Abstract
Poliovirus disrupts nucleocytoplasmic trafficking and results in the cleavage of two nuclear pore complex (NPC) proteins, Nup153 and Nup62. The NPC is a 125-MDa complex composed of multiple copies of 30 different proteins. Here we have extended the analysis of the NPC in infected cells by examining the status of Nup98, an interferon-induced NPC protein with a major role in mRNA export. Our results indicate that Nup98 is targeted for cleavage after infection but that this occurs much more rapidly than it does for Nup153 and Nup62. In addition, we find that cleavage of these NPC proteins displays differential sensitivity to the viral RNA synthesis inhibitor guanidine hydrochloride. Inhibition of nuclear import and relocalization of host nuclear proteins to the cytoplasm were only apparent at later times after infection when all three nucleoporins (Nups) were cleaved. Surprisingly, analysis of the distribution of mRNA in infected cells revealed that proteolysis of Nup98 did not result in an inhibition of mRNA export. Cleavage of Nup98 could be reconstituted by the addition of purified rhinovirus type 2 2Apro to whole-cell lysates prepared from uninfected cells, suggesting that the 2A protease has a role in this process in vivo. These results indicate that poliovirus differentially targets subsets of NPC proteins at early and late times postinfection. In addition, targeting of interferon-inducible NPC proteins, such as Nup98, may be an additional weapon in the arsenal of poliovirus and perhaps other picornaviruses to overcome host defense mechanisms.
Poliovirus is a positive-strand RNA virus belonging to the family Picornaviridae. Poliovirus, like other members of this family, encodes a single large polyprotein that is co- and posttranslationally processed by virus-encoded proteases to produce the individual viral gene products (reviewed in reference 40). After the production of viral proteins, RNA synthesis ensues on virus-induced vesicles in the cell cytoplasm. Despite the fact that viral translation, RNA synthesis, and assembly occur in the cytoplasm, a number of host nuclear proteins have been attributed roles in the viral life cycle (3, 6, 27, 32, 33, 51). Consistent with their having a role in the replication of poliovirus in the cytoplasm, several host nuclear factors have been shown to redistribute to the cytoplasm after infection (3, 5, 32, 33, 51).
For nuclear factors to move from the nucleus to the cytoplasm they must transit the nuclear pore complex (NPC), a large protein channel found embedded in the nuclear envelope. The vertebrate NPC has a molecular mass of 125 MDa and is composed of multiple copies of roughly 30 different proteins that are collectively called nucleoporins (Nups; reviewed in reference 18). A number of findings have suggested that the redistribution of host nuclear factors to the cytoplasm in poliovirus-infected cells is due to alterations of the NPC and a disruption in nucleocytoplasmic trafficking. Previous work has shown that two NPC proteins, Nup153 and Nup62, are targeted for degradation in poliovirus-infected cells and that this correlates temporally with relocalization of host-nuclear proteins and an inhibition of the classical nuclear import pathway (7, 24). Consistent with these findings, Belov et al. (8) showed that staining of NPCs in thin sections prepared from infected cells for electron microscopy was considerably reduced. That study also demonstrated that poliovirus induces an increase in the permeability barrier of the NPC that allows certain proteins to diffuse freely across the nuclear envelope (8). Despite these alterations to the NPC, certain transport pathways remain functional in infected cells, suggesting that the NPC retains some functional and structural integrity (24). This suggests a model whereby poliovirus targets certain NPC proteins for cleavage, leading to the inhibition of specific transport pathways and increased diffusion of macromolecules across the nuclear envelope, thus resulting in cytoplasmic redistribution of host nuclear factors. The finding that relocalization of host nuclear factors could be recapitulated in uninfected cells expressing the viral 2A protease (2Apro) (8) suggests that 2A may also be responsible for the degradation of NPC proteins and the inhibition of nuclear import.
Here we have extended the analysis of the NPC in infected cells by examining the status of Nup98. In uninfected cells Nup98 is localized to both the cytoplasmic and the nuclear sides of the NPC and has been implicated in the process of mRNA export (23, 39). In addition, Nup98 is upregulated in response to interferon treatment, suggesting that it may have a role in the host response to viral infection (16). Our results demonstrate that Nup98 is targeted for cleavage after infection, but that this occurs much more rapidly than it does for Nup153 and Nup62. In addition, we find that cleavage of these NPC proteins displays differential sensitivity to the RNA synthesis inhibitor guanidine hydrochloride (GuHCl). Inhibition of nuclear import and relocalization of host nuclear proteins to the cytoplasm were only apparent at later times after infection when all three Nups were cleaved. Finally, an in vitro cleavage assay is used to show that purified 2Apro can cause cleavage of Nup98. The potential significance of differential targeting of NPC proteins by poliovirus is discussed.
MATERIALS AND METHODS
Cell culture and virus.
HeLa cells were grown in monolayers in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and 2 mM l-glutamine and penicillin-streptomycin at 37°C in 5% CO2. Mahoney type 1 poliovirus stocks were prepared as described previously (51). Subconfluent HeLa cells were either mock infected or infected with poliovirus at a multiplicity of infection of 50. Virus was adsorbed for 30 min at 37°C in phosphate-buffered saline (PBS) supplemented with 10 mg of MgCl2 and 10 mg of CaCl2/ml. After adsorption, residual virus was removed, and Dulbecco modified Eagle medium with 10% fetal bovine serum and 2 mM l-glutamine and penicillin-streptomycin was added. Where indicated, cultures were incubated in the presence of 2 mM guanidine or 100 μg of cycloheximide/ml.
Fluorescence microscopy.
HeLa cells were seeded onto 12-mm-diameter coverslips, and 48 h later the cells were either mock infected or infected with poliovirus at a multiplicity of infection of 50. Coverslips were removed at the indicated times and fixed and permeabilized by incubation in methanol-acetic acid (3:1) for 10 min at 25°C. Coverslips were then washed three times in PBS, incubated in blocking solution (PBS containing 2% bovine serum albumin and 0.05% Triton X-100) for 30 min at 25°C, and incubated overnight at 4°C in MS3 antibody to nucleolin (34). Coverslips where then washed three times in blocking solution, incubated for 1 h at 25°C in secondary antibody conjugated to Alexa Fluor 488 (Invitrogen), washed two times in PBS, stained with Hoechst 33258 (0.2 μg/ml in PBS), and mounted on glass slides with Vectashield mounting medium. Cells were viewed on an Olympus BX-60 fluorescence microscope with a ×40 magnification, and images were acquired by using a 35-mm camera.
Immunoblotting.
HeLa cell lysates were prepared by washing cells once in PBS, followed by a 20-min incubation on ice in Tx buffer (50 mM triethanolamine [pH 7.4], 500 mM NaCl, 0.5% Triton X-100, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and chymostatin, leupeptin, antipain, and pepstatin [10 μg/ml each]) (35). Lysates were cleared by centrifugation at 16,000 × g for 5 min, and protein was quantified by using the Bio-Rad protein assay kit. Equal quantities of protein were separated by sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis, followed by transfer to a polyvinylidene difluoride membrane (Millipore Corp.). Nup153 and p62 were detected by using mouse monoclonal antibody 414 (Covance, Inc.). Nup98 was detected by using rabbit polyclonal antisera (22, 52). Mouse monoclonal antibody MS3 was used to detect nucleolin (34). eIF4GI was detected by using rabbit polyclonal antisera (2). Antibody-antigen complexes were detected by using an horseradish peroxidase-conjugated secondary antibody and chemiluminescence.
In vitro import assay.
Rabbit reticulocyte lysate (Promega) and GST-NLS-EGFP fusion protein (42) were prepared as described previously (24). For import assays, HeLa cells that had been seeded onto 12-mm glass coverslips 2 days prior were mock infected or infected with poliovirus for the indicated times. The permeabilization and assay conditions were as described previously (24). Cells were viewed by using a Nikon E1000M fluorescence microscope with a ×60 objective, and images were acquired by using a Hamamatsu Orca digital camera and Metamorph software.
Fluorescent in situ hybridizations.
HeLa cells that had been seeded onto 12-mm glass coverslips 2 days prior were mock infected or infected with poliovirus for the indicated times. Where indicated, cells were transfected with expression vectors encoding wild-type and mutant VSV M proteins (M51R) fused to green fluorescent protein [GFP]) (36) using the TransIT-HeLaMONSTER transfection reagent (Mirus Bio Corp.). Cells were fixed by incubating at 25°C for 10 min in PBS containing 3% formaldehyde, washed in PBS, permeabilized in PBS containing 0.5% Triton X-100 at 25°C for 10 min, and washed in PBS. Subsequently, cells were incubated for 15 min at 37°C in prewarmed hybridization solution (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 20% formamide, 0.2% bovine serum albumin, 1 μg of yeast tRNA/μl), transferred to prewarmed hybridization solution containing 0.05 pmol of fluorescein isothiocyanate (FITC) or Alexa Fluor 555-conjugated oligo(dT) probe (40mer; Operon Technologies)/μl, and incubated in a humidified chamber at 37°C for 16 h. The next day, cells were washed twice for 5 min at 37°C with prewarmed 2× SSC-20% formamide, twice for 5 min at 37°C with prewarmed 2× SSC, once for 5 min at 25°C with 1× SSC, and once for 5 min at 25°C with PBS. Cells were fixed in 3% formaldehyde for 10 min at 25°C, washed three times in PBS, stained with Hoechst 33258 (0.2 μg/ml in PBS), and mounted on glass slides with Vectashield mounting medium. Cells were viewed by using an Olympus BX-60 fluorescence microscope with a ×60 objective lens, and images were acquired by using a Hamamatsu Orca digital camera and Image Pro Plus or Metamorph software.
In vitro cleavage assay.
Uninfected HeLa cell lysates were prepared essentially as described above except that the Tx buffer lacked protease inhibitors. Lysates were immediately divided into aliquots and stored at −20°C. Human rhinovirus type 2 was purified from bacteria as described previously (31). For cleavage assays, 25 μg of whole-cell lysates was incubated with the indicated amount of human rhinovirus type 2 2Apro at 30°C for 4 h in reaction buffer (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 5 mM dithiothreitol, 1 mM EDTA) (46). After this incubation, reactions were analyzed by immunoblotting as described above.
RESULTS
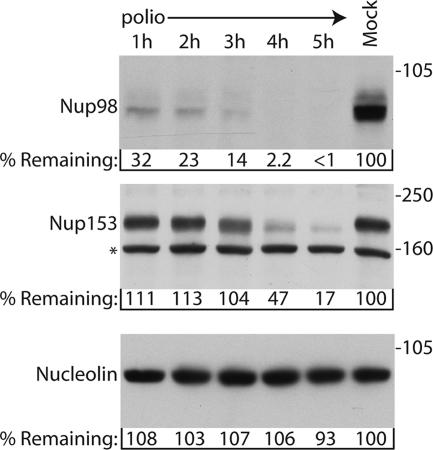
To determine whether poliovirus targets Nup98 for cleavage during infection, the levels of Nup98 were examined in infected cells by immunoblotting. Figure 1 shows that steady-state levels of Nup98 are reduced more than 65% within 1 h of infection compared to mock-infected controls and that by 4 h postinfection the levels of Nup98 were below the level of detection. In contrast, the levels of Nup153 and Nup62 did not begin to decline until after 3 h postinfection (Fig. 1 and data not shown). As shown previously, the decrease in the levels of NPC proteins was not due to a general degradation of proteins within the infected cell since the levels of nucleolin remained unchanged (Fig. 1) (24). Since the relative abundance of Nup153 and Nup98 in the NPC is the same (13) and we obtained similar results with multiple Nup98, Nup153, and Nup62 antibodies (data not shown), it seems unlikely that these results were due simply to differences in protein abundance or antibody affinities. These results indicate that Nup98 is targeted for cleavage in poliovirus-infected cells and that this occurs more rapidly than what is seen for Nup153 and Nup62.
FIG. 1.
Nup98 is targeted for cleavage in infected cells. Portions (50 μg) of whole-cell lysates prepared from mock-infected cells or cells that had been infected with poliovirus for the indicated length of time were analyzed by immunoblotting with monoclonal antibody 414 to detect Nup153 or monoclonal antibody MS3 to detect nucleolin or a polyclonal antisera to Nup98. All antibodies were used at 1:5,000 dilutions, and the exposure times were 10 min, 1 min, and 10 s for the Nup98, Nup153, and nucleolin immunoblots, respectively. For the percent remaining values, band intensities were quantitated by densitometry, and the amount relative to mock-infected cells are indicated. *, Cross-reactive band that is often detected with this antibody. The positions of molecular mass markers are indicated in kilodaltons.
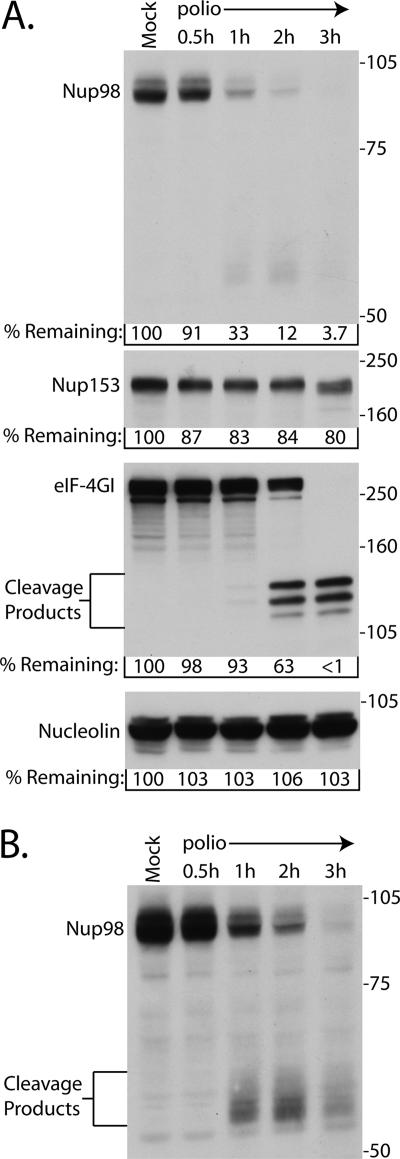
A number of host proteins are proteolyzed after infection with poliovirus (reviewed in reference 45), but most are not targeted until later times in the infectious cycle. In contrast, eIF4GI is cleaved very quickly after infection (17). Figure 2 compares the cleavage of Nup98 with that of eIF4GI and Nup153 in poliovirus-infected cells. The results indicate that levels of Nup98 began a significant decline at between 30 min and 1 h postinfection (33% of control) and that by 2 h postinfection the levels of Nup98 were 12% of the amount detected in mock-infected controls. As reported by others (10), cleavage of eIF4GI appeared to occur slightly more slowly than this, with levels remaining relatively unchanged through 1 h postinfection and then dropping to 63% of the control levels by 2 h postinfection (Fig. 2). As expected, cleavage of Nup153 was just beginning by 3 h postinfection, and the levels of nucleolin remained constant throughout the course of the infection. These results indicate that the cleavage of Nup98 occurs very rapidly after the initiation of infection.
FIG. 2.
Nup98 is cleaved very rapidly after infection. (A) Portions (50 μg) of whole-cell lysates prepared from mock-infected cells or cells that had been infected with poliovirus for the indicated lengths of time were analyzed by immunoblotting with polyclonal antisera to Nup98 or eIF4GI or monoclonal antibodies to detect Nup153 or nucleolin. The positions of the N-terminal eIF4GI cleavage products and molecular mass markers are indicated in kilodaltons. All antibodies were used at 1:5,000 dilutions, and the exposure times were 5 min for Nup98 and 10 s for Nup153, eIF4GI, and nucleolin. For the percent remaining values, band intensities were quantitated by densitometry, and the amounts relative to mock-infected cells are indicated. (B) A 30-min exposure of the immunoblot shown in panel A, with the putative Nup98 cleavage products indicated.
Longer exposures of Nup98 immunoblots revealed the appearance of reactive protein species with molecular masses ranging from ∼55 to ∼65 kDa (Fig. 2B). The appearance of these reactive bands coincided with the onset of Nup98 cleavage, suggesting that they may be proteolytic products. Interestingly, these products were not very abundant and began to disappear by 3 h postinfection (Fig. 2B), suggesting that they may be further proteolyzed. At later times after infection these reactive bands were virtually undetectable (data not shown).
The early cleavage of Nup98 suggested that viral genome amplification and high levels of viral protein synthesis were not required for this process to occur. Conversely, the later cleavage of Nup153 and Nup62 suggested a potential requirement for higher levels of viral proteins. To determine whether either of these possibilities was indeed the case, we examined the status of Nup98, Nup153, and Nup62 in lysates prepared from cells infected in the presence of GuHCl. GuHCl is a potent inhibitor of viral RNA synthesis with sensitivity mapping to the viral 2C protein (37). Cells infected in the presence of GuHCl will not amplify the viral genome, and consequently only input genomes are translated, and low levels of viral proteins are produced. In cells infected in the absence of GuHCl, the cleavage of Nup98 was essentially complete by 2 h postinfection, whereas proteolysis of Nup153, Nup62, and eIF4GI was not complete until 5 h postinfection (Fig. 3, lanes 1 to 3). Infection of cells in the presence of GuHCl had no effect on the cleavage of Nup98 (Fig. 3, compare lanes 1 and 4). Similarly, the addition of GuHCl did not prevent degradation of eIF4GI by 5 h postinfection (Fig. 3, compare lanes 2 and 5) as has been reported elsewhere (10). In contrast, infection in the presence of GuHCl completely prevented the cleavage of Nup153 and Nup62 (Fig. 3, compare lanes 2 and 5). Importantly, infection in the presence of cycloheximide, an inhibitor of viral and cellular protein synthesis, completely prevented the proteolysis of Nup153, Nup62, and Nup98 (Fig. 3, lanes 7 and 8), indicating that new protein synthesis was required for proteolytic targeting of all of these NPC proteins. In addition, the finding that treatment of uninfected cells with cycloheximide did not effect the levels of Nup153, Nup 62, or Nup98 indicates that the loss of these proteins in poliovirus-infected cells is an active process and not simply due to inhibition of protein synthesis and normal turnover of these proteins. These results indicate that although only a low level of viral protein synthesis is required for cleavage of Nup98, the cleavage of Nup153 and Nup62 requires RNA synthesis and subsequently higher levels of viral proteins.
FIG. 3.
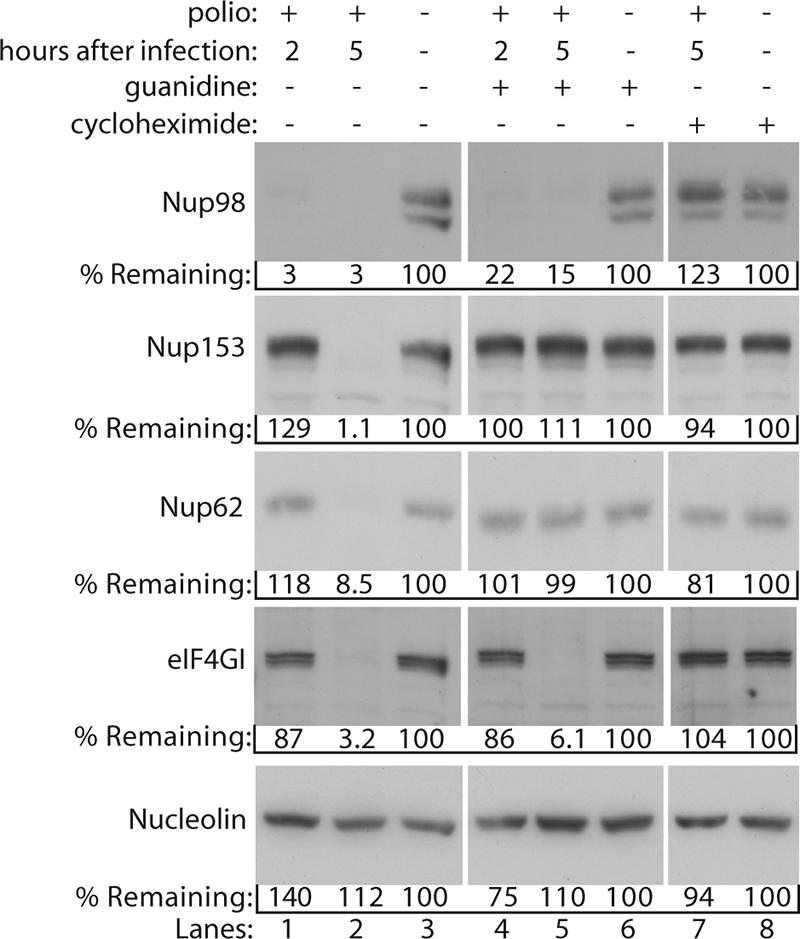
Effect of guanidine on cleavage of NPC proteins. Portions (50 μg) of whole-cell lysates prepared from mock-infected cells or cells that had been infected with poliovirus for 2 or 5 h were analyzed by immunoblotting with the indicated antibodies. “Guanidine” refers to cells treated with 2 mM GuHCl. “Cycloheximide” refers to cells treated with 100 μg of cycloheximide/ml. Mock-infected cells treated with cycloheximide or guanidine were exposed to the reagent for 5 h. All antibodies were used at 1:5,000 dilutions, and exposure times were 1 min for Nup98, Nup153, and Nup62 and 10 s for eIF4GI and nucleolin. For the percent remaining values, band intensities were quantitated by densitometry, and the amounts relative to mock-infected cells are indicated.
Using an in vitro nuclear import assay, we have shown that the classical nuclear import pathway is inhibited in cells that have been infected with poliovirus for 4 h (24). This assay relies on permeabilized cells to provide a functional NPC and a cell extract to provide the soluble factors essential for nuclear import such as transport receptors and the small GTPase Ran. The apparent proteolysis of Nup98 by 2 h postinfection prompted us to determine whether the loss of Nup98 or other early alterations to the NPC might be sufficient to inhibit nuclear import. Figure 4 shows the results of an in vitro import assay using permeabilized mock-infected cells or cells that had been infected with polio for 2 or 4 h. As expected, upon the addition of rabbit reticulocyte lysate as a source for soluble transport factors, mock-infected cells supported robust nuclear import of an NLS-GFP cargo, while cells that had been infected with poliovirus for 4 h did not (Fig. 4A). A similar analysis of cells infected for only 2 h revealed strong nuclear import that was indistinguishable from mock-infected cells (Fig. 4A). As expected, nuclear import in this assay required the addition of rabbit reticulocyte lysate (Fig. 4B), energy, and a functional nuclear localization signal (data not shown). These results suggested that alterations to the NPC that occurred later during infection, such as cleavage of Nup153 and Nup62, were necessary to inhibit the classical nuclear import pathway. However, it was possible that the cleavage of Nup98 could trigger events that by 4 h postinfection might result in an inhibition of import and that this was independent of the loss of Nup153 and Nup62. To determine whether this was the case, we infected cells in the presence of GuHCl. As shown above, GuHCl allows cleavage of Nup98 to proceed normally but prevents the loss of Nup153 and Nup62 (Fig. 3). When mock-infected cells were incubated in the presence of GuHCl, good levels of nuclear import were observed, showing that GuHCl did not inhibit this cellular process. As expected, cells infected with poliovirus for 2 h in the presence of GuHCl supported levels of nuclear import similar to mock-infected cells (Fig. 4C). Infection with poliovirus for 4 h in the presence of GuHCl completely prevented the inhibition of import normally apparent by this time (compare Fig. 4C and A). These results provide further evidence that inhibition of import in poliovirus-infected cells requires the cleavage of Nup153, p62, and/or other events that occur a later times after infection.
FIG. 4.
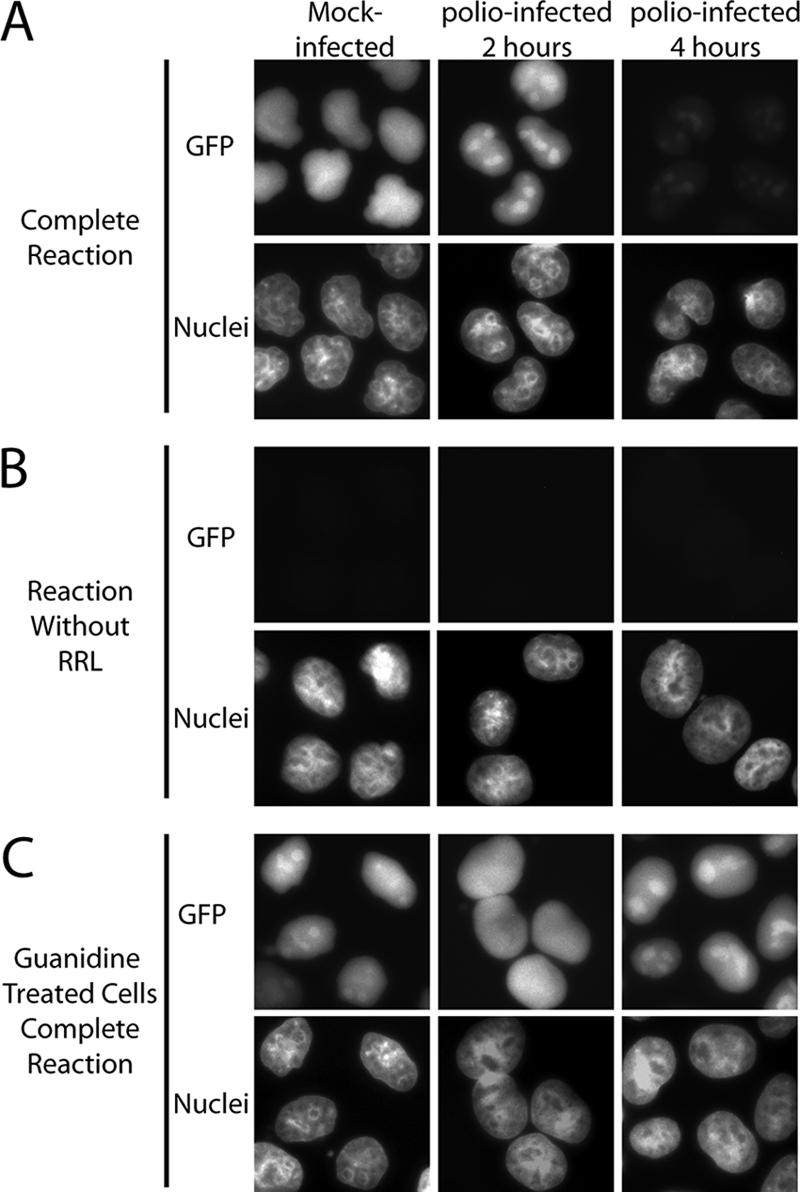
Cell-free nuclear import assays. (A) Uninfected cells (mock infected) or cells that had been infected with poliovirus for the indicated amount of time were permeabilized and used in an in vitro nuclear import assay. Assays were carried out in the presence of rabbit reticulocyte lysate (RRL) as a source of cytosolic factors. The top panels show GFP using an FITC filter, and the bottom panels show Hoechst staining of DNA using a UV filter. (B) Same as in panel A except rabbit reticulocyte lysate was omitted from the reactions. (C) Same as in panel A except that after virus adsorption the cells were incubated in the presence of 2 mM GuHCl. Mock-infected cells were exposed to GuHCl for 4 h. All GFP images were acquired by using identical exposure times and digital manipulations.
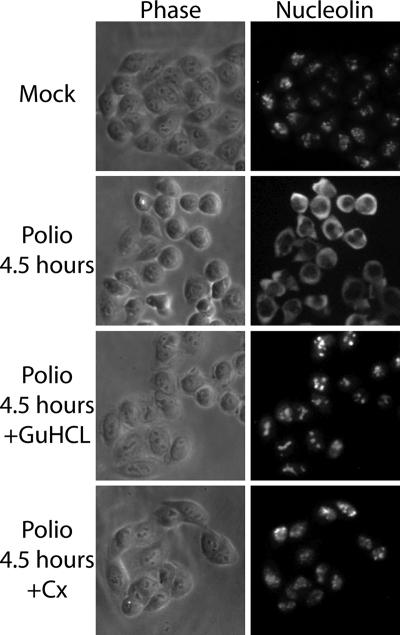
Previously, we found that relocalization of host nuclear proteins to the cytoplasm after infection correlated temporally with the cleavage of Nup153 and Nup62 and the inhibition of nuclear import (24). The observation that GuHCl prevented cleavage of Nup153 and p62, along with inhibition of nuclear import, prompted us to test the effect of GuHCl on the relocalization of host nuclear proteins. Nucleolin is an RNA-binding protein that localizes to the nucleolus in uninfected cells and has been implicated in a number of cellular functions, most prominently rRNA biogenesis (21). In poliovirus-infected cells nucleolin interacts with the viral 3′ noncoding region and relocalizes to the cytoplasm (51). Indirect immunofluorescence analysis of nucleolin in uninfected cells revealed an intranuclear distribution, with localization predominantly to the nucleolus (Fig. 5). As reported previously, by 4.5 h postinfection nucleolin had relocalized from the nucleus to the cytoplasm (Fig. 5) (51). Infection in the presence of GuHCl completely prevented the cytoplasmic accumulation of nucleolin. Similarly, the inhibition of protein synthesis by the addition of cycloheximide 1 h after virus adsorption also prevented the relocalization of nucleolin to the cytoplasm (Fig. 5). Nucleolin binds to the poliovirus RNA 3′ untranslated region (UTR), and this could contribute to relocalization by trapping nucleolin in the cytoplasm (51). However, cells infected with a poliovirus mutant bearing a nearly complete deletion of the 3′ UTR (48) still accumulate nucleolin in the cytoplasm (K. E. Gustin, unpublished data), indicating that interaction with the 3′ UTR is not required for nucleolin relocalization. These results indicate that high levels of viral proteins are needed to cause relocalization of nucleolin in infected cells and suggest that the alterations to the NPC that occur at later times after infection are necessary for redistribution of host nuclear proteins to the cytoplasm.
FIG. 5.
Cytoplasmic redistribution of host nuclear proteins. Uninfected cells (mock) or HeLa cells infected with poliovirus for 4.5 h were fixed and stained with an antibody directed against nucleolin. The left-hand panels show phase-contrast images of the cells (phase), while the right-hand panels show nucleolin staining of the same field (nucleolin). +GuHCl, cells treated with 2 mM GuHCl. Cx, cells treated with 100 μg of cycloheximide/ml. GuHCl and cycloheximide were added 1 h after virus adsorption.
The successful export of cellular mRNAs from the nucleus to the cytoplasm requires interactions with transport factors and subsequent targeting to the NPC (reviewed in reference 15). One NPC protein that has been implicated in the process of mRNA export is Nup98 (22). In addition, the vesicular stomatitis virus (VSV) matrix (M) protein inhibits mRNA export, and this is dependent upon complex formation between the M protein, the cellular transport receptor RAE1, and Nup98 (19, 28, 50). Thus, it was possible that targeting of Nup98 by poliovirus might also result in an mRNA export block. To determine whether this was the case, we examined the distribution of mRNA in infected cells by fluorescence in situ hybridization. Analysis of uninfected HeLa cells using a fluorescently labeled oligo(dT) probe revealed that mRNA localized to brightly staining foci in the nucleus that were excluded from nucleoli, along with a more diffuse staining throughout the cytoplasm (Fig. 6A). Treatment of cells with RNase prior to hybridization with the oligo(dT) probe or the use of an oligo(dA) probe resulted in only background staining, while treatment with DNase had no effect on staining (data not shown). After infection with poliovirus, the number and intensity of the nuclear foci were reduced, and brightly staining foci appeared in the cytoplasm (Fig. 6). This is in contrast to what was seen when the distribution of mRNA in cells expressing the VSV M protein was examined (Fig. 6B). In agreement with earlier reports, cells expressing wild-type M protein exhibited a notable increase in nuclear fluorescence compared to untransfected cells or cells expressing a mutant form of M that is deficient in its ability to inhibit mRNA export (36, 50). The observation that total nuclear fluorescence did not increase in poliovirus-infected cells suggested that mRNA export was not inhibited. However, the reduced nuclear staining in infected cells, most likely due to the inhibition of transcription that occurs during poliovirus infection (4), as well as the possible detection of poliovirus plus-strand RNA in the cytoplasm (9), makes other interpretations of these results possible. To avoid these complications and to examine cells where only Nup98 is cleaved and no inhibition of nuclear import is seen, a similar analysis was done on cells infected in the presence of GuHCl. Under these conditions, infected cells did not show reduced nuclear staining or the appearance of cytoplasmic foci and exhibited a staining pattern that was very similar to that seen in uninfected cells (Fig. 6A). These results indicate that in poliovirus-infected cells mRNA does not accumulate in the nucleus and suggest that cleavage of Nup98 does not result in a block in mRNA export.
FIG. 6.
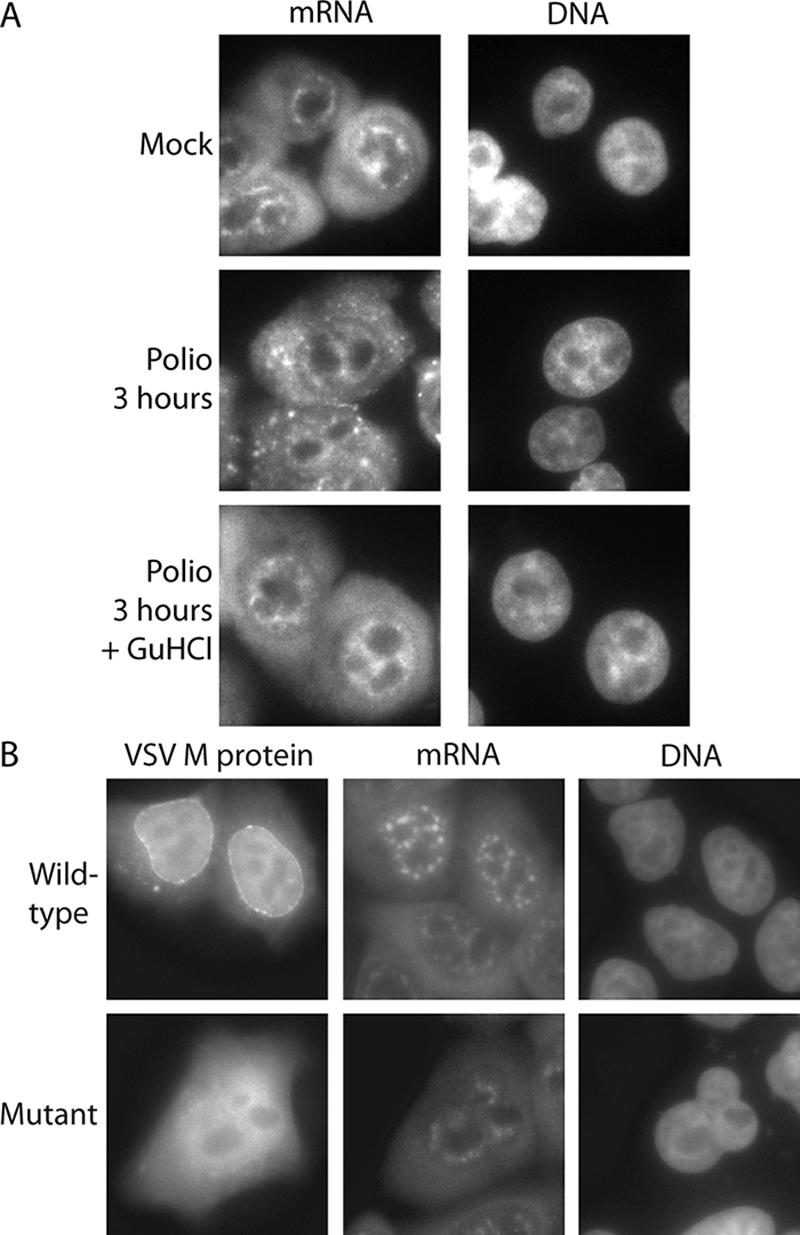
Intracellular distribution of mRNA in poliovirus-infected cells. (A) Uninfected cells (mock) or cells that had been infected with poliovirus for 3 h were analyzed by fluorescence in situ hybridization using a FITC-conjugated oligo(dT) probe. The left-hand panels show oligo(dT) staining of mRNA using an FITC filter. The right-hand panels show Hoechst staining of DNA using a UV filter. +GuHCl, infections carried out in the presence of GuHCl. (B) HeLa cells were transfected with expression vectors encoding GFP fused to wild-type VSV M protein (top panels) or the M51R mutant (bottom panels), followed by in situ hybridization using an oligo(dT) probe conjugated to Alexa Fluor 555. The GFP distribution is shown using an FITC filter, mRNA distribution is shown using a TRITC (tetramethyl rhodamine isothiocyanate) filter, and DNA is shown using a UV filter.
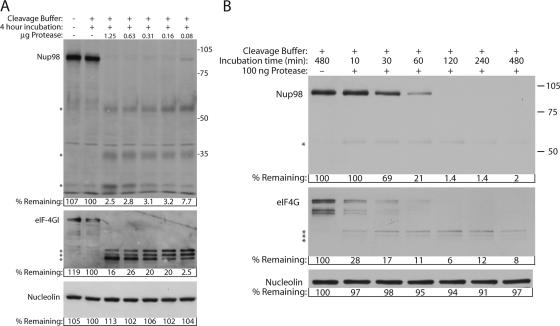
Previous work has shown that the viral 2A protease (2Apro) can cause redistribution of host proteins to the cytoplasm, although the effect of 2Apro on NPC proteins has not been examined (8). To determine whether 2Apro was responsible for cleavage of Nup98, we performed an in vitro cleavage assay using uninfected whole-cell lysates and purified bacterially expressed 2Apro. We were unable to obtain active, purified poliovirus 2A and therefore used the well-characterized picornavirus 2A protease from human rhinovirus 2 for our experiments. We have observed that Nup98 is cleaved in cells infected with rhinovirus type 2 (data not shown). Figure 7 shows that a 4-h incubation of cell lysates in reaction buffer lacking 2Apro had no effect on the levels of Nup98, eIF4GI, or nucleolin. However, when as little as 80 ng of purified 2Apro was added to the reactions, the amount of full-length Nup98 was reduced by >90%. As expected, eIF4GI was proteolyzed efficiently in these reactions, giving rise to the anticipated series of N-terminal cleavage products (53). In contrast, even at the highest concentration of 2Apro, the levels of nucleolin were unaffected, indicating that the cleavage of Nup98 was specific. Incubation with purified RV14 2Apro also resulted in cleavage of Nup98, although the kinetics of cleavage were somewhat slower (data not shown). Interestingly, several potential cleavage products were observed in reactions probed with anti-Nup98 antisera. One of these reactive bands migrated at about 55 kDa and appeared to diminish with increasing concentration or longer incubation with 2Apro (Fig. 7 and data not shown). The size and gradual diminishment of this band intensity raises the possibility that it may correspond to the band seen in lysates prepared from poliovirus-infected cells (Fig. 2). In addition, bands of approximately 35 and 30 kDa were observed in this analysis that appeared to increase in abundance with increasing concentration of protease and thus may arise due to subsequent proteolysis of the 55-kDa band. Of course, it is possible that these reactive bands are not derived from Nup98 but instead represent novel reactivities with unrelated cellular proteins that appear after incubation with 2Apro. These results indicate that 2Apro is responsible for the cleavage of Nup98, although subsequent experiments will be needed to determine whether 2Apro directly cleaves this NPC protein or whether cellular proteases are involved.
FIG. 7.
In vitro cleavage reactions using purified rhinovirus type 2 2Apro. (A) Uninfected HeLa whole-cell lysates were incubated in the presence or absence of the indicated amount of purified 2A protease for 4 h. Cleavage buffer refers to lysates incubated in 2Apro reaction buffer. After incubation, lysates were analyzed by immunoblotting with the indicated antibodies. Nup98, eIF4GI, and nucleolin antibodies were used at 1:2,500, 1:3,000, and 1:5,000, respectively. Exposure times were 2 min for Nup98, 10 min for eIF4GI, and 30 s for nucleolin. For the percent remaining values, band intensities were quantitated by densitometry, and the amounts relative to mock-infected cells are indicated. (B) Uninfected cell lysates were incubated with or without 0.1 μg of purified 2A protease for the indicated times and analyzed by immunoblotting as described above. The exposure times were 10 min for Nup98 and eIF4GI and 1 min for nucleolin. *, Putative Nup98 cleavage products. The positions of molecular mass markers are indicated in kilodaltons.
To examine the relative efficiencies with which 2Apro induces cleavage of Nup98 and eIF4G, we incubated cell lysates with 100 ng of protease for increasing amounts of time. Figure 4 shows that levels of Nup98 remained unchanged for the first 10 min, declined to 21% of control levels within 60 min, and were essentially undetectable by 120 min. In contrast, levels of eIF4GI declined more rapidly, being reduced to 28% of controls within the first 10 min of incubation. As expected, the levels of nucleolin remained relatively unchanged throughout the time course. It is not clear why Nup98 appears to be cleaved more rapidly in vivo (Fig. 2), while eIF4GI is cleaved more rapidly in vitro. Although this could reflect differences between poliovirus and rhinovirus 2Apro, it may also be due to differences in the accessibility or folding of these two substrates in lysis buffer compared to what 2Apro would encounter in vivo.
DISCUSSION
The NPC is composed of at least 30 different proteins and is required for all transport between the nucleus and the cytoplasm. Previously, we had shown that two NPC proteins, Nup153 and Nup62, were degraded during poliovirus and rhinovirus infection and that this correlated with inhibition of nuclear import and relocalization of host-nuclear proteins to the cytoplasm (24, 25). The data presented here demonstrate that at least one other NPC protein, Nup98, is targeted for cleavage after infection with poliovirus and suggest that this is mediated by the viral 2A protease. In contrast to what was seen for Nup153 and Nup62, however, cleavage of Nup98 occurred very quickly after infection and was insensitive to the addition of guanidine, indicating that only very low levels of viral proteins were required. The differential proteolysis of Nup98 compared to Nup153 and Nup62 after infection raises the possibility that poliovirus may selectively target different subsets of NPC proteins for different reasons. Although we do not know the status of the other 27 NPC proteins, our finding that certain transport pathways are functional in infected cells indicates that the NPCs are functional and suggests that not all NPC proteins are degraded.
After infection, several potential cleavage products were observed that migrated in the 55- to 65-kDa range. The presence of multiple bands could be due to cleavage at multiple sites within Nup98, cleavage of different Nup98 isoforms or a combination of both of these possibilities. Significantly, cleavage of Nup98 could be induced by the addition of purified rhinovirus type 2 2Apro to whole-cell lysates, indicating that 2Apro has a role in this process. By using the nomenclature of Schechter and Berger to describe proteolytic cleavage sites (Pn…P2-P1-P1′-P2′…Pn′), where the scissile bond is located between P1 and P1′, 2Apro has a strong preference for glycine at P1′, with threonine found at P2 and a hydrophobic amino acid such as isoleucine or leucine at P4 (26, 43, 47). Analysis of the Nup98 amino acid sequence reveals two sequences that conform to this consensus (370LTTFG374 and 548LQTTG552). Cleavage at G374 would give rise to N- and C-terminal products with estimated molecular masses of 37 and 53 kDa, respectively. Alternatively, cleavage at G552 would result in N- and C-terminal products with predicted molecular masses of 55 and 35 kDa, respectively. Since Nup98 is glycosylated, cleavage at either of these sites could give rise to fragments that correspond to the bands seen in infected cells or whole-cell lysates after the addition of purified 2Apro. Further work will be needed to determine whether one or both of these sites in Nup98 is actually recognized by 2Apro in infected cells.
The rapid targeting of Nup98 and its insensitivity to guanidine compared to the degradation of Nup153 and Nup62 prompted us to determine whether early or late alterations to the NPC were responsible for inhibition of nuclear import and relocalization of host nuclear factors to the cytoplasm. Our results revealed that relocalization of host nuclear factors to the cytoplasm and inhibition of nuclear import occurred only in cells where all three NPC proteins were cleaved. This indicates that early events, including cleavage of Nup98, are not sufficient to cause these effects and suggests that later events, such as the degradation of Nup153 and Nup62, are required. At present, it is not clear whether the cleavage of Nup98 is unrelated to, or is a prerequisite for, the degradation of Nup153/Nup62 and or the inhibition of nuclear import. Our preliminary work indicates that 2Apro also plays a major role in the later stage cleavage of Nup153 and Nup62 (N. Park and K. E. Gustin, unpublished data). Thus, it appears that 2Apro may be responsible for many, if not all of the alterations to the NPC that occur in infected cells. Efforts are currently under way to determine whether 2Apro is both necessary and sufficient to induce the cleavage of Nup98 or whether other viral or cellular proteases have a role in this process.
Our finding that relocalization of host nuclear factors to the cytoplasm is sensitive to the addition of guanidine is in apparent contrast to previous reports. Belov et al. (7, 8) found that the cytoplasmic accumulation of host nuclear factors occurred very rapidly after infection and was not prevented by guanidine. The reason for this discrepancy is unknown but may be due differences between the HeLa cells used in the different studies. Belov and coworkers used a subclone of HeLa cells called HeLa B cells that were found to develop apoptotic markers faster than other HeLa subclones examined (49). Since apoptosis is known to alter the NPC and increase nuclear envelope permeability (11, 20) and since poliovirus has been shown to induce apoptotic markers (1), it is possible that HeLa B cells manifest the poliovirus-induced alterations in nucleocytoplasmic trafficking faster than other cells. Of course, which of these cell lines more closely reflects the in vivo consequences of poliovirus infection is not known.
Cardioviruses also disrupt nucleocytoplasmic trafficking and cause the cytoplasmic accumulation of host nuclear factors in the cytoplasm but appear to do this via a different mechanism than that used by enteroviruses and rhinoviruses (14, 30). In contrast to what is seen with poliovirus, rhinovirus, and coxsackievirus, cardiovirus infection does not result in the degradation of Nup153 and Nup62 (30). In addition, deletion of the cardiovirus 2A coding region does not prevent cytoplasmic accumulation of nuclear proteins (30). In contrast, mutations in the leader (L) protein of encephalomyocarditis virus, mengovirus, or Theiler's murine encephalomyocarditis virus completely prevents the redistribution of cellular proteins to the cytoplasm (14, 30). How encephalomyocarditis virus mediates these effects was discovered recently with the finding that the L protein interacts with and inhibits the activity of the small GTPase Ran, a critical factor for many nuclear import and export pathways (38). It will be interesting to learn whether cardioviruses also disrupt Nup98 function and whether this is related to the inhibition of Ran activity reported for the L protein.
Although the reason why poliovirus targets Nup98 for cleavage is not known, it is interesting that Nup98 is also targeted by the matrix (M) protein of VSV. The VSV matrix (M) protein forms a complex with Nup98 and RAE1, an mRNA transport factor resulting in an inhibition of mRNA export (19, 28, 50). As mentioned above, Nup98 is also an interferon-responsive gene and may thus have a role in defending the cell against viral infection (16). In agreement with this, induction of Nup98 by interferon treatment prevents the block in mRNA export caused by the VSV M protein (16). Our finding that poliovirus infection does not result in an inhibition of mRNA export, however, indicates that the consequences of 2Apro-mediated cleavage of Nup98 and VSV M interaction with Nup98 and are qualitatively different. One intriguing possibility for the rapid cleavage of Nup98 is suggested by recent reports that have implicated the NPC in transcriptional regulation (12, 29, 41, 44). Thus, by removing Nup98, poliovirus may attenuate the host transcriptional response necessary to mount an effective antiviral response. The finding that both a positive- and negative-strand RNA viruses have evolved mechanisms to interfere with Nup98 function raises the possibility that this may be a more common feature of viral pathogenesis than is currently appreciated.
Acknowledgments
This study was initiated in the laboratory of Peter Sarnow, and we are grateful for all his contributions, including critical reading of the manuscript. We also thank Bert Semler for the Δ3′NCR poliovirus mutant, Maureen Powers and Jan Van Deursen for providing Nup98 antibodies, and Jim Dahlberg for the VSV M expression vectors.
This study was supported by grants from the American Cancer Society (RSG 109705), the National Institutes of Health (AI 059467 and AI 064432), and the National Center for Research Resources COBRE (P20 RR15587) and INBRE (P20 RR016454) Program to K.E.G and by National Institutes of Health grant AI 25105 to Peter Sarnow. T.S. was supported by grant P17988 from the Austrian Science Fund.
Footnotes
Published ahead of print on 28 November 2007.
REFERENCES
- 1.Agol, V. I., G. A. Belov, K. Bienz, D. Egger, M. S. Kolesnikova, N. T. Raikhlin, L. I. Romanova, E. A. Smirnova, and E. A. Tolskaya. 1998. Two types of death of poliovirus-infected cells: caspase involvement in the apoptosis but not cytopathic effect. Virology 252343-353. [DOI] [PubMed] [Google Scholar]
- 2.Aldabe, R., E. Feduchi, I. Novoa, and L. Carrasco. 1995. Efficient cleavage of p220 by poliovirus 2Apro expression in mammalian cells: effects on vaccinia virus. Biochem. Biophys. Res. Commun. 215928-936. [DOI] [PubMed] [Google Scholar]
- 3.Almstead, L. L., and P. Sarnow. 2007. Inhibition of U snRNP assembly by a virus-encoded proteinase. Genes Dev. 211086-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bablanian, R., H. J. Eggers, and I. Tamm. 1965. Studies on the mechanism of poliovirus-induced cell damage. I. The relation between poliovirus-induced metabolic and morphological alterations in cultured cells. Virology 26100-113. [DOI] [PubMed] [Google Scholar]
- 5.Back, S. H., S. Shin, and S. K. Jang. 2002. Polypyrimidine tract-binding proteins are cleaved by caspase-3 during apoptosis. J. Biol. Chem. 27727200-27209. [DOI] [PubMed] [Google Scholar]
- 6.Bedard, K. M., S. Daijogo, and B. L. Semler. 2007. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 26459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belov, G. A., A. G. Evstafieva, Y. P. Rubtsov, O. V. Mikitas, A. B. Vartapetian, and V. I. Agol. 2000. Early alteration of nucleocytoplasmic traffic induced by some RNA viruses. Virology 275244-248. [DOI] [PubMed] [Google Scholar]
- 8.Belov, G. A., P. V. Lidsky, O. V. Mikitas, D. Egger, K. A. Lukyanov, K. Bienz, and V. I. Agol. 2004. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J. Virol. 7810166-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolten, R., D. Egger, R. Gosert, G. Schaub, L. Landmann, and K. Bienz. 1998. Intracellular localization of poliovirus plus- and minus-strand RNA visualized by strand-specific fluorescent In situ hybridization. J. Virol. 728578-8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bovee, M. L., W. E. Marissen, M. Zamora, and R. E. Lloyd. 1998. The predominant elF4G-specific cleavage activity in poliovirus-infected HeLa cells is distinct from 2A protease. Virology 245229-240. [DOI] [PubMed] [Google Scholar]
- 11.Buendia, B., A. Santa-Maria, and J. C. Courvalin. 1999. Caspase-dependent proteolysis of integral and peripheral proteins of nuclear membranes and nuclear pore complex proteins during apoptosis. J. Cell Science 1121743-1753. [DOI] [PubMed] [Google Scholar]
- 12.Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus, and P. A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117427-439. [DOI] [PubMed] [Google Scholar]
- 13.Cronshaw, J. M., A. N. Krutchinsky, W. Zhang, B. T. Chait, and M. J. Matunis. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delhaye, S., V. van Pesch, and T. Michiels. 2004. The leader protein of Theiler's virus interferes with nucleocytoplasmic trafficking of cellular proteins. J. Virol. 784357-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dimaano, C., and K. S. Ullman. 2004. Nucleocytoplasmic transport: integrating mRNA production and turnover with export through the nuclear pore. Mol. Cell. Biol. 243069-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enninga, J., D. E. Levy, G. Blobel, and B. M. Fontoura. 2002. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science 2951523-1525. [DOI] [PubMed] [Google Scholar]
- 17.Etchison, D., S. C. Milburn, I. Edery, N. Sonenberg, and J. W. B. Hershey. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000 dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 25714806-14810. [PubMed] [Google Scholar]
- 18.Fahrenkrog, B., and U. Aebi. 2003. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell. Biol. 4757-766. [DOI] [PubMed] [Google Scholar]
- 19.Faria, P. A., P. Chakraborty, A. Levay, G. N. Barber, H. J. Ezelle, J. Enninga, C. Arana, J. van Deursen, and B. M. Fontoura. 2005. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol. Cell 1793-102. [DOI] [PubMed] [Google Scholar]
- 20.Ferrando-May, E., V. Cordes, I. Biller-Ckovric, J. Mirkovic, D. Gorlich, and P. Nicotera. 2001. Caspases mediate nucleoporin cleavage, but not early redistribution of nuclear transport factors and modulation of nuclear permeability in apoptosis. Cell Death Differ. 8495-505. [DOI] [PubMed] [Google Scholar]
- 21.Ginisty, H., H. Sicard, B. Roger, and P. Bouvet. 1999. Structure and functions of nucleolin. J. Cell Sci. 112(Pt. 6)761-772. [DOI] [PubMed] [Google Scholar]
- 22.Griffis, E. R., N. Altan, J. Lippincott-Schwartz, and M. A. Powers. 2002. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell 131282-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffis, E. R., S. Xu, and M. A. Powers. 2003. Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol. Biol. Cell 14600-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gustin, K. E., and P. Sarnow. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gustin, K. E., and P. Sarnow. 2002. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J. Virol. 768787-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hellen, C. U., C. K. Lee, and E. Wimmer. 1992. Determinants of substrate recognition by poliovirus 2A proteinase. J. Virol. 663330-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hellen, C. U., G. W. Witherell, M. Schmid, S. H. Shin, T. V. Pestova, A. Gil, and E. Wimmer. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 907642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Her, L. S., E. Lund, and J. E. Dahlberg. 1997. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science 2761845-1848. [DOI] [PubMed] [Google Scholar]
- 29.Ishii, K., G. Arib, C. Lin, G. Van Houwe, and U. K. Laemmli. 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109551-562. [DOI] [PubMed] [Google Scholar]
- 30.Lidsky, P. V., S. Hato, M. V. Bardina, A. G. Aminev, A. C. Palmenberg, E. V. Sheval, V. Y. Polyakov, F. J. van Kuppeveld, and V. I. Agol. 2006. Nucleocytoplasmic traffic disorder induced by cardioviruses. J. Virol. 802705-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liebig, H. D., E. Ziegler, R. Yan, K. Hartmuth, H. Klump, H. Kowalski, D. Blaas, W. Sommergruber, L. Frasel, B. Lamphear, et al. 1993. Purification of two picornaviral 2A proteinases: interaction with eIF-4 gamma and influence on in vitro translation. Biochemistry 327581-7588. [DOI] [PubMed] [Google Scholar]
- 32.McBride, A. E., A. Schlegel, and K. Kirkegaard. 1996. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. USA 932296-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowicz, D. J. Kenan, E. K. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 673798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson, M. O., K. Guetzow, and H. Busch. 1981. Localization of phosphoprotein C23 in nucleoli by immunological methods. Exp. Cell Res. 135259-265. [DOI] [PubMed] [Google Scholar]
- 35.Pante, N., R. Bastos, I. McMorrow, B. Burke, and U. Aebi. 1994. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J. Cell Biol. 126603-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen, J. M., L. S. Her, V. Varvel, E. Lund, and J. E. Dahlberg. 2000. The matrix protein of vesicular stomatitis virus inhibits nucleocytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Mol. Cell. Biol. 208590-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pincus, S. E., D. C. Diamond, E. A. Emini, and E. Wimmer. 1986. Guanidine-selected mutants of poliovirus: mapping of point mutations to polypeptide 2C. J. Virol. 57638-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter, F. W., Y. A. Bochkov, A. J. Albee, C. Wiese, and A. C. Palmenberg. 2006. A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 10312417-12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powers, M. A., D. J. Forbes, J. E. Dahlberg, and E. Lund. 1997. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J. Cell Biol. 136241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/Raven, Philadelphia, PA.
- 41.Rodriguez-Navarro, S., T. Fischer, M. J. Luo, O. Antunez, S. Brettschneider, J. Lechner, J. E. Perez-Ortin, R. Reed, and E. Hurt. 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 11675-86. [DOI] [PubMed] [Google Scholar]
- 42.Rosorius, O., P. Heger, G. Stelz, N. Hirschmann, J. Hauber, and R. H. Stauber. 1999. Direct observation of nucleocytoplasmic transport by microinjection of GFP-tagged proteins in living cells. BioTechniques 27350-355. [DOI] [PubMed] [Google Scholar]
- 43.Schechter, I., and A. Berger. 1967. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27157-162. [DOI] [PubMed] [Google Scholar]
- 44.Schmid, M., G. Arib, C. Laemmli, J. Nishikawa, T. Durussel, and U. K. Laemmli. 2006. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell 21379-391. [DOI] [PubMed] [Google Scholar]
- 45.Skern, S., B. Hampolz, A. Guarne, I. Fita, E. Bergmann, J. Petersen, and M. N. G. James. 2002. Structure and function of picornavirus proteinases, p. 199-212. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 46.Skern, T., and H. D. Liebig. 1994. Picornains 2A and 3C. Methods Enzymol. 244583-595. [DOI] [PubMed] [Google Scholar]
- 47.Sommergruber, W., H. Ahorn, A. Zophel, I. Maurer-Fogy, F. Fessl, G. Schnorrenberg, H. D. Liebig, D. Blaas, E. Kuechler, and T. Skern. 1992. Cleavage specificity on synthetic peptide substrates of human rhinovirus 2 proteinase 2A. J. Biol. Chem. 26722639-22644. [PubMed] [Google Scholar]
- 48.Todd, S., J. S. Towner, D. M. Brown, and B. L. Semler. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 718868-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tolskaya, E. A., L. I. Romanova, M. S. Kolesnikova, T. A. Ivannikova, E. A. Smirnova, N. T. Raikhlin, and V. I. Agol. 1995. Apoptosis-inducing and apoptosis-preventing functions of poliovirus. J. Virol. 691181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Kobbe, C., J. M. A. van Deursen, J. P. Rodrigues, D. Sitterlin, A. Bachi, X. Wu, M. Wilm, M. Carno-Fonseca, and E. Izaurralde. 2000. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell 61243-1252. [DOI] [PubMed] [Google Scholar]
- 51.Waggoner, S., and P. Sarnow. 1998. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 726699-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, X., L. H. Kasper, R. T. Mantcheva, G. T. Mantchev, M. J. Springett, and J. M. A. van Deursen. 2001. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc. Natl. Acad. Sci. USA 983191-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan, R., W. Rychlik, D. Etchison, and R. E. Rhoads. 1992. Amino acid sequence of the human protein synthesis initiation factor eIF-4 gamma. J. Biol. Chem. 26723226-23231. [PubMed] [Google Scholar]