Abstract
In Saccharomyces cerevisiae, the evolutionarily conserved nucleocytoplasmic shuttling protein Nap1 is a cofactor for the import of histones H2A and H2B, a chromatin assembly factor and a mitotic factor involved in regulation of bud formation. To understand the mechanism by which Nap1 function is regulated, Nap1-interacting factors were isolated and identified by mass spectrometry. We identified several kinases among these proteins, including casein kinase 2 (CK2), and a new bud neck-associated protein, Nba1. Consistent with our identification of the Nap1-interacting kinases, we showed that Nap1 is phosphorylated in vivo at 11 sites and that Nap1 is phosphorylated by CK2 at three substrate serines. Phosphorylation of these serines was not necessary for normal bud formation, but mutation of these serines to either alanine or aspartic acid resulted in cell cycle changes, including a prolonged S phase, suggesting that reversible phosphorylation by CK2 is important for cell cycle regulation. Nap1 can shuttle between the nucleus and cytoplasm, and we also showed that CK2 phosphorylation promotes the import of Nap1 into the nucleus. In conclusion, our data show that Nap1 phosphorylation by CK2 appears to regulate Nap1 localization and is required for normal progression through S phase.
Nap1, or nucleosome assembly protein 1, is a member of the Nap1/SET family of proteins and is highly conserved among eukaryotes (23). The first human homolog to be cloned was purified from HeLa cell extracts and characterized by its ability to assemble chromatin in vitro (21, 22). In mice, deletion of Nap1L2 is embryonic lethal due to hyperproliferation of neuronal precursor cells. Mutations in and chromosomal translocations involving Nap1/SET family members are associated with a variety of human cancers (1, 2, 13, 14, 30, 55, 59). These disorders may be due to Nap1/SET-associated defects in cell cycle regulation or could be the result of transcriptional misregulation, since in a genome-wide expression study in Saccharomyces cerevisiae, about 10% of all genes showed altered transcription in a nap1Δ strain (43).
In Saccharomyces cerevisiae, Nap1 was originally identified as a cyclin B2 (Clb2)-interacting protein. At entry into mitosis, the cyclin-dependent kinase complex Clb2/Cdc28 is activated and, in complex with a number of other proteins, initiates the switch from polar to isotropic bud growth (28). The Swe1 kinase is a negative regulator of this complex, and its inactivation is required for bud growth to proceed appropriately. Nap1 is also involved in the regulation of this switch, and in nap1Δ mutants, a mild elongated bud phenotype is observed in 10 to 30% cells (5). This is thought to be due to the role of Nap1 in regulating phosphorylation and activation of the Nim1-related kinase Gin4, which is a prerequisite for the normal assembly of a septin complex at the bud neck (5). Although Nap1 and Gin4 remain in complex throughout the cell cycle, at the G2/M transition, Gin4 undergoes Nap1-dependent hyperphosphorylation that is required for its localization to the bud neck and association with the septin cortex (5, 34, 36, 38, 44, 45).
At steady state, Nap1 is primarily cytoplasmic, but mutation of a leucine-rich nuclear export sequence (NES) sequesters Nap1 in the nucleus, suggesting it is a nucleocytoplasmic shuttling protein (40). In vivo, Nap1 binds to histones H2A and H2B in the cytoplasm, acts to promote the formation of the Kap114-H2A-H2B complex, and facilitates histone import into the nucleus via the karyopherin Kap114 (40). Once in the nucleus, Nap1 is presumably released from Kap114 and is able to assemble histones H2A and H2B onto DNA. The assembled chromatin then recruits the Ran nucleotide exchange factor Rcc1 (Prp20 in yeast) leading to the production of RanGTP at the chromatin surface (6, 8, 29, 42). Kap114 directly inhibits chromatin assembly by Nap1 in vitro, suggesting the chromatin assembly activity of Nap1 is not inherently coupled to import of the H2A-H2B dimer and that Nap1 may undergo additional regulation inside the nucleus (39).
Nap1 appears to have two quite distinct roles in key cellular processes, namely, regulation of bud formation and nucleosome assembly, and within the cell these functions are temporally and spatially separate. We wanted to gain insight into how the apparently pleiotropic functions of Nap1 might be regulated. NAP1 mRNA levels remain stable throughout the cell cycle, suggesting the regulatory mechanism is independent of transcription (54). We hypothesized that the function of different cellular pools of Nap1 might be determined by different Nap1-interacting proteins and set out to identify and characterize these binding partners. As a validation of this approach, we showed these partners included the previously uncharacterized Yol070c, designated here Nba1, which localizes to the septin cortex prior to bud emergence and to the bud neck. Additionally, we found several kinases including Cka2, the protein encoding the α′ subunit of casein kinase 2 (CK2). Analysis of Nap1 phosphorylation determined that Nap1 is phosphorylated at 11 sites in vivo. We show that CK2 associates with Nap1 and that Nap1 is a substrate for phosphorylation by CK2 at three serines in vitro. We demonstrate that reversible phosphorylation of Nap1 is required for normal S phase progression and that phosphorylation of Nap1 by CK2 appears to promote its import into the nucleus. This suggests that in S. cerevisiae Nap1 function is regulated by phosphorylation.
MATERIALS AND METHODS
Yeast strains and plasmids.
S. cerevisiae strains used in this study were derived from strain DF5 or BY4741 as noted, and construction of mutant nap1Δ and kap114Δ strains have been described previously (39, 49). The Nap1-protein A (PrA), kap114Δ strain was constructed by integration of a PrA tag at the C-terminal end of Nap1 in the kap114Δ strain as described previously (3). nap1Δ double mutants were made by mating BY4741 deletion strains from Open Biosystems with the nap1Δ::cloNAT strain (derived from strain Y2454 provided by C. Boone, MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 MFA1pr-HIS3 can1Δ0) and performing tetrad dissections. Strains DK212 and DK213 were gifts from D. Kellogg (5). Expression of green fluorescent protein (GFP)-tagged proteins was performed using the pGFP2-C-FUS vector (40). Amino acid substitutions were made using oligonucleotide site-directed mutagenesis of a double-stranded DNA plasmid template and verified by sequencing. For plasmid rescue experiments, the NAP1 promoter (400 bp upstream) and terminator (296 bp downstream) fragments were cloned into pRS316 with an intervening BamHI site. The NAP1 and mutant derivatives were inserted into this site. For recombinant protein expression, NAP1 and mutant derivatives were cloned into pGEX4T1, and glutathione S-transferase (GST) fusion proteins were purified as previously described (41).
Purification of Nap1 binding partners and identification of phosphorylation sites.
Postribosomal whole-cell extract was prepared from 1 liter of Nap1-PrA-tagged or untagged, exponentially growing yeast. Cultures were harvested by centrifugation and pretreated with protease and phosphatase inhibitor buffer (1 mM peroxovanadate, 10 nM calyculin, 1 mM phenylmethylsulfonyl fluoride, and 1 μg/ml pepstatin in 100 mM Tris [pH 8.0]) at 30°C for 30 min, followed by the addition of 10 mM dithiothreitol for 10 min. Cells were lysed in 2% polyvinylpyrollidone using the French press. Lysates were cleared by centrifugation to remove ribosomes and incubated with immunoglobulin G (IgG)-Sepharose. After Nap1-PrA and copurifying proteins were washed with 50 mM MgCl2, Nap1-PrA-interacting proteins were eluted with 1 M MgCl2 and identified by mass spectrometry (MS) as described previously (12, 18). Proteins from which a minimum of four unique peptides were isolated were included in the analysis. Proteins present in both experimental and control fractions were included only if the total number of peptides present in the experimental fraction was at least 50% higher than the total number in the control fraction. To detect Nap1 phosphopeptides, the Nap1-PrA bound to Sepharose beads was subjected to proteolytic digestion by trypsin, Glu C, and Asp N. The resulting peptides were separated using a C18 column and esterified in acetyl chloride and methanol. Phosphopeptides were enriched from the sample using an immobilized metal affinity column and analyzed by reverse-phase high-performance liquid chromatography coupled to electrospray ionization tandem MS with a chromatograph interfaced with a Finnigan LTQ-FT ion trap mass spectrometer, and identified using SEQUEST. For the coimmunoprecipitation of Nap1 binding partners, lysates from 500 ml of each tagged strain were prepared as described above, and 50 mg of total protein was incubated with IgG-Sepharose. Coprecipitating proteins were eluted with a MgCl2 gradient, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and visualized by Western blotting using a Nap1 antibody purchased from Santa Cruz Biotechnology Inc.
In vitro binding assays.
Recombinant proteins were purified according to the manufacturer's instructions (GE Healthcare, NEB). Nba1 and Nap1 binding assays were performed using either GST-Nap1 or GST-Nap1 mutants immobilized on glutathione-Sepharose or maltose binding protein (MBP)-Nba1 immobilized on amylose resin. In each case, beads were preblocked with 10% bovine serum albumin in buffer containing 1% Tween 20 and then incubated with recombinant proteins at the concentrations described in the figure legends. For the histone binding assay, glutathione-Sepharose-immobilized Nap1 or mutant protein was incubated with chicken erythrocyte core histones. Beads were washed extensively, and bound material was separated by SDS-PAGE and visualized as indicated. For quantification of Nap1 bound to MBP-Kap114, the membrane was incubated with an anti-Nap1 antibody, followed by anti-MBP antibody, and binding was detected using the Odyssey infrared imaging system (LI-COR, Biosciences). Binding was quantified by comparing the amount of Nap1 or mutant signal relative to the amount of input MBP-Kap114 signal detected by the imaging system.
In vitro kinase assay.
One microgram of GST-Nap1 or Nap1 mutant was incubated with 0.2 mM ATP, 1 μCi of [γ-32P]ATP, and 0.2 μl human recombinant CK2 (NEB, Beverley, MA) at 30°C for 30 min or as indicated. The reaction was stopped by the addition of SDS-PAGE sample buffer. The proteins were separated by SDS-PAGE and stained with Coomassie blue, and 32P-labeled proteins were detected by autoradiography on a PhosphorImager.
Cell culture and microscopy.
Cell culture and microscopy were performed as described previously (41) using a Nikon Microphot-SA microscope (Melville, NY), and images were captured using OpenLab software (Improvision, Lexington, MA) with a 100× objective. Strains containing reporter GFP constructs were induced in synthetic complete medium without uracil and methionine (SC-Ura-Met) either overnight [Nap1(L99S S159A S177A S397A)-GFP2] or for 2 h (all other constructs). Cells were photographed using identical exposure settings, and all image manipulation was performed identically using Adobe Photoshop. For the benomyl sensitivity assay, 10-fold serial dilutions of each strain were spotted onto yeast extract-peptone-dextrose (YPD), YPD plus 20 μg/ml benomyl in 0.4% dimethyl sulfoxide, or YPD plus 0.4% dimethyl sulfoxide and incubated at 30°C for 4 days. For the quantification of elongated buds, 250 budded cells were counted from exponentially growing cultures of each strain, and buds whose length from neck to bud tip was greater than twice the width of the mother cell were scored as elongated. The experiment was performed in triplicate, and a two-way chi-squared test was performed comparing values calculated for each double mutant to those obtained from nap1Δ cells.
Cell cycle analysis.
Exponentially growing DK213 cells carrying pRS316 constructs were cultured in SC-Ura medium; DK212 control cells were cultured in complete supplement mixture. Cells were counted, fixed in 70% ethanol and 30% sorbitol, and stained with 1 μM Sytox green (Molecular Probes, Eugene, OR). Ten thousand cells per sample were acquired on an ImageStream imaging flow cytometer (Amnis Corporation, Seattle, WA) using INSPIRE software. The Sytox green signal and corresponding bright-field images were also collected. Spectral overlap of Sytox green into the bright-field channel was calculated and subtracted using the manufacturer's image analysis software. Random cell images were collected from within a cross-section of the 4N peak on the histogram. Cell cycle analysis was performed on exponentially growing cultures; 500 Hoechst-labeled cells from each sample were assigned to one of three morphological categories on the basis of microscopic analysis: no bud (G1), budded cell with a single nucleus (S phase), or budded cell with two nuclei (G2). Cells of abnormal morphology were excluded from the analysis. A two-way chi-squared test was performed comparing values calculated for each mutant to the expected values obtained from wild-type cells.
RESULTS
Identification of Nap1-interacting proteins.
To identify proteins involved in the regulation of Nap1 function, Nap1-PrA was first immunoprecipitated from whole-cell lysates prepared from exponentially growing yeast. Nap1 binding proteins were eluted, and MS was used to identify the protein constituents. Nonspecific interactions were excluded by comparison with a control experiment using an untagged strain. The remaining Nap1-interacting proteins were evaluated based upon the number of total and unique peptides identified, resulting in the identification of 22 Nap1-interacting proteins (Table 1). Several heat shock proteins, a ribosomal protein, and proteins involved in amino acid biosynthesis were present after filtering (Table 1). These proteins are highly abundant in cellular lysates and commonly regarded as contaminants in proteomic screens (17). The Nap1-interacting proteins identified in our screen included known binding partners of Nap1: H2A, H2B, Kap114, H2A.Z, and the kinase Gin4, as well as some previously unreported interacting proteins and two proteins of unknown function.
TABLE 1.
Nap1-interacting proteins
| Function | Protein name | No. of unique peptidesa
|
No. of total peptidesa
|
Protein localization | ||
|---|---|---|---|---|---|---|
| Nap1 IP | Ctrl. | Nap1 IP | Ctrl. | |||
| Chromatin associated | Nap1 | 19 | 6 | 425 | 11 | Cytoplasm, nucleus, and bud neck |
| Htb2 | 7 | 3 | 95 | 6 | Nucleus | |
| Kap114 | 6 | 0 | 7 | 0 | Cytoplasm and nucleus | |
| Hta2 | 4 | 4 | 108 | 5 | Nucleus | |
| Htz1 | 4 | 0 | 26 | 0 | Nucleus | |
| Kinases | Gin4 | 32 | 0 | 81 | 0 | Cytoplasm, bud neck, and bud |
| Cki1 | 23 | 0 | 52 | 0 | Cytoplasm | |
| Kcc4 | 21 | 0 | 39 | 0 | Bud neck, cell periphery, and bud | |
| Cka2 | 5 | 0 | 7 | 0 | Cytoplasm and nucleus | |
| Protein synthesis/metabolism | Tef2 | 11 | 11 | 40 | 18 | Cytoplasm |
| Rpl18b | 7 | 0 | 14 | 0 | Cytoplasm | |
| Shm1 | 18 | 0 | 38 | 0 | Mitochondria | |
| Sip5 | 7 | 0 | 8 | 0 | Cytoplasm | |
| Tco89 | 6 | 0 | 6 | 0 | Vacuolar membrane | |
| Fol1 | 5 | 0 | 6 | 0 | Cytoplasm and mitochondria | |
| Heat shock | Ssa1 | 35 | 20 | 115 | 37 | Cytoplasm |
| Ssb1 | 28 | 0 | 92 | 0 | Cytoplasm | |
| Ssa2 | 8 | 7 | 15 | 8 | Cytoplasm and nucleus | |
| Ssc1 | 6 | 2 | 6 | 2 | Cytoplasm and mitochondria | |
| Hsc82 | 4 | 0 | 6 | 0 | Cytoplasm | |
| Unknown function | Nis1 | 2 | 0 | 2 | 0 | Bud neck and cell periphery |
| Yol070c/Nba1 | 26 | 0 | 111 | 0 | Cytoplasm, bud neck, and cell periphery | |
Nap1-PrA-interacting proteins were identified by mass spectrometry. The number of peptides identified from a control (Ctrl.) experiment using an untagged strain is shown for comparison. Nap1 IP, Nap1-interacting protein.
Among the Nap1-interacting proteins found in the screen, four kinases were identified: the Nim1-related kinases Gin4 and Kcc4, one of the catalytic subunits of CK2, Cka2, and choline kinase, Cki1. Phosphorylation of Nap1 by Nap1-interacting kinases presents a simple and reversible mechanism by which its distinct functions could be regulated. Additionally, two largely uncharacterized proteins were identified, Nis1 and Yol070c. Nis1 (neck protein interacting with septins) has been shown by yeast two-hybrid screening to interact with septins, Nim1 kinases Gin4 and Kcc4, and Nap1, and it localizes to the bud neck in a septin-dependent manner (24). Yol070c is the product of an uncharacterized open reading frame identified in a screen for potential Cdc28 substrates (57). On the basis of these data, we decided to further investigate the roles of Gin4, Kcc4, Cka2, Cki1, Nis1, and Yol070c in Nap1 function.
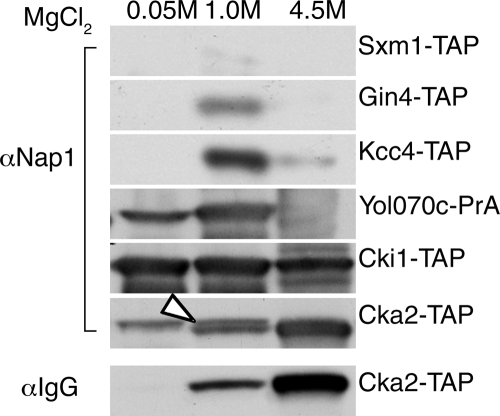
To verify these interactions in vivo, we evaluated the ability of Nap1 to coprecipitate with each protein. Nis1 and Gin4 have previously been shown to interact with Nap1 by coimmunoprecipitation (5, 24). The tandem affinity purification (TAP) or PrA-tagged fusion of each protein of interest was purified from whole-cell extracts, and coprecipitating proteins were eluted with MgCl2 and Western blotted for Nap1 (Fig. 1). We found that Nap1 coprecipitated with Gin4 as expected, and it also coprecipitated with Kcc4, Cka2, Cki1, and Yol070c, but not with the unrelated karyopherin, Sxm1/Kap108. Since Nap1 migrates at a similar size as the Cka2-TAP fusion protein, this membrane was initially probed with IgG to identify Cka2-TAP, followed by a Nap1 antibody. Only Nap1 is visible in the 50 mM MgCl2 fraction, whereas both Nap1 and Cka2-TAP are detected in the 1 M MgCl2 fraction (Fig. 1). These results validated our MS data and showed that these interactions occur either directly or within a shared complex in yeast lysates. The interaction of Nap1 with these proteins suggests that these proteins may be involved in the regulation of Nap1 function.
FIG. 1.
Nap1-interacting proteins. PrA- or TAP-tagged proteins as indicated were isolated from whole-cell lysates using IgG-Sepharose and eluted with MgCl2. The coprecipitation of Nap1 with these proteins was analyzed by Western blotting with an anti-Nap1 antibody (αNap1). For Cka2-TAP, the blot was probed with rabbit IgG antibody (αIgG) to detect Cka2-TAP followed by Nap1 antibody. The white arrowhead indicates Nap1 band.
YOL070c encodes a new bud neck-associated protein, Nba1.
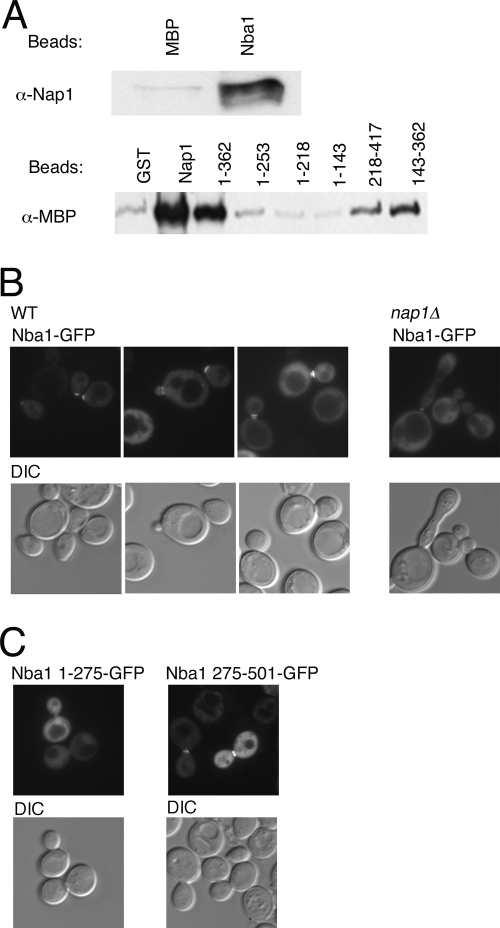
To further validate our candidate Nap1-interacting proteins, we decided to characterize the new Nap1 partner encoded by YOL070c. Yol070c has a predicted molecular mass of 56 kDa. The protein shares no significant homology with other known protein families and is conserved within fungi, although close homologs are not found in higher eukaryotes. The most extensive homology within fungi is observed in the C-terminal domain of Yol070c, and a lysine-rich region (residues 405 to 421) may represent a classical nuclear localization signal. Various global proteomic screens have identified potential partners for Yol070c, including Nap1, Nis1, Clb2, Cdc28, Sxm1/Kap108, and Sua7 (http://www.yeastgenome.org/). We observed that recombinant Nap1 and Yol070c interact directly (Fig. 2A), and we propose that Yol070c be designated Nba1 (Nap1 and bud neck-associated protein). We determined that Nba1 interacts with the central domain of Nap1 (amino acids 143 to 362 [Fig. 2A]). NBA1 does not encode an essential gene, and nba1Δ deletion strains have no obvious growth defects (see Fig. 3). Interestingly, when we expressed Nba1 as a GFP fusion protein, its localization appeared identical to that of Nap1, primarily cytoplasmic and excluded from the nucleus in many cells (Fig. 2B). In addition, bud neck localization was observed in G2/M cells, as well as localization to the actin cap in unbudded cells; this localization was unaltered in nap1Δ cells (Fig. 2B). We also observed that the C-terminal half of Nba1 (amino acids 275 to 501) contains determinants necessary for the normal cellular distribution and bud neck targeting of Nba1, and an amino-terminal fragment (amino acids 1 to 275) did not contain this activity (Fig. 2C). We hypothesize that Nba1 functions with Nap1 at the septin scaffold to regulate G2/M progression. Future experiments will be necessary to determine the relevance of Nba1 to Nap1 function. This characterization of Nba1, together with our identification of known Nap1-interacting proteins, suggests that our screen identified real Nap1 partners.
FIG. 2.
Nap1 interacts directly with the bud neck-associated protein, Nba1. (A) MBP-Nba1 (200 nM), MBP (200 nM), GST (1 μM), GST-Nap1 (1 μM), and GST-Nap1 fragments (indicated by amino acid numbers above the lanes; 1 μM) were immobilized and incubated with 250 nM GST-Nap1 (top blot) or 500 nM MBP-Nba1 (bottom blot). Bound proteins were visualized by Western blotting with anti-Nap1 (α-Nap1) or anti-MBP (α-MBP) (for Nba1) antibodies. (B) Nba1-GFP2 fusion protein was expressed in wild-type (WT) or nap1Δ cells and visualized by fluorescence microscopy. The coincident differential interference contrast (DIC) image is also shown. (C) Nba1 fragments (indicated by amino acid number) were expressed as GFP2 fusion proteins in wild-type yeast and visualized as described above for panel B.
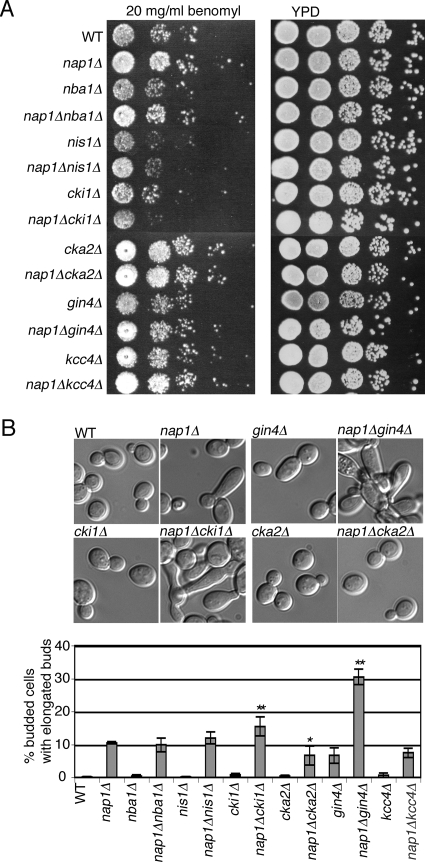
FIG. 3.
Genetic interactions of NAP1. (A) Strains of the indicated genotype were equalized, spotted at 10-fold serial dilutions, and grown on YPD plates with and without benomyl. (B) Strains of the indicated genotypes were examined by differential interference contrast (DIC) microscopy, and the percentage of budded cells exhibiting elongated buds was quantified. The error bars indicate the standard error of the means for three independent experiments. Statistically significant differences between nap1Δ and the double deletion strains are indicated as follows: *, P < 0.05; **, P < 0.01. DIC images of selected strains as indicated are shown above the bar graph. WT. wild type.
NAP1 interacts genetically with several kinases.
In order to understand the relationship between the Nap1-interacting proteins and Nap1 in vivo, we examined strains in which both genes were deleted. None of the double deletion strains showed any obvious growth defect on YPD (Fig. 3A). In S. cerevisiae, nap1Δ mutants have increased resistance to benomyl, a microtubule-destabilizing drug that causes mitotic arrest at high concentrations, demonstrating a role for Nap1 in regulating microtubule dynamics (25). As expected, nap1Δ cells grew better than wild-type cells on plates containing 20 μg/ml benomyl (Fig. 3A). Strains lacking NBA1, GIN4, or KCC4 grew similarly to wild-type cells, which suggests that they are not resistant to benomyl. When these mutations were combined with the NAP1 deletion, nap1Δ nba1Δ, nap1Δ gin4Δ, and nap1Δ kcc4Δ deletion strains grew similarly to the nap1Δ mutant, suggesting that the resistance to benomyl observed in nap1Δ strains is not dependent on these three genes. Both nis1Δ and cki1Δ strains showed increased benomyl sensitivity compared to the wild type. However, unlike the nap1Δ nis1Δ strain, deletion of NAP1 in combination with CKI1 caused increased sensitivity to benomyl, indicating a genetic interaction. Last, the cka2Δ strain showed increased resistance to benomyl in the presence and absence of NAP1, suggesting that these mutations may be epistatic and that both proteins function in microtubule stability (Fig. 3A).
A proportion of nap1Δ cells exhibit an elongated bud phenotype, indicative of a delayed switch from polar to isotropic bud growth (25). We therefore examined the morphology of the single and double deletion strains and quantified the percentage of budded cells exhibiting elongated buds. We observed elongated buds in nap1Δ and gin4Δ cells (∼10% in this strain background), but not in other strains bearing a single deletion (Fig. 3B). The percentage of elongated buds was significantly increased in both the nap1Δ gin4Δ and nap1Δ cki1Δ strains (31% and 15%, respectively) compared to the nap1Δ strain, further suggestive of a genetic interaction (Fig. 3B). No morphological defect was seen in cka2Δ cells, and we did not observe an increase in the percentage of elongated buds in the nap1Δ cka2Δ mutant or in the other double deletion mutants (Fig. 3B). Interestingly, a small decrease in the percentage of cells with elongated buds was observed in the nap1Δ cka2Δ mutant compared to the nap1Δ mutant (Fig. 3B). CK2 contains two catalytic subunits encoded by CKA1 and CKA2, and although disruption of both genes is lethal, depletion of CK2 using conditional alleles is known to cause defects in cell cycle progression, cell polarity, and elongated bud morphology. Whether NAP1 interacts genetically with conditional strains remains to be tested. Collectively, these results suggested that NAP1 interacts genetically with GIN4, CKI1, and CKA2 and may have an overlapping role with CKA2 in regulating microtubule stability. This also suggests that the elongated bud phenotype and the resistance of nap1Δ mutants to benomyl are independent phenotypes. It was of particular interest that NAP1 appeared to be epistatic with CKA2, since CK2 may phosphorylate Nap1 homologs in other species (9, 16, 31, 51). Therefore, because cka2Δ and nap1Δ mutants exhibit overlapping phenotypes, CK2 and Nap1 are present in both the cytoplasm and nucleus, and Nap1 has been implicated as a CK2 substrate in other species, we decided to investigate CK2-mediated regulation of Nap1 function.
Nap1 is a phosphoprotein in vivo.
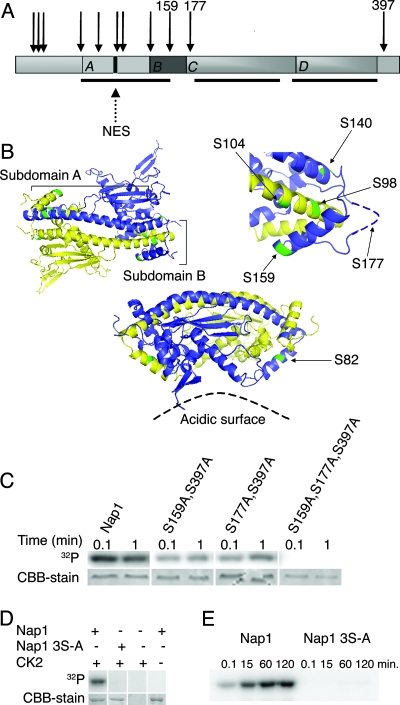
Phosphorylation of Nap1 has not been demonstrated in S. cerevisiae, so we utilized MS to determine whether yeast Nap1 is a phosphoprotein. Nap1-PrA was purified from exponentially growing cells, and after protease digestion, phosphopeptides were enriched using an immobilized metal affinity column and sequenced by MS/MS. Eleven phosphorylated serines and threonines were identified, and all but one are clustered in the amino-terminal half of the protein (Fig. 4A). Only five of these sites are visible on the published crystal structure, and another site (S177) is contained in an unstructured, flexible loop (48). The remaining sites were not included in the fragment that was crystallized. In the published structure of Nap1, four distinct regions of the protein, labeled subdomains A to D, were characterized (Fig. 4A) (48). Nap1 forms a homodimer via subdomain A, and this region also contains the previously characterized NES (40). Subdomain A consists of two alpha helices, of which the shorter α1 helix extends toward the acidic surface of the Nap1 dimer (Fig. 4B). One of the phosphorylated residues, S82, is exposed in the crystal structure and lies at one end of α1, near the loop that joins α1 with α2, and is oriented toward the acidic surface of Nap1 (Fig. 4B). The longer α2 helix couples with the corresponding α2 helix of the opposite subunit by close antiparallel pairing along the entire length of the helix (Fig. 4B). Subdomain B forms a clamp across the α2 helix of the opposing subunit. The clamp may contribute to formation or stabilization of the dimer and also appears to mask the NES of the opposing subunit. Two of the phosphorylated serines we identified, S98 and S104, are located within or adjacent to the NES, and are one and two-third helical turns apart from each other at one end of subdomain A (Fig. 4B). Subdomain B containing the clamp of the opposing subunit appears to bury S98 and partially occlude S104. Two other phosphorylation sites identified in our analysis, S140 and S159, face outward from each end of the clamp region formed between the base of α2 in subdomain A and subdomain B (Fig. 4B). Though not represented in the structure, S177 is located on the flexible loop that forms the hinge of this clamp (Fig. 4B). Due to antiparallel subunit pairing, S98 and S104 of one subunit are in close proximity to S140 and S159 and probably S177 from the other subunit. It is possible that phosphorylation of S140, S159, and S177 in the clamp domain alters the conformation of this domain. In addition, phosphorylation of the occluded residues (S98 and S104) probably requires a change in the clamp domain conformation. Taken together, this suggests that phosphorylation may regulate nuclear transport of the protein and the conformation of the dimer.
FIG. 4.
Nap1 is phosphorylated. (A) Schematic of Nap1. Arrows illustrate the relative positions of the 11 identified phosphosites, with the amino acids of the CK2 sites indicated numerically. Black bars beneath the schematic show the regions contained within the solved structure (48). (B) Tertiary structure of the Nap1 dimer (left). The subunits are shown in purple and yellow, and phosphorylated residues on both subunits are green. The labeled subdomains A and B refer to the purple subunit. Close-up view (right) of the clamp region indicates phosphorylated residues in this domain, and the likely position of S177 is shown. An alternate projection of the dimer, illustrating S82, is shown for clarity (bottom view) (48). (C) Recombinant GST-Nap1 and the indicated Nap1 mutants were incubated with purified CK2 and [γ-32P]ATP for the time indicated. The proteins were separated by SDS-PAGE, Nap1 was visualized by Coomassie blue staining (CBB), and phosphorylation was measured by 32P incorporation detected using a PhosphorImager. (D) In vitro kinase assay of GST-Nap1 or GST-Nap1(S159S177AS397A) (labeled Nap1 3S-A) was carried out for 30 min as described in Materials and Methods, and proteins included in the reaction mixture are as indicated. (E) In vitro kinase assay of GST-Nap1 or GST-Nap1 3S-A was carried out as described in Materials and Methods for the times indicated.
Nap1 is phosphorylated by CK2 in vitro.
CK2 has a consensus substrate recognition motif, and analysis of Nap1 phosphosites revealed that three phosphoserines, S159, S177 and S397, exist within a CK2 consensus sequence (Fig. 4A). Phosphoserine S159 aligns with Drosophila Nap1 S118, which is phosphorylated by CK2 in vitro (31). A fourth phosphorylated residue, S140, that is also within a minimal CK2 consensus sequence was identified; however. this site was not predicted to be a CK2 target site by NetPhosK or KinasePhos prediction tools (7). On the basis of this evidence, we predicted that serines 159, 177, and 397 within Nap1 were CK2 target residues.
To determine whether Nap1 is a substrate for CK2, we assessed the ability of CK2 to phosphorylate Nap1 in vitro. Recombinant GST-tagged Nap1 was incubated with recombinant CK2 and 32P-labeled ATP, and CK2 was able to phosphorylate GST-Nap1 in vitro (Fig. 4C). In order to confirm the CK2 target sites, mutants were constructed in which two or three of the CK2 target serines were mutated to uncharged, unphosphorylatable alanine residues. As predicted, mutation of two of the three serines to alanines significantly reduced phosphorylation of Nap1 as measured by 32P incorporation, whereas mutation of all three serines, creating Nap1(S159A S177A S397A) completely inhibited phosphorylation by CK2 (Fig. 4C). This suggested that all three sites were recognized by CK2. We also demonstrated that phosphorylation of Nap1 in this reaction was specifically dependent upon the presence of both the substrate and CK2 (Fig. 4D). A time course experiment determined that the in vitro phosphorylation progresses to saturation by 60 min, whereas Nap1(S159A S177A S397A) was not detectably phosphorylated even after 120 min (Fig. 4E). MS of recombinant GST-Nap1, phosphorylated by CK2 in vitro, confirmed the presence of phosphorylated S177 and S397 (data not shown). Taken together, these results show that Nap1 contains three substrate serines for phosphorylation by CK2.
Phosphorylation of Nap1 by CK2 is not required for CLB2-dependent bud formation.
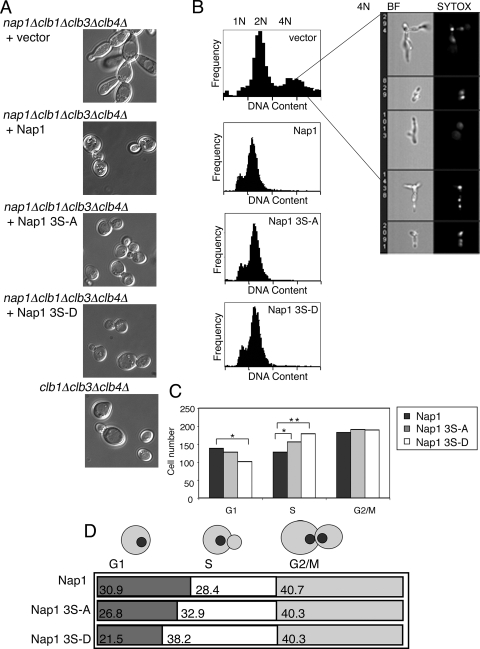
To investigate how phosphorylation of Nap1 by CK2 might regulate its various cellular functions, we constructed plasmids expressing wild-type Nap1 or mutants in which the three CK2 target serines were mutated to alanine, Nap1(S159A S177A S397A), or to negatively charged aspartic acid, Nap1(S159D S177D S397D) to mimic constitutively unphosphorylated or phosphorylated Nap1, respectively. Each was expressed under the control of the endogenous NAP1 promoter and terminator, and Western blotting confirmed that the wild type and mutants were expressed at similar levels (data not shown). In order to determine whether phosphorylation of Nap1 by CK2 was necessary for the regulation of correct bud formation, we expressed Nap1(S159A S177A S397A) and Nap1(S159D S177D S397D) in a CLB2-dependent strain, in which the CLB1, CLB3, CLB4, and NAP1 genes were deleted (DK213). In this background, the nap1Δ phenotype is exacerbated, resulting in cells with highly elongated buds and the formation of large, interconnected clumps of cells (25). In this strain, expression of Nap1 or either of the Nap1 mutants, Nap1(S159A S177A S397A) and Nap1(S159D S177D S397D), was able to rescue normal bud shape (Fig. 5A). This suggests that CK2 phosphorylation of Nap1 is not required for correct bud formation.
FIG. 5.
Reversible phosphorylation of Nap1 by CK2 may regulate normal progression through S phase. (A) Nap1, Nap1(S159A S177A S397A) (labeled Nap1 3S-A), or Nap1(S159D S177D S397D) (labeled Nap1 3S-D) were expressed in the Clb2-dependent strain and examined by differential interference contrast (DIC) microscopy. (B) The same strains were fixed, and the DNA was stained with Sytox green. Cells were analyzed for DNA content using the Amnis ImageStream instrument. Random images of cells within the peak corresponding to 4N DNA content (vector control sample only) are displayed on the right. BF, bright-field image, SYTOX, Sytox green-stained DNA. (C) Strains as in panel A were stained with Hoechst and observed by fluorescence and DIC microscopy and scored morphologically for their cell cycle distribution. The numbers of cells with no bud (G1), cells with a small bud and one nucleus (S phase), and cells with a large bud and two nuclei (G2/M) are indicated. Comparison of strains showed statistically significant differences (*, P < 0.05; **, P < 0.01). (D) The numbers in the boxes indicate the percentage of cells in each phase of the cell cycle derived from panel C.
Reversible phosphorylation of Nap1 by CK2 is required for normal cell cycle progression.
In order to determine whether Nap1 phosphorylation by CK2 regulates cell cycle progression, we observed the cell cycle profile of the Nap1 phosphomutants in the CLB2-dependent strain. Cells were analyzed using Amnis ImageStream allowing the correlation of DNA content with cell morphology of individual cells within the population. In unsynchronized cells, nap1Δ yeast had a higher ratio of cells with 2N DNA to those with 1N DNA content compared to the NAP1 strain, which is indicative of the mitotic delay described previously (25) (Fig. 5B). In this strain, nap1Δ cells exhibited a large proportion of multinucleate cells, as evidenced by the presence of an additional peak representing 4N DNA content. Morphologically, this peak consisted of interconnected buds representing failed cytokinetic events, and Sytox green staining revealed that these interconnected buds had several nuclei (Fig. 5B). In this strain, expression of Nap1, Nap1(S159A S177A S397A), or Nap1(S159D S177D S397D) rescued the cytokinetic defect, resulting in a significant reduction in the number of 2N+ cells and a loss of the peak corresponding to 4N DNA content, further suggesting that phosphorylation of Nap1 by CK2 is not required for its role in regulating bud formation (Fig. 5B). However, analysis of the cell cycle profile for these strains suggested that there was a small increase in the percentage of cells in G1/S in the Nap1(S159A S177A S397A) and Nap1(S159D S177D S397D) strains relative to cells expressing Nap1. To confirm this observation, we used microscopy to characterize the cell cycle profile of each strain on the basis of cell morphology. Random fields were scored for the number of cells in G1, S, and G2/M. Although the doubling times for strains expressing the different forms of Nap1 were equivalent, expression of Nap1(S159A S177A S397A) or Nap1(S159D S177D S397D) led to a prolonged S phase and a shortened passage through G1 relative to the wild type, a result which was more pronounced with Nap1(S159D S177D S397D) (Fig. 5C and D). As we observed a similar effect with both Nap1(S159A S177A S397A) and Nap1(S159D S177D S397D), these data suggest that reversible phosphorylation of Nap1 by CK2 is required for normal progression through S phase.
Phosphorylation by CK2 is not necessary for histone binding.
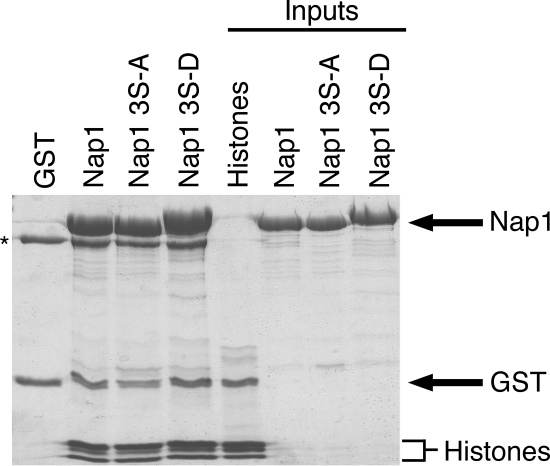
During S phase, Nap1 likely plays an important role loading histones onto DNA following replication. Therefore, we examined the interaction of the Nap1 phosphomutants with histones to determine whether phosphorylation of Nap1 by CK2 might regulate this interaction. Among Nap1 family members, the central NAP1 domain (containing S159 and S177 in yeast Nap1) is the minimal region required for histone binding, and the acidic C-terminal region of Nap1 (containing S397) has been proposed to enhance its histone binding activity (15). Core chicken erythrocyte histones were incubated with either immobilized GST-Nap1 or Nap1(S159A S177A S397A) (both representing unphosphorylated Nap1) or Nap1(S159D S177D S397D) (mimicking phosphorylated Nap1). All three isoforms of Nap1 interacted with the core histones. As has been shown in previous in vitro studies, Nap1 interacted with H3 and H4 as well as H2A and H2B (Fig. 6); Nap1(S159D S177D S397D) consistently has slightly reduced mobility. These data suggested that the Nap1-histone interaction occurs regardless of the phosphorylation status of Nap1.
FIG. 6.
Phosphorylation of Nap1 by CK2 does not prevent histone binding by Nap1. GST, GST-Nap1, GST-Nap1(S159A S177A S397A) (labeled Nap1 3S-A), or GST-Nap1(S159D S177D S397D) (labeled Nap1 3S-D) (all at 1 μM) was immobilized and incubated with purified core histones (1 μM). Bound proteins were visualized by Coomassie blue staining. The black asterisk indicates a band corresponding to bovine serum albumin. Inputs of each protein (25 μmol) are shown.
Phosphorylation of Nap1 by CK2 regulates nuclear import.
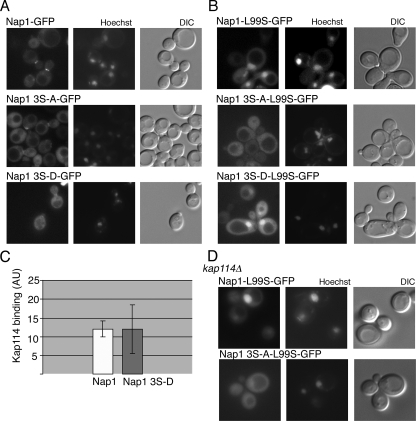
Nap1 has been demonstrated to shuttle between the nucleus and cytoplasm in a NES-dependent manner (40). In yeast, Nap1 is predominantly cytoplasmic with no obvious cell cycle-dependent change in nuclear localization (data not shown). However, it is possible that CK2 phosphorylation may regulate the import or export of Nap1, thus affecting the availability of histones in the nucleus. As two of the phosphorylation sites are located near the NES on the basis of the crystal structure, we hypothesized that CK2 may regulate Nap1 export. GFP2 fusions of Nap1, Nap1(S159A S177A S397A), and Nap1(S159D S177D S397D) were each expressed from a plasmid in a nap1Δ strain to prevent dimerization or competition with endogenous Nap1. In exponentially growing cells, wild-type and mutant Nap1 GFP fusions were predominantly cytoplasmic and able to localize to the bud neck in G2/M cells (Fig. 7A). This suggested that the loss of CK2 phosphorylation was not specifically blocking export. Since in this assay Nap1 is almost undetectable in the nucleus at steady state, we decided to assess the ability of the mutants to be imported in the context of an export-deficient reporter. In this way, the steady-state localization of Nap1 would be shifted to the nucleus, making it possible to reveal an import defect. It has previously been demonstrated that mutation of two of the leucine residues in the NES, L99 and L102, is sufficient to make Nap1 predominantly nuclear (37). We constructed a minimal export-deficient mutant, in which a single leucine (L99) within the NES was mutated to a serine, redistributing Nap1-GFP2 to the nucleus. The Nap1(L99S) mutation was incorporated into the GFP reporter constructs described above. Nap1(L99S)-GFP2 and the phosphomimic Nap1(L99S S159D S177D S397D)-GFP2 fusion proteins localized to the nucleus (Fig. 7B). However, the unphosphorylatable Nap1(L99S S159A S177A S397A)-GFP2 mutant was predominantly cytoplasmic, although it was still detectable in the nucleus at low levels. This indicated that nuclear import of the unphosphorylatable mutant was greatly reduced relative to the wild-type and phosphomimic forms and supported the idea that CK2 phosphorylation promotes the nuclear import of Nap1.
FIG. 7.
Phosphorylation of Nap1 by CK2 regulates nuclear import. (A) Nap1, Nap1(S159A S177A S397A) (labeled Nap1 3S-A), or Nap1(S159D S177D S397D) (labeled Nap1 3S-D) were expressed as GFP2 fusion in nap1Δ cells and visualized by fluorescence microscopy. The coincident differential interference contrast (DIC) and Hoechst-stained images are shown. (B) Export-deficient Nap1-L99S, Nap1 3S-A-L99S, and Nap1 3S-D-L99S GFP2 fusion proteins were expressed and visualized as described above for panel A. (C) MBP-Kap114 (500 nM) was immobilized and incubated with recombinant Nap1 or Nap1 3S-D (both 250 nM). Bound proteins were visualized by Western blotting and quantitated using the Odyssey infrared imaging system as described in Materials and Methods. Relative binding of Nap1 to Kap114 is expressed in arbitrary units (AU). (D) The indicated GFP fusions were expressed in kap114Δ cells and visualized as described above for panel A.
Nap1 can be imported by the karyopherin Kap114 in a Kap114-Nap1-histone complex. Indeed, the presence of Nap1 in the Kap114-histone complex increases the binding of Kap114 to the complex (40). We tested whether phosphorylation may promote binding of Nap1 to Kap114, as this might explain the observed mislocalization of Nap1(L99S S159A S177A S397A)-GFP2. We performed an in vitro binding assay with unphosphorylated recombinant Nap1 or the Nap1(S159D S177D S397D) phosphomimic and quantified the amount of Nap1 bound to immobilized Kap114. No difference was detected between the two recombinant forms of Nap1, suggesting that phosphorylation at these sites does not regulate the interaction of Nap1 with Kap114 (Fig. 7C). We also examined the localization of these mutants in a Δkap114 strain. In this background, Nap1(L99S)-GFP2 is still predominantly nuclear, suggesting that Nap1 import is not solely dependent upon Kap114. In the absence of Kap114, as in wild-type cells, low levels of Nap1(L99S S159A S177A S397A)-GFP2 were observed in the nucleus (Fig. 7D). Taken together, these results imply that the decreased import of Nap1(L99S S159A S177A S397A)-GFP2 was not due to a decreased affinity for Kap114. It is possible, however, that CK2 may regulate the import of Nap1 through a Kap114-independent import pathway. In summary, our results suggest that CK2 phosphorylation of Nap1 regulates its nuclear import. In addition to the regulation of import, reversible phosphorylation is required for timely S phase progression, raising the possibility that decreased import negatively affects the availability of histones during replication.
DISCUSSION
We have shown that Nap1 function is regulated by phosphorylation. We observed that Nap1 interacts with several kinases, including CK2. Here, we present evidence that Nap1 is phosphorylated on several sites and is a substrate for CK2. CK2 phosphorylation appears to be necessary for efficient nuclear import of Nap1. This is the first report on the mechanism of yeast Nap1 regulation by phosphorylation. CK2 is ubiquitously expressed in the cytoplasm and nucleus, and thus, the phosphorylation of Nap1 is likely reversible, regulated by the combined action of this kinase and a phosphatase. The importance of regulated phosphorylation and dephosphorylation events is further emphasized by the fact that yeast strains expressing either the phosphorylation-defective Nap1(S159A S177A S397A) or constitutively charged Nap1(S159D S177D S397D) mutant had alterations in the cell cycle consistent with a defect in S phase progression and replication.
Functionally, members of the Nap1/SET superfamily have in common their interaction with the mitotic B-type cyclins and the ability to assemble nucleosomes onto chromatin (47). Whether these two functions are interconnected or represent an example of a single protein performing multiple, unrelated functions is not yet understood. In higher eukaryotes, there are several members of the Nap1 superfamily to which different functions in chromatin metabolism, transcription, and cell cycle regulation have been ascribed. S. cerevisiae has only two members of this protein family, Nap1 and Vps75, and we set out to use this simple model organism to elucidate the regulatory mechanisms governing Nap1's pleiotropic functions (52). We identified many Nap1-interacting proteins, including most of the expected and previously characterized partners, such as histones, Kap114, and Gin4. In addition, some new partners were identified; these included the kinases CK2, Kcc4, and Cki1. Many members of the Nap1/SET superfamily in higher eukaryotes are known phosphoproteins; we show here the first evidence that yeast Nap1 is also a target for phosphorylation in vivo.
Our data show that Nap1 is a specific substrate for CK2. CK2 is a ubiquitous kinase that is highly conserved throughout eukaryotes, has a wide range of cellular targets, and is required for viability (for a review, see reference 32). It is composed of four subunits, two catalytic α subunits and two regulatory β subunits. Interestingly, the catalytic subunits are not entirely functionally redundant. In strains carrying a temperature-sensitive allele of a single-subunit disruption of CKA1, encoding the α subunit, prevents polarization of the actin cytoskeleton and causes a loss of cell polarity (50). In contrast, loss of CKA2 (encoding the α′ subunit) is reported to result in elongated buds in 10 to 30% of cells and cell cycle arrests in both G1 and G2/M (19, 46). Both Cka2 and Cka1 were detected among our Nap1-interacting proteins. After stringent filtering, Cka1 was removed from the list but may also represent a relevant physiological interaction. The cka2Δ strain used in this study has the CKA1 locus intact, but this strain, like the nap1Δ strain, showed robust resistance to benomyl. Whether this phenotype is specific to the CKA2 isoform is not known. CK2 has been found in association with tubulin, centrosomes, and the mitotic spindle, offering further evidence for a role in the regulation of microtubule stability during mitosis (10, 11, 26, 60). Surprisingly, deletion of both CKA2 and NAP1 in our strain background led to a slight decrease in the number of cells with abnormal buds compared to the nap1Δ strain, suggesting a synthetic genetic interaction. These results strengthen the idea that the delayed switch from polar to isotropic growth and the increased microtubule stability observed in nap1Δ are independent phenotypes.
It has previously been shown that Drosophila Nap1 binds to and is phosphorylated by human CK2 holoenzyme in vitro (31, 51). The Nap1 homolog in the nematode Steinernema feltiae interacts with endogenous CK2 by a yeast two-hybrid (16), and the homolog in the rice Oryza sativa is phosphorylated by recombinant CK2 in vitro (9). Human TSPY (testis-specific protein, Y encoded) predominantly occurs in a phosphorylated form, and a putative CK2 phosphorylation site is required for nuclear import of this protein (27). These studies have led to speculation that Nap1 is a substrate for phosphorylation in vivo and that phosphorylation may regulate Nap1 localization. Our studies have demonstrated for the first time that this is indeed the case in S. cerevisiae. In addition to CK2, we identified Gin4 and other kinases that associate with Nap1. Gin4 is thought to regulate bud formation through a pathway parallel to that of Nap1, since in this study and others, the deletion of both enhances the elongated bud phenotype seen in both single deletion strains (5). Another Nim1-like kinase, Kcc4, was found to associate with Nap1, although the two proteins did not appear to interact genetically. All three proteins, Nap1, Kcc4, and Gin4, cause an elongated bud phenotype when overexpressed (4, 44; also data not shown). Taken together, these results suggest that these proteins function in overlapping but nonidentical pathways in the regulation of bud formation. We do not yet know whether Nap1 is a substrate for these kinases. The fourth kinase we identified in our screen for Nap1-interacting proteins was Cki1. Cki1 is involved in the synthesis of the membrane lipid phosphatidylcholine in the Kennedy pathway (20). Cki1 and Nap1 interact genetically, as indicated by the fact that deletion of NAP1 greatly increases the benomyl sensitivity observed in the cki1Δ mutant and exacerbates the elongated bud phenotype compared to either single deletion strain. Phosphorylation of Nap1 itself or a Nap1-interacting substrate by Cki1 may be required for normal bud formation.
Two other Nap1-interacting proteins identified were Nis1 and the newly described Nba1. Nap1 has previously been shown to bind to Nis1, whose specific function is not well understood, although it is proposed to be involved in the mitotic signaling network (24). The nis1Δ strain was very sensitive to benomyl relative to the wild type, implying a role for Nis1 in regulating microtubule stability. The bud neck-associated protein Nba1 is a substrate for phosphorylation by Cdc28 in vitro (57). Global analysis of NBA1 mRNA levels during the cell cycle demonstrated that its expression, like that of Clb2, is periodic and peaks during mitosis (54). Taken together, these data imply that Nba1 may have a cell cycle-dependent function. We show that, like Nap1, Nba1 is primarily cytoplasmic and localizes to the bud neck at G2/M. We hypothesize that Nap1 and Nba1 interact at the bud neck and function in the regulation of G2/M progression. As yet we have no evidence that Nba1, like Nap1, can be imported into the nucleus. However, proteomic experiments have indicated that the yeast karyopherin Kap108/Sxm1 interacts with Nba1, suggesting Kap108 may function in Nba1 nuclear import and raising the possibility that Nba1 may also shuttle between the nucleus and cytoplasm (58).
Of the 11 phosphorylated residues identified on Nap1 in vivo, 3 were targets for CK2, and we have not yet determined which kinases phosphorylate the other sites. The only remaining sites that fit a consensus sequence are two in the amino terminus that are within the minimal consensus motif for phosphorylation by Cdc28. It is plausible that Nap1 is a substrate for this kinase, given the fact that Nap1 binds Clb2, the mitotic cyclin subunit for Cdc28 (25). Future studies will focus on the functional significance of Nap1 phosphorylation at each site, and it remains possible that there are other phosphorylation sites not identified in this study.
We analyzed the potential impact of phosphorylation on Nap1 conformation using the published crystal structure (48). Serine 82 represents the most conserved phosphorylation site identified. This site is conserved in all four human Nap1 isoforms, Nap1L1 to Nap1L4. Interestingly, comparison of the sequence surrounding this site from different species allowed us to identify a potentially novel consensus sequence. The sequence, S/T-X-Y/F-V/I, is shared from yeast to humans, including mouse, Xenopus laevis and Schizosaccharomyces pombe. Although we do not know as yet which kinase phosphorylates S82, this conserved sequence may constitute a novel recognition motif. S82 is in the α1 domain that is proposed to play a role in orienting the C-terminal acidic domain of the opposite subunit (48). The acidic domain is also important for histone binding, suggesting that S82 phosphorylation may play a role in dimer stabilization or regulate the interaction of Nap1 with chromatin.
Several of the identified phosphorylated residues are located adjacent to the NES on the dimerization α2 helix (subdomain A) and the clamp domain (subdomain B). Nap1 has been predicted to be an obligate dimer; however, it is possible that phosphorylation of residues in the clamp domain leads to a change in the conformation of the clamp. As the clamp likely contributes to the formation or stability of the dimer, this may alter the interaction of the two subunits in some way (48). In addition, two phosphorylation sites on the α2 helix appeared to be occluded by the clamp domain, suggesting that this domain must move to allow phosphorylation to occur and further supporting the notion that the clamp has more than one conformation. Park and Luger also noted three potential CK2 targets on the basis of the kinase consensus sequence (S140, S159, and S177) (48). As S140 and S159 are located on the clamp domain, adjacent to the NES of the opposing molecule, they predicted that phosphorylation of these residues by CK2 could affect localization by increasing the accessibility of the NES. We demonstrated that S159 and S177 are phosphorylated by CK2, as well as a third residue, S397, and so far we have no evidence that S140 is phosphorylated by CK2. Serine 397 is of particular interest, since in both Oryza and Drosophila Nap1 homologs, a C-terminal serine is proposed as a CK2 target (9, 31), and in human TSPY phosphorylation of a C-terminal serine is required for nuclear import (27). As noted above, the location of the CK2 phosphorylation sites suggested that phosphorylation would impact export of Nap1 by altering the availability of the NES to transport factors (48). However, we found that export was seemingly unaffected when phosphorylation was prevented, but rather nuclear import was inhibited.
The mechanism by which CK2 phosphorylation promotes nuclear import of Nap1 is not yet understood. We examined whether phosphorylation could enhance the affinity of Nap1 for its known karyopherin Kap114; however, in this study the association of recombinant Nap1 with Kap114 was unaffected by mutation of the phosphorylated residues or by in vitro phosphorylation of Nap1 (data not shown). This suggests that phosphorylation does not regulate the Kap114-Nap1 interaction. Because the Nap1 L99S mutant was not significantly redistributed to the cytoplasm in strains lacking KAP114, Nap1 likely has additional routes to the nucleus. These routes may be mediated by other karyopherins or indirectly by piggybacking onto other nuclear proteins, and it is possible that phosphorylation could promote these interactions. It is also possible that CK2 phosphorylation modulates the Nap1-histone interaction and hence regulates the formation of a stable histone-Kap114-Nap1 complex. However, we did not observe an obvious difference between the binding of the Nap1(S159A S177A S397A) or Nap1(S159D S177D S397D) to histones in an in vitro binding assay, suggesting that this was not the case.
Interestingly, we did not observe steady-state changes in the nuclear to cytoplasmic distribution of Nap1 during the cell cycle, as has been shown in higher eukaryotic homologs, although we cannot rule out the cell cycle-dependent relocalization of a small pool of Nap1 (data not shown). However, both Nap1(S159A S177A S397A) and Nap1(S159D S177D S397D) exhibited a shortened G1 and prolonged S phase relative to the wild type, with Nap1(S159D S177D S397D), which mimics constitutive phosphorylation by CK2, showing a more pronounced defect. The fact that we saw the defect with both mutants suggests that regulated cycles of phosphorylation and dephosphorylation are important for correct cell cycle progression. Loss of Nap1 is known to lead to an elongated bud phenotype and delayed G2/M progression. In contrast, the defect we observed with the Nap1-phosphomutants was a prolonged S phase, a phenotype associated with replication stressors, such as impaired deoxynucleoside triphosphate synthesis and defects in DNA damage repair (33). As Nap1 is a chromatin assembly and histone import factor, the observed S phase defect may be due to an alteration in the availability of histones during replication as a result of the phosphosite mutations. A cell cycle defect is also seen in yeast lacking the chromatin assembly factor, Asf1. asf1Δ strains exhibit delays in both S phase and G2/M and are hypersensitive to DNA double-stranded break-inducing agents, suggesting that the chromatin assembly activity of Asf1 is required during both replication and repair (56). No role in DNA repair has yet been defined for Nap1, but it was recently shown that Nap1 greatly enhances the disassembly of nucleosomes from chromatin by RSC in vitro (35). The RSC complex is specifically recruited to regions of double-stranded breaks, where remodeling activity makes chromatin more accessible to the repair machinery (53). Therefore, the cell cycle defect exhibited by the Nap1 phosphomutants may in part be due to a defect in chromatin remodeling during DNA repair.
In summary, this report examines the regulation of Nap1 function, and we determine that reversible phosphorylation of Nap1 by CK2 may be involved in cell cycle regulation and the regulation of Nap1 nuclear import. The ability of Nap1(S159A S177A S397A) to rescue normal bud morphology in a nap1Δ strain implies that Nap1 phosphorylation is not required for its role in bud formation. Mechanistically, we predict that phosphorylation of Nap1 occurs in the cytoplasm prior to import, though nuclear phosphorylation is also possible, since kinase and substrate are abundant in both compartments. We propose that once Nap1 is inside the nucleus, dephosphorylation occurs, as reversible phosphorylation is necessary for timely S-phase progression, with the phosphomimic showing a greater cell cycle defect. This suggests that cycles of phosphorylation and dephosphorylation may occur in the nucleus, and it is tempting to speculate that they are necessary for Nap1 to efficiently function in chromatin assembly during S phase. Assembly factors, such as Nap1, function by binding histones and titrating them slowly onto the DNA. This requires the assembly factor to act as both histone donor and acceptor during chromatin assembly, and cycles of phosphorylation and dephosphorylation could regulate this function. In conclusion, our data show for the first time that Nap1 phosphorylation by CK2 appears to regulate Nap1 localization and is required for normal progression through S phase.
Acknowledgments
We thank Douglas R. Kellogg and Charles Boone for providing strains. We thank Brian Del Rosario, Jeffrey Blackwell, and David Wotton for suggestions and comments. We also thank Joanne Lannigan and Michael Solga in the Flow Cytometry Core Facility and Ken Victor in the Department of Chemistry at the University of Virginia for technical help.
This work was supported by NIH research grants R01 GM65385 and R01 GM037537.
Footnotes
Published ahead of print on 17 December 2007.
REFERENCES
- 1.Adachi, Y., G. N. Pavlakis, and T. D. Copeland. 1994. Identification and characterization of SET, a nuclear phosphoprotein encoded by the translocation break point in acute undifferentiated leukemia. J. Biol. Chem. 2692258-2262. [PubMed] [Google Scholar]
- 2.Adachi, Y., G. N. Pavlakis, and T. D. Copeland. 1994. Identification of in vivo phosphorylation sites of SET, a nuclear phosphoprotein encoded by the translocation breakpoint in acute undifferentiated leukemia. FEBS Lett. 340231-235. [DOI] [PubMed] [Google Scholar]
- 3.Aitchison, J. D., M. P. Rout, M. Marelli, G. Blobel, and R. W. Wozniak. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 1311133-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akada, R., J. Yamamoto, and I. Yamashita. 1997. Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol. Gen. Genet. 254267-274. [DOI] [PubMed] [Google Scholar]
- 5.Altman, R., and D. Kellogg. 1997. Control of mitotic events by Nap1 and the Gin4 kinase. J. Cell Biol. 138119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnaoutov, A., and M. Dasso. 2003. The Ran GTPase regulates kinetochore function. Dev. Cell 599-111. [DOI] [PubMed] [Google Scholar]
- 7.Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 2941351-1362. [DOI] [PubMed] [Google Scholar]
- 8.Carazo-Salas, R. E., G. Guarguaglini, O. J. Gruss, A. Segref, E. Karsenti, and I. W. Mattaj. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400178-181. [DOI] [PubMed] [Google Scholar]
- 9.Dong, A., Z. Liu, Y. Zhu, F. Yu, Z. Li, K. Cao, and W. H. Shen. 2005. Interacting proteins and differences in nuclear transport reveal specific functions for the NAP1 family proteins in plants. Plant Physiol. 1381446-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faust, M., J. Gunther, E. Morgenstern, M. Montenarh, and C. Gotz. 2002. Specific localization of the catalytic subunits of protein kinase CK2 at the centrosomes. Cell. Mol. Life Sci. 592155-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faust, M., N. Schuster, and M. Montenarh. 1999. Specific binding of protein kinase CK2 catalytic subunits to tubulin. FEBS Lett. 46251-56. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez, J., L. Andrews, and S. M. Mische. 1994. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal. Biochem. 218112-117. [DOI] [PubMed] [Google Scholar]
- 13.Fornerod, M., J. Boer, S. van Baal, M. Jaegle, M. von Lindern, K. G. Murti, D. Davis, J. Bonten, A. Buijs, and G. Grosveld. 1995. Relocation of the carboxyterminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene 101739-1748. [PubMed] [Google Scholar]
- 14.Fornerod, M., J. Boer, S. van Baal, H. Morreau, and G. Grosveld. 1996. Interaction of cellular proteins with the leukemia specific fusion proteins DEK-CAN and SET-CAN and their normal counterpart, the nucleoporin CAN. Oncogene 131801-1808. [PubMed] [Google Scholar]
- 15.Fujii-Nakata, T., Y. Ishimi, A. Okuda, and A. Kikuchi. 1992. Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. J. Biol. Chem. 26720980-20986. [PubMed] [Google Scholar]
- 16.Gal, T. Z., I. Glazer, A. Sherman, and H. Koltai. 2005. Protein interaction of nucleosome assembly protein 1 and casein kinase 2 during desiccation response in the insect-killing nematode Steinernema feltiae IS-6. J. Parasitol. 91691-693. [DOI] [PubMed] [Google Scholar]
- 17.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415141-147. [DOI] [PubMed] [Google Scholar]
- 18.Gharahdaghi, F., M. Kirchner, J. Fernandez, and S. M. Mische. 1996. Peptide-mass profiles of polyvinylidene difluoride-bound proteins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in the presence of nonionic detergents. Anal. Biochem. 23394-99. [DOI] [PubMed] [Google Scholar]
- 19.Hanna, D. E., A. Rethinaswamy, and C. V. Glover. 1995. Casein kinase II is required for cell cycle progression during G1 and G2/M in Saccharomyces cerevisiae. J. Biol. Chem. 27025905-25914. [DOI] [PubMed] [Google Scholar]
- 20.Hosaka, K., T. Kodaki, and S. Yamashita. 1989. Cloning and characterization of the yeast CKI gene encoding choline kinase and its expression in Escherichia coli. J. Biol. Chem. 2642053-2059. [PubMed] [Google Scholar]
- 21.Ishimi, Y., J. Hirosumi, W. Sato, K. Sugasawa, S. Yokota, F. Hanaoka, and M. Yamada. 1984. Purification and initial characterization of a protein which facilitates assembly of nucleosome-like structure from mammalian cells. Eur. J. Biochem. 142431-439. [DOI] [PubMed] [Google Scholar]
- 22.Ishimi, Y., W. Sato, M. Kojima, K. Sugasawa, F. Hanaoka, and M. Yamada. 1985. Rapid purification of nucleosome assembly protein (AP-I) and production of monoclonal antibodies against it. Cell Struct. Funct. 10373-382. [DOI] [PubMed] [Google Scholar]
- 23.Ito, T., M. Bulger, R. Kobayashi, and J. T. Kadonaga. 1996. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 163112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwase, M., and A. Toh-e. 2001. Nis1 encoded by YNL078W: a new neck protein of Saccharomyces cerevisiae. Genes Genet. Syst. 76335-343. [DOI] [PubMed] [Google Scholar]
- 25.Kellogg, D. R., and A. W. Murray. 1995. NAP1 acts with Clb2 to perform mitotic functions and to suppress polar bud growth in budding yeast. J. Cell Biol. 130675-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krek, W., G. Maridor, and E. A. Nigg. 1992. Casein kinase II is a predominantly nuclear enzyme. J. Cell Biol. 11643-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krick, R., A. Aschrafi, D. Hasgun, and J. Arnemann. 2006. CK2-dependent C-terminal phosphorylation at T300 directs the nuclear transport of TSPY protein. Biochem. Biophys. Res. Commun. 341343-350. [DOI] [PubMed] [Google Scholar]
- 28.Lew, D. J., and S. I. Reed. 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 1201305-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, H. Y., and Y. Zheng. 2004. Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes Dev. 18512-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, M., A. Makkinje, and Z. Damuni. 1996. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J. Biol. Chem. 27111059-11062. [DOI] [PubMed] [Google Scholar]
- 31.Li, M., D. Strand, A. Krehan, W. Pyerin, H. Heid, B. Neumann, and B. M. Mechler. 1999. Casein kinase 2 binds and phosphorylates the nucleosome assembly protein-1 (NAP1) in Drosophila melanogaster. J. Mol. Biol. 2931067-1084. [DOI] [PubMed] [Google Scholar]
- 32.Litchfield, D. W. 2003. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 3691-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longhese, M. P., M. Clerici, and G. Lucchini. 2003. The S-phase checkpoint and its regulation in Saccharomyces cerevisiae. Mutat. Res. 53241-58. [DOI] [PubMed] [Google Scholar]
- 34.Longtine, M. S., C. L. Theesfeld, J. N. McMillan, E. Weaver, J. R. Pringle, and D. J. Lew. 2000. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 204049-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorch, Y., B. Maier-Davis, and R. D. Kornberg. 2006. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl. Acad. Sci. USA 1033090-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMillan, J. N., M. S. Longtine, R. A. Sia, C. L. Theesfeld, E. S. Bardes, J. R. Pringle, and D. J. Lew. 1999. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol. Cell. Biol. 196929-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyaji-Yamaguchi, M., K. Kato, R. Nakano, T. Akashi, A. Kikuchi, and K. Nagata. 2003. Involvement of nucleocytoplasmic shuttling of yeast Nap1 in mitotic progression. Mol. Cell. Biol. 236672-6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mortensen, E. M., H. McDonald, J. Yates III, and D. R. Kellogg. 2002. Cell cycle-dependent assembly of a Gin4-septin complex. Mol. Biol. Cell 132091-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosammaparast, N., B. D. del Rosario, and L. F. Pemberton. 2005. Modulation of histone deposition by the karyopherin Kap114p. Mol. Cell. Biol. 251764-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosammaparast, N., C. S. Ewart, and L. F. Pemberton. 2002. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 216527-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mosammaparast, N., K. R. Jackson, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and L. F. Pemberton. 2001. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J. Cell Biol. 153251-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nemergut, M. E., C. A. Mizzen, T. Stukenberg, C. D. Allis, and I. G. Macara. 2001. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 2921540-1543. [DOI] [PubMed] [Google Scholar]
- 43.Ohkuni, K., K. Shirahige, and A. Kikuchi. 2003. Genome-wide expression analysis of NAP1 in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 3065-9. [DOI] [PubMed] [Google Scholar]
- 44.Okuzaki, D., and H. Nojima. 2001. Kcc4 associates with septin proteins of Saccharomyces cerevisiae. FEBS Lett. 489197-201. [DOI] [PubMed] [Google Scholar]
- 45.Okuzaki, D., S. Tanaka, H. Kanazawa, and H. Nojima. 1997. Gin4 of S. cerevisiae is a bud neck protein that interacts with the Cdc28 complex. Genes Cells 2753-770. [DOI] [PubMed] [Google Scholar]
- 46.Padmanabha, R., J. L. Chen-Wu, D. E. Hanna, and C. V. Glover. 1990. Isolation, sequencing, and disruption of the yeast CKA2 gene: casein kinase II is essential for viability in Saccharomyces cerevisiae. Mol. Cell. Biol. 104089-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park, Y. J., and K. Luger. 2006. Structure and function of nucleosome assembly proteins. Biochem. Cell Biol. 84549-558. [DOI] [PubMed] [Google Scholar]
- 48.Park, Y. J., and K. Luger. 2006. The structure of nucleosome assembly protein 1. Proc. Natl. Acad. Sci. USA 1031248-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pemberton, L. F., M. P. Rout, and G. Blobel. 1995. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc. Natl. Acad. Sci. USA 921187-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rethinaswamy, A., M. J. Birnbaum, and C. V. Glover. 1998. Temperature-sensitive mutations of the CKA1 gene reveal a role for casein kinase II in maintenance of cell polarity in Saccharomyces cerevisiae. J. Biol. Chem. 2735869-5877. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez, P., J. Pelletier, G. B. Price, and M. Zannis-Hadjopoulos. 2000. NAP-2: histone chaperone function and phosphorylation state through the cell cycle. J. Mol. Biol. 298225-238. [DOI] [PubMed] [Google Scholar]
- 52.Selth, L., and J. Q. Svejstrup. 2007. Vps75, a new yeast member of the NAP histone chaperone family. J. Biol. Chem. 28212358-12362. [DOI] [PubMed] [Google Scholar]
- 53.Shim, E. Y., S. J. Hong, J. H. Oum, Y. Yanez, Y. Zhang, and S. E. Lee. 2007. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 271602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 93273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsuchiya, K., R. Reijo, D. C. Page, and C. M. Disteche. 1995. Gonadoblastoma: molecular definition of the susceptibility region on the Y chromosome. Am. J. Hum. Genet. 571400-1407. [PMC free article] [PubMed] [Google Scholar]
- 56.Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka, and J. T. Kadonaga. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402555-560. [DOI] [PubMed] [Google Scholar]
- 57.Ubersax, J. A., E. L. Woodbury, P. N. Quang, M. Paraz, J. D. Blethrow, K. Shah, K. M. Shokat, and D. O. Morgan. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425859-864. [DOI] [PubMed] [Google Scholar]
- 58.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403623-627. [DOI] [PubMed] [Google Scholar]
- 59.von Lindern, M., S. van Baal, J. Wiegant, A. Raap, A. Hagemeijer, and G. Grosveld. 1992. can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol. Cell. Biol. 123346-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu, I. J., D. L. Spector, Y. S. Bae, and D. R. Marshak. 1991. Immunocytochemical localization of casein kinase II during interphase and mitosis. J. Cell Biol. 1141217-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]