Abstract
The male-specific lethal (MSL) protein-RNA complex is required for X chromosome dosage compensation in Drosophila melanogaster. The MSL2 and MSL1 proteins form a complex and are essential for X chromosome binding. In addition, the MSL complex must integrate at least one of the noncoding roX RNAs for normal X chromosome binding. Here we find the amino-terminal RING finger domain of MSL2 binds as a complex with MSL1 to the heterochromatic chromocenter and a few sites on the chromosome arms. This binding required the same amino-terminal basic motif of MSL1 previously shown to be essential for binding to high-affinity sites on the X chromosome. While the RING finger domain of MSL2 is sufficient to increase the expression of roX1 in females, activation of roX2 requires motifs in the carboxyl-terminal domain. Binding to hundreds of sites on the X chromosome and efficient incorporation of the roX RNAs into the MSL complex require proline-rich and basic motifs in the carboxyl-terminal domain of MSL2. We suggest that incorporation of the roX RNAs into the MSL complex alters the binding specificity of the chromatin-binding module formed by the amino-terminal domains of MSL1 and MSL2.
In Drosophila melanogaster, X chromosome dosage compensation is achieved by doubling the expression of most X-linked genes (5, 16, 17, 48). The male-specific lethal (MSL) ribonucleoprotein complex is required for this process. The core of the MSL complex consists of MSL1, MSL2, MSL3, the males absent on the first (MOF) and maleless (MLE) proteins, and at least one of the roX1 and roX2 noncoding RNAs (9, 26, 41, 47). Several other proteins have been shown to copurify with the core MSL complex, including components of the nuclear pore (35). In addition, the MSL complex interacts with proteins such as HP1 and ISWI that facilitate chromatin compaction (7, 46) and with the DNA supercoiling factor that is important for hypertranscription of X-linked genes and may counteract ISWI (12). The MSL complex binds to hundreds of sites along the X chromosome in male larval salivary gland nuclei (21, 26). High-resolution chromatin immunoprecipitation-chip studies have shown that the MSL complex binds within most actively expressed X-linked genes (2, 13, 23).
MSL1 and MSL2 have been shown to directly form a complex by yeast two-hybrid analysis and by coimmunoprecipitation of in vitro translated proteins (6, 24). Complex formation is mediated by the amino-terminal domains of both proteins (6, 44). An apolar motif in MSL1, which immediately precedes a leucine zipper-like motif, has been shown to be essential for binding to MSL2 (24). Amino acid residues in the RING finger domain of MSL2 and in the immediately preceding predicted coiled-coil motif, are essential for binding to MSL1 in yeast (6).
MSL1 and MSL2 are essential for binding of the MSL complex to the male X chromosome (27). Neither MSL1 nor MSL2 binds to the X chromosome in the absence of the other protein (27). However, in mle, msl3, or mof mutant larvae, the MSL1/MSL2 complex binds to approximately 30 high-affinity sites on the X chromosome (14, 27). Two of these sites are within the roX1 and roX2 genes. roX2 is specifically expressed in males from 6 h onward (31). roX1 is initially expressed in both male and female embryos but is male specific from midembryogenesis onward (31). In roX1 roX2 double-mutant males, there is a dramatic relocation of most of the MSL complex from the X chromosome to a few autosomal sites, the fourth chromosome, and the pericentromeric heterochromatin (33). There is also some residual binding to several sites on the X chromosome.
With the long-term aim of understanding how the MSL complex selectively binds to the male X chromosome, we have previously identified motifs in the MSL1 protein that are essential for X chromosome binding (24). In addition to the apolar motif that is essential for binding to MSL2, an amino-terminal basic motif was essential for binding to the high-affinity sites. The basic motif is highly conserved among the MSL1 proteins predicted from the genome sequences of several Drosophila species. The aim of this study was to identify motifs in the MSL2 protein that are required for X chromosome binding.
MATERIALS AND METHODS
Construction of truncated msl2 mutant plasmids.
The pBluescript-msl2 cDNA plasmid was used as a template for PCR to amplify the truncated MSL2 mutants with respective primers containing an EcoRI site in the forward primer and an XbaI site in the reverse primer. The PCR products were digested with EcoRI and XbaI and cloned into P transformation vector pCaSpeR-hs. The MSL2(1-193)F construct with a C-terminal FLAG tag was made by using a reverse primer encoding the FLAG peptide (5′-TCTAGATTACTTGTCATCGTCGTCCTTGTAGTCGTGATCATAGGGCAGTTCCGG-3′). MSL2(490-773)F with an N-terminal FLAG tag was created by PCR with a forward primer containing FLAG sequences (5′-GAATTCAACATGGACTCAAGGACGACGATGACAAGGAGGTCAAGACTAAAGTGCAA-3′).
Fly transgenesis and genetic crosses.
Maintenance of Drosophila cultures and generation of P transformant lines were done as previously described (24). To generate homozygous msl1L60 female larvae that express MSL2(1-193)F, recombinant y w/Y; P[MSL2(1-193)F w+] msl1L60/Bc males were crossed to w; msl1L60 females. To create homozygous msl1L60 female larvae that express MSL1NHA and MSL2(1-193)F, recombinant y w/Y; P[MSL2(1-193)F w+] msl1L60/CyO males were crossed to P[MSL1NHA w+] y w; Bc/CyO females. Subsequently, the P[MSL1NHA w+] y w/Y; P[MSL2(1-193)F w+] msl1L60/Bc male offspring were crossed to w; msl1L60 females. Homozygous msl1L60 female larvae were distinguished by the absence of a black cell marker. To create homozygous msl1L60 female larvae that express MSL1(27-265)HA and MSL2(1-193)F, recombinant y w; P[MSL1(27-265)HA w+] msl1L60 females were crossed to recombinant y w/Y; P[MSL2(1-193)F w+] msl1L60/Bc males. Homozygous msl1L60 female larvae were distinguished by the absence of a black cell marker.
To determine if expression of truncated versions of MSL2 could rescue msl2 mutant males, w; msl21/CyO; P[HSP-MSL2 w+] males and females were crossed, raised at 25°C, and given a daily heat shock of 37°C for 30 min. Without the daily heat shock, msl2 males were not rescued by expression of the hsp-msl2 transgenes (not shown).
Immunofluorescent polytene chromosome staining and in situ hybridization.
Transgenic female larvae were grown at 25°C. For heat shock, the larvae were treated at 37°C for 20 min and then left to recover at 25°C for 4 h. Polytene chromosome squashes and immunostaining were carried out according to the procedures of Lyman et al. (27) and Li et al. (24). Rat anti-hemagglutinin (HA; Roche), rabbit anti-MSL2, rabbit anti-MSL1, goat anti-MSL3, rabbit anti-MLE, rabbit anti-MOF, rabbit anti-H4K16ac (Upstate), and rabbit anti-HP1 (Abcam) were used as primary antibodies. Fluorescein isothiocyanate (FITC)-conjugated rabbit anti-rat (Sigma), Alexa Fluor 594-conjugated donkey anti-rabbit (Molecular Probes), and FITC-conjugated rabbit anti-goat (Sigma) were used as secondary antibodies. DNA was stained by 4′,6′-diamidino-2-phenylindole (DAPI).
Fluorescence in situ hybridizations of polytene chromosome were performed as previously described (34). Briefly, single-stranded antisense roX1 probe was labeled by in vitro transcription with biotin RNA labeling mix (Roche) with roX1 1.6-kb cDNA (nucleotides 1536 to 3110) in pGEM as the template. Chromosome spreads were fixed in 4% formaldehyde, treated with proteinase K, washed in glycine, and prehybridized at 42°C for 3 h. Hybridizations containing biotinylated riboprobe were carried out overnight at 42°C. The biotin-avidin system (Vector Laboratories) was used for detecting hybridized biotin-labeled probes under conditions recommended by the manufacturer. Briefly, after hybridization, chromosome spreads were blocked for 30 min in 1× ISH blocking solution at 37°C and then incubated with fluorescein-avidin DCS solution (5 μg/ml) for 30 min at room temperature. After washing, the chromosome spreads were incubated with biotinylated anti-avidin (5 μg/ml) solution for 30 min. Slides were washed and then incubated again with fluorescein-avidin DCS solution for 30 min. The final wash was in 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% Tween 20. Slides were mounted with Vectashield mounting medium containing DAPI.
RNA expression analysis.
Total RNA was extracted from adult flies with TRIzol reagent (Life Technologies/Invitrogen) and subsequently treated with Turbo DNase (Ambion). For Northern blot hybridization analysis, 20 μg of total RNA was loaded per lane and the membrane was hybridized with a 32P-labeled roX1 cDNA fragment derived from a 262-bp PCR product (nucleotides 1535 to 1797). Blots were stripped and reprobed with a fragment from the rp49 gene as a loading control. For quantitative real-time reverse transcription (RT)-PCR, 1.5 μg of total RNA was used to synthesize cDNA with Expand reverse transcriptase (Roche) under conditions recommended by the manufacturer. Real-time PCR was performed with a LightCycler 480 real-time PCR system and a LightCycler 480 SYBR green I master kit (Roche Applied Science). A standard curve of amplification efficiency for each set of primers was generated with a serial dilution of cDNA. The primers used for real-time PCR were roX1 forward (5′-ATGCGAGCGAGACAATGATACT-3′) and reverse (5′-GACTTGCAGTCCGCCCTATG-3′), roX2 forward (5′-AGCTCGGATGCCATCGAAA-3′) and reverse (5′-CGTTACTCTTGCTTGATTTTGCTTCG-3′), and pka forward (5′-TTCTCGGAGCCGCACTCGCGCTTCTAC-3′) and reverse (5′-CAATCAGCAGATTCTCCGGCT-3′) (4). Samples were analyzed in triplicate in a 10-μl final volume containing 1 μl of cDNA, 1× LightCycler 480 SYBR green I master kit (Roche Applied Science), and 500 nM each primer and subjected to 45 cycles of PCR (initial denaturation for 10 min at 95°C, 95°C for 10 s, 57°C for 10 s, and 72°C for 20 s). Melting curve analysis was performed to eliminate nonspecific products from the reaction. pka RNA levels were used for normalization. Calibrator-normalized relative quantification analysis was performed with the Roche LightCycler 480 Relative Quantification Software (version 1.2.9.11). Control quantitative RT-PCR was also performed without reverse transcriptase. No significant amplification was observed with any of the samples and primer sets used (cycle threshold [CT], >38).
Immunoprecipitation and Western blotting.
Immunoprecipitation of FLAG-tagged versions of MSL2 and of full-length MSL2 from female extracts and Western blotting were carried out as described previously (24), except that the immunoprecipitation and wash buffers used contained 150 mM NaCl since association of MLE with the MSL complex is reduced if high-salt buffers are used for immunoprecipitation analysis (45). For the RNA coimmunoprecipitation experiments, female adult flies from indicated MSL2 mutants were heat shocked at 37°C for 1 h and then transferred to 25°C for 4 h. Flies were frozen in liquid nitrogen and store at −70°C. Approximately 100 flies were homogenized in 2 ml of cold lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% Nonidet P-40, 10 μM dithiothreitol) containing RNasin proteinase inhibitor (Roche). The homogenate was put on ice for 30 min and then centrifuged at 20,800 × g for 10 min at 4°C. The supernatant was preincubated with 50 μl of a 50% slurry of protein A-Sepharose beads (Sigma) for 1 h at 4°C with rocking. After a brief (30-s) centrifugation, the protein concentration was determined by the Bradford method. A 1.2-mg sample of whole lysate proteins was either incubated with 100 μl of a 50% slurry of protein A-Sepharose beads prebound with anti-MSL2 antibody or no antibody (mock) in 800 μl of lysis buffer containing RNasin proteinase inhibitor for 4 h at 4°C with rocking. The beads were then washed four times with lysis buffer containing RNasin and proteinase inhibitor and then once with TE buffer. Total RNA in the pellet was treated with Turbo DNase (Ambion), extracted with phenol-chloroform, and precipitated with ethanol. The RNA pellet was dissolved in 10 μl of diethyl pyrocarbonate-treated water. A 5-μl volume of immunoprecipitated RNA was used for RT. Quantitative RT-PCR was performed with roX1, roX2, and pka primers as described above. Fold differences were determined by raising 2 to the CT power, where CT = [CT (pka) − CT (roX)]MSL2 IP[CT (pka) − CT (roX)]mock IP, where IP is immunoprecipitation.
RESULTS
Binding patterns of truncated versions of MSL2 to polytene chromosomes.
Previous comparisons of Drosophila melanogaster MSL2 amino acid sequence with orthologous sequences from invertebrates and vertebrates identified two well-conserved motifs, a RING finger domain at the amino end and a cysteine-rich “CXC” domain toward the carboxyl end (29). The function of the CXC domain is unknown. However, the RING finger domain (amino acids [aa] 1 to 190) is sufficient to bind to MSL1 in yeast (6). To determine which domains of MSL2 are important for X chromosome binding, we generated transgenic Drosophila lines that express truncated versions of MSL2 (Fig. 1A and B). MSL2 expression was controlled with the heat-inducible hsp70 promoter, which has a low basal activity. As overexpression of some of the MSL proteins can lead to nonspecific chromatin binding (42, 44), larvae were not heat treated in this study unless otherwise indicated. Since the endogenous MSL2 protein is only present in males (50), salivary gland nuclei from transgenic female larvae were stained with anti-MSL2 antibody. The binding pattern was determined in at least two independent transgenic lines for each gene construct.
FIG. 1.
A carboxyl-terminal region of MSL2 that contains proline-rich and basic motifs is important for binding of the MSL complex to the X chromosome. (A) Schematic representation of the msl2 gene constructs used in this study. The numbers in parentheses correspond to the amino acid numbers in full-length MSL2. All constructs were controlled by the Drosophila hsp70 promoter. MSL2(1-193)F has a C-terminal and MSL2(490-773)F has an N-terminal FLAG tag. (B) Protein expression in adult flies was confirmed by Western blotting with anti-MSL2 antibody. (C to N) Female larval salivary gland nuclei from transgenic lines were stained with anti-MSL2 antibody (red) and counterstained with DAPI (blue). All MSL2 protein is expressed from the transgene, as there is no production of MSL2 protein from the endogenous gene in female nuclei. MSL2(1-193)F bound to heterochromatin at the chromocenter (arrowhead) and fourth chromosome (4) (C and D) and to a few other sites, including 8D (asterisk) on the X chromosome (C and E) and 21B on the second chromosome (F). MSL2(1-524) (G), MSL2(1-649) (H), and MSL2(1-684) (I) bound in a similar pattern to MSL2(1-193)F (C), although MSL2(1-684) bound more weakly to chromatin. MSL2(1-743), which contains a proline-rich motif and basic motifs, bound to many sites on the X chromosome (K), as did the control of full-length MSL2 expressed from the hsp70 promoter (J). MSL2(1-743) does not bind to the chromocenter (L) or autosomal sites such as 21B (M). No significant binding to either the X chromosome or chromocenter was detected with just the carboxyl-terminal region of MSL2(490-773)F (N).
The RING finger domain of MSL2 bound to a few sites on the X chromosome and autosomes, to sites on the fourth chromosome, and to the chromocenter (Fig. 1C and D). The strongly stained site on the X chromosome was mapped to 8D (Fig. 1E), which does not contain one of the high-affinity sites. The nearest high-affinity sites are at 8A and 8E (8, 27). Weaker but consistent binding was detected at the high-affinity sites at 3F and 10C (see below). Binding to other sites on the X chromosome was generally not observed, although in some nuclei binding was seen at 2B. Binding to autosomal sites was highly variable, with the notable exception that strong staining was consistently seen at 21B, toward the tip of the left arm of chromosome 2 (Fig. 1F). A longer version of MSL2(1-524) that did not include the CXC domain showed a similar binding pattern (Fig. 1G). Surprisingly, inclusion of the conserved CXC motif [MSL2(1-649)] (Fig. 1H) did not substantially change the binding profile. Thus, normal binding to the X chromosome must require motifs between the CXC domain and the carboxyl terminus. We reasoned that motifs important in X chromosome binding would be well conserved among Drosophila MSL2 homologs. Alignment of the MSL2 carboxyl-terminal domain sequences from nine Drosophila species identified three regions where the amino acid composition is well conserved (see Fig. S1 in the supplemental material). They were an acid-rich motif that immediately followed the CXC domain, a proline-rich motif (aa 685 to 713 in D. melanogaster MSL2), and a 12-aa base-rich motif that follows the proline-rich motif. In addition, a 66-aa region that immediately precedes the proline-rich motif is generally well conserved across Drosophila species (28/66 aa identical in all species). A truncated version of MSL2 that includes this conserved region but not the proline-rich motif (aa 1 to 683) showed a binding pattern similar to that of aa 1 to 649, although binding was consistently weaker (Fig. 1I). Inclusion of the proline-rich and basic motifs (1 to 743) (Fig. 1K), however, resulted in binding to many sites on the X chromosome in a pattern very similar to that seen with full-length MSL2 (Fig. 1J). Binding to the chromocenter was lost (Fig. 1L), as was binding to some autosomal sites, including 21B (Fig. 1M). The carboxyl-terminal domain of MSL2, which cannot form a complex with MSL1, did not bind significantly to sites on the X chromosome or chromocenter (Fig. 1N).
Expression of any of the truncated versions of MSL2 did not disrupt the normal X chromosome binding of MSL1 in males (not shown). However, in MSL2(1-193)F, MSL2(1-524), MSL2(1-649), and MSL2(1-683) males there was abnormal binding of MSL1 to 21B, which is presumably due to expression of the truncated form of MSL2. Further, overexpression of MSL2(1-193)F, induced by administering a daily heat shock of 1 h at 37°C, had no effect on male viability (241 males, 204 females). The level of MSL1 protein in females is lower than in males, as MSL1 is stabilized by MSL2 (37). Since MSL2 does not bind to the male X chromosome without MSL1 (27), it is possible that differences in the chromatin-binding profile of the truncated versions of MSL2 could, in part, be due to variation in the level of MSL1 protein. However, similar levels of MSL1 protein were present in females that express any version of MSL2 that contains the amino-terminal RING finger domain (not shown). Further, the level of MSL1 was comparable to that of males. We next asked if the expression of any of the truncated versions of MSL2 could rescue msl2 mutant males. We found that homozygous msl2 males were viable if either full-length MSL2 or MSL2(1-743) was expressed (Table 1). That is, only the truncated version of MSL2 that bound normally to the X chromosome could rescue msl2 males.
TABLE 1.
Expression of MSL2(1-743), but not shorter versions of MSL2, can rescue msl2 males
| Transgene | No. of:
|
|||
|---|---|---|---|---|
| msl21/+ males | msl21/+ females | msl21 males | msl21 females | |
| hsp-msl2 | 146 | 72 | 34 | 59 |
| hsp-msl2(1-743) | 213 | 87 | 32 | 59 |
| hsp-msl2(1-649) | 110 | 45 | 0 | 64 |
| hsp-msl2(1-524) | 187 | 220 | 0 | 46 |
| hsp-msl2(1-193)F | 346 | 423 | 0 | 107 |
The amino-terminal domain of MSL2 binds to chromatin complexed with MSL1.
The binding pattern of the amino-terminal domain was similar to that of MSL2 in roX1 roX2 mutant male nuclei where MSL2 bound to the chromocenter, the fourth chromosome, to several sites on the X chromosome and a few sites on the autosomes, including 21B (33). The intensity of binding to sites on the X chromosome diminished with increasing severity of the roX1 allele (11). The other core protein components of the MSL complex colocalized with MSL2 to the chromocenter and other sites (10, 36). Thus, we next investigated if the other MSL proteins colocalized with the amino-terminal domain of MSL2. In female nuclei that express MSL2(1-193)F, MSL1 bound to the chromocenter and to a few sites on the X chromosome and autosomes (Fig. 2A). In particular, consistent binding was seen at 8D and 10C on the X chromosome and 21B on chromosome two. Furthermore, the other MSL proteins, MSL3, MOF, and MLE, showed a similar binding pattern, although MLE bound to more sites than MSL1 (Fig. 2B to D). There was a significant enrichment for histone H4 acetylated at lysine 16 at the chromocenter, indicating that MOF bound to the pericentric heterochromatin is active (Fig. 2E). These results suggest that expression of the amino-terminal domain of MSL2 is sufficient for the protein components of the core complex to assemble into a complex.
FIG. 2.
MSL proteins colocalize with the amino-terminal domain of MSL2 on polytene chromosomes. Female polytene nuclei that express truncated MSL2(1-193) were immunostained with (A) anti-MSL1, (B) anti-MSL3, (C) anti-MLE, (D) anti-MOF, (E) anti-H4K16ac, and (F) anti-HP1 antibodies. MSL1, MLE, MOF, H4K16ac, and HP1 were detected by anti-rabbit Alexa Fluor 594 (red). MSL3 was detected by anti-goat-FITC (green). MSL1, MSL3, MLE, and MOF colocalize with MSL2(1-193) to the chromocenter (arrowhead), the fourth chromosome (4), and sites on the X chromosome (X). H4K16ac staining appears to be largely confined to the chromocenter (E).
To determine if MSL1 was required for binding of the RING finger domain of MSL2 to chromatin, a line carrying the MSL2(1-193)F transgene was crossed into an msl1L60 null mutant background. There was no significant binding of MSL2(1-193)F to chromatin in homozygous msl1L60 female nuclei (Fig. 3A). We previously found that the amino-terminal domain of MSL1 is sufficient for the MSL1/MSL2 complex to bind to high-affinity sites on the X chromosome (24). To determine if this was also the case for binding to the chromocenter, a recombinant line that carried both the MSL2(1-193)F and MSL1(1-265)HA transgenes was crossed into an msl1L60 null mutant background. It was necessary to cross into an msl1 mutant background as the amino-terminal domain of MSL1 contains a glycine-rich motif that mediates self-association of MSL1 (24). The amino-terminal domains of MSL2 and MSL1 colocalize to the chromocenter and other sites in homozygous msl1L60 female nuclei (Fig. 3B). A well-conserved 26-aa basic motif at the amino end of MSL1 was essential for binding of the amino-terminal domain to high-affinity sites on the X chromosome (24). To determine if the same motif is essential for binding to the chromocenter, homozygous msl1L60 female nuclei that coexpress MSL1(27-265)HA and MSL2(1-193)F were immunostained with anti-HA and anti-MSL2 antibodies. No significant binding to the chromocenter or other sites was observed (Fig. 3C). Thus, MSL2(1-193)F binds to the chromocenter and other sites complexed with MSL1 and binding requires the same N-terminal basic motif in MSL1 that is essential for binding to the X chromosome.
FIG. 3.
The N-terminal X chromosome-binding domain of MSL1 is essential for binding of the N-terminal domain of MSL2 to chromatin. (A) Salivary gland nuclei from female larvae that express MSL2(1-193)F in an msl1L60 null background were stained with anti-MSL2 (red) and counterstained with DAPI (blue). No binding to chromatin was observed. (B and C) Salivary gland nuclei from female larvae that coexpress the amino-terminal domains of MSL2(1-193)F and either MSL1(1-265)HA (B) or MSL1(27-265)HA (C) in an msl1L60 background were stained with anti-HA antibody (green) and anti-MSL2 antibody (red) and counterstained with DAPI (blue). The amino-terminal domain of MSL1 colocalizes with MSL2(1-193)F (B) to the chromocenter (arrowhead), but the N-terminal basic motif of MSL1 is essential for the binding of both domains to chromatin (C).
Co-overexpression of MSL1 and MSL2 in roX1 roX2 mutant male larvae led to a significant increase in binding of the proteins to sites on the X chromosome (36). To determine if overexpression of the amino-terminal domains of MSL1 and MSL2 altered the chromatin-binding pattern, female larvae that contain both the MSL2(1-193)F and MSL1(1-265)HA transgenes were briefly heat shocked at 37°C and then left to recover for 4 h. Expression of both transgenes was controlled with the heat-inducible hsp70 promoter. The heat treatment significantly increased protein expression and resulted in stronger immunostaining of the chromocenter and 8D (Fig. 4B to D). Weaker binding was seen at a few sites on the X chromosome (e.g., near the tip) in addition to 3F and 10C. However, in general the binding pattern was very similar to that of non-heat-treated larvae (Fig. 4A). Thus, overexpression does not significantly change the chromatin-binding specificity of the amino-terminal domains of MSL1/MSL2.
FIG. 4.
Overexpression of the amino-terminal domains of MSL1 and MSL2 does not significantly change the chromatin-binding pattern. (A and B) Female salivary gland nuclei from larvae that coexpress the amino-terminal domains of MSL1(1-265)HA and MSL2(1-193)F were stained with anti-HA antibody (green) and anti-MSL2 antibody (red) and counterstained with DAPI (blue). Larvae were raised at 25°C and either given no heat treatment (A) or heat shocked at 37°C for 1 h and then left to recover at 25°C for 4 h (B). Western blot assays with anti-HA (top of panel C), anti-FLAG (top of panel D), or antitubulin (bottom of panels C and D) of protein extracts from untreated (lane 1) or heat-shocked (lane 2) flies. Heat treatment led to a significant increase in the levels of the MSL1NHA and MSL2(1-193)F proteins (arrows).
roX gene expression is elevated in females that express truncated versions of MSL2.
The chromatin-binding pattern of truncated versions of MSL2 that lack the carboxyl-terminal domain is similar to that of the MSL complex in roX1 roX2 double mutants. roX RNAs are highly enriched in males, as the MSL complex stabilizes the RNAs (32) and binding of the MSL complex to the roX genes overcomes constitutive repression by the NURF complex (4). Thus, a possible explanation for the MSL2 binding patterns shown in Fig. 1 is that the roX RNA levels are low in female nuclei that express the shorter versions of MSL2 but high in females that express the longer version that binds to the X chromosome. However, MSL2(1-193)F, MSL2(1-524), and MSL2(1-649) consistently bound to 3F and 10C in female nuclei, the locations of the roX1 and roX2 genes, respectively (Fig. 5A). Although MSL2(1-683) bound weakly to chromatin, binding to 10C was seen in most nuclei. Thus, it is possible that these shorter versions of MSL2 could be either binding and stabilizing nascent roX transcripts or binding to the roX genes and increasing expression in female nuclei.
FIG. 5.
roX RNA levels in hsp-msl2 lines. (A) Salivary gland nuclei from female larvae that express MSL2(1-193)F were stained with anti-MSL2 (top) and counterstained with DAPI (lower). MSL2(1-193)F bound to 3F and 10C, the locations of the roX1 and roX2 genes, respectively. (B) Northern blot hybridization analysis of RNA isolated from transgenic females and wild-type flies. The membrane was hybridized with 32P-labeled roX1 probe (top) and reprobed with an rp49 probe (lower) as a loading control. Low levels of roX1 RNA were detected in all lines except MSL2(490-773)F females. (C and D) Quantitative real-time RT-PCR was used to measure the levels of the roX1 (C) and roX2 (D) RNAs in transgenic females and y w (parental strain “wild-type”) flies. The RNAs were isolated from females from the following strains: y w (sample 1), MSL2(1-193)F (sample 2), MSL2(1-524) (sample 3), MSL2(1-649) (sample 4), MSL2(1-683) (sample 5), MSL2(1-743) (sample 6), full-length MSL2 (sample 7), and MSL2(490-773)F (sample 8). Sample 9 was from y w males. Each sample was measured in triplicate. (E) To determine if the amino-terminal domain of MSL2 is sufficient to induce roX1 transcription, RT-PCR was performed with RNA from females that express MSL2(1-193)F and were either homozygous for msl1L60 (lanes 2 and 3), heterozygous for msl1L60 (lanes 4 and 5), or wild type for msl1 (lane 6 and 7). RT(+) indicates the presence of reverse transcriptase in cDNA synthesis reaction; RT(−) indicates its absence. M is a molecular weight marker (lane 1). The lower part of the panel shows RT-PCR products with a primer pair for pgd transcripts as a cDNA synthesis control. (F) Real-time quantitative RT-PCR of the RNA samples analyzed in panel E. roX1 transcript was readily detected in msl1+ females but was not above background levels in msl1L60 homozygotes. (G) Quantitative RT-PCR of roX RNA that coimmunoprecipitated (co-IP) with MSL2 antibody relative to control immunoprecipitation with no added antibody (mock). Each RNA sample was analyzed in triplicate, and the average ± the standard deviation is shown. (H) Western blot assay with MSL2 antibody of immunoprecipitated (IP) extracts from which the RNA quantified in panel G was extracted. Bands corresponding to the expected sizes for MSL2(1-683) (lane 1), MSL2(1-743) (lane 2), and MSL2(1-773) (lane 3) were detected in extract immunoprecipitated with MSL2 antibody but not in the controls (lanes 4 to 6).
We next measured roX RNA levels in females that express truncated versions of MSL2. Low levels of roX1 RNA were detected by Northern blot hybridization analysis in transgenic females that express any version of MSL2 that contains the amino-terminal domain (Fig. 5B). Further, the levels of roX1 RNA appeared to be similar in the different strains. Quantitative real-time RT-PCR assays were performed to more accurately determine the level of the roX RNAs. We found that there was a three- to fivefold increase in the roX1 RNA level in females that express any truncated version of MSL2 that contains the amino-terminal domain relative to wild-type females (Fig. 5C). Similarly, there was a threefold increase in roX1 in females that express full-length MSL2. roX1 RNA levels were significantly lower than in wild-type males, possibly because of the low basal expression levels of the hsp-msl2 transgenes. Thus, roX1 RNA levels are not significantly higher in females that express versions of MSL2 that bind to the X chromosome. Surprisingly, although MSL2(1-193)F, MSL2(1-524), and MSL2(1-649) bound to 10C, roX2 RNA levels were similar to those of wild-type females (Fig. 5D). There was an 8- to 10-fold increase in MSL2(1-683)- and MSL2(1-743)-expressing females and a 70-fold increase in females that express full-length MSL2. These results suggest that the carboxyl-terminal domain of MSL2 is required for roX2 RNA stabilization and/or activation of roX2 gene expression. Further, aa 649 to 683 and 743 to 773 appear to be particularly important. Both regions are well conserved across nine Drosophila species, with the exception that the 743-to-773 motif appears to be absent in D. grimshawi MSL2 (see Fig. S1, in the supplemental material). The acidic nature of the 743-to-773 motif may be significant, as some transcription activators contain acidic activation domains (28). Significantly, roX2 RNA levels were similar in females that express MSL2(1-683) and MSL2(1-743), although only the latter binds normally to the X chromosome. The results shown in Fig. 5C and D were obtained with the roX1 and roX2 primer pairs used by Bai et al. (4); however, similar results were obtained with different roX1 and roX2 primer pairs (data not shown). Further, comparable results were obtained with independently isolated RNA preparations (not shown). In total, our results indicate that the different binding patterns of the truncated versions of MSL2 are not due to differences in the levels of the roX RNAs.
Since MSL1 is required for chromatin binding of the amino-terminal domain of MSL2, we next sought to determine if MSL1 was essential for roX gene expression. roX1 RNA was readily detected by RT-PCR in females that express MSL2(1-193)F (Fig. 5E and F). However, in homozygous msl1L60 females, roX1 RNA levels were not significantly above that of y w females. Thus, MSL1 is required for roX1 gene expression in females that express the amino-terminal domain of MSL2. These results are more consistent with the results of reference 3 than with those of reference 40.
As roX RNAs coimmunoprecipitate with the MSL proteins (45), we next investigated if the roX1 and roX2 RNAs coimmunoprecipitated with MSL2(1-683), MSL2(1-743), and full-length MSL2(1-773) from female extracts. In females that express these versions of MSL2, roX1 and roX2 RNA levels are significantly higher than in the control y w females (Fig. 5C and D, lanes 5 to 7). Immunoprecipitation experiments were carried out with MSL2 antibody under conditions that preserve RNA integrity. With each protein extract, a mock immunoprecipitation was also performed without added antibody (control). Considerably more roX1 and roX2 RNA coimmunoprecipitated with MSL2(1-743) and full-length MSL2(1-773) relative to the control (Fig. 5G). In contrast, there was little difference in the level of either roX1 or roX2 RNA that coimmunoprecipitated with MSL2(1-683) compared to the mock immunoprecipitation. Similar levels of MSL2 protein were immunoprecipitated from the transgenic strains, but no MSL2 protein was detected in the matched controls (Fig. 5H). Similar results were obtained with an independent set of immunoprecipitation experiments (not shown).
The carboxyl-terminal domain of MSL2 is required for efficient incorporation of the roX RNAs into the MSL complex.
The coimmunoprecipitation experiments suggested that the roX RNAs are only incorporated into the MSL complex that contains the longer versions of MSL2 that bind normally to the X chromosome. To confirm this suggestion, we next performed in situ hybridizations with a labeled roX1 RNA probe on female polytene chromosomes. roX1 was not associated with either the X chromosome or chromocenter in females that express MSL2(1-193)F or MSL2(1-649) (Fig. 6A and B). In contrast, the roX1 RNA hybridized to hundreds of sites along the X chromosomes in females that express MSL2(1-743) (Fig. 6C). Since similar low levels of roX1 RNA are present in females of all three lines (Fig. 5C), these results suggest that the carboxyl-terminal domain of MSL2 is required for efficient incorporation of roX1 RNA.
FIG. 6.
Association of roX1 RNA with polytene chromosomes in hsp70-msl2 lines. (A to H) Single-stranded biotinylated roX1 probe was hybridized to salivary gland nuclei from female larvae that express the indicated version of MSL2 and were detected with fluorescein-avidin (green) and counterstained with DAPI (blue). (D to H) The female nuclei coexpress roX1 RNA from an autosomal hsp83-roX1 transgene. (I to K) Female nuclei that coexpress roX1 RNA and MSL2(1-193)F (I and J) or MSL2(1-649) (K) were immunostained with MSL2 antibody (red) and counterstained with DAPI (blue). The chromocenter (arrowhead), X chromosome (X), and hybridization/binding sites on the X chromosome (arrows) are indicated. (I to K, insets) Binding to sites at the base of the X chromosome, but not the chromocenter, was detected in most nuclei. (L) Quantitative real-time RT-PCR of roX1 RNA levels in females that coexpress roX1 and the indicated truncated version of MSL2. The average of two independent experiments is shown.
The core MSL complex contains three proteins, MLE, MSL3, and MOF, that contain RNA-binding domains (1, 22). Since these proteins colocalize with the amino-terminal domain of MSL2 to the chromocenter and other sites (Fig. 2), it might be anticipated that one or more of these proteins could mediate incorporation of roX1 RNA. We wondered if this did not occur in MSL2(1-193)F female nuclei because of the low level of roX1 RNA. To test this idea, we crossed the MSL2(1-193)F, MSL2(1-649), and MSL2(1-743) lines with a strain that contains an hsp83-roX1 transgene that constitutively expresses roX1 RNA (32). As anticipated, there was an about 50- to 150-fold increase in roX1 levels in female offspring of these crosses relative to the y w strain (wild type) (Fig. 6L), which was at least 20-fold higher than in females of the parental hsp70-msl2 strains (Fig. 5C). In the salivary gland nuclei of females that coexpress roX1 and MSL2(1-193)F or MSL2(1-649), we observed strong signals of hybridization to many sites at the base of the X chromosome in 80 to 90% of the nuclei (Fig. 6D to G). In approximately 30% of the nuclei, we observed hybridization to sites spanning several numbered divisions at the base of the X chromosome and also to sites near the tip of the X chromosome (Fig. 6F). Hybridization signals were also detected at other sites on the X chromosome and autosomes, but these where very inconsistent. Interestingly, in most nuclei we did not detect hybridization to the chromocenter or 8D, although weak diffuse staining was seen over the chromocenter in a few nuclei (Fig. 6F). In females that coexpress roX1 and MSL2(1-743), hybridization signals were detected at many sites along the length of the X chromosome (Fig. 6H). To determine if elevated levels of roX1 had altered the chromatin binding of the shorter versions of MSL2, nuclei were stained with MSL2 antibody. Many binding sites were detected at the base of the X chromosome but not at the chromocenter or 8D (Fig. 6I to K). In some nuclei, complex binding spanned several numbered divisions (Fig. 6I) and in a few nuclei binding was also detected at sites close to the tip of the X chromosome (Fig. 6K). These results suggested that some roX1 has been incorporated into the MSL complex containing truncated MSL2 and this appears to have led to local spreading from the chromocenter along the X chromosome.
The carboxyl-terminal domain of MSL2 could either bind roX RNA directly or associate with one or more of the proteins in the complex that binds RNA. To address the latter possibility, we tested if any of the MSL proteins coimmunoprecipitate with the FLAG-tagged MSL2 C-terminal domain from extracts from adult females. Neither MSL1, nor MSL3, nor MOF coimmunoprecipitated with MSL2(490-773)F (Fig. 7A, B, and D). However, a small amount MLE coimmunoprecipitated with MSL2(490-773)F in three independent experiments (Fig. 7C). MLE did not coimmunoprecipitate with the amino-terminal domain of MSL2 (Fig. 7E). However, a small amount of MLE coimmunoprecipitated with full-length MSL2 from female extracts (Fig. 7F). Thus, the carboxyl-terminal domain of MSL2 does not bind strongly to any of the other MSL proteins but does appear to weakly associate with MLE.
FIG. 7.
MLE weakly coimmunoprecipitates with the carboxyl-terminal domain of MSL2. Protein extracts from transformant female flies that co-overexpressed MSL2(490-773)F and MSL1 (A), MSL3 (B), MLE (C), or MOF (D) were immunoprecipitated (IP) with anti-FLAG affinity matrix and detected by Western blotting (WB) with the indicated antibodies. Immunoprecipitated extracts (Ip) are shown in the right lane of each panel, and 10% of the corresponding input (Input) is shown in the left lane. Coimmunoprecipitation experiments were also performed with extracts from females that overexpress MLE and either MSL2(1-193)F (E) or full-length MSL2 (F). A small amount of MLE consistently coimmunoprecipitated with the carboxyl-terminal domain of MSL2 or full-length MSL2 but not with the amino-terminal domain.
DISCUSSION
It is has recently become apparent that a large fraction of complex genomes is devoted to the production of noncoding RNAs (38, 39). While it is clear that some of these noncoding RNAs regulate gene expression, the underlying mechanisms are poorly understood. The Drosophila roX1 and roX2 noncoding RNAs are required for the normal localization of the MSL complex to sites on the male X chromosome. The question is: how do the roX RNAs direct the MSL complex? One possibility is that the roX RNAs invade X chromosomal DNA and form RNA-DNA heteroduplexes (19). In support of this model, MLE is an ATP-dependent RNA-DNA helicase and this activity is required for binding of the MSL complex to most sites on the X chromosome, except the high-affinity sites (15). This mechanism is similar to one of the models proposed for how small interfering RNAs guide the transcription-silencing RITS complex to homologous centromeric repetitive sequences in Schizosaccharomyces pombe (49). Alternatively, incorporation of the roX RNAs could alter the chromatin-binding specificity of the MSL complex. Our results provide some support for this model. We found that the amino-terminal domains of MSL1 and MSL2 (MSL1N/MSL2N) bind as a complex to the chromocenter and a few other sites. Although a low level of roX1 RNA was present, it was not incorporated into the MSL1N/MSL2N complex. Binding to the chromocenter required the same 26-aa N-terminal basic motif in MSL1 that was previously shown to be essential for binding to the high-affinity sites on the X chromosome (24). Thus, the amino-terminal domains of MSL1 and MSL2 form a chromatin-binding module that is required for binding of the complex to pericentromeric heterochromatin (no roX RNA in complex) and to the high-affinity sites on the X chromosome. MSL3, MLE, and MOF colocalized with MSL1 and MSL2N to the pericentromeric heterochromatin and other sites (Fig. 2 and 8A). If considerably higher levels of roX1 RNA were supplied by a constitutively active transgene, then MSL2N no longer bound to the chromocenter. Instead, binding was seen over many bands at the base of the X chromosome, suggesting local spreading of the complex from the chromocenter (Fig. 8B). In situ hybridization data indicated that some of the roX1 was incorporated into the complex, presumably by interaction with one or more of the MSL proteins that contain an RNA-binding domain. Thus, it appears that incorporation of roX1 RNA has altered the binding specificity of the MSL1/MSL2 complex to recognize features on the X chromosome. Conceivably, incorporation of roX RNA could induce a conformational change in the MSL proteins or could combine with the proteins to create a novel chromatin-binding module. Normal binding to hundreds of sites on the X chromosome required motifs in the carboxyl-terminal domain of MSL2 (Fig. 8C). Further, although levels of the roX RNAs were low, the RNAs were incorporated into the complex. Thus, the role of the carboxyl-terminal domain of MSL2 could be to promote X chromosome binding by facilitating efficient incorporation of the roX RNAs into the complex. It is also possible that motifs (e.g., basic) in the carboxyl-terminal domain may contribute directly to X chromosome recognition. This could explain why shorter versions of MSL2 lacking the carboxyl-terminal domain only bound to a limited number of sites on the X chromosome in the presence of high levels of roX1 RNA.
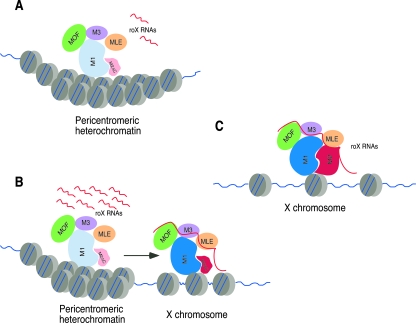
FIG. 8.
Proposed model. (A) In female nuclei that express a truncated version of MSL2 that lacks the carboxyl-terminal domain (M2ΔC) and contain a low level of roX RNA, the MSL complex assembles and binds to pericentromeric heterochromatin. roX RNA is not incorporated into the protein complex. Chromatin binding is mediated by the amino-terminal domains of MSL1 (M1) and M2ΔC. (B) However, if roX RNA levels are high, some RNA is incorporated into the complex, which then spreads from the pericentromeric heterochromatin to sites at the base of the X chromosome. (C) Longer versions of MSL2 that contain proline-rich and basic motifs in the C-terminal domain efficiently incorporate low levels of roX RNA into the complex and bind to hundreds of sites along the length of the X chromosome. We suggest that incorporation of roX RNA alters the chromatin-binding specificity of the MSL1/MSL2 complex to recognize features on the X chromosome (B and C). Most of the interactions shown are known from this or previous studies, but it is not known how MLE would associate with the MSL complex that contains M2ΔC and lacks roX RNA (A).
The proline-rich and basic motifs in the carboxyl-terminal domain of MSL2 could mediate incorporation of the roX RNAs either directly or indirectly. With regard to the latter, there are numerous examples where proline-rich motifs participate in protein-protein interactions. For example, both SH3 and WW domains recognize proline-rich docking sites (25). We found that MLE, which contains an RNA-binding domain, weakly associated with the carboxyl-terminal domain of MSL2. Since MLE is required for incorporation of the roX RNAs into the MSL complex (32), the carboxyl-terminal domain of MSL2 may mediate incorporation of the roX RNAs indirectly via association with MLE. However, while MLE colocalized with the amino-terminal domain of MSL2 to the chromocenter, roX1 RNA did not. Since similar levels of roX1 RNA were found in MSL2(1-193)F and MSL2(1-743) females, MLE does not appear to be sufficient to mediate efficient incorporation of roX1 RNA. One possibility is that the proline- and base-rich motifs of MSL2 combine with the MLE to incorporate the roX RNAs. The herpes simplex virus type 1 US11 protein binds RNA via an arginine- and proline-rich motif (20). The first double-stranded RNA-binding motif (dsRBM) in human RNA helicase A combines with a proline-rich motif to form a novel RNA-binding module (18). RNA helicase A is the human ortholog of MLE. While the dsRBM domains are well conserved between RNA helicase A and MLE, the proline-rich motif is not conserved. Perhaps in Drosophila the proline-rich motif in MSL2 combines with the dsRBM of MLE to form a module that specifically binds the roX RNAs.
In Sciara ocellaris, a very distant dipteran relative of drosophilid species, the MSL proteins bind to all chromosomes and thus appear to have no role in X chromosome dosage compensation (43). The MSL complex does bind specifically to the male X chromosome in Drosophila species spanning four genera (30). It will be of interest to determine if a key step in the adaptation of the MSL complex for X chromosome dosage compensation was acquisition of the carboxyl-terminal domain of MSL2 that is required for normal binding to the X chromosome and efficient incorporation of roX RNAs into the complex.
Supplementary Material
Acknowledgments
We are grateful to Esther Belikoff for excellent technical assistance and to Corey Laverty for comments on the manuscript. We thank Mitzi Kuroda, Rick Kelley, and John Lucchesi for generously sending plasmid DNAs, antibodies, and fly strains.
This research was funded by grant MAU204 from the Royal Society of New Zealand Marsden Fund.
Footnotes
Published ahead of print on 17 December 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akhtar, A., D. Zink, and P. B. Becker. 2000. Chromodomains are protein-RNA interaction modules. Nature 407405-409. [DOI] [PubMed] [Google Scholar]
- 2.Alekseyenko, A. A., E. Larschan, W. R. Lai, P. J. Park, and M. I. Kuroda. 2006. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 20848-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai, X., A. A. Alekseyenko, and M. I. Kuroda. 2004. Sequence-specific targeting of MSL complex regulates transcription of the roX RNA genes. EMBO J. 232853-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai, X., E. Larschan, S. Y. Kwon, P. Badenhorst, and M. I. Kuroda. 2007. Regional control of chromatin organization by noncoding roX RNAs and the NURF remodeling complex in Drosophila melanogaster. Genetics 1761491-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belote, J. M., and J. C. Lucchesi. 1980. Control of X chromosome transcription by the maleless gene in Drosophila. Nature 285573-575. [DOI] [PubMed] [Google Scholar]
- 6.Copps, K., R. Richman, L. M. Lyman, K. A. Chang, J. Rampersad-Ammons, and M. I. Kuroda. 1998. Complex formation by the Drosophila MSL proteins: role of the MSL2 RING finger in protein complex assembly. EMBO J. 175409-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corona, D. F., C. R. Clapier, P. B. Becker, and J. W. Tamkun. 2002. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 3242-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demakova, O. V., I. V. Kotlikova, P. R. Gordadze, A. A. Alekseyenko, M. I. Kuroda, and I. F. Zhimulev. 2003. The MSL complex levels are critical for its correct targeting to the chromosomes in Drosophila melanogaster. Chromosoma 112103-115. [DOI] [PubMed] [Google Scholar]
- 9.Deng, X., and V. H. Meller. 2006. Non-coding RNA in fly dosage compensation. Trends Biochem. Sci. 31526-532. [DOI] [PubMed] [Google Scholar]
- 10.Deng, X., and V. H. Meller. 2006. roX RNAs are required for increased expression of X-linked genes in Drosophila melanogaster males. Genetics 1741859-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, X., B. P. Rattner, S. Souter, and V. H. Meller. 2005. The severity of roX1 mutations is predicted by MSL localization on the X chromosome. Mech. Dev. 1221094-1105. [DOI] [PubMed] [Google Scholar]
- 12.Furuhashi, H., M. Nakajima, and S. Hirose. 2006. DNA supercoiling factor contributes to dosage compensation in Drosophila. Development 1334475-4483. [DOI] [PubMed] [Google Scholar]
- 13.Gilfillan, G. D., T. Straub, E. de Wit, F. Greil, R. Lamm, B. van Steensel, and P. B. Becker. 2006. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev. 20858-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu, W., P. Szauter, and J. C. Lucchesi. 1998. Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Dev. Genet. 2256-64. [DOI] [PubMed] [Google Scholar]
- 15.Gu, W., X. Wei, A. Pannuti, and J. C. Lucchesi. 2000. Targeting the chromatin-remodeling MSL complex of Drosophila to its sites of action on the X chromosome requires both acetyl transferase and ATPase activities. EMBO J. 195202-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamada, F. N., P. J. Park, P. R. Gordadze, and M. I. Kuroda. 2005. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 192289-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry, R. A., B. Tews, X. Li, and M. J. Scott. 2001. Recruitment of the male-specific lethal (MSL) dosage compensation complex to an autosomally integrated roX chromatin entry site correlates with an increased expression of an adjacent reporter gene in male Drosophila. J. Biol. Chem. 27631953-31958. [DOI] [PubMed] [Google Scholar]
- 18.Hung, M. L., P. Chao, and K. Y. Chang. 2003. dsRBM1 and a proline-rich domain of RNA helicase A can form a composite binder to recognize a specific dsDNA. Nucleic Acids Res. 315741-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelley, R. L. 2004. Path to equality strewn with roX. Dev. Biol. 26918-25. [DOI] [PubMed] [Google Scholar]
- 20.Khoo, D., C. Perez, and I. Mohr. 2002. Characterization of RNA determinants recognized by the arginine- and proline-rich region of Us11, a herpes simplex virus type 1-encoded double-stranded RNA binding protein that prevents PKR activation. J. Virol. 7611971-11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuroda, M. I., M. J. Kernan, R. Kreber, B. Ganetzky, and B. S. Baker. 1991. The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell 66935-947. [DOI] [PubMed] [Google Scholar]
- 22.Lee, C. G., K. A. Chang, M. I. Kuroda, and J. Hurwitz. 1997. The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J. 162671-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legube, G., S. K. McWeeney, M. J. Lercher, and A. Akhtar. 2006. X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev. 20871-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, F., D. A. Parry, and M. J. Scott. 2005. The amino-terminal region of Drosophila MSL1 contains basic, glycine-rich, and leucine zipper-like motifs that promote X chromosome binding, self-association, and MSL2 binding, respectively. Mol. Cell. Biol. 258913-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, S. S. 2005. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem. J. 390641-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucchesi, J. C., and M. I. Kuroda. 2007. Dosage compensation in Drosophila, p. 307-319. In C. D. Allis, T. Jenuwein, and D. Reinberg (ed.), Epigenetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Lyman, L. M., K. Copps, L. Rastelli, R. L. Kelley, and M. I. Kuroda. 1997. Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics 1471743-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma, J., and M. Ptashne. 1987. A new class of yeast transcriptional activators. Cell 51113-119. [DOI] [PubMed] [Google Scholar]
- 29.Marín, I. 2003. Evolution of chromatin-remodeling complexes: comparative genomics reveals the ancient origin of “novel” compensasome genes. J. Mol. Evol. 56527-539. [DOI] [PubMed] [Google Scholar]
- 30.Marín, I., A. Franke, G. J. Bashaw, and B. S. Baker. 1996. The dosage compensation system of Drosophila is co-opted by newly evolved X chromosomes. Nature 383160-163. [DOI] [PubMed] [Google Scholar]
- 31.Meller, V. H. 2003. Initiation of dosage compensation in Drosophila embryos depends on expression of the roX RNAs. Mech. Dev. 120759-767. [DOI] [PubMed] [Google Scholar]
- 32.Meller, V. H., P. R. Gordadze, Y. Park, X. Chu, C. Stuckenholz, R. L. Kelley, and M. I. Kuroda. 2000. Ordered assembly of roX RNAs into MSL complexes on the dosage-compensated X chromosome in Drosophila. Curr. Biol. 10136-143. [DOI] [PubMed] [Google Scholar]
- 33.Meller, V. H., and B. P. Rattner. 2002. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 211084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meller, V. H., K. H. Wu, G. Roman, M. I. Kuroda, and R. L. Davis. 1997. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88445-457. [DOI] [PubMed] [Google Scholar]
- 35.Mendjan, S., M. Taipale, J. Kind, H. Holz, P. Gebhardt, M. Schelder, M. Vermeulen, A. Buscaino, K. Duncan, J. Mueller, M. Wilm, H. G. Stunnenberg, H. Saumweber, and A. Akhtar. 2006. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell 21811-823. [DOI] [PubMed] [Google Scholar]
- 36.Oh, H., Y. Park, and M. I. Kuroda. 2003. Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes Dev. 171334-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmer, M. J., R. Richman, L. Richter, and M. I. Kuroda. 1994. Sex-specific regulation of the male-specific lethal-1 dosage compensation gene in Drosophila. Genes Dev. 8698-706. [DOI] [PubMed] [Google Scholar]
- 38.Pang, K. C., M. C. Frith, and J. S. Mattick. 2006. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 221-5. [DOI] [PubMed] [Google Scholar]
- 39.Prasanth, K. V., and D. L. Spector. 2007. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2111-42. [DOI] [PubMed] [Google Scholar]
- 40.Rattner, B. P., and V. H. Meller. 2004. Drosophila male-specific lethal 2 protein controls sex-specific expression of the roX genes. Genetics 1661825-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rea, S., and A. Akhtar. 2006. MSL proteins and the regulation of gene expression. Curr. Top. Microbiol. Immunol. 310117-140. [DOI] [PubMed] [Google Scholar]
- 42.Richter, L., J. R. Bone, and M. I. Kuroda. 1996. RNA-dependent association of the Drosophila maleless protein with the male X chromosome. Genes Cells 1325-336. [DOI] [PubMed] [Google Scholar]
- 43.Ruiz, M. F., M. R. Esteban, C. Donoro, C. Goday, and L. Sanchez. 2000. Evolution of dosage compensation in Diptera: the gene maleless implements dosage compensation in Drosophila (Brachycera suborder) but its homolog in Sciara (Nematocera suborder) appears to play no role in dosage compensation. Genetics 1561853-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott, M. J., L. L. Pan, S. B. Cleland, A. L. Knox, and J. Heinrich. 2000. MSL1 plays a central role in assembly of the MSL complex, essential for dosage compensation in Drosophila. EMBO J. 19144-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, E. R., A. Pannuti, W. Gu, A. Steurnagel, R. G. Cook, C. D. Allis, and J. C. Lucchesi. 2000. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20312-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spierer, A., C. Seum, M. Delattre, and P. Spierer. 2005. Loss of the modifiers of variegation Su(var)3-7 or HP1 impacts male X polytene chromosome morphology and dosage compensation. J. Cell Sci. 1185047-5057. [DOI] [PubMed] [Google Scholar]
- 47.Straub, T., and P. B. Becker. 2007. Dosage compensation: the beginning and end of generalization. Nat. Rev. Genet. 847-57. [DOI] [PubMed] [Google Scholar]
- 48.Straub, T., G. D. Gilfillan, V. K. Maier, and P. B. Becker. 2005. The Drosophila MSL complex activates the transcription of target genes. Genes Dev. 192284-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verdel, A., S. Jia, S. Gerber, T. Sugiyama, S. Gygi, S. I. Grewal, and D. Moazed. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou, S., Y. Yang, M. J. Scott, A. Pannuti, K. C. Fehr, A. Eisen, E. V. Koonin, D. L. Fouts, R. Wrightsman, J. E. Manning, and J. C. Lucchesi. 1995. Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. EMBO J. 142884-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.