Abstract
The broadly conserved Sir2 NAD+-dependent deacetylase is required for chromatin silencing. Here we report the discovery of physical and functional links between Sir2 and Slx5 (Hex3), a RING domain protein and subunit of the Slx5/8 complex column, which is a ubiquitin E3 ligase that targets sumoylated proteins. Slx5 interacted with Sir2 by two-hybrid and glutathione S-transferase-binding assays and was found to promote silencing of genes at telomeric or ribosomal DNA (rDNA) loci. However, deletion of SLX5 had no detectable effect on the distribution of silent chromatin components and only slightly altered the deacetylation of histone H4 lysine 16 at the telomere. In vivo assays indicated that Sir2-dependent silencing was functionally intact in the absence of Slx5. Although no previous reports suggest that Sir2 contributes to the fitness of yeast populations, we found that Sir2 was required for maximal growth in slx5Δ mutant cells. A similar requirement was observed for mutants of the SUMO isopeptidase Ulp2/Smt4. The contribution of Sir2 to optimal growth was not due to known Sir2 roles in mating-type determination or rDNA maintenance but was connected to a role of sumoylation in transcriptional silencing. These results indicate that Sir2 and Slx5 jointly contribute to transcriptional silencing and robust cellular growth.
In the budding yeast Saccharomyces cerevisiae, transcriptional silencing is defined as constitutive, chromatin-mediated repression of transcription that occurs at HM loci, ribosomal DNA (rDNA), and telomeres. The Sir2 protein, essential for S. cerevisiae transcriptional silencing, is of particular interest as the founding member of the sirtuin family of NAD-dependent protein deacetylases that is conserved throughout eukaryotes. Increased expression of SIR2 or its closest homologs increases the life span in yeast and animals, respectively (reviewed in references 31 and 44). Although these studies suggest the conservation of an aging pathway, molecular mechanisms underlying the conserved function of sirtuins in aging remain unclear. In yeast, the only reported targets of Sir2 activity are histones. By contrast, the mammalian sirtuin SIRT1 deacetylates many different targets, including p53 (reviewed in reference 55). In yeast, Sir2 binds the telomeres and HM loci in complex with Sir3 and Sir4. Deacetylation of histone H4 lysine 16 (H4K16) promotes binding of Sir3 to chromatin and spreading of transcriptional silencing (reviewed in reference 54). However, Sir3 and Sir4 have no known metazoan homologs. Thus, it seems likely that some Sir2 molecular interactions remain undiscovered. Indeed, molecular mechanisms that contribute to sir2Δ phenotypes, such as suppression of replication onset (47) and unequal inheritance of oxidative damage between mother and daughter cells (1), are undefined. To identify novel Sir2-interacting proteins that might participate in these and other uncharacterized molecular pathways, we performed a screen for Sir2 physical interactions.
We identified Slx5 as a novel Sir2 interactor by two-hybrid analysis with Sir2 fused to the Gal4 DNA-binding domain (GBD). Slx5 is a RING domain protein that was first identified as essential for viability in the absence of Sgs1 (45). Sgs1 is the sole S. cerevisiae representative of the RecQ helicase family that includes human WRN and BLM. Slx5 forms a complex with another RING domain protein, Slx8 (45, 76). Mutants lacking Slx5 or Slx8 show an increase in global sumoylation (6, 28, 70). SUMO is a ubiquitin-like protein moiety that is covalently attached to lysine residues. The Slx5/8 complex may function as a SUMO E3 ligase (28). Recently, it was discovered that Slx5 contains SUMO interaction motifs and that Slx8 is a ubiquitin E3 ligase (49, 67, 73). The Slx5/8 complex is thus a member of a newly defined conserved family of factors, known as STUbLs (SUMO-targeted ubiquitin ligases), present in humans as a single protein, RNF4 (49, 62). A function shared by some members of this protein family may be ubiquitination of polysumoylated proteins. The polysumoylated and ubiquitinated proteins are then degraded by the proteasome (67), providing a previously unsuspected mechanism for down-regulation.
Although Sir3 and Sir4, but not Sir2, are known to be sumoylated (11, 72), the Sir2-Slx5 two-hybrid interaction required neither protein and the GAD-Slx5 interactor lacked the N-terminal SUMO interaction motifs. Furthermore, we recapitulated the physical interaction in vitro by affinity precipitation under conditions in which protein sumoylation is not preserved. Thus, the physical interaction was not likely to be bridged by sumoylation. The proteins have a functional overlap defined by several independent assays. Yeast lacking the SLX5 gene grew poorly, and additionally deleting SIR2, SIR3, or SIR4 exacerbated this defect, indicating that Slx5 and the Sir2/3/4 complex may have parallel roles in a vital process. Deletion of the SLX5 gene caused a previously unsuspected loss of telomeric and rDNA silencing, as does sir2Δ. Furthermore, ulp2Δ mutant cells, which share the slx5Δ phenotype of increased global sumoylation, also shared the silencing defect and loss of viability in combination with the sir2Δ or sir4Δ mutation. Therefore, the role of Slx5 in silencing appears to be linked to its role in regulating sumoylation, such that excess accumulation of sumoylated proteins is toxic in the absence of silenced chromatin.
MATERIALS AND METHODS
Yeast methods.
For the yeast strains used in this study, see Table S1 in the supplemental material. Strains were grown at 30°C and standard manipulations were performed as previously described (2). Yeast media were prepared as previously described (56). Yeast cells were grown on yeast-peptone-dextrose (YPD) unless otherwise noted. 5-Fluoroorotic acid (5FOA; U.S. Biological, Marblehead, MA) was added to supplemented minimal medium at 0.1% to test for URA3 reporter expression. Telomeric and rDNA silencing assays were performed as described previously (22, 59). In serial-dilution assays, fivefold dilutions were plated starting at an optical density greater than 1 and grown at a temperature of 30°C. The growth rate of yeast strains was measured by spectrophotometer in logarithmic phase (A600 readings between 0.03 and 0.3). The A600 readings were fitted to the exponential curve A = A0ekt, and then the calculated A0 values were used to normalize and combine data from two to four independent cultures. Experiments with slx5Δ haploid strains were performed immediately after counterselection against an SLX5 URA3 plasmid.
Microscopy.
GFP microscopic imaging was performed on an Axiovert 200 M system (Carl Zeiss MicroImaging) with a monochrome digital camera and software provided by the manufacturer. Immunofluorescence microscopy was performed on an Applied Precision optical sectioning microscope to collect images spaced at 0.2-μm increments. The images were deconvolved with the Delta Vision deconvolution software as previously described (48). For immunofluorescence microscopy, fluorescein-conjugated anti-rabbit and Texas Red-conjugated anti-mouse secondary antibodies (Jackson ImmunoResearch) were preadsorbed against spheroplasted mutant and wild-type yeast cells.
Plasmid construction.
For the plasmids used in this study, see Table S2 in the supplemental material. Inserting a BclI-NruI SIR2 fragment, with Klenow-filled ends, from plasmid pLP285 into the SmaI site of pLP956 yielded plasmid Gbd-coreSir2 (pLP1073). Plasmid pLP1074 contains the ClaI fragment of SIR2 from pLP285 inserted into pLP958. Insertion of SacI-NcoI fragments of pLP1102, pLP1110, and pLP1112 (17) into pLP1074 created plasmids pLP1369, pLP1370, and pLP1371, respectively. SLX5 was cloned by molecular amplification with oligonucleotides 404 and 415 (see Table S3 in the supplemental material), inserted into pBluescript digested with ClaI and SmaI, and then subcloned into pRS315 cut with SalI and XbaI to make pLP1739.
Two-hybrid screen.
Details of the Gad-C1 library construction and PJ694-A two-hybrid reporter strain are described elsewhere (29). To test the bait constructs for in vivo Sir2 activity, MATa sir2Δ strain LPY11 was transformed with GBD, SIR2 (pLP983), GBD-coreSIR2, and two different GBD-Δ73NSIR2 constructs and tested for mating ability. Plasmids recovered from yeast cells capable of growth on medium lacking histidine and adenine were digested with HindIII as a diagnostic to distinguish bait from prey plasmids. To test for autoactivation, the prey plasmids were transformed into strain PJ694-A, lacking a bait construct, and these yeast cells were plated on medium lacking leucine, histidine, and adenine. For directed two-hybrid interactions, yeast transformants were selected on medium lacking leucine, tryptophan, histidine, and adenine and grown at 30°C for 6 days.
To identify the inserts of the candidate interactors, sequencing of both ends was performed with two plasmid-specific primers. For the 5′ end of the inserts, an oligonucleotide was used that hybridizes to the activation domain of Gal4 456 (5′-CGATGAGAAGATACCCC-3′). Sequencing of the 3′ end of the insert was performed with oligonucleotide 457 (5′-ATAGATCTCTGCAGGTCG-3′) and an Applied Biosystems automated facility at the UCSD Cancer Center and Center for Molecular Genetics (La Jolla, CA). Sequences obtained were aligned with the S. cerevisiae complete genomic sequence by using the Saccharomyces Genome Database BLAST program (http://genome-www2.stanford.edu/cgi-bin/SGD/nph-blast2sgd). The GAD-Slx5 interactor lacked only the first 182 residues of the Slx5 protein and contained the entire Slx5 RING domain.
GST-binding assays.
Glutathione S-transferase (GST; pLP1302), GST-Sir2 (pLP1275), and GST-sir2-R139K (pLP1335) fusion proteins were expressed in Escherichia coli BL21DE3 during a 4-h induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside at room temperature. Cells were resuspended in lysis buffer (20 mM Tris [pH 8]; 1 mM EDTA; 1 mM EGTA; 1× protease inhibitors tosylsulfonyl phenylalanyl chloromethyl ketone [TPCK], phenylmethylsulfonyl fluoride [PMSF], benzamidine, leupeptin, and pepstatin; 200 mg/ml lysozyme; 1% NP-40; 350 mM NaCl; 10 mM dithiothreitol) and lysed on ice for 30 min, followed by sonication. Proteins were purified from the E. coli extracts on glutathione-agarose beads as directed by the manufacturer (GE Healthcare, Piscataway, NJ). Whole-cell extracts were prepared from yeast strains LPY5, LPY9187, LPY12490, LPY12628, and LPY12630 in Sir2 IP lysis buffer (50 mM HEPES [pH 7.5]; 0.5 M NaCl; 0.5% NP-40; 10% glycerol; 1 mM EDTA; 1× PMSF, TPCK, leupeptin, pepstatin, and benzamidine). Whole-cell extracts (approximately 45 A600 cell equivalents) were mixed with ∼10 μg GST, GST-Sir2, and GST-sir2-R139K bound to glutathione-agarose beads and incubated at 4°C for 1 h. The beads were washed once with Sir2 lysis buffer and twice with wash buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA) and then boiled in 50 μl 1× sodium dodecyl sulfate (SDS) sample loading buffer. For hemagglutinin (HA) immunoblot analysis, 40 μl of sample was loaded onto 9% SDS-polyacrylamide gels (10 by 12 cm). Immunoblots were probed with a 1:5,000 dilution of mouse anti-HA monoclonal antiserum (Covance Research Products, Berkeley, CA). A horseradish peroxidase-coupled anti-mouse secondary antibody (Promega, Madison, WI) was used at 1:10,000 and detected with enhanced chemiluminescence reagents (Perkin-Elmer, Waltham, MA). For GST immunoblot analysis, 5 μl of sample was loaded onto a 10% SDS-polyacrylamide gel (8 by 10 cm). Immunoblots were probed with a 1:5,000 dilution of rabbit anti-GST antibody (Sigma-Aldrich, St. Louis, MO). A horseradish peroxidase-coupled anti-rabbit secondary antibody (Promega, Madison, WI) was used at 1:10,000 and detected with enhanced chemiluminescence reagents (Perkin-Elmer, Waltham, MA).
Generation of deletion mutants.
The Saccharomyces genome deletion project strain collection (71) obtained from Research Genetics was used to build most of the null strains constructed in this study. Additional information on this strain collection can be found at http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html. Strain 23709 was used for the construction of the slx5 deletion strains. Oligonucleotides 404 and 415 were used to amplify DNA containing the SLX5 flanking sequence and a Geneticin resistance cassette (kanMX). Strains LPY5, LPY408, LPY2446, and LPY4931 were transformed by an optimized lithium acetate method (50) with the following modifications. Twenty microliters of PCR product was mixed with 150 μl of cells and 20 μl of herring sperm DNA (10 mg/ml). Immediately following a heat shock at 42°C for 8 min, cells were resuspended in 100 μl 1 M sorbitol and 900 μl YPD and grown at 30°C for a 2- to 3-h recovery period. Geneticin (G418)-resistant transformants were selected by plating the recovered cultures onto YPD plates containing 100 μg/ml G418 (Mediatech Inc., Herndon, VA). Transformants were replica plated onto plates containing 200 μg/ml G418, and these colonies were subsequently streaked for singles onto plates containing 200 μg/ml G418. Correct deletion of the target gene at each specific genomic locus was verified by analytical PCR. The same procedure was used to make the slx8 deletion strains used. The ulp2Δ mutant with a silencing reporter and sir2Δ ulp2Δ double-mutant strains were derived partly from MHY1380, provided by M. Hochstrasser (38).
The ura3Δ0 strains used for reverse transcription-PCR analysis (see Fig. 4) were derived from DR1726 (51) crossed to isogenic sir2Δ or slx5Δ strains with the telomeric adh4::URA3-UAS reporter. Deletion of the nontelomeric URA3 gene was confirmed in candidate segregants by PCR with primers oLP#382, oLP#383, oLP#387, and oLP#389 (see Table S3 in the supplemental material).
FIG. 4.
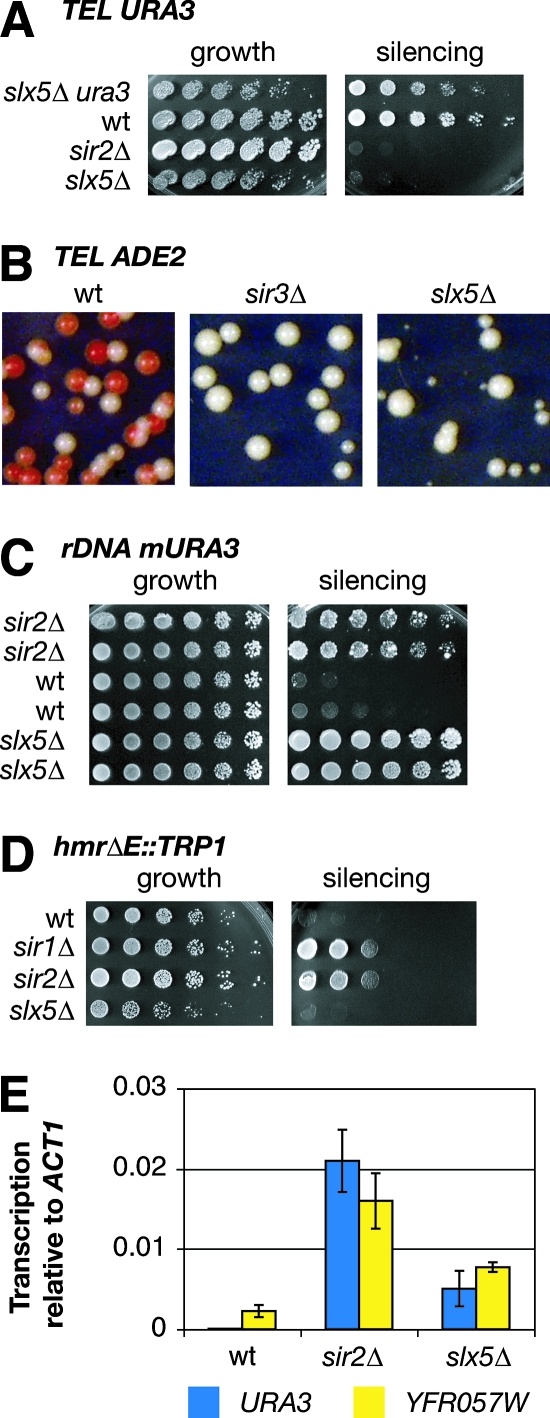
Slx5 contributes to transcriptional silencing. (A) Deletion of SLX5 disrupted telomeric silencing. URA3 gene expression is toxic on 5FOA. Wild-type (wt) yeast silenced the telomere VR-proximal URA3 reporter and grew on 5FOA (silencing), whereas silencing mutants such as the sir2Δ mutant did not. The slx5Δ mutant grew on 5FOA when it lacked the URA3 gene (top) but not in the presence of the telomeric URA3 reporter. (B) Red indicates silencing at the telomeric VR-proximal ADE2 reporter gene. Wild-type cells formed both red and white colonies; both the sir3Δ and slx5Δ mutants formed uniformly white colonies. Note the colony size heterogeneity of slx5Δ cells, a characteristic of the mutants that has been described previously (45). (C) Deletion of SLX5 disrupted rDNA silencing. Growth on medium lacking uracil (silencing) indicates the expression of an mURA3 reporter located in nontranscribed spacer 1 (NTS1) of one unit of the rDNA array. Two independent isolates each of the sir2Δ mutant, the wild type, and the slx5Δ mutant are shown. (D) Deletion of SLX5 did not disrupt mating-type silencing even in the presence of the hmrΔE mutation, which weakens silencing at the HMR locus. Growth on medium lacking tryptophan (silencing) indicates loss of silencing of a TRP1 reporter integrated at HMR, as in the sir1Δ and sir2Δ mutants but not the slx5Δ mutant. (E) Quantification of mRNA by reverse transcription and real-time molecular amplification. Experiments were performed with strains with a telomeric URA3 reporter gene and a chromosomal deletion of the ura3-1 locus. Bars represent the URA3 or YFR057W cDNA signal minus control reaction mixtures not containing reverse transcriptase, divided by ACT1 cDNA. No enrichment of URA3 signal was detected in the wild type. For the primer sequences, see Table S3 in the supplemental material. Error bars are standard deviations from two independent experiments.
ChIP.
Chromatin immunoprecipitations (ChIP) were performed as previously described (18). Cultures were grown to an A600 of 0.25 to 1.0, and protein-DNA complexes were cross-linked for 45 min with 0.86% formaldehyde. After quenching with 125 mM glycine and 0.2% ammonium hydroxide, cells were lysed with glass beads in 50 mM HEPES (pH 7.5)-0.5 M sodium chloride-0.5% Nonidet P-40-10% glycerol-1 mM EDTA-1 mM PMSF-2 mM benzamidine-1 mM leupeptin-1 mM pepstatin-1 mM TPCK and then DNA was broken by sonication into fragments approximately 500 bp long. Immunoprecipitation mixtures were incubated overnight at 4°C with polyclonal antiserum (2916/8) raised to a C-terminal peptide of Sir2 (60) or purified antibody to acetylated H4K16 peptide (Upstate, Charlottesville, VA). Efficiency of immunoprecipitations was assayed by Western blot assay. DNA in input and immunoprecipitated (IP) samples was quantified by real-time PCR on a DNA Engine Opticon 2 (MJ Research, Waltham MA). For the sequences of the primers designed for these experiments, see Table S3 in the supplemental material; primers for TEL6R-200, 25S, and 5S have been used previously (13). For each strain, the values reported are IP at each locus divided by the input average of all loci, normalized to the average IP/input at the nonspecific loci. Each datum point is the average of two or more experiments.
mRNA quantification.
For expression analysis, RNA was prepared by RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions, with the following modifications. Cells were grown in 25 ml rich medium to an A600 of 1 before harvest, suspended in 600 μl guanidine buffer with 1% β-mercaptoethanol, and then flash-frozen at −20°C; cells were lysed in 300-μl aliquots by 3 min of vortexing with glass beads. Reverse transcriptase reactions were performed with a TaqMan kit (Applied Biosystems, Foster City, CA) with oligo(dT) to prime the reactions. The cDNA preparations were then diluted 100-fold prior to real-time PCRs, performed as described for ChIP (see above), with the primers for ACT1, URA3, and YFR057W (see Table S3 in the supplemental material).
RESULTS
Interactions between Sir2 and Slx5 proteins.
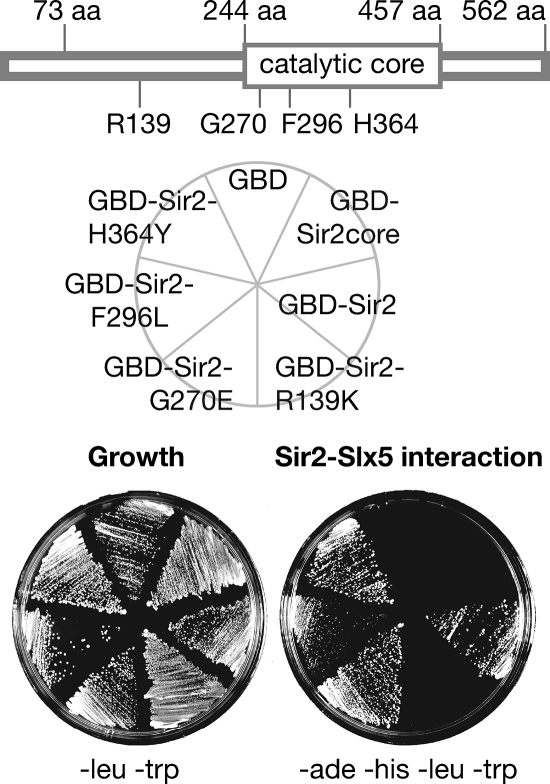
In search of proteins that contribute to specific cellular functions of Sir2, two different GBD (GBD1-147)-Sir2 fusion proteins were designed for two-hybrid assay analysis. GBD-Sir2core contains residues 244 to 457 of Sir2 and constitutes the sirtuin family conserved catalytic domain. GBD-Sir2 contains residues 74 to 562 of Sir2. Both fusion proteins were expressed and had the expected molecular weights, as determined by protein immunoblotting (not shown). The Δ73N GBD-Sir2 fusion rescued the mating defect of a sir2Δ haploid, although GBD-Sir2core did not (not shown), indicating that Δ73N GBD-Sir2 has biological activity that GBD-Sir2core lacks. A two-hybrid screen performed with Δ73N GBD-Sir2 produced a single interactor, the Slx5 protein.
In a directed test, the GAD-Slx5 fusion recovered from the screen did not interact with GBD-Sir2core (Fig. 1), nor did it interact with other members of the sirtuin family (data not shown). These data suggested that the sirtuin catalytic core is not sufficient for the interaction with Slx5. To determine whether Sir2 enzymatic activity is necessary for association with Slx5, four point mutations previously shown to impair Sir2 histone deacetylase function (17, 65) were tested for two-hybrid interactions in the Δ73N GBD-Sir2 construct. All GBD-Sir2 fusions were fully expressed, as determined by Western blot assay, except Sir2-H364Y, the expression of which was slightly reduced (not shown). The only point mutation found to disrupt the association, sir2-R139K, was outside the core domain (Fig. 1). Perhaps when mutated to lysine, the residue may be subject to some posttranslational modification that interferes with Slx5 association or with the transcriptional readout of the interaction. Thus, although Sir2 catalytic activity was dispensable for association with Slx5, the amino acid sequence N terminal to the core domain of Sir2 appeared to be critical for the two-hybrid interaction.
FIG. 1.
GAD-Slx5 interaction with GBD-Sir2. Sir2 is diagrammed above. The core contains residues 244 to 457 of Sir2. GBD-Sir2 contains residues 74 to 562. All sir2 point mutants were tested in this construct. GBD fusions were expressed from a TRP1-marked plasmid. GAD-Slx5 fusion was expressed from a LEU2-marked plasmid. The growth plate lacks leucine and tryptophan. The interaction plate additionally lacks adenine and histidine to assay simultaneously for activation of the reporter genes GAL1-HIS3 and GAL2-ADE2, which share minimal promoter sequence similarity yet are highly induced by the same activator, Gal4, thus eliminating promoter-specific false positives (29). Growth on this plate indicates a physical association between the GBD-Sir2 fusion and GAD-Slx5. Point mutants demonstrate that the interaction is not solely dependent on catalytic activity but is dependent on at least one residue outside the Sir2 catalytic domain.
Because Sir2 participates in multicomponent protein complexes in silent chromatin, it was possible that known Sir2 interactors mediated the Slx5 interaction. To address this idea, the two-hybrid assay was repeated with two strains in which nonessential Sir2 complex members had been deleted. Absence of SIR3 or SIR4 did not disrupt the Sir2-Slx5 two-hybrid interaction (data not shown). Thus, the two-hybrid interaction was independent of the partner Sir3 or Sir4 protein. Because the NET1 gene is essential, it was not included in this analysis. Both the Sir2/3/4 and Sir2-Net1 complexes have been characterized biochemically, and Slx5 was not reported to be a stable member of either complex (20, 26, 52). Therefore, Slx5 appears to be a new molecular partner for Sir2.
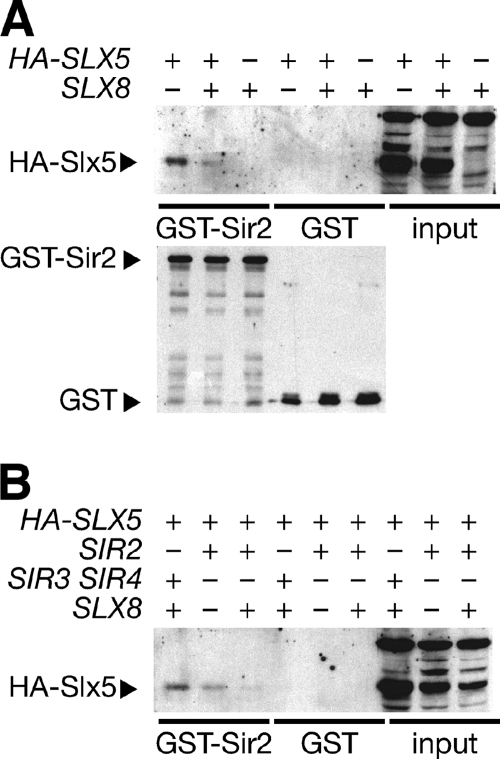
To determine whether the two-hybrid Sir2-Slx5 interaction could be validated independently, affinity binding experiments were performed with the GST moiety that binds glutathione-conjugated medium. Lysate of yeast cells expressing Slx5 with or without an HA epitope tag was mixed with GST or a GST-Sir2 fusion protein purified from bacteria. Western blotting for the HA epitope revealed that the GST-Sir2 fusion, but not GST alone, bound the HA-Slx5 fusion (Fig. 2A). Binding of Slx5 to the Sir2-R139K mutant protein that did not support the two-hybrid interaction was slightly diminished (not shown). Sir2 R139K is defective for interaction with Net1 (17), suggesting that Slx5 and Net1 may interact with the same region of Sir2.
FIG. 2.
Evaluating Sir2 and Slx5 interaction in vitro. (A) GST and GST-Sir2 were purified from bacteria with GST-Sepharose. Purified proteins were incubated with whole-cell extracts from yeast with or without Slx8 or chromosomally tagged HA-Slx5. Bound protein was subjected to SDS-polyacrylamide gel electrophoresis. Immunoblot analysis of HA-Slx5 was performed with anti-HA monoclonal antibody. Note the enhanced interaction in the slx8Δ mutant, perhaps reflecting decreased competition for binding. (B) The same experiment was performed in the absence of Sir3 and Sir4, Slx8, or Sir2. As before, deletion of SLX8 enhances the interaction, even in the absence of Sir3 and Sir4. Although the interaction appeared to be enhanced in the absence of Sir2, more Slx5 was apparent in the input as well. Immunoblot analysis of GST was performed with anti-GST polyclonal antibody.
We noted that only a small amount of Sir2 and Slx5 interacted, consistent with the presence of most Sir2 in the RENT and TEL complexes that do not appear to contain Slx5. When extracts were prepared from sir2Δ or slx8Δ strains, Slx5 binding was increased, although in the sir2Δ mutant this may have been due to a modestly increased Slx5 abundance. Sir3 and Sir4 were not required for the interaction (Fig. 2B), consistent with the lack of a requirement for Sir3 or Sir4 in the two-hybrid assay. We note that these results do not necessarily indicate direct binding of Sir2 to Slx5, as other proteins present in yeast lysate may bridge the interaction.
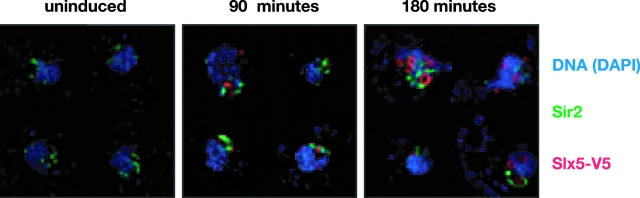
Slx5 has been reported to have a relatively uniform nuclear localization when evaluated by indirect immunofluorescence (76). By comparison, we observed that when epitope-tagged SLX5 expression was turned on in a null mutant background, at early times of expression, the protein appeared not to be evenly distributed but rather to occupy distinct foci. By 3 h, the foci became ring-like structures that did not coincide with the Sir2-associated nucleolus or the telomeres (Fig. 3). Thus, the Sir2-Slx5 interaction is not stoichiometric or fully overlapping, yet it is likely to be functionally significant, as detailed below.
FIG. 3.
Epitope-tagged Slx5 protein resided in the nucleus in structures distinct from the telomeres or nucleolus. Strain LPY8908 contains plasmid pLP1737 with the SLX5-V5 construct on a GAL1 promoter (45). Cells were grown in 2% raffinose to late log phase. To induce, galactose was added to 2% and cultures were incubated at 30°C for the indicated times. Indirect immunofluorescence was performed as previously described (14), with antiserum to Sir2 (fluorescein isothiocyanate-conjugated secondary antibody, in green) and antiserum to the V5 epitope (Texas Red-conjugated secondary antibody, in red). DNA is stained with DAPI (blue).
SLX5 has a distinct role in transcriptional silencing.
The interaction with Sir2 suggested that Slx5 might influence transcriptional silencing, although this function had not previously been ascribed to it, nor did any previous screens for silencing genes uncover SLX5. Silencing in slx5Δ mutants was assayed with well-characterized reporter genes at each of the major silenced loci. To assess telomeric silencing, ADE2 and URA3 reporter genes were used (reviewed in reference 68). Cells with a URA3 reporter gene located at telomere VR grow on 5FOA only if they have functional silencing. Otherwise, expression of the URA3 gene product converts 5FOA into a toxic metabolite. As a control, growth of slx5Δ mutants lacking the URA3 reporter on plates containing 5FOA demonstrated that slx5Δ mutants are not inherently sensitive to the drug (Fig. 4A). However, in the presence of the telomeric URA3 reporter, deletion of SLX5 caused extreme 5FOA sensitivity comparable to that seen in sir2Δ mutants. In an independent assay, loss of silencing in slx5Δ strains was also seen when monitoring an ADE2 reporter gene integrated at telomere VR. Absence of the ADE2 product results in accumulation of red pigmentation. Normal telomeric position effect causes variegation of wild-type cells, resulting in white and sectored colonies. Defective telomeric silencing, as in sir3Δ mutants, gives rise to uniformly white colonies. On rich medium, slx5Δ mutant colonies were fully defective for telomeric ADE2 silencing, as indicated by their white color (Fig. 4B).
In addition to telomeric silencing, chromatin-mediated transcriptional silencing also occurs at the rDNA and the HM mating-type loci. To assay rDNA silencing, a URA3 reporter was again used. The mURA3 gene with a weakened promoter has been integrated into nontranscribed spacer 1 (NTS1) of a single rDNA repeat (59). Wild-type yeast did not express mURA3 and did not grow on medium lacking uracil, but both sir2Δ and slx5Δ mutants grew robustly (Fig. 4C). This indicated that, as at telomeres, slx5Δ mutants were defective in transcriptional silencing at the rDNA. However, unlike sir1Δ or sir2Δ mutants, slx5Δ mutants had no expression of a TRP1 gene integrated at HMR (Fig. 4D) or any lack of mating competence (not shown). Together, these results indicate that Slx5 was required for transcriptional repression at some, but not all, loci subject to Sir2-dependent silencing.
The silencing defects above represent endpoint assays with reporter genes. To test the degree to which deletion of SLX5 altered the expression of an endogenously silenced gene, the transcription of one such gene was studied by real-time transcriptional analysis. The gene encoding the unnamed open reading frame YFR057W, located approximately 1 kb from telomere VIR, was previously reported to be regulated by SIR2 (75). By using the same technique of reverse transcription and quantitative molecular amplification, we also observed that deletion of SIR2 increased the transcription of YFR057W (Fig. 4E). Likewise, the slx5Δ mutant showed an increase in YFR057W transcription, although the magnitude of this effect was less than that seen in sir2Δ mutant cells. YFR057W transcription in the sir2Δ slx5Δ double mutant equaled 96.9% ± 21.6% of that in the sir2Δ single mutant (not shown). Thus, it appears that the contributions of Sir2 and Slx5 to telomeric silencing are not synergistic.
Relative to the transcription of the ACT1 control, telomeric silencing was diminished in the slx5Δ mutant. However, the degree of the effect observed indicated that the Sir2-mediated silencing pathway remained partly functional, not as expected from the URA3 and ADE2 silencing assays. To determine whether slx5Δ mutants had the same degree of effect on silencing at the telomeric URA3 reporter, transcriptional analysis was performed with strains bearing a complete deletion of the endogenous chromosomal URA3 locus. Consistent with the effect on YFR057W, slx5Δ strains had intermediate expression of the telomeric URA3 gene (Fig. 4E).
A similar result was obtained for rDNA silencing. Sir2-mediated silencing in the rDNA suppresses unequal rDNA recombination (33). An assay for unequal rDNA recombination is the frequency of loss of an ADE2 gene inserted into a single copy of the rDNA repeats (21). By this assay, a sir2Δ mutant had a 20-fold increase in the rate of loss of an individual rDNA repeat (Table 1), consistent with previous reports (5, 21, 59). However, the slx5Δ mutant had a less-than-twofold increase in the rate of marker loss. The double-mutant marker loss rate was comparable to that of the sir2Δ mutant. Thus, the modest defect of slx5Δ mutants is consistent with a previous report that they do not accumulate rDNA circles faster than the wild type (30) and indicates that Sir2 function in the rDNA is largely intact in the absence of Slx5.
TABLE 1.
rDNA gene ADE2 marker loss
| Strain | Genotype | % Sectoreda | Probability equal to wild type |
|---|---|---|---|
| LPY4931 | Wild type | 0.30 | 1 |
| LPY4981 | sir2Δ | 6.33 | <0.0001 |
| LPY8790 | slx5Δ | 0.56 | 0.0074 |
| LPY10564 | sir2Δ slx5Δ | 5.09 | <0.0001 |
Molecular hallmarks of silencing are intact in slx5Δ mutants.
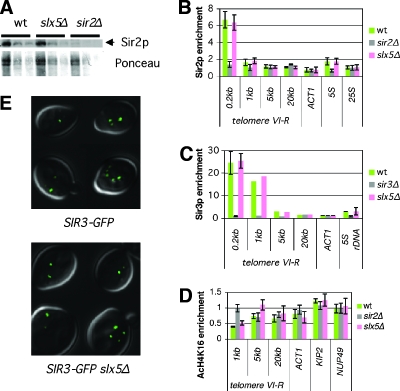
One conjecture from the physical interaction data to explain the SLX5 dependence of silencing is that Slx5 has a global effect on Sir2 abundance or activity and different silenced loci and assays differ in sensitivity to Sir2. Protein immunoblotting with antisera to Sir2 revealed no change in the Sir2 protein level in the absence of Slx5 (Fig. 4A), so any global effect of Slx5 on Sir2 did not alter protein abundance. Another model is that Slx5 promotes Sir2 recruitment or activity at some genomic loci but not others. To test this idea, Sir2 abundance at silenced loci was assayed by ChIP.
Previous observations have shown modest changes in Sir2-silenced chromatin composition at telomere VIR associated with clear differences in silencing (32, 61). Therefore, primers were designed to study Sir2 ChIP at telomere VIR. The slx5Δ mutant displayed neither an increase in the distance from the telomere over which Sir2 associated nor a significant decrease in Sir2 abundance near the telomere (Fig. 5B). The same results were seen for Sir3 (Fig. 5C) and Sir4 (not shown). In addition, Sir2 itself remained associated with the rDNA in the absence of Slx5 (Fig. 5B). Therefore, Slx5 did not significantly alter Sir2 occupancy at either of the Slx5-silenced loci.
FIG. 5.
Molecular hallmarks of telomeric silencing are largely intact in slx5Δ mutants. (A) Sir2 was expressed at the wild-type (wt) level in slx5Δ cells. The top part of the panel is an anti-Sir2 protein immunoblot assay showing Sir2 in lysates produced from four, two, or one relative amount of cells; the bottom part of the panel is Ponceau stain as a loading control. (B) Silencing protein occupancy at telomere VIR was evaluated by ChIP with antiserum to Sir2. Sir2-immunoprecipitated (IP) DNA was quantified by real-time molecular amplification. Error bars equal 1 standard deviation of three to five independent experiments at each locus. (C) ChIP with antiserum to Sir3. Error bars, where shown, equal 1 standard deviation from two independent experiments. (D) Histone modification state was evaluated by ChIP with antiserum to acetylated histone H4 lysine 16. Error bars equal 1 standard deviation from two independent experiments. The primers for 0.2 kb from telomere VIR were not included because the AcH4K16 immunoprecipitation was less efficient at that locus in all of the strains tested, suggesting altered nucleosome occupancy at that position. (E) Sir3GFP fusion protein, expressed from an in-frame integration of the GFP coding sequence into the chromosomal SIR3 locus (27), was visualized in live cells. Three-dimensional deconvolution was used to resolve telomeric Sir3GFP foci. Each image is a collage of four representative z sections superimposed on their corresponding differential interference contrast images. In addition to the apparently normal cells shown here, the slx5Δ population included many dead cells and misshapen cells that did not have clear Sir3GFP foci.
Because Sir2 recruitment appeared intact, another possibility was that Sir2 catalytic activity was regulated by Slx5 in the absence of any change in Sir2 abundance. However, deacetylation of telomeric H4K16, a mark of Sir2-dependent silencing that is thought to be inherently repressive to transcription, was not significantly affected in slx5Δ mutants (Fig. 5D). Thus, known molecular markers of silent chromatin were intact in the absence of Slx5, even as silencing was functionally disrupted.
Cytological markers of silencing are telomeric foci, where telomeres cluster at the nuclear periphery (19). To determine if these were intact in slx5Δ mutants, immunofluorescence was performed for Sir2 (not shown), and both Sir3GFP (Fig. 5E) and Sir4GFP (not shown) fusion proteins were imaged in live cells. A large fraction of slx5Δ mutant populations consists of dead and inviable, morphologically abnormal cells (discussed below), and in these cells, no foci were observed. However, in slx5Δ cells of normal morphology, telomeric foci were observed. Thus, loss of Slx5 function does not perturb large-scale telomeric structure in viable cells. This result was in keeping with reports that deletion of SLX5 does not have a severe effect on telomeric integrity (3; see Fig. S1 in the supplemental material). Together, these findings indicate that traditionally defined telomeric silenced chromatin was largely intact in the absence of Slx5, although silencing was not.
Enhancing Sir2-dependent telomeric silencing in slx5Δ mutants.
Sir2 deacetylase activity is limiting for silencing (60). Although slx5Δ mutants had moderate silencing defects, slx5Δ mutants had no defect in Sir2 function in silencing, as assayed by ChIP. This discrepancy suggested that in an slx5Δ mutant, some factor other than Sir2 might become limiting for silencing. To test this prediction, three genetic interventions that promote Sir2-dependent silencing were performed in slx5Δ mutants.
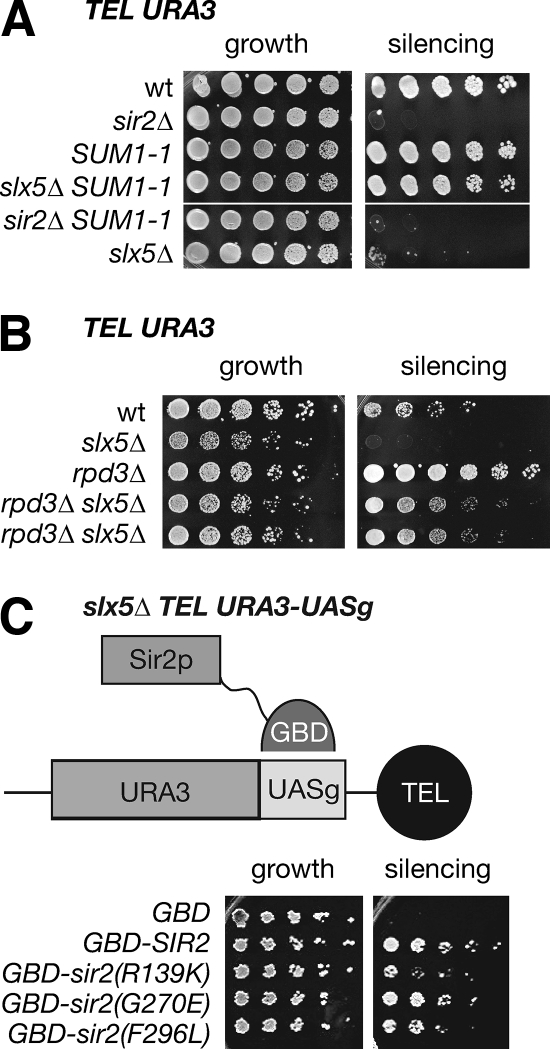
First, the dominant mutation SUM1-1 has been shown to enhance telomeric silencing in SIR2 but not sir2Δ yeast (8). Sum1 is a promoter-specific repressor that functions with the sirtuin Hst1. When mutated to SUM1-1, the Sum1/Hst1 complex behaves like the Sir2/3/4 complex, binding deacetylated H4 tails and spreading on chromatin (42, 64). As shown in Fig. 6A, SUM1-1 caused strengthened silencing in slx5Δ mutants, as well as wild-type yeast, but not in the absence of SIR2.
FIG. 6.
Restoring silencing in slx5Δ mutant strains. (A) Although deletion of SLX5 caused expression of the telomeric URA3 reporter and toxicity on 5FOA-containing medium, this phenotype was fully suppressed by SUM1-1. wt, wild type. (B) Deletion of RPD3 improves telomeric silencing, as previously reported (63), and bypasses the requirement for SLX5. Single mutants and two independently derived rpd3Δ slx5Δ double mutants are shown. (C) Tethering Sir2 near the telomere restores silencing to slx5Δ mutants. The GBD-Sir2 fusion binds the UASg sequence placed downstream of the URA3 reporter as diagrammed above. Presence of the GBD-Sir2 fusion construct restored silencing (growth on 5FOA) in the slx5Δ mutant, even by Sir2 mutant proteins with reduced catalytic activity (17).
Second, deletion of RPD3 also has a positive effect on silencing (12, 53, 69). Although RPD3 encodes a histone deacetylase, its deletion causes enhanced silencing, which requires the Sir2/3/4 complex (63). The rpd3Δ slx5Δ double mutant displayed strong silencing of the telomeric URA3 reporter (Fig. 6B), indicating that the Sir2 and Sir2 complex functions remained intact.
Finally, we found that Sir2 artificially recruited to a telomeric reporter gene could bypass the requirement for Slx5 in silencing. In this case, the GBD-Sir2 fusion was used in conjunction with a previously described telomeric URA3 gene with a 3′-proximal Gal4 DNA-binding site (UASg) (9). As before, in the absence of SLX5, the URA3 gene was expressed and cells were inviable on 5FOA (Fig. 6C). However, tethering full-length Sir2 to the UASg element restored silencing of the URA3 gene. Together, these three experiments indicated that, despite the reduced responsiveness of slx5Δ mutants to Sir2-mediated silencing, strength of telomeric silencing remained a function of Sir2 availability.
SIR2 and SLX5 contribute to maximal growth.
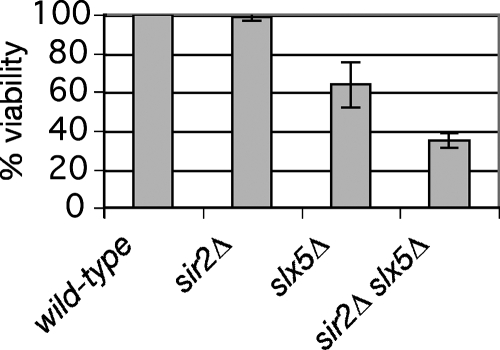
Although slx5Δ mutants have an obvious growth defect (6, 45), no previous observations suggest a role for Sir2 in promoting optimal growth of populations of cells. To test for functional interactions with SIR2, sir2Δ slx5Δ double mutants were constructed. Because the double-mutant strains had a more severe growth defect than slx5Δ single mutants on plates, quantitative analyses were performed to determine single-cell viability and growth rates in culture.
Micromanipulation of normal-looking G1- or S-phase (unbudded) cells taken from cultures undergoing logarithmic growth was used to determine the fraction of freshly divided cells that are competent for growth. Of the wild-type cells selected, all formed colonies (Fig. 7). However, only 97 of 152 slx5Δ mutant G1/S founder cells formed visible colonies. Those that failed to grow into visible colonies typically formed microscopic colonies or clumps of abnormal cells, often incompletely divided or lysed. Of 192 double-mutant founder cells examined, only 67 formed visible colonies. Furthermore, the double mutants displayed markedly slower growth in culture: a 3.98-h doubling time for the homozygous sir2Δ slx5Δ diploid, compared to 2.63 h for the slx5Δ homozygote (Table 2; see Fig. S2A in the supplemental material). Because sir2Δ slx5Δ populations do undergo logarithmic growth, the rate at which freshly divided cells become inviable cannot be 50% or more a generation; therefore, the unbudded fraction of a double-mutant population must include many that arrested as G1/S cells in a previous generation.
FIG. 7.
G1/S-phase cells mutated for SLX5 have a reduced ability to form colonies. Individual cells were taken from a liquid culture growing logarithmically and positioned on rich medium by micromanipulation. Resulting colony growth from these founder cells was monitored microscopically after incubation at 30°C. Error bars are based on two or three replicates from independent cultures.
TABLE 2.
Growth rates in rich medium at 30°C
| Strain | Genotype | Time (h)a | R2b |
|---|---|---|---|
| LPY11031c | Wild-type haploid | 1.58 | 0.9925 |
| LPY94 | sir1Δ haploid | 1.56 | 0.9987 |
| LPY11034c | sir2Δ haploid | 1.58 | 0.9884 |
| LPY10 | sir3Δ haploid | 1.72 | 0.9951 |
| LPY9 | sir4Δ haploid | 1.45 | 0.9981 |
| LPY11033c | slx5Δ haploid | 2.10 | 0.9847 |
| LPY11738c | sir1Δ slx5Δ haploid | 2.14 | 0.9998 |
| LPY11032c | sir2Δ slx5Δ haploid | 3.12 | 0.9823 |
| LPY11343c | sir3Δ slx5Δ haploid | 3.10 | 0.9234 |
| LPY10547c | sir4Δ slx5Δ haploid | 3.34 | 0.9527 |
| LPY1552 | Wild-type diploid | 1.81 | 0.9869 |
| LPY10370 | sir2Δ diploid | 1.75 | 0.9972 |
| LPY10545 | slx5Δ diploid | 2.63 | 0.9868 |
| LPY10546 | sir2Δ slx5Δ diploid | 3.98 | 0.9827 |
| LPY4931 | ADE2 haploid | 1.39 | 0.9968 |
| LPY8790 | ADE2 slx5Δ haploid | 1.93 | 0.9986 |
| LPY12073 | ADE2 SUM1-1 haploid | 2.96 | 0.9949 |
| LPY12076 | ADE2 slx5Δ SUM1-1 haploid | 2.69 | 0.9863 |
| LPY12074 | ADE2 sir2Δ slx5Δ SUM1-1 haploid | 2.82 | 0.9929 |
| LPY11980 | Wild-type cir0 haploid | 1.43 | 0.9948 |
| LPY11981 | sir2Δ slx5Δ cir0 haploid | 1.99 | 0.9994 |
| LPY11982 | slx5Δ cir0 haploid | 1.70 | 0.9953 |
| LPY12569 | slx8Δ haploid | 2.18 | 0.9992 |
| LPY12571 | sir2Δ slx8Δ haploid | 2.51 | 0.9984 |
| LPY12633 | siz1Δ siz2Δ haploid | 1.92 | 0.9973 |
| LPY12634 | sir2Δ siz1Δ siz2Δ haploid | 2.18 | 0.98 |
Calculated generation time is shown. See Fig. S2 in the supplemental material for a graphical representation.
Square of correlation coefficient of growth curve fitted to data.
After selection on 5FOA.
Molecular basis for SIR2-SLX5 genetic interaction.
To better define the functional overlap between Sir2 and Slx5, we evaluated slx5Δ mutants in the context of sir2Δ phenotypes. Sir2 is essential for transcriptional silencing, thereby enabling mating and promoting longevity, and it contributes to nonhomologous end-joining double-strand break repair (36), sequestration of oxidative damage in mother rather than daughter cells (1), and timing of replication initiation (47). In the case of HM silencing, there are several ways to mimic the defect of sir2Δ mutants and ask whether slx5Δ mutant growth is thereby impaired.
Diploid yeast expresses both MATa and MATα mating-type information, yet the sir2Δ slx5Δ double mutant was as sick in homozygous diploids as in haploids (Table 2). Heterologous expression of both mating-type cassettes did not impede the growth of slx5Δ haploids relative to that of SLX5 haploids (not shown), nor did deletion of HML α information enhance the growth of MATa sir2Δ slx5Δ haploids (not shown). Furthermore, deletion of SIR1, which is required for HM silencing but not telomeric or rDNA silencing, had no detrimental effect on the growth of slx5Δ mutants (Table 2; see Fig. S2B in the supplemental material). Therefore, the contribution of Sir2 to growth in the absence of SLX5 is not HM silencing.
The role of Sir2 in the rDNA has been well characterized and is known to be independent of the Sir3 and Sir4 proteins. In fact, Sir4 opposes Sir2 function in the rDNA by recruiting it to other loci, such as the telomeres (60). If Sir2 activity in the rDNA were important for growth in the absence of SLX5, deletion of SIR3 or SIR4 should not have a negative effect on slx5Δ mutant growth. Instead, both double mutants were synthetic sick, just as the sir2Δ slx5Δ double mutant was (Table 2; see Fig. S2C in the supplemental material). The Sir2 contribution to growth in the absence of SLX5 thus is not likely to be solely rDNA silencing but is likely in a pathway shared with Sir3 and Sir4, such as telomeric silencing.
Since SUM1-1 restores telomeric silencing in the absence of SLX5, it was possible that the same mutation would restore maximal growth to sir2Δ slx5Δ double mutants, although SUM1-1 sir2Δ double mutants lack telomeric silencing. SUM1-1 mutants themselves have a growth defect that is more severe than that of slx5Δ mutants (Table 2; see Fig. S2D in the supplemental material) and share the slx5Δ phenotypes of defective chromosomal maintenance and an increased rate of cell death (8). In fact, SUM1-1 mutants grew at the same rate as sir2Δ slx5Δ double mutants (Table 2; see Fig. S2D in the supplemental material). Moreover, deleting SLX5 had no effect on the SUM1-1 mutant growth rate, even in combination with deletion of SIR2 (not shown). Therefore, the growth defect of sir2Δ slx5Δ double mutants may be due to a process similar to that which is defective in SUM1-1 mutants.
Linking regulators of sumoylation to silencing and Sir2-dependent growth.
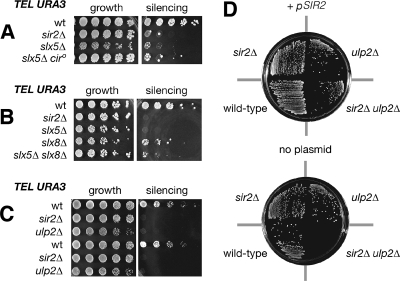
We also evaluated sir2Δ mutants in the context of published slx5Δ phenotypes. Deletion of SLX5 causes sensitivity to hydroxyurea and is synthetic lethal with deletion of SGS1 (45). However, the sir2Δ sgs1Δ double mutant was not synthetic lethal (not shown) and sir2Δ mutants grew as wild type in the presence of 100 mM hydroxyurea (not shown). As previously reported (45), slx5Δ mutants grow heterogeneously on plates (Fig. 4B, compare the slx5Δ mutant to the wild type). This heterogeneous growth is common to mutants with changes in several genes affecting sumoylation and is due to a defect in 2μm plasmid maintenance (6). To test whether the slx5Δ mutant phenotypes that we discovered were due to aberrant 2μm plasmid maintenance, we cured slx5Δ and sir2Δ slx5Δ strains of endogenous 2μm plasmids by the method of Tsalik and Gartenberg (66). Neither slx5Δ mutant telomeric silencing (Fig. 8A) nor the synthetic growth defect of sir2Δ slx5Δ mutants (Table 2) was rescued by elimination of endogenous 2μm plasmids.
FIG. 8.
Silencing defect of slx5Δ mutants is linked to the SUMO pathway. (A) The 2μm plasmid is not required for the silencing defect of the slx5Δ mutant. A 5FOA silencing assay was performed as described in the legend to Fig. 4. As shown in the left part of the panel, curing slx5Δ mutants of the 2μm plasmid (cir0) rescued clonal lethality; however, the right part of the panel indicates that silencing was not restored. (B) The slx8Δ mutant has a moderate silencing defect. The slx5Δ slx8Δ double mutant also has a silencing defect, indicating that loss of silencing is not due to unregulated activity of one complex member in the absence of the other. (C) Silencing of a telomeric URA3 reporter is lost in ulp2Δ mutants. (D) Mutants of SIR2 require ULP2 for viability. Haploid products of a heterozygous diploid (SIR2/sir2Δ::TRP1 ULP2/ulp2Δ::HIS3 + URA3 SIR2 plasmid) with the indicated genotypes and the URA3 SIR2 plasmid pLP37 were grown in medium lacking uracil (+ pSIR2) to maintain the plasmid or in 5FOA (no plasmid) to select against the plasmid. Little to no viability was observed for multiple independent double mutants.
A large fraction of Slx5 occurs in a complex with Slx8 (76). Because Slx8 competed with the Sir2-Slx5 affinity precipitation interaction, it seemed possible that slx5Δ phenotypes relating to Sir2 function might be independent of Slx8. We therefore constructed slx8Δ mutant strains either with silencing reporters or as sir2Δ double mutants. As assayed by URA3 reporter genes at both telomeres and rDNA, the slx8Δ mutants had silencing defects similar to the slx5Δ silencing defects (Fig. 8B). Thus, both subunits of the Slx5/8 complex were required for full silencing at telomeres and rDNA. By contrast, no synthetic sickness was observed for sir2Δ slx8Δ double mutants on plates (not shown). In liquid culture, slx8Δ mutants grew slightly slower when SIR2 was also deleted (Table 2). Although SIR2 was required for robust growth in the absence of either subunit of the Slx5/8 complex, SIR2 had a greater effect on the growth of slx5Δ mutants than on that of slx8Δ mutants. Therefore, some contribution of SLX5 to sir2Δ growth may be independent of SLX8.
Recent studies report that the Slx5/8 complex promotes ubiquitination of sumoylated proteins, leading to their degradation in the proteasome (67, 73). Mutants lacking SLX5 or SLX8 have increased global levels of sumoylated proteins (6, 28, 70). Because both slx5Δ and slx8Δ mutants had silencing defects, we hypothesized that the silencing defect and perhaps the genetic interaction with sir2Δ mutants could be due to an excess of a target sumoylated protein. Siz1 and Siz2 are the two best-characterized SUMO E3 ligases in yeast. We therefore deleted SIR2 in a siz1Δ siz2Δ double-mutant strain to determine whether the sir2Δ mutation was deleterious in combination with any interference with the SUMO conjugation pathway. The sir2Δ siz1Δ siz2Δ triple mutant grew similarly to the siz1Δ siz2Δ double mutant, which has a slight growth defect (Table 2).
ULP2/SMT4 encodes a SUMO isopeptidase, and ulp2Δ mutants have elevated levels of sumoylated proteins, similar to slx5Δ mutants (38, 70). Therefore, we tested whether ulp2Δ mutants shared the phenotypes of defective telomeric silencing or genetic interaction with the sir2Δ mutation. As assayed by a URA3 reporter, telomeric silencing was defective in an ulp2Δ mutant (Fig. 8C). Thus, both slx5Δ and ulp2Δ silencing defects may be due to excessive sumoylation of some target protein.
Similar to slx5Δ mutants, ulp2Δ mutants had a growth defect in liquid culture, with doubling times ranging from 2.89 to 3.98 h. An even more severe growth defect was observed in the sir2Δ ulp2Δ double mutant (Fig. 8D). In standard plasmid shuffle experiments, the double mutant appeared inviable, with only rare colonies arising that we interpreted as suppressors. When attempting to perform growth rate measurements for ulp2Δ or ulp2Δ sir2Δ mutant yeast as in Table 2, we found significant variability between cultures, perhaps reflecting the suppressors. The rare sir2Δ ulp2Δ double-mutant survivors from selection on solid medium had doubling times ranging from 4.13 to 5.46 h in liquid culture. In liquid medium, the sir2Δ slx5Δ ulp2Δ triple mutant had doubling times of 5.78 to 7.22 h. These findings supported the hypothesis that sir2Δ mutants depend on the Slx5/8 ubiquitination pathway for maximal growth.
The other SIR genes were deleted in combination with ulp2Δ to see whether ULP2 had the same pattern of interactions as SLX5. Growth of sir1Δ ulp2Δ and sir4Δ ulp2Δ double mutants was not obviously defective on solid medium (not shown). No sir1Δ ulp2Δ double mutant grew more slowly than the slowest ulp2Δ single mutants. However, the doubling times of the sir4Δ ulp2Δ double mutants ranged from 3.50 to 5.78 h. Thus, the sir4Δ ulp2Δ growth defect was potentially as severe as the sir2Δ ulp2Δ growth defect. These data were consistent with the model in which Ulp2 and Slx5 act in a pathway that promotes telomeric silencing and, in the absence of this pathway, all components of the Sir2/3/4 complex are required for optimal growth.
DISCUSSION
Sir2 and other members of the sirtuin protein family are the subject of intense study because of their enzymatic activity and reports that their roles in regulation of metabolism and aging may be broadly conserved. Long-standing interest in the SIR2 gene itself originally came from its identification as a key player in transcriptional silencing (reviewed in reference 41).
For several decades, studies with yeast have provided important insight for uncovering broader mechanisms of transcriptional silencing, including a basic model whereby Sir2 deacetylates H4K16, which is followed by recruitment of other silencing factors. Yet recent results, along with those presented in this study, now challenge the model in which Sir2-mediated H4K16 deacetylation alone is sufficient for transcriptional silencing. For example, the O-acetyl ADP ribose product of the NAD-dependent deacetylation reaction appears to facilitate Sir3 oligomerization (39). Furthermore, bypass of Sir2's catalytic activity in silencing through constitutively deacetylated nucleosomes fails to provide complete silencing in the presence of a catalytically inactive Sir2 mutant protein, suggesting an additional role for Sir2 (75). There is growing evidence that other posttranslational histone modifications, such as ubiquitination, methylation, sumoylation (reviewed in reference 4), and Sir2-regulated H3K56 deacetylation (74), are positively or negatively associated with silenced loci. These modifications may affect H4K16 deacetylation, affect the transduction of histone deacetylation into inhibition of transcription, or both. Certainly, it is true that the molecular picture of Sir2-silenced chromatin is incomplete.
At an organismal level, yeast cells receive multiple benefits from silent chromatin. These include mating capacity as haploids and suppression of unequal recombination that, if uncontrolled, can lead to toxic rDNA circle accumulation. Transcriptional silencing occurs at telomeres in both yeast and animals, suggesting that it serves a conserved function. SIR2 is essential for telomeric silencing yet has little reported effect on telomere integrity. Sir2 is involved, however, in DNA repair by nonhomologous end joining, as are other telomere-associated proteins (reviewed in reference 37). This suggests that there may be additional conserved functions of telomeric silencing for which the molecular basis is not established. Insight into both the molecular mechanism of silencing and its conservation is provided by the identification of SLX5 as a gene necessary for full transcriptional silencing and growth potential in the absence of SIR2.
A new role for SLX5 in transcriptional silencing.
In vivo assays revealed defective telomeric and rDNA silencing in slx5Δ mutants. Curiously, the most well-defined molecular hallmarks of silencing dependent on SIR2 were intact. Genetic experiments and ChIP showed that Sir2 was present and active at silenced loci and that silencing was responsive to conditions that enhance Sir2 recruitment. The question of how Slx5 affects silencing is not yet resolved. Slx5 may subtly promote Sir2 activity or affect silencing at previously undefined steps downstream of Sir2-mediated histone deacetylation. Given the natural variation in wild-type silencing observed in sectoring assays (reviewed in reference 68) and the decreased viability of slx5Δ mutants, another possibility is that there is selection against strong silencing in slx5Δ populations.
Linking sumoylation and silencing through Sir2.
Multiple lines of evidence suggest that Slx5 affects silencing through its contributions to sumoylation, ubiquitination, and protein degradation as part of the STUbL complex with Slx8. In fission yeast, sumoylation of heterochromatic proteins is required for full transcriptional silencing (57). Sumoylation of histones is enriched at the telomeres in budding yeast (46). We have now shown that Slx5, Slx8, and the SUMO isopeptidase Ulp2 all contribute to telomeric silencing. Although sumoylation has been correlated with transcriptional repression (24, 43), biochemical evidence indicates that Slx5, Slx8, and Ulp2 all negatively regulate sumoylation (6, 67, 70, 73). Slx5 and Slx8 promote ubiquitination (67, 73), and deubiquitination of histone H2B by Ubp10 promotes telomeric silencing (13). We did not find an effect of Slx5 on global ubiquitinated H2B levels (not shown). Formation of SUMO chains has been shown to be a key requirement for Slx5/8-dependent ubiquitination (67). Although polysumoylation of histones has been reported (46), further studies are required to understand its mechanistic significance.
Another group of potential targets for Slx5/8 and Ulp2 activity are the Sir proteins. Both Sir3 and Sir4 are sumoylated (11, 72). Whether Sir2 is sumoylated is not clear. Although it was retrieved from the same proteomic screens that identified Sir3 and Sir4, Sir2 was a low-scoring candidate and did not appear to be sumoylated in a targeted biochemical assay (11). However, the human sirtuin SIRT1 is sumoylated and this modification affects its activity in vitro (77). Because the Slx5/8 complex is now known to promote ubiquitination and subsequent degradation of sumoylated proteins, assessing the proteomic and targeted biochemical sumoylation assays in the absence of Slx5 or under inhibition of the proteasome should provide a more complete atlas of proteins modified by sumoylation.
Slx5 defines an unusual nuclear structure when induced.
It has been reported that constitutively expressed Slx5 has a relatively uniform nuclear localization when evaluated by indirect immunofluorescence (76). By comparison, we observed that when SLX5 expression was turned on in a null mutant background, at early times of expression, the protein appeared not to be evenly distributed but rather to occupy distinct foci. By 3 h, the foci became ring like (Fig. 3) and did not coincide with the Sir2-associated nucleolus or the telomeres. Instead, the Slx5 pattern of staining appears to define a new nuclear structure or compartment.
To date, only a few distinct patterns of subnuclear localization have been described in yeast, such as Rad52 and Rad53 foci, which appear in S phase (15, 40). Mammals, on the other hand, have numerous subnuclear bodies, such as promyelocytic leukemia (PML) and Cajal bodies. A number of subnuclear body components, including the PML protein itself, have RING domains, as does Slx5, the integrity of which is required for function (70). One RING domain protein associated with PML bodies is RNF4 (23), which has recently been identified as a human member of the STUbL family, to which Slx5 and Slx8 also belong (49, 62). Intriguingly, Schizosaccharomyces pombe STUbL protein Rfp1, thought to be an Slx5 homolog, also forms distinct nuclear foci when overexpressed ectopically (49). It has also been observed that a yeast SUMO ligase complex localizes to a subset of nuclear pore complexes (78). This complex contains several RING domain proteins and influences transcriptional silencing and is thus another example, like Slx5, of a telomeric silencing factor the localization of which is not restricted to telomeres.
A new role for silencing factors in promoting growth.
It had previously been reported that high levels of Sir2 could be lethal (25), although no essential requirement for SIR2 in mitotic cells had been observed. We show here that both Sir2 and Slx5 contribute to maximal growth, yet the contribution of Sir2 is ordinarily masked in the presence of Slx5. The growth defects observed in the double mutants are manifested as both slowed growth rates of populations (Table 2) and failure of individual cells to divide (Fig. 7). As noted above, and because the defects are observed in both haploid and homozygous diploid double mutants, it is unlikely that this interaction is due to a synthetic defect in rDNA or that it is related to the role of Sir2 in HM silencing or nonhomologous end joining. Instead, it is likely that a distinct aspect of Sir2 function has an unsuspected role in promoting growth. Candidate Sir2 functions include suppression of initiation of replication (47), promotion of unequal inheritance of oxidative damage (1), and other aspects of telomere function.
By what mechanism does loss of Slx5 make Sir2 more important for growth? Because sir2Δ ulp2Δ double mutants were even more synthetic sick than sir2Δ slx5Δ double mutants, it seems likely that the sir2Δ slx5Δ defects are related to a defect in protein sumoylation. For example, it is possible that Ulp2 opposes Slx5/8-dependent ubiquitination by desumoylating potential Slx5/8 targets or that it promotes Slx5/8-dependent protein degradation by desumoylating Slx5/8 targets subsequent to Slx5/8 activity and prior to degradation in the proteasome (7, 67). Thus, the growth defects observed in sir2Δ slx5Δ and sir2Δ ulp2Δ double mutants could be linked to excessive sumoylation of one or more target proteins or to aberrant target protein accumulation due to insufficient degradation.
In previous screens, a number of genes have been identified that interfere with silencing when overexpressed (10, 35, 58). An attractive possibility is that the proteins found in these screens may interfere not only with silencing but also with viability when control of sumoylation is disrupted. Future combined proteomic and genomic studies should resolve the means by which STUbL and sumoylation factors such as Ulp2 and the silencing factors Sir2, Sir3, and Sir4 jointly contribute to viability.
Finally, it is of note that the significant functional interactions observed among SIR2, other silent chromatin components, and SLX5 and ULP2/SMT4 have not been reported from previous genome-wide screens for synthetic lethal or critical interactions. This is for the technically simple yet biologically critical reason that the sir2-4Δ mutants do not mate and are therefore invisible in the majority of current genomic screens that require successful diploid formation as part of the query process. The link defined here between silent chromatin and optimal cellular growth therefore raises the possibility that there are other critical as-yet-undiscovered cellular components and processes that are dependent on intact silent chromatin.
ADDENDUM
In further support of the conclusions presented in this paper, it should be noted that an additional, independent report of the SUMO-directed ubiquitin ligase activity of the Slx5-Slx8 complex was published by Ii and colleagues (27a).
Supplementary Material
Acknowledgments
We are indebted to S. Berger, S. Brill, S. Forsburg, M. Gartenberg, D. Gottschling, V. Guacci, M. Hochstrasser, P. James, E. Johnson, D. Moazed, D. Rivier, K. Runge, L. Rusche, D. Shore, J. Smith, L. Sompayrac, and X. Zhao for reagents, assistance, and advice. M. Grunstein, S. Jacobson, and J. Wilson provided sequences for several of the primers used. We are grateful to S. Jacobson, P. Laurenson, and J. Wilson for comments on the manuscript. We thank E. Stone and M. Sharp for assistance with immunofluorescence and A. Babour for assistance with GFP microscopy. V. Lundblad and E. Ford assisted with telomeric blotting.
Research in the lab has been supported by funding from the National Institutes of Health. R.P.D. was supported in part by a Kirschstein Individual NRSA, and M.R.K. was supported in part by the Molecular Endocrinology training grant (M. G. Rosenfeld, principal investigator).
Footnotes
Published ahead of print on 17 December 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aguilaniu, H., L. Gustafsson, M. Rigoulet, and T. Nystrom. 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 2991751-1753. [DOI] [PubMed] [Google Scholar]
- 2.Amberg, D., D. Burke, and J. Strathern. 2005. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Azam, M., J. Y. Lee, V. Abraham, R. Chanoux, K. A. Schoenly, and F. B. Johnson. 2006. Evidence that the S. cerevisiae Sgs1 protein facilitates recombinational repair of telomeres during senescence. Nucleic Acids Res. 34506-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. [DOI] [PubMed] [Google Scholar]
- 5.Bryk, M., M. Banerjee, M. Murphy, K. E. Knudsen, D. J. Garfinkel, and M. J. Curcio. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11255-269. [DOI] [PubMed] [Google Scholar]
- 6.Burgess, R. C., S. Rahman, M. Lisby, R. Rothstein, and X. Zhao. 2007. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell. Biol. 276153-6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bylebyl, G. R., I. Belichenko, and E. S. Johnson. 2003. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 27844113-44120. [DOI] [PubMed] [Google Scholar]
- 8.Chi, M.-H., and D. Shore. 1996. SUM1-1, a dominant suppressor of SIR mutations in Saccharomyces cerevisiae, increases transcriptional silencing at telomeres and HM mating-type loci and decreases chromosome stability. Mol. Cell. Biol. 164281-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien, C. T., S. Buck, R. Sternglanz, and D. Shore. 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75531-541. [DOI] [PubMed] [Google Scholar]
- 10.Cockell, M., H. Renauld, P. Watt, and S. M. Gasser. 1998. Sif2p interacts with Sir4p amino-terminal domain and antagonizes telomeric silencing in yeast. Curr. Biol. 8787-790. [DOI] [PubMed] [Google Scholar]
- 11.Denison, C., A. D. Rudner, S. A. Gerber, C. E. Bakalarski, D. Moazed, and S. P. Gygi. 2005. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell. Proteomics 4246-254. [DOI] [PubMed] [Google Scholar]
- 12.De Rubertis, F., D. Kadosh, S. Henchoz, D. Pauli, G. Reuter, K. Struhl, and P. Spierer. 1996. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature 384589-591. [DOI] [PubMed] [Google Scholar]
- 13.Emre, N. C., K. Ingvarsdottir, A. Wyce, A. Wood, N. J. Krogan, K. W. Henry, K. Li, R. Marmorstein, J. F. Greenblatt, A. Shilatifard, and S. L. Berger. 2005. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol. Cell 17585-594. [DOI] [PubMed] [Google Scholar]
- 14.Ersfeld, K., and E. M. Stone. 1999. Simultaneous in situ detection of DNA and proteins, p. 51-66. In V. Allan (ed.), The practical approach series. Oxford University Press, New York, NY.
- 15.Frei, C., and S. M. Gasser. 2000. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 1481-96. [PMC free article] [PubMed] [Google Scholar]
- 16.Fritze, C. E., K. Verschueren, R. Strich, and R. Easton Esposito. 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 166495-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia, S. N., and L. Pillus. 2002. A unique class of conditional sir2 mutants displays distinct silencing defects in Saccharomyces cerevisiae. Genetics 162721-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner, K. A., and C. A. Fox. 2001. The Sir1 protein's association with a silenced chromosome domain. Genes Dev. 15147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasser, S. M., M. Gotta, H. Renauld, T. Laroche, and M. Cockell. 1998. Nuclear organization and silencing: trafficking of Sir proteins. Novartis Found. Symp. 214114-132. [DOI] [PubMed] [Google Scholar]
- 20.Ghidelli, S., D. Donze, N. Dhillon, and R. T. Kamakaka. 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 204522-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb, S., and R. E. Esposito. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56771-776. [DOI] [PubMed] [Google Scholar]
- 22.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63751-762. [DOI] [PubMed] [Google Scholar]
- 23.Häkli, M., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2005. SUMO-1 promotes association of SNURF (RNF4) with PML nuclear bodies. Exp. Cell Res. 304224-233. [DOI] [PubMed] [Google Scholar]
- 24.Hay, R. T. 2005. SUMO a history of modification. Mol. Cell 181-12. [DOI] [PubMed] [Google Scholar]
- 25.Holmes, S. G., A. B. Rose, K. Steuerle, E. Saez, S. Sayegh, Y. M. Lee, and J. R. Broach. 1997. Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics 145605-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, J., I. L. Brito, J. Villen, S. P. Gygi, A. Amon, and D. Moazed. 2006. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 202887-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425686-691. [DOI] [PubMed] [Google Scholar]
- 27a.Ii, T., J. Fung, J. R. Mullen, and S. J. Brill. 2007. The yeast Slx5-Slx8 DNA integrity complex displays ubiquitin ligase activity. Cell Cycle 62800-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ii, T., J. R. Mullen, C. E. Slagle, and S. J. Brill. 2007. Stimulation of in vitro sumoylation by Slx5-Slx8: evidence for a functional interaction with the SUMO pathway. DNA Repair 61679-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1441425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaliraman, V., and S. J. Brill. 2002. Role of SGS1 and SLX4 in maintaining rDNA structure in Saccharomyces cerevisiae. Curr. Genet. 41389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy, B. K., E. D. Smith, and M. Kaeberlein. 2005. The enigmatic role of Sir2 in aging. Cell 123548-550. [DOI] [PubMed] [Google Scholar]
- 32.Kimura, A., T. Umehara, and M. Horikoshi. 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32370-377. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi, T., T. Horiuchi, P. Tongaonkar, L. Vu, and M. Nomura. 2004. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rDNAs in yeast. Cell 117441-453. [DOI] [PubMed] [Google Scholar]
- 34.Koshland, D., and P. Hieter. 1987. Visual assay for chromosome ploidy. Methods Enzymol. 155351-372. [DOI] [PubMed] [Google Scholar]
- 35.Le, S., C. Davis, J. B. Konopka, and R. Sternglanz. 1997. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 131029-1042. [DOI] [PubMed] [Google Scholar]
- 36.Lee, S. E., F. Paques, J. Sylvan, and J. E. Haber. 1999. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 9767-770. [DOI] [PubMed] [Google Scholar]
- 37.Lewis, L. K., and M. A. Resnick. 2000. Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutat. Res. 45171-89. [DOI] [PubMed] [Google Scholar]
- 38.Li, S. J., and M. Hochstrasser. 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 202367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liou, G.-G., J. C. Tanny, R. G. Kruger, T. Walz, and D. Moazed. 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121515-527. [DOI] [PubMed] [Google Scholar]
- 40.Lisby, M., R. Rothstein, and U. H. Mortensen. 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 988276-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Longo, V. D., and B. K. Kennedy. 2006. Sirtuins in aging and age-related disease. Cell 126257-268. [DOI] [PubMed] [Google Scholar]
- 42.Lynch, P. J., H. B. Fraser, E. Sevastopoulos, J. Rine, and L. N. Rusche. 2005. Sum1p, the origin recognition complex, and the spreading of a promoter-specific repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 255920-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marx, J. 2005. Cell biology. SUMO wrestles its way to prominence in the cell. Science 307836-839. [DOI] [PubMed] [Google Scholar]
- 44.Michan, S., and D. Sinclair. 2007. Sirtuins in mammals: insights into their biological function. Biochem. J. 4041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullen, J. R., V. Kaliraman, S. S. Ibrahim, and S. J. Brill. 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157103-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nathan, D., K. Ingvarsdottir, D. E. Sterner, G. R. Bylebyl, M. Dokmanovic, J. A. Dorsey, K. A. Whelan, M. Krsmanovic, W. S. Lane, P. B. Meluh, E. S. Johnson, and S. L. Berger. 2006. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 20966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pappas, D. L., Jr., R. Frisch, and M. Weinreich. 2004. The NAD+-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 18769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pogliano, J., N. Osborne, M. D. Sharp, A. Abanes-De Mello, A. Perez, Y. L. Sun, and K. Pogliano. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 311149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prudden, J., S. Pebernard, G. Raffa, D. A. Slavin, J. J. Perry, J. A. Tainer, C. H. McGowan, and M. N. Boddy. 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 264089-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reifsnyder, C., J. Lowell, A. Clarke, and L. Pillus. 1996. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat. Genet. 1442-49. [DOI] [PubMed] [Google Scholar]
- 51.Replogle, K., L. Hovland, and D. H. Rivier. 1999. Designer deletion and prototrophic strains derived from Saccharomyces cerevisiae strain W303-1a. Yeast 151141-1149. [DOI] [PubMed] [Google Scholar]
- 52.Rudner, A. D., B. E. Hall, T. Ellenberger, and D. Moazed. 2005. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell. Biol. 254514-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner, and M. Grunstein. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 9314503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72481-516. [DOI] [PubMed] [Google Scholar]
- 55.Sauve, A. A., C. Wolberger, V. L. Schramm, and J. D. Boeke. 2006. The biochemistry of sirtuins. Annu. Rev. Biochem. 75435-465. [DOI] [PubMed] [Google Scholar]
- 56.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 1943-21. [DOI] [PubMed] [Google Scholar]
- 57.Shin, J. A., E. S. Choi, H. S. Kim, J. C. Ho, F. Z. Watts, S. D. Park, and Y. K. Jang. 2005. SUMO modification is involved in the maintenance of heterochromatin stability in fission yeast. Mol. Cell 19817-828. [DOI] [PubMed] [Google Scholar]
- 58.Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson, C. Goggin, M. Mahowald, and D. E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11241-254. [DOI] [PubMed] [Google Scholar]
- 60.Smith, J. S., C. B. Brachmann, L. Pillus, and J. D. Boeke. 1998. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics 1491205-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suka, N., K. Luo, and M. Grunstein. 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32378-383. [DOI] [PubMed] [Google Scholar]
- 62.Sun, H., J. D. Leverson, and T. Hunter. 2007. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 264102-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun, Z. W., and M. Hampsey. 1999. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics 152921-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sutton, A., R. C. Heller, J. Landry, J. S. Choy, A. Sirko, and R. Sternglanz. 2001. A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol. Cell. Biol. 213514-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tanny, J. C., G. J. Dowd, J. Huang, H. Hilz, and D. Moazed. 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99735-745. [DOI] [PubMed] [Google Scholar]
- 66.Tsalik, E. L., and M. R. Gartenberg. 1998. Curing Saccharomyces cerevisiae of the 2 micron plasmid by targeted DNA damage. Yeast 14847-852. [DOI] [PubMed] [Google Scholar]
- 67.Uzunova, K., K. Gottsche, M. Miteva, S. R. Weisshaar, C. Glanemann, M. Schnellhardt, M. Niessen, H. Scheel, K. Hofmann, E. S. Johnson, G. J. Praefcke, and R. J. Dohmen. 2007. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 28234167-34175. [DOI] [PubMed] [Google Scholar]
- 68.van Leeuwen, F., and D. E. Gottschling. 2002. Assays for gene silencing in yeast. Methods Enzymol. 350165-186. [DOI] [PubMed] [Google Scholar]
- 69.Vannier, D., D. Balderes, and D. Shore. 1996. Evidence that the transcriptional regulators SIN3 and RPD3, and a novel gene (SDS3) with similar functions, are involved in transcriptional silencing in S. cerevisiae. Genetics 1441343-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, Z., G. M. Jones, and G. Prelich. 2006. Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics 1721499-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Winzeler, E. A., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285901-906. [DOI] [PubMed] [Google Scholar]
- 72.Wohlschlegel, J. A., E. S. Johnson, S. I. Reed, and J. R. Yates III. 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 27945662-45668. [DOI] [PubMed] [Google Scholar]
- 73.Xie, Y., O. Kerscher, M. B. Kroetz, H. F. McConchie, P. Sung, and M. Hochstrasser. 2007. The yeast HEX3·SLX8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 28234176-34184. [DOI] [PubMed] [Google Scholar]
- 74.Xu, F., Q. Zhang, K. Zhang, W. Xie, and M. Grunstein. 2007. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell 27890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang, B., and A. L. Kirchmaier. 2006. Bypassing the catalytic activity of SIR2 for SIR protein spreading in Saccharomyces cerevisiae. Mol. Biol. Cell 175287-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang, L., J. R. Mullen, and S. J. Brill. 2006. Purification of the yeast Slx5-Slx8 protein complex and characterization of its DNA-binding activity. Nucleic Acids Res. 345541-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang, Y., W. Fu, J. Chen, N. Olashaw, X. Zhang, S. V. Nicosia, K. Bhalla, and W. Bai. 2007. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat. Cell Biol. 91253-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao, X., and G. Blobel. 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 1024777-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.