Abstract
The early autologous neutralizing antibody response in human immunodeficiency virus type 1 (HIV-1) subtype C infections is often characterized by high titers, but the response is type specific with little to no cross-neutralizing activity. The specificities of these early neutralizing antibodies are not known; however, the type specificity suggests that they may target the variable regions of the envelope. Here, we show that cross-reactive anti-V3 antibodies developed within 3 to 12 weeks in six individuals but did not mediate autologous neutralization. Using a series of chimeric viruses, we found that antibodies directed at the V1V2, V4, and V5 regions contributed to autologous neutralization in some individuals, with V1V2 playing a more substantial role. However, these antibodies did not account for the total neutralizing capacity of these sera against the early autologous virus. Antibodies directed against the C3-V4 region were involved in autologous neutralization in all four sera studied. In two sera, transfer of the C3-V4 region rendered the chimera as sensitive to antibody neutralization as the parental virus. Although the C3 region, which contains the highly variable α2-helix was not a direct target in most cases, it contributed to the formation of neutralization epitopes as substitution of this region resulted in neutralization resistance. These data suggest that the C3 and V4 regions combine to form important structural motifs and that epitopes in this region are major targets of the early autologous neutralizing response in HIV-1 subtype C infection.
The envelope glycoprotein (Env) of human immunodeficiency virus type 1 (HIV-1) is the target of neutralizing antibodies (NAbs). Almost all individuals develop NAbs to their own virus (autologous neutralization) within a few months of infection (12, 21, 32, 37). In subtype C, these antibodies develop to high titer and are type specific with little or no cross-neutralizing activity within the first year of infection (12, 21). The target(s) of these early antibodies are unknown, but their type specificity suggests that they might recognize the most variable regions of Env, namely V1V2, V3, V4, and V5. Characterization of the targets of NAbs in early infection will allow a better understanding of the epitopes involved in early neutralization.
Anti-V3 antibodies play a minimal role in neutralization of primary viruses (1, 22) because the V3 loop is occluded on the trimeric Env (13, 19, 30). Comparisons of the V3 regions of subtype B and subtype C viruses suggest that there are subtype-specific selection pressures applied to this region. Among subtype B viruses, a high nonsynonymous-to-synonymous substitution ratio typifies the V3 region, whereas in subtype C this region remains relatively conserved, with a much higher nonsynonymous-to-synonymous substitution ratio in the C3 region downstream of V3 (8, 17). The highly conserved nature of V3 in subtype C suggests that it is unlikely to play a role in type-specific neutralization. However, anti-V3 antibodies have been implicated in autologous neutralization of South African subtype C viruses (2).
The V1V2 region regulates neutralization sensitivity by occluding conserved epitopes such as the coreceptor binding site (3, 18-20, 30, 35, 39). Variable regions (V1 to V4) have also been implicated in shielding neutralization determinants in subtype C viruses, where infection may be mediated by viruses with relatively short variable loops and correspondingly high sensitivity to neutralization by donor sera (5). We have previously shown that the V1V2 length of subtype C viruses correlated with resistance to broadly cross-neutralizing serum (12) and that a corresponding relationship exists between the length of the variable loops (V1V2 and V1 to V4) of subtype C viruses and their ability to induce antibodies which cross-neutralize heterologous subtype C viruses (31).
In contrast to the role of V1V2 in shielding conserved neutralization epitopes, V1V2 may also act as a neutralization target in laboratory-adapted isolates (6) and primary isolates (10, 15, 24, 25, 29, 38). In subtype C viruses, the role of V1V2 in neutralization resistance was examined by generating chimeric viruses within four transmission pairs (33). In most cases longer V1V2 loops conferred neutralization resistance while viruses with shorter loops were generally more sensitive, in accordance with the idea that V1V2 masks neutralization-sensitive sites. However, in some viruses, longer loops conferred neutralization sensitivity, possibly because they contained neutralization epitopes. We have also previously suggested that V1V2 may be a potential target of autologous NAbs in subtype C infection (12).
Unlike V1V2, the role of V4 and V5 in neutralization resistance is not clear, although these regions are likely to impact Env conformation and glycan packing (16, 36, 37), thereby sterically limiting accessibility to neutralization determinants. The V4 region in subtype C viruses is generally shorter and significantly more positively charged than in subtype B viruses (9, 21, 23). Increased length in the V4 region in subtype C viruses correlated with resistance to neutralization in linked donor plasma but less so for heterologous plasma, suggesting that changes in V4 may be driven by type-specific NAbs (34).
As with the V3 region of HIV-1, there are distinct structural differences between subtypes B and C in the α2-helix of C3, located in the outer domain of gp120 slightly upstream of the V3 loop. As mentioned, the C3 region of subtype C viruses appears to be under strong diversifying pressure (8). Furthermore, the α2-helix of subtype C viruses is more solvated and shows greater amphipathicity than its subtype B counterpart (9), despite high sequence entropy at the polar faces. Although it is not clear whether these structural and diversity differences in subtype C reflect subtype-specific properties of NAbs, unique mutations within the α2-helix were linked with resistance to neutralization by donor plasma (34). However, viruses chimeric for the α2-helix did not show altered susceptibility to neutralization by donor plasma, implying that another region(s) of Env together with the α2-helix plays a role in resistance to neutralization.
Here, we have attempted to dissect the specificities of antibodies responsible for autologous neutralization in the first year of infection with HIV-1 subtype C, focusing on gp120, where the regions of greatest variability are located.
MATERIALS AND METHODS
Participants.
Participants were from the CAPRISA (Centre for the AIDS Programme of Research in South Africa) 002 Acute Infection study, a cohort of 245 high-risk HIV-negative women which was established in 2004 in Durban, South Africa, for follow-up and subsequent identification of HIV seroconversion. We previously reported that 14 of these individuals had potent but type-specific autologous neutralizing activity (12). In this study, six of these individuals were selected for in-depth analysis on the basis of potent autologous neutralizing responses but absence of cross-neutralizing capacity at 12 months postinfection. Written informed consent was obtained from all participants. This study received ethical approval from the University of the Witwatersrand, University of KwaZulu-Natal, and University of Cape Town.
Cell lines.
The JC53-bl cell line, engineered by J. Kappes and X. Wu, was obtained from the NIH AIDS Research and Reference Reagent Program. 293T cells were obtained from George Shaw (University of Alabama, Birmingham, AL). Both cell lines were cultured in Dulbecco's modified Eagle's medium (Gibco BRL Life Technologies) containing 10% heat-inactivated fetal bovine serum and 50 μg/ml gentamicin (Sigma). Cell monolayers were disrupted at confluence by treatment with 0.25% trypsin in 1 mM EDTA.
Peptide ELISAs.
Peptides (33-mer) representing the V3 region of six subjects were purchased from NMI peptides (Reutlingen, Germany). Autologous peptides spanning the V1V2 region (15-mer peptides overlapping by 5 amino acids) of the gp160 were purchased through New England Peptides (Gardner, MA). Peptides from the Ebola surface proteins were used as irrelevant peptide controls. Peptides were coated onto high-binding 96-well enzyme-linked immunosorbent assay (ELISA) plates at a concentration of 2.5 μg/ml in sodium bicarbonate buffer (pH 8.5), 50 μl/well, overnight at 4°C. Unbound peptide was removed by washing four times with phosphate-buffered saline containing 0.3% Tween 20 (wash solution), and plates were blocked for 1 h at room temperature with 200 μl of phosphate-buffered saline, 0.3% Tween 20, and 5% nonfat milk (block solution). Serum samples were diluted 1:500 in block solution, and 100 μl per well was added, followed by incubation at room temperature for 1 h. Plates were washed four times with wash solution before the addition of 100 μl of secondary antibody (alkaline phosphatase-labeled goat anti-human [Fc-specific] antibody [Sigma-Aldrich, St. Louis, MO] diluted 1:1,000 in blocking solution) for 1 h at 37°C. After four washes with wash solution, bound antibody was detected using p-nitrophenyl phosphate substrate and stopped by the addition of 25 μl of 3 M NaOH stop solution. Samples were scored positive when the optical density at 405 nm (OD405) exceeded 2.5 times the negative control.
Viruses.
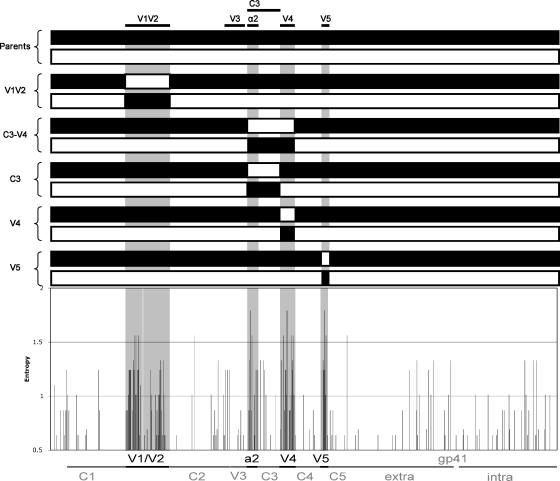
Envelope clones CAP45.2.00.G3J, CAP63.2.00.A9J, CAP84.2.00.32J, CAP88.2.00.B5J, CAP206.2.00.08J, and CAP239.2.00.G3J have been previously described (12). The gp120 sequences and the lengths and numbers of predicted N-linked glycosylation sites of the V1V2, C3-V4, V4, and V5 regions are shown in Fig. 3 and Table 1, respectively. Env-pseudotyped viruses were obtained by cotransfecting the Env plasmid with pSG3ΔEnv (37) using Fugene transfection reagent (Roche) as previously described (12).
FIG. 3.
Amino acid alignment of gp120 from the six parental envelope clones used to construct chimeric viruses. Regions exchanged between viruses are annotated, and within the C3 region, the α2-helix and β-14 and β-15 strands are shown. PNGS are highlighted in gray, and amino acids involved in CD4 binding (40) are indicated by asterisks.
TABLE 1.
Characteristics of parental viruses
| Participant identifier | Env clone name | Accession no. | Length (aa) and no. of PNGS of the indicated regiona
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| V1V2
|
α2-V4
|
V4 alone
|
V5
|
|||||||
| Length | PNGS | Length | PNGS | Length | PNGS | Length | PNGS | |||
| CAP45 | CAP45.2.00.G3J | EF203960 | 66 | 5 | 75 | 7 | 22 | 3 | 17 | 1 |
| CAP84 | CAP84.2.00.32J | EF203963 | 66 | 5 | 77 | 6 | 24 | 4 | 16 | 1 |
| CAP63 | CAP63.2.00A9J | EF203973 | 72 | 6 | 83 | 5 | 30 | 2 | 17 | 3 |
| CAP88 | CAP88.2.00.B5J | EF203972 | 73 | 5 | 76 | 5 | 22 | 3 | 18 | 2 |
| CAP206 | CAP206.2.00.08J | EF203967 | 70 | 6 | 78 | 6 | 25 | 4 | 18 | 1 |
| CAP239 | CAP239.2.00.G3J | EF203983 | 64 | 5 | 84 | 9 | 31 | 6 | 20 | 2 |
aa, amino acids.
Generation of chimeras.
Chimeric gp160 proteins were created using an overlapping PCR strategy with the inserts and flanking regions amplified in separate reactions (primer sequences are available on request from the authors). After linkage, the 3-kb chimeric PCR fragments, generated using env A and env M primers (7), were cloned into the pCDNA 3.1D-TOPO vector (Invitrogen) and screened for function as previously described (11). Chimerism was confirmed by sequence analysis.
gp160 sequencing and analysis.
Cloned env genes were sequenced using an ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA) and resolved on an ABI 3100 automated genetic analyzer. The full-length gp160 sequences were assembled and edited using Sequencher software, version 4.0 (Genecodes, Ann Arbor, MI). The number of potential N-linked glycosylation sites (PNGS) was determined using N-glycosite (http:/www.hiv.lanl.gov/content/hiv-db/GLYCOSITE/glycosite.html). Multiple sequence alignments were performed using Clustal X (version 1.83) and edited with BioEdit (version 5.0.9). Entropy plots were generated using the software Entropy-One (http://www.hiv.lanl.gov/content/hiv-db/ENTROPY/entropy.html).
Neutralization assays.
Neutralization was measured as described previously (12) by a reduction in luciferase gene expression after a single round infection of JC53-bl cells with Env-pseudotyped viruses (26). Titers were calculated as the inhibitor concentration (IC50) or reciprocal plasma/serum dilution (ID50) causing 50% reduction of relative light units. Peptide adsorption neutralization assays were performed by preincubating serum with autologous peptides at a final concentration (after addition of cells) of 50 μg/ml for 1 h at 37°C before continuing with the standard neutralization assay.
RESULTS
Anti-V3 antibodies are present early in subtype C infection but do not mediate autologous neutralization.
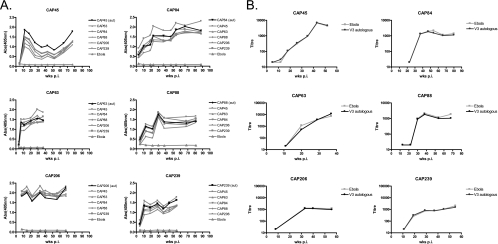
Autologous V3 peptides (33-mer) were synthesized and used to measure anti-V3 binding antibodies in sera from six HIV-1-infected individuals during the first year of infection. Sera were tested against both autologous and heterologous V3 peptides. In all individuals, anti-V3 binding antibodies developed early in infection, with antibodies first detected between 3 and 12 weeks postinfection (p.i). (Fig. 1A). In one case, anti-V3 antibodies preferentially recognized the autologous peptide (participant CAP45) although there was still good recognition of heterologous peptides. In the remaining individuals, there was a high level of cross-reactivity between peptides, with binding to the autologous peptide no stronger than to heterologous peptides.
FIG. 1.
(A) Development of anti-V3 binding antibodies in six individuals (absorbance at 405 nm [Abs405] versus weeks postinfection). Binding of each serum to the autologous peptides is shown in black, with binding to heterologous peptides shown in gray. (B) Peptide adsorption neutralization assays. Sera were preincubated with autologous V3 peptides (black) or irrelevant peptides (gray) prior to performing neutralization assays against the autologous enrollment virus. Neutralization titer (ID50) is plotted against the number of weeks p.i.
Although the extensive cross-reactivity of anti-V3 antibodies makes it unlikely such antibodies mediate type-specific autologous neutralization, anti-V3 antibodies have been previously implicated in autologous neutralization (2). We therefore performed neutralization assays in which serum was preincubated with peptides to adsorb anti-V3 antibodies. Binding ELISAs where serum was preincubated with peptides confirmed that adsorption of anti-V3 antibodies was efficient (data not shown). However, in all six subjects, we observed no change in autologous NAb titers in the presence of autologous peptides (Fig. 1B), suggesting that these binding antibodies do not mediate autologous neutralization.
What are the likely targets of the autologous neutralizing response?
The type specificity of the autologous response suggests that these antibodies may target the most variable regions of the Env. Entropy analysis of gp160 sequences from six individuals at enrollment showed that the V1V2, V4, and V5 regions as well as the C3 region (specifically the α2-helix) were the most variable between viruses (Fig. 2, gray regions). There were numerous substitutions, deletions, insertions, and shifts in glycosylation sites in these regions (Fig. 3). The V3 region showed minimal diversity, as has been reported previously for subtype C viruses (8, 17). The absence of variability in the V3 region supports our data, suggesting that this region is not the target of the potent neutralizing responses in these individuals. We therefore turned our attention to the remaining variable loops of the HIV-1 envelope, namely, V1V2, V4, and V5. Using a series of chimeric viruses, schematically shown in Fig. 2, we attempted to transfer sensitivity to NAbs by exchanging variable loops between pairs of viruses that did not share neutralization determinants.
FIG. 2.
Entropy analysis of six amino acid Env (gp160) sequences. Regions of greatest variability (V1V2, C3, V4, and V5) are highlighted in gray. Above the entropy plot is a schematic depicting pairs of chimeric viruses constructed for this study.
Anti-V1V2 antibodies targeting conformational epitopes mediate autologous neutralization in some individuals. (i) Chimeras.
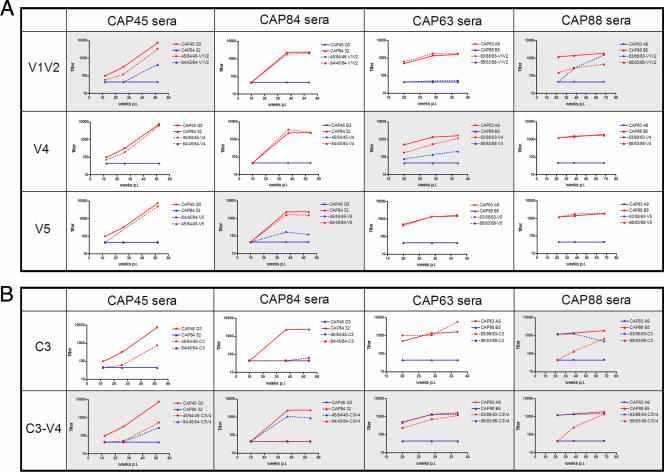
We generated three pairs of viruses chimeric for the V1V2 region using an overlapping PCR strategy. Pairs of chimeras were tested, along with both parental viruses, in neutralization assays against autologous sera at various time points up to 1 year p.i. (excepting participant CAP63, who progressed rapidly to AIDS and entered a treatment program at 37 weeks p.i.).
Results of autologous neutralization assays are shown in Fig. 4A (data not shown for CAP206 and CAP239 chimeras). Each graph represents neutralization by serum from one individual of two chimeras and their parental viruses. In four of six participants (CAP45, CAP88, CAP206, and CAP239) a proportion of autologous neutralization appeared to be mediated by anti-V1V2 antibodies, as reflected by acquisition of partial or complete sensitivity in chimeric viruses in which the V1V2 was swapped in and by loss of some sensitivity in viruses where the autologous V1V2 had been swapped out. For example, serum from participant CAP45 neutralized the parental virus CAP45 (IC50 of 6,377 by 12 months p.i.) (Fig. 4A, solid red line) but showed no activity against CAP84 (solid blue line) even after 1 year of infection. However, when the V1V2 region from CAP45 was inserted into the CAP84 backbone (yielding the chimera designated 84/45/84-V1V2) (Fig. 4A, dotted blue line), this chimera acquired partial sensitivity to serum from CAP45 (IC50 of 392). Conversely, the reverse chimera (45/84/45-V1V2; dotted red line) was slightly less sensitive than the parental CAP45 virus to neutralization by CAP45 serum. Collectively, these data suggest that some of the NAbs in serum from CAP45 are directed at the V1V2 region. Chimera 63/88/63-V1V2 was especially striking, and acquired nearly complete sensitivity to serum from CAP88, with titers similar to those measured against the autologous parental viruses at 12 months p.i. In two other sera, CAP206 (IC50 of 102 against 239/206/239-V1V2 compared to autologous IC50 of 1,843) and CAP239 (IC50 of 1,252 against 206/239/206-V1V2 compared to autologous IC50 of 3,535), anti-V1V2 NAbs were also implicated in autologous neutralization (data not shown). In two participants (CAP63 and CAP84), there was no evidence that neutralization was mediated by anti-V1V2 antibodies as parental and chimeric viruses were equally sensitive to neutralization.
FIG. 4.
Neutralization data for chimeric and parental viruses. ID50 titers are shown on the y axis and the number of weeks p.i. are on the x axis. Solid lines indicate parent viruses; dotted lines indicate chimeras, with the color matching the backbone of the parental viruses. Each graph represents neutralization data using serum from a single individual, indicated above each column. Shaded boxes indicate transfer of sensitivity in chimeras. (A) Neutralization data for viruses chimeric for the V1V2, V4, and V5 regions. (B) Neutralization data for viruses chimeric for the C3 and C3-V4 regions.
(ii) Peptides.
In order to determine whether the neutralization epitopes in V1V2 were linear, we performed adsorption neutralization assays using autologous V1V2 peptides for all six individuals (15-mer peptides overlapping by 5-mer). However, none of these peptides was able to inhibit neutralization of parental viruses by matched sera. Thus, while the use of chimeric viruses suggested the presence of neutralization targets in V1V2 in four cases, these data suggested that these epitopes are likely to be of a conformational nature. In contrast to V3 binding antibodies, only one of six individuals had detectable binding antibodies to these V1V2 peptides (ELISA data not shown). Binding antibodies to the peptide TNKSMNGEIDMKEEM in the V1 region were detected at approximately 47 weeks p.i in CAP88 serum. Interestingly, this response was considerably later than the autologous NAb response in CAP88, which was detectable at 22 weeks p.i. (12), although it was similar to the timing of anti-V1V2 NAbs detected using the chimera 63/88/63-V1V2.
Antibodies targeting the V4 and V5 regions do not play a major role in autologous neutralization.
Viruses chimeric for the V4 and V5 regions were created from six parental env clones (however, chimeras between CAP206 and CAP239 for the V4, V5, C3, and C3-V4 regions were nonfunctional) and were tested in neutralization assays against parental sera from multiple time points (Fig. 4A). NAbs targeting the V4 region were detected only in CAP63, where insertion of the CAP63 V4 region into a CAP88 backbone (88/63/88-V4) showed some sensitivity to neutralization by CAP63 serum (IC50 of 212 at 1 yr p.i. compared to an autologous titer of 1,633). The reverse chimera, 63/88/63-V4, showed some reduction in sensitivity, dropping from the parental titer of 1,633 to 1,142 at 12 months p.i., suggesting that in CAP63, NAbs targeting V4 were present. In the remaining three sera no anti-V4 NAbs were observed.
V5 chimeras indicated that anti-V5 NAbs were undetectable in sera from participants CAP45, CAP63, and CAP88. CAP84 serum contained low levels of NAbs directed at the V5 region, with insertion of the V5 region of CAP84 into the resistant CAP45 backbone allowing transfer of some sensitivity (IC50 of 120 compared to the autologous titer of 2,447 against CAP84) and a reciprocal drop in titer when the V5 region was swapped out (IC50 dropping from 2,447 to 1,551). In general, the role of antibodies targeting the V4 and V5 regions in autologous neutralization appeared to be minor.
The C3 region in subtype C is involved in the formation of NAb epitopes.
The α2-helix of the C3 region, which showed high entropy in the results shown in Fig. 2, has been implicated in neutralization resistance in subtype C viruses (34). We therefore also tested four viruses chimeric for the C3 region against parental sera. As with the other chimeras, we considered that autologous neutralization may be mediated by antibodies directed at the C3 region if there is acquisition of partial or complete sensitivity in heterologous viruses in which the autologous C3 has been introduced.
Neutralization assays using sera from CAP45, CAP84, and CAP63 against constructs carrying the autologous C3 regions showed no sensitivity to neutralization, indicating that C3 alone was not a target of NAbs in these three sera (Fig. 4B, dotted blue lines). However, interestingly, in CAP45 and CAP84, replacement of the autologous C3 with an unrelated C3 resulted in a dramatic drop in the titer in both chimeras. Taken together, these data suggest that C3 in CAP45 and CAP84 may be involved in the formation of neutralization epitope(s), as has been proposed by other investigators (34), though it is not a direct target of NAbs. Replacement of the CAP63 C3 with CAP88 C3 did not result in a reduction in titer compared to the parent virus. CAP88 was unique in that 63/88/63-C3 acquired considerable sensitivity to CAP88 serum, with a concomitant reduction in the titer of the reverse chimera, 88/63/88-C3, suggesting that in CAP88, C3 may itself be a direct target of circulating NAbs in this individual.
The C3-V4 region is a major target of autologous neutralization in subtype C infection.
In addition to the C3 chimeras, we constructed viruses chimeric for the C3 through V4 regions, which have been proposed to be in close proximity and to interact with one another (9). Significantly, in all four individuals, the introduction of the autologous C3-V4 into a heterologous parent virus resulted in an increase in sensitivity to neutralization, with two of the chimeras (63/88/63-C3-V4 and 88/63/88-C3-V4) reaching titers observed in autologous neutralization of the parent virus (Fig. 4B). In all sera tested, the chimera in which the autologous C3-V4 was swapped out also had a partial (CAP45, CAP63, and CAP88) or complete (CAP84) reduction in the sensitivity to parental sera, further confirming the role of the C3-V4 region as a target of autologous neutralization in these individuals.
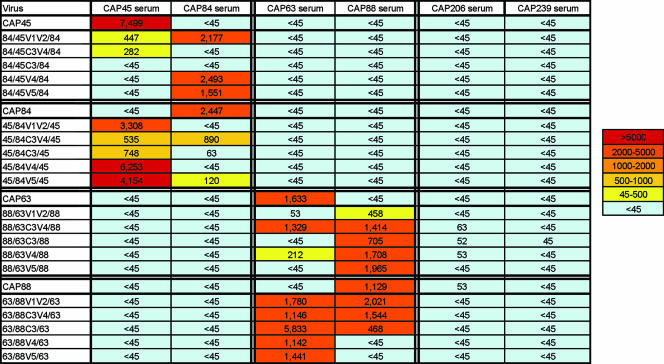
Chimeric viruses do not display a generally neutralization-sensitive phenotype.
In order to assess whether the acquisition of neutralization sensitivity was due to these chimeras' acquiring a more generally neutralization-sensitive phenotype, perhaps through more open conformations, we tested them against heterologous CAPRISA sera obtained 12 months p.i. We have previously reported that sera from these six individuals, while potently neutralizing the autologous enrollment virus, showed no cross-reactivity at this time point (12). No neutralization of chimeras by sera other than the parental sera was observed, suggesting that acquisition of neutralization sensitivity was specific to the region transferred (Fig. 5). Furthermore, we showed above that all six individuals developed high-titer anti-V3 binding antibodies. These antibodies, although unable to neutralize parental viruses, were able to potently block viral entry of an HIV-2 scaffold into which either subtype B or C consensus V3 regions have been introduced (K. Davis et al., unpublished data). The lack of cross-neutralization, despite high titers of anti-V3 antibodies, indicated that the V3 region of these chimeras was not exposed.
FIG. 5.
Chimeras do not acquire a generally neutralization-sensitive phenotype. Summary of neutralization data for all chimeras and parental viruses tested against heterologous sera. Data are shown as neutralization titer (ID50) and are color coded.
DISCUSSION
This study aimed to determine the specificities of NAbs which mediate potent but type-specific autologous neutralization in early HIV-1 subtype C infection. Although high levels of cross-reactive binding anti-V3 antibodies were detected, these did not mediate neutralization of primary viruses. NAbs targeting the V1V2, V4, and V5 regions contributed to autologous neutralization in some individuals to various degrees, with V1V2 playing a more significant role than V4 or V5. In contrast, antibodies directed against the C3-V4 region were involved in autologous neutralization in all sera tested, suggesting that epitopes in this region are major targets of the early autologous neutralizing response in subtype C infection.
The V3 region is known to be highly immunogenic, with most individuals developing antibodies soon after infection (27). Anti-V3 antibodies developed in these six individuals within 3 to 12 (median, 5 weeks) weeks, which in five of six subjects corresponded to the earliest time point tested. This is considerably earlier than the autologous neutralizing response in these individuals, which became detectable within a median of 19 weeks (range, 12 to 37 weeks) (12). These anti-V3 binding antibodies were characterized by substantial cross-reactivity, which is not surprising, given that the V3 region in subtype C viruses is remarkably well conserved (8). Indeed, within the CAPRISA cohort, two viruses had identical V3 loops (CAP8 and CAP244) but no neutralization cross-reactivity (12). It is possible that, in a similar manner as has been suggested for CD4-induced antibodies (4), anti-V3 antibodies play a role in constraining the virus to shield the V3 loop, thereby limiting the accessibility of the V3 region. However, the high degree of conservation in the subtype C V3 suggests that anti-V3 antibodies in subtype C infection are not of great biological relevance in driving viral escape from the neutralization response, a suggestion which is borne out in this study where we detected no role for anti-V3 antibodies in autologous neutralization using peptide adsorption studies.
The role of V1V2 in shielding neutralization determinants is well recognized (3, 18-20, 30, 35, 39). However, we along with others have proposed that the V1V2 may also serve as the neutralization target in some cases (12, 33). These data suggest that in some individuals a proportion of autologous neutralization is indeed directed at conformational epitopes contained within the V1V2 region. The role of V4 and V5 was less significant. Although we did observe some activity directed against V4 alone (CAP63) and V5 (CAP84), in both these cases the contribution of antibodies targeting these regions as a proportion of total autologous neutralization was minor.
Although it was evident that all sera contained some activity against variable loops, this activity did not account for the total neutralizing capacity of these sera against the autologous enrollment virus. This prompted us to look at other regions potentially targeted by the early autologous neutralizing response.
The C3 region of the HIV-1 subtype C envelope and, in particular, unique mutational patterns in the α2-helix have been implicated in neutralization resistance (34). However, exchange of the α2-helix between resistant donor Env and sensitive recipient Env did not result in transfer of neutralization phenotypes (34), an observation that is generally consistent with our data, where in three of four individuals (Fig. 4B) viruses containing heterologous C3 regions (including the α2-helix) did not acquire sensitivity to the matching heterologous serum. However, of these three sera, it was noticeable that two of the three reverse chimeras, where the C3 region was removed and replaced with an unrelated C3 region, were much less sensitive to neutralization than the parental viruses, suggesting that removal of the C3 altered NAb epitopes. This was not the case in the study by Rong et al. (34), where the replacement of the α2-helix did not result in substantial loss of sensitivity to neutralization. Given the strong interaction between the β-14 strand of C3 and the V4 region (9), it is possible that replacement of the α2-helix in conjunction with the β-14 strand in the C3 chimeras as described in this study had a deleterious effect on the conformation of the V4 region, whereas in the α2-helix chimeras described by Rong and coworkers, the interaction of V4 with the autologous β-14 strand was intact, which afforded some stability to the conformation of the V4 region. Furthermore, the parent viruses of chimeras were transmission pairs, with less variation across the entire envelope.
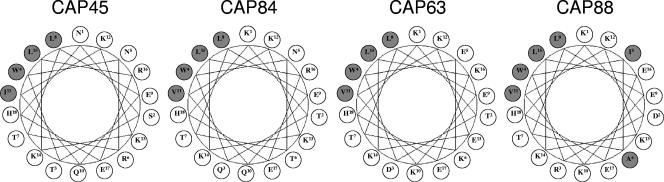
In general, the α2-helices of subtype B viruses are more hydrophobic than the subtype C α2-helices, which have more defined polar and nonpolar faces (9). This increased amphipathicity, which is more characteristic of a surface helix, in subtype C may indicate that the α2-helix is more exposed. The high levels of switching between oppositely charged residues, a feature of the subtype C α2-helix and not of subtype B, where variable positions maintain similar charges, may therefore be a mechanism of immune escape (9). It has therefore been proposed that NAbs directly target the α2-helix in subtype C viruses. However, in this study, we observed only one of four individuals (CAP88) where transfer of the C3 region implicated antibodies targeting this region, although it is not clear whether these antibodies were specifically directed against the α2-helix or against other regions in C3. It is noteworthy that the α2-helix of CAP88 contained two hydrophobic residues which replaced residues that are normally hydrophilic (5I and 6A), and the polar and nonpolar faces have been perturbed (Fig. 6). It is therefore possible that the anti-C3 activity observed in CAP88 is the result of an unusual conformation of the α2-helix, possibly altering the exposure of the α2-helix. As noted above, the C3 region also contains the highly variable β-14 strand, as well as the conserved β-15 strand and α3-helix which contain elements of the CD4 binding site, so it is also possible the C3-directed activity in CAP88 is mediated by anti-CD4 binding site (CD4bs) antibodies or others of unknown specificities. Anti-CD4bs antibodies mediating autologous neutralization would be interesting to investigate since they are clearly type specific. Type-specific anti-CD4bs antibodies may not be entirely surprising as even the monoclonal CD4bs NAb immunoglobulin G1b12, which is characterized as broadly neutralizing, neutralizes only approximately half of primary isolates (1). Furthermore, there is evidence that the functional CD4 binding region is smaller than the structural epitope of immunoglobulin G1b12 (14), perhaps allowing some flexibility in secondary binding sites to other nonconserved residues (28).
FIG. 6.
Helical wheel plot of the α2-helices of viruses from CAP45, CAP84, CAP63, and CAP88. Hydrophilic amino acid residues are shown in clear circles, and hydrophobic residues are shown in gray circles.
In contrast to C3 or V4 in isolation, both of which seem to be only sporadically involved in autologous neutralization, chimeras containing the C3 region through to V4, when transferred as a single unit, showed much more dramatic shifts in their neutralization phenotypes. In all four cases, insertion of C3-V4 into an insensitive backbone resulted in increased sensitivity, reaching levels at or close to those seen in the autologous (parental) virus, with a corresponding decrease in sensitivity observed in reverse chimeras. Molecular dynamics simulations have indicated that the N-terminal half of the α2-helix of C3 interacts strongly with the C-terminal portion of V4 (9). Furthermore, it has been proposed that the α2-helix, though not a direct target of the neutralizing response, may contribute to the quaternary structure of the trimer and so affect neutralization epitopes, possibly via the strong interactions this region has with the V4 loop or the with β-10, β-11, β-14, and β-24 strands (34). Our study suggests that the α2-helix and the V4 loop of subtype C viruses may coevolve during antibody escape, with the V4 region undergoing mutations and the α2-helix undergoing concomitant compensatory mutations which therefore track with neutralization resistance. The conformation of V4 may therefore be dependent on the proper context provided by the α2-helix of the C3 region. The transfer of sensitivity observed in this study suggests the possibility that NAbs in all four sera may target the V4 loop but that the conformation of the epitopes in the V4 region is informed by the underlying C3 region. This model is further supported by the C3 chimera data, where in three individuals, and regardless of the presence of the autologous V4 region, the removal of the C3 region destroyed the epitope recognized by circulating NAbs.
The use of chimeras in the study has proven helpful in the identification of regions which are likely targeted by autologous NAbs early in infection. However, a caveat to these data is that variations in the length and glycosylation of the heterologous insert may impact neutralization sensitivity by exposure of epitopes. Furthermore, the exchange of fairly large portions of the envelope may result in conformational changes which are hard to predict. Even in cases such as the transfer of V1V2 between CAP45 and CAP84 (in chimeras 84/45/84-V1V2 and 45/84/45-V1V2) where the exchanged region was of equal length and with the same number of predicted glycosylation sites (Table 1), the effect of residues with various charges and properties on the overall conformation may change. We have shown that in all cases, the chimeras used in this study did not acquire a universally sensitive phenotype, with acquisition of neutralization sensitivity related only to sera from one or another parent virus. These data suggest that where we observed acquisition of neutralization sensitivity in chimeras, this is likely through transfer of epitopes rather than through gross conformational changes.
In general, results obtained using reverse chimeras mirrored the constructs where regions of interest were swapped in, although this was not always the case. For example, the chimera 88/63/88-C3-V4 acquired complete sensitivity to neutralization by CAP63 sera, but in the reverse chimera, 63/88/63-C3-V4, the titers did not drop substantially (Fig. 4B). This may be explained by the presence of other antibody specificities in CAP63 sera which are magnified in the absence of the autologous C3-V4 epitope. The contribution of each specificity to the total neutralizing capacity, as measured by the chimeras, is, however, unlikely to be additive. Chimeras may have subtle conformational changes which could alter the efficiency of entry compared to parental viruses, leading to over- or underestimates of the proportions of other antibody specificities. Although neutralization assays using gp120-gp41 chimeras suggest that autologous neutralization is not mediated by anti-gp41 antibodies (data not shown), there are almost certainly other antibody specificities present in these sera, which could perhaps only be measured using chimeras where larger subregions are transferred together. Indeed, in serum from CAP45, the role of both anti-V1V2 and anti-C3-V4 antibodies seems relatively minor, suggesting that as yet unidentified specificities likely contribute to autologous neutralization in this serum.
Our results indicate that autologous neutralization may be targeted to multiple regions of the gp120. However, a common and striking feature of autologous neutralization in these sera is a high level of antibodies targeting the C3-V4 region, which includes the α2-helix and some determinants of the CD4bs. These data support available bioinformatics data implicating the α2-helix in neutralization resistance in subtype C and further supporting suggestions that the C3 region in subtype C has distinct structural features from that of subtype B. It is perhaps surprising, given the type-specific nature of the autologous response, that the same region was targeted in all four individuals. Clearly, the antibody specificities are distinct, and future work will aim to precisely define the epitopes in this region. Elucidation of the specificities of early autologous NAbs in a globally prevalent subtype will enable studies to understand the mechanism whereby neutralization breadth develops and will provide information regarding accessible epitopes on the transmitted early virus which may inform vaccine design.
Acknowledgments
We thank the clinical and laboratory staff at CAPRISA for providing specimens, Mary Phoswa and Sarah Cohen for sample and data management, and Natasha Taylor for assistance with peptide ELISA. We are grateful to Katie Davis and George Shaw for sharing their data on V3 NAbs. We also thank S. Gnanakaran and J. Mascola for useful discussions.
This work was funded by CAPRISA and CHAVI and by a grant to P.L.M. from the South African Medical Research Council. CAPRISA was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S. Department of Health and Human Services (grant U19 AI51794). P.L.M. and E.S.G. are partially salaried by SAAVI.
Footnotes
Published ahead of print on 5 December 2007.
REFERENCES
- 1.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 7813232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bures, R., L. Morris, C. Williamson, G. Ramjee, M. Deers, S. A. Fiscus, S. Abdool-Karim, and D. C. Montefiori. 2002. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J. Virol. 762233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 717719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decker, J. M., F. Bibollet-Ruche, X. Wei, S. Wang, D. N. Levy, W. Wang, E. Delaporte, M. Peeters, C. A. Derdeyn, S. Allen, E. Hunter, M. S. Saag, J. A. Hoxie, B. H. Hahn, P. D. Kwong, J. E. Robinson, and G. M. Shaw. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 2011407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derdeyn, C. A., J. M. Decker, F. Bibollet-Ruche, J. L. Mokili, M. Muldoon, S. A. Denham, M. L. Heil, F. Kasolo, R. Musonda, B. H. Hahn, G. M. Shaw, B. T. Korber, S. Allen, and E. Hunter. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 3032019-2022. [DOI] [PubMed] [Google Scholar]
- 6.Fung, M. S., C. R. Sun, W. L. Gordon, R. S. Liou, T. W. Chang, W. N. Sun, E. S. Daar, and D. D. Ho. 1992. Identification and characterization of a neutralization site within the second variable region of human immunodeficiency virus type 1 gp120. J. Virol. 66848-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao, F., S. G. Morrison, D. L. Robertson, C. L. Thornton, S. Craig, G. Karlsson, J. Sodroski, M. Morgado, B. Galvao-Castro, H. von Briesen, S. Beddows, J. Weber, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID Networks for HIV Isolation and Characterization. J. Virol. 701651-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 2962354-2360. [DOI] [PubMed] [Google Scholar]
- 9.Gnanakaran, S., D. Lang, M. Daniels, T. Bhattacharya, C. A. Derdeyn, and B. Korber. 2007. Clade-specific differences between human immunodeficiency virus type 1 clades B and C: diversity and correlations in C3-V4 regions of gp120. J. Virol. 814886-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorny, M. K., J. P. Moore, A. J. Conley, S. Karwowska, J. Sodroski, C. Williams, S. Burda, L. J. Boots, and S. Zolla-Pazner. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J. Virol. 688312-8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray, E. S., T. Meyers, G. Gray, D. C. Montefiori, and L. Morris. 2006. Insensitivity of paediatric HIV-1 subtype C viruses to broadly neutralising monoclonal antibodies raised against subtype B. PLoS Med. 3e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray, E. S., P. L. Moore, I. A. Choge, J. M. Decker, F. Bibollet-Ruche, H. Li, N. Leseka, F. Treurnicht, K. Mlisana, G. M. Shaw, S. S. Karim, C. Williamson, and L. Morris. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 816187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartley, O., P. J. Klasse, Q. J. Sattentau, and J. P. Moore. 2005. V3: HIV's switch-hitter. AIDS Res. Hum. Retrovir. 21171-189. [DOI] [PubMed] [Google Scholar]
- 14.Huang, C. C., F. Stricher, L. Martin, J. M. Decker, S. Majeed, P. Barthe, W. A. Hendrickson, J. Robinson, C. Roumestand, J. Sodroski, R. Wyatt, G. M. Shaw, C. Vita, and P. D. Kwong. 2005. Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth. Structure 13755-768. [DOI] [PubMed] [Google Scholar]
- 15.Kayman, S. C., Z. Wu, K. Revesz, H. Chen, R. Kopelman, and A. Pinter. 1994. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J. Virol. 68400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinsey, N. E., M. G. Anderson, T. J. Unangst, S. V. Joag, O. Narayan, M. C. Zink, and J. E. Clements. 1996. Antigenic variation of SIV: mutations in V4 alter the neutralization profile. Virology 22114-21. [DOI] [PubMed] [Google Scholar]
- 17.Korber, B. T., K. MacInnes, R. F. Smith, and G. Myers. 1994. Mutational trends in V3 loop protein sequences observed in different genetic lineages of human immunodeficiency virus type 1. J. Virol. 686730-6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krachmarov, C., A. Pinter, W. J. Honnen, M. K. Gorny, P. N. Nyambi, S. Zolla-Pazner, and S. C. Kayman. 2005. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade A and clade B V3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J. Virol. 79780-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krachmarov, C. P., W. J. Honnen, S. C. Kayman, M. K. Gorny, S. Zolla-Pazner, and A. Pinter. 2006. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J. Virol. 807127-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, B., J. M. Decker, R. W. Johnson, F. Bibollet-Ruche, X. Wei, J. Mulenga, S. Allen, E. Hunter, B. H. Hahn, G. M. Shaw, J. L. Blackwell, and C. A. Derdeyn. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J. Virol. 805211-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, M., F. Gao, J. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 7910108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, M., J. F. Salazar-Gonzalez, C. A. Derdeyn, L. Morris, C. Williamson, J. E. Robinson, J. M. Decker, Y. Li, M. G. Salazar, V. R. Polonis, K. Mlisana, S. A. Karim, K. Hong, K. M. Greene, M. Bilska, J. Zhou, S. Allen, E. Chomba, J. Mulenga, C. Vwalika, F. Gao, M. Zhang, B. T. Korber, E. Hunter, B. H. Hahn, and D. C. Montefiori. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol. 8011776-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Y., K. Svehla, N. L. Mathy, G. Voss, J. R. Mascola, and R. Wyatt. 2006. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J. Virol. 801414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKeating, J. A., C. Shotton, J. Cordell, S. Graham, P. Balfe, N. Sullivan, M. Charles, M. Page, A. Bolmstedt, S. Olofsson, et al. 1993. Characterization of neutralizing monoclonal antibodies to linear and conformation-dependent epitopes within the first and second variable domains of human immunodeficiency virus type 1 gp120. J. Virol. 674932-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montefiori, D. 2004. Evaluating neutralizing antibodies against HIV, SIV and SHIV in luciferase reporter gene assays, p. 12.11.1-12.11.15. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley and Sons, New York, NY. [DOI] [PubMed]
- 27.Moore, J. P., and D. D. Ho. 1993. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J. Virol. 67863-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pantophlet, R., and D. R. Burton. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24739-769. [DOI] [PubMed] [Google Scholar]
- 29.Pinter, A., W. J. Honnen, P. D'Agostino, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2005. The C108g epitope in the V2 domain of gp120 functions as a potent neutralization target when introduced into envelope proteins derived from human immunodeficiency virus type 1 primary isolates. J. Virol. 796909-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 785205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rademeyer, C., P. L. Moore, N. Taylor, D. Martin, I. Choge, E. S. Gray, H. W. Sheppard, C. M. Gray, L. Morris, and C. Williamson. 2007. Genetic characteristics of HIV-1 subtype C envelopes inducing cross-neutralizing antibodies. Virology 368172-181. [DOI] [PubMed] [Google Scholar]
- 32.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 1004144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rong, R., F. Bibollet-Ruche, J. Mulenga, S. Allen, J. L. Blackwell, and C. A. Derdeyn. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J. Virol. 811350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rong, R., S. Gnanakaran, J. M. Decker, F. Bibollet-Ruche, J. Taylor, J. N. Sfakianos, J. L. Mokili, M. Muldoon, J. Mulenga, S. Allen, B. H. Hahn, G. M. Shaw, J. L. Blackwell, B. T. Korber, E. Hunter, and C. A. Derdeyn. 2007. Unique mutational patterns in the envelope α2 amphipathic helix and acquisition of length in gp120 hypervariable domains are associated with resistance to autologous neutralization of subtype C human immunodeficiency virus type 1. J. Virol. 815658-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sagar, M., X. Wu, S. Lee, and J. Overbaugh. 2006. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J. Virol. 809586-9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teeraputon, S., S. Louisirirojchanakul, and P. Auewarakul. 2005. N-linked glycosylation in C2 region of HIV-1 envelope reduces sensitivity to neutralizing antibodies. Viral Immunol. 18343-353. [DOI] [PubMed] [Google Scholar]
- 37.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422307-312. [DOI] [PubMed] [Google Scholar]
- 38.Wu, L., Z. Y. Yang, L. Xu, B. Welcher, S. Winfrey, Y. Shao, J. R. Mascola, and G. J. Nabel. 2006. Cross-clade recognition and neutralization by the V3 region from clade C human immunodeficiency virus-1 envelope. Vaccine 244995-5002. [DOI] [PubMed] [Google Scholar]
- 39.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 695723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. Nabel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]