Abstract
The proteasome homeostasis in Saccharomyces cerevisiae is regulated by a negative feedback circuit in which the Rpn4 transcription factor upregulates the proteasome genes and is rapidly degraded by the proteasome. Previous work has identified Ubr2 and Rad6 as the cognate E3 and E2 enzymes for Rpn4 ubiquitylation. However, our recent attempts to ubiquitylate Rpn4 using purified Ubr2 and Rad6 proteins in a reconstitution system have been unsuccessful, suggesting that an additional factor is required for Rpn4 ubiquitylation. Here, we screened the entire collection of the single-gene-deletion yeast mutants generated by the Saccharomyces Genome Deletion Project and identified the mub1Δ mutant defective in ubiquitin-dependent degradation of Rpn4. An in vitro reconstitution ubiquitylation assay confirms that Mub1 is the missing factor for Rpn4 ubiquitylation. We further show that Mub1 directly interacts with Ubr2 and Rpn4. The MYND domain of Mub1 may play an important role in Rpn4 ubiquitylation. Interestingly, Mub1 itself is a short-lived protein and its degradation is dependent on the Ubr2/Rad6 ubiquitin ligase. Together, these data suggest that Mub1 and Ubr2 cooperate to transfer ubiquitin to Rpn4 from Rad6 and that Mub1 may switch from a partner to a substrate of the Ubr2/Rad6 ubiquitin ligase.
The ubiquitin (Ub) system selects abnormal and regulatory proteins for degradation by the proteasome (13, 38, 46). Protein ubiquitylation is a multistep process involving three enzymes. Ub is first activated by the Ub-activating enzyme E1, forming an E1∼Ub thioester. The Ub moiety of the E1∼Ub thioester is thereafter transferred to a Ub-conjugating enzyme, E2, forming an E2∼Ub thioester. With the participation of an E3 enzyme, the Ub moiety of the E2∼Ub thioester is transferred to the target substrate. An E2/E3 complex is also referred to as a Ub ligase. Most E3s are grouped into either the HECT or RING E3 family, based on their catalytic modules and features of sequence and structure. A HECT E3 can accept the Ub moiety from an associated E2∼Ub thioester, forming an E3∼Ub thioester intermediate and acting as a proximal Ub donor to the selected substrate. By contrast, the formation of a thioester between RING E3s and Ub has not been detected. The mechanism by which RING E3s catalyze Ub transfer from the E2∼Ub thioester to the substrate remains speculative. The current hypothesis is that RING E3s act as adaptors to optimize the orientation of the bound substrate to the E2∼Ub thioester (14, 35, 43, 49, 52, 59). The specificity of protein ubiquitylation is mainly controlled by the degradation signal of the substrate and the activity of the cognate Ub ligase. E3-substrate interaction is often modulated through covalent modification of the substrate's degradation signal. The known modifications include phosphorylation, acetylation, hydroxylation, and glycosylation (17, 23, 26, 37, 45, 58).
The Saccharomyces cerevisiae RPN4 gene (also named SON1 and UFD5) was originally isolated as a suppressor of mutant gene sec63-101, a temperature-sensitive mutant of SEC63 which encodes an essential component of the endoplasmic reticulum translocation channel (7, 8, 18, 32). Recent studies demonstrated that the RPN4-encoded protein, Rpn4, is a transcription factor that activates the proteasome genes (16, 29, 53). Interestingly, Rpn4 is an extremely short-lived protein (half-life, ≤2 min) and is degraded by the proteasome (53). Moreover, stabilization of Rpn4 due to inhibition of the proteasome activity results in an upregulation of the proteasome genes (22, 27). Together, these observations led to the formation of a negative-feedback model for the regulation of proteasome homeostasis in S. cerevisiae (22). On the one hand, Rpn4 upregulates proteasome expression; on the other hand, Rpn4 is rapidly destroyed by the proteasome. It has recently become clear that the Rpn4-proteasome negative-feedback circuit plays an important role in a wide range of cellular processes (3, 11, 22, 27, 33, 36, 39, 40, 57). Interestingly, a similar negative-feedback mechanism also exists in higher eukaryotes, including humans, even though the functional homolog of Rpn4 has not yet been identified (28, 31, 51, 56).
Rpn4 degradation is the key element of the Rpn4-proteasome negative-feedback circuit. Our recent work has shown that the proteasomal degradation of Rpn4 can be mediated by two distinct pathways (19). One is Ub dependent, whereas the other is Ub independent. While the Ub-independent degradation pathway remains largely unclear, progress has been made in understanding the Ub-dependent degradation of Rpn4. Ubr2 and Rad6 have been identified as the cognate E3 and E2 enzymes for Rpn4 ubiquitylation (48). It has been shown that Rpn4 carries six different ubiquitylation sites, of which K187 is the preferred one (20). The degradation signal of Rpn4 has also been mapped to the N-terminal acidic domain, including amino acids 211 to 229 (20). We have further demonstrated that the degradation signal of Rpn4 is modulated by phosphorylation of Ser 220 (21). In spite of this progress, our recent attempts to ubiquitylate Rpn4 using purified Ubr2 and Rad6 proteins in a reconstitution system have been unsuccessful. This suggests that a factor other than Ubr2 and Rad6 is required for Rpn4 ubiquitylation.
In this study, we screened the entire collection of the single-gene-deletion yeast mutants generated by the Saccharomyces Genome Deletion Project (50) and isolated the mub1Δ mutant that is defective in Ub-dependent degradation of Rpn4. An in vitro reconstitution ubiquitylation assay confirmed that Mub1, a MYND domain-containing protein, is the sought factor essential for Rpn4 ubiquitylation. We further showed that Mub1 directly interacts with Ubr2 and Rpn4. Whereas the MYND domain is largely dispensable for the binding of Mub1 to Ubr2 and Rpn4, the deletion of this domain virtually abolishes the activity of Mub1 in supporting the Ub-dependent degradation of Rpn4. Interestingly, Mub1 itself is a short-lived protein and its degradation is dependent on the Ubr2/Rad6 Ub ligase. Together, these data suggest a model in which Mub1 cooperates with Ubr2 to ubiquitylate Rpn4, and in the meantime, Mub1 may switch from a partner to a substrate of the Ubr2/Rad6 Ub ligase.
MATERIALS AND METHODS
Strains, plasmids, and yeast manipulation.
The yeast strains used were JD52 (MATa his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52), JD53 (MATα his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52), YXY36 (a ufd4Δ::LEU2 derivative of JD53), YXY78 (an rad6Δ::URA3 derivative of JD52), YXY274 (a ubr2Δ::HIS3 derivative of JD52), YXY346 (an mub1Δ::URA3 derivative of JD52), YXY352 (an mub1Δ::URA3 ubr2Δ::HIS3 double mutant derived from JD52), Y791 (MATa cim5-1 ura3-52 his3-Δ200 leu2Δ1), BY4741 (MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0), and the collection of single-gene-deletion strains (50). Manipulation of the single-gene-deletion mutants was done according to the protocols provided by the Consortium of Deletion Yeast Strains (see the Yeast Deletion Project website, http://www-sequence.stanford.edu/group/yeast_deletion_project). Escherichia coli strain BL21(DE3) was used to express glutathione S-transferase (GST) fusion proteins. The details of plasmid construction are available upon request.
β-Gal assay.
The enzymatic activity of β-galactosidase (β-gal) in liquid yeast culture was determined as described in reference 1, using the chromogenic substrate o-nitrophenyl-β-d-galactopyranoside (ONPG). For induction of the CUP1 promoter, CuSO4 was added to a final concentration of 0.1 mM.
Pulse-chase and immunoprecipitation analysis.
S. cerevisiae cells from 10-ml cultures (optical density at 600 nm of 0.8 to 1.0) in synthetic dextrose medium containing 0.1 mM CuSO4 or in galactose medium supplemented with essential amino acids were harvested. The cells were resuspended in 0.3 ml of the same medium supplemented with 0.15 mCi of [35S]methionine-cysteine (EXPRESS [35S] protein labeling mix; Perkin-Elmer) and incubated at 30°C for 5 min. The cells were then pelleted and resuspended in the same medium with cycloheximide (0.2 mg/ml) and excessive cold l-methionine-l-cysteine (2 mg/ml l-methionine and 0.4 mg/ml l-cysteine) and chased at 30°C. An equal volume of the sample was withdrawn at each time point. Labeled cells were harvested and lysed in equal volumes of 2× sodium dodecyl sulfate (SDS) buffer (2% SDS, 30 mM dithiothreitol, 90 mM Na-HEPES, pH 7.5) by incubation at 100°C for 3 min. The supernatants were diluted 20-fold with buffer A (1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 50 mM Na-HEPES, pH 7.5) before being applied to immunoprecipitation with antihemagglutinin (anti-ha) antibody (Sigma), anti-FLAG antibody (Sigma), or anti-β-Gal antibody (Promega) combined with protein A agarose (Calbiochem). The volumes of supernatants used in immunoprecipitation were adjusted to equalize the amounts of 10% trichloroacetic acid-insoluble 35S. The immunoprecipitates were washed three times with buffer A and resolved by SDS-polyacrylamide gel electrophoresis (PAGE), followed by autoradiography and quantitation with a PhosphorImager (Molecular Dynamics).
GST pulldown-immunoblotting assay.
The ubr2Δ mub1Δ cells expressing N-terminally FLAG-tagged Ubr2 (FUbr2) or N-terminally myc-tagged Mub1 (mMub1) from the GAL1 promoter in a high-copy-number vector were grown to an optical density at 600 nm of 1.8 in synthetic selective medium containing 2% galactose. The cells were spun down and manually ground to a fine powder with a pestle. Cell extracts were prepared by incubation of the powder in buffer B (150 mM NaCl, 50 mM HEPES, pH 7.5, 0.2% Triton X-100) plus protease inhibitor mix (Roche Diagnostics). For each pulldown, approximately 100 μg of yeast extract or 0.1 μg of purified protein was incubated with glutathione-agarose beads preloaded with ∼1 μg GST fusion protein at 4°C for 2 h. The beads were then washed three times with buffer B, and the retained proteins were separated by SDS-PAGE, followed by immunoblotting with anti-FLAG (Sigma) and antimyc (Covance) antibodies and detection with an Odyssey infrared imaging system, according to the manufacturer's instructions (Li-Cor Biosciences).
In vitro reconstitution ubiquitylation system.
Yeast extracts from ubr2Δ mub1Δ cells expressing FUbr2 or C-terminally Tev-FLAG-tagged Mub1 (Mub1TF) were prepared as described above. For the preparation of purified FUbr2 and Mub1TF proteins, supernatants were incubated with a one-fifth volume of anti-FLAG M2 affinity agarose (Sigma) at 4°C for 3 h. The beads were washed three times with buffer B and eluted with buffer B plus 150 μg/ml FLAG peptide at 4°C for 2 h. The eluted proteins were then passed through a YM-100 (for FUbr2) or YM-50 (for Mub1TF) column (Millipore) to remove the FLAG peptides, and the buffer was exchanged for 25 mM HEPES (pH 7.5). The preparation of purified Rad6 and Uba1 was previously described (48). Bovine Ub was purchased from BostonBiochem (Cambridge, MA). The phosphorylated GST-Rpn4172-229 (a GST fusion with residues 172-229 of Rpn4 which contain the Ub-dependent degradation signal of Rpn4) substrates were prepared as follows. GST-Rpn4172-229 proteins preloaded on glutathione-agarose were phosphorylated by recombinant casein kinase 2 (CK2) (New England BioLabs). After being washed with buffer B, the fusion proteins were eluted with buffer C (50 mM Tris Cl, pH 8.06, 150 mM NaCl, 20 mM glutathione). The eluted proteins were then passed through a YM-30 column to remove glutathione, and the buffer was exchanged for 25 mM HEPES (pH 7.5). A typical 5-μl ubiquitylation reaction mixture consisted of 100 nM FUbr2, 1 μM Rad6, 100 nM Uba1, 7.5 μM Ub, 2 mM ATP, 5 mM MgCl2, 0.5 μM substrate, and various amounts of Mub1TF in 25 mM HEPES, pH 7.5. The reactions were stopped by the addition of SDS loading buffer and resolved by SDS-PAGE, followed by immunoblotting analysis with anti-GST antibody (Santa Cruz).
RESULTS
A systematic screen for mutants defective in Ub-dependent degradation of Rpn4.
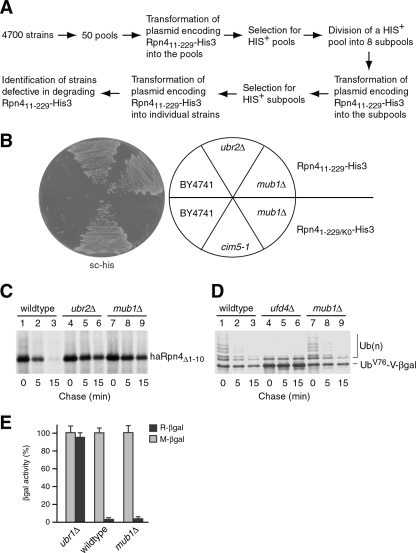
Taking advantage of the collection of ∼4,700 single-gene-deletion strains generated by the Saccharomyces Genome Deletion Project (50), we set up a genome-wide screen for genes involved in the Ub-dependent degradation of Rpn4. Different from previous screens of this yeast deletion library (30, 42), we designed an approach that enables us to screen the entire collection with a manageable number of yeast transformations (Fig. 1A). Specifically, the deletion mutants were divided into 50 pools, each containing ∼100 strains that have a similar growth rate. The 50 pools of mutants were transformed with a plasmid encoding Rpn411-229-His3, a His3 fusion protein bearing the Ub-dependent degradation signal of Rpn4 (20), and selected on histidine-lacking plates. Each of the pools that came up with HIS+ transformants was further divided into eight subpools and transformed with the Rpn411-229-His3 plasmid. Individual strains from the HIS+ subpools were then tested for defects in degrading Rpn411-229-His3. Note that the parental strain (BY4741) used to construct the deletion mutants has a his3Δ background.
FIG. 1.
Mub1 is essential for Ub-dependent degradation of Rpn4. (A) Schematic outline of the genome-wide search for yeast mutants defective in Ub-dependent degradation of Rpn4. Yeast cells transformed with the plasmid encoding Rpn411-229-His3 from the CUP1 promoter in a CEN-URA3-based vector were selected on plates (of synthetic complete medium lacking Ura and His) containing 10 μM CuSO4 and 5 mM 3-amino-1,2,4-triazole. (B) Wild-type (BY4741), ubr2Δ, mub1Δ, and cim5-1 cells expressing Rpn411-229-His3 or Rpn41-229/K0-His3 from the CUP1 promoter in a CEN-URA3-based vector were selected on a histidine-free plate as described for panel A. sc-his, synthetic complete medium lacking His. (C) Mub1 is required for Ub-dependent degradation of Rpn4. The metabolic stability of haRpn4Δ1-10 in wild-type, ubr2Δ, and mub1Δ strains was assessed by pulse-chase analysis. (D) Pulse-chase analysis was conducted to measure the degradation of UbV76-V-β-Gal in wild-type, ufd4Δ, and mub1Δ strains. Ub(n), polyubiquitylated species. (E) Deletion of MUB1 does not affect the N-end rule pathway. β-Gal enzymatic assay was performed to compare the turnover of R-β-Gal in wild-type, ubr1Δ, and mub1Δ strains. The stable substrate M-β-Gal served as a control. Error bars show standard deviations.
Four deletion strains were isolated from this screen, including ubr2Δ, ump1Δ, cvt9Δ, and mub1Δ mutants (Fig. 1B and data not shown). Ump1 is essential for proper maturation of the 20S proteasome and Ub-dependent protein degradation (41). The isolation of the ump1Δ and ubr2Δ strains essentially validated our screen. The cvt9Δ mutant was not further pursued because we found that this mutant contains a point mutation in the PRE2 gene, resulting in an Asp-to-Gly mutation at codon 243 of Pre2, a catalytic subunit of the 20S proteasome (12), and that deletion of CVT9 per se did not stabilize Rpn411-229-His3 (data not shown). In this study, we focused on the mub1Δ mutant. The MUB1 gene, encoding a 620-amino-acid protein of ∼72 kDa calculated molecular mass, was originally identified as an S. cerevisiae homolog of the samB gene of Aspergillus nidulans, deletion of which resulted in a multibudding phenotype (25). The biochemical function of Mub1, however, remains unexplored. Here, we studied the involvement of Mub1 in Rpn4 ubiquitylation.
Mub1 is required for Ub-dependent degradation of Rpn4.
To verify the outcome of the genetic screen, we used pulse-chase analysis to compare the Ub-dependent degradation of Rpn4 in the presence and absence of Mub1. Since BY4741 is an met strain, which is not suitable for pulse-chase analysis with [35S]methionine, we generated an mub1Δ strain (YXY346) by replacing MUB1 with the URA3 selection marker in the JD52 strain background. A plasmid expressing haRpn4Δ1-10 (an Rpn4 derivative with the N-terminal 10 residues replaced by an ha tag) from the CUP1 promoter in a low-copy-number vector was transformed in YXY346, JD52, and YXY274. Our early work has shown that this replacement substantially inhibits the Ub-independent degradation of Rpn4 but has no effect on the Ub-dependent degradation (19, 48). YXY274 is a congenic ubr2Δ strain of JD52. As shown in Fig. 1C, haRpn4Δ1-10 was stabilized in the ubr2Δ and mub1Δ mutants but degraded rapidly in JD52. Thus, Mub1 is essential for Ub-dependent degradation of Rpn4.
To test if deletion of MUB1 would have a general effect on protein degradation mediated by the Ub-proteasome system, we examined the degradation of two model substrates, UbV76-V-β-Gal and R-β-Gal, in the mub1Δ mutant. UbV76-V-β-Gal is a model substrate of the Ub fusion degradation pathway (18, 54), whereas R-β-Gal is a model substrate of the N-end rule pathway (2). Note that the E3 component of the N-end rule pathway, Ubr1, shares the E2 enzyme (Rad6) with Ubr2 (55). We found that both model substrates were degraded in mub1Δ cells as rapidly as in the wild-type cells but stabilized in the ufd4Δ and ubr1Δ E3 mutants (Fig. 1D and E). Consistent with these results, a cell survival assay showed that Rpn41-229/K0-His3, a substrate carrying the Ub-independent degradation signal of Rpn4 and lacking all Rpn4 ubiquitylation sites (19, 20), was efficiently degraded in mub1Δ but stabilized in the cim5-1 proteasome mutant (10) (Fig. 1B). Together, these results lead to the conclusion that Mub1 is specifically required for the Ub-dependent degradation of Rpn4.
Mub1 is an essential factor for Rpn4 ubiquitylation.
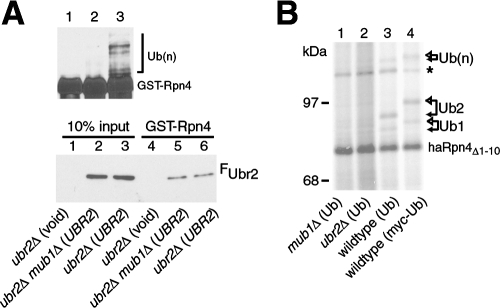
The current study originated from the hypothesis that a protein other than the Ubr2/Rad6 Ub ligase is required for Rpn4 ubiquitylation. To examine the involvement of Mub1 in Rpn4 ubiquitylation, we first performed a pulldown ubiquitylation assay. Cell extracts prepared from the ubr2Δ and ubr2Δ mub1Δ mutants overexpressing FUbr2 were incubated with agarose beads preloaded with GST-Rpn4. Extract from the ubr2Δ mutant containing a void vector was used as a negative control. After a standard pulldown, the beads were further incubated with purified Rad6 and Uba1 in the presence of Ub and ATP. The GST-Rpn4 fusions were then separated by SDS-PAGE and subjected to immunoblotting analysis with anti-GST antibody (Fig. 2A, upper panel). Consistent with our previous observation (48), GST-Rpn4 was ubiquitylated by FUbr2 from the ubr2Δ mutant (compare lanes 3 and 1). By contrast, FUbr2 pulled down from the ubr2Δ mub1Δ double mutant barely ubiquitylated GST-Rpn4 (lane 2). Thus, Mub1 is essential for Ubr2/Rad6-mediated ubiquitylation of Rpn4. It is noteworthy that the extracts prepared from the ubr2Δ and ubr2Δ mub1Δ transformants had similar levels of FUbr2 (Fig. 2A, lower panel, lanes 2 and 3) and that comparable amounts of FUbr2 were pulled down from these two extracts by GST-Rpn4 (lanes 5 and 6). This pulldown analysis suggests that Mub1 is not required for the binding of Rpn4 to Ubr2.
FIG. 2.
Mub1 is required for Rpn4 ubiquitylation. (A) Pulldown ubiquitylation assay. Upper panel shows the results of a pulldown ubiquitylation assay. Agarose beads preloaded with GST-Rpn4 were incubated with extracts from ubr2Δ (lane 3) and ubr2Δ mub1Δ (lane 2) cells expressing FUbr2 from the GAL1 promoter on a high-copy-number vector. Extract from ubr2Δ cells bearing a void vector served as a control (lane 1). After the pulldown, the beads were further incubated with purified Rad6 and Uba1 in the presence of Ub and ATP at 30°C for 30 min. GST fusions were then separated by SDS-PAGE and analyzed by immunoblotting with anti-GST antibody. Ubiquitylated species are marked by half of a square bracket. The lower panel shows the result of a pulldown assay to compare the binding of FUbr2 from different extracts to GST-Rpn4 (lanes 4 to 6). Ten percent of the input extracts was included, to estimate the pulldown efficiency (lanes 1 to 3). (B) Mub1 is required for in vivo ubiquitylation of Rpn4. Wild-type, mub1Δ, and ubr2Δ cells coexpressing haRpn4Δ1-10 with Ub were metabolically labeled with [35S]methionine for 5 min (lanes 1 to 3). Wild-type cells coexpressing haRpn4Δ1-10 and myc-Ub were used as a control (lane 4). Cell extracts were immunoprecipitated with anti-ha antibody and separated by SDS-PAGE (6% gel). Filled and open arrows mark conjugates with Ub and myc-Ub, respectively. The asterisk shows a cross-reactive band. Ub(n), polyubiquitylated species.
To validate the pulldown ubiquitylation result, we compared the in vivo ubiquitylation of haRpn4Δ1-10 in the presence and absence of Mub1. Wild-type, mub1Δ, and ubr2Δ cells coexpressing haRpn4Δ1-10 and Ub were metabolically labeled with [35S]methionine. To facilitate the assessment of ubiquitylated substrates, wild-type cells coexpressing haRpn4Δ1-10 and N-terminally myc-tagged Ub (myc-Ub) were used as the control. It has been shown that myc-Ub conjugates of Rpn4 characteristically migrate more slowly than their Ub counterparts (19). haRpn4Δ1-10 proteins were immunoprecipitated with anti-ha antibody and separated by SDS-PAGE. Whereas ubiquitylated species of haRpn4Δ1-10 were observed in the wild-type cells (Fig. 2B, lanes 3 and 4), no ubiquitylation of haRpn4Δ1-10 was detected in either ubr2Δ or mub1Δ cells (lanes 1 and 2), indicating that Mub1 is required for Rpn4 ubiquitylation in vivo.
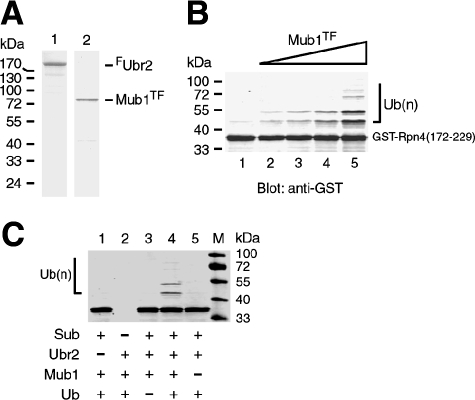
To more directly assess the involvement of Mub1 in Rpn4 ubiquitylation, we established an in vitro reconstitution ubiquitylation system using purified Ubr2 and Mub1 proteins. Mub1TF was overexpressed and purified from the ubr2Δ mub1Δ double mutant by using an anti-FLAG affinity column (Fig. 3A, lane 2). The attachment of the Tev-FLAG tag to Mub1 did not noticeably impair its activity in supporting the Ub-dependent degradation of Rpn4 (data not shown). FUbr2 was also overexpressed and purified from the ubr2Δ mub1Δ double mutant by using an anti-FLAG affinity column (Fig. 3A, lane 1). The substrate used in this assay was GST-Rpn4172-229 (20). Our early work has shown that Ub is selectively conjugated to K187, the preferred ubiquitylation site of Rpn4 (20). GST-Rpn4172-229 purified from E. coli was phosphorylated by CK2 before being applied to the ubiquitylation reactions. It has been shown that phosphorylation of S220 by CK2 facilitates Rpn4 ubiquitylation (21). We compared the ubiquitylation of GST-Rpn4172-229 in the absence and in the presence of increasing amounts of Mub1TF, with all other components included in the reaction mixtures (Fig. 3B). In the absence of Mub1TF, a very small amount of monoubiquitylated species was generated by the Ubr2/Rad6 Ub ligase (lane 1). In contrast, the ubiquitylation efficiency markedly increased, in a dose-dependent manner, with the input of Mub1TF (lanes 2 to 5). The ubiquitylation did not take place when Ubr2, Ub, or the substrate was removed from the reaction mixture (Fig. 3C). The in vitro reconstitution ubiquitylation assay clearly demonstrates that Mub1 is the sought factor required for Rpn4 ubiquitylation.
FIG. 3.
In vitro ubiquitylation of Rpn4 by a reconstitution system. (A) Coomassie blue staining of purified FUbr2 and Mub1TF proteins. Approximately 0.4 μg FUbr2 (lane 1) and 0.2 μg Mub1TF (lane 2) were run on an SDS-PAGE gel (10% gel). (B) In vitro reconstitution ubiquitylation assays. Phosphorylated GST-Rpn4172-229 was incubated with purified FUbr2, Rad6, Uba1, Ub, and ATP in the absence (lane 1) or presence of increasing amounts (8, 20, 80, and 160 nM) of Mub1TF (lanes 2 to 5) at 30°C for 90 min. The reaction products were separated by SDS-PAGE (10% gel), followed by immunoblotting analysis with anti-GST antibody. (C) In vitro ubiquitylation reaction mixtures similar to those described for panel B, except that individual components were omitted as indicated, were set up. Rad6 and Uba1 were included in all reaction mixtures. Ubiquitylated species are marked by half of a square bracket. Sub, substrate; Ub(n), polyubiquitylated species.
Mub1 interacts with Ubr2 and Rpn4.
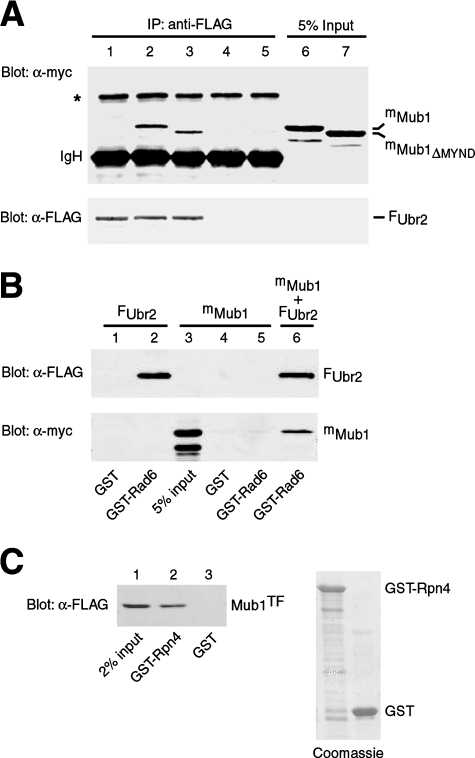
To gain insight into the mechanism of Mub1 in Rpn4 ubiquitylation, we examined whether Mub1 forms a complex with the Ubr2/Rad6 Ub ligase. We first tested whether Mub1 interacts with Ubr2 by using a coimmunoprecipitation assay. Since Mub1 was rapidly degraded in a Ubr2-dependent manner (see below), we expressed mMub1 and FUbr2, separately, in the ubr2Δ mub1Δ double mutant. The cell extracts from these two transformants were immunoprecipitated, either individually or in combination, by anti-FLAG antibody, followed by Western analysis with antimyc and anti-FLAG antibodies (Fig. 4A, lanes 1, 2, and 4). Clearly, mMub1 was specifically coprecipitated with FUbr2 (lane 2). We went on to examine whether Mub1 also interacts with Rad6 by using a GST pulldown assay (Fig. 4B). The same cell extracts used in the coimmunoprecipitation assay were incubated with agarose beads preloaded with GST-Rad6 or GST. As expected, FUbr2 was specifically pulled down by GST-Rad6 (Fig. 4B, lane 2). In contrast, mMub1 did not bind to GST-Rad6 (lane 5). Interestingly, GST-Rad6 was able to pull down mMub1 from the mixed extracts containing both mMub1 and FUbr2 (Fig. 4B, lane 6). These results indicate that Ubr2 mediates the formation of an Mub1-Ubr2-Rad6 ternary complex. The early observed interaction of Mub1 with Rad6 in a genome-wide affinity capture-mass spectrometry analysis likely resulted from coimmunoprecipitation of Rad6 with Ubr2 by the tagged Mub1 protein (24).
FIG. 4.
Mub1 interacts with Ubr2 and Rpn4. (A) Mub1 was coimmunoprecipitated with Ubr2. FUbr2, mMub1, and mMub1ΔMYND were expressed in the ubr2Δ mub1Δ double mutant. The cell extracts were immunoprecipitated (IP) with anti-FLAG antibody, either individually or in combination, followed by Western analysis with anti-myc (upper panel) and anti-FLAG (lower panel). Lanes 1, 4, and 5, individual extracts containing FUbr2, mMub1, and mMub1ΔMYND; lanes 2 and 3, mixed extracts containing FUbr2 with mMub1 and mMub1ΔMYND. Five percent of the mMub1 and mMub1ΔMYND input extracts was included in the immunoblotting analysis (lanes 6 and 7). The asterisk marks a nonspecific band. IgH, immunoglobulin H. (B) Ubr2 mediates the formation of an Mub1-Ubr2-Rad6 complex. Cell extracts containing FUbr2 (lanes 1 and 2), mMub1 (lanes 4 and 5), or mixed extracts with FUbr2 and mMub1 (lane 6) were pulled down by GST-Rad6 or GST, followed by Western analysis with anti-FLAG (upper panel) and anti-myc (lower panel) antibodies. Five percent of the mMub1 input extracts was included in the Western analysis (lane 3). (C) Mub1 directly interacts with Rpn4. Purified Mub1TF protein was applied in a pulldown assay with GST-Rpn4 (lane 2) and GST (lane 3). Two percent of the Mub1TF input was included in the Western analysis with anti-FLAG antibody (lane 1). Comparable amounts of GST (right lane) and GST-Rpn4 (left lane) were used in the pulldown assay (right panel). α, anti.
We have shown that Ubr2 directly binds Rpn4 in the absence of Mub1 (Fig. 2A) (48). However, the physical association between Ubr2 and Rpn4 in the absence of Mub1 was not productive in Rpn4 ubiquitylation. We decided to examine whether Mub1 is also an Rpn4-binding protein by using a GST pulldown assay. As shown in Fig. 4C, purified Mub1TF was indeed pulled down by GST-Rpn4, indicating that Mub1 directly binds to Rpn4.
The MYND domain of Mub1 is required for Ub-dependent degradation of Rpn4.
Sequence analysis shows that Mub1 carries a 44-amino-acid MYND domain near the C terminus (Fig. 5A). The MYND domain, named after myeloid translocation protein 8, Nervy, and DEAF-1, is a zinc-binding domain defined by seven conserved Cys residues and a single His residue in a C4-C2HC format (44). Somewhat different from the cross-braced RING domain, the C4-C2HC MYND motif forms two sequential zinc binding sites, comprising a small β hairpin and two short α helices, respectively (4, 44). MYND domains have been shown to mediate protein-protein interactions involved in transcriptional regulation and signal transduction (44). To examine whether the MYND domain of Mub1 is also required for the interactions with Ubr2 and Rpn4, we generated an mMub1 mutant that lacks the MYND domain (mMub1ΔMYND). Cell extracts from the ubr2Δ mub1Δ double mutant expressing FUbr2 and from mMub1ΔMYND cells, respectively, were immunoprecipitated by anti-FLAG antibody either individually or in combination, followed by Western analysis with anti-myc and anti-FLAG antibodies. As shown in Fig. 4A, mMub1ΔMYND was specifically coimmunoprecipitated with FUbr2 (compare lanes 3 and 5). The binding of mMub1ΔMYND to FUbr2 was modestly weaker than that of mMub1 (compare lanes 2 and 3). Similarly, deletion of the MYND domain slightly decreased the binding of Mub1 to Rpn4 in the GST pulldown assay (Fig. 5C). Interestingly, in spite of the relatively mild effect on the binding of Mub1 to Ubr2 and Rpn4, deletion of the MYND domain virtually abolished the activity of Mub1 in supporting the degradation of Rpn4172-229-β-Gal, a β-Gal fusion with the Ub-dependent degradation signal of Rpn4 (Fig. 5B). These results suggest that the function of the Mub1 MYND domain in Rpn4 ubiquitylation may involve a novel mechanism.
FIG. 5.
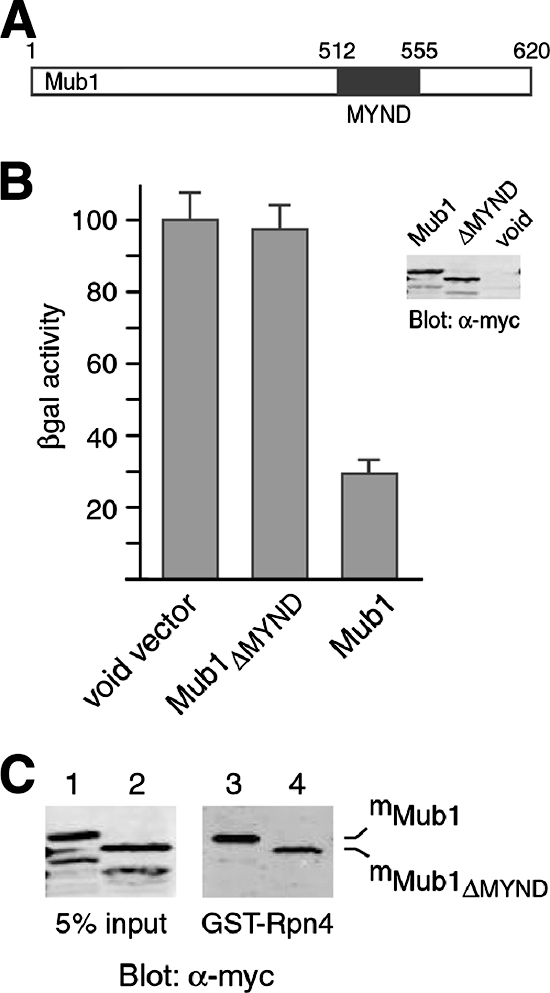
The MYND domain of Mub1 is essential for Ub-dependent degradation of Rpn4. (A) Diagram of the Mub1 protein. The MYND domain consists of amino acids 512 to 555. (B) Ub-dependent degradation of Rpn4 is inhibited by deletion of the MYND domain of Mub1. Plasmids expressing mMub1, mMub1ΔMYND, or a void vector were transformed into the mub1Δ cells coexpressing Rpn4172-229-β-Gal. The degradation of Rpn4172-229-β-Gal was measured by β-Gal enzymatic assay as previously described (20). The β-Gal activity in the mub1Δ cells bearing the void vector was set at 100%. Error bars show standard deviations. The expression levels of mMub1 and mMub1ΔMYND were comparable, as confirmed by Western analysis with anti-myc antibody. (C) The MYND domain of Mub1 is not required for interaction with Rpn4. Cell extracts from the ubr2Δ mub1Δ double mutant expressing mMub1 or mMub1ΔMYND were applied in a pulldown assay with GST-Rpn4 (lanes 3 and 4). Five percent of the input extracts was included in the Western analysis with anti-myc antibody (lanes 1 and 2). α, anti.
Mub1 is a short-lived protein whose degradation is dependent on the Ubr2/Rad6 Ub ligase.
Several F-box substrate adaptor proteins of the Cul1-Rbx1 Ub ligase have been found to be short-lived substrates of the same Ub ligase (9, 34, 37, 60). The binding of Mub1 to the Ubr2/Rad6 Ub ligase prompted us to assess whether Mub1 is a short-lived protein as well. Moreover, we wondered if Mub1 is a substrate of the Ubr2/Rad6 Ub ligase. To test these possibilities, we compared the stability of Mub1TF in wild-type, ubr2Δ, and rad6Δ cells by pulse-chase analysis. Indeed, Mub1TF was short-lived in the wild type (half-life, ∼15 min) but was stabilized in the ubr2Δ and rad6Δ mutants (Fig. 6). Thus, Mub1 is a short-lived protein and its degradation is dependent on the Ubr2/Rad6 Ub ligase.
FIG. 6.
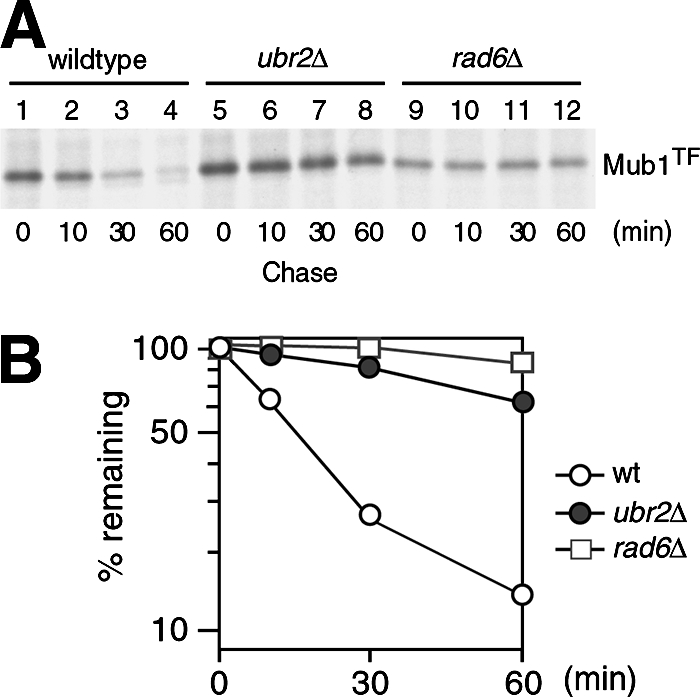
In vivo degradation of Mub1 is dependent on the Ubr2/Rad6 Ub ligase. (A) The stability of Mub1TF in wild-type, ubr2Δ, and rad6Δ cells was measured by pulse-chase assay. Mub1TF was expressed from the CUP1 promoter in a low-copy-number vector. Anti-FLAG antibody was used in the immunoprecipitation. (B) Quantitation with PhosphorImager of data shown in panel A, to show the decay curves. wt, wild type.
DISCUSSION
In this study, we conducted a genome-wide analysis and identified Mub1 as an essential factor for Rpn4 ubiquitylation catalyzed by the Ubr2/Rad6 Ub ligase. In the course of this study, a systematic analysis of genetic interactions between proteins unveiled a potential functional relationship between Mub1 and Ubr2 (6), which is in line with our finding. Our data show that Mub1 forms a complex with the Ubr2/Rad6 Ub ligase via an interaction with Ubr2. This is in contrast to the results of a previous report showing that Mub1 physically interacts with Rad6 in a genome-wide affinity capture-mass spectrometry analysis (24). Whereas we cannot rule out exclusively the possibility that the sensitivity of the GST pulldown assay in our study was not sufficient to detect a weak Mub1-Rad6 binding, it is very likely that the early observation resulted from coimmunoprecipitation of Rad6 with Ubr2 by the tagged Mub1 protein rather than a direct Rad6-Mub1 interaction. Nevertheless, Ubr2 mediates the formation of an Mub1-Ubr2-Rad6 complex.
Our data demonstrate that Mub1 directly interacts with Rpn4, suggesting that it may act as an adaptor to recruit Rpn4 to the Ubr2/Rad6 Ub ligase. However, we have also shown that Ubr2 directly binds Rpn4 in the absence of Mub1 (Fig. 2) (48). Interestingly, the Ubr2-Rpn4 association without the involvement of Mub1 is unable to catalyze Rpn4 ubiquitylation. What is the mechanism of Mub1 in Rpn4 ubiquitylation? One possibility is that Mub1 may activate the intrinsic activity of the Ubr2/Rad6 Ub ligase. We found, however, that the kinetics of Ubr2 autoubiquitylation was similar in the presence or absence of Mub1, indicating that Mub1 does not affect the intrinsic catalytic activity of the Ubr2/Rad6 Ub ligase (data not shown). We propose that Mub1 may play a role in directing the transfer of Ub from Rad6 to Rpn4. A tentative model that explains how Mub1 and Ubr2 may cooperate in ubiquitylating Rpn4 is postulated (Fig. 7). Although Ubr2 can bind Rpn4 on its own, this interaction is not productive, perhaps because the ubiquitylation site of Rpn4 is not close enough or not in the right orientation to the Ub∼Rad6 thioester (Ub donor) to receive Ub. The entry of Mub1 may rotate the Rpn4 substrate through the Mub1-Ubr2 and Mub1-Rpn4 protein-protein interactions so as to position the ubiquitylation site of Rpn4 proximal to the Ub∼Rad6 thioester associated with Ubr2 (Fig. 7). This model is in line with our recent finding that an E3-binding site does not necessarily function as a ubiquitylation signal, regardless of the binding stability (21). The position of the E3-binding site on the substrate (e.g., relative to the ubiquitylation site) may be a more critical element that determines whether or not the E3-binding site is a legitimate ubiquitylation signal.
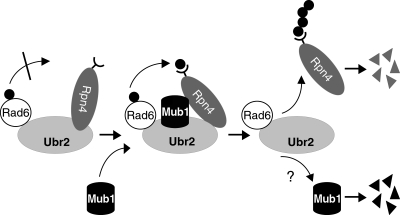
FIG. 7.
A model in which Mub1 and Ubr2 cooperate to ubiquitylate Rpn4. Binding of Rpn4 to the Ubr2 E3 does not concurrently confer access for the E3-bound Ub∼Rad6 thioester (Ub donor) to the ubiquitylation site of Rpn4. The entry of Mub1 may rotate the Rpn4 substrate through the Mub1-Ubr2 and Mub1-Rpn4 protein-protein interactions, positioning the Rpn4 ubiquitylation site proximal to the Ub∼Rad6 thioester and allowing the transfer of Ub (black dots) from Rad6 to Rpn4. After Rpn4 is ubiquitylated, Mub1 may be released from Ubr2 and is rapidly degraded. The details of the Ubr2/Rad6-dependent degradation of Mub1 remain to be elucidated.
It is interesting to note that Mub1 is a short-lived substrate in vivo and that the Ubr2/Rad6 Ub ligase is required for Mub1 degradation. Our attempts to detect Mub1 ubiquitylation by Ubr2/Rad6 in vitro were unsuccessful (data not shown). It is likely that the in vitro assay conditions were not optimal for Mub1 ubiquitylation. Alternatively, another factor, in addition to the Ubr2/Rad6 Ub ligase, may be required for Mub1 ubiquitylation, just as Mub1 is required for Rpn4 ubiquitylation. Nevertheless, the Ubr2/Rad6-dependent degradation of Mub1 suggests that Mub1 may switch from a partner to a substrate of the Ubr2/Rad6 Ub ligase in the course of Rpn4 ubiquitylation. The degradation of Mub1 may facilitate the reentrance of Ubr2 into the next cycle of Rpn4 ubiquitylation, i.e., recruitment of new Rpn4 molecules. A more intriguing possibility is that the removal of Mub1 from Ubr2/Rad6 may be necessary to leave room for the extension of the Ub chain on Rpn4. It is also possible that the degradation of Mub1 may provide a mechanism to minimize the competition for a limited amount of Ubr2 in vivo, as Ubr2 may target other substrates in addition to Rpn4. It is not uncommon that the same Ub ligase ubiquitylates different substrates.
Another interesting observation from this study is that the MYND domain of Mub1 may play an important role in Rpn4 ubiquitylation. Whereas deletion of the MYND domain only modestly weakens the binding of Mub1 to Ubr2 and Rpn4, the MYND-less Mub1 protein is unable to support the Ub-dependent degradation of Rpn4. Both the MYND and RING domains contain two zinc-binding sites capable of interacting with two zinc ions. The RING domain is a well-known hallmark of the RING E3 family. Could the MYND domain represent a signature for yet another group of proteins involved in protein ubiquitylation? Our current study and several recent reports suggest that this possibility may exist. The prolyl-hydroxylase PHD2, which specifically hydroxylates HIF-1α, a prerequisite step for HIF-1α ubiquitylation by the VHL Ub ligase, contains an MYND domain (5). The MYND domain protein BS69 interacts with the TRAF6 E3, which plays an important role in the latent membrane protein 1-mediated c-Jun N-terminal kinase pathway (47). Interestingly, BS69 is also required for inhibition of the ubiquitylation of the adenovirus E1A protein (15). Further delineation of the exact function of these MYND domains may provide important information on the potential involvement of MYND domains in protein ubiquitylation.
Acknowledgments
We thank G. Brush for yeast strains and X. Mao for assistance in the early phase of this work.
This study was supported by grants to Y.X. from the Barbara Ann Karmanos Cancer Institute and the American Cancer Society (RGS-0506401-GMC).
Xiaogang Wang is a Ph.D. candidate of Fudan University, Shanghai, China.
Footnotes
Published ahead of print on 10 December 2007.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl (ed.). 1998. Current protocols in molecular biology. Wiley-Interscience, New York, NY.
- 2.Bachmair, A., D. Finley, and A. Varshavsky. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234179-186. [DOI] [PubMed] [Google Scholar]
- 3.Cai, H., S. Kauffman, F. Naider, and J. M. Becker. 2006. Genomewide screen reveals a wide regulatory network for di/tripeptide utilization in Saccharomyces cerevisiae. Genetics 1721459-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capili, A. D., D. C. Schultz, I. F. Rauscher, and K. L. Borden. 2001. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, K.-O., T. Lee, N. Lee, J.-H. Kim, E. G. Yang, J. M. Yoon, J. H. Kim, T. G. Lee, and H. Park. 2005. Inhibition of the catalytic activity of hypoxia-inducible factor-1α-prolyl-hydroxylase 2 by a MYND-type zinc finger. Mol. Pharmacol. 681803-1809. [DOI] [PubMed] [Google Scholar]
- 6.Collins, S. R., K. M. Miller, N. L. Maas, A. Roguev, J. Fillingham, C. S. Chu, M. Schuldiner, M. Gebbia, J. Recht, M. Shales, et al. 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446806-810. [DOI] [PubMed] [Google Scholar]
- 7.Finley, D., K. Tanaka, C. Mann, H. Feldmann, M. Hochstrasser, R. Vierstra, S. Johnston, R. Hampton, J. Haber, J. McCusker, et al. 1998. Unified nomenclature for subunits of the Saccharomyces cerevisiae proteasome regulatory particle. Trends Biochem. Sci. 23244-245. [DOI] [PubMed] [Google Scholar]
- 8.Fujimuro, M., K. Tanaka, H. Yokosawa, and A. Toh-e. 1998. Son1p is a component of the 26S proteasome of the yeast Saccharomyces cerevisiae. FEBS Lett. 423149-154. [DOI] [PubMed] [Google Scholar]
- 9.Galan, J. M., and M. Peter. 1998. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA 969124-9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghislain, M., A. Udvardy, and C. Mann. 1993. S. cerevisiae 26S proteasome mutants arrest cell division in G2/metaphase. Nature 366358-362. [DOI] [PubMed] [Google Scholar]
- 11.Hahn, J.-S., D. W. Neef, and D. J. Thiele. 2006. A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Mol. Microbiol. 60240-251. [DOI] [PubMed] [Google Scholar]
- 12.Heinemeyer, W., A. Gruhler, V. Möhrle, Y. Mahé, and D. H. Wolf. 1993. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J. Biol. Chem. 2685115-5120. [PubMed] [Google Scholar]
- 13.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67425-479. [DOI] [PubMed] [Google Scholar]
- 14.Hochstrasser, M. 2006. Lingering mysteries of ubiquitin-chain assembly. Cell 12427-34. [DOI] [PubMed] [Google Scholar]
- 15.Isobe, T., C. Uchida, T. Hattori, K. Kitagawa, T. Oda, and M. Kitagawa. 2006. Ubiquitin-dependent degradation of adenovirus E1A protein is inhibited by BS69. Biochem. Biophys. Res. Commun. 339367-374. [DOI] [PubMed] [Google Scholar]
- 16.Jelinsky, S. A., P. Estep, G. M. Church, and L. D. Samson. 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 208157-8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong, J.-W., M.-K. Bae, M.-Y. Ahn, S.-H. Kim, T.-K. Sohn, M.-H. Bae, M.-Q. Yoo, E. J. Song, K.-J. Lee, and K.-W. Kim. 2002. Regulation and destabilization of HIF-1α by ARD1-mediated acetylation. Cell 111709-720. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, E. S., P. C. Ma, I. M. Ota, and A. Varshavsky. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 27017442-17456. [DOI] [PubMed] [Google Scholar]
- 19.Ju, D., and Y. Xie. 2004. Proteasomal degradation of RPN4 via two distinct mechanisms: ubiquitin-dependent and -independent. J. Biol. Chem. 27923851-23854. [DOI] [PubMed] [Google Scholar]
- 20.Ju, D., and Y. Xie. 2006. Identification of the preferential ubiquitination site and ubiquitin-dependent degradation signal of Rpn4. J. Biol. Chem. 28110657-10662. [DOI] [PubMed] [Google Scholar]
- 21.Ju, D., H. Xu, X. Wang, and Y. Xie. 2007. Ubiquitin-mediated degradation of Rpn4 is controlled by a phosphorylation-dependent ubiquitylation signal. Biochim. Biophys. Acta 17731672-1680. [DOI] [PubMed] [Google Scholar]
- 22.Ju, D., L. Wang, X. Mao, and Y. Xie. 2004. Homeostatic regulation of the proteasome via an Rpn4-dependent feedback circuit. Biochem. Biophys. Res. Commun. 32151-57. [DOI] [PubMed] [Google Scholar]
- 23.Kaelin, W. G. 2005. Proline hydroxylation and gene expression. Annu. Rev. Biochem. 74115-128. [DOI] [PubMed] [Google Scholar]
- 24.Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A. P. Tikuisis, et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440637-643. [DOI] [PubMed] [Google Scholar]
- 25.Krüger, M., and R. Fischer. 1998. Integrity of a Zn finger-like domain in SamB is crucial for morphogenesis in ascomycetous fungi. EMBO J. 17204-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laney, J. D., and M. Hochstrasser. 1999. Substrate targeting in the ubiquitin system. Cell 97427-430. [DOI] [PubMed] [Google Scholar]
- 27.London, M., B. I. Keck, P. C. Ramos, and R. J. Dohmen. 2004. Regulatory mechanisms controlling biogenesis of ubiquitin and the proteasome. FEBS Lett. 567259-264. [DOI] [PubMed] [Google Scholar]
- 28.Lundgren, J., P. Masson, C. A. Realini, and P. Young. 2003. Use of RNA interference and complementation to study the function of the Drosophila and human 26S proteasome subunit S13. Mol. Cell. Biol. 235320-5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mannhaupt, G., R. Schnall, V. Karpov, I. Vetter, and H. Feldmann. 1999. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 45027-34. [DOI] [PubMed] [Google Scholar]
- 30.Medicherla, B., Z. Kostova, A. Schaefer, and D. H. Wolf. 2004. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 5692-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meiners, S., D. Heyken, A. Weller, A. Ludwig, K. Stangl, P.-M. Kloetzel, and E. Krüger. 2003. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of mammalian proteasomes. J. Biol. Chem. 27821517-21525. [DOI] [PubMed] [Google Scholar]
- 32.Nelson, M. K., T. Kurihara, and P. A. Silver. 1993. Extragenic suppressors of mutations in the cytoplasmic C terminus of SEC63 define five genes in Saccharomyces cerevisiae. Genetics 134159-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng, D. T. W., E. D. Spear, and P. Walter. 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 15077-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohta, T., J. J. Michel, A. J. Schottelius, and Y. Xiong. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3535-541. [DOI] [PubMed] [Google Scholar]
- 35.Orlicky, S., X. Tang, A. Willems, M. Tyers, and F. Sicheri. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112243-256. [DOI] [PubMed] [Google Scholar]
- 36.Owsianik, G., E. Balzi, and M. Ghislain. 2002. Control of 26S proteasome expression by transcription factors regulating multidrug resistance in Saccharomyces cerevisiae. Mol. Microbiol. 431295-1308. [DOI] [PubMed] [Google Scholar]
- 37.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-ring ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 69-20. [DOI] [PubMed] [Google Scholar]
- 38.Pickart, C. M., and M. J. Eddins. 2004. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 169555-72. [DOI] [PubMed] [Google Scholar]
- 39.Prinz, S., I. Avila-Campillo, C. Aldridge, A. Srinivasan, K. Dimitrov, A. F. Siegel, and T. Galitski. 2004. Control of yeast filamentous-form growth by modules in an integrated molecular network. Genome Res. 14380-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pritsker, M., Y.-C. Liu, M. A. Beer, and S. Tavazoie. 2004. Whole-genome discovery of transcription factor binding sites by network-level conservation. Genome Res. 1499-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramos, P. C., J. Höckendorff, E. S. Johnson, A. Varshavsky, and R. J. Dohmen. 1998. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell 92489-499. [DOI] [PubMed] [Google Scholar]
- 42.Ravid, T., S. G. Kreft, and M. Hochstrasser. 2006. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 25533-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulman, B. A., A. C. Carrano, P. D. Jeffrey, Z. Bowen, E. R. E. Kinnucan, M. S. Finnin, S. J. Elledge, J. W. Harper, M. Pagano, and N. Pavletich. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408381-386. [DOI] [PubMed] [Google Scholar]
- 44.Spadaccini, R., H. Perrin, M. J. Bottomley, S. Ansieau, and M. Sattler. 2006. Structure and functional analysis of the MYND domain. J. Mol. Biol. 358498-508.16527309 [Google Scholar]
- 45.Tyers, M., and P. Jorgensen. 2000. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr. Opin. Genet. Dev. 1054-64. [DOI] [PubMed] [Google Scholar]
- 46.Varshavsky, A. 2005. Regulated protein degradation. Trends Biochem. Sci. 30282-286. [DOI] [PubMed] [Google Scholar]
- 47.Wan, J., W. Zhang, L. Wu, T. Bai, M. Zhang, K.-W. Lo, Y.-L. Chui, Y. Cui, Q. Tao, M. Yamamoto, et al. 2006. BS69, a specific adaptor in the latent membrane protein 1-mediated c-Jun N-terminal kinase pathway. Mol. Cell. Biol. 26448-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, L., X. Mao, D. Ju, and Y. Xie. 2004. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J. Biol. Chem. 27955218-55223. [DOI] [PubMed] [Google Scholar]
- 49.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2169-178. [DOI] [PubMed] [Google Scholar]
- 50.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285901-906. [DOI] [PubMed] [Google Scholar]
- 51.Wójcik, C., and G. N. DeMartino. 2002. RNA interference of valosin-containing protein (VCP/p97) reveals multiple cellular roles linked to ubiquitin/proteasome-dependent proteolysis. J. Biol. Chem. 2776188-6197.11739392 [Google Scholar]
- 52.Wu, G., G. Xu, B. A. Schulman, P. D. Jeffrey, J. W. Harper, and N. P. Pavletich. 2003. Structure of β-TrCP1-Skp1-β-catenin complex: destruction motif binding and lysine specificity of the SCF (β-TrCP1) ubiquitin ligase. Mol. Cell 111445-1456. [DOI] [PubMed] [Google Scholar]
- 53.Xie, Y., and A. Varshavsky. 2001. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. USA 983056-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie, Y., and A. Varshavsky. 2002. UFD4 lacking the proteasome-binding region catalyses ubiquitination but is impaired in proteolysis. Nat. Cell Biol. 41003-1007. [DOI] [PubMed] [Google Scholar]
- 55.Xie, Y., and A. Varshavsky. 1999. The E2-E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 186832-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu, H., D. Ju, T. Jarois, and Y. Xie. 2008. Diminished feedback regulation of proteasome expression and resistance to proteasome inhibitors in breast cancer cells. Breast Cancer Res. Treat. 107267-274. [DOI] [PubMed] [Google Scholar]
- 57.Yokoyama, H., M. Mizunuma, M. Okamoto, J. Yamamoto, D. Hirata, and T. Miyakawa. 2006. Involvement of calcineurin-dependent degradation of Yap1p in Ca2+-induced G2 cell-cycle regulation in Saccharomyces cerevisiae. EMBO Rep. 7519-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshida, Y., T. Chiba, F. Tokunaga, H. Kawasaki, K. Iwai, T. Suzuki, Y. Ito, K. Matsuoka, K. Yoshida, K. Tanaka, and T. Tadashi. 2002. An E3 ubiquitin ligase that recognizes sugar chains. Nature 418438-472. [DOI] [PubMed] [Google Scholar]
- 59.Zheng, N., B. A. Schulman, L. Song, J. J. Miller, P. D. Jeffrey, P. Wang, C. Chu, D. M. Koepp, S. J. Elledge, M. Pagano, C. R. Conaway, J. W. Conaway, J. W. Harper, and N. P. Pavletich. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416703-709. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, P., and P. M. Howley. 1998. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell 2571-580. [DOI] [PubMed] [Google Scholar]