Abstract
The mechanism of CD4+ T-cell depletion during chronic human immunodeficiency virus type 1 (HIV-1) infection remains unknown. Many studies suggest a significant role for chronic CD4+ T-cell activation. We assumed that the pathogenic process of excessive CD4+ T-cell activation would be reflected in the transcriptional profiles of activated CD4+ T cells. Here we demonstrate that the transcriptional programs of in vivo-activated CD4+ T cells from untreated HIV-positive (HIV+) individuals are clearly different from those of activated CD4+ T cells from HIV-negative (HIV−) individuals. We observed a dramatic up-regulation of cell cycle-associated and interferon-stimulated transcripts in activated CD4+ T cells of untreated HIV+ individuals. Furthermore, we find an enrichment of proliferative and type I interferon-responsive transcription factor binding sites in the promoters of genes that are differentially expressed in activated CD4+ T cells of untreated HIV+ individuals compared to those of HIV− individuals. We confirm these findings by examination of in vivo-activated CD4+ T cells. Taken together, these results suggest that activated CD4+ T cells from untreated HIV+ individuals are in a hyperproliferative state that is modulated by type I interferons. From these results, we propose a new model for CD4+ T-cell depletion during chronic HIV-1 infection.
Human immunodeficiency virus type 1 (HIV-1) is the etiologic agent of AIDS, which is a manifestation of the progressive CD4+ T-cell depletion that occurs in the setting of HIV-1 infection (19, 59). During acute infection by HIV-1, the process of CD4+ T-cell depletion is initiated by unchecked viral replication that occurs before an adaptive immune response is mounted (9, 19, 21, 45, 51, 59, 71, 77). Concomitant with development of a cytotoxic T-lymphocyte (CTL) response and the consequent reduction in viral replication, however, the CD4 count rebounds to a quasi-steady-state level that is below the baseline CD4 count of HIV-negative (HIV−) individuals and that decreases on the time scale of years (19, 59). In the gut-associated lymphoid tissue, where the majority of the body's lymphocytes reside, there is a dramatic depletion of CD4+ T cells during acute infection (9, 45, 51, 71, 77). However, AIDS develops only when CD4+ T cells have been sufficiently depleted that opportunistic infections occur (reflected by a CD4 count below 200 cells/μl). It is unclear how and why CD4+ T cells are depleted in the setting of chronic HIV-1 infection (19, 25, 28, 50, 59).
The “tap and drain” model of CD4+ depletion during HIV-1 infection developed by Ho and colleagues suggests that the rate of CD4+ T-cell destruction by HIV-1 during chronic infection is too fast to be compensated for by natural CD4+ T-cell production (34). Consistent with the idea of increased destruction, CD4+ T cells from simian immunodeficiency virus (SIV)-infected macaques have been shown to have a substantially higher turnover rate than CD4+ T cells from uninfected macaques (30, 41, 52, 65). While more-recent studies continue to indicate a role for viral destruction (56) in CD4+ T-cell depletion, several important experimental observations suggest that factors in addition to destruction by viral infection are responsible for CD4+ T-cell depletion in the setting of HIV-1 infection. In particular, in both SIV-infected macaques and HIV-1-infected individuals, the majority of apoptotic CD4+ T cells are “bystander” cells rather than productively infected cells (20). Furthermore, the frequency of HIV-1-infected CD4+ T cells in humans is, on average, very low (14) and likely is too low if virus-induced death subsequent to infection is solely responsible for HIV-1-mediated CD4+ T-cell depletion (20), given known parameters of CD4+ T-cell dynamics in the setting of HIV-1 infection (2). Consistent with this observation, natural hosts of SIV, such as the sooty mangabey or African green monkey, do not exhibit a dramatic decline in the number of CD4+ T cells despite high viral loads similar to those observed in AIDS-susceptible nonhuman primate species, such as the rhesus macaque (38, 70).
Chronic HIV-1 infection is characterized by massive and chronic activation of CD4+ T cells (29, 41, 63). This activation is reduced to near-normal levels by treatment with highly active antiretroviral therapy (HAART) (29) or in long-term nonprogressors (13). Interestingly, the level of CD4+ T-cell activation correlates better than the viral load with the degree of reduction in CD4 count in both HIV-1- and HIV-2-infected individuals (43, 54, 74). As with HIV-1, SIV induces excessive CD4+ T-cell activation in AIDS-susceptible species of nonhuman primates (e.g., rhesus macaques) but not in its natural hosts (e.g., the sooty mangabey or African green monkey) (70). These studies strongly suggest that massive and chronic CD4+ T-cell activation induced by HIV-1 infection plays a role in CD4+ T-cell depletion (25, 28, 50). However, several important questions remain unanswered. In particular, what is the mechanism through which chronic CD4+ T-cell activation leads to CD4+ T-cell depletion? Furthermore, what signals are acting on CD4+ T cells in the setting of HIV-1 infection (i) to induce such a substantial increase in activated CD4+ T cells and (ii) to modulate CD4+ T-cell depletion?
In the present report, we describe a novel approach aimed at elucidating the molecular mechanisms through which chronic CD4+ T-cell activation in the setting of HIV-1 infection leads to CD4+ T-cell depletion. We hypothesized that if chronic CD4+ T-cell activation contributes to CD4+ T-cell depletion, then this pathogenic process should be reflected at the molecular level in the transcriptional profiles of activated CD4+ T cells of chronically infected HIV-positive (HIV+) individuals. Specifically, we wanted to determine whether there were qualitative differences in the activated CD4+ T cells from HIV-1-infected individuals compared to the activated CD4+ T cells from uninfected individuals. With this idea in mind, we carried out a microarray study of the gene expression profiles of activated CD4+ T cells from untreated, chronically infected HIV+ individuals with a comparison to activated CD4+ T cells from healthy HIV− individuals. We observed dramatic differences in patterns of gene expression in activated CD4+ T cells from infected versus uninfected individuals that lead to a new model for the role of chronic CD4+ T-cell activation in CD4+ T-cell depletion in the setting of HIV-1 infection.
MATERIALS AND METHODS
Study subjects.
Seventeen HIV− donors and 28 HIV+ donors (21 untreated and viremic and 7 treated and aviremic; unpublished data) were enrolled in this study after signed, informed consent approved by the Johns Hopkins Medical Institutions Institutional Review Board was obtained. Inclusion criteria for viremic HIV+ donors were as follows: (i) lack of prior antiretroviral therapy or lack of antiretroviral therapy for more than 1 year prior to study entry and (ii) viremia of >1,000 copies/ml for more than 1 year, as well as (iii) CD4 count of >200 cells/μl (in order to ensure that enough CD4+ T cells could be isolated for experiments). Inclusion criteria for aviremic HIV+ donors were as follows: (i) on HAART for more than 1 year, (ii) suppression of viremia to <50 copies/ml for more than 1 year, and (iii) CD4 count of >200 cells/μl (in order to ensure that enough CD4+ T cells could be isolated for experiments). Exclusion criteria for all blood donors were as follows: (i) chronic hepatitis B or hepatitis C infection and (ii) AIDS defining illness within 1 year of blood draw.
Cell isolation.
CD4+ T cells were positively selected to >98% purity from freshly isolated peripheral blood mononuclear cells, using magnetic microbeads and subsequent column purification according to the manufacturer's protocol (Miltenyi Biotec, Auburn, CA). Activated CD4+ T cells were similarly isolated by positive selection from CD4+ T cells using CD25 microbeads (Miltenyi Biotec, Auburn, CA). In order to ensure isolation of CD25lo cells, the manufacturer's protocol was altered such that 2 μl of CD25 microbeads and 8 μl of wash medium (phosphate-buffered saline [pH 7.2 to 7.4], 2% newborn calf serum, 0.1% glucose, 1% penicillin/streptomycin, 12 mM HEPES [pH 7.2]) were used per million CD4+ T cells. The purity of isolated CD25+ cells was almost always greater than 95%.
Microarray.
Total RNA was isolated from cells using the RNeasy minikit (Qiagen). RNA from control and experimental tissue was processed using the two-round RNA amplification protocol described by Affymetrix (Affymetrix GeneChip eukaryotic small-sample target labeling assay, version II). Briefly, 100 ng of total RNA was used to synthesize first-strand cDNA, using the SuperScript Choice system (Invitrogen, Carlsbad, CA). The 24 oligo(dT)-plus-T7 promoter was used as a primer (Proligo LLC, Boulder, CO). Following double-stranded-cDNA synthesis, the product was purified by phenol-chloroform extraction and unlabeled ribonucleotides were used in a first round of in vitro transcription cRNA amplification (MegaScript, Ambion, Austin, TX). The following cycle of cDNA synthesis was started with random primers, and the oligo(dT)-with-T7 promoter was again used as a primer at the second-strand cDNA synthesis step. The double-stranded cDNA product was then purified again by phenol-chloroform extraction. Consequently, biotinylated antisense cRNA was generated through in vitro transcription using the BioArray RNA high-yield transcript labeling Kit (ENZO Life Sciences Inc., Farmingdale, NY). Fifteen micrograms of the biotinylated labeled cRNA was fragmented at 94°C for 35 min (100 mM Tris-acetate, pH 8.2, 500 mM potassium acetate, 150 mM magnesium acetate), and 10 μg of total fragmented cRNA was hybridized to the Affymetrix Human Genome GeneChip array U133Plus 2.0 for 16 h at 45°C with constant rotation (60 rpm). The Affymetrix Fluidics Station 450 was then used to wash and stain the chips, removing the nonhybridized target and incubating with a streptavidin-phycoerythrin (PE) conjugate to stain the biotinylated cRNA. The staining was then amplified using goat immunoglobulin G (IgG) as a blocking reagent and biotinylated antistreptavidin antibody (goat), followed by a second staining step with a streptavidin-PE conjugate. Fluorescence was detected using the Affymetrix-GS3000 GeneArray scanner, and image analysis of each GeneChip was done through the GeneChip Operating System software from Affymetrix (GCOS1.3), using the standard default settings. For a comparison between different chips, global scaling was used, scaling all arrays to a user-defined target intensity of 150.
Microarray analysis.
The quality of the microarray experiment was assessed with affyPLM and Affy, two BioConductor packages for statistical analysis of microarray data. To estimate the gene expression signals, data analysis was conducted with the chips' CEL file probe signal values at the Affymetrix probe pair (perfect match probe and mismatch probe) level using the statistical algorithm RMA (robust multiarray expression measure) (37) with Affy. This probe-level data processing includes a normalization procedure utilizing a quantile normalization method (7) to reduce the obscuring variation between microarrays which might be introduced during the processes of sample preparation, manufacture, fluorescence labeling, hybridization, and/or scanning. With these signal estimates, principal component analysis and multidimensional scaling analyses were performed using the R software program to assess sample variability. The quality assessment, principal component analysis, and multidimensional scaling analysis identified and disqualified a discordant control sample chip, C9.
With the signal data in a log-transformed format, differential gene expression between controls and HIV+ patients was also assessed by statistical linear model analysis using the BioConductor package limma, in which an empirical Bayes method is used to moderate the standard errors of the estimated log-fold changes of gene expression, resulting in more-stable inference and improved power, especially for experiments with small numbers of microarrays (www.bioconductor.org) (72). The moderated t-statistic P values derived from the linear-model analysis described above were further adjusted for multiple testing by Benjamini and Hochberg's method to control the false discovery rate (FDR). The FDR was used to obtain the list of differentially expressed genes.
All Bioconductor packages are available at http://www.bioconductor.org. All computation was performed using the R software program (http://www.r-project.org) (36), a highly extensible language and environment that provides a wide variety of statistical (linear and nonlinear modeling, classical statistical tests, time-series analysis, classification, clustering, etc.) and graphical techniques.
We performed hierarchical clustering, gene ontology analysis, and graphical representation (volcano plots) using the Spotfire analytic platform with its DecisionSite for Functional Genomics extension. All Affymetrix expression data from the 20 retained chips, representing 11 HIV+ specimens and 9 HIV− controls, had been previously normalized by RMA, as described above, before data were imported into Spotfire.
Hierarchical clustering was performed on a subset of 3,742 probe sets, representing 2,669 genes, that were determined to be significantly differentially expressed at a threshold of an FDR of ≤0.01. The pairwise clustering algorithm employed the unweighted pair-group method with arithmetic mean method and defined the similarity between clusters by their Pearson's r correlation. This measure rewards positive correlation, i.e., the most similar expression profiles in terms of both the magnitude and direction of changes. Both probe sets (rows) and chips (columns) were blindly clustered, with the final order determined by average values and not by input order.
We analyzed gene functionality by employing the classifications and vocabulary of the Gene Ontology Consortium (GO) (4). The GO project annotates the function and cellular localization of gene products based on evidence from manual literature-based annotation, structural or sequence similarity, and/or computational analysis, and every GO annotation contains both evidence and source attribution. Since GO annotates gene products, i.e., proteins, it is necessary to map the Affymetrix GeneChips' probe sets first to genes and thence to their GO protein annotations. Although Affymetrix provides GO annotation directly on their NetAffx website, they include inferred annotations. We therefore elected to map these relationships ourselves in order to restrict our analysis to explicit annotation. In brief, we used the Entrez gene identifiers (IDs) provided by Affymetrix to establish one-to-one relationships with gene ontology IDs, based on the NCBI gene2go file, downloaded from their FTP site (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA). We used Spotfire's Gene Ontology browser to examine the full depth of gene ontology annotation for selected sets of probe sets/genes.
Promoter analysis.
We used a previously described computational approach (62) in order to identify enrichment of transcription factor (TF) binding sites in the promoters of genes we found to be up-regulated in activated CD4+ T cells of HIV+ individuals. Regulatory signatures, defined as the set of all genes whose promoters contain at least one binding site for a specific TF, were identified as follows. We identified all probe sets present on the Affymetrix Human Genome GeneChip array U133Plus 2.0 that had Entrez gene IDs, using the file HG-U133_Plus_2.na21.annot.csv, available at http://www.affy.com. We identified the RefSeq IDs associated with each Entrez gene ID using the gene2refseq file available at ftp://ftp.ncbi.nlm.nih.gov/gene/DATA. The genomic coordinate of each RefSeq ID's start transcription site was determined using the refgene table available at the UCSC Genome Bioinformatics website (http://www.genome.ucsc.edu/goldenPath/gbdDescriptions.html). A promoter sequence corresponding to each RefSeq ID was identified as the 1-kb sequence upstream of the start transcription site associated with that RefSeq ID, using chromosomal genomic sequences available at ftp://hgdownload.cse.ucsc.edu/goldenPath/hg18/chromosomes. Individual promoter sequences were submitted to TRANSFAC Professional, version 10.4 (48), an online database of 533 position weight matrices (an algorithm for scoring DNA sequence to identify a particular motif, in this case a TF binding site) for TF sites and scanned for TF binding sites using the program MATCH v10.4 with the settings “group of matrices” set to “vertebrates,” with “high quality matrices” selected. Our analysis was performed five separate ways for “cutoff selection,” set to “minimize false positives” and “minimize the sum of both error rates,” with “mat. sim.” and “core sim.” set (respectively) to 0.8 and 0.85, 0.9 and 0.9, and 0.95 and 0.95. Analysis for enrichment of TF binding sites was performed with P values calculated using a simple binomial test, and Q values were calculated (significance taken to be a Q value of <0.1) to correct for the FDR, exactly as previously described (62).
We repeated our analysis by defining promoters as the 1.5-kb or 2-kb sequences upstream of each gene and found no qualitative differences.
Flow cytometry.
Freshly isolated CD4+ T cells were resuspended in wash medium (106 to 2 × 106 cells in 100 μl). Cells were incubated with antibodies to cell surface markers on ice for 25 min. The following antibodies were used for surface staining of cells: PE-conjugated anti-CD25, PE anti-HLA-DR, PE mouse IgG1 isotype, PE mouse IgG2a isotype, and fluorescein isothiocyanate (FITC)-conjugated CD4.
For intracellular staining after we stained for the appropriate extracellular markers, cells were fixed and permeabilized with 50% ethanol overnight at −20°C. The following antibodies were used for intracellular staining as previously described (23): FITC cyclin A (clone BF683) and FITC cyclin B1 (clone GNS-1). 7-Aminoactinomycin was used to stain DNA. All antibodies and 7-aminoactinomycin were purchased from BD Biosciences. Data were collected by routinely acquiring >300,000 total events (based on forward- and side-scatter properties) with BD FACScalibur and CellQuest software. Fluorescence-activated cell sorting data were analyzed with CellQuest. P values were calculated using Welch's t test, comparing HIV+ samples to HIV− samples.
Microarray data accession number.
All microarray results have been deposited in the Gene Expression Omnibus database (accession number GSE9927).
RESULTS
Microarray analysis of in vivo-activated CD4+ T cells.
All individuals, regardless of their HIV status, have activated CD4+ T cells in their peripheral blood and in secondary lymphoid and peripheral tissues. However, HIV− individuals do not exhibit depletion of CD4+ T cells. If chronic activation of CD4+ T cells in the setting of HIV-1 infection drives a pathogenic process, then activated CD4+ T cells from untreated, HIV+ individuals should reflect this process. To gain insight into the changes that may arise in activated CD4+ T cells in the setting of HIV-1 infection, we performed a microarray study of transcriptional differences between activated CD4+ T cells from HIV+ and HIV− individuals. To gain the clearest understanding of transcriptional changes induced by chronic HIV-1 infection, we used strict inclusion criteria for both HIV+ and HIV− blood donors. HIV+ donors used for our microarray study met the following criteria: (i) lack of prior antiretroviral therapy or lack of antiretroviral therapy for more than 1 year prior to study entry, (ii) HIV+ viremia of >1,000 copies/ml for more than 1 year, and (iii) CD4 count of >200 cells/μl. Exclusion criteria for HIV+ donors were as follows: (i) chronic hepatitis B or hepatitis C infection and (ii) AIDS defining illness within 1 year of blood draw. For our microarray study, blood donors were also chosen to closely age match the HIV+ cohort with the HIV− cohort in order to avoid any confounding effects of age on immune function.
Furthermore, the transcriptional state of an activated CD4+ T cell is dynamic and changes with time, beginning at the initial activation event (76). Activated CD4+ T cells in the peripheral blood include cells that were activated over a wide temporal spectrum. In order to minimize the confounding effects of different temporal distributions of activated CD4+ T cells between the patient populations, we used CD25 as our marker of activation. CD25, the alpha chain of the interleukin 2 (IL-2) receptor, is up-regulated within 2 days of activation and is down-regulated several days later (16). Therefore, CD4+ CD25+ T cells represent a temporally discrete and relatively homogeneous population of activated CD4+ T cells. Although high-level expression of CD25 has also been used as a marker of regulatory T cells (Tregs), Tregs compose a very small fraction of the total activated CD4+ CD25+ T-cell population in human subjects (<5% in mice and considerably less in humans [61]). Additionally, by taking additional steps in cell purification to isolate all CD4+ CD25+ cells rather than just CD4+ CD25hi cells, we enriched for activated CD4+ CD25+ cells and reduced the contribution of these Tregs to our study. Finally, the results of our analyses do not show differential expression of any Treg-specific markers, such as FOXP3, CTLA4, and GITR, which strongly suggests that differential Treg composition of CD4+ CD25+ T cells from HIV− and untreated HIV+ individuals did not contribute to our observed results. Therefore, the contribution of these Tregs to our overall results is most likely insignificant.
For our microarray study, we used Affymetrix U133Plus 2.0 GeneChips, which probe for 54,630 different transcripts, representing 20,254 different Entrez genes. Raw microarray signals were preprocessed using the RMA method and analyzed to obtain P values for each probe set, with corresponding FDRs, from which we generated our list of differentially expressed genes.
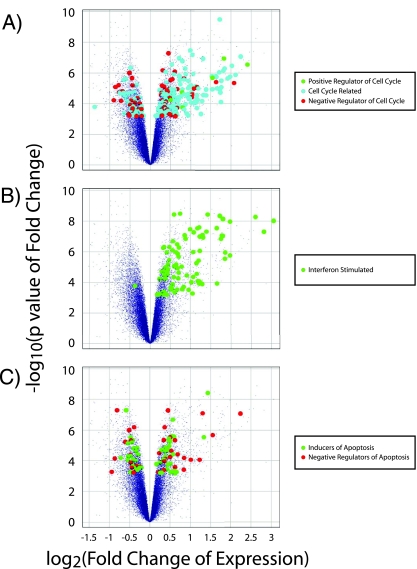
The results of the microarray study demonstrated that clear transcriptional differences exist between the CD4+ CD25+ T cells of HIV+ individuals and those of HIV− individuals (Fig. 1A). For a 1% FDR, analysis of microarray results yielded 2,110 transcripts that were 1.10- to 2.85-fold down-regulated and 1,633 transcripts that were 1.09- to 8.33-fold up-regulated in the CD4+ CD25+ T cells of chronically infected, untreated HIV+ individuals (Fig. 1B). When categorized with respect to the change in expression, the number of transcripts that were differentially expressed in CD4+ CD25+ T cells from HIV+ versus HIV− individuals decreased with increasing degree of change. This was true for both down- and up-regulated genes, but the transcripts showing the largest change (ranging from 3- to 8.33-fold differentially expressed) were transcripts that were all up-regulated in CD4+ CD25+ T cells from HIV+ versus HIV− individuals (Fig. 1B). There was no difference in CD25 expression between the HIV− and untreated HIV+ individuals, confirming that the purified cells were relatively homogeneous with respect to this activation marker. Genes that were differentially expressed in the activated CD4+ T cells of HIV+ individuals compared to expression for HIV− individuals belong to many different gene categories, including those involving the cell cycle, growth, cytokine responses, apoptosis, and survival, as well as metabolic genes (all with an unadjusted P value of <10−8 according to gene ontology [75]). Quite strikingly, the most highly up-regulated transcripts could almost entirely be categorized as related to the cell cycle or interferon signaling. As is discussed below, of the differentially expressed genes, cell cycle genes and interferon-stimulated genes stood out with respect to consistency of differential expression, number of differentially expressed genes, and overall change.
FIG. 1.
Differential expression of transcripts in CD4+ CD25+ T cells from untreated HIV+ individuals compared to expression in those from HIV− individuals. (A) Heat map of 3,743 differentially expressed transcripts in CD4+ CD25+ T cells from untreated HIV+ individuals compared to those in cells from HIV− individuals. Rows and columns are arranged by hierarchical clustering. Coloring was based on the log2 change over the row mean (mean expression across all samples). (B) The breakdown of the magnitudes of change for significantly up- and down-regulated transcripts in CD4+ CD25+ T cells from untreated HIV+ individuals compared to those in cells from HIV− individuals.
Cell cycle genes are up-regulated in untreated HIV+ patients.
Based on gene ontology annotations, we found 255 (out of 3,742 total) differentially expressed transcripts that were classified as involved in the cell cycle (without including transcripts of genes directly involved in signal transduction pathways) (unpublished data). Of these, 190 (out of 1,632 total) were up-regulated, while only 65 (out of 2,110 total) were down-regulated (Fig. 2A).
FIG. 2.
Differential expression profile of transcripts categorized as cell cycle associated, interferon stimulated, or regulators of apoptosis. Volcano plots of microarray results (small purple dots) with identification of differentially expressed positive regulators of the cell cycle (green circles), cell cycle-associated transcripts (blue circles), or negative regulators of the cell cycle (red circles) (A); interferon-stimulated genes (B), or inducers of apoptosis (green circles) or negative regulators of apoptosis (red circles) (C). The vertical axis represents significance [−log10(P value)], and the horizontal axis represents the change in gene expression for HIV+ individuals compared to that for HIV− individuals [−log2(fold change)].
The 190 up-regulated transcripts represented 150 different genes. These genes include regulators of the cell cycle as well as cell cycle-regulated genes (genes whose temporal expression is regulated by the cell cycle). Transcription of cell cycle-regulated genes from all phases of the cell cycle was found to be up-regulated. For example, transcription of the primary cyclins (cyclins D2, E, A, and B) from each phase of the cell cycle was up-regulated. Interestingly, the change in expression of up-regulated cell cycle-regulated transcripts increased for genes expressed later in the cell cycle. Differentially expressed G1-phase-associated genes (such as the cyclin D2 gene [CCND2], E2F1, E2F2, E2F3, and the Ki67 gene [MKI67]) were generally up-regulated between 1.5- and 2-fold, while differentially expressed S-phase-associated genes, such as cyclin A (CCNA2) and cyclin E (CCNE1 and CCNE2), were generally up-regulated between 2- and 3-fold. Differentially expressed G2/M-phase-associated genes (such as those encoding cdc2 [CDC2], cyclin B1 [CCNB1], cyclin B2 [CCNB2], BIRC5/survivin, and aurora kinase [AURKA, AURKB, and AURKC]) are mostly up-regulated by more than threefold. Furthermore, all differentially expressed positive regulators of the cell cycle (as defined by gene ontology) are up-regulated, while only a minority of differentially expressed negative regulators of the cell cycle (as defined by gene ontology) are up-regulated.
Some of the up-regulated cell cycle-related transcripts in CD4+ CD25+ cells from HIV+ individuals are associated with entry into the G1 phase. These include the TFs E2F1, E2F2, and E2F3 (55). Interestingly, these are genes whose transcription is associated specifically with entry into the cell cycle from quiescence or G0.
We observed 65 cell cycle-associated transcripts, representing 52 different genes, to be down-regulated from ∼1.2- to 2.5-fold in CD4+ CD25+ cells from HIV+ individuals compared to results for HIV− individuals. For some of these genes, the mechanistic link to the cell cycle is unclear (including the most highly down-regulated cell cycle-associated gene—the septin 10 gene [SEPT10]). Of the other down-regulated transcripts categorized as cell cycle related, most are known negative regulators of the cell cycle (or proliferation), such as that encoding cyclin L2 (CCLN2), HEXIM2, LZTS1, GADD45B, TSC1, and TUSC4, or mediators of cell cycle arrest, such as HBP1 and the sestrin 1 gene (SESN1). Interestingly, two cell cycle genes (MDM4 and RB1CC1) that we have identified to be down-regulated in CD4+ CD25+ T cells of HIV+ individuals are also known to be associated with cellular quiescence and the G0 phase of the cell cycle.
Interferon-related genes are up-regulated in untreated HIV+ patients.
The other major category of highly up-regulated transcripts in CD4+ CD25+ cells from HIV+ individuals compared to cells from HIV− individuals comprises interferon-stimulated genes. Importantly, almost all of the differentially expressed, interferon-stimulated transcripts were up-regulated rather than down-regulated (Fig. 2B). We found 94 up-regulated interferon-stimulated transcripts (from 1.13- to 8.33-fold), including those known to be up-regulated in response to type I interferons (e.g., alpha interferon and beta interferon), type II interferons (i.e., gamma interferon), or type I and II interferons (unpublished data). An extensive literature search revealed that the majority of interferon-stimulated genes up-regulated in CD4+ CD25+ T cells from untreated HIV+ individuals were type I responsive. Up-regulated interferon-stimulated genes included IFI44L, IFIT3, the gene for 2′,5′-oligoadenylate synthetase 1 (OAS1), OAS2, OAS3, OASL, IFI27, IFIT1, IFI44, G1P2 (ISG15), MxA (MX1), G1P3 (IFI6) (6-16), IFITM1 (9-27), STAT1, IL-12-RB, and IRF7. Of the most highly up-regulated interferon-stimulated transcripts, the majority represented genes known to be regulated specifically by type I interferons (alpha and beta interferons) (e.g., OAS1, 6-16, MxA, and IRF7) or primarily by type I interferons (e.g., STAT1 and 9-27) (17). Although up-regulated to a lesser extent, some type II interferon-regulated transcripts were also identified among the differentially expressed genes (e.g., IFI16, AIM2, and HLA-DRA). Interestingly, in activated CD4+ CD25+ T cells from untreated HIV+ individuals, we observed up-regulation of T-bet (TBX21), IL-12-RB, and PIM2, which are all induced by type I interferons and promote TH1 effector differentiation (1, 33). The up-regulation of these genes is consistent with previously published reports of a TH1 skewed immune response during chronic HIV-1 infection (15) and may be due to induction by type I interferons (33). These findings suggest that CD4+ T cells in untreated HIV+ individuals are activated in the presence of type I interferons.
Apoptosis-regulating genes are not differentially expressed in a consistent pattern in activated CD4+ T cells from HIV+ versus HIV− donors.
Interestingly, 93 of the differentially expressed transcripts (unpublished data), representing 68 different genes, were categorized by gene ontology as “induction of programmed cell death” and “negative regulation of programmed cell death” (Fig. 2C). However, there was no clear pattern suggesting a pro- or antiapoptotic phenotype. Of the differentially expressed apoptosis-related transcripts, 37 are negative regulators of apoptosis. Of these, 22 transcripts, including those of BIRC5/survivin, PIM2, and MYBL2, were up-regulated, while 15 transcripts including those for c-Myc (MYC), apoptosis inhibitor 5 (API5), and BIRC1 (NAIP), were down-regulated. We identified 61 differentially expressed transcripts that are considered to be inducers of programmed cell death. Of these, 32 transcripts, including those of BID, the BAK gene (BAK1), the p53 gene (TP53), the caspase 3 gene (CASP3), the caspase 7 gene (CASP7), and CD38 were up-regulated, while 24 transcripts, including PDCD5, PDCD6, FOXO3, and BCLAF1, were down-regulated.
Interestingly, the majority of these differentially expressed, apoptosis-related transcripts demonstrated small changes in CD4+ CD25+ T cells from HIV+ individuals compared to results for HIV− individuals. Furthermore, there is no obvious directionality of differential gene expression for either negative regulators or positive inducers of programmed cell death. This is clear from the intermingling of both groups of differentially expressed transcripts on the volcano plot (Fig. 2C). Therefore, the observed transcriptional changes do not suggest an obvious trend toward either increased apoptosis/decreased survival or decreased apoptosis/increased survival in CD4+ CD25+ T cells from HIV+ individuals compared to results for HIV− individuals.
Enrichment of proliferative and interferon signaling-related TF binding sites in the promoters of up-regulated genes.
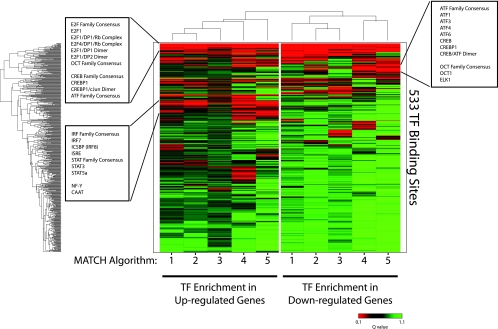
Having characterized the major differences in gene expression between activated CD4+ T cells from untreated HIV+ individuals and those from HIV− individuals, we sought to understand the proximal causes of these differences. We assumed that differential expression of sets of genes was the result of differential activation of transcriptional programs regulated by specific TFs. We hypothesized that the promoters of differentially expressed genes would be enriched in binding sites of TFs that are differentially activated in CD4+ CD25+ T cells from untreated HIV+ individuals compared to those from HIV− individuals. From an enrichment of TF binding sites, we could potentially trace the signals that drive the observed differences between activated CD4+ T cells from untreated HIV+ individuals and those from HIV− individuals. To determine whether particular TF binding sites were enriched in the promoters of genes that were differentially expressed between CD4+ CD25+ T cells from untreated HIV+ individuals compared to those from HIV− individuals, we performed a variation of a previously described promoter analysis (62). This method of analysis was previously used to predict activated TFs and signaling pathways in various cancers and to predict E2F-regulated transcripts (62). We used TRANSFAC Professional (48), a database of 533 position weight matrices for TF binding sites, to analyze the promoter (defined as 1 kb upstream of the start transcription site) of each transcript represented on the Affymetrix U133Plus 2.0 GeneChip with the program MATCH (39). For robustness, we performed our analysis with only high-quality position weight matrices in TRANSFAC and for five different core and matrix similarity score cutoffs (described in Materials and Methods) in MATCH. Having defined regulatory signatures (the set of all promoters that contain at least one binding site specified by a particular high-quality position weight matrix) for each TF binding site on the background of the U133Plus 2.0 GeneChip, we used a simple binomial test with an FDR (calculated as a Q value) cutoff of 0.10 to determine which TF binding sites were enriched in the set of genes that were up-regulated in CD4+ CD25+ T cells of untreated HIV+ individuals compared to results for HIV− individuals. For each MATCH matrix and similarity score cutoff, our analysis detected enrichment of particular TF binding sites in the promoters of genes whose transcripts are differentially expressed between CD4+ CD25+ T cells of HIV+ and HIV− individuals. Interestingly, we found that the TF binding sites enriched in the promoters of both up- and down-regulated genes were binding sites of TFs related to proliferation and type I interferon signaling. We observed three clusters of significantly enriched TF binding sites: (i) enriched in the promoters of all differentially expressed transcripts, (ii) enriched in only the promoters of down-regulated transcripts, and (iii) enriched in only the promoters of up-regulated transcripts (Fig. 3).
FIG. 3.
Enrichment of TF binding sites in the promoters of differentially expressed genes. Hierarchical clustering and heat map of Q values calculated for the enrichment of TF binding sites in the promoters of differentially expressed genes in CD4+ CD25+ T cells from untreated HIV+ individuals compared to those from HIV− individuals. The color represents the degree of significance for the Q value (green = low significance; red = high significance). Each row represents a specific TF binding site specified by a high-quality position weight matrix in TRANSFAC, while each column reflects a different algorithm (user-defined cutoff) for significance in identifying a TF binding site in MATCH: 1, minimize the sum of both error rates; 2, mat. sim. = 0.90; core sim. = 0.90; 3, mat. sim. = 0.80; core sim. = 0.85; 4, minimize false positives; 5, mat. sim. = 0.95; core sim. = 0.95.
We found that TF binding sites related to proliferation, in particular, binding sites for E2F-mediated transcription (e.g., general E2F family, E2F1, E2F1/DP2 dimer, and E2F1/DP1/Rb complex), as well as CREB/ATF-mediated transcription (general CREB family, general ATF family, CREBP1/c-Jun dimer, and CREBP1), were enriched in the promoters of all differentially expressed transcripts (Fig. 3). This is consistent with these TFs having dual roles as transcriptional activators and repressors during proliferation (49, 55). Interestingly, we found a small cluster of ATF family member and CREB family binding sites that are enriched in only the promoters of transcripts that are down-regulated in CD4+ CD25+ T cells from untreated HIV+ individuals compared to those from HIV− individuals (Fig. 3), which suggests that these TFs may be involved with down-regulation of certain genes in activated CD4+ T cells during HIV infection. However, because we found other binding sites for these same TFs enriched in the promoters of up-regulated transcripts (within the same cluster as E2F family members), it may be that other factors are regulating the binding of CREB and ATF family members at binding sites in the promoters of down-regulated transcripts (26). Finally, we observed a cluster of TF binding sites enriched in only the promoters of transcripts that are up-regulated in CD4+ CD25+ T cells from untreated HIV+ individuals compared to those from HIV− individuals. This cluster contains primarily binding sites for TFs that are downstream of type I interferon signaling, including IRF7, ISRE, and STAT family members (Fig. 3). These findings are consistent with the most dramatic transcriptional changes that we identified in our microarray study, i.e., cell cycle-related and type I-stimulated transcripts. Furthermore, these results suggest that active processes, involving type I interferon signaling and proliferative stimuli, are mediating the transcriptional changes in CD4+ CD25+ T cells of untreated HIV+ individuals compared to those of HIV− individuals. These results confirm our gene expression results that cell cycle-associated and type I interferon-stimulated genes comprise the most highly up-regulated genes in CD4+ CD25+ T cells of untreated HIV+ individuals compared to results for HIV− individuals.
Cell cycle distribution and protein expression in in vivo-activated CD4+ T cells from HIV+ individuals corroborate gene expression profiling and promoter analysis.
To test whether our transcript-level microarray findings reflect functional effects in vivo, we studied cell cycle parameters in CD4+ T cells activated in vivo from HIV− donors, from untreated chronically infected HIV+ patients, and from HIV+ patients on HAART. Using flow cytometry, we measured the expression of cyclin A (CCNA2) and cyclin B1 (CCNB1), whose two corresponding genes we have found to be up-regulated in CD4+ CD25+ cells from untreated HIV+ individuals, at the protein level in relation to cell cycle phase (reflected by a flow cytometric measurement of DNA quantity). Cyclin A is up-regulated late in G1 and begins to accumulate in the S phase of the cell cycle, mostly due to the action of E2F TFs. Interacting with both CDK2 and CDC2, cyclin A functions in both the S and G2/M phases of the cell cycle (79). Cyclin B1 is up-regulated late in S phase and accumulates through the G2/M phase of the cell cycle, due in part to regulatory effects of cyclin A, and is a critical component of the mitosis promoting factor complex (40). Cyclins A and B are both degraded during mitosis.
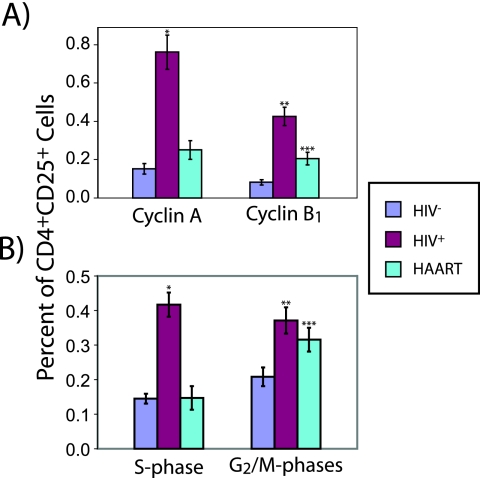
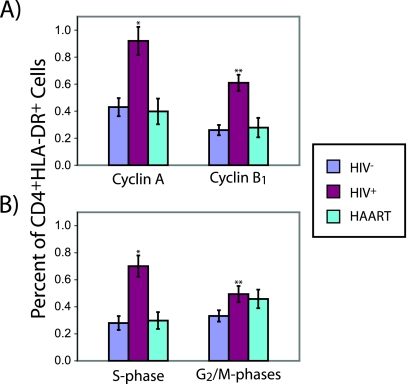
We found that a significantly larger fraction of CD4+ CD25+ T cells from untreated HIV+ individuals expressed cyclin A and/or cyclin B1 than was the case with those from HIV− individuals (Fig. 4A). The frequencies of CD4+ CD25+ cyclin A+ T cells and CD4+ CD25+ cyclin B1+ T cells in HIV+ individuals on HAART approached those for HIV− individuals. As expected, the increased fraction of CD4+ CD25+ cells expressing cyclin A and/or cyclin B1 was reflected by a significant shift toward post-G0/G1 phases of the cell cycle in CD4+ CD25+ cells from untreated HIV+ individuals. This shift was much less prominent for patients on HAART (Fig. 4B). In order to check whether these cell cycle findings were specific to CD4+ CD25+ T cells, we examined activated CD4+ T cells using a different activation marker: HLA-DR, which is up-regulated later than CD25 during the process of activation. We found an increase in the frequency of cells expressing cyclin A and/or cyclin B1 among CD4+ T cells positive for the late activation marker HLA-DR+ for untreated HIV+ individuals similar to that for HIV− individuals (Fig. 5A). This increase was much less prominent for patients on HAART. As expected, we also found a shift toward post-G0/G1 phases in the cell cycle distribution of CD4+ HLA-DR+ cells from untreated HIV+ individuals (Fig. 5B). This shift was also much less prominent for patients on HAART. Interestingly, we found that the percentages of cells expressing cyclin A and cyclin B1 were much higher among CD4+ CD25+ T cells from untreated HIV+ individuals than among CD4+ HLA-DR+ cells from HIV− donors (P = 0.014 and P = 0.013, respectively). This is particularly surprising since HLA-DR+ cells should be considerably further along in the cell cycle than CD4+ CD25+ cells. While the observed fraction of post-G0/G1 or cyclin A/B1-expressing activated CD4+ T cells in our study is low, these results are consistent with previous measurements of these parameters in CD4+ T cells from the peripheral blood, where cycling cells are much less common than in peripheral lymphoid organs (69). However, these results most likely reflect the state of activated CD4+ T cells in the lymph nodes of HIV+ individuals compared to those of HIV− individuals qualitatively, if not quantitatively.
FIG. 4.
Cell cycle parameters of in vivo-activated T cells (CD4+ CD25+). Percentage of in vivo-activated T cells (CD4+ CD25+) from HIV− (n = 11) or untreated HIV+ individuals (n = 15) or from HIV+ individuals on HAART (n = 7) expressing either cyclin A or B1 (P value for comparison to HIV− samples: *, <2 × 10−5; **, <5 × 10−6; ***, <0.01) (A) or in either the S or G2/M phase of the cell cycle (P value for comparison to HIV− samples: *, <5 × 10−7; **, <0.002; ***, <0.03) (B). Error bars represent one standard error.
FIG. 5.
Cell cycle parameters of in vivo-activated T cells (CD4+ HLA-DR+). Percentage of in vivo CD4+ HLA-DR+ T cells from HIV− (n = 9) or untreated HIV+ individuals (n = 14) or from HIV+ individuals on HAART (n = 6) expressing either cyclin A or B1 (P value for comparison to HIV− samples: *, <9 × 10−4; **, <9 × 10−5 (A) or in either the S or G2/M phase of the cell cycle (P value for comparison to HIV− samples: *, <2 × 10−4; **, <0.04) (B). Error bars represent one standard error.
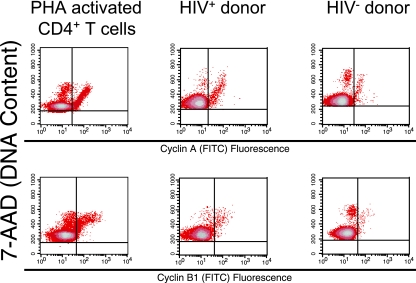
Finally, because we simultaneously measured CD25 and cyclin expression, as well as DNA content, in freshly isolated CD4+ T cells, we were able to examine the temporal pattern of cyclin expression. We found that cyclin A and cyclin B1 were expressed at appropriate phases of the cell cycle in CD4+ T cells activated in vivo from untreated HIV+ individuals. Indeed, the temporal pattern of cyclin A and cyclin B1 protein expression in CD4+ CD25+ T cells of untreated HIV+ individuals was not different from expression in CD4+ CD25+ T cells of HIV− individuals or phytohemagglutinin (PHA)-activated CD4+ T cells (Fig. 6). Specifically, we observe that in all samples, cyclin A expression increases from late G1 phase to the G2/M phases while cyclin B1 expression increases from S phase to the G2/M phases. We also observed appropriate temporal expression patterns for cyclin A and cyclin B1 in CD4+ HLA-DR+ T cells of untreated HIV+ individuals (data not shown).
FIG. 6.
Cell cycle phase expression of cyclins A and B1 in CD4+ T cells. Cell cycle phase expression of cyclin A and cyclin B1 in PHA-activated CD4+ T cells and CD4+ CD25+ T cells from HIV− or untreated HIV+ individuals.
DISCUSSION
A preponderance of data now support the hypothesis that chronic CD4+ T-cell activation plays a role in CD4+ T-cell depletion during HIV-1 infection (13, 22, 25, 28, 29, 41, 43, 50, 52, 54, 63, 65, 74). Despite numerous suggestive and correlative studies, a mechanism for the effect of chronic CD4+ T-cell activation on CD4 depletion has not yet been established. Proposed mechanisms include accelerated CD4+ T-cell senescence (3, 25, 34, 52, 63, 65), CD4+ T-cell exhaustion (25, 28, 34, 52, 63, 65), and activation-induced cell death (18, 22, 24, 25).
In this study, we present results that support a novel mechanism for the role of chronic CD4+ T-cell activation in CD4+ T-cell depletion. Our approach was based on the assumption that the pathogenic process leading to chronic CD4+ T-cell activation would be reflected by transcriptional changes in CD4+ T cells activated in vivo. We studied and compared in vivo-activated CD4+ T cells from untreated HIV+ individuals to in vivo-activated CD4+ T cells from healthy individuals. Using transcriptional profiling and analysis of the promoters of differentially expressed genes together with functional cell cycle analysis, we show that activated CD4+ T cells in untreated HIV+ individuals are in a hyperproliferative state under the influence of type I interferons. The power of our approach and our ability to see such clear differences is largely a result of our focus on a homogeneous population of activated CD4+ CD25+ T cells, rather than total CD4+ T cells, which are a heterogeneous population of cells, the composition of which varies from person to person. Because of the relatively small fraction of CD4+ CD25+ T cells that are Tregs and our experimental steps to enrich for activated CD4+ CD25+ T cells, as well as the fact that we did not observe differential Treg-specific gene expression (e.g., FOXP3, CTLA4, and GITR), we believe that our results reflect true transcriptional differences between activated CD4+ CD25+ T cells from untreated HIV+ individuals and in vivo-activated CD4+ CD25+ T cells from healthy individuals. Nonetheless, our findings also apply to activated CD4+ T cells defined in another way, by expression of HLA-DR. Although chronically infected HIV+ individuals have a significantly larger fraction of memory CD4+ T cells than do HIV− individuals (19), we do not believe that this was a significant factor contributing to the observed hyperproliferative state of CD4+ CD25+ T cells in untreated HIV+ individuals. In our microarray screen, we observed no significant difference in the expression levels of CD62L, CD40L, CD11a, or CD27 (all markers that can distinguish naive from memory CD4+ T cells) between the HIV− and untreated HIV+ groups. Furthermore, we found that a larger fraction of CD4+ CD25+ cells from untreated HIV+ individuals express cyclin A and cyclin B1 and are also beyond the G0/G1 phases of the cell cycle than was the case even for CD4+ HLA-DR+ cells from HIV− individuals. These observations lead us to believe that the hyperproliferative state of CD4+ CD25+ T cells in HIV+ individuals is a reflection of the pathophysiologic changes induced by HIV-1 infection.
Many previous studies have demonstrated increases in apoptotic markers, such as Fas and FasL, on total CD4+ T cells in HIV-1 infection (18, 24, 54). These increases correlated with increased CD4+ T-cell activation (18, 24, 54), suggesting a mechanism for HIV-induced CD4+ T-cell depletion. However, in focusing on a relatively homogeneous population of cells, we observed only a modest increase in the transcription of some apoptotic genes in activated CD4+ T cells from HIV+ individuals compared to results for HIV− individuals. We do not discount the modest transcriptional changes in proapoptotic gene products observed in our screen, since some of these genes are known mediators of death in the setting of viral infections and type I interferon signaling (12a, 74a). However, our results suggest that bystander death by apoptosis of activated CD4+ T cells in HIV+ individuals likely occurs at later stages of activation.
It has also been reported that CD4+ T cells from viremic HIV+ individuals exhibit cell cycle dysregulation, which resolves upon treatment with HAART (11, 57). Similarly, cell cycle dysregulation has been observed in CD4+ T cells of SIV-infected rhesus macaques (an AIDS-susceptible primate species), the degree of which correlates with immune activation and T-cell apoptosis, while cell cycle dysregulation is not observed in the CD4+ T cells of SIV-infected sooty mangabeys (a natural host of SIV) (58). Because these reports are based on correlative studies of the cell cycle markers that are confounded by focus on total CD4+ T-cell populations, it is difficult to compare these results to those of the present study. Here, we used a direct experimental approach with flow cytometry to visualize simultaneously the temporal expression patterns of cyclins A and B1 in specific activated subsets (CD25+ or HLA-DR+) of CD4+ T cells and found no difference between the temporal expression patterns of cyclins A and B1 in activated CD4+ T cells from untreated, viremic HIV+ individuals and those of cells from HIV− individuals or of PHA-activated CD4+ T cells.
Our finding that activated CD4+ T cells are under the influence of type I interferons in the setting of HIV-1 infection is consistent with findings of previous studies documenting type I interferon effects in CD4+ T cells from the secondary lymphoid tissues of HIV+ individuals (5, 32, 35, 44, 53, 81). However, we extend these previous studies by specifically studying the subset of activated (CD25+) CD4+ T cells from HIV+ individuals with a comparison to activated cells from HIV− individuals. Without purification of these activated cells, it is not possible to know whether previously observed interferon-stimulated transcriptional changes represent simply a change in the proportion of activated cells in the overall CD4+ T-cell population or a real qualitative change in the character of the activated cells. Recent work has implicated type I interferons as important mediators of CD4+ T-cell death in the setting of HIV-1 infection (31, 32), in particular through the up-regulation of TRAIL and Fas. These in vitro findings are consistent with in vitro studies suggesting a negative regulatory role for type I interferons in CD4+ T-cell survival and proliferation (82). However, the consequences of interferon signaling for CD4+ T cells are clearly dependent on the integration of context-specific environmental signals. Previous in vivo studies have demonstrated that type I interferons not only are necessary for a proper antiviral CD4+ T-cell response (27) but may also impart strong proliferative and survival signals to CD4+ T cells (46, 47). Furthermore, type I interferons enhance in vitro and in vivo TH1 effector differentiation through various direct and indirect mechanisms, including up-regulation of the IL-12 receptor and T-bet (60, 64, 73), which is consistent with our observation that IL-12 receptor beta chain 2 and the TH1-specific TF T-bet are up-regulated in activated CD4+ T cells from HIV+ individuals compared to expression in cells from HIV− individuals. In the context of these in vivo studies, our results suggest that type I interferons may be responsible for many of the major differences in gene expression (e.g., up-regulation of cell cycle-related and interferon-stimulated transcripts) between activated CD4+ T cells from HIV+ individuals and those from HIV− individuals.
Based on our findings and previously published reports on CD4+ T-cell dynamics, we propose a model for the role of chronic CD4+ T-cell activation in HIV-1-mediated CD4+ T-cell depletion and the development of AIDS. Recent studies have shown that CD4+ T cells differentiate along a branching, rather than linear, pathway from the naive to effector to memory states (12, 68, 78). The process of activation and subsequent proliferation leads to asymmetric production of long-lived memory cells and short-lived effector cells. In this way, normal CD4+ T-cell dynamics maintain a roughly steady-state level of naive, memory, and effector cells in healthy individuals (Fig. 7A). Furthermore, consistent with our transcriptional profiling, type I interferons may affect this steady state by shifting the balance toward TH1 effectors (42) through up-regulation of IL-12RB2 (33), which also leads to subsequent up-regulation of T-bet by enhancing sensitivity to IL-12. In the context of this straightforward model of CD4+ T-cell dynamics, it is easy to see how type I interferons could decrease the steady-state number of CD4+ T cells by shifting a greater fraction of activated cells to the short-lived effector compartment (Fig. 7B). Our model is consistent with the recent findings of Okoye et al. with the SIV model (56). Their results indicate that while effector memory cells are rapidly depleted in SIV-infected macaques, depletion of central memory cells, which give rise to effector memory cells, is the underlying cause of CD4+ T-cell depletion in SIV infection. In this elegant study, Okoye et al. suggest that viral infection and direct killing are the underlying cause of central memory T-cell depletion in SIV infection (56). Our model likewise predicts a gradual depletion of the memory compartment. Our results are consistent with the idea that in HIV-1 infection, depletion of memory T cells may also occur through the type I interferon-mediated skewing of T-cell differentiation, which results in an increased number of cells that are susceptible to destruction by activation-induced cell death or by direct infection. While we cannot rule out direct viral killing in the setting of HIV-1 infection, we believe that our model may describe a primary mechanism for CD4+ T-cell depletion in chronic infection, as supported by the findings that the frequency of infected CD4+ T cells in both peripheral blood and the lymphoid tissue appears to be very low during chronic HIV-1 infection (14, 20).
FIG. 7.
A model of CD4+ T-cell dynamics. A model of CD4+ T-cell dynamics in a healthy individual (A) or in an untreated HIV+ individual (B), where HIV-1 antigens enhance the rate of activation of naive and memory cells while type I interferons skew the activated T-cell differentiation toward short-lived effector cells instead of long-lived memory cells.
This model is consistent with recent work demonstrating an increased frequency of effector CD4+ T cells in the lymph nodes of untreated HIV+ individuals (6). In this model, the fraction of activated CD4+ T cells differentiating into short-lived effectors (reflected by the number of activated CD4+ T cells) governs the degree of CD4+ T-cell depletion, since an increase in this parameter pushes a larger fraction of CD4+ T cells toward the short-lived effector phenotype rather than the long-lived memory phenotype. Furthermore, because the vast majority of activated CD4+ T cells in HIV+ individuals are not HIV-1 antigen specific (19, 25), this model demonstrates how type I interferons could cause an expansion of short-lived activated CD4+ T cells independently of HIV-1 antigens. During chronic HIV-1 infection, the steady-state number of CD4+ T cells may gradually decrease as increasing levels of type I interferons drive more activated CD4+ T cells toward effector differentiation (43, 74). In the setting of HAART, when there is significantly less viral antigen, the rate at which activated CD4+ T cells differentiate into effector cells is lower and likely does not increase with time. Therefore, the CD4+ T-cell count reaches a steady-state level that depends on the degree of local type I interferon secretion and fibrosis of lymph nodes, preventing repopulation by CD4+ T cells (66, 67).
Interestingly, type I interferons are secreted in response to many viral infections that do not cause CD4+ T-cell depletion. We propose that the main difference between the beneficial effects of type I interferons during other viral infections and the potentially harmful effects of type I interferons in the setting of HIV-1 infection lies in recruitment of plasmacytoid dendritic cells (pDCs), the major type I interferon-producing cell type, to the location of active viral replication. In the setting of HIV-1 infection, pDCs are recruited to the major sites of active viral replication which lie in the secondary lymphoid tissues (32). At these sites, resting CD4+ T cells are activated, and therefore, resting CD4+ T cells going through the early stages of the activation are more likely to be exposed to and affected by higher levels of type I interferons than in other viral infections not localized in the secondary lymphoid tissues. The dominant and concentration-dependent effects of type I interferons during the early stages of T-cell activation may be the difference between other viral infections and HIV infection, where a dramatic shift in normal CD4+ T-cell dynamics may be manifested as a reduced CD4+ T-cell count. CD8+ T cells are also activated in the secondary lymphoid tissues. However, CD8+ T cells, unlike CD4+ T cells, appear to have a linear pathway of differentiation (68). This linear pathway may prevent cytokines, such as type I interferons, from skewing differentiation of activated CD8+ T cells from the long-lived memory pool to the short-lived effector pool. Therefore, the differentiation pathway of CD8+ T cells may render them relatively immune to depletion by type I interferons.
Finally, the TH1 immune response against HIV-1 primarily controls viral replication by promoting a CTL response. Because our results are strongly suggestive that type I interferons may play a major role in stimulation and maintenance of the TH1 phenotype during chronic infection, the transition to AIDS and subsequent uncontrolled viral replication may be due to the failure of the innate immune system to produce type I interferons, which is consistent with recent observations that pDCs are depleted in the lymph nodes of SIV-infected macaques with AIDS (10). This suggests that type I interferons are important for maintaining some control over viral replication.
It appears, then, that the complex interaction of a greater-than-normal innate response (mediated by type I interferons) and the adaptive response (mediated by CD4+ and CD8+ T cells) in the environment of secondary lymphoid tissues acts as a double-edged sword that both controls viremia and causes CD4+ T-cell depletion. Therefore, we suggest that CD4+ T-cell depletion during chronic and end-stage (AIDS) HIV-1 infection are mediated by two separate mechanisms that may be linked by the actions of type I interferons. In the case of chronic infection, CD4+ T-cell depletion occurs as a result of preferential differentiation of CD4+ T cells under the influence of type I interferons into short-lived effectors. CD4+ T-cell depletion during end-stage HIV-1 infection and AIDS is a consequence of the failure of type I interferon action on CD4+ and CD8+ T cells (from either decreased pDCs or perhaps even decreased T-cell responsiveness), resulting in a loss of effective control of viral replication. There remain, however, several unanswered questions. In particular, what is the mechanism that slowly increases type I interferon secretion and thus drives down the CD4+ T-cell count during chronic infection (e.g., viral evolution [80] or microbial translocation across compromised mucosal barriers [8])? Also, what is the mechanism for type I interferon failure that would mark the dramatic changes observed during the transition from chronic HIV-1 infection to AIDS (e.g., T-cell exhaustion/senescence [3, 28] or depletion of pDCs [10])? We hope that future work can be directed at modulating these mechanisms for therapeutic applications.
Acknowledgments
We acknowledge the generous contribution of all blood donors who participated in this study.
This work was supported by NIH grants AI43222 and AI51178, by a grant from the Doris Duke Charitable Foundation, and by the Howard Hughes Medical Institute.
We declare that we have no competing financial interests.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Aho, T. L., R. J. Lund, E. K. Ylikoski, S. Matikainen, R. Lahesmaa, and P. J. Koskinen. 2005. Expression of human pim family genes is selectively up-regulated by cytokines promoting T helper type 1, but not T helper type 2, cell differentiation. Immunology 11682-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, R. W., M. S. Ascher, and H. W. Sheppard. 1998. Direct HIV cytopathicity cannot account for CD4 decline in AIDS in the presence of homeostasis: a worst-case dynamic analysis. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 17245-252. [DOI] [PubMed] [Google Scholar]
- 3.Appay, V., J. R. Almeida, D. Sauce, B. Autran, and L. Papagno. 2007. Accelerated immune senescence and HIV-1 infection. Exp. Gerontol. 42432-437. [DOI] [PubMed] [Google Scholar]
- 4.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, G. Sherlock, et al. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 2525-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Audige, A., M. Urosevic, E. Schlaepfer, R. Walker, D. Powell, S. Hallenberger, H. Joller, H. U. Simon, R. Dummer, and R. F. Speck. 2006. Anti-HIV state but not apoptosis depends on IFN signature in CD4+ T cells. J. Immunol. 1776227-6237. [DOI] [PubMed] [Google Scholar]
- 6.Biancotto, A., J. C. Grivel, S. J. Iglehart, C. Vanpouille, A. Lisco, S. F. Sieg, R. Debernardo, K. Garate, B. Rodriguez, L. B. Margolis, and M. M. Lederman. 2007. Abnormal activation and cytokine spectra in lymph nodes of persons chronically infected with HIV-1. Blood 1094272-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19185-193. [DOI] [PubMed] [Google Scholar]
- 8.Brenchley, J. M., D. A. Price, T. W. Schacker, T. E. Asher, G. Silvestri, S. Rao, Z. Kazzaz, E. Bornstein, O. Lambotte, D. Altmann, B. R. Blazar, B. Rodriguez, L. Teixeira-Johnson, A. Landay, J. N. Martin, F. M. Hecht, L. J. Picker, M. M. Lederman, S. G. Deeks, and D. C. Douek. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 121365-1371. [DOI] [PubMed] [Google Scholar]
- 9.Brenchley, J. M., T. W. Schacker, L. E. Ruff, D. A. Price, J. H. Taylor, G. J. Beilman, P. L. Nguyen, A. Khoruts, M. Larson, A. T. Haase, and D. C. Douek. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, K. N., A. Trichel, and S. M. Barratt-Boyes. 2007. Parallel loss of myeloid and plasmacytoid dendritic cells from blood and lymphoid tissue in simian AIDS. J. Immunol. 1786958-6967. [DOI] [PubMed] [Google Scholar]
- 11.Cannavo', G., M. Paiardini, D. Galati, B. Cervasi, M. Montroni, G. De Vico, D. Guetard, M. L. Bocchino, I. Picerno, M. Magnani, G. Silvestri, and G. Piedimonte. 2001. Abnormal intracellular kinetics of cell-cycle-dependent proteins in lymphocytes from patients infected with human immunodeficiency virus: a novel biologic link between immune activation, accelerated T-cell turnover, and high levels of apoptosis. Blood 971756-1764. [DOI] [PubMed] [Google Scholar]
- 12.Chang, J. T., V. R. Palanivel, I. Kinjyo, F. Schambach, A. M. Intlekofer, A. Banerjee, S. A. Longworth, K. E. Vinup, P. Mrass, J. Oliaro, N. Killeen, J. S. Orange, S. M. Russell, W. Weninger, and S. L. Reiner. 2007. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 3151687-1691. [DOI] [PubMed] [Google Scholar]
- 12a.Chawla-Sarkar, M., D. J. Lindner, Y. F. Liu, B. R. Williams, G. C. Sen, R. H. Silverman, and E. C. Borden. 2003. Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis 8237-249. [DOI] [PubMed] [Google Scholar]
- 13.Choudhary, S. K., N. Vrisekoop, C. A. Jansen, S. A. Otto, H. Schuitemaker, F. Miedema, and D. Camerini. 2007. Low immune activation despite high levels of pathogenic human immunodeficiency virus type 1 results in long-term asymptomatic disease. J. Virol. 818838-8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387183-188. [DOI] [PubMed] [Google Scholar]
- 15.Clerici, M., and G. M. Shearer. 1993. A TH1→TH2 switch is a critical step in the etiology of HIV infection. Immunol. Today 14107-111. [DOI] [PubMed] [Google Scholar]
- 16.Depper, J. M., W. J. Leonard, M. Kronke, P. D. Noguchi, R. E. Cunningham, T. A. Waldmann, and W. C. Greene. 1984. Regulation of interleukin 2 receptor expression: effects of phorbol diester, phospholipase C, and reexposure to lectin or antigen. J. Immunol. 1333054-3061. [PubMed] [Google Scholar]
- 17.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 9515623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dockrell, D. H., A. D. Badley, A. Geciras-Schimnich, M. Simpson, R. Schut, D. H. Lynch, and C. V. Paya. 1999. Activation-induced CD4+ T cell death in HIV-positive individuals correlates with Fas susceptibility, CD4+ T cell count, and HIV plasma viral copy number. AIDS Res. Hum. Retrovir. 151509-1518. [DOI] [PubMed] [Google Scholar]
- 19.Douek, D. C., L. J. Picker, and R. A. Koup. 2003. T cell dynamics in HIV-1 infection. Annu. Rev. Immunol. 21265-304. [DOI] [PubMed] [Google Scholar]
- 20.Finkel, T. H., G. Tudor-Williams, N. K. Banda, M. F. Cotton, T. Curiel, C. Monks, T. W. Baba, R. M. Ruprecht, and A. Kupfer. 1995. Apoptosis occurs predominantly in bystander cells and not in productively infected cells of HIV- and SIV-infected lymph nodes. Nat. Med. 1129-134. [DOI] [PubMed] [Google Scholar]
- 21.Finzi, D., and R. F. Siliciano. 1998. Viral dynamics in HIV-1 infection. Cell 93665-671. [DOI] [PubMed] [Google Scholar]
- 22.Giorgi, J. V., L. E. Hultin, J. A. McKeating, T. D. Johnson, B. Owens, L. P. Jacobson, R. Shih, J. Lewis, D. J. Wiley, J. P. Phair, S. M. Wolinsky, and R. Detels. 1999. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J. Infect. Dis. 179859-870. [DOI] [PubMed] [Google Scholar]
- 23.Gong, J., F. Traganos, and Z. Darzynkiewicz. 1995. Growth imbalance and altered expression of cyclins B1, A, E, and D3 in MOLT-4 cells synchronized in the cell cycle by inhibitors of DNA replication. Cell Growth Differ. 61485-1493. [PubMed] [Google Scholar]
- 24.Gougeon, M. L. 2003. Apoptosis as an HIV strategy to escape immune attack. Nat. Rev. Immunol. 3392-404. [DOI] [PubMed] [Google Scholar]
- 25.Grossman, Z., M. Meier-Schellersheim, A. E. Sousa, R. M. Victorino, and W. E. Paul. 2002. CD4+ T-cell depletion in HIV infection: are we closer to understanding the cause? Nat. Med. 8319-323. [DOI] [PubMed] [Google Scholar]
- 26.Haus-Seuffert, P., and M. Meisterernst. 2000. Mechanisms of transcriptional activation of cAMP-responsive element-binding protein CREB. Mol. Cell Biochem. 2125-9. [PubMed] [Google Scholar]
- 27.Havenar-Daughton, C., G. A. Kolumam, and K. Murali-Krishna. 2006. Cutting edge: the direct action of type I IFN on CD4 T cells is critical for sustaining clonal expansion in response to a viral but not a bacterial infection. J. Immunol. 1763315-3319. [DOI] [PubMed] [Google Scholar]
- 28.Hazenberg, M. D., D. Hamann, H. Schuitemaker, and F. Miedema. 2000. T cell depletion in HIV-1 infection: how CD4+ T cells go out of stock. Nat. Immunol. 1285-289. [DOI] [PubMed] [Google Scholar]
- 29.Hazenberg, M. D., J. W. Stuart, S. A. Otto, J. C. Borleffs, C. A. Boucher, R. J. De Boer, F. Miedema, and D. Hamann. 2000. T-cell division in human immunodeficiency virus (HIV)-1 infection is mainly due to immune activation: a longitudinal analysis in patients before and during highly active antiretroviral therapy (HAART). Blood 95249-255. [PubMed] [Google Scholar]
- 30.Hellerstein, M. K., R. A. Hoh, M. B. Hanley, D. Cesar, D. Lee, R. A. Neese, and J. M. McCune. 2003. Subpopulations of long-lived and short-lived T cells in advanced HIV-1 infection. J. Clin. Investig. 112956-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herbeuval, J. P., J. C. Grivel, A. Boasso, A. W. Hardy, C. Chougnet, M. J. Dolan, H. Yagita, J. D. Lifson, and G. M. Shearer. 2005. CD4+ T-cell death induced by infectious and noninfectious HIV-1: role of type 1 interferon-dependent, TRAIL/DR5-mediated apoptosis. Blood 1063524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbeuval, J. P., A. W. Hardy, A. Boasso, S. A. Anderson, M. J. Dolan, M. Dy, and G. M. Shearer. 2005. Regulation of TNF-related apoptosis-inducing ligand on primary CD4+ T cells by HIV-1: role of type I IFN-producing plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. USA 10213974-13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hibbert, L., S. Pflanz, M. R. De Waal, and R. A. Kastelein. 2003. IL-27 and IFN-alpha signal via Stat1 and Stat3 and induce T-Bet and IL-12Rbeta2 in naive T cells. J. Interferon Cytokine Res. 23513-522. [DOI] [PubMed] [Google Scholar]
- 34.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373123-126. [DOI] [PubMed] [Google Scholar]
- 35.Hyrcza, M. D., C. Kovacs, M. Loutfy, R. Halpenny, L. Heisler, S. Yang, O. Wilkins, M. Ostrowski, and S. D. Der. 2007. Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells. J. Virol. 813477-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ihaka, R., and R. Gentleman. 1996. A language for data analysis and graphics. J. Comput. Graph. Stat. 5299-314. [Google Scholar]
- 37.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4249-264. [DOI] [PubMed] [Google Scholar]
- 38.Kaur, A., R. M. Grant, R. E. Means, H. McClure, M. Feinberg, and R. P. Johnson. 1998. Diverse host responses and outcomes following simian immunodeficiency virus SIVmac239 infection in sooty mangabeys and rhesus macaques. J. Virol. 729597-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kel, A. E., E. Gossling, I. Reuter, E. Cheremushkin, O. V. Kel-Margoulis, and E. Wingender. 2003. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 313576-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kishimoto, T., and E. Okumura. 1997. In vivo regulation of the entry into M-phase: initial activation and nuclear translocation of cyclin B/Cdc2. Prog. Cell Cycle Res. 3241-249. [DOI] [PubMed] [Google Scholar]
- 41.Kovacs, J. A., R. A. Lempicki, I. A. Sidorov, J. W. Adelsberger, B. Herpin, J. A. Metcalf, I. Sereti, M. A. Polis, R. T. Davey, J. Tavel, J. Falloon, R. Stevens, L. Lambert, R. Dewar, D. J. Schwartzentruber, M. R. Anver, M. W. Baseler, H. Masur, D. S. Dimitrov, and H. C. Lane. 2001. Identification of dynamically distinct subpopulations of T lymphocytes that are differentially affected by HIV. J. Exp. Med. 1941731-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krug, A., R. Veeraswamy, A. Pekosz, O. Kanagawa, E. R. Unanue, M. Colonna, and M. Cella. 2003. Interferon-producing cells fail to induce proliferation of naive T cells but can promote expansion and T helper 1 differentiation of antigen-experienced unpolarized T cells. J. Exp. Med. 197899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leng, Q., G. Borkow, Z. Weisman, M. Stein, A. Kalinkovich, and Z. Bentwich. 2001. Immune activation correlates better than HIV plasma viral load with CD4 T-cell decline during HIV infection. J. Acquir. Immune Defic. Syndr. 27389-397. [DOI] [PubMed] [Google Scholar]
- 44.Li, Q., T. Schacker, J. Carlis, G. Beilman, P. Nguyen, and A. T. Haase. 2004. Functional genomic analysis of the response of HIV-1-infected lymphatic tissue to antiretroviral therapy. J. Infect. Dis. 189572-582. [DOI] [PubMed] [Google Scholar]
- 45.Lim, S. G., A. Condez, C. A. Lee, M. A. Johnson, C. Elia, and L. W. Poulter. 1993. Loss of mucosal CD4 lymphocytes is an early feature of HIV infection. Clin. Exp. Immunol. 92448-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marrack, P., J. Kappler, and T. Mitchell. 1999. Type I interferons keep activated T cells alive. J. Exp. Med. 189521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matikainen, S., T. Sareneva, T. Ronni, A. Lehtonen, P. J. Koskinen, and I. Julkunen. 1999. Interferon-alpha activates multiple STAT proteins and upregulates proliferation-associated IL-2Ralpha, c-myc, and pim-1 genes in human T cells. Blood 931980-1991. [PubMed] [Google Scholar]
- 48.Matys, V., E. Fricke, R. Geffers, E. Gossling, M. Haubrock, R. Hehl, K. Hornischer, D. Karas, A. E. Kel, O. V. Kel-Margoulis, D. U. Kloos, S. Land, B. Lewicki-Potapov, H. Michael, R. Munch, I. Reuter, S. Rotert, H. Saxel, M. Scheer, S. Thiele, and E. Wingender. 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31374-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayr, B., and M. Montminy. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2599-609. [DOI] [PubMed] [Google Scholar]
- 50.McCune, J. M. 2001. The dynamics of CD4+ T-cell depletion in HIV disease. Nature 410974-979. [DOI] [PubMed] [Google Scholar]
- 51.Mehandru, S., M. A. Poles, K. Tenner-Racz, A. Horowitz, A. Hurley, C. Hogan, D. Boden, P. Racz, and M. Markowitz. 2004. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 200761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohri, H., S. Bonhoeffer, S. Monard, A. S. Perelson, and D. D. Ho. 1998. Rapid turnover of T lymphocytes in SIV-infected rhesus macaques. Science 2791223-1227. [DOI] [PubMed] [Google Scholar]
- 53.Moir, S., A. Malaspina, O. K. Pickeral, E. T. Donoghue, J. Vasquez, N. J. Miller, S. R. Krishnan, M. A. Planta, J. F. Turney, J. S. Justement, S. Kottilil, M. Dybul, J. M. Mican, C. Kovacs, T. W. Chun, C. E. Birse, and A. S. Fauci. 2004. Decreased survival of B cells of HIV-viremic patients mediated by altered expression of receptors of the TNF superfamily. J. Exp. Med. 200587-599. [PubMed] [Google Scholar]
- 54.Muro-Cacho, C. A., G. Pantaleo, and A. S. Fauci. 1995. Analysis of apoptosis in lymph nodes of HIV-infected persons. Intensity of apoptosis correlates with the general state of activation of the lymphoid tissue and not with stage of disease or viral burden. J. Immunol. 1545555-5566. [PubMed] [Google Scholar]
- 55.Nevins, J. R. 1998. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 9585-593. [PubMed] [Google Scholar]
- 56.Okoye, A., M. Meier-Schellersheim, J. M. Brenchley, S. I. Hagen, J. M. Walker, M. Rohankhedkar, R. Lum, J. B. Edgar, S. L. Planer, A. Legasse, A. W. Sylwester, M. Piatak, Jr., J. D. Lifson, V. C. Maino, D. L. Sodora, D. C. Douek, M. K. Axthelm, Z. Grossman, and L. J. Picker. 2007. Progressive CD4+ central memory T cell decline results in CD4+ effector memory insufficiency and overt disease in chronic SIV infection. J. Exp. Med. 2042171-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paiardini, M., B. Cervasi, D. Galati, S. Dominici, H. Albrecht, A. Sfacteria, M. Magnani, G. Silvestri, and G. Piedimonte. 2004. Early correction of cell cycle perturbations predicts the immunological response to therapy in HIV-infected patients. AIDS 18393-402. [DOI] [PubMed] [Google Scholar]
- 58.Paiardini, M., B. Cervasi, B. Sumpter, H. M. McClure, D. L. Sodora, M. Magnani, S. I. Staprans, G. Piedimonte, and G. Silvestri. 2006. Perturbations of cell cycle control in T cells contribute to the different outcomes of simian immunodeficiency virus infection in rhesus macaques and sooty mangabeys. J. Virol. 80634-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pantaleo, G., and A. S. Fauci. 1995. New concepts in the immunopathogenesis of HIV infection. Annu. Rev. Immunol. 13487-512. [DOI] [PubMed] [Google Scholar]
- 60.Parronchi, P., S. Mohapatra, S. Sampognaro, L. Giannarini, U. Wahn, P. Chong, S. Mohapatra, E. Maggi, H. Renz, and S. Romagnani. 1996. Effects of interferon-alpha on cytokine profile, T cell receptor repertoire and peptide reactivity of human allergen-specific T cells. Eur. J. Immunol. 26697-703. [DOI] [PubMed] [Google Scholar]
- 61.Piccirillo, C. A., and E. M. Shevach. 2004. Naturally-occurring CD4+ CD25+ immunoregulatory T cells: central players in the arena of peripheral tolerance. Semin. Immunol. 1681-88. [DOI] [PubMed] [Google Scholar]
- 62.Rhodes, D. R., S. Kalyana-Sundaram, V. Mahavisno, T. R. Barrette, D. Ghosh, and A. M. Chinnaiyan. 2005. Mining for regulatory programs in the cancer transcriptome. Nat. Genet. 37579-583. [DOI] [PubMed] [Google Scholar]
- 63.Ribeiro, R. M., H. Mohri, D. D. Ho, and A. S. Perelson. 2002. In vivo dynamics of T cell activation, proliferation, and death in HIV-1 infection: why are CD4+ but not CD8+ T cells depleted? Proc. Natl. Acad. Sci. USA 9915572-15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rogge, L., D. D'Ambrosio, M. Biffi, G. Penna, L. J. Minetti, D. H. Presky, L. Adorini, and F. Sinigaglia. 1998. The role of Stat4 in species-specific regulation of Th cell development by type I IFNs. J. Immunol. 1616567-6574. [PubMed] [Google Scholar]
- 65.Rosenzweig, M., M. A. DeMaria, D. M. Harper, S. Friedrich, R. K. Jain, and R. P. Johnson. 1998. Increased rates of CD4(+) and CD8(+) T lymphocyte turnover in simian immunodeficiency virus-infected macaques. Proc. Natl. Acad. Sci. USA 956388-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schacker, T. W., J. M. Brenchley, G. J. Beilman, C. Reilly, S. E. Pambuccian, J. Taylor, D. Skarda, M. Larson, D. C. Douek, and A. T. Haase. 2006. Lymphatic tissue fibrosis is associated with reduced numbers of naive CD4+ T cells in human immunodeficiency virus type 1 infection. Clin. Vaccine Immunol. 13556-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schacker, T. W., P. L. Nguyen, G. J. Beilman, S. Wolinsky, M. Larson, C. Reilly, and A. T. Haase. 2002. Collagen deposition in HIV-1 infected lymphatic tissues and T cell homeostasis. J. Clin. Investig. 1101133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seder, R. A., and R. Ahmed. 2003. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat. Immunol. 4835-842. [DOI] [PubMed] [Google Scholar]
- 69.Sieg, S. F., B. Rodriguez, R. Asaad, W. Jiang, D. A. Bazdar, and M. M. Lederman. 2005. Peripheral S-phase T cells in HIV disease have a central memory phenotype and rarely have evidence of recent T cell receptor engagement. J. Infect. Dis. 19262-70. [DOI] [PubMed] [Google Scholar]
- 70.Silvestri, G., D. L. Sodora, R. A. Koup, M. Paiardini, S. P. O'Neil, H. M. McClure, S. I. Staprans, and M. B. Feinberg. 2003. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity 18441-452. [DOI] [PubMed] [Google Scholar]
- 71.Smit-McBride, Z., J. J. Mattapallil, M. McChesney, D. Ferrick, and S. Dandekar. 1998. Gastrointestinal T lymphocytes retain high potential for cytokine responses but have severe CD4(+) T-cell depletion at all stages of simian immunodeficiency virus infection compared to peripheral lymphocytes. J. Virol. 726646-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 [DOI] [PubMed]
- 73.So, E. Y., H. H. Park, and C. E. Lee. 2000. IFN-gamma and IFN-alpha posttranscriptionally down-regulate the IL-4-induced IL-4 receptor gene expression. J. Immunol. 1655472-5479. [DOI] [PubMed] [Google Scholar]
- 74.Sousa, A. E., J. Carneiro, M. Meier-Schellersheim, Z. Grossman, and R. M. Victorino. 2002. CD4 T cell depletion is linked directly to immune activation in the pathogenesis of HIV-1 and HIV-2 but only indirectly to the viral load. J. Immunol. 1693400-3406. [DOI] [PubMed] [Google Scholar]
- 74a.Takaoka, A., S. Hayakawa, H. Yanai, D. Stoiber, H. Negishi, H. Kikuchi, S. Sasaki, K. Imai, T. Shibue, K. Honda, and T. Taniguchi. 2003. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 424516-523. [DOI] [PubMed] [Google Scholar]
- 75.Tavazoie, S., J. D. Hughes, M. J. Campbell, R. J. Cho, and G. M. Church. 1999. Systematic determination of genetic network architecture. Nat. Genet. 22281-285. [DOI] [PubMed] [Google Scholar]
- 76.Ullman, K. S., J. P. Northrop, C. L. Verweij, and G. R. Crabtree. 1990. Transmission of signals from the T lymphocyte antigen receptor to the genes responsible for cell proliferation and immune function: the missing link. Annu. Rev. Immunol. 8421-452. [DOI] [PubMed] [Google Scholar]
- 77.Veazey, R. S., M. DeMaria, L. V. Chalifoux, D. E. Shvetz, D. R. Pauley, H. L. Knight, M. Rosenzweig, R. P. Johnson, R. C. Desrosiers, and A. A. Lackner. 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280427-431. [DOI] [PubMed] [Google Scholar]
- 78.Wu, C. Y., J. R. Kirman, M. J. Rotte, D. F. Davey, S. P. Perfetto, E. G. Rhee, B. L. Freidag, B. J. Hill, D. C. Douek, and R. A. Seder. 2002. Distinct lineages of T(H)1 cells have differential capacities for memory cell generation in vivo. Nat. Immunol. 3852-858. [DOI] [PubMed] [Google Scholar]
- 79.Yam, C. H., T. K. Fung, and R. Y. Poon. 2002. Cyclin A in cell cycle control and cancer. Cell Mol. Life Sci. 591317-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yates, A., J. Stark, N. Klein, R. Antia, and R. Callard. 2007. Understanding the slow depletion of memory CD4+ T cells in HIV infection. PLoS Med. 4e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zagury, D., A. Lachgar, V. Chams, L. S. Fall, J. Bernard, J. F. Zagury, B. Bizzini, A. Gringeri, E. Santagostino, J. Rappaport, M. Feldman, A. Burny, and R. C. Gallo. 1998. Interferon alpha and Tat involvement in the immunosuppression of uninfected T cells and C-C chemokine decline in AIDS. Proc. Natl. Acad. Sci. USA 953851-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zella, D., F. Romerio, S. Curreli, P. Secchiero, C. Cicala, D. Zagury, and R. C. Gallo. 2000. IFN-alpha 2b reduces IL-2 production and IL-2 receptor function in primary CD4+ T cells. J. Immunol. 1642296-2302. [DOI] [PubMed] [Google Scholar]