Abstract
The stress response in yeast cells is regulated by at least two classes of transcription activators—HSF and Msn2/4, which differentially affect promoter chromatin remodeling. We demonstrate that the deletion of SNF2, an ATPase activity-containing subunit of the chromatin remodeling SWI/SNF complex, eliminates histone displacement, RNA polymerase II recruitment, and heat shock factor (HSF) binding at the HSP12 promoter while delaying these processes at the HSP82 and SSA4 promoters. Out of the three promoters, the double deletion of MSN2 and MSN4 eliminates both chromatin remodeling and HSF binding only at the HSP12 promoter, suggesting that Msn2/4 activators are primary determinants of chromatin disassembly at the HSP12 promoter. Unexpectedly, during heat shock the level of Msn2/4 at the HSP12 promoter declines. This is likely a result of promoter-targeted Msn2/4 degradation associated with transcription complex assembly. While histone displacement kinetic profiles bear clear promoter specificity, the kinetic profiles of recovery from heat shock for all analyzed genes display an equal or even higher nucleosome return rate, which is to some extent delayed by the deletion of SNF2.
The stress response in yeast cells is regulated by at least two types of transcriptional activators: heat shock factor (HSF) and the partially redundant Msn2 and Msn4 (Msn2/4) activators (7, 19). The HSF system is highly conserved in its overall composition and function from yeast to humans (61). HSF binds to the major groove of DNA in heat shock promoter elements (HSEs), which are also conserved from yeast to humans (61). The activity of HSF is regulated via several distinct pathways. These include a monomer-trimer transition (45), phosphorylation, and other posttranslational modifications (30, 31, 54), as well as repression by molecular chaperones interacting with HSF, thus blocking their own production (45, 60) and forming a self-regulatory loop.
The Msn2/4 system is more specific to Saccharomyces cerevisiae. The Msn2 and Msn4 factors recognize and bind stress response element (STRE) sequences (44) in promoter regions of a large array of genes partially overlapping the HSF-regulated array (3, 7, 22, 46). The Msn2 factor seems to have a more pronounced role, since mutants lacking only Msn2 have an already distinct decrease in STRE-regulated transcription; however, only MSN2 and MSN4 double deletions exhibit pleiotropic stress sensitivity (19). Under stress, Msn2/4 accumulate in the nucleus within a few minutes (24, 32). The Msn2/4 factors are regulated by efficient and oscillatory nuclear transport (32), hyperphosphorylation upon stress (21, 24), and degradation associated with transcription initiation (37, 38). While the HSF system is actively involved in chromatin remodeling events at gene promoters, the role of the Msn2/4 system in these processes is poorly understood.
Chromatin remodeling varies in intensity and intermediate states between genes. The most evident and intense examples of chromatin remodeling are the changes taking place at the promoters of certain heat shock genes during heat induction. Chromatin remodeling at the HSP12, HSP82, and SSA4 promoters (14) surpasses in rate and intensity such canonical models as the PHO5 and Gal promoters. At the HSP82 promoter, a noticeable two- to fourfold nucleosome diminishment is observed in the first seconds of heat shock which by 8 to 16 min can reach 15- to 20-fold (14, 65). The kinetic profiles and the intensities of nucleosome displacement vary considerably even between the highly related and coregulated heat shock genes, indicating the possibility of different chromatin remodeling mechanisms. The involvement of the SWI/SNF complex in chromatin remodeling at the HSP82 promoter has been reported previously (65), but the deletion of the SNF2 activity has not been shown to affect nucleosome displacement. Therefore, the function of the SWI/SNF complex at heat shock gene promoters remains unclear.
The SWI/SNF complex is one of the major ATP-dependent chromatin remodeling protein complexes in eukaryotic cells directly affecting expression of a large number of genes. Consequently, there has been a longstanding interest in investigations of the mechanisms of recruitment and action of this complex. The SWI/SNF complex is composed of 12 subunits (40, 52), with the Swi2/Snf2 subunit serving as an ATPase, required for providing energy for disruption and/or mobilization of nucleosomes (8, 11, 28, 36, 47). The mechanistic aspects of SWI/SNF action in vivo are not well defined. Some early data suggest a nontargeted mode of action determining the accessibility of promoter cis elements for transcription activators (11, 36, 59), while more recent studies argue for the targeted recruitment of ATP-dependent chromatin remodeling complexes by activation domains of activators causing nucleosome translocations (4, 28, 42). The prevailing point of view is that due to the low abundance of the SWI/SNF complex in the cell and its interaction with nucleosomal DNA without sequence specificity, the SWI/SNF complex needs to be targeted prior to stimulating local nucleosome sliding or displacement in trans.
In the present study, we compared events taking place at three different heat shock gene promoters during the time course of heat induction and found that the involvement of the SWI/SNF complex in chromatin remodeling at these promoters differs drastically. This involvement is exhibited in the effects caused by the inactivation of the SNF2 subunit, which either eliminates (HSP12) or delays (HSP82 and SSA4) chromatin remodeling at the studied heat shock gene promoters. While the kinetic profiles of nucleosome displacement are promoter specific, the rate of reoccupation appears to be the same for all promoters analyzed and is delayed to the same degree in the ΔSNF2 strain. Additionally we show that Msn2/4 activators, which dominate the HSP12 promoter, are likely under the control of targeted degradation associated with transcription initiation complex assembly. This regulatory mechanism is likely responsible for the attenuation of transcription of Msn2/4-regulated genes during the loss of induction stimuli.
MATERIALS AND METHODS
Strains and cultivation conditions.
Strains utilized in this study are indicated in Table 1. Strain TYE007 was constructed by substitution of the −300 to +351 HSP12 region (relative to the translation start codon) with the KanMX cassette PCR amplified from the pUG6 plasmid (27) according to a previously described procedure. Correct chromosomal integration was confirmed by a diagnostic PCR using HSP12 (−337 to −304) and pUG6 primers. Positions of three Msn2/4 binding sites in the HSP12 promoter are −435, −413, and −376.
TABLE 1.
Yeast strains used
| Strain | Genotype | Reference or source |
|---|---|---|
| FY1350 | MATα leu2Δ0 lys2Δ0 ura3Δ0 | 43 |
| FY1360 | MATα leu2Δ1 snf2::LEU2 his3D200 ura3-52 lys2-173R2 | 43 |
| FY2470 | MATα ura3Δ0 lus2Δ0 leu2Δ0 his3Δ200 trp1Δ63 SNF2-C18myc::TRP1 CHA4-flag::kanMX | 43 |
| JT670 | MATα ade2-101 leu2-3,112 ura3-52 his3 trp1 | 57 |
| JT652 | MATα ade2-101 leu2-3,112 ura3-52 his3 msn2Δ3::HIS3 msn4Δ2::URA3 | 57 |
| YSC1178-7502107 | Open Biosystems | |
| HS1004 | MATaade-can1-100 cyh2r his3-11,15 leu2-3,112 trp1-1 ura3 leu::hsp82-ΔHSE1-lacz::leu2::KanMX | 51 |
| HS1006 | HS1004; ump1 | |
| TYE007 | FY1350; hsp12Δ::KanMX | This study |
S. cerevisiae strains were cultivated at 30°C to early log phase in rich yeast extract-peptone-dextrose (YPD) broth supplemented with 0.04 mg/ml adenine. For kinetics experiments, instantaneous upshift was achieved by rapidly mixing equal volumes of 30°C culture with prewarmed 52°C medium and then incubating with shaking at 39°C for the times indicated. For the recovery experiments, cultures were heat shocked for 16 min at 39°C and then ice cold medium was added to immediately downshift the temperature to 30°C. Samples were taken at various time intervals.
ChIP analysis.
Chromatin immunoprecipitation (ChIP) was performed essentially as previously described (16), with the exception that protein A magnetic beads were used instead of protein A-Sepharose beads to precipitate antigen-antibody complexes. Special attention was paid to the consistency of sonication levels of cell lysates. Before immunoprecipitation, all samples were tested for the level of DNA fragmentation, and the mean size of DNA fragments was always 500 bp. Antibodies specific for the following epitopes were used: histone H3, total (ab1791; AbCam), on RNA polymerase II (Pol II)-YSPT[pS]PS repeat of Pol II C-terminal domain (4H8 monoclonal antibody from Upstate Biotechnology; this antibody recognizes both phospho- and nonphospho-Pol II according to the manufacturer's data); HSF (rabbit antibody raised and characterized previously [15]); Msn2 (sc-33631 and sc-15615; Santa Cruz Biotechnology); Msn4 (sc-15550; Santa Cruz Biotechnology); Myc tag (sc-40; Santa Cruz Biotechnology); and TAP tag (CAB1001; Open Biosystems). Immunoprecipitated DNA samples were used for real-time PCR with Sybr green dye, and signals for individual gene promoters were normalized against the corresponding signal derived from the PHO5 promoter (for histone ChIPs) or the chromosome V intergenic region (in the case of Pol II and HSF ChIPs) and to the input DNA sample. For each DNA sample, at least three consecutive dilutions of DNA were analyzed, making certain that the amplification rate was always optimal and the change in amplification signal was proportional to the change in the amount of DNA. In addition, controls without DNA were always included, making certain that primer dimer formation was not detectable or comparable to the amplification from experimental samples. Experiments were typically repeated three times or more.
Primers for real-time PCRs were selected among a significant number of primers based on PCR efficiency. Only those primer pairs that gave an amplification rate of at least 1.9 per PCR cycle during the linear amplification and did not produce primer dimers were used. The sequences of PCR primers used in this study were as follows (coordinates are relative to ATG): PHO5, −214 to −192 and −20 to −48; HSP12, −304 to −279 or −337 to 304 and −82 to −107; HSP82, −193 to −167 and −37 to −69; SSA4, −307 to −279 and −70 to −98; chromosome V intergenic region, GCAATCAACATCTGAAGAAAAGAAAGTAGT and CATAATCTGCTGAAAAATGGCGTAAAT.
RESULTS
In this study, we compared changes at the promoters of the three highly inducible and coregulated heat shock genes over the time course of induction and repression. The main methodological approach we utilized was ChIP coupled with real-time PCR. This method allows side-by-side comparison of events taking place simultaneously at different promoters by employing promoter-specific primers. Comparison of different gene promoters to each other using the same DNA preparations helped us to quantitate gene-specific effects and to eliminate differences due to assay variability.
The SWI/SNF complex is critical for chromatin remodeling at the HSP12 promoter, and its absence delays nucleosome displacement at the HSP82 and SSA4 promoters.
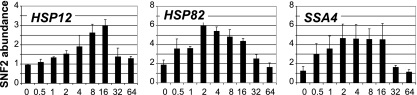
The three genes analyzed in this study, HSP12, HSP82, and SSA4, represent typical genes that are greatly induced upon heat shock. Since all three heat shock gene promoters undergo extensive chromatin remodeling albeit with different kinetic profiles (14), we wanted to test if SWI/SNF, as one of the major chromatin remodeling complexes, is involved. The results, depicted in Fig. 1, show that the HSP12, HSP82, and SSA4 promoters exhibit an initial increase in the SNF2 level during heat induction, followed by a decline during the attenuation period. These results are consistent with the dynamics of chromatin remodeling observed previously by us (14) and with the involvement of the SWI/SNF complex at the HSP82 promoter (65).
FIG. 1.
Kinetics of SNF2 recruitment to heat shock gene promoters. ChIPs were performed using an antibody that recognizes the Myc epitope on SNF2 (S. cerevisiae strain FY2470). The promoter analyzed is indicated in the upper left corner of each panel. y axis, abundance of Myc-tagged SNF2; x axis, time after heat shock (0 to 64 min). All real-time PCR values were normalized relative to the input and to the background values of the intergenic region of chromosome V. Values represent means ± standard deviations (n ≥ 3).
To monitor chromatin remodeling events at the indicated promoters, we used anti-H3 antibody raised against the C-terminal region of H3 (amino acids 125 to 135). This region is not known to be posttranslationally modified, allowing a measure of total histone H3 abundance. Utilization of this antibody for quantification of a change in total H3 has been demonstrated previously by others (34, 39). The results of such experiments are traditionally reported as a drop (1 to 0 or 100% to 0%) in histone content during gene activation. This form of presentation restricts analysis of the data, since the closer the values are to 0 the more difficult it is to see fine differences. Therefore, we have chosen to present the results as an inverse value, which represents the degree of histone displacement and changes from 1 to infinity. This presentation format better conveys fine differential chromatin remodeling and has been utilized by us previously (14).
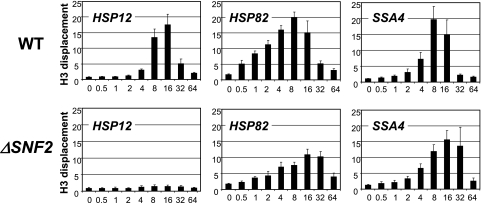
To confirm the involvement of the SWI/SNF complex and to further investigate its role in the regulation of these genes, we utilized the ΔSNF2 strain. The deletion of this key enzymatic activity of the SWI/SNF complex influenced the promoters in different ways. At the HSP12 promoter, the displacement of nucleosomes was almost entirely eliminated, while the effect of this mutation on the HSP82 and SSA4 promoters was only a moderate decrease in the intensity of histone displacement and, more obviously, a delay in reaching the maximum histone loss (Fig. 2).
FIG. 2.
Chromatin remodeling is eliminated at the HSP12 promoter and delayed at the HSP82 and SSA4 promoters in the strain lacking SNF2. ChIPs using an antibody raised against the C terminus of histone H3 were performed with the ΔSNF2 (FY1360) or isogenic WT strain (FY1350). The promoter analyzed is indicated in the upper left corner of each panel. y axis, n-fold displacement of histone H3; x axis, time after heat shock (0 to 64 min). All real-time PCR values were normalized relative to the input and to the signal from the PHO5 promoter, which is known to contain positioned nucleosomes that do not change during heat shock (16). Values represent means ± standard deviations (n ≥ 3). Note: displacement value is an inverse value of abundance.
Deletion of SNF2 eliminates HSF binding and Pol II recruitment at the HSP12 promoter while delaying these processes at the HSP82 and SSA4 promoters.
In the next set of experiments, we utilized antibodies against RNA Pol II that recognize both phosphorylated and unphosphorylated forms of the Pol II C-terminal domain. Our ChIPs therefore are able to provide information about transcription initiation events at specific promoters. This approach has the advantage of not having to deal with cross-reactivity issues of probes in canonical Northern blot experiments or reverse transcription-PCR and not having to deal with the specific mRNA half-life. The restriction of this approach is that it does not necessarily give a measure of completed transcription but rather is a measure of the transcription initiation complex assembly.
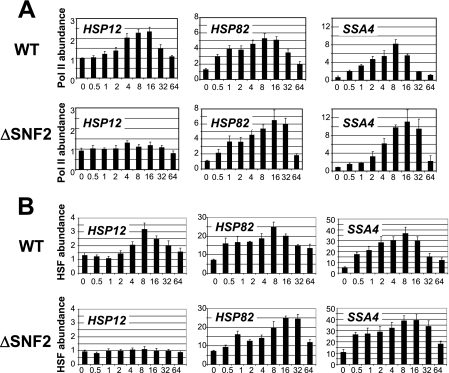
Since nucleosome displacement is not necessarily associated with nucleosome removal from the whole promoter during transcription initiation (34), chromatin remodeling observed at the three promoters may or may not correlate with Pol II loading. However, our ChIPs performed with anti-Pol II antibody (Fig. 3A) indicated that for both the wild-type (WT) and ΔSNF2 strains, there was a good correlation between Pol II loading and the histone H3 loss profiles. Notably, the ΔSNF2 strain showed no Pol II loading at the HSP12 promoter, and both the HSP82 and SSA4 promoters revealed a delay in maximum Pol II loading. These results are consistent with the histone displacement profiles (Fig. 2), suggesting that at least for the three promoters tested, histone displacement is a prerequisite for the initiation of transcription or at least for the initiation of complex assembly.
FIG. 3.
Deletion of SNF2 eliminates HSF binding and Pol II recruitment at the HSP12 promoter while delaying these processes at the HSP82 and SSA4 promoters. (A) ChIPs utilizing antibody against Pol II, performed with the ΔSNF2 or isogenic WT strain. The promoter analyzed is indicated in the upper left corner of each panel. y axis, abundance of Pol II; x axis, time after heat shock (0 to 64 min). All real-time PCR values were normalized relative to the input and to the background values of the intergenic region of chromosome V. Values represent means ± standard deviations (n ≥ 3). (B) Same as panel A except that the antibody utilized was against HSF.
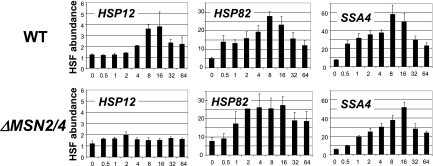
The master regulator for heat shock genes is the HSF, although other transcription factors, such as Msn2/4, are known to be involved in regulation of some heat shock genes, including HSP12 (3, 7). Although involvement of the SWI/SNF complex in regulation of heat shock genes has previously been suggested, a mechanistic role for this coactivator in chromatin remodeling is not clear, since the deletion of SNF2 does not have a significant effect on the HSP82 chromatin remodeling (65). Our previous results (14) indicate that the deletion of both activation domains in HSF drastically affects chromatin remodeling at the HSP12 promoter, indicating a significant role for HSF in HSP12 regulation. In the following experiments, we wanted to test if the deletion of SNF2 has any effect on regulation of HSP12 by HSF. Utilization of antibody raised against HSF in ChIPs (Fig. 3B) revealed that the deletion of SNF2 abolished HSF binding to the HSP12 promoter. The effect of this deletion on the HSP82 and SSA4 promoters was a delay of maximal HSF binding. Most importantly, both of the latter promoters have significant HSF binding before heat shock (5- to 10-fold above the background level as indicated for the zero time point in Fig. 3B). Since HSF cannot bind to the HSE DNA sequence within the nucleosome (56) and chromatin failed to be remodeled in the ΔSNF2 strain at the HSP12 promoter (Fig. 2), it seems plausible to suggest that HSF binding at the HSP12 promoter requires preliminary action by the SWI/SNF complex. It is also possible that SWI/SNF might be initially recruited by Msn2/4 at the HSP12 promoter.
Msn2/4 are required for HSF binding and chromatin remodeling at the HSP12 promoter.
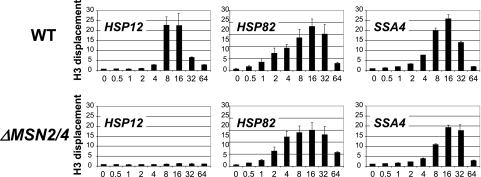
While it is known that the transcription of certain heat shock genes is under dual regulation of HSF and Msn2/4 systems (3, 7), the input of each activator in chromatin remodeling is not clear. To address this issue, we performed histone H3 ChIPs with a strain bearing a double deletion of Msn2 and Msn4 and compared the results to those for the isogenic WT strain (Fig. 4). These experiments revealed that there is no significant effect of the ΔMsn2/4 deletion on nucleosome displacement at the HSP82 and SSA4 promoters. However, histone H3 displacement at the HSP12 promoter is eliminated. These data suggest that Msn2/4 activators play a critical role in chromatin remodeling events at the HSP12 promoter but not at the HSP82 and SSA4 promoters, which is consistent with previously published expression data for these genes (7) and with the lack of a change in the levels of the HSP82 and SSA4 mRNA in the ΔMsn2 ΔMsn4 strain (57). Considering the dynamic recruitment of HSF to the HSP12 promoter (Fig. 3B) and our previously published data indicating strong effects of the deletion of HSF activation domains on chromatin remodeling events at the HSP12 promoter (14), we conclude that HSF and Msn2/4 cooperate in promoting nucleosome displacement at this promoter. However, it is not clear which activator plays the primary role in chromatin remodeling events.
FIG. 4.
MSN2/4 are critical for chromatin remodeling at the HSP12 promoter. ChIPs using antibody against the C terminus of histone H3 were performed with the ΔMSN2/4 or isogenic WT strain. The analyzed promoter is indicated in the upper left corner of each panel. The axis designation is the same as in Fig. 2.
Comparison of the results of HSF ChIPs for the WT and ΔMSN2 ΔMSN4 strains reveals that the MSN2 and MSN4 double deletion eliminates HSF binding to the HSP12 promoter (Fig. 5), while it has no significant effect on the binding of HSF to the HSP82 and SSA4 promoters. This is consistent with the critical role Msn2/4 play in regulation of HSP12 expression (3, 7). Our results for HSF ChIPs lead to the conclusion that HSF plays a primary role in chromatin remodeling at the HSP82 and SSA4 promoters while the Msn2/4 activators are the primary factors facilitating HSF binding at the HSP12 promoter.
FIG. 5.
MSN2/4 deletion prevents HSF from binding to the HSP12 promoter. ChIPs using antibody against HSF performed in the ΔMSN2/4 or isogenic WT strain. The promoter analyzed is indicated in the upper left corner of each panel. The axis designation is the same as in Fig. 3B.
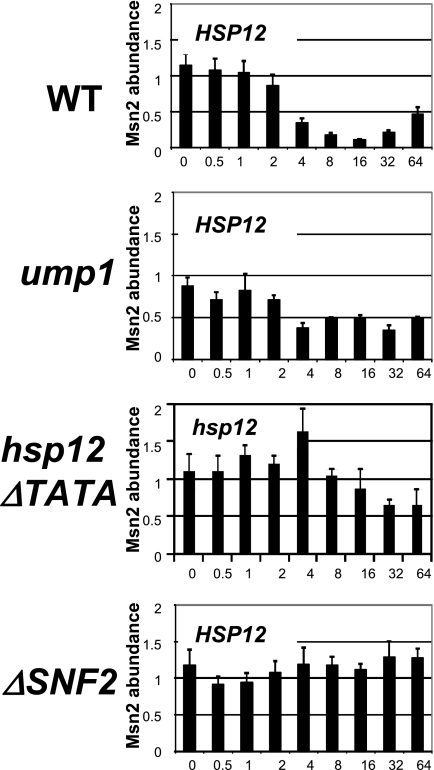
To investigate if Msn2/4 action at the HSP12 promoter is SWI/SNF dependent, we performed ChIPs using anti-Msn2 antibody in the heat shock time course format using WT and ΔSNF2 strains (Fig. 6). Unexpectedly, we found that in the WT strain, the Msn2 level at the HSP12 promoter declined below the background level starting at the fourth minute of heat shock, reached a minimal level at 16 min, and recovered to some degree during the attenuation phase at 32 and 64 min. We were profoundly perplexed by these results and repeated the experiments with the WT strain, using antibody raised against a different epitope of Msn2 or against Msn4. In another modification, a yeast strain with TAP-tagged Msn2 and anti-TAP-tag antibody (see Materials and Methods) were utilized for ChIPs. Consistently in each case, we obtained results (data not shown) similar to those shown in Fig. 6, namely, that the level of Msn2 at the HSP12 promoter declined during heat shock. Changing normalization to the signal from the chromosome V intergenic region (see Materials and Methods) or to the signal from the PHO5 coding region, neither of which has any accidental Msn2/4 STRE sequence, also did not change the outcome. Our conclusion is that the level of Msn2/4 at the HSP12 promoter indeed decreases during heat shock. The Msn2/4 factors are known to be proteolytically degraded in the nucleus in a heat shock-dependent manner (6, 37, 38). Moreover, this degradation seems to occur specifically in the nucleus upon binding to the DNA and is dependent on Srb10 (38) and Gal11 (38), which both are components of the transcription initiation complex. These data might explain our results and will be considered in more detail in the Discussion.
FIG. 6.
Msn2 abundance at the HSP12 promoter during heat shock depends on Snf2, proteasome assembly component Ump1, and intactness of core promoter elements. ChIPs using antibody against yeast Msn2 were performed with the WT, ump1, hsp12ΔTATA, or DSNF2 strain. y axis, Msn2 abundance relative to the PHO5 promoter. x axis, time after heat shock (0 to 64 min).
Msn2 loss at the HSP12 promoter depends on transcription initiation complex assembly.
To test if Msn2 proteolysis is involved in regulation of the HSP12 promoter, we performed the Msn2 ChIPs with the ump1 mutant strain isolated previously by the D. S. Gross group during the genetic screen for mutations affecting expression of HSP genes. Ump1 is a proteasome assembly component, and its inactivation leads to a deficiency in proteasome function (49). Our data (Fig. 6) indicate that mutation of ump1 diminishes the decline in the Msn2 abundance during the time course of heat shock in comparison to results for the WT strain. These data suggest that indeed Msn2 proteolysis might be involved in HSP12 promoter regulation.
To address the possibility of association of Msn2 loss at the HSP12 promoter with transcription initiation, we constructed a strain where the entire HSP12 core promoter, including the TATA box but not the upstream activation sequence (UAS) encompassing the Msn2/4 binding sites, was replaced with the KanMX cassette, thus significantly decreasing the possibility of transcription initiation complex assembly. The results of Msn2 ChIPs with this strain (Fig. 6, hsp12 ΔTATA) indicate that indeed the effect of Msn2 loss was significantly abolished. Moreover, during the fourth minute of heat shock, which coincides with the beginning of chromatin remodeling and Pol II recruitment to the HSP12 promoter (Fig. 2 and 3A), we observed minor increase in Msn2 abundance. These results suggest an association of the Msn2 loss at the HSP12 promoter with transcription initiation. The minor decline of the Msn2 abundance after 32 and 64 min of heat shock we attribute to the possibility of promiscuous transcription mediated by the degenerate TATA box sequences in proximity to the Msn2 UAS. The possibility and the relative effectiveness of such promiscuous transcription initiation were demonstrated recently in experiments, when yeast Gal4 UAS was artificially distanced from the core promoter elements (13).
Since we observed strong SNF2 dependence of HSP12 promoter regulation (Fig. 2, 3, and 4), we wanted to test if Msn2 loss, at least at this promoter, is SWI/SNF dependent. Importantly, the SNF2 deletion eliminated the heat shock-dependent loss of Msn2 at the HSP12 promoter (Fig. 6). This experiment suggests that similar to HSF (Fig. 3B), Msn2/4 might require prior action of the SWI/SNF complex.
The rates of reoccupation of HSP promoters with nucleosomes during recovery from heat shock are the same for all three promoters and are delayed in the ΔSNF2 strain.
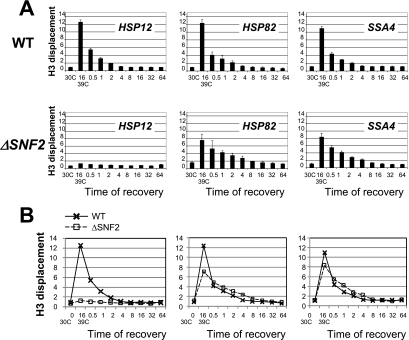
The results described above all show clear gene-specific characteristics. This can be understood when it is considered that the SWI/SNF chromatin remodeling complex is recruited to promoters by different combinations of promoter-specific cis- and trans-acting factors. Therefore, we next wanted to test if the kinetics of recovery from heat shock for the analyzed promoters also have gene-specific characteristics. If SWI/SNF is recruited by different factors or combinations of those factors, then the retention of the SWI/SNF complex during heat shock and release during the return to normal temperature may also be promoter specific. Surprisingly, all three promoters in the WT strain exhibited similar profiles associated with histone return during recovery from heat shock (Fig. 7). Notably, the return of nucleosomes at all three promoters started immediately after the shift of the temperature to normal and was completed in 4 to 8 min, which is similar to or even greater than the rate of histone displacement during heat shock. The rapid and equivalent rate of histone return at all three promoters suggests that the return of nucleosomes is not directed by promoter elements and trans-acting factors but rather is determined by the general translocation rate of nucleosomes.
FIG. 7.
The rate of reoccupation of HSP promoters with nucleosomes during recovery from heat shock is the same for all three promoters and is delayed in the ΔSNF2 strain. (A) ChIPs using antibody against the C terminus of histone H3 were performed with the ΔSNF2 or isogenic WT strain. The promoter analyzed is indicated in each panel. y axis, n-fold displacement of histone H3. For the x axis, the first value is for the initial 30°C culture, the second value is for the culture incubated for 16 min at 39°C, and the following time points (0.5 to 64 min) are after a downshift of the temperature from 39°C to 30°C. All real-time PCR values were normalized relative to the input and to the signal from the PHO5 promoter, similar to results in Fig. 2. (B) Superimposure of data from panel A for WT and ΔSNF2 strains is shown.
Analysis of the ΔSNF2 strain revealed more promoter-specific effects. The HSP12 promoter showed no chromatin changes during the time course of heat shock, consistent with strong dependence of chromatin remodeling at this promoter on SWI/SNF (Fig. 2). The HSP82 and SSA4 promoters exhibit return rates similar to each other, with a delay in nucleosome return. This delay is evident considering the significantly lower change in histone H3 displacement values in the first 30 s of recovery kinetics for the ΔSNF2 strain than for the WT strain (Fig. 7B) and persistently higher H3 displacement values for the ΔSNF2 strain than for the WT. These data indicate a delay of nucleosome return in the ΔSNF2 strain and might be a result of direct or indirect action of the SWI/SNF complex, which will be discussed below.
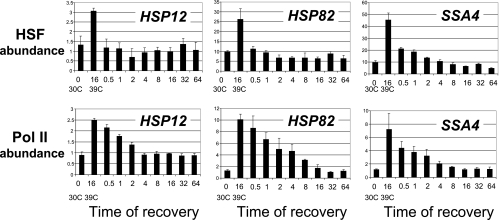
We extended further the analysis of recovery kinetics and asked whether there is any correlation between retention of activators and RNA polymerase occupation of the promoters. Figure 8 indicates that there was a fast drop in the occupation levels of HSF at all three promoters in the first seconds after the temperature downshift. In fact, for the HSP12 and HSP82 promoters, the non-heat-shock level of HSF abundance was restored in the first seconds. The comparison of these recovery kinetics with the Pol II abundance levels indicates that Pol II was delayed in its restoration to the non-heat-shock level, correlating more with the kinetics of histone reoccupation of promoters (Fig. 7A, WT).
FIG. 8.
HSF leaves promoters faster than Pol II. The heat shock recovery experiments were done essentially as described for Fig. 7. ChIPs using antibody against HSF and Pol II were performed with the WT strain.
DISCUSSION
In this study we have investigated three highly inducible heat shock gene promoters which all display an extremely rapid and significant loss of histones upon heat induction. All three promoters exhibit a similar degree of SWI/SNF complex recruitment during heat shock in the WT strain (Fig. 1). Despite these similarities, there is a notable difference in the effect of inactivation of SNF2, the major enzymatic subunit of this chromatin remodeling complex. While SNF2 inactivation causes a minor delay and a decrease of intensity in chromatin remodeling for the HSP82 and SSA4 promoters, the effect on the HSP12 promoter is the elimination of chromatin remodeling and Pol II recruitment. To better understand this phenomenon, we need to consider the composition of the cis-acting elements in these promoters and the actions of regulatory factors involved.
Interaction of HSF with perfect and mismatched HSEs in context of chromatin.
All three promoters are regulated to some degree by HSF, which binds to HSE sequences in the promoter. A typical HSE contains three or more sequential inverted repeats of the sequence nGAAn. This consensus sequence is rarely preserved in natural promoters. Currently HSEs are separated into three groups: perfect, gapped, and stepped. A perfect HSE has all three inverted repeats in a contiguous array (nTTCnnGAAnnTTC) (2, 48, 62, 63). Gapped HSEs have two consecutive inverted sequences, with the third sequence separated by 5 bp (50). Stepped HSEs have 5-bp gaps separating all three modules (64). The HSP12 promoter has a stepped HSE with one mismatched nucleotide, while the HSP82 and SSA4 promoters both have a perfect match to the gapped HSE consensus and additional adjacent HSE sequences of gapped and/or stepped nature with some mismatches (5, 18). In the HSP82 promoter, this mismatched region has the designation HSE2/3 (17, 18), while it has no designation in SSA4. The HSEs with deviations from the consensus are known to be bound by HSF in an inducible manner (17, 23, 29) and require the cooperative action of other activators (3; for a recent review, see reference 58), while perfect and stepped HSEs without mismatches are usually constitutively occupied in yeast by HSF (26, 33, 53). These observations are consistent with our results regarding the HSF occupancy of these promoters (Fig. 6). Normalized to the background signal from the PHO5 promoter in the WT strain, HSF does not exhibit any occupation of the HSP12 promoter before heat shock, while both the HSP82 and SSA4 promoters show a seven- to eightfold enrichment (14; this report) (Fig. 3B, 0-min time point). Upon heat shock, all three promoters show a gradual increase of HSF occupation, consistent with the inducible mode of HSF binding to the mismatched HSE sequences present in all three promoters. These data are also consistent with genomic footprinting results, which indicate that the HSE2/3 region of the HSP82 promoter is bound by HSF in an inducible manner (17, 23).
Since HSF cannot bind to any HSE sequence if DNA is incorporated into a nucleosome (56; our unpublished data) and there is no histone displacement at the HSP12 promoter in the ΔSNF2 strain, it seems plausible to suggest that HSF requires preliminary action of the SWI/SNF complex. Such a requirement has recently been demonstrated at the PHO5 promoter during conditions of lower activator nuclear localization and higher nucleosome occupancy (12). It is also possible that SWI/SNF might initially be recruited by Msn2/4 activators. This assumption is supported by our current results showing that there is no HSF binding and no chromatin remodeling at the HSP12 promoter in the ΔMsn2/4 strain.
Function of Msn2/4.
To elucidate the role of the Msn2/4 activators in chromatin remodeling at the HSP12 promoter, we need to consider the anomalous behavior of Msn2/4 at the HSP12 promoter, exhibited by the decline in Msn2 abundance during heat shock (Fig. 6). The Msn2/4 activators are known to interact with their target sequences in promoters in an inducible manner, translocating from the cytoplasm to the nucleus in a matter of minutes (24, 25, 32). There have also been several reports indicating a tight association between Msn2 degradation and transcriptional induction caused by this activator. This association fits well with the “black widow” model of transcription activation, whereby, to prevent unnecessary continuation of transcription, the transcription activator is destroyed specifically at the promoter right after initiation of transcription (10, 55). The degradation rate of the Msn2 activator is significantly increased upon heat shock. Moreover, it has been demonstrated that heat shock-induced Msn2 degradation occurs in the nucleus, depends on DNA binding (38), and is influenced by Gal11 (37) and Srb10 (6, 9, 38), both of which are components of Mediator complex. Considering that heat shock causes a significant increase in the total Msn2 degradation and considering indications that this degradation is regulated by components of the transcription initiation complex, it is logical to assume that the Msn2 degradation rate at cognate promoters is significantly higher than the total Msn2 degradation rate. Targeting Msn2 degradation to the cognate promoters might be the reason of the heat shock-induced decline in Msn2 abundance at the HSP12 promoter observed in this study (Fig. 6 WT). Importantly, this effect is eliminated by the SNF2 deletion, which also abolishes Pol II recruitment at the HSP12 promoter (Fig. 3A) and presumably transcription initiation complex assembly. The involvement of Msn2/4 degradation in regulation of the HSP12 promoter is also supported by our data indicating that the decline of Msn2 abundance is significantly diminished in the strain bearing the ump1 mutation, which impairs function of the proteasome. The association of Msn2 loss at the HSP12 promoter with transcription initiation complex assembly is farther supported by our experiments utilizing a strain in which HSP12 core promoter elements are eliminated by replacing them with the KanMX cassette. In this situation (Fig. 6, hsp12ΔTATA), Msn2 loss during heat shock is significantly reduced. The remaining loss of Msn2 might be associated with promiscuous transcription, which, as demonstrated recently (13), is relatively efficiently initiated near UAS regions even in the absence of a canonical TATA box and other promoter elements.
In addition to suggesting a connection between Msn2 transcription activation and Msn2 stability, our data indicate that similar to the case with HSF, the Msn2/4-dependent activation at the HSP12 promoter is SWI/SNF dependent. Moreover, considering that the level of Msn2 abundance at the HSP12 promoter before heat shock is indistinguishable from the background level measured at the PHO5 promoter or the intergenic region of chromosome V, our data suggest that there is very little if any Msn2/4 present at the HSP12 promoter before heat shock. This is consistent with previously published data indicating that the Msn2/4 activators interact with their target sequences in promoters in an inducible manner, translocating from the cytoplasm to the nucleus in a matter of minutes (24, 25, 32). There have been no reports regarding the ability of Msn2/4 activators to bind to nucleosomal DNA, but our data (Fig. 6) suggest that Msn2/4 activators are highly SWI/SNF dependent.
Function of SWI/SNF complex.
In our attempt to understand the differential requirement of the SWI/SNF complex for chromatin remodeling between the HSP12 promoter on one hand and the HSP82 and SSA4 promoters on the other, we consider two possibilities. First, it is possible that there is a difference in the abilities to recruit the SWI/SNF complex of two different activators: Msn2/4, which plays a dominant role in HSP12 regulation, and HSF, which plays a dominant role in HSP82 and SSA4 regulation. The second possibility is that the SWI/SNF complex paves the way for both activators at all three promoters, but at the HSP82 and SSA4 promoters HSF is already prebound and the SWI/SNF action is not as critical.
Our preliminary data argue against the first explanation, because the inactivation of the SWI/SNF complex abolishes the Msn2/4 loss at the HSP12 promoter (Fig. 6) and because Snf2 is recruited to all three promoters equally during heat shock (Fig. 1) despite differences in HSF or Msn2/4 domination. We favor the alternative explanation, which considers a possibility of SWI/SNF affecting directly or indirectly the general translocation rate of nucleosomes at least in the context of the heat shock promoters studied here. In this consideration, the dynamic translocation of nucleosomes may create a window of opportunity for activators to bind to the cognate cis-acting promoter element sequences. At the HSP82 and SSA4 promoters, prebound HSF facilitates additional binding to the mismatched HSE consensus regions, while at the HSP12 promoter, there is no such preexisting platform and SWI/SNF action is more important. Although we favor this explanation, formally we cannot exclude the possibility of direct recruitment of SWI/SNF complex by Msn2/4 activators, because the presence of the Msn2 activator at the HSP12 promoter is very transient. This dynamic situation makes it difficult to establish the sequence of actions between Msn2/4 and the SWI/SNF complex.
The assumption regarding the possibility of the SWI/SNF complex affecting the general mobility of the nucleosomes mentioned above is supported by our recovery experiments. While histone displacement is clearly directed by the cooperative action of activators and coactivators, the return of nucleosomes is likely a result of general nucleosome dynamics, since there are no factors identified that specifically attract nucleosomes back to the naked DNA promoter regions. A novel implication of our results is the indication that the rate of nucleosome return is equal to or higher than the rate of nucleosome displacement, at least at the three analyzed heat shock gene promoters. Also, we have demonstrated that the histone displacement at the HSP82 and SSA4 promoters in the ΔSNF2 strain (Fig. 2) is delayed to the same extent as the return during the recovery (Fig. 7). We have been unable to make a similar conclusion regarding the HSP12 promoter, simply because chromatin remodeling is practically eliminated at this promoter when SNF2 is deleted.
The relatively small but reproducible effect of SNF2 deletion on the return rate of nucleosomes at the HSP82 and SSA4 promoters can be explained by considering at least two possibilities. First, there are a significant number of ATP-dependent chromatin remodeling complexes operating in the cell, and their activities often overlap. These protein complexes are divided (20) into families by homology of their protein subunits: the SWI/SNF family (SWI/SNF and RSC), the ISWI family (ISWI1 and ISWI2), the CHD family (Chd1), and the INO80 family (INO80 and SWR1). This redundancy is important, considering the vital necessity of chromatin remodeling for proper gene expression, and may be behind the weak effect of the SNF2 deletion that we observe for the HSP82 and SSA4 promoters. A second possibility is that nucleosome reassembly is a multistep process which may involve ATP-dependent chromatin changes, histone modifications, histone chaperone-assisted assembly/disassembly steps and others, and the impairment of SWI/SNF complex function affects a step which is not rate limiting. The redundancy and multistep nature of nucleosome dynamics become more obvious considering recent publications indicating functional interactions of the SWI/SNF and ISW1 complexes in expression of GAL genes (35) and the important function that ISW1 activity plays in repression of stress-induced genes, including HSP12 (41).
Another interesting observation from the recovery experiments is that the dynamics of Pol II retention at the HSP82 and SSA4 promoters during the recovery after the abrupt downshift of the temperature to normal (Fig. 8) is correlated with the dynamics of the histone reoccupation and not with the change in HSF abundance. In fact, HSF leaves promoters faster than Pol II, while profiles of histone reoccupation and Pol II departure parallel each other (Fig. 7 and 8). That observation signifies a greater importance of a nucleosome-free environment for Pol II than a direct role of recruitment by the activator. This situation is similar to the one described recently for the PHO5 promoter in the absence of the Spt6 histone chaperone, when restoration of promoter chromatin structure and shutdown of the promoter but not the departure of the Pho4 activator were abolished (1).
Acknowledgments
We thank David Gross for discussion of unpublished data, Jocelyn Krebs, Robin Miskimins, and Robert Noiva for critical reading of the manuscript and helpful suggestions, and Fred Winston, Jessica Tyler, Kevin Morano, and David Gross for plasmids and yeast strains.
This work was supported by grants awarded to A.M.E. from NSF (MCB-0352042) and from NIH (P20 RR016479) from the INBRE Program of the National Center for Research Resources.
Footnotes
Published ahead of print on 10 December 2007.
REFERENCES
- 1.Adkins, M. W., and J. K. Tyler. 2006. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell 21405-416. [DOI] [PubMed] [Google Scholar]
- 2.Amin, J., J. Ananthan, and R. Voellmy. 1988. Key features of heat shock regulatory elements. Mol. Cell. Biol. 83761-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amoros, M., and F. Estruch. 2001. Hsf1p and Msn2/4p cooperate in the expression of Saccharomyces cerevisiae genes HSP26 and HSP104 in a gene- and stress type-dependent manner. Mol. Microbiol. 391523-1532. [DOI] [PubMed] [Google Scholar]
- 4.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71247-273. [DOI] [PubMed] [Google Scholar]
- 5.Boorstein, W. R., and E. A. Craig. 1990. Structure and regulation of the SSA4 HSP70 gene of Saccharomyces cerevisiae. J. Biol. Chem. 26518912-18921. [PubMed] [Google Scholar]
- 6.Bose, S., J. A. Dutko, and R. S. Zitomer. 2005. Genetic factors that regulate the attenuation of the general stress response of yeast. Genetics 1691215-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boy-Marcotte, E., G. Lagniel, M. Perrot, F. Bussereau, A. Boudsocq, M. Jacquet, and J. Labarre. 1999. The heat shock response in yeast: differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol. Microbiol. 33274-283. [DOI] [PubMed] [Google Scholar]
- 8.Cairns, B. R., Y. J. Kim, M. H. Sayre, B. C. Laurent, and R. D. Kornberg. 1994. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl. Acad. Sci. USA 911950-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi, Y., M. J. Huddleston, X. Zhang, R. A. Young, R. S. Annan, S. A. Carr, and R. J. Deshaies. 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 151078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, G. A., and W. P. Tansey. 2006. The proteasome: a utility tool for transcription? Curr. Opin. Genet. Dev. 16197-202. [DOI] [PubMed] [Google Scholar]
- 11.Cote, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 26553-60. [DOI] [PubMed] [Google Scholar]
- 12.Dhasarathy, A., and M. P. Kladde. 2005. Promoter occupancy is a major determinant of chromatin remodeling enzyme requirements. Mol. Cell. Biol. 252698-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobi, K. C., and F. Winston. 2007. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol. Cell. Biol. 275575-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erkina, T. Y., and A. M. Erkine. 2006. Displacement of histones at promoters of Saccharomyces cerevisiae heat shock genes is differentially associated with histone H3 acetylation. Mol. Cell. Biol. 267587-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erkine, A. M., C. C. Adams, T. Diken, and D. S. Gross. 1996. Heat shock factor gains access to the yeast HSC82 promoter independently of other sequence-specific factors and antagonizes nucleosomal repression of basal and induced transcription. Mol. Cell. Biol. 167004-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erkine, A. M., and D. S. Gross. 2003. Dynamic chromatin alterations triggered by natural and synthetic activation domains. J. Biol. Chem. 2787755-7764. [DOI] [PubMed] [Google Scholar]
- 17.Erkine, A. M., S. F. Magrogan, E. A. Sekinger, and D. S. Gross. 1999. Cooperative binding of heat shock factor to the yeast HSP82 promoter in vivo and in vitro. Mol. Cell. Biol. 191627-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erkine, A. M., C. Szent-Gyorgyi, S. F. Simmons, and D. S. Gross. 1995. The upstream sequences of the HSP82 and HSC82 genes of Saccharomyces cerevisiae: regulatory elements and nucleosome positioning motifs. Yeast 11573-580. [DOI] [PubMed] [Google Scholar]
- 19.Estruch, F. 2000. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24469-486. [DOI] [PubMed] [Google Scholar]
- 20.Gangaraju, V. K., and B. Bartholomew. 2007. Mechanisms of ATP dependent chromatin remodeling. Mutat. Res. 6183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garreau, H., R. N. Hasan, G. Renault, F. Estruch, E. Boy-Marcotte, and M. Jacquet. 2000. Hyperphosphorylation of Msn2p and Msn4p in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology 1462113-2120. [DOI] [PubMed] [Google Scholar]
- 22.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 114241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giardina, C., and J. T. Lis. 1995. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol. Cell. Biol. 152737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schuller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorner, W., E. Durchschlag, J. Wolf, E. L. Brown, G. Ammerer, H. Ruis, and C. Schuller. 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gross, D. S., K. E. English, K. W. Collins, and S. W. Lee. 1990. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J. Mol. Biol. 216611-631. [DOI] [PubMed] [Google Scholar]
- 27.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 242519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutierrez, J. L., M. Chandy, M. J. Carrozza, and J. L. Workman. 2007. Activation domains drive nucleosome eviction by SWI/SNF. EMBO J. 26730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahn, J. S., Z. Hu, D. J. Thiele, and V. R. Iyer. 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 245249-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hashikawa, N., and H. Sakurai. 2004. Phosphorylation of the yeast heat shock transcription factor is implicated in gene-specific activation dependent on the architecture of the heat shock element. Mol. Cell. Biol. 243648-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoj, A., and B. K. Jakobsen. 1994. A short element required for turning off heat shock transcription factor: evidence that phosphorylation enhances deactivation. EMBO J. 132617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacquet, M., G. Renault, S. Lallet, J. De Mey, and A. Goldbeter. 2003. Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. J. Cell Biol. 161497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakobsen, B. K., and H. R. Pelham. 1988. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol. Cell. Biol. 85040-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristjuhan, A., and J. Q. Svejstrup. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 234243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kundu, S., P. J. Horn, and C. L. Peterson. 2007. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 21997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon, H., A. N. Imbalzano, P. A. Khavari, R. E. Kingston, and M. R. Green. 1994. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370477-481. [DOI] [PubMed] [Google Scholar]
- 37.Lallet, S., H. Garreau, C. Garmendia-Torres, D. Szestakowska, E. Boy-Marcotte, S. Quevillon-Cheruel, and M. Jacquet. 2006. Role of Gal11, a component of the RNA polymerase II mediator in stress-induced hyperphosphorylation of Msn2 in Saccharomyces cerevisiae. Mol. Microbiol. 62438-452. [DOI] [PubMed] [Google Scholar]
- 38.Lallet, S., H. Garreau, C. Poisier, E. Boy-Marcotte, and M. Jacquet. 2004. Heat shock-induced degradation of Msn2p, a Saccharomyces cerevisiae transcription factor, occurs in the nucleus. Mol. Genet. Genomics 272353-362. [DOI] [PubMed] [Google Scholar]
- 39.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36900-905. [DOI] [PubMed] [Google Scholar]
- 40.Lee, K. K., P. Prochasson, L. Florens, S. K. Swanson, M. P. Washburn, and J. L. Workman. 2004. Proteomic analysis of chromatin-modifying complexes in Saccharomyces cerevisiae identifies novel subunits. Biochem. Soc. Trans. 32899-903. [DOI] [PubMed] [Google Scholar]
- 41.Lindstrom, K. C., J. C. Vary, Jr., M. R. Parthun, J. Delrow, and T. Tsukiyama. 2006. Isw1 functions in parallel with the NuA4 and Swr1 complexes in stress-induced gene repression. Mol. Cell. Biol. 266117-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13136-142. [DOI] [PubMed] [Google Scholar]
- 43.Martens, J. A., P. Y. Wu, and F. Winston. 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 192695-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez-Pastor, M. T., G. Marchler, C. Schuller, A. Marchler-Bauer, H. Ruis, and F. Estruch. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 152227-2235. [PMC free article] [PubMed] [Google Scholar]
- 45.Morimoto, R. I. 1998. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 123788-3796. [DOI] [PubMed] [Google Scholar]
- 46.Moskvina, E., C. Schuller, C. T. Maurer, W. H. Mager, and H. Ruis. 1998. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast 141041-1050. [DOI] [PubMed] [Google Scholar]
- 47.Owen-Hughes, T., and J. L. Workman. 1996. Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J. 154702-4712. [PMC free article] [PubMed] [Google Scholar]
- 48.Perisic, O., H. Xiao, and J. T. Lis. 1989. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell 59797-806. [DOI] [PubMed] [Google Scholar]
- 49.Ramos, P. C., J. Hockendorff, E. S. Johnson, A. Varshavsky, and R. J. Dohmen. 1998. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell 92489-499. [DOI] [PubMed] [Google Scholar]
- 50.Santoro, N., N. Johansson, and D. J. Thiele. 1998. Heat shock element architecture is an important determinant in the temperature and transactivation domain requirements for heat shock transcription factor. Mol. Cell. Biol. 186340-6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh, H., A. M. Erkine, S. B. Kremer, H. M. Duttweiler, D. A. Davis, J. Iqbal, R. R. Gross, and D. S. Gross. 2006. A functional module of yeast mediator that governs the dynamic range of heat-shock gene expression. Genetics 1722169-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, C. L., R. Horowitz-Scherer, J. F. Flanagan, C. L. Woodcock, and C. L. Peterson. 2003. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat. Struct. Biol 10141-145. [DOI] [PubMed] [Google Scholar]
- 53.Sorger, P. K., M. J. Lewis, and H. R. Pelham. 1987. Heat shock factor is regulated differently in yeast and HeLa cells. Nature 32981-84. [DOI] [PubMed] [Google Scholar]
- 54.Sorger, P. K., and H. R. Pelham. 1988. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54855-864. [DOI] [PubMed] [Google Scholar]
- 55.Tansey, W. P. 2001. Transcriptional activation: risky business. Genes Dev. 151045-1050. [DOI] [PubMed] [Google Scholar]
- 56.Taylor, I. C., J. L. Workman, T. J. Schuetz, and R. E. Kingston. 1991. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 51285-1298. [DOI] [PubMed] [Google Scholar]
- 57.Treger, J. M., A. P. Schmitt, J. R. Simon, and K. McEntee. 1998. Transcriptional factor mutations reveal regulatory complexities of heat shock and newly identified stress genes in Saccharomyces cerevisiae. J. Biol. Chem. 27326875-26879. [DOI] [PubMed] [Google Scholar]
- 58.Uffenbeck, S. R., and J. E. Krebs. 2006. The role of chromatin structure in regulating stress-induced transcription in Saccharomyces cerevisiae. Biochem. Cell Biol. 84477-489. [DOI] [PubMed] [Google Scholar]
- 59.Utley, R. T., J. Cote, T. Owen-Hughes, and J. L. Workman. 1997. SWI/SNF stimulates the formation of disparate activator-nucleosome complexes but is partially redundant with cooperative binding. J. Biol. Chem. 27212642-12649. [DOI] [PubMed] [Google Scholar]
- 60.Voellmy, R. 2004. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones 9122-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu, C. 1995. Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 11441-469. [DOI] [PubMed] [Google Scholar]
- 62.Xiao, H., and J. T. Lis. 1988. Germline transformation used to define key features of heat-shock response elements. Science 2391139-1142. [DOI] [PubMed] [Google Scholar]
- 63.Xiao, H., O. Perisic, and J. T. Lis. 1991. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell 64585-593. [DOI] [PubMed] [Google Scholar]
- 64.Yamamoto, A., Y. Mizukami, and H. Sakurai. 2005. Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 28011911-11919. [DOI] [PubMed] [Google Scholar]
- 65.Zhao, J., J. Herrera-Diaz, and D. S. Gross. 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 258985-8999. [DOI] [PMC free article] [PubMed] [Google Scholar]