Abstract
The packaging of eukaryotic DNA into chromatin can create an impediment to transcription by hindering binding of essential factors required for transcription. The mammalian SWI/SNF remodeling complex has been shown to alter local chromatin structure and facilitate recruitment of transcription factors. BRG1 (or hBrm), the central ATPase of the human SWI/SNF complex, is a critical factor for the functional activity of nuclear receptor complexes. Analysis using BRG1/SNF2h chimeras suggests BRG1 may contain previously uncharacterized functional motifs important for SWI/SNF. To identify these regions, BRG1 truncation and deletion mutants were designed, characterized, and utilized in a series of assays to evaluate transcriptional activation and chromatin remodeling by the glucocorticoid receptor. We identified a domain within the N terminus of BRG1 that mediates critical protein interactions within SWI/SNF. We find the HSA domain of BRG1 is required to mediate the interaction with BAF250a/ARID1A and show this association is necessary for transcriptional activation from chromatin mouse mammary tumor virus or endogenous promoters in vivo. These studies suggest BAF250a is a necessary facilitator of BRG1-mediated chromatin remodeling required for SWI/SNF-dependent transcriptional activation.
In the eukaryotic nucleus, genetic information is packaged with histone and nonhistone proteins to form a highly organized chromatin structure which tends to have a repressive effect on gene expression by inhibiting access of the transcriptional machinery and gene-specific regulators to recognition sequences within target promoters (4, 34). This structural assembly can represent an additional level of control in numerous nuclear processes, such as transcriptional regulation, replication, recombination, and DNA damage repair. The modulations of chromatin structure that accompany transcriptional regulation often require multiprotein complexes that can manipulate the nucleosomal architecture. At least two highly conserved chromosome-modifying enzymatic activities have been described that alter chromatin structure through ATP-dependent chromatin remodeling complexes or by covalent modification of histone tails by means of acetylation, methylation, phosphorylation, ubiquitination, sumoylation, and/or ADP ribosylation (20, 31).
Several multisubunit complexes have been identified that utilize the energy derived from ATP hydrolysis to alter the arrangement and stability of nucleosomes in a noncovalent manner to regulate nuclear processes. Generally, these ATP-dependent remodeling machines are divided into four major classes according to the identity of their central catalytic subunit which include SWI/SNF, ISWI, Mi-2/NuRD, and INO80 (4, 8, 29). The SWI/SNF family of chromatin remodeling complexes, which is highly divergent and can exist in multiple forms, has been well characterized with regard to structure, function, and enzymatic activity. Mammalian SWI/SNF is a large multiprotein complex that contains either BRG1 (Brahma-related gene 1) or hBrm (human Brahma) as the catalytic ATPase as well as 10 to 12 BAF (BRG1-associated factor) proteins, most of which are orthologous to those found in Saccharomyces cerevisiae SWI/SNF and RSC (31). Human SWI/SNF is composed of a heterogeneous mixture of subunits with most purified complexes containing accessory subunits BAF170, BAF155, BAF60, BAF57, BAF53, and BAF47 (35). Although BRG1 alone is sufficient to stimulate nucleosome remodeling in vitro, the addition of the core BAF proteins, BAF170, BAF155, and BAF47, has been shown to reconstitute chromatin remodeling to near optimal levels (28). Human SWI/SNF can be further grouped into two subfamilies classified as BAF and PBAF complexes (22, 25). Members of these two groups share similar subunit compositions but are distinguished by the presence of specific subunits where BAF250 has been found exclusively in the BAF complex, while BAF200 and BAF180 are exclusively present in PBAF (40). This differential makeup of the various BRG1/Brm-based complexes suggest BAF subunits may play important roles for promoter-specific targeting or to stabilize nucleosomal structure in a particular conformation which is favorable for SWI/SNF remodeling (4, 24). Studies of the human β-globin promoter support this concept by demonstrating that mammalian SWI/SNF-regulated transcription is a selective process involving direct interactions between distinct zinc finger DNA-binding domain structures and individual SWI/SNF subunits, BRG1 and BAF155 or BAF170, to achieve targeted chromatin remodeling and transcriptional activation (19).
The targeting of SWI/SNF to gene-specific promoters is thought to take place through the binding of transcription factors, coactivators, or members of the general transcriptional machinery (4). Interestingly, BAF subunits with bromodomains are known to target acetylated histone tails; likewise, different SWI/SNF components, including BRG1, BAF250, BAF60a, and BAF57, have been reported to mediate critical interactions between steroid hormone receptors and remodeling complexes (2, 12, 14, 18). Within the context of nuclear receptor (NR)-mediated transcriptional activation, multiple interactions are thought to be involved in recruitment and stabilization of the SWI/SNF remodeling complex at target promoters and this targeting may be mediated through direct or indirect interactions involving one or more BAF subunits (4).
The SWI/SNF chromatin remodeling complex has been implicated in transcriptional regulation by glucocorticoid receptor (GR) based on the published interaction of GR with the BRG1 complex in GR-mediated chromatin remodeling at the steroid hormone-responsive mouse mammary tumor virus (MMTV) promoter (32). The MMTV promoter has proven to be a model system for the study of the molecular details involved in NR-mediated chromatin remodeling and transcriptional activation by SWI/SNF.
Studies using the BRG1/hBrm null SW-13 cell line established the requirement for BRG1 in GR-mediated chromatin remodeling and transcriptional activation from gene-specific promoters (32). Interestingly, SW-13 cells also lack the PBAF-specific BAF180 subunit, suggesting that the BAF180 subunit is not required for GR-mediated remodeling at chromatin MMTV because the remodeling process is supported upon reintroduction of BRG1 (22). Although present in SW-13 cells, remodeling activities of ISWI- or Mi-2-based complexes are also unable to substitute for GR-dependent BRG1 actions, such as transcriptional activation, induced transcription factor binding, and chromatin remodeling of the nucleosomal architecture (32).
In this present study, we use various BRG1/SNF2h chimeric proteins to show a dependence of the BRG1 N-terminal region, in conjunction with the active ATPase domain, on GR-induced BRG1-dependent transcriptional activation of chromatin MMTV. The N-terminal domain of BRG1 contains several conserved regions identified based on sequence homology, whose function and biological importance remain unclear. To further analyze these regions and their importance in vivo, we generated and characterized a series of N-terminal BRG1 truncation and deletion mutants specifically targeting the proline-rich region, the helicase/SANT-associated (HSA) domain (7), or the BRM and KIS (BRK) domain (6). Interestingly, deletion of the HSA region within BRG1 greatly reduced GR-mediated transcriptional activation from chromatin MMTV and endogenous promoters. Computational analyses and in vitro binding studies suggest this region may comprise a protein interaction surface that may mediate critical interactions required for GR-dependent SWI/SNF transcriptional regulation (18). We find that the N-terminal HSA domain of BRG1 is necessary to mediate the association with the ARID domain (AT-rich interacting domain) containing SWI/SNF subunit, BAF250a (ARID1A), and that this interaction is required for GR-mediated BRG1-dependent transcriptional activation of chromatin MMTV and endogenous promoters.
MATERIALS AND METHODS
Cell culture, transient transfections, and luciferase assays.
Human SW-13 adrenal carcinoma cells were maintained as previously described (32). SW-13/MMTV cells, which contain 20 stably integrated copies of MMTV fused to a luciferase reporter, were cultured under the same conditions as those used for SW-13 cells but supplemented with 250 μg/ml G418. Transfections were carried out in serum-free Dulbecco modified Eagle medium using Lipofectamine Plus reagent or in antibiotic-free medium using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were assayed for luciferase activity, and relative light units were normalized to the total protein measured as previously described (32).
Generation of BRG1/SNF2h chimeric and BRG1 N-terminal mutant constructs.
Constructs expressing human FLAG-tagged BRG1/SNF2h chimeric proteins were generated by PCR as previously described (11). Mammalian expression vectors for V5-tagged human BRG1 and BRG1 mutants were constructed by directional TOPO cloning of PCR fragment containing the entire coding region or truncated products into pcDNA3.1D/V5-His plasmid according to the manufacturer's suggested protocol (Invitrogen). BRG1 deletion mutant expression plasmids (ΔHSA, ΔBRK, and Δ475-656) were generated by first creating shuttle vectors of TOPO cloned BRG1 PCR fragments encoding amino acids 532 to 1648 (for ΔHSA) or 656 to 1648 (for ΔBRK and Δ475-656) using specific primers containing unique restriction endonuclease sites. These plasmids were digested and used in ligation reaction mixtures containing BRG1-specific PCR products encoding amino acids 1 to 475 (for ΔHSA and Δ475-656) or 1 to 612 (for ΔBRK). The authenticity of each PCR-generated sequence was verified by DNA sequencing. Primer sequences used to generate these BRG1 pcDNA3.1D/V5-His expression vectors are available upon request.
Immunoblotting and immunoprecipitation analysis.
Cells from subconfluent cultures were washed and scraped into phosphate-buffered saline and pelleted by centrifugation. For immunoprecipitation analysis and immunoblotting, whole-cell or nuclear lysates were prepared as previously described using indicated antibodies (32).
Nucleosome assembly.
A 202-bp DNA fragment, containing two 20-bp GT phasing sequences located at one end, was assembled into mononucleosomes with recombinant human histones using step gradient salt dialysis. DNA fragments used for assembly were generated by PCR and labeled with [α-32P]dATP. Recombinant histones were prepared as described previously (23). Mononucleosomes were purified on a 10 to 30% glycerol gradient (10).
Protein purification and ATP-dependent remodeling assays.
DNA encoding BRG1 N-terminus-truncated proteins were transfected into 293T cells using Lipofectamine 2000. Forty hours posttransfection, cells were harvested and lysed in a hypotonic buffer. BRG1 mutant proteins were purified from nuclear extracts using V5 beads, and these bead-bound BRG1 mutant proteins were assayed for their remodeling activity.
Remodeling reactions were performed in 12 mM HEPES (pH 7.9), 10 mM Tris-HCl (pH 7.5), 60 mM KCl, 8% glycerol, 4 mM MgCl2, 2 mM ATP·Mg, and 0.02% NP-40 at 30°C. The mononucleosomal substrate was used at less than 1 nM. Reactions were terminated by adding ADP to a final concentration of 10 mM and DNA to 100 ng/μl as an unlabeled competitor. The remodeled products were resolved on 5% native polyacrylamide gels (0.5× Tris-borate-EDTA).
ChIP.
SW-13/MMTV cells were transiently transfected with GR and wild-type or BRG1 mutant expression plasmids followed by treatment with dexamethasone (Dex) (10−7 M) or an equal volume of vehicle for 1 h. Cell monolayers were fixed for 5 min with 1% formaldehyde and washed with phosphate-buffered saline and then analyzed as previously described (32). Quantitative chromatin immunoprecipitation (ChIP) PCR was performed with Stratagene Mx3000P and Brilliant SYBR green quantitative PCR (QPCR) master mix. Average cycle threshold amplification values were calculated and presented as percentages of sample input.
Restriction enzyme hypersensitivity assay.
SW-13/MMTV cells were transiently transfected with expression plasmids for GR and either pcDNA3.1D (empty vector) or wild-type or mutant BRG1. Cells were treated with Dex (10−7 M) or vehicle for 1 h, followed by nucleus isolation and digestion with SstI (in vivo) and HaeIII (in vitro) as previously described (1). Equal amounts of purified digestion products were analyzed by linear Taq polymerase amplification with a 32P-labeled oligonucleotide specific for MMTV (32). Purified amplification products were resolved on polyacrylamide denaturing gels.
Reverse transcription-QPCR.
Total RNA was isolated from parental SW-13 cells transfected with expression plasmids for GR and/or BRG1, K798R, ΔHSA, or ΔBRK. Forty-eight hours posttransfection, cells were scraped or treated with Dex (10−7 M) or vehicle for 4 h, and total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's protocol. DNase I-treated total RNA was reverse transcribed into single-stranded cDNA using oligo(dT) and SuperScript II (Invitrogen). An equal amount of cDNA, from each experimental condition, was amplified by real-time PCR using the Stratagene Mx3000P and Brilliant Sybr green QPCR master mix with indicated gene-specific primer sets. For reverse transcription-QPCR, the average cycle threshold values for promyelocytic leukemia zinc finger protein (PLZF) and hydroxysteroid 11-β dehydrogenase (HSD-11β) type 2 primer sets were calculated and normalized the cycle threshold values obtained from glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific primers. Optimized conditions for PCR amplification and primer sequences are available upon request.
siRNA transfection.
Small interfering RNA (siRNA) duplexes targeting the mRNA coding region of BAF250a (NCBI Nucleotide Database accession number NM_006015) were created using Ambion or Invitrogen Web-based software and synthesized by Dharmacon. siRNAs were also synthesized targeting BAF155 and BAF170 (used as a positive control of transcriptional activation knock-down) or BAF250b and nontargeting control (NTC) from Dharmacon (used as a negative control of transcriptional activation knockdown). Sequence information regarding chemically synthesized siRNA duplexes used in this study follows: BAF250-1, 5′-GGGCCAGACUCCAUAUUACdTdT-3′; BAF250-2, 5′-UUGCCCAAGAUCGAGGUUAdTdT-3′; BAF250-3, 5′-CGACAUGAUUCCUAUGGCAdTdT-3′ (40); BAF250-4, 5′-CGCCUGGAGAAGUUGUAUAdTdT-3′; BAF155, 5′-CAAGGAUGAUUGGAACAAAdTdT-3′; BAF170, 5′-CCUCUCACUUCCAUGUCUUdTdT-3′; BAF250b, 5′-UCUGUGAUGUACUGUUUCAdTdT-3′; and NTC, siCONTROL nontargeting siRNA 1 (catalog number D-001212-01-20; Dharmacon).
SW-13 cells were seeded into six-well plates, containing normal growth media (minus antibiotics), at a density of 0.3 × 106 cells/well. Cells were cotransfected with GR and wild-type BRG1 expression plasmid (1.0 μg/well) and 50 μM duplex siRNA using Lipofectamine 2000 in Opti-MEM (Invitrogen). Forty-eight hours posttransfection, cells were treated with Dex (10−7M) or vehicle for 16 h in fresh normal growth media (minus antibiotics). The extents of BAF250a protein knockdown and expression of transfected plasmids were determined by immunoblotting at 48 h posttransfection. The effect of BAF250a protein knock-down on MMTV transcriptional initiation was evaluated by luciferase assay.
RESULTS
The N-terminal region of BRG1 is required for SWI/SNF function.
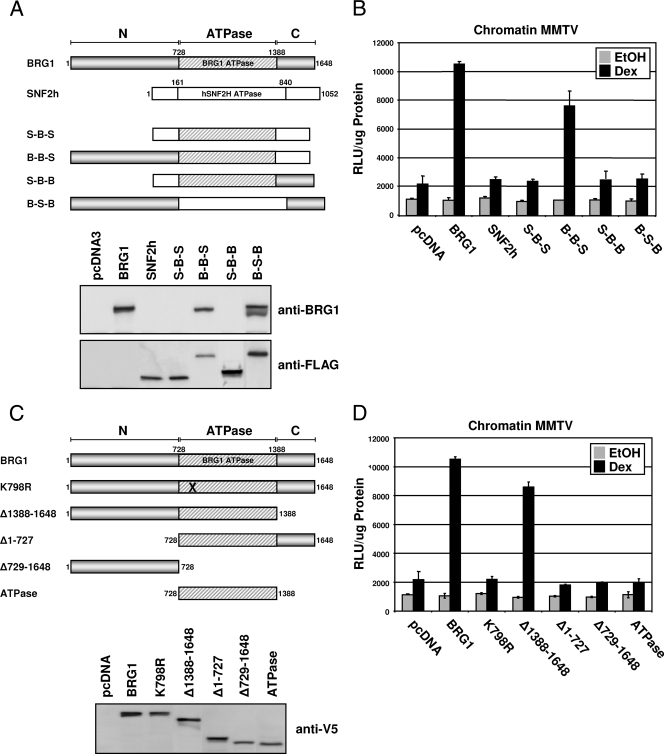
The SWI/SNF and ISWI families of ATP-dependent chromatin remodeling complexes show conserved homology from yeast to humans. BRG1 and SNF2h represent the motor proteins of the major mammalian remodeling complexes, and genetic analysis has indicated these ATPase activities do not complement each other (9). In order to identify domains within the BRG1 remodeling protein that are required for SWI/SNF function, we utilized a series of BRG1/SNF2h chimeric proteins generated through domain swapping between the N-terminal, ATPase, and C-terminal domains of the two chromatin remodeling proteins to create four BRG1/SNF2h chimeric proteins, S-B-S, B-B-S, S-B-B, and B-S-B (Fig. 1A). Each chimeric protein was characterized and evaluated using a series of in vitro assays to determine functional activities within the context of the remodeling complex as previously described (11). In the present study, these BRG1/SNF2h chimeras were used to evaluate GR-dependent transcriptional activation by SWI/SNF.
FIG. 1.
Characterization of BRG1/SNF2h chimeric proteins and BRG1 domain deletion mutants in SW-13 cells. (A) Schematic representation of BRG1/SNF2h chimeric proteins used in this study. BRG1 and SNF2h are divided into three regions: a homologous ATPase domain (ATPase), a nonhomologous N-terminal region (N), and a C-terminal region (C). (B) SW-13 cells, containing stably integrated MMTV reporter, were cotransfected with expression plasmids for GR and pcDNA (empty vector), BRG1, SNF2h, or BRG1/SNF2h chimeric proteins, treated with Dex, and assayed for luciferase activity. Luciferase activity was normalized to the total protein measured (measured in relative light units [RLU] per microgram of protein). EtOH, ethanol. (C) Diagram of BRG1 ATPase-dead (K798R), N- and C-terminal deletion mutants, and N-terminal or ATPase proteins. (D) GR-mediated transcriptional activation assay as described in Materials and Methods in SW-13/MMTV cells expressing GR and BRG1 or domain deletion proteins. Relative luciferase expression values were normalized to total protein measured. Data represented are from three independent experiments.
Studies have shown that BRG1 is required for GR-dependent activity of chromatin MMTV within SW-13 cells and that this activity cannot be substituted by other chromatin remodeling proteins present, such as SNF2h (32). Therefore, to determine which regions within BRG1 are required for NR-mediated SWI/SNF activity, SW-13 cells containing an integrated MMTV luciferase reporter (designated SW-13/MMTV) were cotransfected with expression vectors for GR and BRG1, SNF2h, or BRG1/SNF2h chimeric proteins, treated with Dex or vehicle, and assayed for reporter activity. Cells expressing BRG1 showed a significant increase in reporter activity from the chromatin promoter upon Dex treatment. Interestingly, GR-mediated activity was observed only in the presence of BRG1 and the B-B-S chimera, while activity resulting from the other chimeric proteins was equivalent to that observed with the empty vector control (Fig. 1B). To verify GR-dependent transcription initiation from chromatin MMTV requires an intact N terminus and BRG1 ATPase; we performed reporter studies in the presence of BRG1 deletion mutants. Mutant BRG1 proteins were generated by removing the N or C terminus of BRG1, denoted Δ1-727 or Δ1388-1648, respectively, or by specifically expressing the N-terminal domain (Δ729-1648) or ATPase domain of BRG1 (Fig. 1C). A similar transcriptional dependence was observed in SW-13/MMTV cells expressing these BRG1 domain deletion mutants. Interestingly, the C terminus of BRG1 does not appear to be necessary for GR-mediated BRG1 activity at the chromatin promoter. Consistent with the BRG1/SNF2h chimeric data, GR-mediated MMTV transcription was observed in cells expressing both the N-terminal and ATPase regions of BRG1, while proteins devoid of both domains displayed significantly diminished activity from the promoter (Fig. 1D). Together, these results suggest the N-terminal region of BRG1 is important, in conjunction with BRG1 ATPase activity, for GR-induced SWI/SNF-dependent transcriptional activation of chromatin MMTV.
Generation and characterization of functional BRG1 N-terminal mutant proteins.
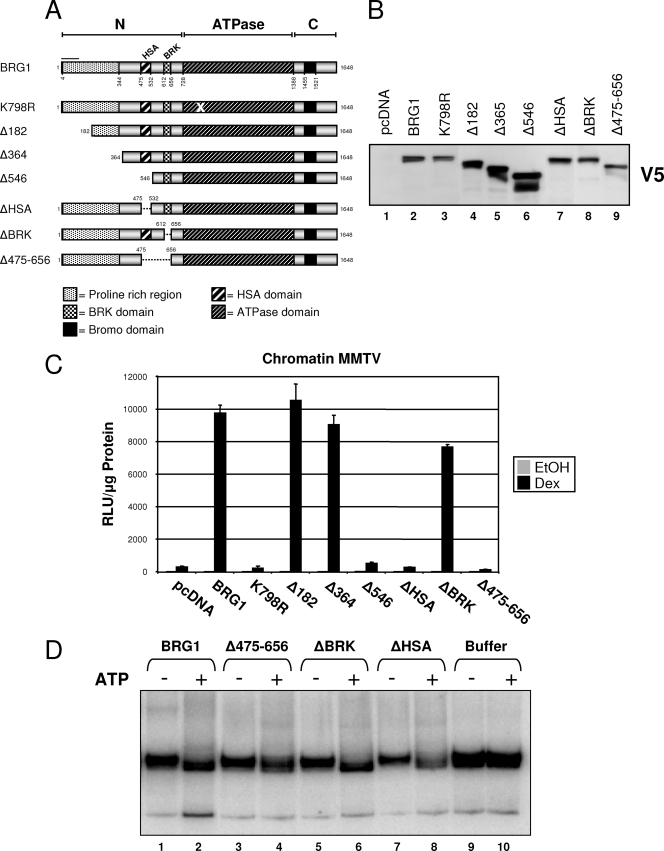
The N-terminal region of BRG1, consisting of amino acids 1 to 728, contain several conserved domains identified based on sequence homology. Studies suggest this region is required for interactions between BRG1 and SWI/SNF subunits as well as other proteins involved in various nuclear processes (15, 30, 33). To investigate the functional importance of the N-terminal region in the context of GR-mediated SWI/SNF activity, we generated and characterized a series of BRG1 N-terminal truncation and deletion mutants. A putative proline-rich region, an HSA domain (domain in helicases and associated with SANT domains), and a BRK domain (domain in transcription and CHROMO domain helicases) were specifically targeted for mutational analysis. Truncation of BRG1 by selectively removing amino acids 1 to 181 or 1 to 363 targeted the putative proline-rich region, while deletion of amino acids 476 to 531 and 611 to 655 targeted the BRG1-HSA and -BRK domains, respectively, while deletion of amino acids 476 to 655 removed both domains along with the linking region (Fig. 2A). The resulting BRG1 N-terminal mutants, each expressing a C-terminal V5 epitope, were expressed in SW-13 cells and found exclusively in the nuclear fraction (Fig. 2B).
FIG. 2.
Characterization of BRG1 N-terminal truncation/deletion mutants. (A) Schematic representation of BRG1 N-terminal mutants. BRG1 mutant expression plasmids were constructed by truncating amino acids or selectively deleting previously identified domains within the amino-terminal (N-terminal) region of BRG1. Each protein contains a C-terminal V5 epitope tag. (B) Protein expression of BRG1 N-terminal mutants in transfected SW-13 cells using nuclear lysate and anti-V5 antibody. (C) SW-13/MMTV cells were transfected with expression plasmids for GR and pcDNA (empty vector), BRG1, or mutant proteins, treated with Dex, and assayed for luciferase activity. Relative luciferase expression values were normalized to total protein measured (measured in relative light units [RLU] per microgram of protein). Data represented are from three independent experiments. EtOH, ethanol. (D) In vitro remodeling activity of BRG1 N-terminal mutant proteins. Nucleosome mobility assay was performed. BRG1 mutant proteins were purified from 293T cell nuclear extracts, immobilized on V5 beads, and assayed for remodeling activity using a 202-bp DNA fragment, assembled in mononucleosomes with recombinant human histones. Remodeled products were resolved on 5% native polyacrylamide gels.
Cells expressing GR and BRG1 showed a significant increase in luciferase activity from the chromatin promoter upon hormone treatment (Fig. 2C) (32). Activity levels detected in the presence of BRG1 mutants, Δ182 and Δ364, were equivalent to that observed with wild-type BRG1, suggesting that the proline-rich region of BRG1 is not required for GR-mediated transcriptional activation of chromatin MMTV. Interestingly, removal of the HSA domain resulted in a complete loss of GR-induced activity compared to empty vector (pcDNA), whereas only a minimal attenuation was observed upon deletion of the BRK region (Fig. 2C). These data suggest the N-terminal HSA domain of BRG1 may be a critical element for GR-mediated BRG1-dependent transcriptional activation.
The inability of ΔHSA to potentiate a GR-mediated transcriptional response may result from impaired ATP-dependent chromatin remodeling activity within the protein. To determine whether the BRG1 deletion mutants contain active nucleosome-remodeling domains, we performed nucleosome mobility analysis. BRG1 mutant proteins were purified from nuclear extracts and assayed for their ability to remodel a 202-bp mononucleosomal substrate. We found that each mutant tested efficiently produced remodeled products in an ATP-dependent manner, enriched for higher-mobility nucleosomes similar to those observed for BRG1 (Fig. 2D). These results indicate deletion of specific domains within the N terminus of BRG1 did not disrupt ATP-dependent remodeling activity of the mutant proteins. Taken together, these results suggest that the HSA domain of BRG1, and not the proline-rich region or the BRK domain, is critical for GR-mediated transcriptional activity of the SWI/SNF remodeling complex.
BRG1-ΔHSA is incorporated into the SWI/SNF complex, targeted to MMTV, but is unable to remodel the chromatin promoter.
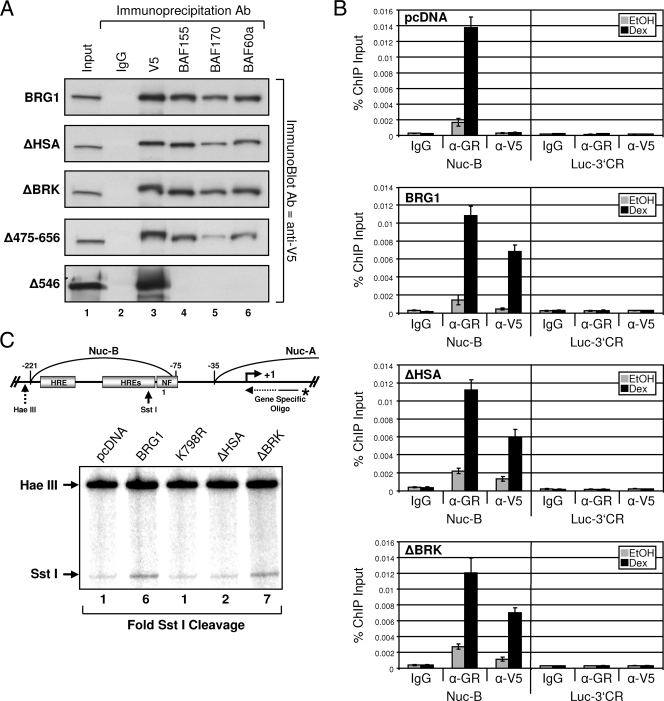
In mammalian cells, BRG1 exists within the context of SWI/SNF as the central catalytic subunit displaying ATPase activity. Multiple complexes have been identified and can be distinguished based on subunit composition; however, BRG1 (or hBrm), BAF170, BAF155, and BAF47 represent the core components of the remodeling complexes (31). The inability of BRG1-ΔHSA to participate in GR-induced activation of chromatin MMTV could be due to its exclusion from the remodeling complex. Therefore, we performed a series of coimmunoprecipitation studies to determine whether the BRG1 N-terminal deletion mutants, ΔHSA, ΔBRK, or Δ475-656 could be incorporated into the SWI/SNF complex. Parental SW-13 cells were transfected with expression plasmids encoding wild-type or mutant BRG1 proteins, and whole-cell lysates were used for immunoprecipitation with antibodies specific for V5 tag, BAF155, BAF170, or BAF60a. Immunoblot analysis reveals BAF170, BAF155, and BAF60a copurified with BRG1 mutants ΔHSA, ΔBRK, and Δ475-656, suggesting the mutant proteins are incorporated into the SWI/SNF complex (Fig. 3A). Interestingly, truncation mutant Δ546 did not copurify with the core BAF subunits, suggesting the BRG1 interactions with BAF170, BAF155, and BAF60a occur within amino acids 1 to 546 of BRG1 (Fig. 3A).
FIG. 3.
BRG1 mutants interact with core SWI/SNF subunits, display GR-dependent promoter binding, and require the HSA domain for remodeling activity. (A) Complexes containing BRG1 and mutant BRG1 were immunopurified from transfected SW-13 cells using antibodies (Ab) specific for V5 epitope, BAF155, BAF170, and BAF60a. Immunoprecipitates were resolved by 6% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting using anti-V5 antibody. Normal IgG was used to detect nonspecific interactions. (B) BRG1-, ΔHSA-, and ΔBRK-containing complexes are recruited to stably integrated MMTV in a GR-dependent manner in SW-13 cells, as demonstrated by ChIP analysis using anti-V5 antibody (α-V5). Purified immunoprecipitated DNA fragments were analyzed by real-time PCR using primer sets covering the MMTV promoter (Nuc-B) or 3′-coding region of the luciferase reporter (Luc-3′CR). Data were displayed as the percentages of ChIP input. Normal IgG was used as a negative control. Data represented are from three independent experiments. EtOH, ethanol. (C) In vivo restriction enzyme accessibility assay to determine MMTV chromatin structure. Nuclei were purified from treated (EtOH or Dex) cells expressing GR and BRG1 or mutant proteins followed by incubation with SstI (in vivo). Purified products were further digested to completion with HaeIII (in vitro) and used as a template for reiterative primer extension with an MMTV-specific primer. Extension products were resolved by 6% denaturing polyacrylamide gel electrophoresis. Change in SstI cleavage is indicated below the gel. HRE, hormone response element; Oligo, oligonucleotide.
To determine whether the SWI/SNF complexes containing BRG1 mutants are targeted to GR-responsive MMTV, we performed ChIP analysis. SW-13/MMTV cells, expressing GR and empty vector, BRG1, ΔHSA, or ΔBRK proteins, were treated with Dex or vehicle and subjected to ChIP analysis. As previously reported, BRG1 recruitment to the MMTV promoter is hormone dependent (32). Interestingly, recruitment of the complexes was shown to be equivalent to that observed for the wild-type protein, suggesting the N-terminal HSA or BRK domains are not required for targeting SWI/SNF to the chromatin promoter (Fig. 3B). When purified ChIP fragments were analyzed using primers specific for the luciferase reporter (∼2,000 bp downstream +1 transcription start site) recruitment/binding of GR, wild-type, or BRG1 mutant proteins were not detected, indicating the specificity of GR and BRG1/mutant recruitment to the MMTV promoter (Fig. 3B).
The nucleosomal structure of integrated MMTV undergoes a conformational change in response to glucocorticoid treatment, inducing the nucleosome B region to become hypersensitive to restriction endonucleases stimulating a remodeled nucleosomal state (21). To determine whether the HSA domain of BRG1 is required for this activity, we performed SstI hypersensitivity assay in the presence of various BRG1 proteins. We observed increased GR-mediated restriction enzyme accessibility in cells expressing wild-type BRG1 compared to treated cells transfected with empty vector or BRG1 ATPase mutant K798R (32). Interestingly, elevated cleavage by SstI was not detected in cells expressing the ΔHSA mutant, indicating this region of BRG1 may be required to induce a remodeled nucleosomal state within the promoter. Restriction enzyme accessibility was restored in cells expressing the ΔBRK protein to levels equivalent to that observed for wild-type BRG1 (Fig. 3C), indicating the HSA domain could be a critical element for GR-mediated SWI/SNF chromatin remodeling.
The HSA domain of BRG1 is required for BRG1-mediated transcriptional activation of endogenous genes.
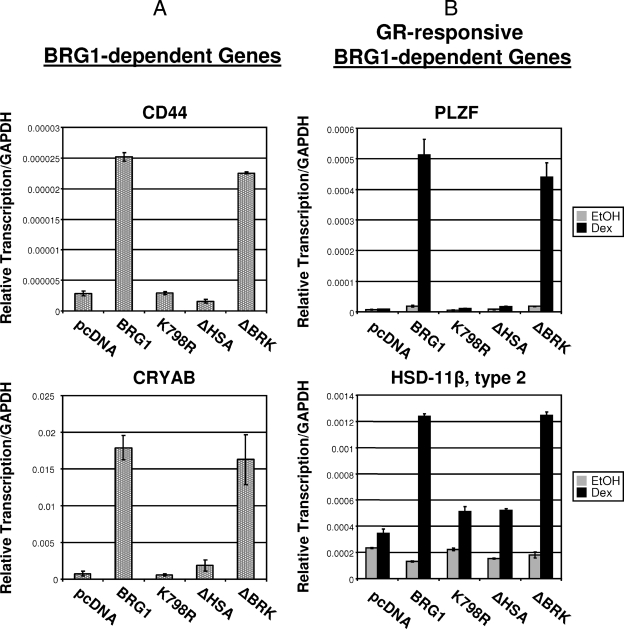
We next analyzed expression of endogenous BRG1-dependent or GR-responsive BRG1-dependent genes, CD44 and crystallin, alpha B (CRYAB) or PLZF and HSD-11β, respectively, within the SW-13 genome in the presence of BRG1, ΔHSA, or ΔBRK proteins. BRG1-dependent expression of both CD44 and CRYAB were increased in the presence of wild-type BRG1. Interestingly, this relative transcription was diminished in cells expressing ΔHSA to levels equivalent to that observed for empty vector or the K798R ATPase dead BRG1 mutant (Fig. 4A). This observation is also seen when evaluating expression of GR-responsive BRG1-dependent genes, PLZF and HSD-11β type 2, in treated SW-13 cells expressing the various BRG1 proteins (Fig. 4A). Consistent with the activation of MMTV, deletion of the BRK domain had no effect on GR-induced BRG1-dependent transcriptional activation from either promoter. As predicted, these data mirror observations made using our MMTV reporter system, confirming the HSA region of BRG1 is required for GR-mediated transcriptional activation of endogenous GR-regulated genes.
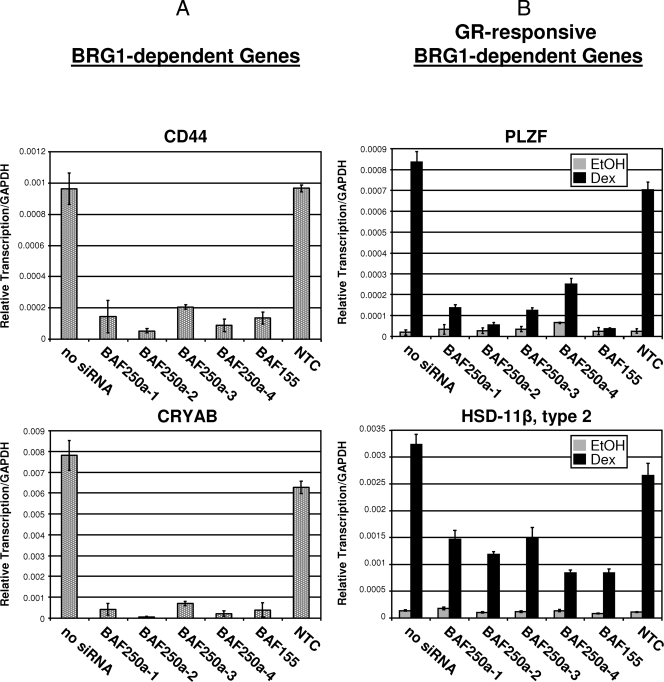
FIG. 4.
Comparison of the abilities of BRG1, ΔHSA, and ΔBRK to activate expression of endogenous BRG1-dependent or GR-responsive BRG1-dependent genes. (A) Real-time reverse transcription-PCR analysis was used to determine the expression level of BRG1-dependent genes, CD44 or CRYAB, in SW-13 wild-type cells expressing BRG1 or mutant proteins. (B) Reverse transcription-PCR expression analysis of GR-responsive BRG1-dependent genes, PLZF or HSD-11β type 2, in treated SW-13 cells expressing GR and BRG1 or mutant proteins. Equal amounts of cDNA were used in real-time PCRs with primers specific for endogenous genes CD44, CRYAB, PLZF, or HSD-11β type 2. Quantitative analysis was performed using Mx3000P software. Data are normalized to GAPDH and displayed as relative transcription. Values are shown as means plus standard deviations (error bars) from three experiments. EtOH, ethanol.
The HSA domain of BRG1 mediates the interaction with BAF250a.
Critical interactions between SWI/SNF subunits BAF170, BAF155, and BAF57 have been shown to take place within the N-terminal region of BRG1 (15). Interactions involving BRG1 with proteins implicated in various cellular processes are also thought to be mediated through the amino terminus of BRG1, such as nuclear receptor estrogen receptor α (17), HP1α (26), FANCA (27), STAT2 (15), and EVI-1 (5).
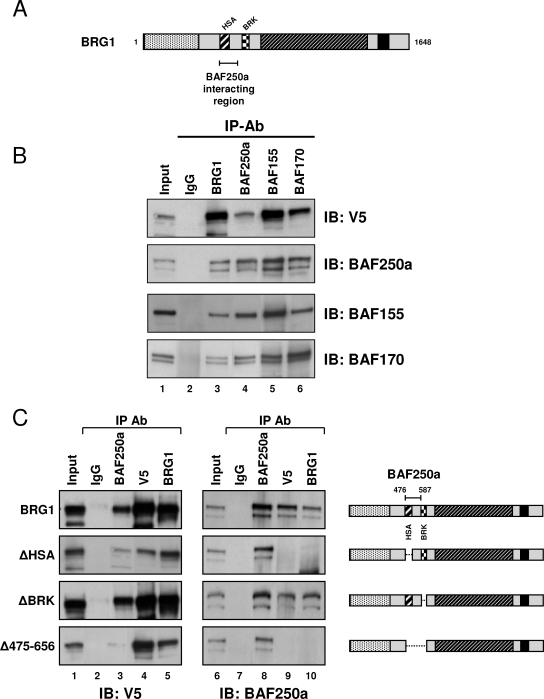
We used the Myriad ProNet database to scan the published literature for reported proteins that bind BRG1 over the HSA domain. Interestingly, a protein was identified from the search that could potentate an interaction utilizing the HSA region of BRG1, BAF250a/ARID1A (16). Therefore, we focused our studies on the BRG1/BAF250a interaction which has been reported to take place between amino acids 380 to 558 of BRG1 encompassing the full HSA domain (Fig. 5A) (18). To verify the interaction between BRG1 and BAF250a, we performed a series of coimmunoprecipitation assays using whole-cell extracts from parental SW-13 cells expressing BRG1 and antibodies specific for normal immunoglobulin G (IgG) (as a negative IP control), BRG1, BAF250a, or BAF155 and BAF170 (as a positive IP control). As previously reported, BAF250a interacts with BRG1 and with core members of the remodeling complex (Fig. 5B) (16).
FIG. 5.
The BAF250a interaction with BRG1 requires the HSA domain. (A) Schematic of wild-type BRG1 indicating the region required for interaction with BAF250a. (B) Wild-type BRG1 was immunopurified from BRG1-transfected SW-13 cells using antibodies specific for proteins reported to interact over the BRG1 HSA region (BAF250a) or core BAF subunits (BAF155 and BAF170). Immunoprecipitates (IP) were resolved by 6% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by immunoblotting (IB) using the indicated antibodies (Ab). Normal IgG was used to detect nonspecific interactions. (C) Complexes containing BRG1 and mutant BRG1 were immunopurified from transfected SW-13 cells using antibodies specific for BAF250a, V5 epitope, and BRG1 (IP Ab). Immunoprecipitates were resolved by 6% SDS-PAGE and analyzed by immunoblotting (IB) using anti-V5 or anti-BAF250a antibodies. Normal IgG was used to detect nonspecific interactions.
To test whether the association of BAF250a and BRG1 requires the full intact HSA domain, we performed coimmunoprecipitation studies in cells expressing wild-type or mutant BRG1 proteins. SWI/SNF complexes were purified using antibodies directed against BAF250a, V5 tag, or BRG1 and detected by immunoblot analysis with anti-V5 or -BAF250a antibodies. As shown by Western blot analysis, BAF250a associates with wild-type BRG1 and the ΔBRK mutant protein, each of which contain intact HSA domains (Fig. 5C). Interestingly, when the HSA region is removed, as in ΔHSA or Δ475-656, the BRG1 interaction with BAF250a is significantly reduced (Fig. 5C). This observation can be seen by immunoblotting using either anti-V5 or anti-BAF250a antibodies, indicating that BAF250a requires the N-terminal HSA domain of BRG1 for association with the SWI/SNF remodeling complex.
The BAF250a subunit of the SWI/SNF remodeling complex is required for transcriptional activation.
To evaluate the requirement for BAF250a in GR-mediated BRG1-dependent transcriptional activation of chromatin MMTV, we used siRNA analysis in conjunction with transcriptional activation assays. SW-13/MMTV cells were cotransfected with expression plasmids for GR, BRG1, and four BAF250a siRNA duplexes 48 h prior to Dex treatment. Western blot analysis was performed with antibodies directed against GR and mammalian SWI/SNF subunits, BRG1, BAF250a, BAF155, or BAF170 to determine the extent of protein expression and to evaluate the level of siRNA protein knockdown within the transfected cells. As observed by immunoblot analysis, transfection of SW-13 cells with four independent BAF250a-specific siRNA duplexes resulted in almost complete knockdown of the BAF250a protein within 48 h (Fig. 6A).
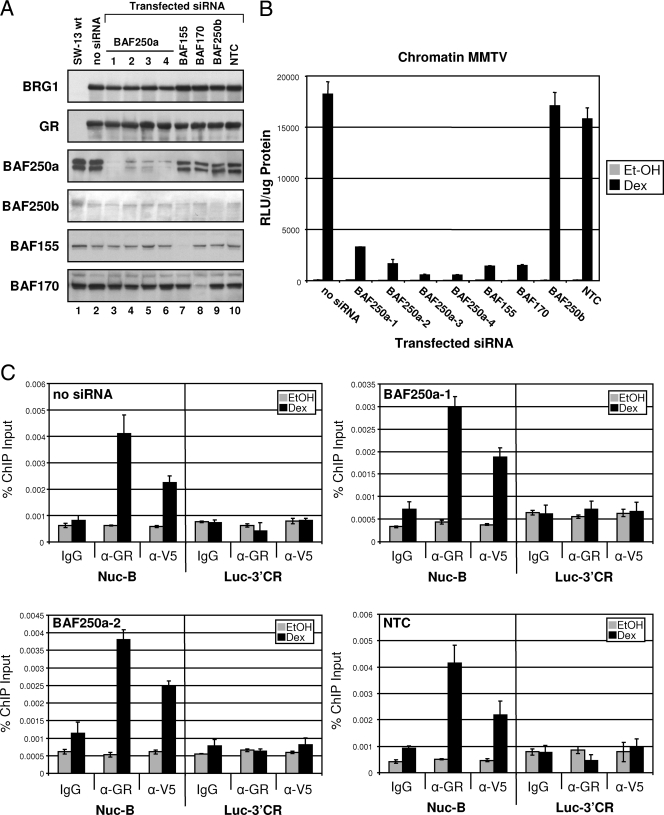
FIG. 6.
BAF250a is required for GR-mediated transcriptional activation of chromatin MMTV. (A) Western blot analysis to monitor protein expression of transfected plasmids and to evaluate the extent of protein knockdown upon introduction of siRNA. Protein expression from SW-13/MMTV cells transfected with GR and BRG1 plasmid and specific siRNAs targeting BAF250a, BAF250b, BAF155, BAF170, or NTC (nontargeting control) was evaluated using the indicated antibodies. wt, wild type. (B) Four siRNA duplexes, specific for BAF250a, were used to evaluate the requirement of BAF250a in GR-mediated BRG1-dependent transcriptional activation of chromatin MMTV. Relative MMTV-luciferase expression levels in treated SW-13/MMTV cells cotransfected with expression plasmids for GR and BRG1 and the indicated siRNAs are shown. Luciferase activity was normalized to the total protein measured (measured in relative light units [RLU] per microgram of protein). NTC siRNA was used as a negative control. Values are shown as means plus standard deviations (error bars) from three experiments. Et-OH, ethanol. (C) BAF250a is not required for recruitment of BRG1-containing SWI/SNF to stably integrated MMTV. BRG1 remodeling complex is recruited to stably integrated MMTV in a GR-dependent manner in SW-13 cells independent of BAF250a protein, as demonstrated by ChIP analysis using anti-V5 antibody (α-V5). Purified immunoprecipitated DNA fragments were analyzed by real-time PCR using primer sets covering the MMTV promoter (Nuc-B) or 3′-coding region of the luciferase reporter (Luc-3′CR). Data are displayed as percentages of ChIP input. Normal IgG was used as a negative control. Data represented are from three independent experiments.
Endogenous expression of core subunits, BAF155 and BAF170, did not change upon BAF250a protein knockdown, suggesting the assembled BRG1 remodeling complex is stable in the absence of BAF250a. Furthermore, expression of both GR and BRG1 proteins remained unchanged in response to BAF250a siRNA. The NTC siRNA duplex did not alter expression from transfected plasmids or change endogenous expression patterns of individual BAF proteins analyzed. Forty-eight hours posttransfection, cells were treated with hormone or vehicle and assayed for MMTV transcriptional activity. Whereas control siRNA had little or no effect on glucocorticoid-stimulated GR/BRG1 activity (compared to no siRNA), treating cells with BAF250a siRNA resulted in a significant decrease in GR-induced BRG1-dependent activity from the chromatin promoter (Fig. 6B). As expected, siRNA knock-down of core SWI/SNF subunits BAF155 and BAF170 also reduced GR-mediated BRG1 activity at the promoter. These findings suggest BAF250a may be a required component of the SWI/SNF chromatin remodeling complex and is necessary for GR-mediated transcriptional activation of MMTV. Interestingly, knockdown of BAF250b (ARID1B) had no effect on GR-mediated BRG1-dependent transcriptional activation from the chromatin promoter, suggesting BAF250b may not be critical for SWI/SNF transcriptional activation of MMTV in this cell system. This result is consistent with previous studies which show both BAF250 proteins compete equally for the BRG1 remodeling complex; however, protein expression levels for BAF250b in various carcinoma cell lines tested were found to be threefold lower compared to BAF250a, implying that three times more BAF250a is associated with the BRG1 complex (36). Therefore, it seems plausible that decreased BAF250a protein would have a more detrimental effect on GR-mediated BRG1-dependent transcriptional activation from chromatin MM TV.
In order to determine whether BAF250a is directly involved in targeting the SWI/SNF remodeling complex to MMTV, we performed ChIP analysis using treated SW-13/MMTV cells expressing GR and wild-type BRG1 in the presence of BAF250a-specific siRNA duplexes. GR-mediated recruitment of wild-type BRG1 to chromatin MMTV does not seem altered upon BAF250a protein knockdown (Fig. 6C). Targeting of BRG1 to the integrated promoter was shown to be equivalent to that observed for no siRNA or the NTC, suggesting that BAF250a is not involved in SWI/SNF recruitment. When purified ChIP fragments were analyzed using primers specific for the 3′ coding region of the luciferase reporter (Luc-3′CR) recruitment/binding of GR or BRG1 mutant proteins was not significantly detected, indicating the specificity of GR and BRG1 recruitment to the chromatin promoter (Fig. 6C).
To test whether BAF250a is required for transcriptional initiation of endogenous BRG1-dependent genes, we performed siRNA-mediated BAF250a protein knocked-down analysis and evaluated relative transcription levels of BRG1-dependent and GR-mediated BRG1-dependent genes CD44 and CRYAB or PLZF and HSD-11β type 2. Expression of each gene tested was significantly reduced in the presence of BAF250a siRNA but not upon treatment with the NTC (Fig. 7). Taken together, these results suggest that BAF250a may play a central role in the process of transcription initiation by the SWI/SNF chromatin remodeling complex. These experiments suggest the function of the BRG1-containing SWI/SNF remodeling complex can be severely compromised through removal of BAF250a protein, resulting in decreased transcription at chromatin MMTV and endogenous genes, including BRG1-responsive CD44 and CRYAB as well as GR-mediated BRG1-responsive genes, such as PLZF and HSD-11β type 2.
FIG. 7.
BAF250a protein is required for expression of endogenous BRG1-dependent and GR-responsive BRG1-dependent genes. (A) Real-time reverse transcription-PCR analysis was used to determine the expression level of endogenous BRG1-dependent genes CD44 or CRYAB in wild-type SW-13 cells cotransfected with BRG1 expression plasmid and BAF250a siRNA duplexes. Equal amounts of cDNA were used for real-time PCR analysis with primers specific for CD44 or CRYAB. (B) Total RNA, extracted from Dex- or vehicle-treated SW-13/MMTV cells cotransfected with GR and BRG1 expression vectors and BAF250a siRNA duplexes, was used as template for cDNA synthesis. Equal amounts of cDNA were used for real-time PCR analysis with primers specific for PLZF or HSD-11β type 2. Quantitative analysis was performed using Mx3000P software (Stratagene). Data were normalized to GAPDH and displayed as relative transcription. Values are shown as means plus standard deviations (error bars) from three experiments. EtOH, ethanol.
DISCUSSION
The SWI/SNF chromatin remodeling complex has been implicated in numerous cellular processes, including transcription, replication, DNA repair, and recombination (32). Protein chimeras, derived from two of the best characterized ATP-dependent remodeling proteins, BRG1 and SNF2h, demonstrate the mechanism of remodeling is specific for each complex, SWI/SNF or ISWI, respectively, and determined by the molecular characteristics provided by the ATPase present (11). These in vitro data also suggest that the ATPase domain is able to confer remodeling activity independent of the flanking N- and C-terminal region of the remodeling proteins. In contrast, in vivo chromatin assays indicate that ATPase activity alone is not sufficient for GR-mediated BRG1 transcriptional activation. Studies using a B-S-B chimera, SNF2h ATPase region flanked by BRG1 N- and C-terminal domains, reveal that the protein is unable to substitute for wild-type BRG1 in GR-dependent transcriptional activity, indicating that the ATPase activity of BRG1 is not functionally interchangeable (11). Taken together, these data suggest BRG1 ATPase is specifically necessary but not sufficient for GR-mediated transcriptional initiation.
The BRG1 protein is composed of multiple domains most of which have been identified by primary sequence analysis. Several protein motifs have been characterized; these include the C-terminal bromodomain, which has been implicated in the recognition of acetylated lysines in histone tails or which may serve as a protein interaction module (24). Further investigation, using a complete series of BRG1/SNF2h chimeras and BRG1 domain deletion proteins, revealed the N-terminal region of BRG1 is necessary, along with a functional ATPase domain for efficient GR-mediated transcriptional activation of chromatin MMTV (Fig. 1). Interestingly, the BRG1 C terminus does not appear necessary for GR-mediated transcriptional activation of the integrated promoter (Fig. 1D).
Conserved motifs within the BRG1 N terminus were identified using the Simple Modular Architecture Research Tool (SMART) and NCBI Conserved Domain (rpsblast) databases. The N-terminal region of BRG1, defined as encompassing amino acids 1 to 728, contains several regions of interest which could prove essential for BRG1 function. A proline-rich domain was identified within amino acids 4 to 344. Although the functional relevance of this area remains unclear, proline-rich domains are often implicated in protein interactions or may contribute to the conformational structure of the protein (37). Also, identified within the N terminus of BRG1 were an HSA domain and a BRK domain located between amino acids 475 to 532 and amino acids 612 to 656, respectively. HSA domains are regions of unknown function found in helicases and other DNA-binding proteins of eukaryotes (7). BRK domains are areas found associated with transcription and CHROMO domain helicases and in DEAD/DEAH box helicase domains (7).
Our results demonstrated that only the HSA domain within the BRG1 N terminus is required for GR-mediated transcriptional activation of BRG1-dependent promoters, PLZF and HSD-11β (type 2) (Fig. 4). Immunoprecipitation studies demonstrate the HSA region is not required for SWI/SNF complex assembly or for association with core BAF subunits, BAF155, BAF170, and GR-associating protein, BAF60a. Targeting of the mutant containing complexes to BRG1-dependent promoters does not seem to require the HSA or BRK regions. As observed by chromatin IP, ligand-induced targeting of BRG1 to MMTV does not appear altered upon removal of either the HSA or BRK domain compared to wild-type BRG1. Interestingly, removal of the HSA region resulted in a significant decrease in the ability of BRG1 to stimulate a remodeled nucleosomal state within the hypersensitive region of MMTV (Fig. 3). Collectively, these observations suggest the HSA domain is necessary, along with the ATPase activity, for GR-mediated BRG1-dependent transcriptional activation of chromatin MMTV and endogenous hormone-responsive promoters.
Previously, mutant BRG1 proteins have been isolated and characterized that exhibit normal ATPase function but display diminished nucleosome remodeling properties (3). A partial loss-of-function BRG1 mutant, designated E1083G, exhibiting normal ATPase activity is recruited to the control region within the β-globin locus but displayed diminished chromatin remodeling and expression, suggesting BRG1-ATPase activity is required but is not sufficient for function. Taken together, these studies, along with the present report, imply SWI/SNF function does not reside exclusively with BRG1-dependent ATP hydrolysis but also require regions within and outside the ATPase domain for optimal nucleosomal remodeling and transcriptional regulation.
To examine the importance of the HSA domain in the context of SWI/SNF function, we scanned the published literature for reported proteins that associate over the HSA region. Several interacting proteins were identified through this screen; however, their minimal BRG1 binding area was reported to encompass the full N-terminal region. A more stringent search was conducted for proteins that interact with BRG1 specifically through the HSA domain. We identified one interacting protein that associates with BRG1 exclusively through the HSA domain, ARID-containing BAF250a (ARID1A/p270/hOsa1) (16). Biochemical studies identified multiple regions within BRG1 that are involved in binding to the C terminus of BAF250a. However, in vitro binding assays suggest the strongest interaction with BAF250a requires residues 380 to 558 of BRG1 (18). Interestingly, amino acids 380 to 558 encompass the complete HSA domain, providing further evidence this region may be essential for SWI/SNF assembly and function. In an attempt to verify this observation, we performed coimmunoprecipitation assays which revealed a significant loss of BAF250a-associated BRG1 upon deletion of the HSA domain (Fig. 5). To assess BAF250a as a critical component of the SWI/SNF complex, which is required for GR-mediated transcriptional activation, the subunit was targeted for siRNA mediated protein knockdown. Removal of BAF250a resulted in a significant decrease in GR-mediated BRG1-dependent transcriptional activation of chromatin MMTV as well as at endogenous PLZF and HSD-11β (type 2) promoters. Taken together, these data suggest BAF250a is a required component of the SWI/SNF chromatin remodeling complex involved in transcriptional initiation of target promoters.
BAF250a is the largest component of the human SWI/SNF complex with an ARID DNA-binding domain as its best-established feature. Members of the ARID protein family have been implicated in numerous cellular processes, such as growth control, differentiation, and development (38). The ARID domain harbors a helix-turn-helix structural motif that has been implicated in sequence-specific and nonspecific DNA binding as seen for the Dead-ringer and BAF250 proteins, respectively (39). It has been suggested that BAF250a may be involved in recruiting SWI/SNF to regulated genes through its ability to stimulate GR-, estrogen receptor-, and androgen receptor-mediated transcriptional activation. Additional motifs within BAF250a include glutamine-rich domains and multiple LXXLL motifs that have been implicated in the association of ligand-bound NRs (13).
Our in vivo mutational analysis suggests a novel and critical role of BAF250a may be to stabilize the SWI/SNF complex at target promoters and/or increase the specific activity of the BRG1 motor protein, via protein-protein interactions that require the BRG1 HSA domain. The HSA deletion proteins are stable, assemble into SWI/SNF-related complexes lacking BAF250a, and are recruited to the promoter in response to hormone. However, GR-mediated chromatin remodeling and target gene expression are significantly diminished. Therefore, while the BRG1 ATPase activity is necessary for SWI/SNF function, our results indicate that it is not sufficient in vivo to alter chromatin architecture and activate gene transcription.
Acknowledgments
We thank the Archer lab members for helpful suggestions and Craig Burd, Brian Keppler, and Paul Wade for critical reading of the manuscript.
This research was supported by Intramural Research Program of NIH, National Institute of Environmental Health Sciences. This work was supported by U.S. National Institutes of Health, National Cancer Institute grant CA-093660 (H.-Y.F).
Footnotes
Published ahead of print on 17 December 2007.
REFERENCES
- 1.Archer, T. K., M. G. Cordingley, R. G. Wolford, and G. L. Hager. 1991. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol. Cell. Biol. 11688-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belandia, B., R. L. Orford, H. C. Hurst, and M. G. Parker. 2002. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 214094-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bultman, S. J., T. C. Gebuhr, and T. Magnuson. 2005. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes Dev. 192849-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, J., H. K. Kinyamu, and T. K. Archer. 2006. Changes in attitude, changes in latitude: nuclear receptors remodeling chromatin to regulate transcription. Mol. Endocrinol. 201-13. [DOI] [PubMed] [Google Scholar]
- 5.Chi, Y., V. Senyuk, S. Chakraborty, and G. Nucifora. 2003. EVI1 promotes cell proliferation by interacting with BRG1 and blocking the repression of BRG1 on E2F1 activity. J. Biol. Chem. 27849806-49811. [DOI] [PubMed] [Google Scholar]
- 6.Daubresse, G., R. Deuring, L. Moore, O. Papoulas, I. Zakrajsek, W. R. Waldrip, M. P. Scott, J. A. Kennison, and J. W. Tamkun. 1999. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development 1261175-1187. [DOI] [PubMed] [Google Scholar]
- 7.Doerks, T., R. R. Copley, J. Schultz, C. P. Ponting, and P. Bork. 2002. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 1247-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eberharter, A., and P. B. Becker. 2004. ATP-dependent nucleosome remodelling: factors and functions. J. Cell Sci. 1173707-3711. [DOI] [PubMed] [Google Scholar]
- 9.Elfring, L. K., R. Deuring, C. M. McCallum, C. L. Peterson, and J. W. Tamkun. 1994. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol. Cell. Biol. 142225-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan, H. Y., X. He, R. E. Kingston, and G. J. Narlikar. 2003. Distinct strategies to make nucleosomal DNA accessible. Mol. Cell 111311-1322. [DOI] [PubMed] [Google Scholar]
- 11.Fan, H. Y., K. W. Trotter, T. K. Archer, and R. E. Kingston. 2005. Swapping function of two chromatin remodeling complexes. Mol. Cell 17805-815. [DOI] [PubMed] [Google Scholar]
- 12.García-Pedrero, J. M., E. Kiskinis, M. G. Parker, and B. Belandia. 2006. The SWI/SNF chromatin remodeling subunit BAF57 is a critical regulator of estrogen receptor function in breast cancer cells. J. Biol. Chem. 28122656-22664. [DOI] [PubMed] [Google Scholar]
- 13.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387733-736. [DOI] [PubMed] [Google Scholar]
- 14.Hsiao, P.-W., C. J. Fryer, K. W. Trotter, W. Wang, and T. K. Archer. 2003. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol. Cell. Biol. 236210-6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, M., F. Qian, Y. Hu, C. Ang, Z. Li, and Z. Wen. 2002. Chromatin-remodelling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes. Nat. Cell Biol. 4774-781. [DOI] [PubMed] [Google Scholar]
- 16.Hurlstone, A. F., I. A. Olave, N. Barker, M. van Noort, and H. Clevers. 2002. Cloning and characterization of hELD/OSA1, a novel BRG1 interacting protein. Biochem. J. 364255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ichinose, H., J. M. Garnier, P. Chambon, and R. Losson. 1997. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene 18895-100. [DOI] [PubMed] [Google Scholar]
- 18.Inoue, H., T. Furukawa, S. Giannakopoulos, S. Zhou, D. S. King, and N. Tanese. 2002. Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J. Biol. Chem. 27741674-41685. [DOI] [PubMed] [Google Scholar]
- 19.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 142441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 21.Lee, H. L., and T. K. Archer. 1994. Nucleosome-mediated disruption of transcription factor-chromatin initiation complexes at the mouse mammary tumor virus long terminal repeat in vivo. Mol. Cell. Biol. 1432-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemon, B., C. Inouye, D. S. King, and R. Tjian. 2001. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414924-928. [DOI] [PubMed] [Google Scholar]
- 23.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 1191-16. [DOI] [PubMed] [Google Scholar]
- 24.Mohrmann, L., and C. P. Verrijzer. 2005. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta 168159-73. [DOI] [PubMed] [Google Scholar]
- 25.Nie, Z., Y. Xue, D. Yang, S. Zhou, B. J. Deroo, T. K. Archer, and W. Wang. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 208879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen, A. L., C. Sanchez, H. Ichinose, M. Cervino, T. Lerouge, P. Chambon, and R. Losson. 2002. Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1alpha. EMBO J. 215797-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otsuki, T., Y. Furukawa, K. Ikeda, H. Endo, T. Yamashita, A. Shinohara, A. Iwamatsu, K. Ozawa, and J. M. Liu. 2001. Fanconi anemia protein, FANCA, associates with BRG1, a component of the human SWI/SNF complex. Hum. Mol. Genet. 102651-2660. [DOI] [PubMed] [Google Scholar]
- 28.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3247-253. [DOI] [PubMed] [Google Scholar]
- 29.Saha, A., J. Wittmeyer, and B. R. Cairns. 2006. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7437-447. [DOI] [PubMed] [Google Scholar]
- 30.Strober, B. E., J. L. Dunaief, S. Guha, and S. P. Goff. 1996. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol. Cell. Biol. 161576-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trotter, K. W., and T. K. Archer. 2007. Nuclear receptors and chromatin remodeling machinery. Mol. Cell. Endocrinol 265-266:162-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trotter, K. W., and T. K. Archer. 2004. Reconstitution of glucocorticoid receptor-dependent transcription in vivo. Mol. Cell. Biol. 243347-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vassallo, M. F., and N. Tanese. 2002. Isoform-specific interaction of HP1 with human TAFII130. Proc. Natl. Acad. Sci. USA 995919-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan, Y., and S. K. Nordeen. 2002. Overlapping but distinct gene regulation profiles by glucocorticoids and progestins in human breast cancer cells. Mol. Endocrinol. 161204-1214. [DOI] [PubMed] [Google Scholar]
- 35.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 155370-5382. [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, X., N. G. Nagl, D. Wilsker, M. Van Scoy, S. Pacchione, P. Yaciuk, P. B. Dallas, and E. Moran. 2004. Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. Biochem. J. 383319-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williamson, M. P. 1994. The structure and function of proline-rich regions in proteins. Biochem. J. 297249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilsker, D., A. Patsialou, P. B. Dallas, and E. Moran. 2002. ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. 1395-106. [PubMed] [Google Scholar]
- 39.Wilsker, D., A. Patsialou, S. D. Zumbrun, S. Kim, Y. Chen, P. B. Dallas, and E. Moran. 2004. The DNA-binding properties of the ARID-containing subunits of yeast and mammalian SWI/SNF complexes. Nucleic Acids Res. 321345-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan, Z., K. Cui, D. M. Murray, C. Ling, Y. Xue, A. Gerstein, R. Parsons, K. Zhao, and W. Wang. 2005. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 191662-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]