Abstract
The transporter Ccc1 imports iron into the vacuole, which is the major site of iron storage in fungi and plants. CCC1 mRNA is destabilized under low-iron conditions by the binding of Cth1 and Cth2 to the 3′ untranslated region (S. Puig, E. Askeland, and D. J. Thiele, Cell 120:99-110, 2005). Here, we show that the transcription of CCC1 is stimulated by iron through a Yap consensus site in the CCC1 promoter. We identified YAP5 as being the iron-sensitive transcription factor and show that a yap5Δ strain is sensitive to high iron. Green fluorescent protein-tagged Yap5 is localized to the nucleus and occupies the CCC1 promoter independent of the iron concentration. Yap5 contains two cysteine-rich domains, and the mutation of the cysteines to alanines in each of the domains affects the transcription of CCC1 but not DNA binding. The fusion of the Yap5 cysteine-containing domains to a GAL4 DNA binding domain results in iron-sensitive GAL1-lacZ expression. Iron affects the sulfhydryl status of Yap5, which is indicative of the generation of intramolecular disulfide bonds. These results show that Yap5 is an iron-sensing transcription factor and that iron regulates transcriptional activation.
Iron is an essential nutrient required by all eukaryotes. In high concentrations, however, iron can be toxic, necessitating tight control over its concentration within cells. Multicellular organisms can transfer iron between cell types; however, multicellular and single-cell eukaryotes do not have an excretory mechanism to dispose of iron. Iron homeostasis results from the ability to regulate iron acquisition or to store iron once it is absorbed. The ability to store iron makes it available for future use while preventing toxicity. In fungi and plants, iron is stored in the vacuole, where it can be exported when needed. In the budding yeast Saccharomyces cerevisiae, high-affinity and low-affinity transporters export iron from the vacuole to the cytosol. The high-affinity vacuolar iron transport complex Fet5/Fth1 is homologous to the high-affinity plasma membrane transport complex Fet3/Ftr1 (22), while the low-affinity vacuolar iron transporter Smf3 is homologous of the cell surface transporter Smf1 (3, 18). The high-affinity vacuolar and cell surface transporters, as well as SMF3, are under the transcriptional control of the iron-sensing transcription factors Aft1 and Aft2 (4, 20). Thus, iron transport into the cytosol from the vacuole and across the cell surface into the cytosol is regulated coordinately.
Much less is known about the regulation of iron transport from the cytosol to the vacuole. The only identified vacuolar iron importer in plants and fungi is Ccc1 (referred to as VIT1 in plants) (14), which can also transport Mn2+ in yeast (13). The overexpression of Ccc1 leads to profound cytosolic iron depletion, implying that this transporter must be regulated to maintain cytosolic iron homeostasis. Recently, Puig et al. showed that the stability of CCC1 mRNA was regulated by Cth1 and Cth2 and that CTH2 was under the transcriptional control of Aft1 (19). Under low-iron conditions, Aft1p induces the expression of Cth2, which then destabilizes CCC1 mRNA, resulting in the decreased expression of the transporter, thus preserving cytosolic iron levels. Iron-dependent changes in CCC1 mRNA levels still occurred in cells with deletions in both CTH1 and CTH2. Based on this observation, we considered that there may be additional modes of regulation for CCC1. We demonstrate that the transcription of CCC1 mRNA is regulated by iron and identify Yap5 as being the iron-responsive transcription factor. YAP5 is one of a family of eight homologous yeast genes that regulate transcriptional responses to a variety of environmental factors including cadmium and hydrogen peroxide. These genes are members of the basic-region leucine zipper transcription factors. We show that Yap5 is the only member of this family that regulates the expression of vacuolar iron transport in response to cytosolic iron.
MATERIALS AND METHODS
Yeast strains and culture conditions.
Wild-type strain DY150 (W303 background) and its deletion strains were used in most experiments. The ccc1Δ strains were described previously (14). Strains with specific deletions (cth1Δ, cth2Δ, cth1Δ cth2Δ, and yap5Δ) were generated in the DY150 background by PCR amplifying the KanMx4 deletion cassettes from specific yeast strains in the homozygous diploid deletion collection (Research Genetics, Stanford, CA). Deletion strains of yap1Δ-yap8Δ were obtained from the BY4741 (S288C background) haploid collection. The glr1Δ and trr1Δ strains were obtained from Dennis Winge (University of Utah). Complete minimal (CM) medium was composed of yeast nitrogen base, dextrose, and the required amino acids. Low-iron medium was made by adding 80 μM bathophenanthroline disulfonate, and high-iron medium resulted from the addition of the designated concentration of FeSO4 or ferrous ammonium sulfate. All experiments were performed a minimum of three times.
Plasmid construction.
The CCC1-lacZ reporter construct was created by cloning 600 bp of the upstream region of CCC1 by PCR. The PCR fragment was placed in a YEp354 lacZ expression vector, which is a high-copy-number vector. For low-copy-number expression studies, we utilized reporter vector YCp pRW95-3LacZ, obtained from D. Stillman (University of Utah). Promoter truncations were performed by using the following PCR primers: 5′-cgcggatccAACAACATCATCGACGAATG-3′ (−555 bp), 5′-cgcggatccCGACACATGCCACTAAGACA-3′ (−496 bp), 5′-cgcggatccGCAGCCGTTAGCAGTTGTTT-3′ (−400 bp), 5′-cgcggatccTTTCGGTCTGGACCAATCGC-3′ (−244 bp), 5′-cgcggatccCTAATATTACTAACATACCC-3′ (−221 bp), 5′-cgcggatccCATACCCTCTTCTCATTGGC-3 (−199 bp), 5′-cgcggatccAGCCCGAATGTCTCACAATG-3′ (−136 bp), and 5′-cccaagcttCATAATATTTGTGTGCACGAG-3′ (−1 bp). The first upstream Yap binding site in the CCC1 promoter region was mutated by site-directed mutagenesis using the QuikChange kit from Stratagene (La Jolla, CA) with two primers: 5′-GCCGCATTTCTCACTAATATTCAGAACATACCCTCTTCTCATTGGC-3′ and 5′-GCCAATGAGAAGAGGGTATGTTCTGAATATTAGTGAGAAATGCGGC-3′. The second Yap binding site in the CCC1 promoter region was mutated by using primer 5′-cgcggatccTTTCGGTCTGGACCAATCGCGCCGCATTTCTCACTAATAGGCAGCCCATACCCTCTTCTCATTGGC-3′. The FLAG-tagged CCC1 construct was described previously by Chen et al. (2).
The YAP5 carboxyl-terminal hemagglutinin (HA) epitope tag was constructed by PCR utilizing a 12× HA-containing plasmid (5). YAP5 amino-terminal green fluorescent protein (GFP)- or His6-tagged plasmids were cloned into a high-copy-number pTF63 vector or a low-copy-number vector (pRS416) obtained from D. Stillman (University of Utah). All these epitope-tagged YAP5 genes were regulated by the YAP5 promoter, as the YAP5 open reading frame and 600 bp upstream of the ATG were cloned by PCR. Mutation of Yap5 cysteines to alanines, or cysteines to aspartic acids, was generated by PCR. A double-fusion PCR technique was used to modify the fragment between the Pst1 site and the stop site. The GAL4 DNA binding domain (DBD) was fused to the YAP5 activation cysteine-rich domain (CRD) by cloning a PCR fragment from YAP5 that had been truncated and that included amino acid 115 to the stop codon into vector pMA424 (16) using the primers 5′-ccg gaa ttc ATG GAA TCG GAA AAT CAT GCC CT-3′ and 5′-cgc gga tcc TCA GTG GAT GAT GGA CCG GA-3′. GAL1-lacZ, constructed by cloning 1,000 bp of the GAL1 promoter into vector YEP353; GAL4-tZAP1, constructed to contain the GAL4 DBD; and the activation domain (amino acids 552 to 800) of ZAP1 (1) were the generous gifts of Dennis Winge (University of Utah).
CHIP.
Chromatin immunoprecipitation (CHIP) was performed as described previously (5). Sheared chromatin was immunoprecipitated using a rabbit anti-GFP antibody (Abcam) against GFP-Yap5 or rabbit anti-HA (Covance, Berkeley, CA) against Aft1-HA, followed by sheep anti-rabbit immunoglobulin G (IgG) Dynabeads (Invitrogen, Carlsbad, CA). The primers, which produced a PCR product of 367 bp including the Yap binding sites in CCC1 promoter region, were 5′-GCAGCCGTTAGCAGTTGTT-3′ and 5′-GAGAGTGATGTCGCTTTAAC-3′. The primers for calmodulin and FET3 as well as the construction of the AFT1-HA plasmid were described previously (5).
β-Galactosidase assay.
β-Galactosidase activity was measured using a 96-well kinetic assay as described previously (5), except that the reaction rate was assayed over 10 min. Specific activity is defined as nmol/min/mg protein.
Northern blotting and S1 nuclease protection assay.
Total RNA was isolated from cells grown at mid-log phase in low-iron or high-iron medium. Northern blotting and S1 assays were performed as described previously (5).
Western blot analysis.
Cell extracts were prepared using a modification of the trichloroacetic acid (TCA) procedure described previously (7). Cells were vortexed with glass beads with either a lysate buffer (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, lysate buffer with protease inhibitors) or a TCA buffer (10% TCA, 1.0 M Tris [pH 8.0], 7.5 M ammonium acetate, 0.5 M EDTA). The TCA extract was dried with acetone and then incubated with 7.5 mM 4-acetamido-4-maleimidylstilbene-2,2′-disulfonic acid (AMS; Sigma, St. Louis, MO) for 1 h at room temperature. Samples, desalted by a G-50 spin column (GE Healthcare), were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). When required, dithiothreitol (DTT) was added to a final concentration of 100 mM. Vacuoles were isolated as described previously (14). Western analysis was performed as described previously (6). A rabbit polyclonal antibody against a glutathione S-transferase fusion construct expressing a peptide containing the terminal 99 amino acids of Ccc1 was made. The polyclonal antiserum, which was immunopurified, could detect endogenous levels of Ccc1 by Western blotting on purified vacuoles and not in whole cells. The antibody could detect overexpressed Ccc1 in the whole-cell extracts. Mouse anti-Gal4 DBD antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-His6 antibody was purchased from Invitrogen (Carlsbad, CA).
Microscopy.
Images were captured on an Olympus BX051 epifluorescence microscope using a 100× 1.4 aperture. DAPI (4′,6′-diamidino-2-phenylindole) staining was performed to identify nuclei. Cells were incubated with 1 μM DAPI for 20 min at room temperature and then mounted onto coverslips precoated with concanavalin A.
All experiments were performed a minimum of two times and on average were performed three to four times.
RESULTS
Ccc1 levels are controlled by mRNA transcript levels.
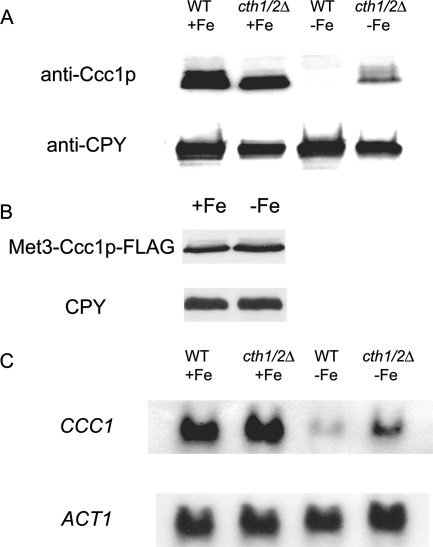
To determine how Ccc1 is regulated, we measured the levels of vacuolar Ccc1 in cells grown in iron-limited or iron-replete medium. Vacuoles isolated from cells grown in iron-limited medium had no detectable Ccc1, while vacuoles isolated from cells grown in iron-replete medium showed abundant Ccc1 (Fig. 1A). Previously, we showed that Ccc1-FLAG could suppress the high-iron sensitivity of ccc1Δ cells, indicating that the epitope-tagged protein is competent to transport iron (14). To determine if Ccc1 is regulated at the protein level, we expressed Ccc1 with a carboxyl-terminal FLAG epitope under the control of a regulated promoter (MET3). The cells were grown in either high- or low-iron medium, without methionine, to permit the expression of Ccc1-FLAG. There was little difference in the amount of Ccc1-FLAG in cells grown under either iron-replete or iron-limited conditions (Fig. 1B), suggesting that iron does not affect protein stability.
FIG. 1.
Regulation of CCC1 protein and mRNA levels by iron. Wild-type (WT) and cth1Δ cth2Δ cells were grown overnight in iron-replete or iron-limited medium. (A) Cells were homogenized, and vacuoles were isolated, solubilized, and examined by Western blot analysis. The Western blots were probed with a polyclonal antibody directed against Ccc1 or a monoclonal antibody against carboxypeptidase Y (CPY), followed by peroxidase-conjugated goat anti-rabbit IgG or peroxidase-conjugated goat anti-mouse IgG. (B) Wild-type cells were transformed with a plasmid containing a MET3-regulated CCC1-FLAG construct. Cells were grown overnight in iron-replete or iron-limited medium in the absence of methionine, which permits the expression of MET3-CCC1. Cells were harvested, and the levels of Ccc1-FLAG or CPY were determined by Western blotting as described above (A). (C) mRNA was isolated from cells grown overnight in iron-replete or iron-limited medium, and CCC1 or ACT1 mRNA was analyzed by Northern blotting.
It may be that the carboxyl-terminal epitope prevented protein degradation or that the protein was not regulated at the level of degradation but at the level of mRNA. To test these possibilities, we examined CCC1 mRNA levels from wild-type and cth1Δ cth2Δ strains grown in iron-replete or iron-limited medium. In iron-replete medium, CCC1 mRNA levels were similar in wild-type and cth1Δ cth2Δ cells (Fig. 1C). Little CCC1 mRNA was detected in wild-type cells grown in iron-limited medium. Puig et al. previously demonstrated that CCC1 mRNA could be regulated by changes in half-life through the binding of Cth1 and Cth2 to the 3′ untranslated region (19). We observed an increase in CCC1 transcripts in cth1Δ cth2Δ cells grown under low-iron conditions compared to wild-type cells grown in low iron. The deletion of both CTH1 and CTH2 did not restore CCC1 mRNA levels to that of wild-type cells grown in iron-replete medium, confirming observations described previously by Puig et al. (19). The inability of the double-deletion strain to restore message levels to that of the iron-replete wild type was also seen at the protein level (c.f. Fig. 1A). These results lead to two conclusions: Ccc1 levels reflect mRNA levels, and CCC1 message levels are not regulated solely by Cth1 and Cth2.
Transcription of CCC1 is iron regulated.
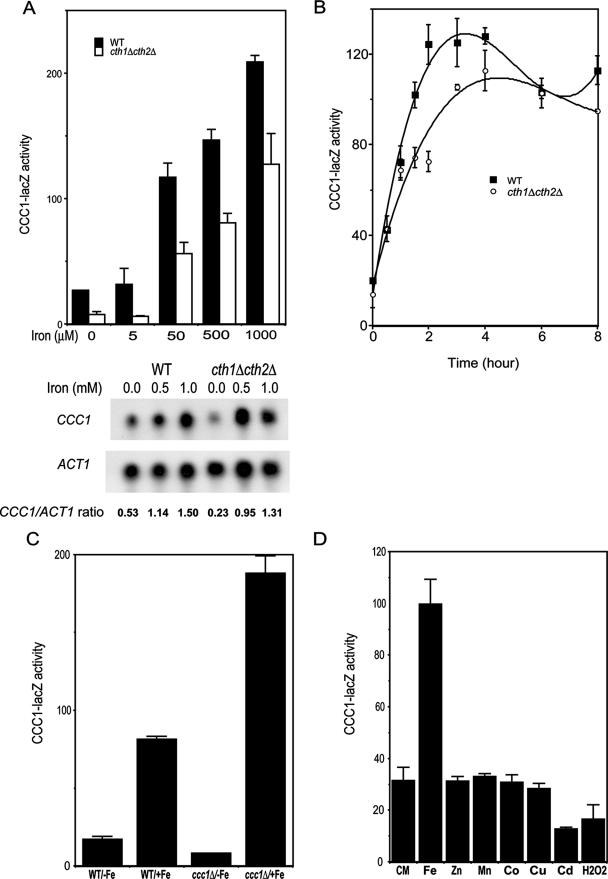
To examine whether the transcription of CCC1 was subject to regulation, we generated a lacZ reporter construct driven by 600 bp of the CCC1 promoter. This region encompasses the complete intergenic region between CCC1 and the 5′-distal gene MSC3. The ability of the reporter construct to respond to iron was measured in wild-type and cth1Δ cth2Δ cells. The addition of iron resulted in the increased expression of CCC1-lacZ (Fig. 2A). The absolute level of reporter expression was lower in cth1Δ cth2Δ cells, but the relative changes in expression were similar to those of wild-type cells. The change in reporter construct activity mirrors changes in mRNA levels, although the absolute amount of the mRNA level depends on the expression of Cth1 and Cth2 (Fig. 2A). We note that cth1Δ cth2Δ cells grown in iron-containing medium have a lower level of CCC1 mRNA than do wild-type cells. If the degradation of CCC1 mRNA was prevented by the deletion of CTH1 and CTH2, then the level of mRNA in cth1Δ cth2Δ cells would be expected to be higher. These results suggest that CCC1 mRNA levels may be responsive to factors other than Cth1 or Cth2. The increase in CCC1 transcripts was rapid, occurring within 2 h of the addition of iron (Fig. 2B). These results indicated that the transcription of CCC1 was affected by iron, independent of the effect of Cth1 and Cth2 on message stability.
FIG. 2.
Expression of a CCC1-lacZ reporter construct is regulated by iron. (A) Cells (wild type [WT] and cth1Δ cth2Δ) were transformed with a reporter plasmid containing 600 bp of the CCC1 promoter region fused with the lacZ gene. Cells were incubated for 3 h in increasing concentrations of iron, harvested, and assayed for β-galactosidase activity and cell protein. For this and all subsequent β-galactosidase measurements, the data are reported as activity in nmol product/min/mg protein. CCC1 and ACT1 mRNAs were analyzed by Northern blot. CCC1 or ACT1 mRNA band intensity was quantified by image analysis, and the ratio of the two is presented. (B) Wild-type and cth1Δ cth2Δ cells were incubated in CM medium supplemented with 500 μM FeSO4. At the selected times, cells were harvested, β-galactosidase activity was determined, and activity was normalized for protein. (C) Wild-type and ccc1Δ cells, transformed with a CCC1-lacZ reporter plasmid, were incubated for 2 h with or without 500 μM FeSO4. Cells were harvested, and β-galactosidase activity was determined. (D) Wild-type cells transformed with a CCC1-lacZ reporter plasmid were incubated in CM medium with 500 μM Fe2+, 1.0 mM Zn2+, 1.0 mM Mn2+, 100 μM Co2+, 200 μM Cu+, 25 μM Cd2+, or 0.0025% H2O2 for 2 h. Cells were harvested, and β-galactosidase activity was determined.
The deletion of CCC1 leads to increased cytosolic iron, as the transport of iron into the vacuole is severely impaired (14). If CCC1 transcription is regulated by cytosolic iron, then an increased level of cytosolic iron should lead to the increased expression of the CCC1-lacZ reporter construct. Wild-type and ccc1Δ cells, transformed with a high-copy reporter construct, were grown in iron-limited or iron-replete medium, and CCC1-lacZ activity was measured. Cells grown in iron-limited medium showed low levels of expression of the reporter construct, and the deletion of CCC1 further reduced that expression (Fig. 2C). Conversely, when ccc1Δ cells were incubated in iron-replete medium, there was enhanced expression of the CCC1-lacZ reporter construct. Similar results were obtained if cells were transformed with a low-copy-number plasmid containing the reporter construct (data not shown). Thus, the lack of Ccc1 results in increased cytosolic iron and consequently increased expression of β-galactosidase.
The overexpression of CCC1 was shown to reduce cytosolic Mn2+, suggesting that Ccc1 may not be specific for iron but that it can transport other transition metals (13). Cells transformed with a high-copy-number CCC1-lacZ plasmid were incubated in CM medium containing different levels of transition metals (Fig. 2D). The concentrations of Cu+, Co2+, and Cd2+ were limited by the toxicity of the metals, but no effect on CCC1-lacZ was seen at concentrations of these metals that ranged from nontoxic to subtoxic. The addition of iron led to the increased expression of CCC1-lacZ; no effect was seen with either Zn2+ or Mn2+. The addition of H2O2 to the medium had no effect on the transcription of the reporter construct. Similar results were obtained with a low-copy-number plasmid containing the reporter construct (data not shown). These results indicate that the transcription of CCC1 is specific for iron and not for oxidant damage, supporting the view that increased cytosolic iron is essential for the transcriptional activation of CCC1.
Yap5 is responsible for the iron-dependent transcription of CCC1.
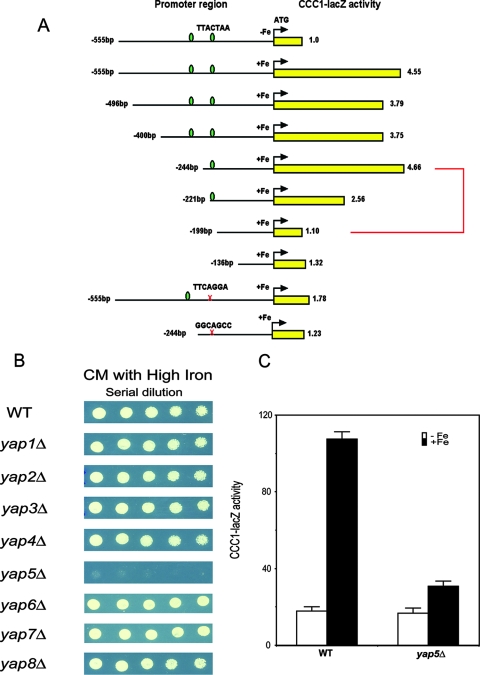
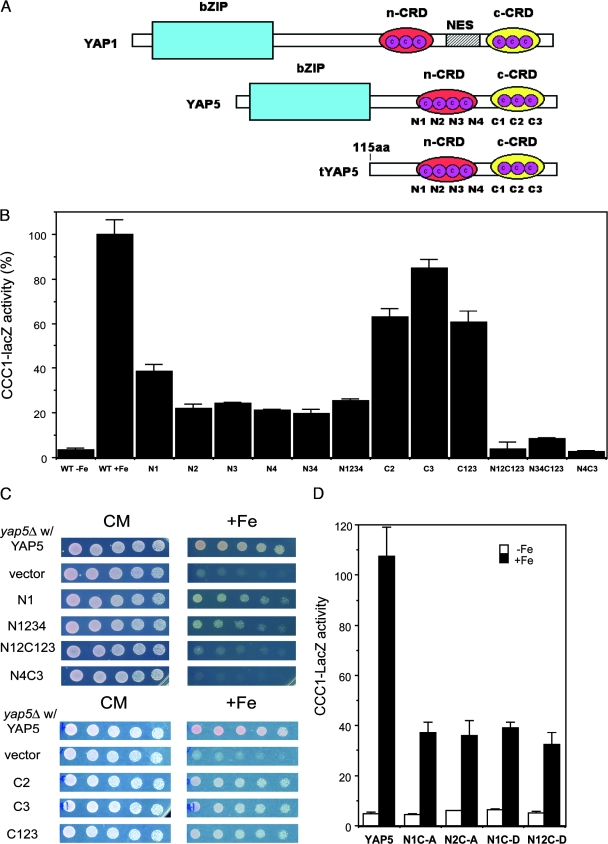
To investigate the transcriptional regulation of CCC1, we subcloned the promoter region to identify cis-acting elements. A 600-bp sequence between the 5′ end of the CCC1 open reading frame and the start of the adjacent gene was placed in front of a lacZ reporter construct, and the plasmid was transformed into wild-type cells. There was little expression of β-galactosidase from a reporter lacking the CCC1 promoter sequence (data not shown). The insertion of the CCC1 promoter sequence, however, resulted in a fivefold increase in CCC1-lacZ expression over background levels upon the addition of iron (Fig. 3A). There was a dramatic decrease in expression in the absence of the 46 bp from the region of residues −244 to −199. Examination of this 46-bp region showed the presence of a Yap consensus binding site (9). Mutation of nucleotides in the Yap site showed that this site is both necessary and sufficient for the iron-regulated expression of the CCC1-lacZ reporter construct. There was a second upstream Yap consensus site, but the truncation of this site had no effect on CCC1-lacZ expression.
FIG. 3.
Yap5 is responsible for the iron-dependent transcription of CCC1. (A) Selected regions of the CCC1 promoter or mutated regions of the CCC1 promoter sequence were cloned into lacZ reporter constructs, and the constructs were transformed into wild-type cells grown in iron-limited medium. Iron (500 μM FeSO4) was added to the medium for 2 h, cells were harvested, β-galactosidase activity was determined, and activity was normalized for protein (yellow bars with associated numbers). The data are presented as the change in β-galactosidase activity. The green ovals represent consensus YAP binding sites, and the × indicates the site of mutations. The 46-bp region is highlighted with a red bracket. (B) Wild-type (WT) and YAP deletion strains in the BY4741 haploid deletion collection were spotted in serial dilutions on medium containing high concentrations of iron. The cells were grown for 2 days at 30°C. (C) Wild-type and yap5Δ cells (haploid W303 background) were transformed with a plasmid containing a CCC1-lacZ reporter construct. Cells grown in iron-limited medium were incubated with (black bars) or without (white bars) 500 μM FeSO4 for 2 h, β-galactosidase activity was determined, and activity was normalized for protein.
Members of the YAP transcription family are responsible for much of the cellular response to oxidative stress. For example, Yap1 is activated by oxidative stresses such as H2O2 (12); however, the addition of 0.0025% H2O2 did not lead to the expression of CCC1-lacZ (Fig. 2D). These results suggest that Yap1 is not responsible for the transcription of CCC1. Previously, we showed that the deletion of CCC1 led to decreased growth on high-iron medium (14). To define which member of the Yap family is involved in the iron regulation of CCC1, we examined the iron sensitivity of BY4741 strains deleted for specific YAP genes (Fig. 3B). In the presence of high iron, only yap5Δ grew poorly. The poor-growth phenotype of the yap5Δ strain could be suppressed by the transformation of cells with a MET3-regulated CCC1 plasmid (data not shown). This result is consistent with the view that the lack of Ccc1 expression was responsible for the iron-sensitive phenotype (14). To determine if YAP5 affected the expression of CCC1, we generated a YAP5 deletion in a W303 background and examined the effect of the deletion on the iron-induced transcription of a CCC1-lacZ reporter construct. The deletion of YAP5 prevented the iron-induced expression of β-galactosidase activity (Fig. 3C). These results indicate that Yap5 is responsible for the iron-induced transcription of CCC1 mRNA.
Yap5 is constitutively localized to the nucleus.
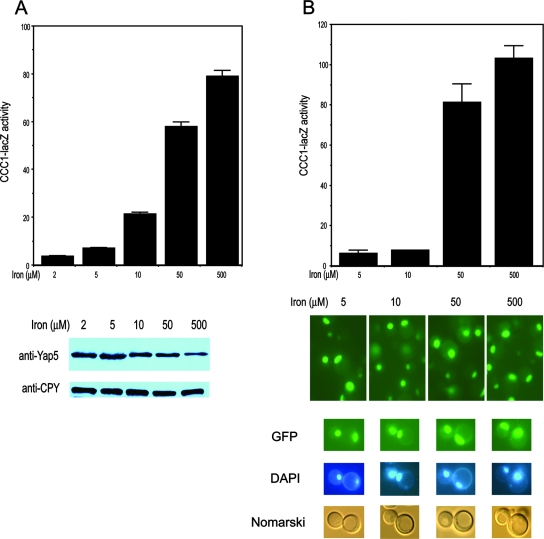
To determine the effect of iron on Yap5, we examined the behavior of epitope-tagged Yap5. Yap5 with a carboxyl-terminal HA epitope was capable of suppressing the iron toxicity of a yap5Δ strain; however, the expression of CCC1-lacZ was constitutive and iron independent (data not shown). Since Yap5-HA was constitutively active, we examined the effect of Yap5 containing other epitopes. Amino-terminal His or amino-terminal GFP-YAP5 constructs complemented the high-iron toxicity of the yap5Δ strain and led to the iron-dependent expression of CCC1-lacZ activity (Fig. 4A and B). There was, however, a slightly lower level of β-galactosidase at 10 μM iron in cells expressing GFP-Yap5 than in those expressing His-Yap5, suggesting that the larger GFP epitope may affect the sensitivity of the protein to iron. Iron increased the transcriptional activity of the His-Yap5 construct but did not increase the amount of His-Yap5 (Fig. 4A). We note a consistent decrease in His-Yap5 protein levels at high iron concentrations. GFP-Yap5 expressed by a high-copy-number plasmid was localized to the nucleus independent of iron levels (Fig. 4B). The fluorescence signal from a low-copy-expressed GFP-Yap5 was too low to be detected. However, as shown below, the data suggest that a low-copy GFP-Yap5 is constitutively localized to the nucleus.
FIG. 4.
Epitope-tagged Yap5 is localized to the nucleus and regulates transcription of CCC1. (A) yap5Δ cells were transformed with a 2μm plasmid containing His-Yap5 under the control of the endogenous YAP5 promoter. The cells also contained a CCC1-lacZ reporter construct. The cells were grown in low-iron medium overnight and then incubated with the specified concentrations of iron for 2 h. Cells were harvested, and β-galactosidase activity was determined. Samples from transformed cells were applied to SDS-PAGE gels, and His-Yap5 was assayed by Western blot analysis using antibodies to the His epitope. The blots were also probed with antibodies to CPY as a control for protein loading. (B) yap5Δ cells containing a CCC1-lacZ reporter construct were transformed with a 2μm plasmid containing GFP-Yap5 under the control of its endogenous promoter. The transformed cells were grown in the specified concentrations of iron, and β-galactosidase activity was determined. The localization of GFP-Yap5 was examined by epifluorescence microscopy. The localization of nuclei was ascertained by DAPI staining.
The best-studied Yap family member is Yap1, which contains two CRDs that surround a nuclear export sequence (Fig. 5A) (for a review, see reference 10). Oxidizing agents directly or indirectly generate disulfides within or between CRDs. The generation of disulfides affects protein structure, resulting in the occlusion of the nuclear export sequence and the nuclear accumulation of Yap1. We have not identified a nuclear export sequence in Yap5, and our data suggest that Yap5 is constitutively localized to the nucleus. The structure of Yap5 is similar to that of Yap1 in that Yap5 contains two CRDs. The amino-terminal CRD of Yap5 contains four cysteines, and the carboxyl-terminal CRD contains three cysteines. The seven cysteine residues within the CRDs of Yap5 are conserved in all species of Saccharomyces. The two CRDs in Yap5 are separated by 37 amino acids, while the two CRDs in Yap1 are separated by 283 amino acids. Mutation of either single or multiple cysteines to alanine in the amino-terminal CRD reduced CCC1-lacZ activity by 70% (Fig. 5B). Mutation of a single cysteine or all of the cysteines to alanine in the carboxyl-terminal CRD lowered CCC1-lacZ activity by approximately 40%. Mutation of all amino- and carboxyl-terminal CRD cysteines resulted in a complete loss of transcription activity. All Yap5 mutants were expressed at wild-type levels and were localized to the nucleus (data not shown). Mutation of the cysteines in the CRD affected the ability of Yap5 to prevent high-iron toxicity (Fig. 5C). Mutation of the cysteines in each domain led to a differential response to iron toxicity when expressed in yap5Δ cells. Amino-terminal Yap5 cysteine mutants supported growth on high iron better than vector alone but not as well as the carboxyl-terminal cysteine Yap5 mutants. The compound Yap5 mutant (N4C3) with mutations in both amino- and carboxyl-terminal CRDs was unable to provide any resistance to high-iron toxicity. These results suggest that the cysteines in the CRD are required for Yap5 function and that the two CRDs may have a modular effect.
FIG. 5.
Expression of CCC1 is dependent on the cysteines in the Yap5 CRD. (A) Comparison of the architectures of Yap1 and Yap5. The architecture of Yap1 is abstracted from data described previously (17, 24). Yap5 has two CRDs in which the cysteines are conserved in all species of Saccharomyces. The cysteines are numbered sequentially starting with the amino-terminal cysteine. A representation of the truncated form of Yap5 lacking the DNA binding domain is shown for comparison. n-CRD, N-terminal CRD; c-CRD, C-terminal CRD; aa, amino acids; NES, nuclear export sequence. (B) yap5Δ cells were transformed with plasmids expressing His-Yap5 in which the designated cysteines were mutated to alanines. The cells also contained a CCC1-lacZ reporter construct. The cells were grown in low-iron medium overnight and then incubated with iron for 2 h. Cells were harvested, β-galactosidase activity was determined, and activity was normalized for cell protein. WT, wild type. (C) yap5Δ cells were transformed with plasmids expressing either HIS-YAP5 or HIS-YAP5 in which the designated cysteines were mutated to alanines. Transformed cells were plated onto medium containing different concentrations of iron, and growth was assayed after 2 days. (D) yap5Δ cells containing a CCC1-lacZ reporter construct were transformed with a plasmid containing His-Yap5 in which cysteines in the amino-terminal CRD were mutated to alanine or aspartic acid. Transformed cells grown in low-iron medium were incubated with 500 μM iron for 2 h, and β-galactosidase activity was determined.
Cysteines are potential metal binding amino acids. No iron was found bound to Yap5 when Yap5 was isolated from 59Fe-labeled cells (data not shown). To further test whether the cysteines bound metals, we mutated the cysteines to aspartic acid, as the carboxyl group should bind positively charged transition metals. These mutations did not lead to increased transcriptional activity compared to the corresponding alanine substitution (Fig. 5D).
Binding of Yap5 to the CCC1 promoter is iron independent, but transcriptional activation is iron dependent.
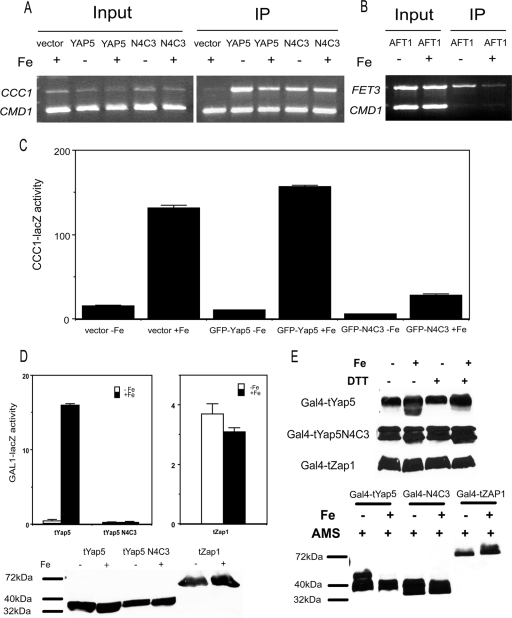
The data suggested that Yap5 constitutively localizes to the nucleus but induces the transcription of CCC1 in an iron-dependent manner only. We determined if iron affected the ability of Yap5 to occupy the CCC1 promoter by CHIP. GFP-Yap5 expressed from a high-copy-number (data not shown) or low-copy-number plasmid occupied the CCC1 promoter in an iron-independent manner, as iron-limited and iron-replete cells showed similar amounts of GFP-Yap5 at the CCC1 promoter, supporting the view that Yap5 is constitutively localized to the nucleus (Fig. 6A). To confirm that the growth conditions employed affected cellular iron, we examined the binding of the low-iron-sensing transcription factor Aft1 to one of its targets, the FET3 promoter. Aft1 binding to the FET3 promoter was reduced under high-iron conditions (Fig. 6B), confirming our previously reported results (5). Since iron regulates CCC1-lacZ expression, we conclude that iron affects the transcriptional activation of Yap5 but does not affect binding to DNA. Based on these data, we examined whether Yap5 cysteine-to-alanine mutants could bind to the CCC1 promoter. The GFP-Yap5(N4C3) mutant with all cysteines mutated (N1234C123 equals N4C3) was chosen because it was unable to induce the expression of CCC1-lacZ. CHIP analysis showed that GFP-Yap5(N4C3) bound to the CCC1 promoter independent of iron.
FIG. 6.
Iron affects Yap5 transcriptional activation but not DNA binding. (A) yap5Δ cells were transformed with a low-copy-number control vector, or low-copy-number plasmids expressing GFP-YAP5 or GFP-YAP5(N4C3), in which all of the cysteines in the CRDs were mutated to alanines. Cells were grown in iron-limited medium overnight and then incubated with or without 500 μM iron for 2 h before being processed for CHIP (IP). (B) Wild-type cells expressing Aft1-HA were incubated as described above (A), and CHIP analysis was performed using probes specific for the FET3 promoter. (C) Wild-type cells containing a CCC1-lacZ reporter construct were transformed with either a control plasmid, a GFP-YAP5 plasmid, or a GFP-YAP5(N4C3) plasmid. Cells grown in low-iron medium were incubated with iron for 2 h, and β-galactosidase and cell protein levels were determined. (D) Cells were transformed with a plasmid expressing a chimeric protein containing the Gal4 binding domain and the Yap5 CRD regions (Gal4-tYap5), a Gal4 binding domain and the Yap5 CRD region from Yap5(N4C3) [Gal4-tYap5(N4C3)], or a Gal4 binding domain and the activation domain (positions 552 to 880) of Zap1 (Gal4-tZap1). Cells were also transformed with a GAL1-lacZ construct. Transformed cells were grown in low-iron medium and then incubated with 500 μM iron for 2 h. Samples were taken for measurement of β-galactosidase activity and Western analysis using antibodies to the Gal4 DBD. (E) Cells transformed with a plasmid expressing Gal4-tYap5, Gal4-tYap5(N4C3), or Gal4-tZap1 were grown in high- or low-iron medium. The cells were extracted with TCA, and the extract was applied to SDS-PAGE gels in either the absence or presence of DTT (top) or after treatment with AMS (bottom).
If GFP-Yap5(N4C3) can occupy the CCC1 promoter but is unable to induce transcription, we predict that it should function as a dominant negative. To test this prediction, we transformed wild-type cells containing the CCC1-lacZ expression plasmid with a control vector, GFP-YAP5 or GFP-YAP5(N4C3), and measured iron-dependent β-galactosidase activity. Wild-type cells transformed with a control vector or GFP-YAP5 expressed similar increased amounts of β-galactosidase in the presence of iron (Fig. 6C). In contrast, wild-type cells transformed with the GFP-YAP5(N4C3) plasmid showed dramatically reduced levels of expression of β-galactosidase in the presence of iron.
If the binding of Yap5 to DNA is iron insensitive but transcription is iron sensitive, then we expect that the Yap5 activation domain would act on heterologous DNA binding proteins. The DNA binding domain of GAL4 was fused to a truncation version of YAP5 lacking the endogenous DNA binding domain but containing the two CRDs. This construct (Gal4-tYap5) was transformed into cells that contained a GAL1-lacZ expression vector, which contained a Gal4 binding site. The addition of iron led to a dose-dependent increase in β-galactosidase activity (Fig. 6D). The expression of the reporter construct was specific for iron, as the addition of Cu, Zn, Mn, or H2O2 had no effect (data not shown). No expression of GAL1-lacZ activity was seen in cells transformed with a construct containing a GAL4 binding domain and the CRDs from Yap5(N4C3). As a further control, we used a fusion protein between Gal4 and the activation domain of the zinc-responsive transcription factor Zap1, which has been shown to transcriptionally activate the GAL1-lacZ reporter construct in response to zinc (1). The Gal4-Zap1 (tZap1) chimeric protein did not induce the transcription of the GAL1-lacZ construct in response to iron. Western analysis of transformed cells showed equivalent levels of chimeric Gal4-tYap5, Gal4-tYap5(N4C3), and Gal4-tZap1. We conclude that iron leads to the transcriptional activation of Yap5 but does not affect its DNA binding activity.
Transcriptional activation is dependent on cysteines in the Yap5 CRD. We attempted to examine the sulfhydryl status of epitope-tagged Yap5 by electrophoretic mobility assays. The migration behavior of epitope-tagged Yap5 in native gels or after cysteine derivitization was difficult to resolve by Western blot analysis (data not shown). We took advantage of the Gal4-tYap5 chimeric protein to determine if iron affected the cysteine status in the CRD. Cells expressing Gal4-tYap5 were grown in high- or low-iron medium, and the migration of Gal4-tYap5 was examined by SDS-PAGE. In the absence of the reducing agent DTT, Gal4-tYap5 extracted from high-iron cells showed two bands, while Gal4-tYap5 extracted from low-iron-grown cells showed one band (Fig. 6E). In contrast, only one band was seen when the samples were treated with high concentrations of DTT. We also examined the effect of DTT on the electrophoretic mobility of Gal4-tYap5(N4C3) and Gal4-tZap1 extracted from control and iron-treated cells. While there is evidence for more than one electrophoretic form of each protein, there was no iron-dependent change in those forms.
To determine if the difference in electrophoretic mobility could be ascribed to disulfide bond formation, we treated samples obtained from high- and low-iron-grown cells with AMS. This cysteine-modifying reagent binds to free sulfhydryls and changes the mobility of the protein. When treated with AMS, samples from both high- and low-iron-grown cells showed decreased mobility by SDS-PAGE. Samples from low-iron-grown cells, however, showed a greater shift in mobility than high-iron-grown cells, suggesting that more of their cysteines were available to be modified by AMS. These results suggest that iron changing the behavior of the cysteines is consistent with iron-induced disulfide formation. AMS treatment did not lead to any change in the Gal4-tYap5(N4C3) protein, suggesting that the cysteines were responsible for the change. The Gal4-tZap1 protein did show a change in AMS response, although the change was seen in the presence and not in the absence of iron, which differs from the Gal4-tYap5 protein. These results indicate that the change in AMS sensitivity cannot be ascribed to the Gal4 domain.
The change in cysteine behavior is not in response to an increase in oxidative stress: the iron-dependent activation of CCC1 transcription occurs anaerobically (data not shown), and the addition of H2O2 does not lead to CCC1 transcription (Fig. 2D). The deletion of TRR1 (thioredoxin reductase) or the deletion of GLR1 (glutathione oxidoreductase) also does not activate CCC1 transcription (data not shown). The deletion of either gene leads to increased oxidation and increased disulfide bond formation (21). Thus, decreased intracellular reductant levels or increased oxidant levels do not lead to the activation of CCC1 transcription, suggesting that the effect of iron is highly specific.
DISCUSSION
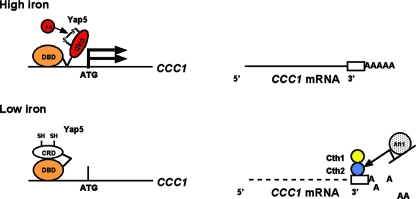
Vacuolar iron homeostasis is a dynamic process in which iron can be transported into and out of the vacuole. Iron transport out of the vacuole is accomplished by two different transport systems, both of which are homologues of cell surface iron transporters. Aft1 transcriptionally regulates the transporters responsible for iron transport into the cell or out of the vacuole in response to low cytosolic iron (23) or defective iron-sulfur cluster synthesis, a measure of low cytosolic iron (2). Iron transport into the vacuole is also a regulated process. The only identified vacuolar iron importer is Ccc1, and two different processes regulate the expression of CCC1 mRNA. Puig et al. previously identified the first process: under low-iron conditions, Cth1 and Cth2 bind to elements in the 3′ untranslated region of CCC1 mRNA, destabilizing the mRNA (19). Aft1 regulates the expression of Cth2, linking a response to low iron and regulation of vacuolar iron transport. Thus, when cells perceive that they are iron limited, transcripts for the vacuolar iron importer are destabilized, preventing reductions in cytosolic iron levels. Simultaneously, the activation of Aft1 leads to the expression of the vacuolar iron exporters Fet5/Fth1 and Smf3. As a result, iron export to the cytosol is increased, while vacuolar iron import is decreased. The second mode of CCC1 regulation is iron-dependent transcriptional activation. The expression of either CCC1 mRNA or a CCC1-lacZ reporter construct is increased in response to iron in both wild-type and cth1Δ cth2Δ cells. The ability to control CCC1 mRNA levels by increased transcription in response to high iron and then to destabilize those mRNA transcripts in response to low iron results in a tight regulation of vacuolar iron import (Fig. 7).
FIG. 7.
Model for the regulation of CCC1 by iron. Yap5 constitutively occupies the CCC1 promoter, and iron, by modifying sulfhydryls, changes the Yap5 conformation, leading to the increased transcriptional activation of CCC1. Reductions in cytosolic iron lead to decreased Yap5 transcriptional activity. As cytosolic iron levels decrease, the transcription factor Aft1 is activated. Aft1 target genes include both cell surface and vacuolar iron transporters, which lead to increased cytosolic iron. Aft1 also induces the expression of Cth2, which binds to the 3′ untranslated region of CCC1 mRNA and destabilizes that mRNA. The lack of transcription of CCC1, in concert with decreased CCC1 mRNA stability, lowers iron import into the vacuole. Thus, cytosolic iron levels are maintained by the coordinated expression of vacuolar and cell surface iron transporters.
Our studies show that Yap5 is responsible for the high-iron induction of CCC1 mRNA, as the deletion of YAP5 results in the decreased expression of CCC1 and increased iron sensitivity. Yap1 is one of eight homologous YAP genes that are a distinct subgroup of the bZIP family of transcription factors similar to that of mammalian AP-1 (9). Yap1, the best-studied member of the yeast YAP family, is a transcription factor that confers resistance to oxidative stresses such as H2O2 and cadmium. Yap2 also controls the transcriptional response to cadmium (11). While Yap1 was shown to bind in vitro to a consensus 7-bp nucleotide sequence, Yap5 did not bind to that sequence in vitro, although genetic studies showed that it could weakly activate a Yap consensus-driven His construct (9). Our data show that one of the two consensus Yap sites in the promoter region of CCC1 responds to Yap5, as the mutation of that site decreases iron-dependent transcription. Comparison of the CCC1 promoter regions of different Saccharomyces species (Saccharomyces Genome Database) showed that the first Yap consensus site is not conserved. In contrast, there is remarkable sequence conservation starting downstream of the first Yap consensus site. This evolutionary conservation of sequence provides support for the function of the downstream Yap sequence and confirms that either the context or nucleotides surrounding the consensus sequence are required for Yap5-dependent expression.
The response of Yap1, the best studied of the Yap family members, to oxidant damage has been well studied. Yap1 is localized in the cytosol, and oxidants promote a change in Yap1 conformation, leading to its accumulation in the nucleus, where it is active as a transcription factor. Yap1 as a result of oxidation forms disulfide bonds, either intramolecular disulfides or disulfides with other proteins such as glutathione peroxidase (Gpx3) (8). These disulfides lead to a change in protein structure, obscuring the nuclear export signal on Yap1, leading to its accumulation in the nucleus and the transcriptional activation of Yap1 target genes. The molecular architecture of Yap5 shows some analogy to Yap1. Yap5, like Yap1, contains two sulfhydryl-rich domains, but these domains do not bracket a nuclear export sequence. Yap5 is constitutively localized to the nucleus even in the absence of iron, and Yap5 protein levels do not change with iron levels. We also observed that Yap5 occupies the CCC1 promoter independent of iron, suggesting that iron regulates transcriptional activation as opposed to DNA binding activity and that the mutation of cysteines in the CRD does not affect promoter binding. The mutation of either one or most of the cysteines in the carboxyl-terminal domain has a milder effect on transcriptional activity than the mutation of cysteines in the amino-terminal domain, suggesting that the CRDs act as independent modules. It is interesting that the incubation of cells with cadmium, a cysteine binding transition metal, resulted in a decrease in Yap5 transcriptional activity (Fig. 2D).
Our data indicate that iron affects the status of the cysteine sulfhydryls. Cysteines can be iron binding residues, and it is possible that iron binding mediates a structural change in Yap5, leading to transcriptional activation. We found no evidence of iron binding to Yap5 and no difference in the transcriptional activity of Yap5 in which the cysteines were mutated to alanine or to aspartic acid. Alanine will not bind metals; however, the metal binding ability of aspartic acid might lead to a constitutive response. Iron affects the migration of the Gal4-Yap5 chimeric protein in SDS-PAGE gels. The change in protein behavior after AMS treatment is consistent with iron-induced disulfide formation. The treatment of cells with H2O2 did not lead to CCC1-lacZ expression, suggesting that transcriptional activation required more than the simple oxidation of sulfhydryls. We recognize that while our data are consistent with disulfide bond formation, it is possible that there could be other modifications of cysteine sulfhydryls. Several experiments indicate that the change in cysteines is not due to the nonspecific oxidation of sulfhydryls, as iron-dependent activation of CCC1 transcription occurs anaerobically, and the addition of H2O2 or deletion of GRl1 or TRR1, which results in increased oxidation, does not activate CCC1 transcription.
The finding that Yap5 can regulate CCC1 expression in response to high iron demonstrates the dynamic response of yeast to the redox-active transition metals iron and copper. Both low and high copper levels result in a transcriptional response regulating copper transport genes and copper storage genes (for a review, see reference 20). High levels of copper result in the transcription of metallothionine genes through the activation of Ace1.
Mac1 and Ace1 are copper binding proteins: the binding of copper activates Ace1, whereas the loss of copper activates Mac1. Our data show that iron homeostasis, like copper homeostasis, is regulated by two different transcription factors, Aft1, which senses low iron and activates the transcription of iron acquisition systems, and Yap5, which senses high iron and activates transporters that affect iron storage. The regulation of vacuolar iron transport is critical for maintaining cytosolic iron levels. Our previous studies showed that the activity of Ccc1 could be affected by changes in mitochondrial iron homeostasis (15). The data presented here indicate that the transcription of CCC1 is responsive to iron. We also note that the level of CCC1 mRNA in cth1Δ cth2Δ cells grown in iron-containing medium is lower than expected if CCC1 mRNA degradation is prevented. This observation suggests that there are other mechanisms that either modify the transcriptional response to iron or regulate the level of CCC1 mRNA.
Acknowledgments
We express our appreciation to the members of the Kaplan laboratory for critical reading of the manuscript.
This work is supported by NIH grant DK30534 to J.K.
Footnotes
Published ahead of print on 10 December 2007.
REFERENCES
- 1.Bird, A. J., H. Zhao, H. Luo, L. T. Jensen, C. Srinivasan, M. Evans-Galea, D. R. Winge, and D. J. Eide. 2000. A dual role for zinc fingers in both DNA binding and zinc sensing by the Zap1 transcriptional activator. EMBO J. 193704-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, O. S., R. J. Crisp, M. Valachovic, M. Bard, D. R. Winge, and J. Kaplan. 2004. Transcription of the yeast iron regulon responds not directly to iron but rather to iron-sulfur cluster biosynthesis. J. Biol. Chem. 27929513-29518. [DOI] [PubMed] [Google Scholar]
- 3.Cohen, A., H. Nelson, and N. Nelson. 2000. The family of SMF metal ion transporters in yeast cells. J. Biol. Chem. 27533388-33394. [DOI] [PubMed] [Google Scholar]
- 4.Courel, M., S. Lallet, J. M. Camadro, and P. L. Blaiseau. 2005. Direct activation of genes involved in intracellular iron use by the yeast iron-responsive transcription factor Aft2 without its paralog Aft1. Mol. Cell. Biol. 256760-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crisp, R. C., E. M. Adkins, E. Kimmel, and J. Kaplan. 2006. Recruitment of Tup1p and Cti6p regulates heme deficient expression of Aft1p target genes. EMBO J. 25512-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis-Kaplan, S. R., D. M. Ward, S. L. Shiflett, and J. Kaplan. 2004. Genome-wide analysis of iron-dependent growth reveals a novel yeast gene required for vacuolar acidification. J. Biol. Chem. 2794322-4329. [DOI] [PubMed] [Google Scholar]
- 7.Delaunay, A., A. D. Isnard, and M. B. Toledano. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 195157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delaunay, A., D. Pflieger, M. B. Barrault, J. Vinh, and M. B. Toledano. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111471-481. [DOI] [PubMed] [Google Scholar]
- 9.Fernandes, L., C. Rodrigues-Pousada, and K. Struhl. 1997. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 176982-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgiou, G. 2002. How to flip the (redox) switch. Cell 111607-610. [DOI] [PubMed] [Google Scholar]
- 11.Hirata, D., K. Yano, and T. Miyakawa. 1994. Stress-induced transcriptional activation mediated by YAP1 and YAP2 genes that encode the Jun family of transcriptional activators in Saccharomyces cerevisiae. Mol. Gen. Genet. 242250-256. [DOI] [PubMed] [Google Scholar]
- 12.Kuge, S., and N. Jones. 1994. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lapinskas, P. J., S. J. Lin, and V. C. Culotta. 1996. The role of the Saccharomyces cerevisiae CCC1 gene in the homeostasis of manganese ions. Mol. Microbiol. 21519-528. [DOI] [PubMed] [Google Scholar]
- 14.Li, L., O. S. Chen, D. M. Ward, and J. Kaplan. 2001. CCC1 is a transporter that mediates vacuolar iron storage in yeast. J. Biol. Chem. 27629515-29519. [DOI] [PubMed] [Google Scholar]
- 15.Li, L., and J. Kaplan. 2004. A mitochondrial-vacuolar signaling pathway in yeast that affects iron and copper metabolism. J. Biol. Chem. 27936339-36348. [DOI] [PubMed] [Google Scholar]
- 16.Ma, J., and M. Ptashne. 1987. A new class of yeast transcriptional activators. Cell 51113-119. [DOI] [PubMed] [Google Scholar]
- 17.Paget, M. S., and M. J. Buttner. 2003. Thiol-based regulatory switches. Annu. Rev. Genet. 3791-121. [DOI] [PubMed] [Google Scholar]
- 18.Portnoy, M. E., X. F. Liu, and V. C. Culotta. 2000. Saccharomyces cerevisiae expresses three functionally distinct homologues of the Nramp family of metal transporters. Mol. Cell. Biol. 207893-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puig, S., E. Askeland, and D. J. Thiele. 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 12099-110. [DOI] [PubMed] [Google Scholar]
- 20.Rutherford, J. C., and A. J. Bird. 2004. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot. Cell 31-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trotter, E. W., and C. M. Grant. 2003. Non-reciprocal regulation of the redox state of the glutathione-glutaredoxin and thioredoxin systems. EMBO Rep. 4184-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urbanowski, J. L., and R. C. Piper. 1999. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 27438061-38070. [DOI] [PubMed] [Google Scholar]
- 23.Van Ho, A., D. M. Ward, and J. Kaplan. 2002. Transition metal transport in yeast. Annu. Rev. Microbiol. 56237-261. [DOI] [PubMed] [Google Scholar]
- 24.Wysocki, R., P. K. Fortier, E. Maciaszczyk, M. Thorsen, A. Leduc, A. Odhagen, G. Owsianik, S. Ulaszewski, D. Ramotar, and M. J. Tamas. 2004. Transcriptional activation of metalloid tolerance genes in Saccharomyces cerevisiae requires the AP-1-like proteins Yap1p and Yap8p. Mol. Biol. Cell 152049-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]