Abstract
Avian encephalomyelitis virus (AEV) is a picornavirus that causes disease in poultry worldwide, and flocks must be vaccinated for protection. AEV is currently classified within the hepatovirus genus, since its proteins are most closely related to those of hepatitis A virus (HAV). We now provide evidence that the 494-nucleotide-long 5′ untranslated region of the AEV genome contains an internal ribosome entry site (IRES) element that functions efficiently in vitro and in mammalian cells. Unlike the HAV IRES, the AEV IRES is relatively short and functions in the presence of cleaved eIF4G and it is also resistant to an inhibitor of eIF4A. These properties are reminiscent of the recently discovered class of IRES elements within certain other picornaviruses, such as porcine teschovirus 1 (PTV-1). Like the PTV-1 IRES, the AEV IRES shows significant similarity to the hepatitis C virus (HCV) IRES in sequence, function, and predicted secondary structure. Furthermore, mutational analysis of the predicted pseudoknot structure at the 3′ end of the AEV IRES lends support to the secondary structure we present. AEV is therefore another example of a picornavirus harboring an HCV-like IRES element within its genome, and thus, its classification within the hepatovirus genus may need to be reassessed in light of these findings.
Translation initiation on the majority of cellular mRNAs is mediated by a cap-dependent mechanism. The cap structure (m7GpppN) found on all cytoplasmic mRNAs is recognized by the translation initiation factor complex eIF4F (reviewed in reference 28). This complex contains three proteins: eIF4E, which is the cap-binding protein; eIF4A, which has RNA helicase activity; and eIF4G, which acts as a protein scaffold between the mRNA and the 40S ribosomal subunit via its interaction with eIF3 (reviewed in reference 14). In contrast, initiation of protein synthesis on some viral mRNAs, for example, from the picornaviruses, occurs by a cap-independent mechanism termed internal initiation. In this case, translation initiation is directed by an internal ribosome entry site (IRES) element located within the 5′ untranslated region (5′UTR) of the viral genome (reviewed in references 2 and 11). These IRES elements are large, typically 450 nucleotides (nt) in length, and contain extensive secondary structure; they have been shown to interact with a variety of cellular proteins (2). Most of these elements work without any requirement for eIF4E and hence can continue to function when cap-dependent protein synthesis is inhibited.
The picornavirus IRES elements are divided into several groups which display distinct secondary structures and biological properties. One group (class I) contains IRES elements from the entero- and rhinoviruses (e.g., poliovirus [PV]), while the second group contains the cardio- and aphthovirus IRES elements (e.g., encephalomyocarditis virus [EMCV]). The cardio-/aphthovirus IRES elements function efficiently in the rabbit reticulocyte lysate (RRL) translation system. However, the PV and rhinovirus IRES elements are inefficient in this system unless the reaction mixture is supplemented with additional proteins, e.g., from HeLa cell extracts (6, 10). The IRES element from hepatitis A virus (HAV) represents a third type of IRES. It is distinct from other picornavirus IRES elements in that it requires an intact eIF4F complex, including eIF4E, for function (1, 4). In contrast, the class I and II picornavirus IRES elements can function efficiently when eIF4G has been cleaved by the expression of an entero-/rhinovirus 2A or aphthovirus L protease (5, 34). This cleavage releases the N terminus of eIF4G including its eIF4E binding site (reviewed in reference 20). The initiation factor eIF4A has also been shown to be required by group I and II IRES elements since dominant-negative mutants of this protein and inhibitors of eIF4A block their activity (9, 26, 38).
Recently, a new group of picornavirus IRES elements has been identified. This group includes the IRES elements from porcine teschovirus 1 (PTV-1) Talfan strain (8, 17, 29), simian virus 2 (SV2), and porcine enterovirus 8 (PEV-8) (9). Strikingly, these IRES elements have many similarities to those from hepatitis C virus (HCV) and classical swine fever virus (CSFV), which both belong to the Flaviviridae family. These recently characterized picornavirus IRES elements are predicted to share a very similar structure to the HCV-type elements, including a pseudoknot near the 3′ end of the IRES that is critical for function (12, 18, 40). These IRES elements are generally shorter than other picornavirus elements, e.g., about 280 nt in the case of the PTV-1 IRES (8). They can also function with cleaved eIF4G (9, 29) and, unlike the group I and II IRES elements, are resistant to both dominant-negative mutants of eIF4A (8, 9) and to hippuristanol, a small molecule inhibitor of eIF4A (3). Like the HCV IRES element, the PTV-1 IRES element does not require any of the eIF4 initiation factors for assembly of 48S initiation complexes on the RNA (27, 29, 30).
Avian encephalomyelitis virus (AEV) is a picornavirus that infects young chickens, quails, pheasants, and turkeys, causing ataxia and rapid tremors, especially in the neck. AEV is a worldwide problem, and almost all flocks are susceptible unless they are vaccinated (7). Around four billion birds are vaccinated worldwide each year to protect them from infection (I. Tarpey, personal communication). The AEV genome is 7,032 nt long (smaller than that of any other picornavirus). It encodes a polyprotein of 2,134 amino acids that is processed to the individual viral proteins which are most closely related to the hepatitis A virus (HAV) proteins. AEV has therefore been assigned to the hepatovirus genus of the picornaviruses (24). The 5′UTR of the AEV RNA is 494 nt long, which is also shorter than most other picornaviruses (24). On the basis of sequence comparisons and secondary structure predictions, it has recently been suggested that the AEV genome contains an HCV-like IRES element (15).
Here, we describe the biological properties of the AEV 5′UTR. We demonstrate for the first time that this region contains an IRES element with clear functional differences from that of HAV. We also show that the AEV IRES has significant functional and structural similarities to the other HCV-like picornavirus IRES elements.
MATERIALS AND METHODS
Reporter plasmids.
DNA preparations and manipulations were performed by standard methods as described previously (36) or as stated in manufacturers' instructions. The reporter plasmids pGEM-CAT/EMC/LUC containing the EMCV IRES cDNA and pGEM-CAT/LUC (lacking any IRES) have been described previously (34). These plasmids express from a T7 promoter dicistronic mRNAs encoding chloramphenicol acetyltransferase (CAT) and firefly luciferase (fLUC). Plasmids containing the HAV and HCV IRES elements between the cyclin and influenza virus nonstructural gene (NS) sequences have also been described previously (4, 32) and were a kind gift from Richard Jackson (University of Cambridge, United Kingdom).
To obtain a single cDNA fragment corresponding to the AEV 5′UTR, overlap PCR was performed. Two separate AEV cDNA clones (a gift from Ian Tarpey [Intervet, United Kingdom] and Dave Cavanagh [Institute for Animal Health, Compton, United Kingdom]) were used as templates to amplify by PCR fragments corresponding to nt 1 to 238 and 238 to 494 of the AEV 5′UTR using primers AEVF1 with AEVR266 and AEVF238 with AEVR494, respectively (Table 1). The two purified products were mixed and used in a further PCR using primers AEVF1 and AEVR494 to create a single fragment corresponding to the full-length AEV 5′UTR (nt 1 to 494) flanked by BamHI sites. The PCR product was ligated into pGEM-T Easy (Promega), and from the resultant plasmid, the AEV cDNA was released by BamHI digestion and then inserted, in both orientations, into similarly digested and phosphatased pGEM-CAT/LUC between the two open reading frames (ORFs). The plasmid containing the AEV 5′UTR cDNA in the sense (genomic) orientation was designated AEVs and that containing the fragment in the antisense orientation was called AEVas (Fig. 1A). A further construct containing cDNA corresponding to the AEV 5′UTR plus 30 nt of coding sequence (AEV+30) was created in a similar way using the reverse primer AEVR524 (Table 1) rather than AEVR494 in the PCRs.
TABLE 1.
Oligonucleotides used for analysis of the AEV IRES
| Primer | Sequencea |
|---|---|
| AEVF1 | ATATGGATCCTTTGAAAGAGGCCTC |
| AEVF238 | TGTAGTATGGGAATCGTGTATGGGGATGA |
| AEVR494 | ATATGGATCCGTTTAAATTGCTACCCT |
| AEVR266 | TCATCCCCATACACGATTCCCATACTACA |
| AEVF100 | ATGGATCCTCCCGCATGGCAAGG |
| AEVF200 | ATGGATCCATCCCTTTGCGTTTC |
| AEVR294 | ATGGATCCACACCTATCCCTCTA |
| AEVR394 | ATGGATCCATACACCGTAACAAT |
| AEVR524 | ATGGATCCCTTGCCTACAGTAGA |
| AEVIIIeF | ATCCTTTCCTACTGCCTAAAAGGGTGGCGTGCCCGG |
| AEVIIIeR | ATCCGGGCACGCCCACCCTTTTAGGCAGTAGGAAAGG |
| AEVS1F | GGGGATGATTAGGATGGGTCGTAGAGGGATAGG |
| AEVS2F | TGATAGGGTGGCGTGGGCGGCCACGAGAGATT |
| AEVS1R | CCTATCCCTCTACGACCCATCCTAATCATCCCC |
| AEVS2R | AATCTCTCGTGGCCGCCCACGCCACCCTATCA |
| AEVS1compF | TGCCCGGCCACGACCCATTAAGGGTAGCAA |
| AEVS2compF | CCACGAGAGATTAAGCCTAGCAATTTAAAC |
| AEVS1compR | TTGCTACCC TTAATGGGTCGTGGCCGGGCA |
| AEVS2compR | GTTTAAATTGCTAGGCTTAATCTCTCGTGG |
| CATForward | ACACCTCCCCCTGAACCTGAAACATAAAAT |
| LUCReverse | CATACTGTTGAGCAATTCAC |
Underlined sequences indicate the restriction sites included in the sequence. Nucleotides in bold italics are those that differ from the nucleotides in the wt sequence.
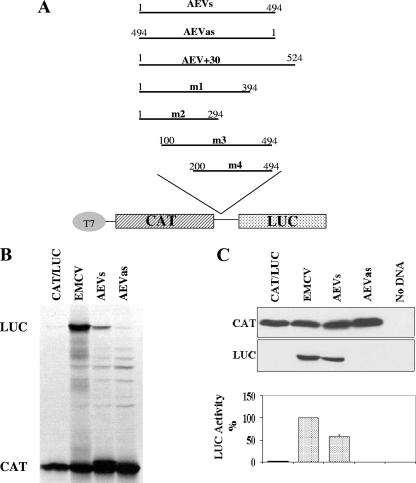
FIG. 1.
The AEV 5′UTR displays IRES activity in vitro and in vivo. (A) The structures of plasmids used in this study are shown. Various fragments of the 5′UTR of the AEV genome were amplified by PCR using primers containing BamHI sites, digested, and inserted between the CAT and LUC ORFs (at the unique BamHI site) in plasmid pGEM-CAT/LUC as described in Materials and Methods. Nucleotide numbers corresponding to the fragments are shown. (B) In vitro translation reactions containing RRL and [35S]methionine were programmed with RNA transcripts derived from the dicistronic plasmids containing the indicated virus sequences. Reaction products were analyzed by SDS-PAGE and autoradiography. The positions of the CAT and LUC proteins are indicated. (C) Transient-expression assay in 293 cells. The dicistronic plasmids (2 μg) containing the indicated virus sequences were transfected into vTF7-3-infected 293 cells. After 20 h, cell lysates were prepared and analyzed for CAT and LUC expression by SDS-PAGE and immunoblotting. LUC assays were performed on cell extracts from three separate transfections, and the results were standardized to the values for LUC expression directed by the EMCV IRES, which was set at 100%. LUC activities were normalized against CAT expression determined using a quantitative CAT enzyme-linked immunosorbent assay kit (Roche). The mean values (plus standard errors of the means [error bars]) are shown.
Four other derivatives (Fig. 1) of the AEVs plasmid with truncated forms of the AEV 5′UTR cDNA were generated as follows: the AEV3′Δ100 fragment (AEVm1) was created using primers AEVF1 and AEVR394, fragment AEV3′Δ200 (AEVm2) was produced using primers AEVF1 and AEVR294, AEV5′Δ100 (AEVm3) was made using primers AEVF100 and AEVR494 while AEV5′Δ200 (AEVm4) was synthesized using primers AEVF200 and AEVR494 (all primer sequences are given in Table 1). The various fragments were cloned, excised with BamHI, and ligated into the pGEM-CAT/LUC dicistronic vector to produce the illustrated plasmids (Fig. 1A). The structures were confirmed by restriction enzyme analysis and sequencing of the inserts.
In vitro translation reactions.
The dicistronic reporter plasmids (1 μg) were assayed in the RRL coupled transcription and translation (TNT) system (Promega) using [35S]methionine as described by the manufacturer. Products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. Alternatively, uncapped mRNA transcripts were produced in vitro using the Ambion MegaScript kit with T7 RNA polymerase, following linearization of the plasmid DNAs with XhoI. Each mRNA was translated in each RRL with [35S]methionine, and products were analyzed by SDS-PAGE and autoradiography.
Transient-expression assays.
The dicistronic reporter plasmids (2 μg) described above were transfected into 293 or HTK-143 cells alone or with the plasmid pGEM3Z/J1 (0.2 μg), which expresses the swine vesicular disease virus (SVDV) 2A protease as previously described (35). Briefly, the plasmids were transfected into cells (35-mm dishes) previously infected with the recombinant vaccinia virus vTF7-3, which expresses T7 RNA polymerase (13), using Lipofectin (8 μl; Invitrogen) and Optimem (192 μl; Gibco BRL). Cell lysates were prepared 20 h after transfection and were analyzed by SDS-PAGE and immunoblotting to determine CAT and LUC expression and eIF4G cleavage. Detection was achieved with anti-CAT (Sigma), anti-fLUC (Promega), or anti-eIF4G (gift from Simon Morley, University of Sussex, United Kingdom) antibodies and peroxidase-labeled anti-rabbit (Amersham) or anti-goat (Dako Cytomation) antibodies, respectively, using chemiluminescence reagents (Pierce). fLUC expression was also quantified using a firefly luciferase assay kit (Promega) with a luminometer.
RNA secondary structure prediction.
AEV 5′UTR sequences (EMBL accession number AJ225173) were aligned with those from HCV (EMBL accession number AB016785) and PTV-1 (EMBL accession number AB038528) using ClustalW and manually edited. Secondary structure elements (other than the pseudoknot) were generated in Mfold (42).
Mutagenesis of the AEV cDNA.
Mutations were introduced into the predicted domain IIIe region in order to change the sequence in the loop region from GAUA to AAAA (nt 446 to 449). The pGEM-CAT/AEVs/LUC plasmid was used as the template for two PCRs, one with each primer set (AEVIIIeF with AEVR494 and AEVIIIeR with AEVF1 [Table 1]). After purification, the products were mixed and a further PCR was performed using AEVF1 and AEVR494 primers. The BamHI-digested product was ligated into pGEM-CAT/LUC as described above, and the resultant plasmid was named pGEM-CAT/AEVIIIemut/LUC. The plasmid was sequenced to verify the presence of the expected mutations.
Mutations within the stem sequences of the predicted pseudoknot were also created (termed S1mut and S2mut). For the S1mut plasmid, nt 273 to 275 (CUC) were changed to GGG, and for the S2mut plasmid, nt 460 to 461 (CC) were changed to GG. The pGEM-CAT/AEVs/LUC plasmid was used as the template for the primary PCRs with the specific mutagenic primers and either CATForward or LUCReverse primer (Table 1) as appropriate. Secondary PCRs used just the latter primers. The final PCR products were purified and digested with BamHI, and the ca. 500-bp fragment was ligated into BamHI-digested and dephosphorylated pGEM-CAT/LUC vector to generate pGEM-CAT/AEVmutS1/LUC and pGEM-CAT/AEVmutS2/LUC. Compensatory mutations were produced in the same way starting with the mutS1 or mutS2 plasmids as templates for the PCRs. The mutagenic primers specified the compensatory mutations (Table 1). The presence of all the expected mutations in the plasmids was confirmed by sequencing.
Translation assays in the presence of hippuristanol.
The requirement of the AEV IRES element for eIF4A was investigated both in vitro and in cells using hippuristanol, a specific inhibitor of eIF4A (3). Dicistronic plasmid DNAs were expressed in the TNT RRL system with or without hippuristanol (10 μM; kind gift from Jerry Pelletier, McGill University, Canada). The products were analyzed by 10% SDS-PAGE and autoradiography. The same plasmids were also assayed in HTK-143 cells with or without the addition of 0.5 μM hippuristanol; cell lysates were prepared after 20 h, and the inhibitor was added for the final 10 h.
RESULTS
Identification of the IRES element within the 5′UTR of AEV RNA.
A dicistronic reporter plasmid was prepared in which cDNA corresponding to the AEV 5′UTR (nt 1 to 494) was inserted between two reporter gene sequences, the first encoding CAT and the second encoding fLUC (AEVs; Fig. 1A). A negative control containing the AEV sequence in the antisense orientation was also constructed (AEVas). Plasmid pGEM-CAT/EMC/LUC, which contains the EMCV IRES, was used as a positive control, and the plasmid pGEM-CAT/LUC, which lacks any IRES sequence, was used as a negative control. RNA transcripts were prepared from each of these constructs using T7 RNA polymerase and were analyzed in translation assays in RRLs. Translation of the first ORF was assessed by the level of CAT expression, and a functional IRES element led to the expression of fLUC. Each of the mRNAs expressed CAT efficiently as expected (Fig. 1B). RNAs containing the EMCV IRES and the AEV 5′UTR sequence in the sense orientation also efficiently expressed fLUC (Fig. 1B), but the AEV 5′UTR was less active in this system than the EMCV IRES was. Only a background level of LUC expression was detected from the AEVas construct or the construct lacking any IRES element. Similar results were obtained in the RRL TNT system (data not shown). No IRES activity from the AEV sequence was detected in the wheat germ TNT system (data not shown). To confirm and extend the results from these in vitro assays, the same dicistronic plasmids were tested in a transient-expression assay in cells. The dicistronic plasmids were transfected into 293 cells, and after 20 h, cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting to detect CAT and LUC expression. As expected, all plasmids expressed CAT efficiently. The AEV and EMCV IRES elements directed efficient fLUC expression (Fig. 1C). LUC assays performed in parallel were consistent with the immunoblotting results, and the AEV 5′UTR generated about 50% of the fLUC expression observed with the EMCV IRES in 293 cells. These results indicated that the AEV 5′UTR contains an IRES element that is functional both in the RRL system and within cells.
The AEV IRES element functions in the presence of an enterovirus 2A protease.
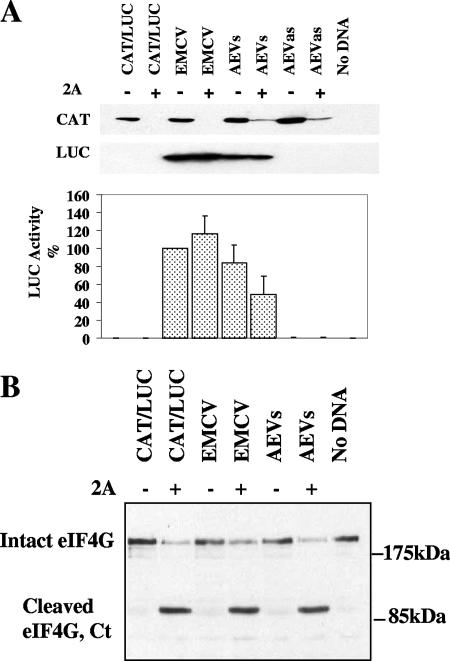
The 2A protease from PV (plus other enteroviruses) and the foot-and-mouth disease virus (FMDV) L protease each inhibit cap-dependent translation by inducing the cleavage of eIF4G, but these proteases have different effects on the various picornavirus IRES elements. Some IRES elements function very efficiently both in the presence or absence of the 2A and L proteases, for example, the EMCV IRES. Other IRES elements, for example, those from PV and other enteroviruses, are stimulated by these proteases within certain cell types, e.g., BHK cells (34). However, the IRES from HAV, the prototype hepatovirus, is strongly inhibited under these conditions, since it requires the intact eIF4F complex (4).We first studied the effect of SVDV 2A on the AEV IRES activity in cells in order to discover any similarity with the HAV IRES. The dicistronic plasmids were transfected into HTK-143 cells either alone or with the pGEM3Z/J1 plasmid which expresses the SVDV 2A protease. After 20 h, cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting to detect CAT and LUC expression. As expected, all plasmids expressed CAT efficiently when transfected into cells alone, but CAT expression was strongly inhibited in the presence of SVDV 2A protease (Fig. 2A) as expected. The AEV and EMCV IRES elements directed efficient fLUC expression in HTK-143 cells (Fig. 2A) in both the presence and absence of the 2A protease, although the AEV IRES did show some inhibition. Note that the AEV IRES displayed higher efficiency compared to the EMCV IRES in this cell type versus the 293 cells (80% versus 50%; Fig. 1). Confirmation of eIF4G cleavage in cells expressing the SVDV 2A protease was achieved by Western blot analysis for eIF4G (Fig. 2B); the C-terminal cleavage product was observed only in the presence of the protease. In addition, we compared the effect of addition of FMDV L protease on the activity of the AEV and HAV IRES elements in vitro. In the presence of FMDV L protease, both the EMCV and AEV IRES elements retained activity, but the HAV IRES was severely inhibited in the presence of the L protease (data not shown), in agreement with previous data (4). These data demonstrate that the AEV IRES directs internal initiation of translation which is cap independent, but it does appear that the intact eIF4F complex is required for optimal activity. This may be due to a direct requirement for binding of a component of eIF4F or possibly to an indirect effect such as a requirement for an eIF4F-dependent factor. These results also indicate that the AEV IRES is different from the IRES element from the other hepatovirus, HAV, as the AEV IRES functions in the presence of cleaved eIF4G.
FIG. 2.
The AEV IRES functions in the presence of cleaved eIF4G. (A) Dicistronic plasmid DNA of the form CAT/IRES/LUC (2 μg) containing the indicated IRES sequences was transfected into HTK-143 cells in the absence (−) or presence (+) of a plasmid encoding SVDV 2A protease (0.2 μg). After 20 h, cell extracts were prepared and analyzed for CAT and LUC expression as described in the legend to Fig. 1. LUC assays were performed on cell extracts from three separate transfections, and the results were standardized to the values for LUC expression directed by the EMCV IRES, which was set at 100%. LUC activities were normalized against CAT expression as described in the legend to Fig. 1. The mean values (plus standard errors of the means [error bars]) are shown. (B) Samples were also analyzed by immunoblotting to analyze the status of eIF4G. The position of the C-terminal cleavage product of eIF4G is indicated (Ct).
The 5′-terminal sequences of the AEV 5′UTR are not required for IRES activity.
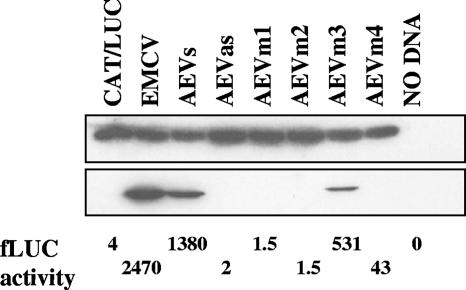
To define the limits of the AEV IRES, four truncated fragments were made by removing sequences from either end of the AEV IRES cDNA. The residual sequences were inserted into the pGEM-CAT/LUC vector as described above. Each plasmid was analyzed within transient-expression assays within HTK-143 cells and in TNT assays as described above. Deletion of 100 nt from the 5′ end of the 5′UTR (mutant AEVm3) had some effect on IRES activity in cells, reducing the activity to 50% of the activity of the wild type (wt) (Fig. 3). However, further deletion to 200 nt from the 5′ end (AEVm4) completely abolished IRES activity (Fig. 3). Deletion of either 100 or 200 nt from the 3′ end of the AEV 5′UTR also completely inhibited IRES activity (AEVm1 and m2), indicating that the 3′ sequences are critical for IRES activity. Similar results were also obtained using TNT assays in vitro (data not shown).
FIG. 3.
Delimitation of the AEV 5′UTR sequences required for IRES activity. Dicistronic plasmids containing the AEV 5′UTR and truncated versions of this sequence, AEVm1 (nt 1 to 294), AEVm2 (nt 1 to 394), AEVm3 (nt 100 to 494), and AEVm4 (nt 1 to 394), were transfected into vTF7-3-infected HTK-143 cells, and cell extracts were analyzed for CAT and LUC expression as described in the legend to Fig. 1. LUC expression was also measured by LUC assay, and the results are shown below the immunoblot in arbitrary units. Similar results were obtained in two independent experiments.
The inclusion of 30 nt of viral coding sequence downstream of the AEV 5′UTR did not result in enhanced IRES activity within cells or in TNT reactions (data not shown), suggesting that these sequences do not play a significant role in AEV IRES function.
Similarity between the AEV IRES and IRES elements from HCV and PTV-1.
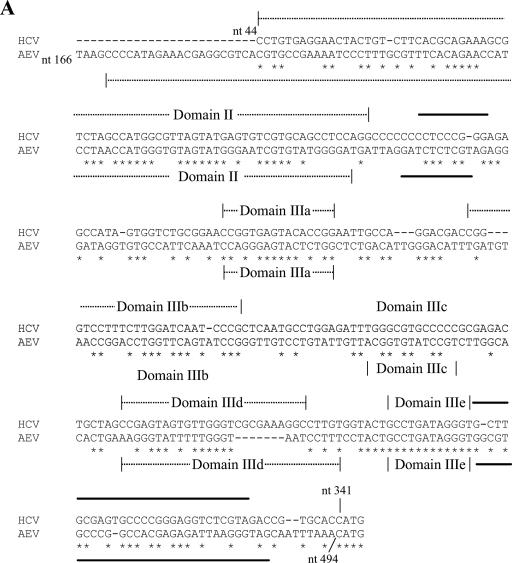
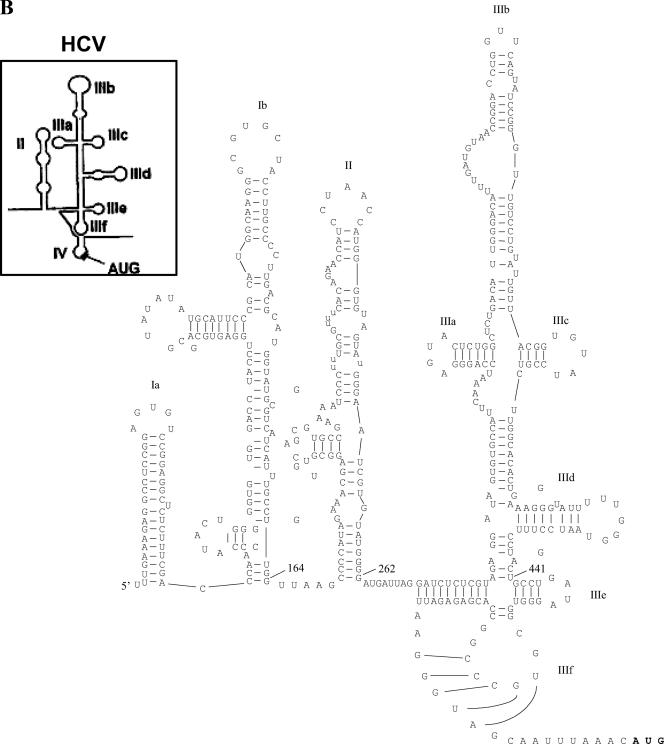
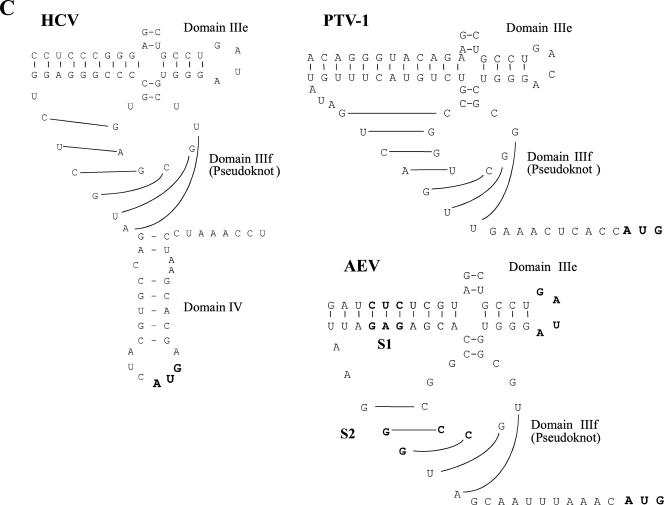
As described above, the functional properties of the AEV IRES are clearly distinct from those of the HAV IRES. To examine the relationship of the AEV IRES to other picornavirus IRES elements, we performed sequence alignments of the AEV sequence using ClustalW. We found that the AEV IRES shares a significant level of identity with the recently characterized PTV-1 IRES (8, 17, 29). The PTV-1 IRES has been shown to resemble the IRES element from HCV, a flavivirus, and the AEV IRES also shares certain critical characteristics with them (Fig. 4A and B). Notably, the HCV domain IIIe is identical to a region of 12 nt within the AEV sequence. Overall, the AEV IRES shares 48% sequence identity with the HCV IRES and 42% identity with the PTV-1 IRES. The similarities are particularly apparent in the regions surrounding and including the pseudoknot found in the HCV and PTV-1 IRES structures (Fig. 4C). We therefore believe that the pseudoknot structure found in these IRES elements is also present in the AEV IRES.
FIG. 4.
Similarity between the HCV and AEV IRES elements. (A) Alignment of the HCV and AEV IRES sequences. Sequences were aligned with ClustalW and manually edited. Identical nucleotides are marked with an asterisk. Individual domains associated with the HCV and AEV IRES elements are indicated above the sequence. Thick black lines indicate regions involved in the formation of the pseudoknot structure within the HCV IRES and AEV IRES elements. The overall sequence identity from this alignment is 48.1%, but note the 100% sequence identity within the domain IIIe region. The gaps introduced to maximize alignment are indicated by dashes. (B) Proposed secondary structure of the entire AEV IRES. Domains are labeled according to corresponding domains of the HCV IRES (inset). The structure was predicted by comparative sequence analysis and using Mfold (42) to predict the most thermodynamically favorable structures. (C) Comparison of predicted secondary structures of the domain IIIe and IIIf regions of the HCV, PTV-1, and AEV IRES elements. The two stems (S1 and S2) and loop regions (L1 and L2) that form the pseudoknot are shown. A domain IV structure is also present in the HCV IRES but not in the PTV-1 or AEV IRES elements. Within the AEV sequence, the nucleotides indicated in bold type are those that were modified in the experiments shown in Fig. 5.
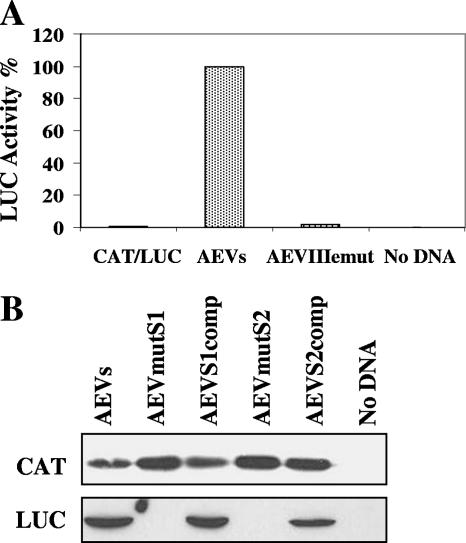
Mutational analysis of the putative domain IIIe region within the AEV IRES.
Studies on domain IIIe of the HCV IRES element have indicated that each of the nucleotides within the highly conserved GAUA tetraloop is crucial for HCV IRES activity (22, 31). This domain, together with domain IIId, plays an essential role in binding the 40S ribosomal subunit (18, 25, 39). A closely related sequence is also present within the PTV-1 IRES element, although the loop sequence (GACA) has a single nucleotide difference from the HCV sequence; mutations in this loop of the PTV-1 IRES also greatly reduced IRES activity (9). To assess the importance of the GAUA sequence within the AEV IRES element, corresponding to the HCV domain IIIe loop, we mutated this motif to AAAA. Consistent with the results from similar mutations within the HCV and PTV-1 IRES elements, these changes resulted in a severely defective AEV IRES as assessed in cells (Fig. 5A).
FIG. 5.
Mutation of the domain IIIe loop or pseudoknot structure results in loss of AEV IRES activity. (A) Dicistronic plasmids containing the wt AEV IRES or the IRES containing the loop IIIe mutation were transfected into vTF7-3-infected HTK-143 cells and analyzed for CAT and LUC expression as described in the legend to Fig. 1. LUC activities (normalized against CAT expression) are shown, and the results are the mean LUC values from three experiments. (B) Dicistronic plasmids containing the indicated mutations within the predicted pseudoknot region were transfected into HTK-143 cells as described above for panel A and analyzed for CAT and LUC expression as described in the legend to Fig. 3. The results are representative of two independent experiments.
Mutational analysis of the predicted pseudoknot region within the AEV IRES.
Evidence for pseudoknot structures within the IRES elements from HCV, CSFV, and GBV-B has been obtained (12, 33, 40). Furthermore, the predicted structure of an analogous pseudoknot within the PTV-1 IRES element has been supported by mutagenesis studies (8). Disruption of base pair interactions within this structure resulted in severely defective HCV (40) and PTV-1 (8) IRES elements. In order to test the structure of this region of the AEV IRES, mutagenesis was carried out to disrupt the predicted base pairing; mutagenesis was followed by the introduction of compensatory mutations to restore the interactions. Two sets of mutations were introduced into the predicted pseudoknot within the base-paired stem regions (Fig. 4B). Mutations in stem 1 (S1) changed nt 273 to 275 (CUC) (which are predicted to base pair with nt 471 to 473 [GAG]) to GGG. These mutations were predicted to disrupt these base pair interactions. This mutant IRES was assayed within the CAT/IRES/LUC vector in cells. As expected (Fig. 5B), the mutations in this region severely inhibited IRES activity. To confirm that it was the disruption of the base pairing in S1 rather than the change in sequence alone that was responsible for the inhibition of IRES activity, compensatory mutations were introduced to restore the predicted base pairing. Nucleotides 471 to 473 (GAG) within the S1 mutant were changed to CCC, resulting in a total of 6 nt differences from the wt sequence; these additional modifications resulted in restoration of IRES activity to about 60 to 80% of wt IRES activity. These results were confirmed in RRLs (data not shown) and strongly suggest that the predicted secondary structure is correct.
Mutations in stem 2 (S2) changed nt 460 and 461 (CC) to GG, these changes were predicted to disrupt the interactions with nt 481 and 482 (GG) and hence destabilize the pseudoknot structure. The results from in vivo (Fig. 5B) and in vitro (data not shown) experiments indicated that these mutations completely abrogated IRES activity as anticipated. Furthermore, compensatory mutations that changed nt 481 and 482 (GG) to CC, predicted to restore base pair interactions, efficiently regenerated IRES activity (about 60% of wt AEV IRES activity in vivo [Fig. 5B]). These results also supported the predicted pseudoknot structure shown in Fig. 4. The fact that full restoration of IRES activity was not achieved may suggest that these nucleotides are also involved in other interactions (such as RNA-protein interactions) as well as forming the pseudoknot structure.
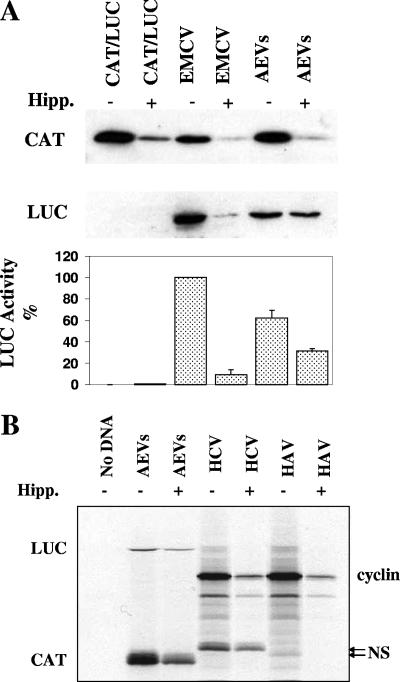
The AEV IRES is resistant to hippuristanol, an inhibitor of eIF4A.
In previous studies, it has been found that the HCV and PTV-1 IRES elements have no requirement for eIF4A for translation initiation (3, 27, 29). As the results presented above indicated that the AEV IRES element resembles these IRES elements, the requirement for eIF4A was studied in vitro and in vivo using hippuristanol, a specific inhibitor of eIF4A. Hippuristanol inhibits cap-dependent translation and the activity of type I and II picornavirus IRES elements. In contrast, the activity of the HCV and PTV-1 IRES elements is resistant to this inhibitor (3). Selected dicistronic reporter plasmids were assayed in cells in the presence and absence of hippuristanol. As expected, the eIF4A inhibitor severely reduced translation of the upstream cistron CAT (Fig. 6A). As seen before (9), the EMCV IRES activity was also reduced to about 10% of its activity in the presence of the eIF4A inhibitor. In contrast, the AEV IRES displayed marked resistance to this inhibitor (Fig. 6A). Similar results were also observed in vitro (Fig. 6B). However, we did note that the AEV IRES was partially inhibited in the presence of this inhibitor in both systems (reduced to about 50% activity in the presence of hippuristanol, which is similar to the reduction seen in the presence of the SVDV 2A protease), although a similar effect was also observed with the HCV IRES element (Fig. 6B). In contrast, the HAV IRES was completely inhibited in the presence of hippuristanol, a previously unreported finding (Fig. 6B). This result is in agreement with the suggestion that the HAV IRES requires the whole eIF4F complex for its function (4).
FIG. 6.
The AEV IRES is resistant to an inhibitor of eIF4A. (A) Dicistronic plasmids containing the indicated IRES sequences were transfected into HTK-143 cells in the absence (−) or presence (+) of 0.5 μM hippuristanol (Hipp.), an inhibitor of eIF4A. Cells were harvested after 20 h, and the inhibitor was added for the last 10 h of the incubation. Cell extracts were analyzed for CAT and LUC expression as described in the legend to Fig. 1. LUC assays were performed on cell extracts from three separate transfections, and the results were standardized to the values for LUC expression directed by the EMCV IRES, which was set at 100%. The mean values (plus standard errors of the means [error bars]) are shown. (B) Dicistronic plasmids containing the indicated IRES elements were also assayed in RRL TNT in the presence of 10 μM hippuristanol. The HAV and HCV IRES elements are within a cyclin/NS dicistronic construct. The positions of all proteins are indicated. Note the slower migration of the HCV IRES-directed NS product compared to the HAV IRES-directed product (arrows) due to the inclusion of 30 nt of the HCV coding sequence in this plasmid (32).
DISCUSSION
The studies presented here demonstrate that the 5′UTR of the AEV genome contains an IRES element that functions efficiently in RRLs and in mammalian cells. Until recently, picornavirus IRES elements were classified into one of three groups. However, a new group of picornavirus IRES elements that includes IRES elements that closely resemble the HCV IRES elements has recently been described; this group includes elements from PTV-1, PEV-8, and SV2 (8, 9, 17, 29). From database searches of sequences, other picornavirus genomes, including AEV, duck hepatitis virus 1, and Seneca valley virus, have recently been predicted to contain HCV-like IRES elements (9, 15). Here, we describe the functional analysis of the IRES from AEV and propose that this IRES element is included within this same group.
We have shown that the functional AEV IRES lies within nt 100 to 494 of the 5′UTR. In contrast to the PV, EMCV, or HAV IRES elements, the AEV IRES lacks a polypyrimidine tract near the 3′ end of the element. It can function when eIF4G is cleaved and is also resistant to hippuristanol, an inhibitor of eIF4A activity. These features distinguish the AEV IRES from these picornavirus IRES elements, but they are shared with the PTV-1, PEV-8, and SV2 IRES elements plus the HCV and CSFV IRES elements. Alignment of the nucleotide sequences of the HCV and AEV IRES elements confirmed that these elements share a striking similarity (about 48% overall identity). Indeed, within the HCV IIIe domain there is 100% sequence identity to a region of the AEV IRES (Fig. 4). The secondary structure models for the HCV-like IRES elements include an important pseudoknot structure. The AEV sequence is also proposed to form this structure (Fig. 4). We have obtained supporting evidence for the formation of this structure in the AEV IRES through mutational analysis of the sequences predicted to form the pseudoknot. Mutations within the S1 or S2 regions that were expected to disrupt the predicted pseudoknot structure inhibited IRES activity, but compensatory mutations designed to restore the base pairing in this structure efficiently rescued activity. Mutation of the GAUA motif within a portion of the AEV sequence that is identical to the domain IIIe of the HCV IRES also disrupted AEV IRES activity. Mutation of this loop region in the HCV and PTV-1 IRES elements also inhibited IRES activity (8, 22, 31). It is known that the IIId and IIIe regions of the HCV IRES domain III interact with the 40S ribosomal subunit (18, 23), while the IIIb region has been shown to interact with eIF3 (16, 18, 37). A recent model of initiation complex formation on the HCV IRES suggests that regions IIId and IIIe of the IRES bind to the 40S ribosomal subunit, and this is followed by the interaction with eIF3 and the ternary complex (eIF2/met-tRNAi/GTP) to form a 48S preinitiation complex (25). Recent data have shown that several proteins of the 40S ribosomal subunit, including p40, S3a, S5, and S16 are positioned close to hairpin IIIe of the HCV IRES element during the early stage of translation initiation (19). It has also recently been shown that domain II of the HCV and CSFV IRES elements plays a role in 80S ribosome assembly on RNA and promotes eIF5-induced GTP hydrolysis and eIF2/GDP release, following 48S initiation complex formation (21).
The AEV IRES does not require any viral coding sequence for function, and secondary structure predictions suggest that there is no region equivalent to HCV IRES domain IV (15; L. O. Roberts, unpublished data). There are some differences between the picornavirus “HCV-like” IRES elements in this respect, as the SV2 IRES is predicted to contain a domain IV region, whereas the PTV-1 and PEV-8 IRES elements do not (9).
The discovery of a number of distinct picornaviruses harboring an HCV-like IRES element in their 5′UTR suggests that recombination between picornavirus and flavivirus genomes has occurred. Previous work has shown that the PV IRES can be replaced with the HCV IRES (41) to produce a viable chimeric virus, suggesting that they are functionally equivalent (although mechanistically very different). However, it is important to note that there are important structural differences between the picornavirus and HCV genomes [e.g., picornaviruses possess a 3′ poly(A) tail] that may be important in translation/replication. There are also differences in the sequence and predicted structures of the domain II regions of the picornavirus “HCV-like” and HCV IRES elements which may have a role in translation and/or replication. It remains to be seen whether the diverse domain II structures found in the picornavirus “HCV-like” IRES elements have the same function or are involved in replication.
AEV has been tentatively classified as being a member of the hepatovirus genus within the Picornaviridae, since it has the highest degree of protein sequence identity to HAV. However, its IRES element is clearly distinct from that of HAV in that it functions well in the presence of cleaved eIF4G, displays resistance to an inhibitor of eIF4A, and shares a striking similarity to the HCV-like IRES elements. Due to these key differences between AEV and HAV, it is suggested that the placement of AEV within the hepatovirus genus should be reconsidered.
Acknowledgments
We thank Ian Tarpey (Intervet, United Kingdom) and Dave Cavanagh (Institute for Animal Health, Compton, United Kingdom) for the AEV cDNA clones, Jerry Pelletier (McGill University, Montreal, Canada) for the kind gift of hippuristanol, Tim Skern (Medical University of Vienna, Austria) for the kind gift of FMDV L protease, Richard Jackson for dicistronic reporter plasmids, and Simon Morley (University of Sussex, United Kingdom) for the gift of eIF4G antisera.
M.B. gratefully acknowledges receipt of a scholarship from the Ministry of Science, Research and Technology and the Ministry of Jihad-Agriculture of Iran.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Ali, I. K., L. McKendrick, S. J. Morley, and R. J. Jackson. 2001. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 757854-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor monograph 39. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Bordeleau, M. E., A. Mori, M. Oberer, L. Lindqvist, L. S. Chard, T. Higa, G. J. Belsham, G. Wagner, J. Tanaka, and J. Pelletier. 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2213-220. [DOI] [PubMed] [Google Scholar]
- 4.Borman, A. M., and K. M. Kean. 1997. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology 237129-136. [DOI] [PubMed] [Google Scholar]
- 5.Borman, A. M., P. LeMercier, M. Girard, and K. M. Kean. 1997. Comparison of picornaviral IRES-driven internal initiation in cultured cells of different origins. Nucleic Acids Res. 25925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, E. A., and E. Ehrenfeld. 1979. Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology 97396-405. [DOI] [PubMed] [Google Scholar]
- 7.Calnek, B. W. 2003. Avian encephalomyelitis, p. 271-281. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), Diseases of poultry, 11th ed. Iowa State University Press, Ames, IA.
- 8.Chard, L. S., Y. Kaku, B. Jones, A. Nayak, and G. J. Belsham. 2006. Functional analyses of RNA structures shared between the internal ribosome entry sites of hepatitis C virus and the picornavirus porcine teschovirus 1 Talfan. J. Virol. 801271-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chard, L. S., M. E. Bordeleau, J. Pelletier, J. Tanaka, and G. J. Belsham. 2006. Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae. J. Gen. Virol. 87927-936. [DOI] [PubMed] [Google Scholar]
- 10.Dorner, A. J., B. L. Semler, R. J. Jackson, R. Hanecak, E. Duprey, and E. Wimmer. 1984. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J. Virol. 50507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doudna, J., and P. Sarnow. 2007. Translation initiation by viral internal ribosome entry sites, p. 129-153. In M. B. Mathews, N. Sonenberg, and J. W. B. Hershey (ed.), Translational control in biology and medicine. Cold Spring Harbor monograph 48. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Fletcher, S. P., and R. J. Jackson. 2002. Pestivirus internal ribosome entry site (IRES) structure and function: elements in the 5′ untranslated region important for IRES function. J. Virol. 765024-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 838122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68913-963. [DOI] [PubMed] [Google Scholar]
- 15.Hellen, C. U. T., and S. de Breyne. 2007. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse Picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 815850-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji, H., C. S. Fraser, Y. Yu, J. Leary, and J. A. Doudna. 2004. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl. Acad. Sci. USA 10116990-16995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaku, Y., L. S. Chard, T. Inoue, and G. J. Belsham. 2002. Unique characteristics of a picornavirus internal ribosome entry site from the porcine teschovirus-1 Talfan. J. Virol. 7611721-11728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieft, J. S., K. Zhou, R. Jubin, and J. A. Doudna. 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7194-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laletina, E., D. Graifer, A. Malygin, A. Ivanov, I. Shatsky, and G. Karpova. 2006. Proteins surrounding hairpin IIIe of the hepatitis C virus internal ribosome entry site on the human 40S ribosomal subunit. Nucleic Acids Res. 342027-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloyd, R. 2006. Translational control by viral proteinases. Virus Res. 11976-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Locker, N., L. E. Easton, and P. J. Lukavsky. 2007. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 26795-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukavsky, P. J., G. A. Otto, A. M. Lancaster, P. Sarnow, and J. D. Puglisi. 2000. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 71105-1110. [DOI] [PubMed] [Google Scholar]
- 23.Lytle, J. R., L. Wu, and H. D. Robertson. 2001. The ribosome binding site of hepatitis C virus mRNA. J. Virol. 757629-7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marvil, P., N. J. Knowles, A. P. Mockett, P. Britton, T. D. Brown, and D. Cavanagh. 1999. Avian encephalomyelitis virus is a picornavirus and is most closely related to hepatitis A virus. J. Gen. Virol. 80653-662. [DOI] [PubMed] [Google Scholar]
- 25.Otto, G. A., and J. D. Puglisi. 2004. The pathway of HCV IRES-mediated translation initiation. Cell 119369-380. [DOI] [PubMed] [Google Scholar]
- 26.Pause, A., N. Methot, Y. V. Svitkin, W. C. Merrick, and N. Sonenberg. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 131205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNA. Genes Dev. 1267-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pestova, T. V., J. R. Lorsch, and C. U. T. Hellen. 2007. The mechanism of translation initiation in eukaryotes, p. 87-128. In M. B. Mathews, N. Sonenberg, and J. W. B. Hershey (ed.), Translational control in biology and medicine. Cold Spring Harbor monograph 48. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Pisarev, A. V., L. S. Chard, Y. Kaku, H. L. Johns, I. N. Shatsky, and G. J. Belsham. 2004. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 784487-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pisarev, A. V., N. E. Shirikiki, and C. U. Hellen. 2005. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C. R. Biol. 328589-605. [DOI] [PubMed] [Google Scholar]
- 31.Psaridi, L., U. Georgopoulou, A. Varaklioti, and O. Mavromara. 1999. Mutational analysis of a conserved tetraloop in the 5′ untranslated region of hepatitis C virus identifies a novel RNA element essential for the internal ribosome entry site function. FEBS Lett. 45349-53. [DOI] [PubMed] [Google Scholar]
- 32.Reynolds, J. E., A. Kaminski, H. J. Kettinen, K. Grace, B. E. Clarke, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 146010-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rijnbrand, R., G. Abell, and S. M. Lemon. 2000. Mutational analysis of the GB virus B internal ribosome entry site. J. Virol. 74773-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts, L. O., R. A. Seamons, and G. J. Belsham. 1998. Recognition of picornavirus internal ribosome entry sites within infected cells: influence of cellular and viral proteins. RNA 4520-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakoda, Y., N. Ross-Smith, T. Inoue, and G. J. Belsham. 2001. An attenuating mutation in the 2A protease of swine vesicular disease virus, a picornavirus, regulates cap- and internal ribosome entry site-dependent protein synthesis. J. Virol. 7510643-10650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2006. The condensed protocols from Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Sizova, D. V., V. G. Kolupaeva, T. V. Pestova, I. N. Shatsky, and C. U. T. Hellen. 1998. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated region of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 724775-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svitkin, Y. V., A. Pause, A. Haghighat, S. Pyronnet, G. Witherell, G. J. Belsham, and N. Sonenberg. 2001. The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is directly proportional to the degree of mRNA 5′ secondary structure. RNA 7382-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tallet-Lopez, B., L. Aldaz-Carroll, S. Chabas, E. Dausse, C. Staedel, and J. J. Toulme. 2003. Antisense oligonucleotides targeted to the domain IIId of the hepatitis C virus IRES compete with 40S ribosomal subunit binding and prevent in vitro translation. Nucleic Acids Res. 31734-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, C., S. Y. Le, N. Ali, and A. Siddiqui. 1995. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1526-537. [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao, W. D., E. Wimmer, and F. C. Lahser. 1999. Poliovirus/hepatitis C virus (internal ribosomal entry site-core) chimeric viruses: improved growth properties through modification of a proteolytic cleavage site and requirement for core RNA sequences but not for core-related polypeptides. J. Virol. 731546-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]