Abstract
The genome components of the Melon chlorotic leaf curl virus (MCLCuV) were cloned from symptomatic cantaloupe leaves collected in Guatemala during 2002. The MCLCuV DNA-A and DNA-B components shared their closest nucleotide identities among begomoviruses, at ∼90 and 81%, respectively, with a papaya isolate of MCLCuV from Costa Rica. The closest relatives at the species level were other members of the Squash leaf curl virus (SLCV) clade, which is endemic in the southwestern United States and Mexico. Biolistic inoculation of cantaloupe seedlings with the MCLCuV DNA-A and -B components resulted in the development of characteristic disease symptoms, providing definitive evidence of causality. MCLCuV experimentally infected species within the Cucurbitaceae, Fabaceae, and Solanaceae. The potential for interspecific reassortment was examined for MCLCuV and its closest relatives, including the bean-restricted Bean calico mosaic virus (BCaMV), and three other cucurbit-infecting species, Cucurbit leaf crumple virus (CuLCrV), SLCV, and SMLCV. The cucurbit viruses have distinct but overlapping host ranges. All possible reassortants were established using heterologous combinations of the DNA-A or DNA-B components. Surprisingly, only certain reassortants arising from MCLCuV and BCaMV, or MCLCuV and CuLCrV, were viable in bean, even though it is a host of all of the “wild-type” (parent) viruses. The bean-restricted BCaMV was differentially assisted in systemically infecting the cucurbit test species by the components of the four cucurbit-adapted begomoviruses. In certain heterologous combinations, the BCaMV DNA-A or -B component was able to infect one or more cucurbit species. Generally, the reassortants were less virulent in the test hosts than the respective wild-type (parent) viruses, strongly implicating adaptive modulation of virulence. This is the first illustration of reassortment resulting in the host range expansion of a host-restricted begomovirus.
The Cucurbitaceae contain economically important crop species of importance to human nutrition in temperate, tropical, and subtropical regions. Plant viruses that infect edible cucurbits pose major constraints to cucurbit production worldwide (49). Among the most important are the circular single-stranded DNA whitefly-transmitted viruses in the family Geminiviridae (5, 11, 45). All cucurbit-infecting begomoviruses reported at present have a bipartite genome (18). In the Eastern Hemisphere, Watermelon chlorotic stunt virus has a native range spanning Sudan, southern Arabia, and Iran (23, 33). Squash leaf curl China virus (25) occurs in China, the Philippines (34), and Vietnam (34), whereas Squash leaf curl Yunnan virus has been reported only in southern China (54). Loofa yellow mosaic virus is host and geographically restricted and is endemic in southern Vietnam (43).
In the Western Hemisphere, cultivated cucurbits are hosts of several bipartite begomoviruses, including Squash leaf curl virus (SLCV) and Squash mild leaf curl virus (SMLCV), which are native to Central America, Mexico, and the southwestern United States (Arizona and California) and Texas (8). Squash leaf curl disease was first described in 1979 and 1980 in California (19) and Arizona (6, 7, 44) and was later shown to be caused by either a single begomovirus or a mixture of two, depending on the host. The viruses are recognized as the separate species SLCV and SMLCV (8, 18), and evidence suggests that the two causal viruses have been present in the southwestern United States and northern Mexico since at least 1977. SLCV was later identified in watermelon plants in the Rio Grande Valley, Texas, during 1993 (32) and has been introduced into Egypt more recently (2004) (30). Based on nucleotide sequence comparisons, SLCV and SMLCV are closely related species (at 79.6%). Despite a sympatric association in nature, they each have a distinct but overlapping host range and they vary with respect to virulence in common hosts (6, 7, 8, 36). A third cucurbit virus, Cucurbit leaf crumple virus (CuLCrV) (also known as Cucurbit leaf curl virus), with distinct biological and molecular characteristics, was identified in squash and/or melon fields in Arizona, Texas, and north-central Mexico (8, 9) and in California (22).
Certain begomoviruses have been shown to exchange noncognate components and produce an infectious reassortant under experimental conditions. To date, no unequivocal, naturally occurring reassortants have been discovered in planta, although compelling evidence for such phenomena has been provided (8, 17, 29). Experimental reassortment has been carried out with test hosts that are susceptible to the parental (wild-type) viruses. In these instances, it has been noted that the replication-associated protein (REP)-binding site, or iteron (1), and key sequences of the REP amino terminus (15, 23, 35, 37) of the viable, noncognate components were either identical or highly similar. For example, heterologous pairs of the CuLCrV DNA-A and -B components for SLCV and SMLCV have been shown to produce viable, and sometimes more virulent, reassortants compared to the parent viruses (8). Even though the latter viruses have an identical iteron (REP binding) sequence and all of them infect pumpkin, not all of the heterologous combinations were viable. This result suggested that the underlying establishment of host range barriers, even among closely related viruses that are otherwise capable of coinfecting the same host, involves levels of complexity beyond that of the successful trans replication of noncognate components.
Recently, we described a fourth Western Hemisphere bipartite begomoviral species that is readily distinguishable from previously studied cucurbit viruses in the tropical Americas by its extreme virulence in cantaloupe, honeydew melon (Cucumis melo L.), and watermelon. The virus was first discovered in the Zacapa Valley (11), a region in Guatemala dedicated to intensive monoculture melon and watermelon production for more than 30 years at the near exclusion of other crop species. In contrast to SLCV and CuLCrV, which infect but are not supervirulent in melon or watermelon, this new virus, Melon chlorotic leaf curl virus (MCLCuV) from Guatemala (MCLCuV-GT), is highly virulent in both melon and watermelon. SLCV, CuLCrV, and MCLCuV-GT are moderately to poorly virulent in bean. In contrast, among cucurbit species (Cucurbita spp.), SMLCV infects only pumpkin (not melon or watermelon) and is highly virulent in bean (8, 11). Bean calico mosaic virus (BCaMV) is restricted or “host adapted” to bean and has a narrow host range that does not include cucurbit species.
Here, we report the identification of MCLCuV-GT, a new begomovirus species of cucurbits in the American Tropics, and its phylogenetic relationship to other well-studied begomoviruses. We further report the potential for and characteristics of interspecific reassortants comprising all viable heterologous combinations of the DNA-A and -B components of MCLCuV-GT and four other begomoviruses in the SLCV clade, BCaMV, Cotton leaf crumple virus (CLCrV), SLCV, and SMLCV. The cucurbit- and bean-infecting viruses in the SLCV clade are unusual among begomoviral species in that they share a highly conserved REP amino-terminal sequence and an identical REP-binding sequence (15, 23, 35), which are predicted to enhance their potential for reassortment. This is the first report of begomoviral reassortants displaying host range promiscuity or “host range shifting” (infecting a nonhost of at least one parent) and adaptive virulence (lesser) of the reassortants compared to wild-type virulence of the respective parent virus cognate pairs. Also reported is the expansion of host range by a reassortant arising from the combined components of a host-restricted and non-host-restricted parent. These results suggest a greater-than-expected role for naturally occurring, interspecific adaptive reassortment in begomovirus diversification and the potential significance of reassortment in narrowing or transcending host range barriers of begomoviruses.
MATERIALS AND METHODS
Virus source.
Six leaf samples were collected from field canteloupe plants (Cucumis melo L.) exhibiting leaf curling and foliar chlorosis (Fig. 1) in Zacapa Valley, Guatemala, during 2000. More than 70% of cantaloupe plants throughout the valley exhibited disease symptoms and heavy infestations of the whitefly Bemisia tabaci B biotype (4; data not shown). Total nucleic acids were extracted from field samples using the cetyltrimethylammonium bromide method (14). Cloning and sequencing of begomoviral diagnostic fragments (28) were undertaken, revealing the presence of a new bipartite begomovirus, MCLCuV-GT (11).
FIG. 1.
Symptoms of experimental test plants inoculated for the host range study of MCLCuV. (A) Common bean (Phaseolus vulgaris); (B) cantaloupe (Cucumis melo); (C) cucumber (Cucumis pepo); (D) Datura stramonium; (E) tobacco (Nicotiana benthamiana); (F) pepper (Capsicum annuum); (G) pumpkin (Cucurbita maxima); (H) sweet basil (Ocimum basilcum); (I) watermelon (Citrullus lanatus).
Cloning and sequencing of MCLCuV-GT.
RNase-treated total DNA preparations extracted from MCLCuV-infected canteloupe were the source of the viral template for cloning of the genomic components of MCLCuV-GT and for amplification of digoxigenin-labeled probes (26). Component-specific probes were amplified by PCR (46) using primers pAV2644 and pAC1154 for the DNA-A component and primers pBV1855 and pBC656 for the DNA-B component (28).
To facilitate cloning of the MCLCuV-GT genome components, unique restriction sites were determined for each component. Purified DNA preparations, extracted from symptomatic pumpkin leaves, were incubated with selected restriction endonucleases, and the resultant products were analyzed by Southern hybridization using a standard protocol (47). Based on hybridization results, ClaI appeared to completely linearize the double-stranded forms of both MCLCuV-GT components (data not shown), rendering this enzyme useful for cloning of full-length viral DNA-A and -B components.
For cloning, total DNA (∼20 μg) from a MCLCuV-GT sample was digested with ClaI. DNA fragments were separated by agarose gel electrophoresis and visualized by ethidium bromide staining to identify linearized viral double-stranded DNA of the expected size at 2.4 to 2.8 kbp. The linearized DNAs were eluted from the gel and purified using the Geneclean II kit (BIO 101, Carlsbad, CA). Size-selected DNAs were ligated to the ClaI-linearized plasmid pGEM7zf+ (Promega, Madison, WI), and the ligation mixture was used to transform Escherichia coli strain DH5α using standard methods (47).
Clones bearing recombinant plasmids containing viral inserts of approximately 2.6 kbp were identified by colony hybridization using digoxigenin-labeled MCLCuV-GT DNA-A or DNA-B probes as described previously (28). Recombinant plasmids containing an apparent full-length insert of the MCLCuV-GT DNA-A (pMCLCuV-C160M) or DNA-B (pMCLCuV-C151M) component were selected for further analysis. The DNA sequence of each of the clones was determined using automated, capillary DNA sequencing at the Genomics Analysis and Technology Center, University of Arizona, Tucson. The DNA sequence was determined in both orientations for each cloned insert using primer walking and overlaps of at least ∼150 to 200 nucleotides (nt) (29). The DNA sequence was determined for five clones for each viral DNA-A and -B component. Sequences were compiled using FAKtory, an online program available through the University of Arizona Biotechnology Computing Facility, which employs Phred, a base-calling algorithm available from the University of Washington (16). The locations and the sizes of viral open reading frames were predicted using Editseq (DNASTAR, Madison, WI).
Infectivity and experimental host range of cloned MCLCuV-GT DNA.
Plasmids containing two tandemly arranged copies of the cloned MCLCuV-GT DNA-A (pMCLCuV-C160D) and DNA-B (pMCLCuV-C151D) were constructed in pGEM7zf+ as described previously (50). Test plants were biolistically inoculated with 0.5 μg of each cloned viral component as described previously (29). Seven “Hales Best Jumbo” cantaloupe seedlings were inoculated in each of three experimental replicates. Seedlings (one to two seedlings per pot) were inoculated with tungsten microprojectiles in water as a negative control (mock inoculation) in each replicate.
Test species included in the host range study represented 10 species and five families (Table 1). Test hosts were bean (Phaseolus vulgaris L. “Top Crop”), cantaloupe (Cucumis melo L. “Hales Best Jumbo”), cotton (Gossypium hirsutum L.), cucumber (Cucumis pepo L. “Bush Champion”), Datura stramonium L., Nicotiana benthamiana L., pepper (Capsicum annuum L. “Anaheim”), pumpkin (Cucurbita maxima Duchnes “Big Max”), sweet basil (Ocimum basilcum “Sweet Dani”), tomato (Lycopersicon esculentum Mill. “Humaya”), and watermelon (Citrullus lanatus L. “Charleston Gray”).
TABLE 1.
Experimental host range of MCLCuV determined by biolistic inoculation and infectivity based on symptom development and PCR amplification of the core CP fragmenta
| Test species | No. of infected plants/no. of inoculated plants | Symptoms phenotypeb | PCR result |
|---|---|---|---|
| Common bean (Phaseolus vulgaris L. “Top Crop”) | 7/17 | Chl, Lc | + |
| Cantaloupe (Cucumis melo L. “Hales Best Jumbo”) | 13/16 | Chl, Chls, Lc, St | + |
| Cotton (Gossypium hirsutum L. “DP 90”) | 0/18 | NS | − |
| Cucumber (Cucumis pepo L. “Bush Champion”) | 8/18 | Lc, Mo, St | + |
| Datura stramonium L. | 9/18 | Lc, Chls | + |
| Nicotiana benthamiana | 18/18 | Lc, Mo, St | + |
| Pepper (Capsicum annuum L. “Anaheim”) | 0/18 | NS | − |
| Pumpkin (Cucurbita maxima Duchnes “Big Max”) | 10/22 | Chl, Lc, St | + |
| Sweet basil (Ocimum basilcum) | 0/18 | NS | − |
| Tomato (Lycopersicum esculentum Mill. “Humaya”) | 0/28 | NS | − |
| Watermelon (Citrullus lanatus L. “Charleston Gray”) | 15/18 | Chl, Chls, Lc, Mo, St | + |
Tests included seven test plants/three replicates.
Chl, interveinal chlorosis; Chls, chlorotic spots; Lc, leaf curling; Mo, mosaic; St, stunting.
Inoculated plants were maintained in an environmentally controlled, insect-free growth chamber (27°C with 12-h day/night cycles) and monitored periodically for symptom development for 3 weeks. Total nucleic acids were extracted from seedlings of symptomatic and asymptomatic test plants inoculated with cloned MCLCuV-GT DNA components 2 to 3 weeks postinoculation. Extracts were subjected to PCR analysis using primers that amplify the core coat protein (CP) gene fragment of 576 bp from Western Hemisphere begomoviruses (53), including MCLCuV-GT, to confirm viable infections in test plants or the lack thereof.
Sequence analysis of MCLCuV-GT.
The nucleotide sequences of the MCLCuV DNA-A and DNA-B components were used to search the GenBank database for closely related viruses. The begomoviruses identified by the BLAST search were aligned with their respective components using the Clustal V option of MegAlign (DNASTAR, Madison, WI). The open reading frames that were greater than 10 kDa in size were identified on the MCLCuV-GT DNA-A and -B components using Editseq (DNASTAR, Madison, WI).
The complete DNA-A and -B component alignments used in the nucleotide comparisons were also used to identify possible regions of intermolecular recombination. To accomplish this, the alignments were analyzed using Genconv (48) and RDP, programs that are both available in the RDP package, version 2.0 (38). The default search parameters for scanning aligned sequences were employed, using a P value of 0.001.
Reassortment experiments.
Three differentially susceptible test species were identified for inoculation with selected combinations of begomoviral DNA-A and -B components based on the following criteria. Watermelon was selected as a test species because MCLCuV-GT is virulent in this host, and it is a symptomatic host of SLCV and CLCrV but not of SMLCV or BCaMV. Bean, a host of all four cucurbit-adapted begomoviruses examined here and of BCaMV, was employed as a test host for reassortment experiments to test the hypothesis that bean could be an important bridge host for begomovirus diversification, particularly in the SLCV clade (3). BCaMV was selected as the only bean-adapted species in the SLCV clade. Pumpkin was included as a test host because SMLCV is highly virulent in pumpkin and in bean but does not infect watermelon or cantaloupe.
Four seedlings of each suite of test species were inoculated with selected heterologous DNA-A and -B components of BCaMV, CuLCrV, MCLCuV-GT, SLCV, and SMLCV in each of three replicates (Table 2). Mock-inoculated (Tris-EDTA [pH 8.0]) negative controls were included for each replicate, and appropriate plant seedlings were inoculated separately with the respective cognate (homologous) pairs of each virus as a positive control (Table 2). Inoculated seedlings were maintained in a growth chamber (27°C with 12-h day/night cycles) and observed for symptom development for 3 weeks. The potential infectivity of reassortants was confirmed visually by inspecting the plants for symptoms and by PCR using primers to amplify the core CP fragment (576 bp) as described previously (53) and restriction fragment length polymorphism (RFLP) for the DNA-A component of each virus. For RFLP analysis, total DNA was extracted from test plants and subjected to PCR amplification of DNA-A using AV2644 and AC1154 degenerate primers (28) to obtain a 1.1-kbp fragment. The PCR fragment was digested with AccI, SalI, or SpeI to confirm the identity of the viral DNA-A included in each experiment (Table 3).
TABLE 2.
Viable reassortants in watermelon, bean, or pumpkin produced from heterologous combinations of the DNA-A or DNA-B component of MCLCuV-GT, BCaMV, CuLCrV-AZ, SLCV, and/or SMLCV and positive controlsa
| Reassortant | DNA-B reassortant | No. of infected plants/no. of inoculated plants | Symptom phenotypeb | PCR result | Test host |
|---|---|---|---|---|---|
| DNA-A | |||||
| BCaMV | MCLCuV | 0/12 | NS | − | Watermelon |
| CuLCrV | MCLCuV | 0/12 | NS | + | Watermelon |
| SLCV | MCLCuV | 3/12 | Chl, Chls | + | Watermelon |
| SMLCV | MCLCuV | 0/12 | NS | − | Watermelon |
| MCLCuV | BCaMV | 0/12 | NS | − | Watermelon |
| MCLCuV | CuLCrV | 4/12 | Chls | + | Watermelon |
| MCLCuV | SLCV | 7/12 | Chl, Lc, St | + | Watermelon |
| MCLCuV | SMLCV | 1/12 | NS | + | Watermelon |
| CuLCrV | MCLCuV | 0/12 | NS | − | Pumpkin |
| MCLCuV | CuLCrV | 5/12 | Chl, Lc | + | Pumpkin |
| MCLCuV | SMLCV | 1/8 | Chl, Lc | + | Pumpkin |
| SMLCV | MCLCuV | 6/8 | Chls, Lc | + | Pumpkin |
| MCLCuV | BCaMV | 4/8 | Chl, Lc | + | Bean |
| BCaMV | MCLCuV | 0/8 | NS | − | Bean |
| Homologous combinations and no-DNA controls | |||||
| BCaMV DNA-A+B | 0/6 | NS | − | Watermelon | |
| CuLCrV DNA-A+B | 6/6 | Chl, Chls, Lc, Mo, St | + | Watermelon | |
| MCLCuV DNA-A+B | 6/6 | Chl, Chls, Lc, Mo, St | + | Watermelon | |
| SLCV DNA-A+B | 6/6 | Chl, Lc, Mo, St | + | Watermelon | |
| SMLCV DNA-A+B | 0/6 | NS | − | Watermelon | |
| No-DNA control | 0/24 | NS | − | Watermelon | |
| No-DNA control | 0/8 | NS | − | Pumpkin | |
| No-DNA control | 0/6 | NS | − | Bean |
Positive controls included wild-type virus or cognate components.
Chl, interveinal chlorosis; Chls, chlorotic spots; Lc, leaf curling; Mo, mosaic; St, stunting; NS, no symptoms.
TABLE 3.
Expected restriction digest fragment sizes for the respective viral DNA-A components used to confirm the identity of parent DNA-A components used to establish reassortants
| Endonuclease | Expected fragment size (bp) after digestion with diagnostic restriction enzymes
|
||||
|---|---|---|---|---|---|
| BCaMV | CuLCrV | MCLCuV | SLCV | SMLCV | |
| AccI | 200 | 138 | 342 | 542 | 542 |
| 342 | 200 | 822 | 621 | 619 | |
| 598 | 342 | ||||
| 489 | |||||
| SalI | 1,140a | 543 | 1,164a | 1,163a | 1,161a |
| 626 | |||||
| SpeI | 1,140a | 1,169a | 1,164a | 1,163a | 128 |
| 1,033 | |||||
Uncut fragment.
Nucleotide sequence accession numbers.
The complete nucleotide sequences for MCLCuV-GT DNA-A and -B have been deposited in the GenBank database under accession numbers AF325497 and AF325498, respectively.
RESULTS
Infectivity of cloned DNA-A and DNA-B components of MCLCuV-GT.
Biolistic inoculation of cantaloupe (C. melo) seedlings with cloned MCLCuV-GT DNA-A (pMCLCuV-C160D) and DNA-B (pMCLCuV-C151D) components resulted in the development of patchy foliar chlorosis and leaf curling, symptoms identical to those observed in field-infected cantaloupe plants (Fig. 1B), thereby fulfilling Koch's postulates. Biolistic inoculation of pumpkin and N. benthamiana seedlings with the DNA-A component of MCLCuV-GT alone failed to establish infectivity (data not shown). The presence or absence of viral DNA in extracts of inoculated test plants was determined by core CP PCR and DNA sequencing of the cloned amplicon (53) (Table 1 and data not shown). When symptoms were observed in test plants, virus was always detected by the PCR analysis, whereas some asymptomatic plants were positive while others were negative in the above-described assays (Table 3). Viral DNA was never detected by PCR in mock-inoculated test plant controls.
Host range.
The inoculation of selected test plant species revealed that MCLCuV-GT experimentally infected species in three plant families, the Cucurbitaceae, the Fabaceae, and the Solanaceae. The results of the host range study and characteristic symptom phenotypes in each test species are summarized in Table 1 and Fig. 1. Test plants of cotton, pepper, sweet basil, and tomato did not develop symptoms in any of the three replicated experiments, nor was viral DNA detected in the latter test species by core CP PCR (Table 1) (Fig. 1A to I). All species that were identified as hosts of MCLCuV-GT developed systemic symptoms and were positive by core CP PCR assay and DNA sequencing (Table 1).
MCLCuV-GT symptoms were severe in cantaloupe and watermelon plants compared to the more mild symptoms in cucumber. MCLCuV-GT infection caused leaf curling and stunting in all four cucurbit host species tested. Unlike the large chlorotic areas noticed in cantaloupe and watermelon, only minute chlorotic spots developed on infected cucumber plants. However, leaf deformation in cucumber was less severe than that in cantaloupe, pumpkin, and watermelon (Fig. 1B, C, G, and I). Symptoms of MCLCuV-infected bean were interveinal chlorosis and mild leaf curling (Fig. 1A). N. benthamiana and Datura stramonium were the only two solanaceous species that were susceptible to MCLCuV-GT infection. The disease symptoms for N. benthamiana were severe and included mosaic, stunting, and mild leaf curling (Fig. 1E). In contrast, D. stramonium developed only mild leaf curling and small chlorotic spots on the leaves (Fig. 1D). Inoculation of N. benthamiana with MCLCuV-GT at 100% infection frequency revealed a high degree of pathogenicity for this host compared to those of the cucurbit species examined: C. melo at 81.3%, C. pepo at 44%, and C. lanatus at 83.3% infectivity (Table 1).
DNA sequence analysis. (i) DNA-A and DNA-B components.
The complete nucleotide sequences for the DNA-A and DNA-B components of MCLCuV-GT were determined to be 2,662 nt and 2,638 nt, respectively. The DNA-A and -B component sequences determined for four field samples of MCLCuV-GT from cantaloupe shared 96.0 to 100% nucleotide identity. The common region (CR) of the DNA-A and DNA-B components of MCLCuV-GT shared 96.9% nucleotide identity. Both components had an identical REP-binding site sequence: GGTGT-CCT-GGTGT, confirming that they were cognate components of the same begomovirus species.
A comparison of MCLCuV-GT DNA-A component nucleotide sequences with other reported begomoviruses indicated that they shared 90.2% and 81.2% nucleotide identities with their closest relative, a strain of MCLCuV from Costa Rica (MCLCuV-CR) (GenBank accession numbers AY064391 and NC_003860). The closest MCLCuV-GT DNA-A component relative at the species level was SLCV, at 86%, whereas the DNA-B component was most closely related to SMLCV, at 70.6%.
(ii) Recombination analysis.
Neither the Genconv (48) nor the RDP (38) program, applied to detect intermolecular recombination between the MCLCuV-GT DNA-A and DNA-B components and selected begomoviruses (data not shown), revealed significant fragments indicative of recombination between these closely related species (data not shown).
(iii) Reassortment experiments.
Watermelon and pumpkin seedlings biolistically inoculated with MCLCuV-GT and selected combinations of heterologous (noncognate) BCaMV, CuLCrV, SLCV, and SMLCV components yielded seven viable reassortants (Table 2 and Fig. 2). Reciprocal MCLCuV-GT and SLCV reassortants developed obvious systemic symptoms, and infection was confirmed by positive PCR in the inoculated test plants (Table 2 and Fig. 2). Symptoms observed in watermelon were milder for the reassortants (Fig. 2I and J) than for infections obtained using the cognate MCLCuV-GT (Fig. 1I and 2E) and were reminiscent of symptoms of SLCV infection (Fig. 2M). Most notable was that chlorotic spots were more or less small and discrete, affecting only portions of the leaf, in contrast to larger chlorotic spots distributed on the entire leaf lamina, which are characteristic of MCLCuV-GT infection. However, pumpkin plants infected with the reassortant MCLCuV-GT A × SLCV B (Fig. 2F) developed symptoms reminiscent of the severe symptoms caused by SLCV (Fig. 2B) instead of the phenotype associated with the other parent virus, MCLCuV-GT, which is characterized by extremely mild symptoms in pumpkin (Fig. 1G and 2A).
FIG. 2.
Symptoms of selected reassortants in test plants. BA, CA, MA, SA, and SMA stand for DNA-A for BCaMV, Cucurbia leaf crumple virus from Arizona (CuLCrV-AZ), MCLCuV-GT, SLCV, and/or SMLCV, respectively. BB, CB, MB, SB, and SMB stand for DNA-B for BCaMV, CuLCrV, MCLCuV-GT, SLCV, SMLCV, respectively.
The MCLCuV-GT DNA-A × SMLCV DNA-B reassortant was viable in watermelon and pumpkin seedlings (Fig. 2G); however, this reassortant was asymptomatic in watermelon (data not shown) but symptomatic in pumpkin. Symptoms of the reassortant in pumpkin were more severe than those characteristically observed for SMLCV (Fig. 2C), and more closely resembled those associated with MCLCuV-GT infection (Fig. 1G and 2A).
The reassortant SMLCV A × MCLCuV-GT B was viable only in pumpkin, which developed systemic symptoms. Viral DNA was confirmed to be present in symptomatic plants (Fig. 2K). Interestingly, symptoms were milder for this reassortant in pumpkin than for those of the reciprocal combination (see above).
Both heterologous combinations of MCLCuV-GT and CuLCrV were viable in watermelon and pumpkin. This reassortant was infectious in watermelon and pumpkin, but CuLCrV A × MCLCuV-GT B caused a symptomless infection of watermelon that was detectable by core CP PCR (Table 2). In contrast, the reciprocal combination MCLCuV-GT DNA-A × CuLCrV DNA-B was highly virulent in watermelon (Fig. 2P) and pumpkin (Fig. 2H). Disease symptoms in watermelon were less severe than those for the respective wild-type parent viruses, MCLCuV and CuLCrV (Fig. 2E and L, respectively). Furthermore, chlorotic spots developed only on the older leaves, together with stunting and leaf curling; collectively, hallmark phenotypes of the wild-type MCLCuV parent were observed for the latter reassortant. In comparison, the symptom phenotypes in pumpkin infected with the reassortant MCLCuV-GT A × CuLCrV B (Fig. 2H) were similar to those of the wild-type parent viruses MCLCuV and CuLCrV (Fig. 2A and D, respectively), particularly with respect to the chlorotic lesion patterns and leaf curling. In contrast, the latter reassortant did not cause severe stunting in pumpkin, which also is characteristic of wild-type parent viruses.
Watermelon (a nonhost of BCaMV and MCLCuV) inoculated with MCLCuV-GT A and BCaMV B reassortants was asymptomatic, and virus was not detected in plant extracts by PCR. However, inoculation of bean (a host of both viruses) with the reassortant MCLCuV-GT DNA-A and BCaMV DNA-B resulted in a viable MCLCuV-GT × BCaMV infection, but the efficiency was notably low (Table 2). The interveinal chlorosis phenotype for this reassortant was extremely mild and less virulent (Fig. 2O) than that of either of the parent viruses MCLCuV-GT and BCaMV (Fig. 1A and 2N, respectively). However, the degree of interveinal chlorosis in bean plants infected by the reassortant MCLCuV-GT DNA-A × BCaMV DNA-B (Fig. 2O) and in those infected by the cognate MCLCuV-GT DNA-A and -B (Fig. 1A) was similar. The latter phenotype was not observed in BCaMV-infected bean, which instead developed green vein banding in the major veins of leaves.
For the most part, the results of PCR and RFLP analyses of viral components were consistent with conclusions based on visual symptom assessment. However, several test species were symptomless hosts for certain reassorted combinations based the ability to detect the inoculated viral components in asymptomatic plants (Table 3).
DISCUSSION
Alignment and phylogenetic analysis of the cloned MCLCuV-GT DNA-A and DNA-B components, respectively, with well-studied begomoviruses revealed that the closest species relative to MCLCuV is SLCV, at 86% nucleotide identity, indicating that MCLCuV-GT is a distinct begomoviral species (11; our unpublished results). At the species level, the next closet relatives of MCLCuV-GT (DNA-A component) are SMLCV at 83% and CuLCrV at ∼80% nucleotide identities. These closest MCLCuV relatives were first identified from cucurbits in irrigated, monoculture cropping systems in the Sonoran Desert (Arizona, California, and Sonora, Mexico) (6, 7, 36). The next closest relative to the latter four cucurbit viruses is BCaMV (78% nucleotide identity with MCLCuV), a bean-adapted begomovirus also from the Sonoran Desert agroecosystem. BCaMV and its cucurbit-infecting relatives in the SLCV clade are similarly divergent from one another with respect to percent nucleotide identity. As would be expected for the less tightly conserved DNA-B components of this genus, MCLCuV-GT diverges from its closest relatives, SLCV, SMLCV, CuLCrV, and BCaMV, at 65, 70, 53%, and 54.5% nucleotide identities, respectively, underscoring the clear species demarcations.
A second isolate, MCLCuV-CR, from papaya, whose DNA-A (GenBank accession number AY064391) and DNA-B (accession number NC_003860) components share 90 and 81% nucleotide identities, respectively, with MCLCuV-GT has been described. By applying the working cutoff of <89% shared nucleotide identity for demarcating begomoviral species (18), the MCLCuV-GT and MCLCuV-CR isolates are considered to be strains of the same species. Interestingly, the CP gene (Cp) for the two strains share only 87% identity, making these isolates somewhat unusual, given the high degree of Cp conservation at the species level that is characteristic of begomoviruses. This suggests either that the MCLCuV-GT and MCLCuV-CR strain has been isolated geographically and/or by the host for an extended period of time or that the Cp of one of the strains was acquired owing to recombination with an unidentified begomovirus. Analysis of the Cp sequence for additional begomoviruses detected in melon and watermelon plants in Guatemala from 1995 to 2005 has revealed the presence of a number of SLCV and SMLCV variants in the vicinity where MCLCuV-GT emerged (our unpublished data). To date, CuLCrV is the only Western Hemisphere cucurbit-infecting begomovirus not yet reported in Central America (8). The distribution of cucurbit-infecting begomoviruses in Mexico, Arizona, California, and Texas in North America and in Guatemala in Central America suggests that this major geographic region is a center of diversity for the cucurbit-BCaMV subclade. Bean (P. vulgaris) and New World cucurbits are native to Mesoamerica and North America, respectively, and a number of cucurbits and legumes are endemic in both locales, suggesting that these virus-host combinations have a history of coevolution.
Intermolecular reassortment experiments with four cucurbit-infecting species, CuLCrV, MCLCuV, SLCV, and SMLCV (Fig. 2 and 3), resulted in symptomatic, systemic infection of watermelon and/or pumpkin, common hosts for three and/or four of the parent species (SMLCV does not infect watermelon; whereas all species infect pumpkin). Interestingly, infectivity (efficiency) and virulence (symptom severity) varied with the particular reassortant-host combination. The REP-binding sites are identical among this suite of cucurbit-infecting begomoviruses (15, 35) and in the bean-adapted relative BCaMV, which does not infect cucurbits (13). The finding that MCLCuV-GT encodes the same REP-binding site as the former three species but shares in common only the hosts of CuLCrV (watermelon), SLCV (watermelon), SMLCV (pumpkin), and BCaMV (bean), with their respective viruses, was unexpected, given the overly simplistic hypothesis examined herein, in which viruses sharing iterons and host range were expected to reassort.
FIG. 3.
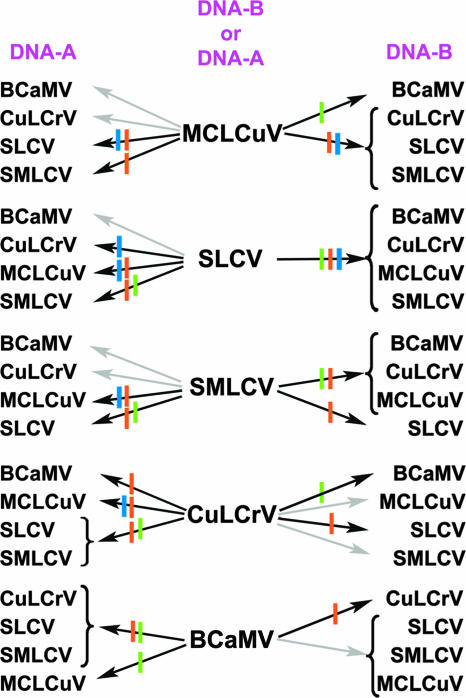
Reassortant experiments illustrating the host range barriers for reassortments of noncognate DNA-A and DNA-B components of five begomoviral species in the SLCV-BCaMV sister clade: BCaMV, CuLCrV-AZ (CuLCrV), MCLCuV, SLCV, and/or SMLCV. Solid line, infectious in bean (green), pumpkin (orange), and watermelon (blue); broken line, not infected.
The biotic and genetic features of the SLCV subclade suggested substantial potential for interspecies reassortment within the sister clade and also generally supported predictions for complementary interactions between certain heterologous DNA-A and -B components. Such molecular interactions are thought, minimally, to involve sequence-specific binding by REP to cognate DNA-A and DNA-B components during the initiation of replication, together with a compatible movement protein to affect the cell-to-cell spread of both components (2, 15, 23, 35). These functions, although interdependent, are encoded on separate components, ensuring codependence and virus survival. Indeed, in certain hosts, proteins encoded by the DNA-A and -B components, e.g., CP, BC1, and BV1, coordinate cell-to-cell spread and affect viral host range and pathogenicity (31, 36, 42). Thus, it was not surprising that bidirectional MCLCuV-GT × SLCV reassortants systemically infected watermelon (Table 2) (6-8; this report). Interestingly, the reassortant MCLCuV-GT A × SMLCV B was viable in pumpkin (Fig. 2 and 3 and Table 2), a host of both parent viruses (7, 8; this report), but infected watermelon in a single direction only, and unexpectedly, infected plants were asymptomatic. In contrast, only the MCLCuV-GT A × CuLCrV B reassortant infected watermelon and pumpkin, even though both hosts supported infection by both wild-type parents. The finding that seemingly host-compatible viruses did not produce viable, bidirectional reassortants suggests intracomponent incompatibility independent of replication and movement functions (Fig. 2 and 3) (Table 2). Furthermore, pumpkin and watermelon infected by MCLCuV-GT × CuLCrV reassortants did not develop severe stunting symptoms, a phenotype associated with both parent viruses. Also, pumpkin and watermelon infected by MCLCuV-GT × CuLCrV reassortants did not develop severe stunting symptoms, a phenotype associated with both parent viruses. These trends suggest that extremely virulent (parental) viruses could become targets of negative selection if extreme virulence becomes detrimental and favors more moderately or mildly pathogenic variants.
As a corollary, certain combinations of reassortment also appeared to favor the adaptive modulation of pathogenicity and virulence. This was borne out in the case of bean systemically infected with the unidirectional reassortant MCLCuV-GT DNA-A × BCaMV, which developed chlorotic spots and leaf curling reminiscent of MCLCuV-GT symptoms in bean. However, reassortants failed to infect pumpkin, a nonhost of BCaMV (12), even though MCLCuV-GT readily infects pumpkin. Here, reassortment was favored by the adaptive modulation of pathogenicity and virulence in which reassortants exhibited a low rate of infectivity and reduced virulence compared to those of parent viruses. This striking phenomenon was underscored by the asymmetric reassortment of seemingly host-compatible parent viruses yielding mildly or moderately virulent hybrids and, secondly, by the more extreme condition in which reassortants established asymptomatic infections, the least virulent phenotype of all.
Finally, a BCaMV component infected a cucurbit host when accompanied by a cucurbit-infecting (parent) component, illustrating the potential for host range expansion, in this case, for both BCaMV components. Given the rather small representation of begomoviruses that are highly host restricted (certain tropical legume viruses and cassava viruses), a state that could limit interactions with non-host-restricted begomoviruses, the expansion of viral host range through such a reassortment would certainly be viewed as a highly adaptive trait. Citing the example of the apparent extinction of Bean golden yellow mosaic virus in Puerto Rico, which followed the establishment of the exotic B biotype vector there (unpublished data), Bean golden yellow mosaic virus would likely have remained successful had it not become host restricted to bean (and several wild legumes). Even so, perhaps adaptive virulence linked to host range shifting provides a common vehicle for multiple directions of diversification through reassortment.
The cucurbit virus-BCaMV subclade of the SLCV clade is an unusual example in the genus Begomovirus. It comprises five closely related species that show little or no evidence of recombination (12, 13). Wild-type parent virus(es) and certain reassortants exhibited differential capacities for host shifting and degrees of virulence, indicating that they are readily responsive to genetic factors associated with virus-host interactions (3). It is notable that the bean-adapted BCaMV, which does not infect cucurbits, appears to be competitive (survived extinction) with other naturally occurring sympatric species in the SLCV clade and also that all neotropical cucurbit viruses known to date infect bean. CLCrV, a basal species in the SLCV clade, is host adapted to cotton and malvaceous species and infects bean, but it has not been found to persistently infect bean from year to year in bean-growing areas (29). These collective observations support the hypothesis that adaptation to bean could be an important diversification vehicle for begomoviruses, implicating bean as a cultivated bridge host for reassortment in this region and perhaps more widely.
The results herein did not support all possible reassortment combinations predicted for otherwise “apparently” genetically and biotically compatible DNA-A and -B components. Perhaps as interestingly, some combinations that would not have been predicted produced viable reassortants. Although not surprising, these results and previous studies support a “higher-complexity” hypothesis that reassortment between bipartite begomovirus components can extend beyond interactions predicted by compatible REP-binding sequences and their REP motifs (1, 8, 10, 15, 20-23, 35, 52). More importantly, the results herein suggest that begomoviruses may undergo greater-than-expected interspecific reassortment and leave open the possibility that reassortment may frequently be employed to modulate virulence and host range shifts. Reports of experimental (8, 10, 11, 20, 21, 24, 27) and natural (10, 29, 41) predicted reassortment in begomoviruses underscore the growing recognition of this genetic mechanism in host range shifting and modulated virulence in diversification. Indeed, there is no evidence for “cross-protection” (silencing) between begomoviruses, and they are commonly reported to establish mixed infections, even to the extent of occupying the same nucleus (39), all scenarios that would be expected to facilitate reassortment-based selection (3).
Footnotes
Published ahead of print on 5 December 2007.
REFERENCES
- 1.Arguello-Astorga, G. R., R. G. Guevara-Gonzalez, L. R. Herrera-Estrella, and R. F. Rivera-Bustamante. 1994. Geminivirus replication origins have a group-specific organization of iterative elements: a model for replication. Virology 20390-100. [DOI] [PubMed] [Google Scholar]
- 2.Bisaro, D. M. 1996. Geminivirus DNA replication, p. 833-854. In M. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Brown, J. K. 2001. The molecular epidemiology of begomoviruses, p. 279-316. In J. A. Khan and J. Dykstra (ed.), Trends in plant virology. Haworth Press, Inc., New York, NY.
- 4.Brown, J. K. 2007. The Bemisia tabaci complex: genetic and phenotypic variability drives begomovirus spread and virus diversification. Plant disease APSNet feature article January 2007. http://www.apsnet.org/online/feature/btabaci/.
- 5.Brown, J. K., and J. Bird. 1992. Whitefly-transmitted geminiviruses in the Americas and the Caribbean Basin: past and present. Plant Dis. 76220-225. [Google Scholar]
- 6.Brown, J. K., and M. R. Nelson. 1989. Characterization of watermelon curly mottle virus, a geminivirus distinct from squash leaf curl virus. Ann. Appl. Biol. 115243-252. [Google Scholar]
- 7.Brown, J. K., and M. R. Nelson. 1986. Whitefly-borne viruses of melons and lettuce in Arizona. Phytopathology 76236-239. [Google Scholar]
- 8.Brown, J. K., A. M. Idris, C. Alteri, and D. C. Stenger. 2002. Cucurbit leaf curl virus, a new emergent begomovirus species able to form viable reassortants with related viruses in the Squash leaf curl virus cluster. Phytopathology 92734-742. [DOI] [PubMed] [Google Scholar]
- 9.Brown, J. K., A. M. Idris, M. Olsen, M. E. Miller, T. Isakeit, and J. Anciso. 2000. Cucurbit leaf curl virus, a new whitefly-transmitted geminivirus in Arizona, Texas, and Mexico. Plant Dis. 84809. [DOI] [PubMed] [Google Scholar]
- 10.Brown, J. K., A. M. Idris, K. M. Ostrow, R. French, and D. C. Stenger. 2005. Genetic and phenotypic variation of three strains of the Pepper golden mosaic virus complex. Phytopathology 951217-1224. [DOI] [PubMed] [Google Scholar]
- 11.Brown, J. K., A. M. Idris, D. Rogan, M. H. Hussein, and M. Palmieri. 2001. Melon chlorotic leaf curl virus, a new begomovirus associated with Bemisia tabaci infestations in Guatemala. Plant Dis. 851027. [DOI] [PubMed] [Google Scholar]
- 12.Brown, J. K., K. M. Ostrow, A. M. Idris, and D. C. Stenger. 1999. Biotic, molecular, and phylogenetic characterization of bean calico mosaic virus, a distinct begomovirus species with affiliation in the squash leaf curl virus cluster. Phytopathology 89273-280. [DOI] [PubMed] [Google Scholar]
- 13.Brown, J. K., K. M. Ostrow, A. M. Idris, and D. C. Stenger. 2000. Chino del tomate virus: relationships to other begomoviruses and identification of A-component variants that affect symptom expression. Phytopathology 90546-552. [DOI] [PubMed] [Google Scholar]
- 14.Doyle, J. J., and J. L. Doyle. 1987. A rapid DNA isolation procedure for small quantities of fresh tissue. Phytochem. Bull. 1911-15. [Google Scholar]
- 15.Eagle, P. A., B. M. Orozco, and L. Hanley-Bowdoin. 1994. A DNA sequence required for geminivirus replication also mediates transcriptional regulation. Plant Cell 1994 61157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8175-185. [DOI] [PubMed] [Google Scholar]
- 17.Faria, J. C., R. L. Gilbertson, S. F. Hanson, F. J. Morales, P. Ahlquist, A. O. Loniello, and D. P. Maxwell. 1994. Bean golden mosaic geminivirus type II isolates from the Dominican Republic and Guatemala: nt sequences, infectious pseudorecombinants, and phylogenetic relationships. Phytopathology 84321-329. [Google Scholar]
- 18.Fauquet, C., M. Mayo, J. Maniloff, U. Desselberger, and L. Ball (ed). 2005. Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA.
- 19.Flock, R. A., and D. E. Mayhew. 1981. Squash leaf curl, a new disease of cucurbits in California. Plant Dis. 6575-76. [Google Scholar]
- 20.Frischmuth, T., M. Engel, S. Lauster, and H. Jeske. 1997. Nucleotide sequence evidence for the occurrence for three distinct whitefly-transmitted, Sida-infecting bipartite geminiviruses in Central America. J. Gen. Virol. 782675-2682. [DOI] [PubMed] [Google Scholar]
- 21.Gilbertson, R. L., S. H. Hidayat, E. J. Paplomatas, M. R. Rojas, Y. M. Hou, and D. P. Maxwell. 1993. Reassortment between infectious cloned DNA components of tomato mottle and bean dwarf mosaic geminiviruses. J. Gen. Virol. 7423-31. [DOI] [PubMed] [Google Scholar]
- 22.Guzman, P., M. R. Sudarshana, Y.-S. Seo, M. R. Rojas, E. Natwick, T. Turini, K. Mayberry, and R. L. Gilbertson. 2000. A new bipartite geminivirus (begomovirus) causing leaf curl and crumpling in cucurbits in the Imperial Valley of California. Plant Dis. 84488. [DOI] [PubMed] [Google Scholar]
- 23.Hanley-Bowdoin, L., S. B. Settlage, B. M. Orozco, S. Nagar, and D. Robertson. 1999. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 1871-106. [PubMed] [Google Scholar]
- 24.Hill, J. E., J. O. Strandberg, E. Hiebert, and S. G. Lazarowitz. 1998. Asymmetric infectivity of pseudorecombinants of cabbage leaf curl virus and squash leaf curl virus: implications for bipartite geminivirus evolution and movement. Virology 250283-292. [DOI] [PubMed] [Google Scholar]
- 25.Holtke, H. J., G. Sagner, C. Kessler, and G. Schmitz. 1992. Sensitive chemiluminescent detection of digoxigenin-labeled nucleic acids: a fast and simple protocol and its application. BioTechniques 12104-113. [PubMed] [Google Scholar]
- 26.Hong, Y., X. Wang, and B. Tian. 1995. Chinese squash leaf curl virus: a new whitefly-transmitted geminivirus. Sci. China 38179-186. [PubMed] [Google Scholar]
- 27.Hou, Y. M., and R. L. Gilbertson. 1996. Increased pathogenicity in a pseudorecombinant bipartite geminivirus correlates with intermolecular recombination. J. Virol. 705430-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Idris, A. M., and J. K. Brown. 1998. Sinaloa tomato leaf curl geminivirus: biological and molecular evidence for a new subgroup III virus. Phytopathology 88648-657. [DOI] [PubMed] [Google Scholar]
- 29.Idris, A. M., and J. K. Brown. 2004. Cotton leaf crumple virus is a distinct western hemisphere begomovirus species with complex evolutionary relationships indicative of recombination and reassortment. Phytopathology 941068-1074. [DOI] [PubMed] [Google Scholar]
- 30.Idris, A. M., A. Abdel-Salam, and J. K. Brown. 2006. Introduction of the New World Squash leaf curl virus to squash (Cucurbita pepo) in Egypt: a potential threat to important food crops. Plant Dis. 901262. [DOI] [PubMed] [Google Scholar]
- 31.Ingham, D. J., E. Pascal, and S. G. Lazarowitz. 1995. Both bipartite geminivirus movement proteins define virus host range, but only BL1 defines pathogenicity. Virology 207191-204. [DOI] [PubMed] [Google Scholar]
- 32.Isakeit, T., N. L. Robertson, J. K. Brown, and R. L. Gilbertson. 1994. First report of Squash leaf curl virus on watermelon in Texas. Plant Dis. 781010. [Google Scholar]
- 33.Kheyr-Pour, A., K. Bananji, G. A. Dafalla, P. Caciagli, E. Noris, A. Ahoomanmesh, H. Lecoq, and B. Gronenborn. 2000. Watermelon chlorotic stunt virus from Sudan and Iran: sequence comparisons and identification of a whitefly-transmission determinant. Phytopathology 90629-635. [DOI] [PubMed] [Google Scholar]
- 34.Kon, T., L. M. Dolores, N. B. Bajet, S. Hase, H. Takahashi, and M. Ikegami. 2003. Molecular characterization of a strain of Squash leaf curl China virus from the Philippines. J. Phytopathol. 151535-539. [Google Scholar]
- 35.Lazarowitz, S. G. 1992. Geminiviruses: genome structure and function. Crit. Rev. Plant Sci. 11327-349. [Google Scholar]
- 36.Lazarowitz, S. G. 1991. Molecular characterization of two bipartite geminiviruses causing squash leaf curl disease: role of viral replication and movement functions in determining host range. Virology 18070-80. [DOI] [PubMed] [Google Scholar]
- 37.Lazarowitz, S. G., L. C. Wu, S. G. Rogers, and J. S. Elmer. 1992. Sequence-specific interaction with the viral AL1 protein identifies a geminivirus DNA replication origin. Plant Cell 4799-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin, D. P., and E. P. Rybicki. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16562-563. [DOI] [PubMed] [Google Scholar]
- 39.Morilla, G., B. Krenz, H. Jeske, E. R. Bejarano, and C. Wege. 2004. Tête à tête of tomato yellow leaf curl virus and tomato yellow leaf curl Sardinia virus in single nuclei. J. Virol. 7810715-10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reference deleted.
- 41.Pita, J. S., V. N. Fondong, A. Sangare, G. W. Otim-Nape, S. Ogwal, and C. M. Fauquet. 2001. Recombination, psuedo-recombination and synergism of geminiviruses are determinant keys to epidemic of the cassava mosaic disease in Uganda. J. Gen. Virol. 82655-661. [DOI] [PubMed] [Google Scholar]
- 42.Qin, S., B. M. Ward, and S. G. Lazarowitz. 1998. The bipartite geminivirus coat protein aids BR1 function in viral movement by affecting the accumulation of single-stranded DNA. J. Virol. 729247-9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Revill, P. A., C. V. Ha, S. C. Prchun, M. T. Vu, and J. L. Dale. 2003. The complete nt sequence of two distinct geminiviruses infecting cucurbits in Vietnam. Arch. Virol. 1481523-1541. [DOI] [PubMed] [Google Scholar]
- 44.Rosemeyer, M. E., J. K. Brown, and M. R. Nelson. 1986. Five viruses isolated from field-grown buffalo gourd, Cucurbita foetidissima HBK, a potential crop for semi-arid lands. Plant Dis. 70405-409. [Google Scholar]
- 45.Rybicki, E. P. 1994. A phylogenetic and evolutionary justification for three genera of Geminiviridae. Arch. Virol. 13949-77. [DOI] [PubMed] [Google Scholar]
- 46.Saiki, R. K., S. Scharf, F. Falcona, K. B. Mullis, G. T. Hom, H. A. Erlich, and N. Arnheim. 1985. Enzymatic amplification of B-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 2301350-1354. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Sawyer, S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6526-538. [DOI] [PubMed] [Google Scholar]
- 49.Schrijnwerkers, C. C. F. M., N. Huijberts, and L. Bos. 1991. Zucchini yellow mosaic virus: two outbreaks in The Netherlands and seed transmissibility. N. J. Plant Pathol. 97187-191. [Google Scholar]
- 50.Stenger, D. C., J. E. Duffus, and B. Viallon. 1990. Biological and genomic properties of a geminivirus isolated from pepper. Phytopathology 80704-709. [Google Scholar]
- 51.Reference deleted.
- 52.Unseld, S., M. Ringel, A. Konrad, S. Lauster, and T. Frischmuth. 2000. Virus-specific adaptations for the production of a pseudorecombinant virus formed by two distinct bipartite geminiviruses from Central America. Virology 274179-2000. [DOI] [PubMed] [Google Scholar]
- 53.Wyatt, S. D., and J. K. Brown. 1996. Detection of subgroup III geminivirus isolates in leaf extracts by degenerate primers and polymerase chain reaction. Phytopathology 861288-1293. [Google Scholar]
- 54.Xei, Y., and X. P. Zhou. 2003. Molecular characterization of squash leaf curl Yunnan virus, a new begomovirus and evidence for recombination. Arch. Virol. 1482047-2054. [DOI] [PubMed] [Google Scholar]