Abstract
We have shown previously that MDM2 promotes the degradation of the cyclin-dependent kinase inhibitor p21 through a ubiquitin-independent proteolytic pathway. Here we report that the MDM2 analog, MDMX, also displays a similar activity. MDMX directly bound to p21 and mediated its proteasomal degradation. Although the MDMX effect was independent of MDM2, they synergistically promoted p21 degradation when coexpressed in cells. This degradation appears to be mediated by the 26S proteasome, as MDMX and p21 bound to S2, one of the subunits of the 19S component of the 26S proteasome, in vivo. Conversely, knockdown of MDMX induced the level of endogenous p21 proteins that no longer cofractionated with 26S proteasome, resulting in G1 arrest. The level of p21 was low at early S phase but markedly induced by knocking down either MDMX or MDM2 in human cells. Ablation of p21 rescued the G1 arrest caused by double depletion of MDM2 and MDMX in p53-null cells. These results demonstrate that MDMX and MDM2 independently and cooperatively regulate the proteasome-mediated degradation of p21 at the G1 and early S phases.
Proteolysis maintains normal homeostasis of cell cycle-regulated proteins and is crucial for the progression of the cell cycle. These proteins include cyclins and cyclin-dependent kinase inhibitors, such as p27 or p21 (9, 45, 51). Proteolytic pathways have been identified and well characterized for many of these cell cycle proteins (34, 36, 38). For instance, ubiquitylation and turnover of cyclin E are implemented by the SCFFbw7 complex during the cell cycle (23, 54). Also, it has been demonstrated that p27 degradation is facilitated by the SCFSkp2 complex-mediated ubiquitylation and ubiquitin-dependent proteasomal system (6, 50). In contrast, the molecular mechanisms by which p21 is degraded during the cell cycle are still under debate (4, 8, 21, 44).
While it is indisputable that p21 is degraded through the proteasome system (3, 12), argument still remains as to whether p21 degradation is mediated through a ubiquitin-dependent or -independent proteasome system (4, 8, 21, 44). It is clear that p21 is ubiquitylated in cells (30, 35). Several studies have suggested that Skp2 may target p21's N terminus for ubiquitylation and p21 degradation (4, 5, 10, 53). However, these notions have been challenged by other studies (2, 7, 21, 28, 44), showing that p21 ubiquitylation is dispensable for its proteasomal degradation in vitro and in vivo (19, 29, 44). Also, the N terminus of p21 was shown to be acetylated and therefore less likely to be ubiquitylated (7). In addition, knocking out Skp2 had no effect on p21 turnover (7). Recently, we found that UV-induced p21 turnover was also independent of ubiquitin and Skp2 (27). Hence, it appears that Skp2 would not be a major player in regulating p21 stability during the cell cycle.
So, the question remains which protein(s) regulates p21's proteasomal degradation during the cell cycle. It is possible that p21 may be degraded directly by the 20S proteasome during the cell cycle, as it has been shown that the 20S proteasome binds to and directly degrades p21 in vitro (29, 49). However, this notion has not been verified in vivo using synchronized cells. Also, one study showed that endogenous p21 cofractionated with the 26S proteasome (43), suggesting that the 26S proteasome may participate in the proteolysis of p21. Thus, it is still uncertain which proteasome complex is in charge of p21 turnover during the cell cycle. The other possible candidate would be MDM2 (also called HDM2 in humans; MDM2 used here for simplicity). MDM2 was originally discovered as an E3 ubiquitin ligase that specifically mediates p53's ubiquitylation and degradation (11, 15, 16, 24). Later studies by others and us have shown that MDM2 also mediates p21 degradation by a ubiquitin-independent mechanism (21, 58). However, whether MDM2 exerts this p21 degradation activity during the cell cycle and whether there are other proteins that may regulate this MDM2 activity remain to be addressed.
In our initial experiments to address these questions, we have observed that knocking down the endogenous MDM2 level partially rescued the low level of p21 at early S phase during the cell cycle. In addition, we have explored the possibility of whether MDMX (also called HDMX or MDM4; MDMX used here for simplicity), an MDM2 analog that has been shown to assist MDM2 in suppressing p53 function in cells and animals (18, 25, 33, 39, 41, 46), also works with MDM2 in degrading p21. Indeed, we found that MDMX not only cooperated with MDM2 in degrading p21 in cells but also mediated p21's degradation independently of MDM2. Like MDM2, MDMX was also an important player in p21 degradation at the G1 and early S phases, as depletion of MDMX by small interfering RNA (siRNA) markedly induced the p21 level specifically at the G1 and early S phases. Interestingly, MDMX cofractionated with p21 and S2, one of the 19S components of 26S proteasome, in the 26S-containing fractions. Also, both MDMX and p21 bound to S2 in cells. Hence, our studies demonstrate that MDMX can, independently of and in cooperation with MDM2, mediate the 26S proteasomal turnover of p21 at the G1 and early S phases of the cell cycle.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney (HEK) epithelial 293 cells, human lung non-small cell carcinoma H1299 cells, and mouse embryonic fibroblast (MEF) cells were cultured as described previously (22, 55).
Buffers.
Lysis buffer consisted of 50 mM Tris-HCl (pH 8.0), 0.5% NP-40, 1 mM EDTA, 150 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride (PMSF). SNNTE buffer consisted of 50 mM Tris-HCl (pH 8.0), 1% NP-40, 5% sucrose, 5 mM EDTA, 500 mM NaCl, and 1 mM PMSF. Buffer C 100 (BC100) included 20 mM Tris-HCl (pH 7.9), 0.1 mM EDTA, 10% glycerol, 100 mM KCl, 4 mM MgCl2, 0.2 mM PMSF, 1 mM dithiothreitol, and 0.25 μg/ml of pepstatin A.
Antibodies and plasmids.
Monoclonal anti-Flag, anti-α-tubulin antibodies and polyclonal anti-V5 antibody were purchased from Sigma. Polyclonal anti-green fluorescent protein (GFP) and anti-p21 antibodies were purchased from Santa Cruz Biotech. Monoclonal anti-MDM2 antibodies 4B11 and 2A10 were described previously (56). Monoclonal anti-p21 antibody (Ab11) was purchased from Neomarker Biotech. Monoclonal anti-myc tag (9E10) and monoclonal anti-His4 antibodies were purchased from Upstate and Qiagen, respectively. pCDNA3-HA-MDM2 and pCMV-p53 plasmids were previously described (22). The monoclonal anti-MDMX antibody (8C6) and pcDNA3-c-myc-MDMX plasmids were kind gifts from Jiandong Chen (H. Lee Moffitt Comprehensive Cancer Center, Florida). pCEP4-p21 and the p21-6KR mutant were described previously (21). The pCDNA3-2X FLAG-p21 and pCDNA3-2X FLAG-p21 6KR plasmids were cloned using BamHI and EcoRI sites. The mammalian expression vectors for Myc-tagged MDMX deletion mutants and V5-tagged 19S subunits were provided by Zhixiong Xiao (Department of Biochemistry and Molecular Biology, Boston University, Boston, MA) and Christine Blattner (Forschungszentrum Karlsruhe, Institute of Toxicology & Genetics, Karlsruhe, Germany), respectively.
Transient transfection and Western blot analyses.
HEK 293, H1299, or MEF cells (70% confluence) were transfected with combinations of plasmids (see figure legends for detail) with TransFectin lipid reagent (Bio-Rad). Forty-eight hours posttransfection, cells were harvested and lysed in lysis buffer. The clarified whole-cell lysates were loaded directly onto a sodium dodecyl sulfate (SDS) gel for Western blot (WB) analysis with antibodies as indicated in each figure.
GST fusion protein-protein association assay.
The fusion proteins were expressed in Escherichia coli and purified on a glutathione S-transferase (GST)-Sepharose 12B column. Protein-protein association assays were conducted as reported previously (56).
siRNA transfection.
siRNA duplexes specific to human MDMX or MDM2 were synthesized by Dharmacon. p21 siRNA (h2) was purchased from Santa Cruz Biotechnology, Inc. (sc-44214). siRNA duplexes were transfected using oligofectAMINE reagent (Invitrogen) as described previously (13, 21). Cells were harvested 48 h posttransfection and lysed for SDS-polyacrylamide gel electrophoresis (PAGE) and WB. Some transfected cells were synchronized by a double-thymidine block and harvested for cell cycle analysis and WB.
FACS analysis.
H1299 cells were transfected with siRNA or siRNA against human MDMX and treated with 150 ng/ml nocodazole for 16 h prior to harvest. Forty hours posttransfection, cells were harvested and resuspended in 100 μl of phosphate-buffered saline (PBS) and transferred to a polystyrene tube for fluorescence-activated cell sorter (FACS) analysis. The cells were stained in staining buffer (100 μg/ml propidium iodide [PI], 30 μg/ml polyethylene glycol 8000, 200 μg/ml RNase A, 0.1% Triton X-100, 0.38 M NaCl) with 200 μl of PI (Sigma) and incubated for at 37°C for 30 min and analyzed for DNA content using a Becton Dickinson FACScan flow cytometer. Data were analyzed with the multicycle software program using a polynomial S-phase algorithm.
In vivo ubiquitylation assay.
In vivo ubiquitylation assays were conducted as described previously (21). Briefly, H1299 or p53−/− MDMX−/− MEF cells in 100-mm plates were transfected with combinations of His6-ubiquitin (2 μg), p21 (2 μg), p21 6KR (2 μg), and MDMX (2 μg) expression plasmids. Prior to harvest, cells were treated with 10 mM MG132 for 8 h and harvested at 48 h after transfection. Each sample was split into two aliquots: one for WB and the other for ubiquitylation assays using Ni-nitrilotriacetic acid (NTA) beads (Qiagen). Cell pellets were lysed in buffer A (6 M guanidinium-HCl, 0.1 M Na2HPO4-NaH2PO4, 10 mM Tris-HCl [pH 8.0], 10 mM β-mercaptoethanol) and incubated with Ni-NTA beads at room temperature for 4 h. Beads were washed once with each of buffer A, buffer B (8 M urea, 0.1 M Na2HPO4-NaH2PO4, 10 mM Tris-HCl [pH 8.0], 10 mM β-mercaptoethanol), and buffer C (8 M urea, 0.1 M Na2HPO4-NaH2PO4, 10 mM Tris-HCl [pH 6.3], 10 mM β-mercaptoethanol). Proteins were eluted from beads with buffer D (200 mM imidazole, 0.15 M Tris-HCl [pH 6.7], 30% glycerol, 0.72 M β-mercaptoethanol, and 5% SDS). The eluted proteins were analyzed by WB for polyubiquitylation of p21 with monoclonal p21 antibodies (Neomarker; Ab11).
Analysis of p21 half-life in cells.
To measure exogenous p21 half-life, H1299 cells were transfected with the p21 plasmid alone or together with Flag-MDMX expression plasmid in 100-mm plates. Forty-eight hours posttransfection, cells were labeled with 100 μCi/ml Easy Expression (PerkinElmer Life Sciences) in 4 ml Dulbecco's modified Eagle's medium with 2% dialyzed methionine-free calf serum at 37°C for 45 min. Cells were then washed with PBS and incubated in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and harvested at different time points. Cell lysates were prepared for immunoprecipitation (IP). Clarified whole-cell lysates with equal counts of 35S were prebound for 2 h using 2 μg antibody (NeoMarker; Ab11) at 4°C, and then 20 μl protein G-agarose bead slurry (Santa Cruz) was added and the samples continued to rotate at 4°C for 2 more h. The samples were then washed once with lysis buffer), twice with SNNTE buffer, and a second time with lysis buffer. The samples were then analyzed by SDS-PAGE followed by autoradiography. To determine how MDMX affects endogenous p21 stability, human retinal pigment epithelial (RPE) cells were transiently transfected with empty vector or plasmid encoding human MDMX. Cycloheximide (CHX; 100 μg/ml) was added to the cultured cells at 40 h posttransfection. Cells were harvested at the indicated time of CHX treatment. Cells were lysed, and 50 μg protein of each sample was used for WB. The level of p21 from the transfected cells was quantified by the intensities of the bands using the Adobe Photoshop program and plotted with Microsoft Excel.
Cell cycle synchronization.
Cells were synchronized by a double-thymidine block. H1299 cells or MEF cells were plated and treated with 2 mM thymidine at about 40% cell confluence. The cells were released by washing with PBS twice at 37°C 16 h later and cultured in fresh medium for 9 h followed by a second arrest-release process. The cells were harvested at the indicated time points after the second release.
RESULTS
MDMX decreased p21 levels in cooperation with and independently of MDM2.
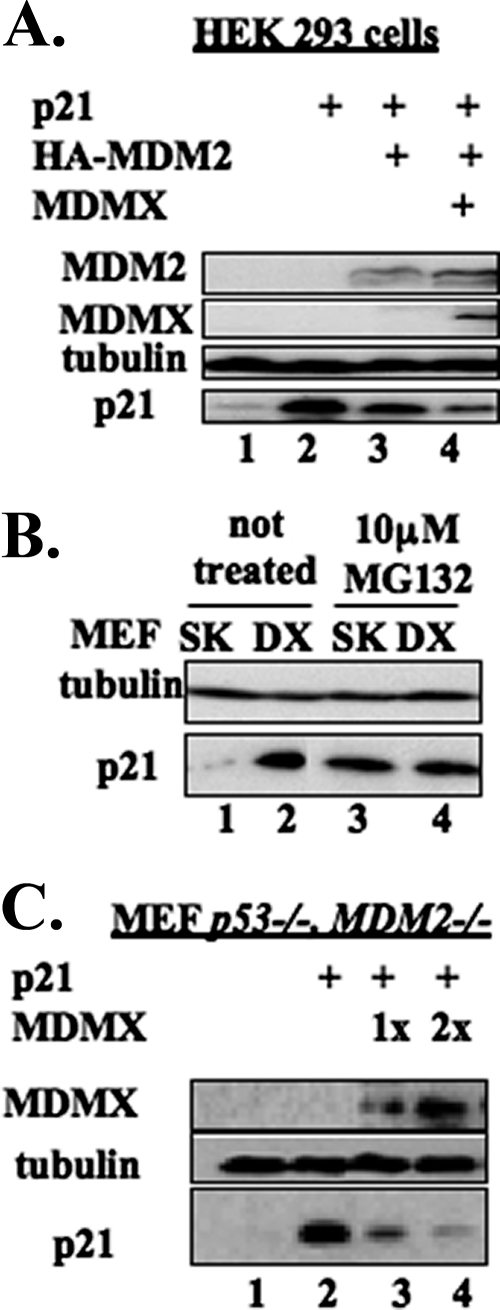
We have reported previously that MDM2 mediates p21 protein turnover independent of both p21 ubiquitylation and MDM2 E3 ligase activity (21). Because MDMX, an MDM2 homolog without demonstrable E3 ubiquitin ligase activity (18, 47), has been shown to assist MDM2 in suppressing p53 activity in vitro and in vivo (31, 32), we wanted to test if MDMX also helps MDM2 degrade p21 in cells by performing transient transfection in HEK 293 cells followed by WB analysis. As expected (21), when coexpressed with MDM2, p21 levels decreased (Fig. 1A). Interestingly, p21 levels further declined when MDMX was coexpressed with MDM2. This result suggests that MDMX may work with MDM2 in down-regulating p21 protein levels in cells.
FIG. 1.
MDMX decreased p21 levels in cooperation with and independently of MDM2. (A) Ectopically expressed MDMX further decreased p21 levels in addition to MDM2 overexpression. HEK 293 cells were transiently transfected with plasmids encoding human p21, HA-MDM2, or Flag-MDMX, as indicated. The transfected cells were harvested 48 h posttransfection and lysed, and the cell lysates (70 μg) were subjected to SDS-PAGE followed by WB with the indicated antibodies. (B) Endogenous p21 level was higher in p53−/− MDMX−/− (DX) MEF cells than in p53−/− (SK) MEF cells. p53−/− MDMX−/− and p53−/− MEF cells were treated with dimethyl sulfoxide (DMSO) or 10 μM MG132 in DMSO for 16 h and harvested at 80% confluence. After lysis, the cell lysates (100 μg) were subjected to SDS-PAGE followed by WB with the indicated antibodies. (C) MDMX decreased the p21 level independently of MDM2. p53−/− MDM2−/− MEF cells were transiently transfected with plasmids encoding human p21 or Flag-MDMX as indicated. The transfected cells were harvested 48 h posttransfection and lysed. The cell lysates (70 μg/sample) were subjected to SDS-PAGE followed by WB with the indicated antibodies.
Our previous report showed that endogenous p21 increased in p53−/− mdm2−/− MEF cells in comparison with p53 single-knockout MEF cells (21). Hence, we wanted to learn whether this would be the case for p53−/− mdmx−/− MEF cells by conducting a straight WB analysis. Indeed, the protein level of p21 in p53−/− mdmx−/− MEF cells was clearly higher than that in p53−/− MEF cells (Fig. 1B). The low level of p21 in p53−/− MEF cells was rescued by the proteasome inhibitor MG132 (lanes 3 and 4), as expected (21). This result suggests that MDMX may also regulate the p21 protein level in cells. To test this idea, we introduced exogenous MDMX and p21 into p53−/− mdm2−/− MEF cells and conducted WB analysis. As shown in Fig. 1C, ectopic MDMX reduced p21 levels in a dose-dependent manner, but independent of MDM2, since these MEF cells are MDM2 deficient. Immunofluorescent staining of p53−/− mdm2−/− MEF cells using an MDMX-specific antibody showed that MDMX resided in both the cytoplasm and the nucleus in the absence of MDM2 (see Fig. S1A in the supplemental material), suggesting that MDMX may regulate p21 levels in the nucleus independent of MDM2.
To address whether MDMX lowered p21 protein levels through inhibition of gene transcription, we tested relative p21 mRNA levels after overexpression or depletion of MDMX by quantitative PCR. As shown in Fig. S1B and S1C in the supplemental material, the MDMX levels had no significant effect on the p21 mRNA level.
Taken together, our results suggest that MDMX also regulates the abundance of p21. This regulation appears to be independent of MDM2, although these two proteins can synergistically reduce p21 levels in cells when coexpressed.
MDMX reduces p21 half-life in cells.
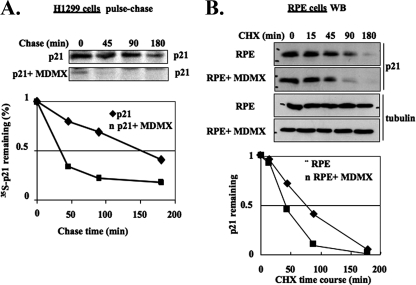
To determine whether MDMX affects the half-life of p21 in cells, we conducted a pulse-chase assay in the p53-deficient human non-small cell lung carcinoma H1299 cells transiently transfected with p21 and/or MDMX. As expected (21), the half-life of p21 was about one and a half hours (Fig. 2A). In striking contrast, this half-life was markedly shortened to less than 40 min in the presence of exogenous MDMX (Fig. 2A). To further confirm this result, we also tested if exogenous MDMX would affect the half-life of endogenous p21 by introducing MDMX into human RPE cells which contain high levels of p21 (27). Protein syntheses in the cells were inhibited by CHX, a protein synthesis inhibitor, to measure the half-life of p21. Again, exogenous MDMX reduced the half-life of endogenous p21 from ∼90 min to ∼45 min (Fig. 2B). These results indicate that MDMX indeed plays a role in destabilizing p21 protein in mammalian cells.
FIG. 2.
Overexpressed MDMX accelerates the turnover of exogenous and endogenous p21. (A) Overexpressed MDMX accelerates the turnover of exogenous p21 in H1299 cells. H1299 cells were transfected with the p21 plasmid alone or together with Flag-MDMX plasmid in 100-mm plates. Forty-eight hours posttransfection, the cells were pulse-chase labeled and harvested at different time points. Cell lysates were prepared for IP with monoclonal anti-p21 antibody. The levels of p21 were quantified from the autoradiograph by Adobe Photoshop software and plotted with Microsoft Excel. (B) MDMX destabilized endogenous p21 in RPE cells. RPE cells were transiently transfected with empty vector or plasmid encoding human MDMX and treated with 100 μg/ml CHX at 40 h posttransfection. The cells were harvested at the indicated time post-CHX treatment. The cell lysates (50 μg/sample) were used for WB. The levels of p21 were quantified as described above.
MDMX associates with p21 in vitro and in cells.
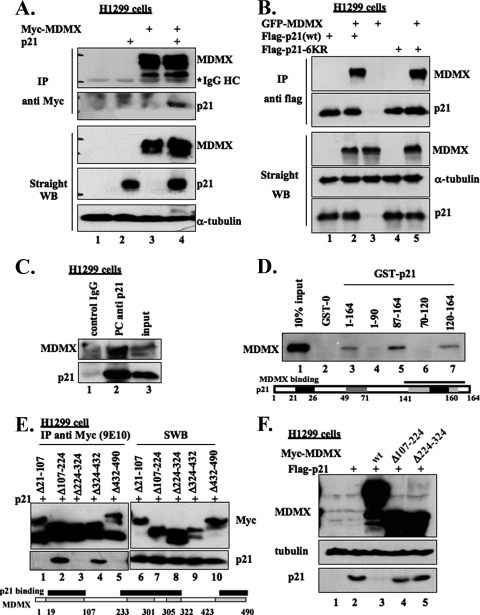
To investigate whether MDMX promotes p21 turnover by associating with the latter, we performed a co-IP-WB analysis following transient transfection of a plasmid encoding Myc-MDMX (Fig. 3A) or GFP-MDMX (Fig. 3B) together with a Flag-p21 expression vector in H1299 cells. Proteins were pulled down by either anti-Myc (Fig. 3A) or anti-Flag antibodies followed by WB, as indicated in the figure legends. As shown in Fig. 3A and B, ectopic p21 and MDMX were reciprocally coimmunoprecipitated with either of these antibodies. Interestingly, MDMX was also coimmunoprecipitated with the p21 mutant (p21-6KR) harboring six lysine-to-arginine mutations by anti-Flag antibodies (lanes 4 and 5 of Fig. 3B). Consistently, endogenous MDMX and p21 also coimmunoprecipitated with anti-p21 antibodies (Fig. 3C) and vice versa with anti-MDMX antibodies (see Fig. S2A in the supplemental material). These results demonstrate that MDMX associates with p21 in cells.
FIG. 3.
p21 and MDMX interact in cells and in vitro. (A) p21 interacts with MDMX in cells. H1299 cells were transfected with the p21 plasmid alone or together with c-myc-MDMX plasmid. The transfected cells were treated with 10 μM MG132 for 16 h and harvested at 48 h posttransfection and lysed. The cell lysates were immunoprecipitated using monoclonal anti-myc (9E10) antibody (300 μg/for each sample) or directly loaded for straight WB (50 μg/sample). The detected proteins are indicated on the right. HC, heavy chain. (B) Reciprocally, MDMX interacts with both wild-type and lysine mutant p21. The Flag-tagged wild-type p21 (wt) or p21-6KR (a p21 mutant in which all lysine residues of p21 were replaced by arginines) was transfected alone or together with GFP-MDMX, in H1299 cells, and treated as in panel A. The cell lysates were immunoprecipitated with monoclonal anti-Flag antibody followed by WB analyses, as indicated on the right. (C) Endogenous p21 complexes with MDMX. H1299 cell lysate (400 μg) was immunoprecipitated with polyclonal anti-p21 antibody or control immunoglobulin G (IgG) and probed for p21 and MDMX. An aliquot of the same cell lysate (80 μg) was used for input. (D) MDMX physically associates with the C terminus of p21 in vitro. Preparation of GST-fusion protein beads and purification of His-MDMX were described previously (20). His-MDMX (100 ng) was incubated with beads conjugating 500 ng GST-0 (GST only) or GST-fused full-length p21 or fragments of p21 in lysis buffer. Thirty minutes after incubation at room temperature, the mixtures were washed with lysis buffer once, SNNTE buffer twice, and lysis buffer again. The samples, together with 10% His-MDMX input, were resolved by SDS-PAGE, followed by straight WB (SWB) against MDMX (8C6). (E) Mapping of p21-binding domains on MDMX in cells. H1299 cells were transfected with Flag-p21 alone or together with myc-tagged wild-type MDMX or MDMX deletion mutants. The cells were treated and lysed as in panel A. The lysates were immunoprecipitated with monoclonal anti-myc antibody (9E10), followed by WB analyses, as indicated on the right. (F) The MDMX deletion mutants that do not bind to p21 lack the ability to decrease p21 levels in cells. H1299 cells were transfected with Flag-p21 alone or together with myc-tagged wild-type MDMX or MDMX deletion mutants. The transfected cells were harvested at 48 h posttransfection and lysed. The cell lysates (50 μg/sample) were used for straight WB, as indicated on the left.
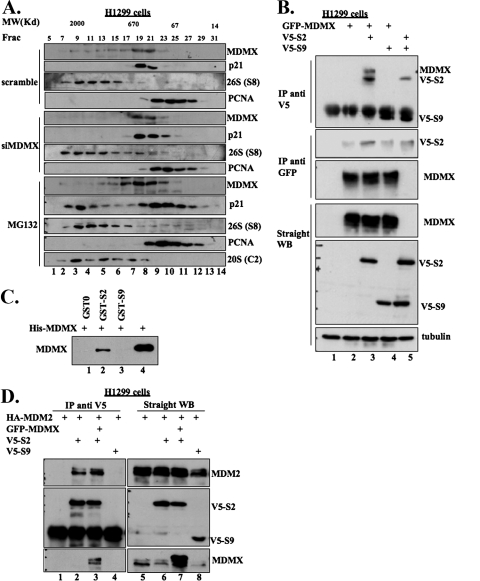
To determine if MDMX associated with p21 directly or through other factors, we conducted a set of GST-fusion protein-protein interaction assays using purified proteins. As shown in Fig. 3D, MDMX bound to the GST-p21 fusion protein (lane 3), but not to GST alone (lane 2). MDMX appeared to specifically bind to the C terminus of p21, as MDMX was pulled down by GST-C-p2187-164 and GST-C-p21120-164, but not by GST-N-p211-90 and GST-N-p2170-120. In a reciprocal pull-down assay, purified His-p21 was also specifically pulled down by GST-MDMX in vitro (see Fig. S2B in the supplemental material).
To map the p21-binding domain(s) of MDMX, H1299 cells were cotransfected with the p21 plasmid and a series of plasmids encoding different MDMX fragments, followed by a co-IP-WB assay. Consistent with our previous mapping of p21-binding domains in MDM2 (21), the central acidic and C-terminal Ring domains were found to be essential for MDMX-p21 binding in cells (Fig. 3E). In addition, we found that the N terminus of MDMX was also likely involved in p21 binding (lane 1 of Fig. 3E), which is different from MDM2 (21). These results demonstrate that MDMX requires three major domains for binding to the C terminus of p21. These p21-binding domains appear to be important for MDMX-mediated p21 degradation, as MDMX mutants that are missing some of these domains were either less potent or unable to degrade p21 in cells (Fig. 3F).
MDMX does not affect p21 ubiquitylation in cells.
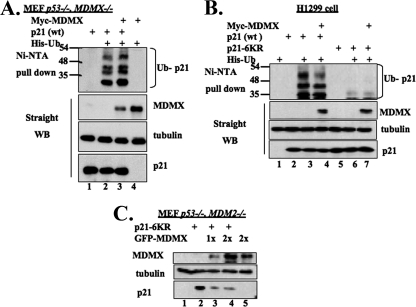
It has been shown that the proteasomal degradation of p21 is mediated through ubiquitin-dependent or -independent mechanisms. Previous studies by our laboratory and others showed that MDM2 mediates the ubiquitin-independent degradation of p21 (21, 58). It would be interesting to determine whether ubiquitylation played a role in MDMX-mediated p21 degradation, although MDMX has not yet been shown to possess an intrinsic E3 ubiquitin ligase activity (18, 47). To address this question, we performed in vivo p21 ubiquitylation assays using p53−/− MDMX−/− MEF or H1299 cells. As shown in Fig. 4A and B, coexpression of Myc-MDMX with p21 did not significantly enhance p21 ubiquitylation in either cell line. This result suggests that MDMX displays no E3 ubiquitin ligase activity toward p21. However, surprisingly, increasing amounts of MDMX further reduced p21 ubiquitylation in either H1299 or p53−/− mdmx−/− MEF cells (see Fig. S2C and S2D in the supplemental material), suggesting that MDMX may block p21 ubiquitylation by binding to it or MDMX may compete with p21 for His-ubiquitin substrates in cells, as MDMX can also be ubiquitinated. Nevertheless, these data show that like MDM2 (21, 58), MDMX may mediate p21 degradation in a ubiquitin-independent fashion.
FIG. 4.
Overexpression of MDMX does not change p21 ubiquitylation in cells. (A and B) Coexpression of MDMX with p21 does not change the p21 ubiquitylation pattern in cells. p53−/− MDMX−/− MEF cells (A) or H1299 cells (B) were transfected with a combination of plasmids encoding His6-ubiquitin (His-Ub), human p21, p21-6KR, or c-myc-MDMX, as indicated. The cells were treated with 10 μM MG132 for 6 h prior to harvest. Cell lysates were incubated with Ni-NTA beads and washed intensively for SDS-PAGE and WB analyses using antibodies against p21 or MDMX. The cell lysates (50 μg/sample) were used for straight WB, as indicated. (C) MDMX mediates the degradation of the ubiquitylation-deficient p21 mutant independent of MDM2. p53−/− MDM2−/− MEF cells were transiently transfected with plasmids encoding p21-6KR or GFP-MDMX. The transfected cells were harvested 48 h posttransfection and lysed. The cell lysates (70 μg/sample) were subjected to SDS-PAGE, followed by WB, as indicated.
To further address the above possibility, we tested whether MDMX also affects the protein level of the p21-6KR mutant, which is lysine free and ubiquitylation deficient (21, 44). As expected, p21-6KR was not polyubiquitylated in cells (Fig. 4B), but its levels were reduced by ectopic MDMX (Fig. 4C), indicating that MDMX also mediates the degradation of p21-6KR independently of ubiquitylation. This degradation was independent of MDM2 because the assay was conducted in p53−/− MDM2−/− MEF cells. Altogether, these results indicate that ubiquitylation is not required for MDMX-mediated p21 proteasomal turnover.
MDMX and MDM2 associate with the 19S component S2 subunit.
Previous studies showed that p21 could be degraded directly by the 20S proteasome (29, 49). However, p21 was also shown to cofractionate with the 26S proteasome when cells were treated with a proteasome inhibitor (43). To understand whether MDMX mediates p21 degradation through the 20S or 26S proteasome, we conducted molecular size exclusion chromatography analyses for the cell lysates from H1299 cells transfected with scramble or MDMX siRNA or treated with MG132. This experiment was to examine if p21 cofractionates with the 20S or 26S proteasome after knocking down endogenous MDMX by siRNA or inhibiting the proteasome activity by MG132. In the absence of MG132, p21 was only detected in the fractions that comigrated with the 600-kDa molecular mass marker while the 26S proteasome eluted with the 2,000-kDa molecular mass marker (upper panel of Fig. 5A). In contrast, in the presence of MG132, p21 cofractionated both with the 26S complex in the high-molecular-weight range and with PCNA in lower-molecular-weight fractions (lower panels of Fig. 5A). The lower-molecular-weight p21 complex contained PCNA, a previously identified p21-interacting protein (52), indicating that p21 in this complex may be functional. Interestingly, unlike the case of MG132 treatment, p21 was only detectable in the 600-kDa fractions, but not in the 26S proteasome fractions, when MDMX was knocked down by siRNA (Fig. 5A). Of note, siRNA appeared to cause the more apparent reduction of MDMX that cofractioned with the 26S proteasome (Fig. 5A). These results suggest two possibilities. First, depletion of endogenous MDMX may prevent the association of p21 with the 19S complex of the 26S proteasome so that p21 no longer associates with this complex, for further degradation. Alternatively, depletion of MDMX may dissociate p21 from the 20S proteasome, which partially comigrated with the 600-kDa complex (43), so that the 20S proteasome is no longer able to degrade p21, perhaps leaving p21 to associate with other proteins such as PNCA in the 600-kDa fractions.
FIG. 5.
MDMX and MDM2 interact with the 19S component of the 26S proteasome. (A) After the size exclusion chromatography, the molecular mass (MW) distribution profile of p21 in the depletion of MDMX by siRNA differs from that in the inhibition of proteasomal activity by MG132. H1299 cells were transfected as indicated or treated with 10 μM MG132 for 16 h and harvested. The cell pellets were lysed and cleared by centrifugation at 13,200 rpm for 15 min. The supernatants were transferred to fresh centrifugation tubes and subjected to a second centrifugation under the same conditions. The clarified cell lysates (300 mg/sample) were applied to a Superose 6 size exclusion chromatographic column (GE Biosience) on a SMART analytical high-performance liquid chromatography system (GE Bioscience) under the following conditions: temperature, 4°C; loading buffer, BC150; flow rate, 50 ml/min; and fraction size, 50 ml/fraction. The column was calibrated under the same conditions, and molecular mass markers (kDa) are shown on the top of all WB lanes. Thirty milliliters of each selected fraction was resolved by 12% SDS-PAGE followed by WB, as indicated on the right. The S8 subunit of the 19S particle was detected for the marker of both the 19S particle and the 26S proteasome. The C2 subunit was used as the marker for the 20S proteasome. (B) MDMX associates with subunit S2 of the 19S particle when ectopically expressed. H1299 cells were transfected as indicated. The transfected cells were treated with 10 mM MG132 for 16 h and harvested at 48 h posttransfection. The cell lysates were used for IP-WB (300 μg/sample) and straight WB (50 μg/sample). (C) Purified His-MDMX binds to S2 in vitro. GST-0 (GST only), GST-S2, GST-S9 beads, and His-MDMX were prepared as described above. One hundred nanograms of His-MDMX was incubated with beads conjugating 500 ng GST-0 or GST fusion proteins in lysis buffer. Thirty minutes after incubation at room temperature, mixtures were washed with lysis buffer once, SNNTE buffer twice, and lysis buffer again. The samples, together with 10% His-MDMX input, were resolved by SDS-PAGE followed by WB against MDMX (8C6). (D) Overexpressed MDM2 and S2 associate in cells. H1299 cells were transfected as indicated and treated as in panel A. The cell lysates were used for IP-WB (300 μg/sample) and straight WB (50 μg/sample), as indicated.
Next, we investigated if MDMX interacts with some of 19S components by conducting transient transfection-IP-WB and GST-pull-down assays. As shown in Fig. 5B, when it was cotransfected with the S2 or the S9 subunit of the 19S complex in H1299 cells, MDMX bound to the S2, but not the S9, subunit. The in vitro GST-fusion protein pull-down assay in Fig. 5C showed the physical interaction between MDMX and the S2 subunit. Interestingly, both C and N termini of MDMX, but not its central domains, were essential for S2 binding (see Fig. S3 in the supplemental material). Similarly, MDM2 bound to the S2, but not the S9, subunit (Fig. 5D). Further transient-transfection studies showed that MDMX also interacted with several other subunits of the 19S complex (data not shown). These results suggest that by associating with some subunits of the 19S complex, MDMX and MDM2 may mediate the recruitment of p21 to the 26S proteasome and promote p21 degradation.
MDMX or MDM2 forms a complex with p21 and the S2 subunit in the 26S proteasome.
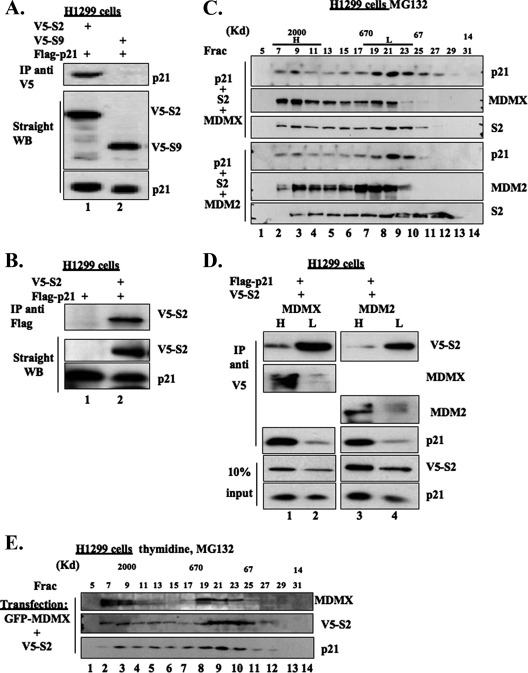
The analyses similar to that in Fig. 5B were conducted to test if p21 complexes with the 19S components, except plasmids encoding p21 or S2 as well as other 19S subunits were used for transfection. As shown in Fig. 6A and B, exogenously, p21 bound to S2, but not S9, in cells. p21 appeared to bind to S2 more strongly than to the other subunits of the 19S component (see Fig. S4A in the supplemental material). Consistent with the result in Fig. 4C, p21-6KR was able to associate with S2 in cells as well (see Fig. S4B in the supplemental material).
FIG. 6.
p21 complexes with the 19S subunit together with MDMX or MDM2 in cells. (A and B) p21 interacts with S2 in cells. H1299 cells were transfected as indicated. Two micrograms of each plasmid was used. The transfected cells were treated with 10 μΜ MG132 for 16 h and harvested at 48 h posttransfection. The cell lysates (300 μg/sample) were immunoprecipitated with polyclonal anti-V5 antibody (A) or monoclonal anti-Flag antibody (B). The cell lysates (50 μg/sample) were also used for WB, as indicated on the right. (C) Molecular mass distribution profiles of overexpressed p21, S2, and MDMX or MDM2 after size exclusion chromatography. H1299 cells were transfected as indicated and treated with 10 μΜ MG132 for 16 h. The size exclusion chromatography was carried out as in Fig. 5A. The selected fractions (Frac; 30 μl/fraction) were revolved by 12% SDS-PAGE followed by WB, as indicated on the right. (D) p21 preferentially associates with S2 in the high-molecular-weight fractions of the Superose 6 column. As indicated in panel C, the Superose 6 column fractions were combined as the high-molecular-weight pool (H, fractions 6 to 12) and the low-molecular-weight pool (L, fractions 18 to 24). One hundred micrograms of proteins per pool was immunoprecipitated with polyclonal anti-V5 antibody and analyzed by WB, as shown on the right. (E) Exogenous MDMX and S2 are cofractionated with p21 in double-thymidine-arrested cells. GFP-MDMX and V5-S2 were ectopically expressed in H1299 cells. The transfected cells were synchronized in G1/S phase by a double-thymidine arrest. A 10 μM concentration of MG132 was added to the culture in the second arrest. The cells were harvested and lysed. The cell lysate (300 μg) was subjected to size exclusion chromatography after clarification as described for Fig. 5A. The selected fractions (30 μl/fraction) were revolved by 12% SDS-PAGE followed by WB, as indicated on the right.
Individually overexpressed p21, MDMX, or MDM2 appeared in a wide range of molecular weight fractions following size exclusion chromatography, including the high-molecular-weight fractions that contain the 26S proteasome (see Fig. S5 in the supplemental material). To determine if p21, MDMX, and S2 associate with one another in the 26S complex, we conducted size exclusion chromatography followed by WB or IP-WB analyses using H1299 cells that expressed exogenous proteins. As shown in Fig. 6C, exogenous p21 and MDMX or MDM2 cofractionated with S2 in the 26S proteasome fractions as well as the 600-kDa fractions. We pooled the high-molecular-weight fractions (H, containing the 26S proteasome) and low-molecular-weight fractions (L) for co-IP-WB analyses. More p21 and MDMX or MDM2 were pulled down by the antibody against V5, which was tagged with S2, along with S2 in the 26S fractions than that in the 600-kDa fractions (Fig. 6D). This result indicates that p21 indeed complexes with MDMX or MDM2 and S2 in the 26S proteasome, although our result does not appear to exclude the possibility that p21 may also associate with MDMX or MDM2 in the 20S complex.
In the H1299 cells transiently transfected with MDMX and S2, synchronized in G1 to early S phase and treated with MG132 (Fig. 6E), similar to that in nonsynchronized cells in Fig. 6C, the exogenous MDMX and S2 cofractionated with p21 in size exclusion chromatography. This implies that MDMX or MDM2 may complex with p21 and the 26S proteasome in this stage of the cell cycle to mediate p21 degradation.
Depletion of MDMX or/and MDM2 by siRNA induces p21 levels and G1 arrest in p53-null cells.
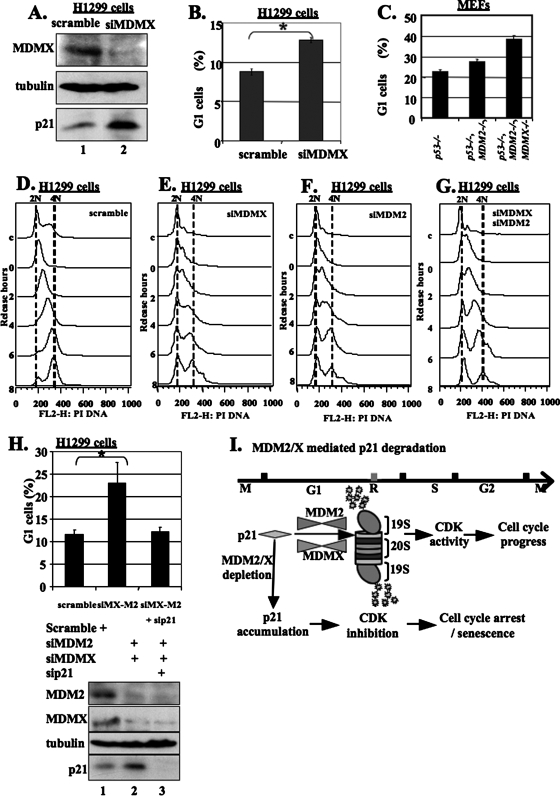
Next, we wanted to know if depletion of endogenous MDMX would stabilize p21 and in turn stall the cell cycle. In doing so, MDMX was knocked down in H1299 cells by an siRNA duplex specific to human MDMX (Fig. 7A). Consistent with the observation in MEF cells (Fig. 1B), this treatment led to a drastic induction of p21 protein. When analyzing the cell cycle of the cells from the same transfection, we found that MDMX siRNA also caused cell cycle arrest at the G1 phase in comparison with the control transfection (Fig. 7B). Cell cycle analyses of MEF cells with different genetic backgrounds indicated that mdmx knockout further caused G1 arrest independently of MDM2 (Fig. 7C). For example, the G1 cells increased from 22% in p53−/− MEF cells to 28% in p53−/− MDM2−/− MEF cells (Fig. 7C). Further knockout of mdmx (p53−/− MDM2−/− and MDMX−/−) (J. A. Barboza, T. Iwakuma, T. Terzian, A. K. El-Naggar, and G. Lozano, submitted for publication) resulted in additional increase of the G1 population to 38% (Fig. 7C). These results suggest that MDMX regulates cell cycle progression by directly monitoring p21 levels independently of p53 and MDM2.
FIG. 7.
Depletion of MDMX and/or MDM2 by siRNA increases the protein level of p21 and causes G1/early S-phase accumulation. (A) siRNA specifically for human MDMX (siMDMX) inhibits MDMX expression and induces p21 in p53-null H1299 cells. H1299 cells were transfected with 300 nM of a scrambled RNA duplex or the anti-MDMX siRNA duplex and harvested 48 h posttransfection. The cell lysates (100 μg/sample) were used to detect MDMX, p21, and tubulin (loading control) by WB, as indicated. (B) Depletion of MDMX by siRNA results in G1 cell accumulation. H1299 cells were transfected as indicated and treated with 150 nM nocodazole for 16 h before harvest. The cells were harvested at 40 h posttransfection and analyzed for DNA content. A total of 50,000 cells were counted for each sample. The error bars represent the standard deviation of results from three independent experiments. Data were analyzed for statistical significance using the Student's t test.*, P < 0.05. (C) Further knockout of MDMX in p53 and MDM2-double-knockout MEF cells leads to more G1 arrest. The cultured p53−/− p53−/− and MDM2−/− or p53−/− MDM2−/− and MDMX−/− MEF cells (Barboza et al., submitted) were treated with 150 nM nocodazole for 16 h. The cells were harvested analyzed for DNA content. A total of 50,000 cells were counted for each sample. (D to G) Depletion of MDMX or/and MDM2 by siRNA causes the accumulation of cells in G1/early S phase accompanied by increased p21 levels. The H1299 cells transfected with scramble siRNA, MDMX siRNA, or MDM2 siRNA were synchronized by a double-thymidine arrest and harvested 0, 2, 4, 6, and 8 h after the second release. The cell pellets were split into two aliquots: one for the cell cycle by FACS analysis (C to E) and the other for WB to determine protein levels (see Fig. S6 in the supplemental material). c, nonsynchronized cells used as a control in both WB and FACS analyses. (H) Depletion of p21 by siRNA (sip21) rescues cells from G1 arrest caused by depletion of MDM2 and MDMX. H1299 cells were transfected as indicated and treated with 150 nM nocodazole for 16 h before harvest. The cells were harvested at 40 h posttransfection and analyzed for DNA content. A total of 50,000 cells were counted for each sample. siMX-M2, MDMX and MDM2 siRNAs. The error bars represent the standard deviations of results from three independent experiments. Data were analyzed for statistical significance using Student's t test. *, P < 0.05. (I) Model for MDMX/MDM2-mediated p21 proteasomal turnover during the cell cycle (see Discussion for details).
To determine at which phase of the cell cycle MDMX regulates p21 levels, we synchronized scramble, MDMX siRNA-, MDM2 siRNA-, or double-siRNA-transfected H1299 cells and conducted FACS and WB analyses. As shown in Fig. 7D to G, the cells expressing scramble siRNA were well synchronized, while the cells expressing MDMX siRNA and/or MDM2 siRNA were mostly arrested at G1/early S phase, consistent with the results in Fig. 7B and C. Interestingly, p21 levels were lower at the G1 and early S phases during the cell cycle of the control cells when MDMX and MDM2 were expressed while MDMX or MDM2 siRNA mainly depleted these two proteins correspondingly and consequently rescued p21 levels at these phases (see Fig. S6A and S6B in the supplemental material). Consistent with this result, p21 levels were also considerably lower at the G1 and S phases in p53-null MEF cells, but were rescued in p53−/− MDMX−/− MEF cells (see the middle panels of Fig. S6C in the supplemental material). Of note, the reduction of p21 levels at the G2/M phase was also rescued in p53−/− MDMX−/− MEF cells (see lane 6 of Fig. S6C in the supplemental material). The lower level of p21 at the G2/M phase was also observed in nontransfected H1299 cells (data not shown), but not in siRNA-transfected H1299 cells (see Fig. S6A and S6B in the supplemental material), implying that siRNA transfection might induce the p21 level at the G2/M phase by an unknown mechanism. Taken together, these results demonstrate that MDMX mainly monitors the p21 level at the G1/S phase. Thus, depletion of MDMX or MDM2 can partially rescue p21 and result in G1 arrest.
Knockdown of p21 by siRNA rescues G1 arrest by depletion of MDMX and MDM2 in p53-null cells.
The above results show that depleting MDMX or MDM2 restores p21 levels at the G1/S phase (see Fig. S6A and S6B in the supplemental material) and leads to G1 arrest (Fig. 7D to G). We then asked whether this G1 arrest depends on p21. To address this question, we performed double- and triple-knockdown experiments using siRNAs against each of the p21, MDM2, and MDMX proteins in p53-deficient H1299 cells followed by FACS analysis. As shown in Fig. 7H, in comparison with the result of depletion of MDMX alone (Fig. 7B), the G1 population further increased when both MDMX and MDM2 were simultaneously depleted, with a net increase of ∼8% after subtracting the control value in H1299 scramble siRNA cells. The increase was remarkably eliminated by additional knockdown of p21 (Fig. 7H). These results demonstrate that p21 can be a target of MDM2 and MDMX in regulating G1-to-S-phase progression in the cell cycle independently of p53.
DISCUSSION
Although several proteins have been identified to potentially regulate p21 proteasomal turnover in cells (21, 28, 49, 53, 58, 59), it remains to be addressed how exactly p21 is degraded during the cell cycle. Our study, as described here, not only identifies MDMX as another regulator of p21 stability in a mechanism similar to that for MDM2-mediated p21 degradation, which is independent of ubiquitylation and p53 (21, 58), but also and more importantly demonstrates that MDMX and MDM2 monitor the p21 level mainly at the G1 and early S phases of the cell cycle. In addition, we provide evidence that suggests that MDMX or MDM2 appears to mediate 26S-catalyzed degradation of p21. Hence, our study uncovers a molecular mechanism underlying p21 turnover at a specific phase of the cell cycle (Fig. 7I).
It is interesting to identify MDMX, like MDM2 (21, 58), as another direct p21 regulator, because biochemical, cellular and genetic studies over the past decade have demonstrated that MDMX acts primarily as a partner of MDM2 in negatively regulating p53 stability and activity (31, 32), although they may have independent functions (17, 20, 26, 37, 40, 46, 47). Our finding is congruent with genetic studies that show that MDMX deficiency causes cell proliferation arrest, which is partially rescued by further knocking out the p21 gene (48). In terms of p21 regulation, MDMX can degrade this protein in cooperation with MDM2 (Fig. 1A), while it can also do so independently of MDM2 (Fig. 1B and C and Fig. 4C). Our previous study showed that MDM2 also degraded p21 independently of MDMX (21). These studies indicate that MDMX and MDM2 can either independently or synergistically promote p21 degradation in cells, although whether the interaction between the two oncoproteins is necessary for their cooperation in degrading p21 during the cell cycle remains to be elucidated.
The role and regulation of MDM2 or MDMX during the cell cycle have been suggested by previous studies (1, 14, 17, 33, 57). However, none of these studies links MDM2 or MDMX directly with p21 regulation. Thus, it is also important to demonstrate that MDMX- or MDM2-mediated p21 degradation occurs in the G1 or early S phase during the cell cycle, as mentioned above. As previously shown, p21 levels were lower in G1 or early S phase than that in other phases of the cell cycle, while MDM2 and MDMX were detected at these phases (Fig. 7 and see Fig. S6 in the supplemental material). However, when MDM2 or MDMX was individually knocked down by siRNA, the level of p21 at G1 and early S phases was induced partially (Fig. 7 and see Fig. S6 in the supplemental material). This result suggests that either of these proteins is required for monitoring p21 turnover at the G1 and early S phases. Consequently cells were arrested at the G1 phase after knocking down either MDM2 or MDMX (Fig. 7 and see Fig. S6 in the supplemental material). In line with these siRNA-mediated knockdown results, p21 levels were also induced specifically at the G1 and early S phases in either p53−/− mdmx−/− or p53−/− mdm2−/− MEF cells compared to p53-null MEF cells (see Fig. S6C in the supplemental material). Consistent with these results, simultaneous deletion of MDM2 and MDMX further increased the cell population at the G1 phase and additional knockdown of p21 rescued this G1 arrest, leading to normal cell proliferation (Fig. 7C and H). These results firmly confirm the role of MDMX and MDM2 in regulating p21 turnover at the G1 and early S phases during the cell cycle.
Then, how does MDMX or MDM2 mediate p21 degradation? Completely addressing this mechanistic question requires more thorough and systematic dissection of the interplay between MDMX or MDM2 and the proteasome system in vitro and in vivo. However, our current study offers some clues that suggest that MDMX may mediate the recruitment of p21 to the 26S proteasome complex. First, we found that p21 cofractionated with the 19S components of the 26S proteasome in the 2,000-kDa fractions when cells were treated with MG132, whereas it disappeared in these fractions and mostly appeared with the 600-kDa fractions in either untreated or siRNA-treated cells (Fig. 5A). Although knocking down MDMX induced p21 (Fig. 7), this induced p21 level appeared merely at the 600-kDa fractions, but not the 2,000-kDa fractions, unlike the case of MG132 treatment (Fig. 5A). This intriguing result suggests that MDMX may play a role in recruitment of p21 to the 26S proteasome, as the lack of MDMX leads to the accumulation of p21 in the non-26S proteasome fractions (Fig. 5A). In accordance with this assumption is the finding that both MDM2 and p21 associate with S2, but not S9, two components of the 19S lid complex of the 26S proteasome (Fig. 5 and 6). Also, the MDM2-p21-S2 complex was detected in the 2,000-kDa fractions when the three proteins were ectopically expressed in cells (Fig. 6C and D). These results suggest that MDMX may mediate p21 turnover that is catalyzed by the 26S proteasome in cells, while MDM2 was previously shown to possibly mediate p21 degradation through the 20S proteasome (58).
In summary, our study uncovers a previously unknown function for MDMX in regulating p21 proteasomal turnover at the G1 and S phases of the cell cycle. This finding further emphasizes the direct role of this oncoprotein in controlling cell proliferation in addition to through suppression of p53 activity. As more evidence shows that MDMX is highly expressed in several human cancers and plays a role in oncogenesis of different tissues (25, 31, 32, 42), our finding also offers one molecular mechanism underlying its oncogenic function. Future studies are necessary to elucidate detailed biochemical mechanisms by which MDMX regulates p21 proteasomal turnover.
Supplementary Material
Acknowledgments
We thank Jiandong Chen, Carol Prives, Aart G. Jochemsen, and Matt Thayer for providing plasmids and antibodies as well as Jayme Gallegos for proofreading. We also thank Guillermina Lozano and Jean-Christophe Marine for kindly providing p53−/−, p53−/− MDM2−/−, p53−/− MDMX−/−, and p53−/− MDM2−/− MDMX−/− MEF cells. Mushui Dai provided technical assistance with FACS analysis.
This work was supported by NCI grants CA93614, CA095441, and CA 079721 to H.L.
Footnotes
Published ahead of print on 17 December 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Argentini, M., N. Barboule, and B. Wasylyk. 2000. The contribution of the RING finger domain of MDM2 to cell cycle progression. Oncogene 193849-3857. [DOI] [PubMed] [Google Scholar]
- 2.Bhat, K. P., K. Itahana, A. Jin, and Y. Zhang. 2004. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 232402-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blagosklonny, M. V., G. S. Wu, S. Omura, and W. S. el-Deiry. 1996. Proteasome-dependent regulation of p21WAF1/CIP1 expression. Biochem. Biophys. Res. Commun. 227564-569. [DOI] [PubMed] [Google Scholar]
- 4.Bloom, J., V. Amador, F. Bartolini, G. DeMartino, and M. Pagano. 2003. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 11571-82. [DOI] [PubMed] [Google Scholar]
- 5.Bornstein, G., J. Bloom, D. Sitry-Shevah, K. Nakayama, M. Pagano, and A. Hershko. 2003. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 27825752-25757. [DOI] [PubMed] [Google Scholar]
- 6.Carrano, A. C., E. Eytan, A. Hershko, and M. Pagano. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1193-199. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X., Y. Chi, A. Bloecher, R. Aebersold, B. E. Clurman, and J. M. Roberts. 2004. N-acetylation and ubiquitin-independent proteasomal degradation of p21(Cip1). Mol. Cell 16839-847. [DOI] [PubMed] [Google Scholar]
- 8.Colledge, M., E. M. Snyder, R. A. Crozier, J. A. Soderling, Y. Jin, L. K. Langeberg, H. Lu, M. F. Bear, and J. D. Scott. 2003. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron 40595-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coqueret, O. 2003. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 1365-70. [DOI] [PubMed] [Google Scholar]
- 10.Coulombe, P., G. Rodier, E. Bonneil, P. Thibault, and S. Meloche. 2004. N-terminal ubiquitination of extracellular signal-regulated kinase 3 and p21 directs their degradation by the proteasome. Mol. Cell. Biol. 246140-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs, S. Y., V. Adler, T. Buschmann, X. Wu, and Z. Ronai. 1998. Mdm2 association with p53 targets its ubiquitination. Oncogene 172543-2547. [DOI] [PubMed] [Google Scholar]
- 12.Fukuchi, K., H. Maruyama, Y. Takagi, and K. Gomi. 1999. Direct proteasome inhibition by clasto-lactacystin beta-lactone permits the detection of ubiquitinated p21(waf1) in ML-1 cells. Biochim. Biophys. Acta 1451206-210. [DOI] [PubMed] [Google Scholar]
- 13.Gu, J., H. Kawai, L. Nie, H. Kitao, D. Wiederschain, A. G. Jochemsen, J. Parant, G. Lozano, and Z. M. Yuan. 2002. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J. Biol. Chem. 27719251-19254. [DOI] [PubMed] [Google Scholar]
- 14.Gu, L., H. Ying, H. Zheng, S. A. Murray, and Z. X. Xiao. 2003. The MDM2 RING finger is required for cell cycle-dependent regulation of its protein expression. FEBS Lett. 544218-222. [DOI] [PubMed] [Google Scholar]
- 15.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387296-299. [DOI] [PubMed] [Google Scholar]
- 16.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 42025-27. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, M. W., and S. J. Berberich. 1999. Constitutive mdmx expression during cell growth, differentiation, and DNA damage. DNA Cell Biol. 18693-700. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, M. W., and S. J. Berberich. 2000. MdmX protects p53 from Mdm2-mediated degradation. Mol. Cell. Biol. 201001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin, S., T. Tong, W. Fan, F. Fan, M. J. Antinore, X. Zhu, L. Mazzacurati, X. Li, K. L. Petrik, B. Rajasekaran, M. Wu, and Q. Zhan. 2002. GADD45-induced cell cycle G2-M arrest associates with altered subcellular distribution of cyclin B1 and is independent of p38 kinase activity. Oncogene 218696-8704. [DOI] [PubMed] [Google Scholar]
- 20.Jin, Y., M. S. Dai, S. Z. Lu, Y. Xu, Z. Luo, Y. Zhao, and H. Lu. 2006. 14-3-3gamma binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J. 251207-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, Y., H. Lee, S. X. Zeng, M. S. Dai, and H. Lu. 2003. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 226365-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin, Y., S. X. Zeng, M. S. Dai, X. J. Yang, and H. Lu. 2002. MDM2 inhibits PCAF (p300/CREB-binding protein-associated factor)-mediated p53 acetylation. J. Biol. Chem. 27730838-30843. [DOI] [PubMed] [Google Scholar]
- 23.Koepp, D. M., L. K. Schaefer, X. Ye, K. Keyomarsi, C. Chu, J. W. Harper, and S. J. Elledge. 2001. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294173-177. [DOI] [PubMed] [Google Scholar]
- 24.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387299-303. [DOI] [PubMed] [Google Scholar]
- 25.Laurie, N. A., S. L. Donovan, C. S. Shih, J. Zhang, N. Mills, C. Fuller, A. Teunisse, S. Lam, Y. Ramos, A. Mohan, D. Johnson, M. Wilson, C. Rodriguez-Galindo, M. Quarto, S. Francoz, S. M. Mendrysa, R. K. Guy, J. C. Marine, A. G. Jochemsen, and M. A. Dyer. 2006. Inactivation of the p53 pathway in retinoblastoma. Nature 44461-66. [DOI] [PubMed] [Google Scholar]
- 26.LeBron, C., L. Chen, D. M. Gilkes, and J. Chen. 2006. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J. 251196-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, H., S. X. Zeng, and H. Lu. 2006. UV induces p21 rapid turnover independently of ubiquitin and Skp2. J. Biol. Chem. 28126876-26883. [DOI] [PubMed] [Google Scholar]
- 28.Lee, J. Y., S. J. Yu, Y. G. Park, J. Kim, and J. Sohn. 2007. Glycogen synthase kinase 3β phosphorylates p21WAF1/CIP1 for proteasomal degradation after UV irradiation. Mol. Cell. Biol. 273187-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, C. W., M. J. Corboy, G. N. DeMartino, and P. J. Thomas. 2003. Endoproteolytic activity of the proteasome. Science 299408-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maki, C. G., and P. M. Howley. 1997. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol. Cell. Biol. 17355-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marine, J. C., and A. G. Jochemsen. 2004. Mdmx and Mdm2: brothers in arms? Cell Cycle 3900-904. [PubMed] [Google Scholar]
- 32.Marine, J. C., and A. G. Jochemsen. 2005. Mdmx as an essential regulator of p53 activity. Biochem. Biophys. Res. Commun. 331750-760. [DOI] [PubMed] [Google Scholar]
- 33.Migliorini, D., E. L. Denchi, D. Danovi, A. Jochemsen, M. Capillo, A. Gobbi, K. Helin, P. G. Pelicci, and J. C. Marine. 2002. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol. Cell. Biol. 225527-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minella, A. C., and B. E. Clurman. 2005. Mechanisms of tumor suppression by the SCF(Fbw7). Cell Cycle 41356-1359. [DOI] [PubMed] [Google Scholar]
- 35.Nakanishi, Y., X. H. Pei, K. Takayama, F. Bai, M. Izumi, K. Kimotsuki, K. Inoue, T. Minami, H. Wataya, and N. Hara. 2000. Polycyclic aromatic hydrocarbon carcinogens increase ubiquitination of p21 protein after the stabilization of p53 and the expression of p21. Am. J. Respir. Cell Mol. Biol. 22747-754. [DOI] [PubMed] [Google Scholar]
- 36.Nakayama, K. I., S. Hatakeyama, and K. Nakayama. 2001. Regulation of the cell cycle at the G1-S transition by proteolysis of cyclin E and p27Kip1. Biochem. Biophys. Res. Commun. 282853-860. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto, K., K. Kashima, Y. Pereg, M. Ishida, S. Yamazaki, A. Nota, A. Teunisse, D. Migliorini, I. Kitabayashi, J.-C. Marine, C. Prives, Y. Shiloh, A. G. Jochemsen, and Y. Taya. 2005. DNA damage-induced phosphorylation of MdmX at serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Mol. Cell. Biol. 259608-9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pagano, M. 2004. Control of DNA synthesis and mitosis by the Skp2-p27-Cdk1/2 axis. Mol. Cell 14414-416. [DOI] [PubMed] [Google Scholar]
- 39.Parant, J., A. Chavez-Reyes, N. A. Little, W. Yan, V. Reinke, A. G. Jochemsen, and G. Lozano. 2001. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat. Genet. 2992-95. [DOI] [PubMed] [Google Scholar]
- 40.Rallapalli, R., G. Strachan, B. Cho, W. E. Mercer, and D. J. Hall. 1999. A novel MDMX transcript expressed in a variety of transformed cell lines encodes a truncated protein with potent p53 repressive activity. J. Biol. Chem. 2748299-8308. [DOI] [PubMed] [Google Scholar]
- 41.Rallapalli, R., G. Strachan, R. S. Tuan, and D. J. Hall. 2003. Identification of a domain within MDMX-S that is responsible for its high affinity interaction with p53 and high-level expression in mammalian cells. J. Cell Biochem. 89563-575. [DOI] [PubMed] [Google Scholar]
- 42.Riemenschneider, M. J., R. Buschges, M. Wolter, J. Reifenberger, J. Bostrom, J. A. Kraus, U. Schlegel, and G. Reifenberger. 1999. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res. 596091-6096. [PubMed] [Google Scholar]
- 43.Sdek, P., H. Ying, D. L. Chang, W. Qiu, H. Zheng, R. Touitou, M. J. Allday, and Z. X. Xiao. 2005. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol. Cell 20699-708. [DOI] [PubMed] [Google Scholar]
- 44.Sheaff, R. J., J. D. Singer, J. Swanger, M. Smitherman, J. M. Roberts, and B. E. Clurman. 2000. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell 5403-410. [DOI] [PubMed] [Google Scholar]
- 45.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 131501-1512. [DOI] [PubMed] [Google Scholar]
- 46.Shvarts, A., W. T. Steegenga, N. Riteco, T. van Laar, P. Dekker, M. Bazuine, R. C. van Ham, W. van der Houven van Oordt, G. Hateboer, A. J. van der Eb, and A. G. Jochemsen. 1996. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 155349-5357. [PMC free article] [PubMed] [Google Scholar]
- 47.Stad, R., Y. F. Ramos, N. Little, S. Grivell, J. Attema, A. J. van Der Eb, and A. G. Jochemsen. 2000. Hdmx stabilizes Mdm2 and p53. J. Biol. Chem. 27528039-28044. [DOI] [PubMed] [Google Scholar]
- 48.Steinman, H. A., H. K. Sluss, A. T. Sands, G. Pihan, and S. N. Jones. 2004. Absence of p21 partially rescues Mdm4 loss and uncovers an antiproliferative effect of Mdm4 on cell growth. Oncogene 23303-306. [DOI] [PubMed] [Google Scholar]
- 49.Touitou, R., J. Richardson, S. Bose, M. Nakanishi, J. Rivett, and M. J. Allday. 2001. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 202367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsvetkov, L. M., K. H. Yeh, S. J. Lee, H. Sun, and H. Zhang. 1999. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr. Biol. 9661-664. [DOI] [PubMed] [Google Scholar]
- 51.Vidal, A., and A. Koff. 2000. Cell-cycle inhibitors: three families united by a common cause. Gene 2471-15. [DOI] [PubMed] [Google Scholar]
- 52.Waga, S., G. J. Hannon, D. Beach, and B. Stillman. 1994. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 369574-578. [DOI] [PubMed] [Google Scholar]
- 53.Wang, W., L. Nacusi, R. J. Sheaff, and X. Liu. 2005. Ubiquitination of p21Cip1/WAF1 by SCFSkp2: substrate requirement and ubiquitination site selection. Biochemistry 4414553-14564. [DOI] [PubMed] [Google Scholar]
- 54.Welcker, M., J. Singer, K. R. Loeb, J. Grim, A. Bloecher, M. Gurien-West, B. E. Clurman, and J. M. Roberts. 2003. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol. Cell 12381-392. [DOI] [PubMed] [Google Scholar]
- 55.Zeng, S. X., M. S. Dai, D. M. Keller, and H. Lu. 2002. SSRP1 functions as a co-activator of the transcriptional activator p63. EMBO J. 215487-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeng, X., L. Chen, C. A. Jost, R. Maya, D. Keller, X. Wang, W. G. Kaelin, Jr., M. Oren, J. Chen, and H. Lu. 1999. MDM2 suppresses p73 function without promoting p73 degradation. Mol. Cell. Biol. 193257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, T., and C. Prives. 2001. Cyclin a-CDK phosphorylation regulates MDM2 protein interactions. J. Biol. Chem. 27629702-29710. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Z., H. Wang, M. Li, S. Agrawal, X. Chen, and R. Zhang. 2004. MDM2 is a negative regulator of p21WAF1/CIP1, independent of p53. J. Biol. Chem. 27916000-16006. [DOI] [PubMed] [Google Scholar]
- 59.Zhou, B. P., Y. Liao, W. Xia, B. Spohn, M. H. Lee, and M. C. Hung. 2001. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat. Cell Biol. 3245-252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.