Abstract
Bats have been identified as the natural reservoir of severe acute respiratory syndrome (SARS)-like and SARS coronaviruses (SLCoV and SCoV). However, previous studies suggested that none of the currently sampled bat SLCoVs is the descendant of the direct ancestor of SCoV, based on their relatively distant phylogenetic relationship. In this study, evidence of the recombinant origin of the genome of a bat SLCoV is demonstrated. We identified a potential recombination breakpoint immediately after the consensus intergenic sequence between open reading frame 1 and the S coding region, suggesting the replication intermediates may participate in the recombination event, as previously speculated for other CoVs. Phylogenetic analysis of its parental regions suggests the presence of an uncharacterized SLCoV lineage that is phylogenetically closer to SCoVs than any of the currently sampled bat SLCoVs. Using various Bayesian molecular-clock models, interspecies transfer of this SLCoV lineage from bats to the amplifying host (e.g., civets) was estimated to have happened a median of 4.08 years before the SARS outbreak. Based on this relatively short window period, we speculate that this uncharacterized SLCoV lineage may contain the direct ancestor of SCoV. This study sheds light on the possible host bat species of the direct ancestor of SCoV, providing valuable information on the scope and focus of surveillance for the origin of SCoV.
Severe acute respiratory syndrome (SARS) is a contagious respiratory disease caused by a newly emerged coronavirus (CoV) named SARS-CoV (SCoV) (10). SCoV is phylogenetically distinct from other CoVs in animals and humans (45). SCoV was also isolated from small mammals, such as civets (Paguma larvata) and raccoon dogs (Nyctereutes procyonoides), in live-animal markets of southern China, suggesting that these mammals may have been the direct sources of the SARS epidemic in early 2003 (11). However, further studies demonstrated the lack of widespread infections in wild or farmed civets, implying that civets might act as only amplifying hosts and not a natural reservoir of SCoV (20). Recently, a group of CoVs that are closely related to SCoVs were identified in various species of horseshoe bats (Rhinolophus spp.) (29, 31). Their genomes share the same organization and an overall 88% to 92% sequence identity with that of the human and civet SCoVs (collectively designated Hu-SCoV), and thus, they are termed bat SARS-like CoVs (Bt-SLCoVs).
Genetic analysis revealed a considerable diversity among Bt-SLCoV genomes, suggesting the presence of a wide spectrum of genetically diverse Bt-SLCoVs in various bat species (43). In addition, previous studies indicated a high seroprevalence against Bt-SLCoVs among various bat populations (29, 31). Therefore, bats were proposed to be the natural reservoir of the lineage of SLCoV and SCoV. Nonetheless, based on the relatively distant phylogenetic relationship between Hu-SCoVs and Bt-SLCoVs, researchers suggested that none of the currently sampled Bt-SLCoVs is the descendant of the direct ancestor of Hu-SCoVs (51). Therefore, the direct ancestor of Hu-SCoVs, as well as its corresponding host species, remains elusive.
In this study, we reanalyzed the available Bt-SLCoV genomes and identified a possible recombination event within the genome of a Bt-SLCoV. Phylogenetic analysis of its parental regions suggests the presence of an uncharacterized SLCoV lineage that is phylogenetically closer to Hu-SCoVs than any of the currently sampled Bt-SLCoVs and is therefore a candidate for the direct ancestor of the Hu-SCoV lineage.
To investigate the time of divergence between Hu-SCoVs and this SLCoV lineage, we analyzed the SCoV and SLCoV genome data under both strict- and relaxed-molecular-clock models. Previous studies demonstrated that the rate variations among lineages can mislead estimation of the divergence date if a strict clock is assumed (54). In contrast, if the data set is clocklike, assumption of a molecular clock increases the precision of rate estimates without compromising accuracy (14). The choice of a molecular-clock model is thus crucial for accurate molecular dating. Therefore, we analyzed our data sets under various Bayesian molecular-clock models, aiming to place a robust time scale on the interspecies transmission of Bt-SLCoVs and to provide insights into the zoonotic origin of Hu-SCoVs.
MATERIALS AND METHODS
Detection of recombination.
Complete genome sequences of Hu-SCoV (n = 10) and Bt-SLCoV (n = 7) were downloaded from GenBank, and these nucleotide sequences were aligned using ClustalX with all gap columns removed. The data set was preliminarily scanned for recombination events by Recombination Detection Program (RDP) 2.0 (35), using MaxChi and Chimaera algorithms with a 0.6 and 0.05 fraction of variable sites per window, respectively. To further investigate the potential recombination event suggested by RDP, similarity plot and bootscan analyses, implemented in Simplot 3.5.1 (33), were performed on the complete genome alignment of selected strains, including Bt-SLCoV strain Rp3 (DQ071615) as the query; Hu-SCoV strains Tor2 (AY274119), SZ3 (AY304486), GD01 (AY278489), ZJ01 (AY297028), GZ04 (AY613947), and PC4 (AY613950) as potential major parents; Bt-SLCoV strain Rm1 (DQ412043) as a potential minor parent; and strain Rf1 (DQ412042) as an outgroup.
Estimation of the potential recombination breakpoint location.
The data set was further analyzed using single-breakpoint estimation algorithms implemented in Genetic Algorithms for Recombination Detection (GARD) and Likelihood Analysis of Recombination in DNA (LARD). Based on the bootscan analysis, only the 2,000 nucleotides (nt) around the open reading frame 1b (ORF1b)/S junction (nt 20150 to 22202; all nucleotide numberings in this study are based on AY274119) were analyzed in order to increase the precision of recombination breakpoint estimation. Based on the RDP results, three selected taxa, Rp3, Tor2, and Rm1, were used in the analyses described below. Briefly, GARD uses a genetic algorithm to search for the best breakpoint locations (23). LARD uses a maximum likelihood (ML) method and a likelihood ratio test (LRT) to access the significance of the inferred breakpoint (15). To demonstrate that the detected recombination event is not likely to be a result of random chance (15), the likelihood ratio (LR) of our data set was evaluated against the null distributions of LRs of 1,000 simulated data sets, assuming no recombination, using Seq-Gen (42).
Investigation of the phylogenetic origin of the potential parents.
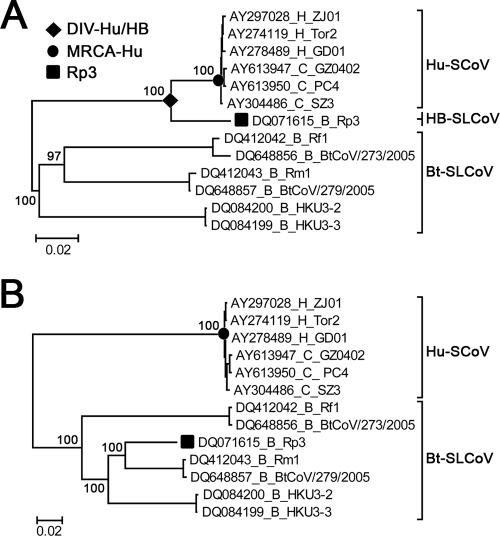
The genome regions 5′ upstream and 3′ downstream of the estimated breakpoint were designated major and minor parental regions, respectively. To investigate the phylogenetic origins of these potential parents, coding sequences of essential ORFs of the major (i.e., ORF1) and minor (i.e., S, E, M, and N genes) parental regions of selected CoV strains (n = 13) were aligned independently using ClustalX based on their codon sequences. The aligned ORFs of the two parental regions were degapped and concatenated separately, generating two alignments of 20,085 bp and 5,778 bp for the major and minor parental regions, respectively. For each of the parental regions, phylogenies were constructed using the Bayesian Markov chain Monte Carlo (BMCMC) method. The BMCMC analyses summarized the majority consensus trees produced by two sets of four tempered MCMC chains of 107 states sampled every 1,000th generation, with the initial 10% of states discarded. The Bayesian phylogenetic analysis was performed with MRBAYES 3 (44) under the best-fit substitution model determined by MRMODELTEST 2 (http://people.scs.fsu.edu/∼nylander/). According to the BMCMC phylogeny (see Fig. 2A), the major parental lineage of Rp3 is designated the human-bat SLCoV (HB-SLCoV) lineage based on its close phylogenetic relationship with the Hu-SCoV lineage.
FIG. 2.
Phylogenetic origins of the major and minor parental regions of Rp3. ML phylogenies were constructed from the concatenated sequences of the essential ORFs of the major (A) and minor (B) parental regions of selected CoVs. For the purposes of display, the phylogenies were midpoint rooted. The taxa were annotated according to their accession numbers and host species—civets (C), humans (H), or bats (B)—and strain names. The numbers on the left of the nodes refer to the BMCMC posterior probabilities. The percentages of support for all other internal nodes within the two lineages were omitted for simplicity. The recombinant strain Rp3, the most recent common ancestor of Hu-SCoVs (MRCA-Hu), and the divergence event between Hu-SCoVs and HB-SLCoVs (DIV-Hu/HB) are indicated. The scale bars are in units of nucleotide substitutions per site.
Estimation of the time of the divergence events.
To estimate the time of the most recent common ancestor (tMRCA) of Hu-SCoVs, as well as the time of divergence events (tDIV) between the Hu-SCoV and HB-SLCoV lineage (designated tMRCA-Hu and tDiv-Hu/HB, respectively) (see Fig. 2), coding sequences of S1 (nt 21492 to 22784; n = 36) and ORF1 (nt 898 to 21479; n = 24) were analyzed under various molecular-clock models in both ML and Bayesian frameworks (details of the taxa in the two data sets are listed as supplementary material at http://evolution.hku.hk/SARS_dating.htm). The sampling times (i.e., the month and year) of the taxa were collected from the literature and used as calibration points in the clock models (41).
First, the strict molecular clock (i.e., a constant rate of evolution) of the two data sets was evaluated in an ML framework using PAML 3.15 as previously described (16, 41, 53). Briefly, the performances of the single-rate dated-tip (SRDT) (i.e., strict-clock) and the different-rate (DR) (i.e., no-clock) models in the data sets were compared using an LRT. Second, the two data sets were analyzed under the strict-clock model (CLOC), as well as the uncorrelated exponentially and lognormally distributed relaxed-clock models (UCED and UCLN) in a Bayesian framework. The CLOC model assumed a constant rate of evolution throughout the tree. The UCED and UCLN models assumed independent rates on different branches, which were drawn from an underlying exponential and lognormal distribution, respectively (6). These clock models are implemented in BEAST 1.4 (8). The MCMC chains were run for 5 × 106 (S1 data set) or 1 × 108 (ORF1 data set) states sampled every 1,000 generations with the initial 10% of burn-in samples discarded (7). For both data sets, the best-fit substitution model was the general time-reversible (GTR) model allowing four categories of gamma-distributed rate heterogeneity distribution and a proportion of invariant sites (GTR + Γ4 + I), as determined by MODELTEST. Since the past population dynamics of the data sets were not the primary interest of our study, we assumed a constant coalescent tree prior for all analyses, with a Jeffreys prior on the constant population size hyperparameter (7). To investigate if this tree prior biased our date estimation, we also analyzed our data sets using a Yule tree prior, which assumes a constant speciation rate per lineage (6). All MCMC chains were independently run twice for the same analysis.
To use information from the S1 data set to improve our estimate of tDIV-Hu/HB from the ORF1 data set, an S1-derived prior distribution was specified on tMRCA-Hu, which is a divergence event shared by the phylogenies of both data sets. This prior distribution was based on the posterior distribution of tMRCA-Hu estimated from the S1 data set under the best-fit clock model. The mode and parameters of this distribution were estimated using distribution-fitting software, EasyFit 3.2 (MathWave Technologies). The MCMC chains for the ORF1 data set were rerun under the same configurations described above, except an S1-derived prior was specified on tMRCA-Hu. For all Bayesian analyses, median and the highest posterior density regions at 95% (HPD) of the parameters were summarized from two identical but independent MCMC chains using TRACER 1.3 (http://beast.bio.ed.ac.uk/). The adequacy of sampling was assessed via effective sample size, which was larger than 200 for all summary statistics investigated (all xml files for BEAST are available as supplementary material at http://evolution.hku.hk/SARS_dating.htm).
Comparison of the performance characteristics of Bayesian clock models.
To compare the performance of any two Bayesian clock models for the same data set, the Bayes factor (BF) was calculated. The BF is the ratio of the marginal likelihoods of the two models. A simple method described by Newton and coworkers (39) computes the BF via importance sampling. A BF of >20, or a ln BF of >2.99, is defined as strong support for the favored model. Clock models of the same data set were compared two by two, and estimates of the best-fit model were taken as the final results.
RESULTS AND DISCUSSION
Detection of recombination and estimation of breakpoint location.
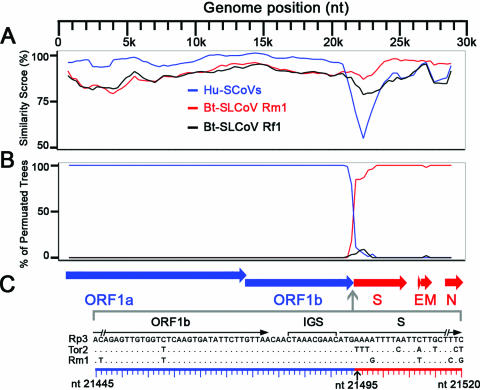
The RDP analysis suggested that Bt-SLCoV Rp3 may be a recombinant of a Bt-SLCoV strain and a strain that is closely related to Hu-SCoVs (data not shown). The similarity plot indicated that the 5′ genomic region of Rp3 shares a substantially higher similarity with the Hu-SCoVs, while its 3′ genomic region is more similar to that of the Bt-SLCoVs (Fig. 1A). Moreover, the bootscan analysis suggested discordance of phylogenetic signals between different genomic regions (Fig. 1B). Taken together, these analyses suggested a single recombination breakpoint located around the junction between the S and ORF1b coding regions (Fig. 1C).
FIG. 1.
Detection of recombination and estimation of a breakpoint within the genome of Rp3. A similarity plot (A) and a bootscan analysis (B) detected a single recombination breakpoint at around the ORF1b/S junction. Both analyses were performed with an F84 distance model, a window size of 1,500 bp, and a step size of 300 bp. The Hu-SCoV group includes strains Tor2 (AY274119), GD01 (AY278489), ZJ01 (AY297028), SZ3 (AY304486), GZ0402 (AY613947), and PC4 (AY613950). (C) Organization of essential ORFs of the SCoV genome and location of the estimated breakpoint. The blue and red horizontal arrows represent the essential ORFs from the major and minor parents, respectively. A sequence alignment of the ORF1b/S junction regions of Rp3, Tor2, and Rm1 is shown below. A consensus IGS and the coding regions of ORF1b and S are annotated above the alignment. The black vertical arrow below the alignment indicates the estimated breakpoint located immediately after the start codon of the S coding region.
To accurately locate the potential recombination breakpoint and to determine the level of its statistical significance, GARD and LARD analyses were performed. Both analyses estimated a potential breakpoint at nt 21495, which is the nucleotide immediately after the start codon of the S coding region (Fig. 1C). The model average support of the breakpoint estimated in GARD analysis was >0.9. In the LARD analysis, the P value of the LRT was <0.0001. Moreover, the LR for this putative breakpoint was greater than any of the LRs of the corresponding simulated data sets (data not shown). These results suggest that the discordance of phylogenetic signals within the genome of Rp3 is not a result of chance and that the recombination breakpoint estimated from both analyses is statistically significant. It should be noted that Rp3 was not plaque isolated, and its genome was obtained by direct sequencing of the PCR products amplified from the field samples (31). Therefore, if the host was infected by multiple strains, we cannot exclude the possibility that the Rp3 genome represents a mosaic sequence of a number of strains. Nonetheless, only one recombination breakpoint was identified within the 29-kb genome, and its parental regions are relatively long (about 21 and 8 kb). Given that the genome was assembled from the sequences of a number of overlapping PCR products, we believe that the probability that the detected recombination breakpoint is an artifact should be negligible.
Genomes of CoVs are reported to have relatively high recombination rates (28). For example, experimental recombination of temperature-sensitive mutants and the wild type of mouse hepatitis virus strains have been studied extensively (21, 27, 34). Moreover, evidence of recombination has also been reported in field isolates of infectious bronchitis virus (19, 25, 30) and feline CoV (13). The occurrence of a high frequency of homologous RNA recombination in CoV genomes is probably related to the unique discontinuous transcription mechanism of its mRNA, in which the nascent RNA transcripts must dissociate from the template and fuse with the leader RNA to a distant mRNA start site (28). Regular dissociation and rejoining of the complex of polymerase and nascent RNA during transcription are similar to the template-switching mechanism in “copy choice” model of recombination in RNA viruses (26). In fact, one of the most utilized recombination sites within the mouse hepatitis virus genome is at the junction between the leader RNA and the remainder of its genome (22). In addition, a previous report suggested that the consensus intergenic sequences (IGS) and the highly conserved sequences around this region may serve as recombination “hot spots” in infectious bronchitis virus (25). In this study, we identifed a potential recombination site immediately after the consensus IGS (17), suggesting that the replication intermediates may participate in the recombination event, as speculated previously in other CoVs. Previous studies suggested that the relatively high rates of recombination and mutation may facilitate the cross-species transmission of CoVs (2, 3), and therefore, CoVs were speculated to be potentially important emerging pathogens (1). A wider surveillance of Bt-SLCoVs may shed light on the possible roles of this observed recombination event in the emergence of SARS.
Phylogenetic origin of the putative parental strains.
To investigate the phylogenetic origin of the putative parents, two BMCMC phylogenies were constructed based on the major (Fig. 2A) and minor (Fig. 2B) parental regions, respectively. The minor parental region of Rp3 was clustered within the Bt-SLCoV lineage and shared monophyly with Rm1 and BtCoV/279/2005 (Fig. 2B). This suggests that the potential minor parent of Rp3 is probably a Bt-SLCoV that shared a close phylogenetic relationship with Rm1 and BtCoV/279/2005. It has been suggested that there is species-specific host restriction of CoVs in bats, since most CoVs from a single bat species grouped together in phylogenetic analyses (48). Moreover, the S protein (which is located within the minor parental region) is the primary determinant of species specificity in CoVs (12, 36), and thus, we speculate that this minor parent may be a Bt-SLCoV residing in Rhinolophus pearsoni, i.e., the host species of Rp3.
On the other hand, the major parental region of Rp3 grouped with, but clustered outside of, the Hu-SCoV lineage (Fig. 2A). Based on this observation, the potential major parent of Rp3 is possibly derived from an uncharacterized lineage that is phylogenetically closely related to Hu-SCoVs. The host species of this speculative parental lineage cannot be ascertained, as it was clustered within neither the Hu-SCoV nor the Bt-SLCoV lineage. Here, we outline three possibilities regarding the host species of this lineage. First, the lineage may originate from an unsampled group of phylogenetically distinct SCoVs residing in live-animal market mammals, like civets or racoon dogs. However, extensive surveillances of various mammalian species over a wide range of geographic locations have been performed, and only CoVs that are highly similar to SCoVs in humans were sampled (20). Thus, this possibility seems unlikely. Second, the lineage may originate from an unknown nonbat intermediate host species, which possibly acquired a SLCoV from bats and transmitted the virus to an amplifying host, such as civets, resulting in spillover in live-animal markets in southern China. However, one of the prerequisites for recombination is coinfection of parental strains within an individual. Therefore, recombination of parental strains residing in different species, i.e., bats and the unknown intermediate host in this case, may be rare due to the relatively strict tropism barrier of CoVs (12, 52). Third, the strain may originate from an unsampled SLCoV lineage residing in a bat species that is phylogenetically closer to Hu-SCoVs than all other currently sampled Bt-SLCoVs. Based on the relatively high genetic diversity among the currently sampled Bt-SLCoVs, the existence of an unsampled phylogenetically distinct lineage of Bt-SLCoV is highly likely, and therefore, the third hypothesis seems to be the most plausible. In the discussions below, this parental lineage is therefore referred to as the HB-SLCoV lineage, while the term “Bt-SLCoV lineage” refers to all other sampled Bt-SLCoVs (Fig. 2). This lineage is proposed to contain the major parent of Rp3 and other closely related strains, and we cannot exclude the possibility that the lineage may also contain the direct ancestor of Hu-SCoVs. To further investigate the time of this interspecies transmission event, tMRCA-Hu and tDIV-Hu/HB (Fig. 2) were estimated under various molecular-clock models in both ML and Bayesian frameworks.
Molecular clock-like behavior of the data sets and choice of Bayesian clock models.
For the ORF1 data set, under the ML framework, LRT analysis suggests that the SRDT model should be rejected in favor of the DR model (Table 1). Moreover, BF analysis suggests that the UCED model fits the ORF1 data set significantly better than the other two models (Table 2), implying that the rate variations among branches of the ORF1 phylogeny are significant and that a strict clock cannot be assumed. The Bt-SLCoV lineage may contribute to the rate variations in the ORF1 data set, since CoVs of different hosts (i.e., bats and humans or civets) may have different substitution rates.
TABLE 1.
Details of the two data sets and results of the ML molecular-clock tests
| Data set | Viral lineages includeda | No. of taxab | dfc | 2Δd | LRT (P)e |
|---|---|---|---|---|---|
| S1 | Hu-SCoVs only | 36 | 34 | 39.29 | 0.24 |
| ORF1 | All Hu-SCoVs and Bt-SLCoVs | 24 | 22 | 87.55 | <0.001 |
S1 and ORF1 data sets were used for estimation of tMRCA-Hu and tDIV-Hu/HB, respectively.
Due to the relatively low variability among Hu-SCoV ORF1 sequences, highly similar Hu-SCoV ORF1 taxa with identical sampling dates were removed (n = 24 after removal).
df.refers to the degree of freedom in the LRT.
2Δ is twice the difference between the log likelihoods for the SRDT and DR models.
The SRDT model cannot be rejected if P is >0.05.
TABLE 2.
Performances of the Bayesian clock models
| Parameter | Value
|
||
|---|---|---|---|
| Clock modelc | S1 data set | ORF1 data set | |
| Marginal likelihooda | CLOC | −3,159.31 | −52,478.47 |
| UCED | −3,155.66 | −52,452.64 | |
| UCLN | −3,157.55 | −52,467.24 | |
| BFb | UCED vs. CLOC | 3.65 | 25.82 |
| UCLN vs. CLOC | 1.76 | 11.23 | |
| UCED vs. UCLN | 1.89 | 14.59 | |
The marginal likelihoods are presented in natural log scale.
The BFs are presented in natural log scale (i.e., ln BF); a ln BF of >2.99 is defined as a strong support for the favored model.
The clock models for the ORF1 data set refer to analyses without the S1-derived lognormal prior on tMRCA-Hu.
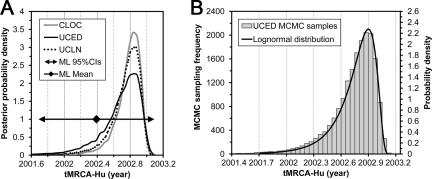
For the S1 data set, LRT analysis suggests the SRDT model cannot be rejected (Table 1). Moreover, the performance of the CLOC model is not significantly worse than that of the UCLN model, implying that the rate variations may not be significant among branches in the S1 phylogeny (Table 2). However, the BF analysis also suggests the UCED model performed slightly better than the CLOC model. Nonetheless, the tMRCAs estimated under the relaxed- and strict-clock models are generally consistent (Fig. 3A), suggesting that these rate variations did not have a significant impact on our estimates of tMRCA-Hu. Based on the marginal likelihoods and the BF analysis (Table 2), the estimates under the UCED model were taken as the final dating results of both data sets.
FIG. 3.
tMRCA-Hu estimated from the S1 data set. (A) tMRCA-Hu estimated from the S1 data set under various Bayesian clock models and the ML SRDT model. (B) Posterior MCMC samples (left y axis) of tMRCA-Hu estimated from the S1 data set under the UCED model and the lognormal distribution (right y axis) fitted using Easyfit. The values of the parameters for the lognormal distribution are as follows: σ = 0.56, μ = −1.00, and γ = 2.04.
tMRCA-Hu.
Based on the analysis of the S1 data set under the UCED model, tMRCA-Hu was estimated to be at a median of 2002.74 (HPD, 2002.18 to 2003.04). This time point refers to the emergence of the common ancestor of all Hu-SCoVs. Under modest assumptions, i.e., that the root had been sampled and the emergence was the result of a single cross-species infection of a single viral lineage, this time point can be considered an estimate of the theoretical onset of the 2003 SARS outbreak. Our estimation of tMRCA-Hu is consistent with previous estimations (5, 47, 55).
tDIV-Hu/HB.
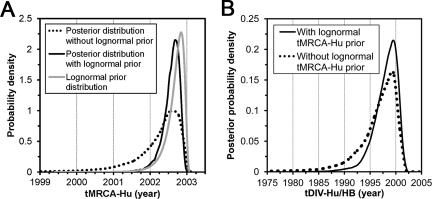
A prior was specified on tMRCA-Hu as a lognormal distribution with parameters chosen to fit the posterior distribution estimated from the S1 data set (Fig. 3B). Bayesian inference specifically provides for the incorporation of prior knowledge, and in this way, we were able to combine information from both data sets in the estimation of tDIV-Hu/HB. Under the UCED model, the medians of tMRCA-Hu estimated from the ORF1 data set with or without the S1-derived tMRCA-Hu prior were similar, and the posterior distribution of tMRCA-Hu was not solely dependent on its prior distribution (Fig. 4A), suggesting that the ORF1 data set was providing additional information in the Bayesian inference. Moreover, tDIV-Hu/HB was consistently estimated at a median around the late 1990s with or without the S1-derived tMRCA-Hu prior (Fig. 4B). It was noted that the specification of S1-derived tMRCA-Hu priors substantially narrowed the HDP of the tDIV-Hu/HB estimate by about 40%, i.e., it decreased from 12.8 to 7.7 years (Table 3). Similar results were observed under the UCLN model (the data are not shown for simplicity).
FIG. 4.
Specification of an S1-derived lognormal tMRCA-Hu prior in the analysis of the ORF1 data set under the UCED model. (A) Prior and posterior distributions of tMRCA-Hu. (B) Effects of the tMRCA-Hu prior on the posterior distribution of tDIV-Hu/HB.
TABLE 3.
Estimates from the ORF1 data set under the Bayesian UCED model
| tMRCA-Hu prior | tMRCA-Hua | tDIV-Hu/HBa | Branch Ab |
|---|---|---|---|
| With S1-derived prior | 2002.63 (2002.14-2002.96) | 1998.51 (1993.55-2001.32) | 4.08 (1.45-8.84) |
| Without S1-derived prior | 2002.40 (2000.69-2003.01) | 1997.44 (1987.68-2001.49) | 4.86 (1.37-13.47) |
Medians with HPD in parentheses.
Length of Branch A (Fig. 5) in years. Similar results were obtained under the UCLN model (data not shown for simplicity).
The relatively low substitution rate of ORF1 and the rate variation among branches in the ORF1 phylogeny may limit the power of the molecular-clock analysis on the ORF1 data set. These factors add uncertainty to our analysis and may widen the credible intervals of our estimates. In contrast, the S1 coding region has been identified as the most variable region among SCoVs (40) and the S1 data set was found to be more clocklike than the ORF1 data set. Therefore, we specified an informative prior on tMRCA-Hu based on the S1 data set, allowing us to combine information across the data sets and to reduce the uncertainty of our divergence time estimates.
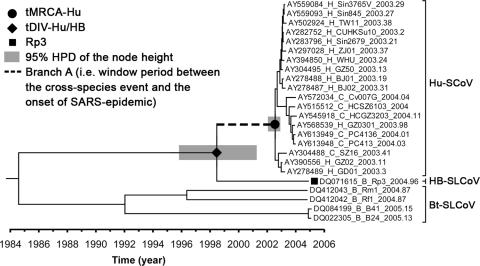
Assuming there was an interspecies transmission of HB-SLCoVs from bats to an amplifying host (e.g., civets), the upper and lower bounds of this event should be theoretically represented by tDIV-Hu/HB and tMRCA-Hu, respectively (Fig. 5). Therefore, the time period between these two events can be considered the most conservative estimation of the period between the cross-species event and the onset of the epidemic. The median and HPD of this period were summarized by sampling the length of a particular branch (i.e., branch A in Fig. 5) of all time-scaled MCMC phylogenies under the UCED model. This period was estimated at a median of 4.08 years (HPD, 1.45 to 8.84 years) (Table 3). The estimated mean substitution rate of the ORF1 data set under the UCED model was 2.79 × 10−3 (HPD, 1.64 × 10−3 to 4.35 × 10−3) substitution per site per year. This estimate is comparable to a previous estimation for the whole genome of Hu-SCoV (i.e., 0.80 × 10−3 to 2.38 × 10−3) (55) and is at the same order of magnitude as in other RNA viruses (4, 9, 18, 37, 38, 50). In addition, the ORF1 data set was reanalyzed under the UCED model with a Yule tree prior assumption, and the estimate is generally consistent with the estimate under the constant coalescent tree prior assumption, suggesting our date estimation is robust for the choice of tree priors.
FIG. 5.
Estimation of the window period between the cross-species event and the onset of the 2003 SARS epidemic. This time-scaled phylogeny was summarized from all MCMC phylogenies of the ORF1 data set analyzed under the UCED model with the S1-derived tMRCA-Hu prior. The heights of the nodes are represented by the median of their estimates. The HPD of tMRCA-Hu and tDIV-Hu/HB are indicated by gray boxes at these nodes. The taxa were labeled in the same style as in Fig. 2, except their sampling dates were annotated.
Implications for the origin of the Hu-SCoV lineage.
Previous studies concluded that none of the currently sampled Bt-SLCoVs is the direct ancestor of the Hu-SCoV lineage based on their relatively distant phylogenetic relationships (43) and molecular-dating results of the putative interspecies transmission event (49). These reports suggest that there may be an unknown intermediate host that acquired a Bt-SLCoV from bats and transmitted it to an amplifying host, such as civets (49), or that Bt-SLCoVs that are phylogenetically closer to the Hu-SCoVs were not sampled (43). However, due to the conflicting phylogenetic relationships between different genomic regions of Rp3 and Hu-SCoV revealed in this work, previous interpretations regarding the closest related Bt-SLCoV strain must be reconsidered. This study demonstrates the recombinant origin of Rp3, emphasizing the presence of an uncharacterized lineage (i.e., the HB-SLCoV lineage) that is phylogenetically closer to Hu-SCoVs than any of the currently sampled Bt-SLCoVs. In addition, our molecular-dating analyses suggest the HB-SLCoV and Hu-SCoV lineage diverged a median of 4.08 years prior to the outbreak. Based on this relatively short window period and their close phylogenetic relationship, we speculate that strains arising from this previously uncharacterized lineage may include the direct ancestor of the SCoVs in live-market animals that contributed to the emergence of SARS in 2003. It is noted that a previous report suggested that the most closely related Bt-SLCoV (i.e., Rp3) and Hu-SCoV diverged a mean of 17 years prior to the outbreak (49). Our credible interval excludes a divergence time this long ago (Table 3). However, due to the relatively large credible interval of the earlier estimate, our estimate falls within its HPD but with improved precision. The choice of genome region for molecular dating, i.e., the HEL gene in the earlier work versus ORF1 in this study, may contribute to the observed differences.
Based on the S protein sequences of the currently sampled Bt-SLCoV, Li and coworkers (32) pointed out that substantial genetic changes in the S protein are likely to be necessary for the virus to infect humans. Due to the fact that the S protein sequence of the direct ancestor of Hu-SCoV is currently unavailable, the genetic factors (e.g., residues under positive selection) that contributed to the switch of species tropism from the bat to the amplifying hosts cannot be determined. We expect that further characterization of the S sequences of the strains of the HB-SLCoV lineage should provide important information regarding the changes that may contribute to cross-species adaptation of the virus.
The observed genetic diversity among currently sampled Bt-SLCoVs strongly suggests bats, in particular, the genus Rhinolophus, are the natural reservoir of SLCoVs and SCoVs. However, among the 69 species of the genus Rhinolophus, the specific species that harbors the direct ancestor of Hu-SCoVs is still unknown (51). One possibility is that there were two phylogenetically distinct lineages of Bt-SLCoV residing in the bat species R. pearsoni that underwent recombination, giving rise to the recombinant strain Rp3. Thus, we suggest a more focused surveillance of SLCoVs in R. pearsoni, which may provide insights into the prevalence and diversity of this recombinant genotype, as well as the possible direct ancestor of Hu-SCoVs.
Another interesting outcome of our analysis is the very young age of the common ancestor of SLCoVs in bats (i.e., the root of the phylogeny in Fig. 5; median, 1982.81; HDP, 1965.75 to 1995.83). It is noted that this estimate refers only to the tMRCA of all currently sampled Bt-SLCoVs, and characterization of more diverged Bt-SLCoVs should extend the age of the lineage. Nonetheless, this estimate precludes codivergence of Bt-SLCoVs with their host bat species. More importantly, it suggests that cross-species transmission of these viruses between different bat species is very common and occurs on an ongoing basis. Interspecies transmissions of CoVs among wildlife and livestock species are well documented (46). With SARS as an example, more comprehensive surveillances of pathogens in wildlife species should make an important contribution to the detection and control of emerging zoonotic infections (24).
Acknowledgments
This work was supported by the Research Fund for the Control of Infectious Diseases (reference number 06060672) from the Hong Kong SAR government.
We thank Susanna K. P. Lau of the Department of Microbiology, Faculty of Medicine, University of Hong Kong, for her valuable comments on the manuscript.
Footnotes
Published ahead of print on 5 December 2007.
REFERENCES
- 1.Baric, R. S., K. Fu, W. Chen, and B. Yount. 1995. High recombination and mutation rates in mouse hepatitis virus suggest that coronaviruses may be potentially important emerging viruses. Adv. Exp. Med. Biol. 380571-576. [DOI] [PubMed] [Google Scholar]
- 2.Baric, R. S., E. Sullivan, L. Hensley, B. Yount, and W. Chen. 1999. Persistent infection promotes cross-species transmissibility of mouse hepatitis virus. J. Virol. 73638-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric, R. S., B. Yount, L. Hensley, S. A. Peel, and W. Chen. 1997. Episodic evolution mediates interspecies transfer of a murine coronavirus. J. Virol. 711946-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, W., and R. S. Baric. 1995. Function of a 5′-end genomic RNA mutation that evolves during persistent mouse hepatitis virus infection in vitro. J. Virol. 697529-7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinese SARS Molecular Epidemiology Consortium. 2004. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 3031666-1669. [DOI] [PubMed] [Google Scholar]
- 6.Drummond, A. J., S. Y. Ho, M. J. Phillips, and A. Rambaut. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drummond, A. J., G. K. Nicholls, A. G. Rodrigo, and W. Solomon. 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 1611307-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drummond, A. J., and A. Rambaut. 2006. BEAST v1.4. http://code.google.com/p/beast-mcmc/.
- 9.Enjuanes, L., C. Sune, F. Gebauer, C. Smerdou, A. Camacho, I. M. Anton, S. Gonzalez, A. Talamillo, A. Mendez, M. L. Ballesteros, et al. 1992. Antigen selection and presentation to protect against transmissible gastroenteritis coronavirus. Vet. Microbiol. 33249-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen, G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302276-278. [DOI] [PubMed] [Google Scholar]
- 12.Haijema, B. J., H. Volders, and P. J. Rottier. 2003. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 774528-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrewegh, A. A., I. Smeenk, M. C. Horzinek, P. J. Rottier, and R. J. de Groot. 1998. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 724508-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho, S. Y., M. J. Phillips, A. J. Drummond, and A. Cooper. 2005. Accuracy of rate estimation using relaxed-clock models with a critical focus on the early metazoan radiation. Mol. Biol. Evol. 221355-1363. [DOI] [PubMed] [Google Scholar]
- 15.Holmes, E. C., M. Worobey, and A. Rambaut. 1999. Phylogenetic evidence for recombination in dengue virus. Mol. Biol. Evol. 16405-409. [DOI] [PubMed] [Google Scholar]
- 16.Hon, C. C., T. Y. Lam, A. Drummond, A. Rambaut, Y. F. Lee, C. W. Yip, F. Zeng, P. Y. Lam, P. T. Ng, and F. C. Leung. 2006. Phylogenetic analysis reveals a correlation between the expansion of very virulent infectious bursal disease virus and reassortment of its genome segment B. J. Virol. 808503-8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussain, S., J. Pan, Y. Chen, Y. Yang, J. Xu, Y. Peng, Y. Wu, Z. Li, Y. Zhu, P. Tien, and D. Guo. 2005. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 795288-5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins, G. M., A. Rambaut, O. G. Pybus, and E. C. Holmes. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 54156-165. [DOI] [PubMed] [Google Scholar]
- 19.Jia, W., K. Karaca, C. R. Parrish, and S. A. Naqi. 1995. A novel variant of avian infectious bronchitis virus resulting from recombination among three different strains. Arch. Virol. 140259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kan, B., M. Wang, H. Jing, H. Xu, X. Jiang, M. Yan, W. Liang, H. Zheng, K. Wan, Q. Liu, B. Cui, Y. Xu, E. Zhang, H. Wang, J. Ye, G. Li, M. Li, Z. Cui, X. Qi, K. Chen, L. Du, K. Gao, Y. T. Zhao, X. Z. Zou, Y. J. Feng, Y. F. Gao, R. Hai, D. Yu, Y. Guan, and J. Xu. 2005. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 7911892-11900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keck, J. G., L. H. Soe, S. Makino, S. A. Stohlman, and M. M. Lai. 1988. RNA recombination of murine coronaviruses: recombination between fusion-positive mouse hepatitis virus A59 and fusion-negative mouse hepatitis virus 2. J. Virol. 621989-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keck, J. G., S. A. Stohlman, L. H. Soe, S. Makino, and M. M. Lai. 1987. Multiple recombination sites at the 5′-end of murine coronavirus RNA. Virology 156331-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosakovsky Pond, S. L., D. Posada, M. B. Gravenor, C. H. Woelk, and S. D. Frost. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 223096-3098. [DOI] [PubMed] [Google Scholar]
- 24.Kuiken, T., F. A. Leighton, R. A. Fouchier, J. W. LeDuc, J. S. Peiris, A. Schudel, K. Stohr, and A. D. Osterhaus. 2005. Public health. Pathogen surveillance in animals. Science 3091680-1681. [DOI] [PubMed] [Google Scholar]
- 25.Kusters, J. G., E. J. Jager, H. G. Niesters, and B. A. van der Zeijst. 1990. Sequence evidence for RNA recombination in field isolates of avian coronavirus infectious bronchitis virus. Vaccine 8605-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai, M. M. 1992. RNA recombination in animal and plant viruses. Microbiol. Rev. 5661-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai, M. M., R. S. Baric, S. Makino, J. G. Keck, J. Egbert, J. L. Leibowitz, and S. A. Stohlman. 1985. Recombination between nonsegmented RNA genomes of murine coronaviruses. J. Virol. 56449-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai, M. M. C. 1996. Recombination in large RNA viruses: coronaviruses. Semin. Virol. 7381-388. [Google Scholar]
- 29.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 10214040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, C. W., and M. W. Jackwood. 2000. Evidence of genetic diversity generated by recombination among avian coronavirus IBV. Arch. Virol. 1452135-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310676-679. [DOI] [PubMed] [Google Scholar]
- 32.Li, W., S. K. Wong, F. Li, J. H. Kuhn, I. C. Huang, H. Choe, and M. Farzan. 2006. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J. Virol. 804211-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makino, S., J. G. Keck, S. A. Stohlman, and M. M. Lai. 1986. High-frequency RNA recombination of murine coronaviruses. J. Virol. 57729-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21260-262. [DOI] [PubMed] [Google Scholar]
- 36.McRoy, W. C., and R. S. Baric. 2006. Spike gene determinants of mouse hepatitis virus host range expansion. Adv. Exp. Med. Biol. 581301-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller-Trutwin, M. C., S. Corbet, M. D. Tavares, V. M. Herve, E. Nerrienet, M. C. Georges-Courbot, W. Saurin, P. Sonigo, and F. Barre-Sinoussi. 1996. The evolutionary rate of nonpathogenic simian immunodeficiency virus (SIVagm) is in agreement with a rapid and continuous replication in vivo. Virology 22389-102. [DOI] [PubMed] [Google Scholar]
- 38.Nakao, H., H. Okamoto, M. Fukuda, F. Tsuda, T. Mitsui, K. Masuko, H. Iizuka, Y. Miyakawa, and M. Mayumi. 1997. Mutation rate of GB virus C/hepatitis G virus over the entire genome and in subgenomic regions. Virology 23343-50. [DOI] [PubMed] [Google Scholar]
- 39.Newton, M. A., A. E. Raftery, A. C. Davison, M. Bacha, G. Celeux, B. P. Carlin, P. Clifford, C. Lu, M. Sherman, M. A. Tanner, A. E. Gelfand, B. K. Mallick, A. Gelman, A. P. Grieve, H. R. Kunsch, T. Leonard, J. S. J. Hsu, J. S. Liu, D. B. Rubin, A. Y. Lo, T. A. Louis, R. M. Neal, A. B. Owen, D. S. Tu, W. R. Gilks, G. Roberts, T. Sweeting, D. Bates, G. Ritter, B. J. Worton, G. A. Barnard, R. Gibbens, and B. Silverman. 1994. Approximate Bayesian-inference with the weighted likelihood bootstrap. J. R. Stat. soc. B 563-48. [Google Scholar]
- 40.Pavlovic-Lazetic, G. M., N. S. Mitic, and M. V. Beljanski. 2004. Bioinformatics analysis of SARS coronavirus genome polymorphism. BMC Bioinform. 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rambaut, A. 2000. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics 16395-399. [DOI] [PubMed] [Google Scholar]
- 42.Rambaut, A., and N. C. Grassly. 1997. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 13235-238. [DOI] [PubMed] [Google Scholar]
- 43.Ren, W., W. Li, M. Yu, P. Hao, Y. Zhang, P. Zhou, S. Zhang, G. Zhao, Y. Zhong, S. Wang, L. F. Wang, and Z. Shi. 2006. Full-length genome sequences of two SARS-like coronaviruses in horseshoe bats and genetic variation analysis. J. Gen. Virol. 873355-3359. [DOI] [PubMed] [Google Scholar]
- 44.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 191572-1574. [DOI] [PubMed] [Google Scholar]
- 45.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 3001394-1399. [DOI] [PubMed] [Google Scholar]
- 46.Saif, L. J. 2004. Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? Rev. Sci. Tech. 23643-660. [DOI] [PubMed] [Google Scholar]
- 47.Song, H. D., C. C. Tu, G. W. Zhang, S. Y. Wang, K. Zheng, L. C. Lei, Q. X. Chen, Y. W. Gao, H. Q. Zhou, H. Xiang, H. J. Zheng, S. W. Chern, F. Cheng, C. M. Pan, H. Xuan, S. J. Chen, H. M. Luo, D. H. Zhou, Y. F. Liu, J. F. He, P. Z. Qin, L. H. Li, Y. Q. Ren, W. J. Liang, Y. D. Yu, L. Anderson, M. Wang, R. H. Xu, X. W. Wu, H. Y. Zheng, J. D. Chen, G. Liang, Y. Gao, M. Liao, L. Fang, L. Y. Jiang, H. Li, F. Chen, B. Di, L. J. He, J. Y. Lin, S. Tong, X. Kong, L. Du, P. Hao, H. Tang, A. Bernini, X. J. Yu, O. Spiga, Z. M. Guo, H. Y. Pan, W. Z. He, J. C. Manuguerra, A. Fontanet, A. Danchin, N. Niccolai, Y. X. Li, C. I. Wu, and G. P. Zhao. 2005. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA 1022430-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang, X. C., J. X. Zhang, S. Y. Zhang, P. Wang, X. H. Fan, L. F. Li, G. Li, B. Q. Dong, W. Liu, C. L. Cheung, K. M. Xu, W. J. Song, D. Vijaykrishna, L. L. Poon, J. S. Peiris, G. J. Smith, H. Chen, and Y. Guan. 2006. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 807481-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vijaykrishna, D., G. J. Smith, J. X. Zhang, J. S. Peiris, H. Chen, and Y. Guan. 2007. Evolutionary insights into the ecology of coronaviruses. J. Virol. 814012-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villaverde, A., M. A. Martinez, F. Sobrino, J. Dopazo, A. Moya, and E. Domingo. 1991. Fixation of mutations at the VP1 gene of foot-and-mouth disease virus. Can quasispecies define a transient molecular clock? Gene 103147-153. [DOI] [PubMed] [Google Scholar]
- 51.Wang, L. F., Z. Shi, S. Zhang, H. Field, P. Daszak, and B. T. Eaton. 2006. Review of bats and SARS. Emerg. Infect. Dis. 121834-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 802535-2543. [DOI] [PubMed] [Google Scholar]
- 53.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13555-556. [DOI] [PubMed] [Google Scholar]
- 54.Yoder, A. D., and Z. Yang. 2000. Estimation of primate speciation dates using local molecular clocks. Mol. Biol. Evol. 171081-1090. [DOI] [PubMed] [Google Scholar]
- 55.Zhao, Z., H. Li, X. Wu, Y. Zhong, K. Zhang, Y. P. Zhang, E. Boerwinkle, and Y. X. Fu. 2004. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol. Biol. 421. [DOI] [PMC free article] [PubMed] [Google Scholar]