Abstract
The SMC protein complexes play important roles in chromosome dynamics. The function of the SMC5-6 complex remains unclear, though it is involved in resolution of different DNA structures by recombination. We have now identified and characterized the four non-SMC components of the human complex and in particular demonstrated that the MAGEG1 protein is part of this complex. MAGE proteins play important but as yet undefined roles in carcinogenesis, apoptosis, and brain development. We show that, with the exception of the SUMO ligase hMMS21/hNSE2, depletion of any of the components results in degradation of all the other components. Depletion also confers sensitivity to methyl methanesulfonate. Several of the components are modified by sumoylation and ubiquitination.
SMC (structural maintenance of chromosomes) proteins form the cores of three essential protein complexes involved in important aspects of chromosome dynamics (see reference 15 for a review). Cohesin, containing Smc1 and Smc3, is the molecular glue that holds sister chromatids together from the time that they are established during DNA replication until they separate in anaphase. Smc2 and Smc4 form the core of condensin, which plays a crucial role in compaction of chromosomes at the beginning of mitosis. The function of the SMC5-6 complex is less clear, but it has an important role in a variety of different DNA repair processes and in resolving recombination structures.
The SMC proteins are conserved throughout eukaryotes, and all have similar structures. Globular domains at the N and C termini are separated by two long coiled-coil domains linked together by a hinge. Each SMC protein is doubled back at the hinge region such that the coiled-coil domains interact with each other in an antiparallel manner and the globular domains are brought together. In this way a Walker A motif in the N-terminal domain and a Walker B motif in the C-terminal domain are juxtaposed to generate a globular ATP binding and hydrolysis “head” domain. The two Smc proteins in each complex interact with each other via the hinge. In cohesin the head domains of Smc1 and Smc3 are also brought together with the non-Smc components to form a ring, which is thought to encircle the two sister chromatids. It is not known if the other Smc complexes also form such a ring structure.
The rad18 gene of Schizosaccharomyces pombe was isolated as a mutant sensitive to both UV and ionizing radiation. It was subsequently found to encode an SMC protein (12) that was redesignated Smc6 as part of the SMC5-6 complex. Further genetic and biochemical studies revealed a role in the repair of double-strand breaks as well as in other repair processes involving repair by homologous recombination (12, 29). The precise function remains unclear. The SMC5-6 complex is essential for cell proliferation, but cells are still able to go through several rounds of division in the absence of Smc6 (9). This suggests that death might result from an accumulation of unrepaired damage. Recent work has shown an accumulation of SMC5-6 at ribosomal DNA and telomeres and has shown also that segregation of ribosomal DNA at anaphase is aberrant in temperature-sensitive mutants of SMC6 in Saccharomyces cerevisiae (28). Furthermore, although gross DNA replication was unaffected in a temperature-sensitive smc6 mutant, the ability to resolve replicated chromatids was defective (13). Following treatment of S. cerevisiae with methyl methanesulfonate (MMS), two-dimensional gel analysis revealed the accumulation of X-like structures in smc5-6 mutants (5). Also, in S. pombe, smc6 mutants are unable to rescue collapsed replication forks (1). Taken together, these data suggest a role for SMC5-6 in the resolution of complex structures by homologous recombination.
Aside from the Smc5 and Smc6 proteins that form the core of the S. pombe SMC5-6 complex, there are four essential non-Smc proteins, designated Nse1 to Nse4 (8, 9, 16, 17, 21, 26), and two further proteins, Nse5 and Nse6, that are not essential for proliferation (22). In contrast in S. cerevisiae all six non-SMC proteins are essential (10, 30).
Nse1 has a RING finger typical of E3 ubiquitin ligases, but evidence for E3 activity has so far not been reported. Nse2 is a Sumo ligase, which binds to Smc5, but in S. pombe it sumoylates Smc6, and this sumoylation is stimulated by DNA damage (2). Mutations that abolish the sumoylation activity confer sensitivity to DNA damage but do not affect the essential function of the SMC5-6 complex (2). Similar results were obtained with Mms21, the ortholog from S. cerevisiae (30), although in S. cerevisiae, the substrate appeared to be Smc5. Nse3 is related to the MAGE family of proteins (7), and Nse4 is a member of the kleisin superfamily (20). The kleisins are thought to bridge the head domains of the Smc proteins (25), and we have shown that Nse4 is indeed able to bind the heads of Smc5 and Smc6 (20). Nse6 contains ARM/HEAT repeats, but there is no sequence conservation between these presumed orthologs from S. cerevisiae and S. pombe. In S. pombe they also appear to bridge the head domains of Smc5 and Smc6 (20, 22). Although all these proteins appear to form a single tight complex inside cells (22, 26, 30), attempts to reassemble the complex from individual polypeptides have identified three subcomplexes, namely, Smc6-Smc5-Nse2, Nse1-Nse3-Nse4, and Nse5-Nse6 (22, 26).
In previous work we characterized the SMC5 and SMC6 proteins from human and mouse cells (27). We showed that they interact in human cells and that the interaction is mediated by the hinge regions and abolished by mutation of a single conserved glycine residue in the hinge region of either human SMC5 (hSMC5) or hSMC6 (26). Mouse SMC5 (mSMC5) and mSMC6 are expressed in all tissues and localize to the sex vesicle during meiosis (27). Others have identified human orthologs of Nse1 (9) and Nse2 (24), and the human NSE2 ortholog (hMMS21) has been extensively characterized (23, 24). However, there are two human homologs of Nse4 in the sequence databases and 55 human proteins containing MAGE domains. In this paper we describe the characterization of the mammalian NSE proteins. In particular, we show that both NSE4 paralogs can form part of the complex but that only one of them is expressed in somatic cells, we identify MAGEG1 as the ortholog of Nse3, and we show that all the components form a tight complex and that some of them are modified by ubiquitylation or sumoylation. Finally, we show that knockdown of hNSE1 or hNSE4a confers sensitivity to MMS.
MATERIALS AND METHODS
Plasmids.
DNA sequences encoding the open reading frames (ORFs) of hNSE1, hNSE4a, MAGEG1, and MAGEF1 were acquired from the IMAGE consortium. hMMS21/hNSE2, hNSE4b, and SUMO1 ORFs were amplified by PCR from human cDNA generated from 1BR.3 primary human fibroblasts using the First Strand cDNA synthesis kit (GE Healthcare). hNSE1 cDNA starting at Met2 (see Fig. 1A) was amplified from the IMAGE clone. To make the longer clone starting at Met1, approximately 110 nucleotides of cDNA upstream of Met2 was amplified by PCR from human genomic DNA and ligated to the construct starting at Met2.
FIG. 1.
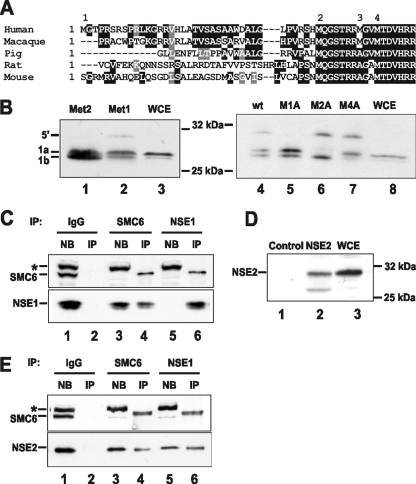
Endogenous hNSE1 and hMMS21/hNSE2 coimmunoprecipitate with SMC6. (A) Alignment of the upstream sequences of hNSE1 from several mammalian species. Dark shading and light shading indicate identical and similar amino acids, respectively. (B) Anti-hNSE1 antibody detects two major translation products (1a and 1b) on Western blots when hNSE1 is expressed in vitro from expression constructs including the complete ORF (Met2) or the ORF plus 5′ untranslated region sequence (Met1) (lanes 1 and 2). Both protein products are also seen in MRC5V1 whole-cell extracts (WCE; lane 3). To confirm the predicted start sites, the indicated methionines were mutated to alanine in the construct including the 5′ untranslated region (right panel). wt, wild type. (C and E) HeLa nuclear extract was incubated with protein A-Sepharose coupled to either anti-hSMC6 or anti-hNSE1 antibodies or control rabbit IgGs. The nonbound (NB) and immunoprecipitated (IP) fractions were subsequently analyzed by Western blotting with the indicated antibodies. The asterisk indicates a nonspecific cross-reacting band. (D) Anti-hMMS21/hNSE2 antibodies detect the hMMS21/hNSE2 protein on Western blots when hMMS21/hNSE2 is expressed in vitro in reticulocyte lysate (lane 2) but not in control reticulocyte lysate (lane 1). An equivalent band is recognized in MRC5V1 cell extracts (lane 3).
Full-length ORFs were subcloned into various vectors for different expression purposes: pEPEX was used for in vitro transcription/translation using the TNT Quick Coupled Transcription/Translation System (Promega); pCI-neo (Promega) incorporating FLAG, Myc, 10-His, and/or green fluorescent protein (GFP) tags was used for expression in mammalian cells; pQE30 (Qiagen) was used for expression of six-His-tagged recombinant protein; and pGEX-KG was used for expression of glutathione S-transferase-tagged recombinant protein in Escherichia coli.
Antibodies.
Full-length hNSE1, hNSE4a, and MAGEG1 were expressed in E. coli as N-terminally hexahistidine-tagged fusion proteins. The proteins were each purified to near homogeneity under denaturing conditions by Ni2+-nitrilotriacetic acid (Ni2+-NTA) affinity chromatography and then used to inoculate two rabbits for antibody generation (Eurogentec). Full-length hMMS21/hNSE2 was expressed in bacteria as a glutathione S-transferase fusion, purified on glutathione Sepharose (GE Healthcare) according to the manufacturer's instructions, and used for antibody production. hNSE1 and hMMS21/hNSE2 antibodies were affinity purified using antigen immobilized with Aminolink Plus coupling gel (Pierce). Antibodies to MAGEG1 and hNSE4a were affinity purified using antigen that had been transferred to nitrocellulose membrane. hSMC5 and hSMC6 antibodies have been described previously (27). Anti-FLAG M2 (Sigma-Aldrich) and antivimentin (Oncogene) commercial antibodies were also used in this study.
Immunoblotting and immunoprecipitation.
For immunoblotting MRC5V1 cells were lysed in sodium dodecyl sulfate (SDS) sample buffer, sonicated, and boiled, and the proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were transferred to a nitrocellulose membrane (GE Healthcare) and blotted with appropriate primary antibodies followed by secondary horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (Dako) or HRP-conjugated rabbit anti-mouse antibody (Dako). Proteins were visualized using Western Lightning chemiluminescent detection reagent (Perkin-Elmer) according to the manufacturer's instructions.
For immunoprecipitation of FLAG-tagged proteins, whole-cell extracts were made by lysing cells in lysis buffer (20 mM HEPES, pH 7.9, 20 mM NaCl, 1 mM MgCl2, 0.5% Triton X-100, 1 μl/ml benzonase [Merck] and 1× complete EDTA-free protease inhibitor mix [Roche]) at 25°C for 10 min followed by incubation on ice for 10 min. One volume of extraction buffer (20 mM HEPES, pH 7.9, 0.5 M NaCl, 0.2 mM EDTA, 20% glycerol, 0.5 mM dithiothreitol [DTT], and 1× complete EDTA-free protease inhibitor mix) was added, and the extract was held on ice for 15 min before the addition of 8 volumes of dilution buffer (20 mM HEPES, pH 7.9, 100 mM NaCl, 0.2 mM EDTA, 10% glycerol, 0.5 mM DTT, and 1× complete EDTA-free protease inhibitor mix). Extracts were cleared by centrifugation at 13,000 rpm for 5 min, and soluble fractions were mixed with anti-FLAG M2 antibody and protein G-Sepharose beads (CRUK, London, United Kingdom) overnight at 4°C. The beads were washed once with wash buffer 1 (20 mM HEPES, pH 7.9, 0.5 M NaCl, 0.2 mM EDTA, 0.5 mM DTT, and 0.1% Triton X-100) and three times with wash buffer 2 (20 mM HEPES, pH 7.9, 1 M NaCl, 0.2 mM EDTA, 0.5 mM DTT, and 0.1% Triton X-100) and then resuspended and boiled in SDS sample buffer.
Endogenous proteins were immunoprecipitated from either commercially prepared HeLa cell nuclear extract (4C, Belgium) or from whole-cell extracts prepared from MRC5V1 cells as described above. Cleared extracts were incubated for 4 h or overnight with protein A-Sepharose beads that had been previously cross-linked to either anti-hSMC6 antibodies, anti-hNSE1 antibodies, or nonspecific rabbit immunoglobulin Gs (IgGs) using dimethylpimelimidate. The beads were washed two times with wash buffer 1 and three times with wash buffer 2, resuspended, and boiled in SDS sample buffer. Immunoprecipitated samples were resolved by SDS-PAGE and analyzed by blotting with appropriate antibodies. For detection of immunoprecipitated proteins a protein A-HRP conjugate (Pierce) was routinely used in place of secondary antibody in order to reduce problems with IgG heavy and light chains masking proteins of interest at the same molecular weight.
Mass spectrometric analysis of SMC5/6-associated proteins.
The hSMC5-6 complex was purified from HeLa cell nuclear extract (4C, Belgium) by anion-exchange chromatography followed by immunoprecipitation with anti-hSMC6 or anti-hNSE1 antibody beads. Briefly, HeLa cell nuclear extract (60 mg) was cleared by centrifugation (18,000 rpm, 30 min) and applied to a Mono Q anion-exchange column. Fractions containing hSMC5/6 following elution with a salt gradient were then diluted twofold with 20 mM HEPES, pH 7.9, 0.2 mM EDTA, 10% glycerol, 0.5 mM DTT, and 1× complete EDTA-free protease inhibitor mix and immunoprecipitated as described above. The hSMC5-6 complex was eluted from the antibody beads using 200 mM glycine (pH 2.5) and then precipitated with 4 volumes of acetone. Following reduction and alkylation, proteins were digested with 25 ng/μl trypsin for 16 h at 37°C. The resulting peptide mixture was diluted in 0.1% trifluoroacetic acid for analysis by nano-liquid chromatography-tandem mass spectrometry at the Sussex Centre for Proteomics using an LTQ linear ion trap (Thermo Fisher). Proteins were identified by automated database searching against the NCBI human protein sequence database using the SEQUEST algorithm as implemented in Bioworks v. 3.1 (Thermo Fisher). Filtering criteria for positive protein identifications were the detection of at least two fully tryptic peptides with Xcorr values greater than 1.9 for +1 spectrum, 2.2 for +2 spectra, and 3.5 for +3 spectra and a delta Cn of greater than 0.1.
Size-exclusion chromatography.
For gel filtration analysis HeLa cell nuclear extract was diluted twofold with buffer A (20 mM HEPES, pH 7.9, 150 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, and 1× complete EDTA-free protease inhibitor mix) and cleared by centrifugation (20,000 rpm, 10 min). Proteins (2 mg) were loaded onto a Superdex 200 column, and 0.5-ml fractions were collected. Proteins in each fraction were acetone precipitated, dissolved in SDS sample buffer, resolved by SDS-PAGE, and analyzed by Western blotting.
Nickel-NTA pull-downs.
For nickel-NTA pull-down experiments extracts were made, 24 h after transfection with the appropriate plasmids, by briefly sonicating cells in Ni2+-NTA lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% SDS, 0.5% Triton X-100, 20 mM imidazole, 4 mM N-ethylmaleimide, and 1× complete EDTA-free protease inhibitor mix). Extracts were cleared by centrifugation (13,000 rpm, 5 min) and diluted with 5 volumes of Ni2+-NTA dilution buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 20 mM imidazole, 4 mM N-ethylmaleimide, and 1× complete EDTA-free protease inhibitor mix) and then mixed with Ni2+-NTA agarose (Qiagen) for 3 h. The beads were washed five times with Ni2+-NTA dilution buffer and resuspended in SDS sample buffer, and bound proteins were analyzed by Western blotting.
Cell culture and transfection.
Simian virus 40-transformed MRC5 cells (designated MRC5V1) were grown in Eagle's minimal essential medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and 100 μg/ml penicillin and streptomycin (Invitrogen). Plasmid transfections were performed using Fugene (Roche) according to the manufacturer's instructions. For stable transfectants, MRC5V1 cells were transfected with pCI-neo-derived plasmids and selection (800 μg/ml G418) was applied 48 h after transfection. Stable transfectants were isolated after 2 weeks.
siRNA techniques.
Short interfering RNA (siRNA) oligonucleotides were obtained from Qiagen. The proteins and siRNA target sequences were as follows: hNSE1, CCGGCTTTGCGTCTTCCACAA; hMMS21/hNSE2, AACTCTGGTATGGACACAGCT; MAGEG1, TAGGATGTGTTTGCAAAGTTT; NSE4a, CTCCTTTGACATGTTAAGATA; and hSMC5, AAGCGAAGAGAGAGGGAAACT, respectively. siRNA transfections were performed at 60 to 70% confluence using HiPerfect transfection reagent (Qiagen) according to the manufacturer's instructions. To determine the efficiency of RNA interference knockdown, siRNA-transfected cells were lysed (3 to 4 days posttransfection) in 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.1% SDS, and 1 μl/ml benzonase at 25°C for 10 min. Extracts were cleared by centrifugation (13,000 rpm, 5 min), boiled in SDS sample buffer, and analyzed by Western blotting. MMS sensitivity was determined 96 h after siRNA transfection by adding MMS at the indicated doses (0 to 0.0075%) for 1 h. The cells were then washed free of the drug and plated for colony formation. Colonies were counted 12 days later.
RESULTS
hNSE1 and hMMS21/hNSE2.
A human ortholog of Nse1 (hNSE1, NMSCE1) has been identified by several groups (8, 9, 24). However, the start site proposed by Fujioka et al. (8) (MQGS…, indicated as “2” in Fig. 1A) is 10 amino acids (aa) upstream of that proposed by Harvey et al. (9) (MTDV…, indicated as “4” in Fig. 1A), and the 10 intervening amino acids are highly conserved in mammals (Fig. 1A). Comparison of the hNSE1 sequences in the expressed sequence tag database with the genomic sequence identifies an intron 4 aa upstream of the MQGS sequence and an ORF extending to a methionine a further 33 aa upstream (indicated as “1” in Fig. 1A). Conservation of the predicted amino acids translated from this upstream exon is poor, with significant sequence identity found only among primates (Fig. 1A). In contrast the rest of the sequence is highly conserved among mammals (not shown). In order to determine the actual translation start site, we generated an antibody against hNSE1. We then cloned hNSE1 cDNA into an expression vector, starting from either methionine 1 (as shown in Fig. 1A) or from methionine 2 and expressed the inserted protein using in vitro transcription-translation. Using the shorter construct, the antibody detected two bands (Fig. 1B, left panel, lane 1), which we interpret to represent initiation from methionines 2 and 4, respectively. With the longer construct, we obtained the same doublet and in addition a faint band of higher molecular weight, which we interpret as protein initiated from methionine 1 (lane 2). To confirm this interpretation, we used the construct starting from methionine 1 and mutated each of methionines 1, 2, and 4 to alanine. We found that the predicted bands disappeared, i.e., the top, middle, and bottom bands disappeared, if we mutated methionines 1, 2, and 4, respectively (Fig. 1B, right panel, lanes 5 to 7). Thus, the major initiating ATGs are methionines 2 and 4, and methionine 1 is rarely used. Consistent with this, when we examined human cell extracts by immunoblotting with the anti-hNSE1 antibody, we again obtained a doublet, corresponding to initiation at methionines 2 and 4, the former being the stronger band (Fig. 1B, lanes 3 and 8). We conclude that the major initiation site is at methionine 2 (MQGS).
To see if hNSE1 interacts with hSMC6, we immunoprecipitated endogenous hSMC6 from MRC5 cell extracts and analyzed the immunoprecipitates. Figure 1C shows that anti-hSMC6 pulls down hNSE1 (lane 4) and conversely immunoprecipitation with anti-hNSE1 pulls down hSMC6 (lane 6). We note that in the immunoprecipitation with anti-hSMC6, some hNSE1 remains unbound (lane 3), whereas anti-hNSE1 quantitatively immunoprecipitates hSMC6 (compare with lane 5). This suggests that some hNSE1 protein is not associated with the complex (see below for further discussion).
hNSE2 (hMMS21, NSMCE2), the human homolog of Nse2 in S. pombe and Mms21 in S. cerevisiae, has been identified and characterized by Potts et al. (23, 24). They showed that hMMS21/hNSE2 coimmunoprecipitated with hSMC5. We have raised an antibody against hMMS21/hNSE2, which is specific as it identifies a band from hMMS21/hNSE2 generated by in vitro transcription-translation (Fig. 1D, lane 2), and a band of identical size from human cell extracts (lane 3). hMMS21/hNSE2 is pulled down in immunoprecipitations with anti-hSMC6 and anti-hNSE1 (Fig. 1E), confirming that it is part of the SMC5-6 complex in human cells.
hNSE4.
We previously showed, as suggested by Nasmyth and Haering (18), that the predicted structure of S. pombe Nse4 was similar to that of the kleisin superfamily, members of which bridge the heads of SMC proteins. Consistent with this idea we found that the C-terminal half of Nse4 interacted with Smc5 and that Nse4 also interacted with Smc6 (20). There are two human homologs of Nse4 in the sequence databases. The first of these (NSMCE4A; C10orf86) is on chromosome 10 (accession no. Q9NXX6). The second is identical to a protein designated E1A-like inhibitor of differentiation 3 (EID3) on chromosome 12 (4) and appears to be a transcriptional repressor.
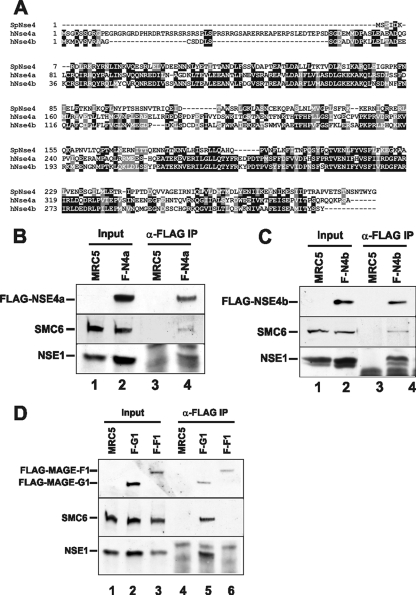
We refer to these as hNSE4a and hNSE4b, respectively. These sequences are approximately 50% identical to each other, and each is approximately 25% identical to S. pombe Nse4 (Fig. 2A). Sequence similarity is highest in the two regions that we previously identified as conserved helix-turn-helix and winged helix domains, respectively (20). hNSE4b/EID3 is expressed exclusively in testis (4), whereas the expressed sequence tags from hNSE4a were derived from many different tissues. We transfected FLAG-tagged constructs of each paralog into MRC5 cells and immunoprecipitated them from cell extracts with anti-FLAG antibody. Immunoblotting revealed the presence of both hSMC6 and hNSE1 in each of the immunoprecipitates (Fig. 2B and C), suggesting that both paralogs could be incorporated into the SMC5-6 complex.
FIG. 2.
FLAG-hNSE4a, FLAG-hNSE4b, and MAGEG1 coimmunoprecipitate with hSMC6 and hNSE1. (A) Alignment of two human hNSE4 paralogs with S. pombe Nse4. (B to D) Anti-FLAG M2 antibody was used to immunoprecipitate FLAG-tagged proteins from whole-cell extracts prepared from MRC5V1 control cells and stably transfected MRC5V1 cells expressing FLAG-hNSE4a (F-N4a) (B), FLAG-hNSE4b (F-N4b) (C), and FLAG-MAGEG1 (F-G1) and FLAG-MAGEF1 (F-F1) (D). hSMC6, hNSE1, and FLAG-tagged proteins were detected in the extracts and in the immunoprecipitated fractions by Western blotting.
MAGEG1.
Nse3 is related to the MAGE family of proteins. There are 55 MAGE genes in the human genome, which have been subdivided into different classes, based on their protein structures (3, 7). Many of them are expressed only in tumor cells or in the germ line. To identify the MAGE protein in the SMC5-6 complex, we immunoprecipitated the complex with antibodies to either hNSE1 or hSMC6. We subjected the two immunoprecipitates, without further fractionation, to mass spectroscopy to identify proteins that were present in both immunoprecipitates. We were able to identify hSMC5, hSMC6, hNSE1, hNSE2, and hNSE4a and one member of the MAGE family, MAGEG1 (Table 1).
TABLE 1.
Mass spectrometric analysis of proteins copurifying with hNSE1 and hSMC6a
| Protein | Purification with:
|
|||
|---|---|---|---|---|
| hNSE1
|
hSMC6
|
|||
| No. of peptides | % Coverage | No. of peptides | % Coverage | |
| hSMC5 | 28 | 19.9 | 7 | 6.6 |
| hSMC6 | 21 | 23.5 | 8 | 8.4 |
| hNSE1 | 5 | 18.6 | ||
| hMMS21/hNSE2 | 5 | 19.0 | ||
| hNSE4a | 6 | 17.1 | 2 | 8.1 |
| MAGEG1 | 4 | 17.9 | 3 | 3.5 |
Anti-hNSE1 or anti-hSMC6 antibodies coupled to protein A-Sepharose were used to immunoprecipitate the indicated proteins from HeLa nuclear extract. Eluted complexes were analyzed by mass spectrometry. The number of independent peptides of each protein recovered and the percentage of primary sequences covered by these peptides are presented.
MAGEG1 (NDNL2) maps to chromosome 15q close to the autistic disorder susceptibility region but is not thought to be involved in these disorders (6). It is expressed in all tissues and is closely related to MAGEF1 (3, 7). We therefore FLAG-tagged both MAGEG1 and MAGEF1 and expressed them in MRC5 cells. Immunoprecipitation with anti-FLAG followed by immunoblotting showed that MAGEG1 was able to coimmunoprecipitate hNSE1 and hSMC6 from cell extracts (Fig. 2D, lane 5), whereas they were not precipitated by MAGEF1 (Fig. 2D, lane 6). These results indicate that MAGEG1 is the human ortholog of Nse3. There is 20% sequence identity between these orthologs.
Composition and stability of the complex.
To confirm the composition of the complex, we immunoprecipitated cell extracts with either anti-hNSE1 or anti-hSMC6 antibodies in 1 M NaCl. Both of the immunoprecipitated complexes contained hSMC5, hSMC6, hNSE4a, MAGEG1, hMMS21/hNSE2, and hNSE1 (Fig. 3A, lanes 2 and 3). These data demonstrate that these six proteins form a stable complex in human cells.
FIG. 3.
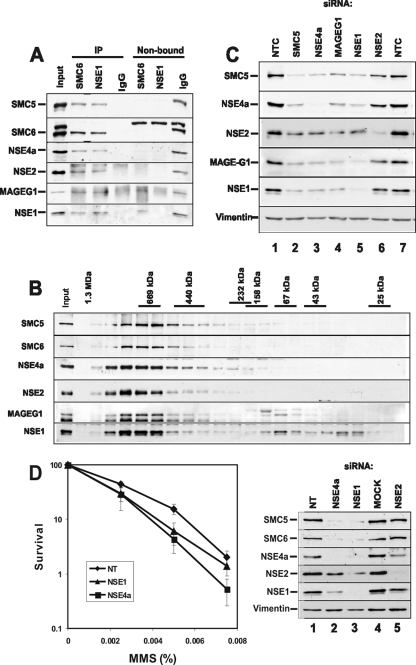
hSMC5 and hSMC6 form a stable complex with hNSE1, hMMS21/hNSE2, hNSE4a, and MAGEG1 that is required for survival following exposure to MMS. (A) Protein A-Sepharose coupled to anti-hSMC6 or anti-hNSE1 antibodies was used to immunoprecipitate the hSMC5-6 complex from HeLa nuclear extract. Nonspecific rabbit IgGs were used as a negative control. Immunoprecipitated complexes were washed in buffer containing 1 M NaCl before Western blot analysis of the immunoprecipitated and nonbound fractions. (B) HeLa nuclear extract was applied to a Superdex 200 gel filtration column, and fractions were analyzed by Western blotting with antibodies to each of the hSMC5-6 complex components. (C) MRC5V1 cells were transfected with a nontargeting control (NTC) siRNA or siRNAs specific for hSMC5, hNSE1, hMMS21/hNSE2, hNSE4a, or MAGEG1. Seventy-two hours after siRNA transfection whole-cell extracts were prepared and protein expression levels were examined by immunoblotting with the antibodies indicated. (D) Ninety-six hours after transfection with hNSE1, hNSE4a, or nontargeting (NT) siRNAs, MRC5V1 cells were treated with the indicated doses of MMS for 1 h. Following removal of the drug cells were counted and plated for colony formation. Colonies were counted after 12 days, and the percent survival was determined relative to untreated cells for each transfected cell line (left panel). Results show means ± standard errors of the means of at least three experiments. Whole-cell extracts were prepared 96 h posttransfection, and protein levels were analyzed by Western blotting with the indicated antibodies (right panel).
Interestingly, the anti-hSMC6 immunoprecipitates contained essentially all of the hSMC5, hMMS21/hNSE2, and hNSE4a in the extracts. None of these proteins remained in the supernatant. MAGEG1 and hNSE1 were found in both unbound and bound fractions. Similarly, the anti-hNSE1 immunoprecipitates quantitatively bound all the other components. These data suggest that, with the exception of MAGEG1 and hNSE1, the hSMC5-6 components are present exclusively in the complex. To obtain independent evidence to support this conclusion, soluble extracts were analyzed by gel filtration and immunoblotting (Fig. 3B). A large proportion of each component was present in fractions corresponding to a globular protein of approximately 800 kDa. In the case of hSMC5, hSMC6, hNSE4a, and hMMS21/hNSE2, almost all of the protein was in this high-molecular-weight complex with a small amount spread through the following few fractions. In the case of MAGEG1 there was a small amount at lower molecular weight, and in the case of hNSE1, there were two smaller protein peaks. Thus, most of the proteins are present in the hSMC5-6 complex with little free protein in the extracts.
To examine if all the proteins were necessary to maintain the stability of the complex, we developed siRNAs against most of the identified components (Materials and Methods). As can be seen from Fig. 3C, transfection of MRC5 human fibroblasts with any of the siRNAs reduced the concentration of the corresponding proteins by >80%. Furthermore, knock-down of hSMC5, hNSE4a, MAGEG1, or hNSE1 (lanes 2 to 5) also reduced the concentration of the other components, with the exception of hMMS21/hNSE2, which was somewhat reduced but not as drastically as the other components. Conversely knock-down of hMMS21/hNSE2 had little effect on the concentration of any of the other components (lane 6). These results suggest that all the components except for hMMS21/hNSE2 are necessary for the stability of the complex, and in the absence of the complex, the protein components are degraded. hMMS21/hNSE2 appears to be the only protein that remains stable in the absence of the others.
DNA damage responses.
To examine the biological effect of reducing the levels of SMC5-6 components, we treated MRC5 cells with the siRNAs described above and then exposed the cells to different DNA-damaging agents. We found no significant effect on colony-forming ability after treatment with UV or gamma irradiation or treatment with mitomycin C. Sensitivity to MMS was observed when either hNSE1 or hNSE4a was knocked down (Fig. 3D).
Posttranslational modifications.
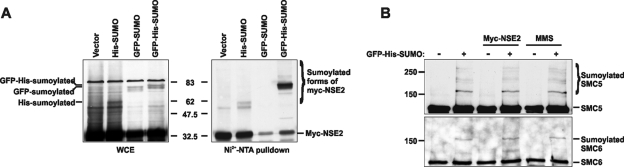
hNSE1 has a RING finger motif characteristic of E3 ubiquitin ligases, and hMMS21/hNSE2 is a SUMO ligase. As a first step in identifying possible targets of these proteins, we checked to see if any of the components are themselves modified by ubiquitin or Sumo. We cotransfected cells with Myc-tagged hMMS21/hNSE2 and SUMO1 tagged with either His, GFP, or GFP-His. Lysates were analyzed by immunoblotting with anti-hMMS21/hNSE2 antibody (Fig. 4A), either directly (lanes 1 to 4) or after extraction on nickel agarose (lanes 5 to 8). In both samples in which the SUMO1 had a His tag, specific bands corresponding to sumoylated hMMS21/hNSE2 were detected (lanes 2, 4, 6, and 8). A much stronger band was obtained with GFP-His-tagged SUMO1 samples (lanes 4 and 8).
FIG. 4.
Sumoylation of hSMC5-6 complex components. (A) MRC5V1 cells were transfected with myc-hMMS21/hNSE2 along with SUMO1 expression constructs (His-SUMO, GFP-SUMO, and GFP-His-SUMO) or empty vector. Twenty-four hours after transfection cells were lysed and the lysates were incubated with Ni2+-NTA agarose for 3 h. Sumoylated forms of myc-hMMS21/hNSE2 were detected in the whole-cell extracts and in the nickel pull-downs by Western blotting with anti-hMMS21/hNSE2 antibody. (B) Twenty-four hours after transfection of MRC5V1 cells with GFP-His-SUMO (or mock transfection) extracts were prepared and His-tagged proteins were captured on Ni2+-NTA agarose. Sumoylated forms of hSMC5 and hSMC6 were detected in the nickel pull-downs by Western blotting with anti-hSMC5 and anti-hSMC6 antibodies. Treatment with 0.0075% MMS for 3 h immediately prior to lysing the cells or cotransfection with myc-hMMS21/hNSE2 had no effect on hSMC5/6 sumoylation. Numbers in the center of panel A and at the left of panel B are molecular masses in kilodaltons.
In a similar experiment with GFP-His-tagged SUMO1, the lysates were analyzed for modified forms of either hSMC5 or hSMC6. Figure 4B shows that high-molecular-weight species representing sumoylated hSMC5 (top panel, lanes 2, 4, and 6) and SMC6 (bottom panel, lanes 2, 4, and 6) were detected specifically in those cells which had been transfected with GFP-His-tagged SUMO1. The intensity of this higher-molecular-weight band was not affected by overexpression of hMMS21/hNSE2 or by treatment of cells with MMS. There was no evidence for sumoylation of hNSE1, MAGEG1, or hNSE4a (data not shown). Note that unexpectedly we found bands corresponding to unmodified hSMC5, SMC6, and Myc-HMMS21/hNSE2 binding to nickel beads. We have found in many experiments that several members of the complex have affinity for nickel beads even without His tags.
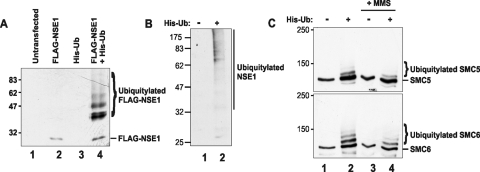
In analogous experiments to look for ubiquitinated species, MRC5 cells were transfected with FLAG-hNSE1 and His-tagged ubiquitin. Cells were lysed under denaturing conditions, and the lysates were extracted with Ni-agarose beads and analyzed by immunoblotting with anti-hNSE1 antibody. Strong bands corresponding to ubiquitinated species of hNSE1 were detected in these extracts (Fig. 5A, lane 4) but not in cells which were transfected with FLAG-tagged hNSE1 (lane 2) or His-tagged ubiquitin (lane 3) alone. In subsequent experiments, ubiquitylation of endogenous hNSE1 was analyzed by transfecting cells with His-tagged ubiquitin, extracting on nickel, and probing with anti-hNSE1 antibody. High-molecular-weight species were detected in cells cotransfected with His-ubiquitin but not in cells transfected with empty vector (Fig. 5B). We were similarly able to identify ubiquitinated species of SMC5 and SMC6 in cells transfected with His-ubiquitin (Fig. 5C). The ubiquitylated species were not affected by treatment of the cells with MMS.
FIG. 5.
Ubiquitylation of SMC5-6 complex components. (A) MRC5V1 cells were transfected with FLAG-hNSE1 and His-ubiquitin expression constructs, either singly or in combination. Twenty-four hours after transfection the cells were lysed and His-tagged proteins were captured on Ni2+-NTA agarose. His-ubiquitylated FLAG-hNSE1 protein was detected on a Western blot using anti-hNSE1 antibody. (B) Nickel pull-down from extracts prepared 24 h posttransfection with His-ubiquitin (or mock transfected). Ubiquitylated forms of hNSE1 were detected by Western blotting with anti-hNSE1 antibody. (C) Twenty-four hours after transfection of MRC5V1 cells with His-ubiquitin (or mock transfection) cells either were left untreated or were treated with 0.0075% MMS for 3 h. Whole-cell extracts were prepared, and His-tagged proteins were captured on Ni2+-NTA agarose. Ubiquitylated forms of hSMC5 and hSMC6 were detected in the nickel pull-downs by Western blotting with anti-hSMC5 and anti-hSMC6 antibodies. Numbers at left of each panel are molecular masses in kilodaltons.
DISCUSSION
The SMC5-6 complexes in S. cerevisiae and S. pombe contain eight components. These are Smc5 and Smc6 and six non-SMC components, Nse1 to Nse6. Human orthologs of Smc5 and Smc6 and of Nse1 and Nse2 have been identified previously by ourselves and others (8, 9, 24, 27). Potts and Yu (24) extensively characterized hNSE2 (hMMS21), showing that, like S. pombe Nse2 (2), it is a SUMO ligase and is able to sumoylate hSMC6 when both hMMS21/hNSE2 and hSMC6 are overexpressed. In apparent contrast, our results in Fig. 4B did not show an increase in sumoylation of hSMC5 and hSMC6 when we overexpressed hMMS21/hNSE2. However, we analyzed sumoylation of endogenous hSMC5 and hSMC6, and we know that these proteins are entirely in the protein complex, including hMMS21/hNSE2. We might anticipate, therefore, that the associated SUMO ligase activity of hMMS21/hNSE2 has carried out the sumoylation of hSMC5 and hSMC6 within the same molecule of the complex. The apparent difference in our results from those of Potts and Yu can therefore be attributed to the analysis of overexpressed hSMC6 in their experiments and endogenous proteins in our experiments.
Potts and Yu also reported that depletion of hMMS21/hNSE2 by siRNA sensitized HeLa cells to the toxic effects of MMS as measured by trypan blue exclusion and apoptosis. In subsequent work the same authors showed that depletion of hMMS21/hNSE2 increased gene targeting and had no effect on nonhomologous end joining but reduced the levels of sister chromatid exchanges induced by camptothecin and reduced the number of long-tract gene conversions. These data suggest that hMMS21/hNSE2 and by implication the hSMC5-6 complex are required for recombination between sisters but not between homologous chromosomes. hMMS21/hNSE2 is loaded onto chromatin at double-strand break sites and is required for loading cohesin at these sites (23). Our data show that not only hMMS21/hNSE2, but also endogenous hSMC5 and hSMC6, are sumoylated in vivo and that hNSE1, hSMC5, and hSMC6 are ubiquitylated (Fig. 4 and 5). The biological roles of these modifications remain to be determined.
Using siRNA knockdowns, we have shown that all members of the complex with the exception of hMMS21/hNSE2 are required to maintain the stability of the complex. This is consistent with our previous finding that S. pombe Nse2 binds to the coiled-coil domain of Smc5, whereas all other components bind at the heads of the SMC5-6 structure (20, 26). In S. pombe, mutations in Smc5-6 components confer sensitivity to a wide range of DNA-damaging agents. We found that depletion of hNSE1 and hNSE4a conferred sensitivity to MMS but not to several other DNA-damaging agents, as measured by colony-forming activity (Fig. 3D). So far, we have not observed similar sensitivity on depletion of other components. Although the levels of depletion appeared to be around 80%, we consider that the most likely reason for this apparent discrepancy is that for some of the depletions, sufficient amounts of the complex remained to carry out its function in response to MMS. In support of this suggestion, the depletions conferred by siRNA for hNSE1 and hNSE4a, which did confer sensitivity to MMS, appeared to be almost complete with residual protein barely detectable. Taken together with the results of Potts et al. (23, 24), we conclude that the hSMC5-6 complex, like that of the yeasts, is involved in the response to DNA damage. It is hazardous to make definitive conclusions when normal responses are obtained using siRNA approaches. We therefore consider it premature at this point to conclude that the sensitivity to a broad range of DNA-damaging agents in yeast and the relatively narrow range of sensitivity that we have found in human cells represent a genuine mechanistic difference rather than a technical issue. Further work is necessary to resolve whether these differences are real or apparent.
We used a proteomic approach to identify the two new components, the orthologs of Nse4 and Nse3, and have shown that all six components form a tight complex in human cells. We previously provided evidence that Nse4 is a member of the kleisin superfamily (25), which bridges the Smc5 and Smc6 globular heads (20). Structure prediction programs indicated that Nse4 and its orthologs had N-terminal helix-turn-helix and C-terminal winged-helix motifs. The amino acids involved in these structural motifs are the most highly conserved sequences in the alignments of Nse4 with the human orthologs, and both orthologs have these motifs (Fig. 2A). Although both orthologs of Nse4 are capable of forming part of the complex, only hNSE4a is found in the endogenous complex in cultured cells. Whereas hNSE4a is expressed in somatic cells, hNSE4b/EID3 is expressed only in testis and is an inhibitor of CBP-dependent coactivation of nuclear receptor-mediated transcription (4). Whether it fulfills this transcriptional function in a testis-specific form of the hSMC5-6 complex and whether more generally there is a meiosis-specific form of the hSMC5-6 complex remain to be determined.
We identified MAGEG1 as the ortholog of Nse3. The MAGE family has received much attention because members of the MAGEA, -B, and -C families (classified as type I MAGE genes) are expressed exclusively in tumor cells and/or germ cells. The 12 members comprising the MAGED, -E, -F, -G, -H, and -L2 proteins and necdin are expressed in all tissues and have been designated type II (3, 7). Individual members have been reported as playing roles in neuronal development, apoptosis, and cell cycle control (reviewed in reference 3). No biochemical function has been identified for any of the MAGE proteins. The MAGEG1/NDNL2 gene has been mapped close to the necdin and MAGEL2 genes near the region deleted in Prader-Willi syndrome (6). It was not subject to imprinting like nearby genes implicated in Prader-Willi syndrome and therefore probably does not play a role in the etiology of this disorder. The gene was expressed in all tissues but, like hSMC5 and hSMC6 (27), most abundantly in testis (11). When transfected into cells, MAGEG1, like necdin, reduced cell proliferation, an effect possibly mediated by its interaction with the E2F1 transcriptional activator. Furthermore like necdin, MAGEG1 interacts with the p75 neurotrophin receptor and may be involved in brain development (11). Intriguingly, recent evidence suggests that only one MAGE gene exists in genomes from plants, nematodes, insects, and even nonmammalian vertebrates (14). Like MAGEG1, the chicken MAGE protein interacts with E2F1 and the p75 neurotrophin receptor, and interestingly the Drosophila melanogaster MAGE protein showed highest sequence identity to MAGEG1 and is highly expressed during postembryonic neurogenesis (19).
We previously found a particularly strong interaction between S. pombe Nse3 and Nse1 (26), and it is likely that there is a similar interaction between hNSE1 and MAGEG1. Interestingly, our gel filtration analysis of Fig. 3A suggests that all the other components of the SMC5-6 complex are located exclusively in the complex, whereas a proportion of hNSE1 and MAGEG1 eluted with a much lower molecular weight. One might speculate that this represents an hNSE1-MAGEG1 heterodimer. It would be interesting to determine if, during brain development, MAGEG1 functioned on its own, as a heterodimer with hNSE1 or as part of the SMC5-6 complex.
Our finding that MAGEG1 and the yeast members of the family are essential components of the hSMC5-6 complex may provide a clue to the biochemical function of the MAGE family and may also point to an involvement of SMC5-6 in brain development. It is possible that, as with the SMC5-6 complex, other mammalian MAGE proteins are part of protein complexes, where they provide interaction surfaces or platforms for other polypeptides. Our current efforts are directed toward resolving these issues.
Acknowledgments
We are indebted to Lucas Bowler of the Sussex Centre for Proteomics for carrying out the mass spectrometry and analyzing the data. We are grateful to Tony Carr for helpful comments on the manuscript.
This work was supported by grants from the MRC and EU.
Footnotes
Published ahead of print on 17 December 2007.
REFERENCES
- 1.Ampatzidou, E., A. Irmisch, M. J. O'Connell, and J. M. Murray. 2006. Smc5/6 is required for repair at collapsed replication forks. Mol. Cell. Biol. 269387-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, E. A., J. Palecek, J. Sergeant, E. Taylor, A. R. Lehmann, and F. Z. Watts. 2005. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 25185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker, P. A., and A. Salehi. 2002. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J. Neurosci. Res. 67705-712. [DOI] [PubMed] [Google Scholar]
- 4.Bavner, A., J. Matthews, S. Sanyal, J. A. Gustafsson, and E. Treuter. 2005. EID3 is a novel EID family member and an inhibitor of CBP-dependent co-activation. Nucleic Acids Res. 333561-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branzei, D., J. Sollier, G. Liberi, X. Zhao, D. Maeda, M. Seki, T. Enomoto, K. Ohta, and M. Foiani. 2006. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127509-522. [DOI] [PubMed] [Google Scholar]
- 6.Chibuk, T. K., J. M. Bischof, and R. Wevrick. 2001. A necdin/MAGE-like gene in the chromosome 15 autism susceptibility region: expression, imprinting, and mapping of the human and mouse orthologues. BMC Genet. 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomez, P., O. De Backer, M. Bertrand, E. De Plaen, T. Boon, and S. Lucas. 2001. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 615544-5551. [PubMed] [Google Scholar]
- 8.Fujioka, Y., Y. Kimata, K. Nomaguchi, K. Watanabe, and K. Kohno. 2002. Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5-SMC6 complex involved in DNA repair. J. Biol. Chem. 27721585-21591. [DOI] [PubMed] [Google Scholar]
- 9.Harvey, S. H., D. M. Sheedy, A. R. Cuddihy, and M. J. O'Connell. 2004. Coordination of DNA damage responses via the Smc5/Smc6 complex. Mol. Cell. Biol. 24662-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazbun, T. R., L. Malmstrom, S. Anderson, B. J. Graczyk, B. Fox, M. Riffle, B. A. Sundin, J. D. Aranda, W. H. McDonald, C. H. Chiu, B. E. Snydsman, P. Bradley, E. G. Muller, S. Fields, D. Baker, J. R. Yates III, and T. N. Davis. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 121353-1365. [DOI] [PubMed] [Google Scholar]
- 11.Kuwako, K., H. Taniura, and K. Yoshikawa. 2004. Necdin-related MAGE proteins differentially interact with the E2F1 transcription factor and the p75 neurotrophin receptor. J. Biol. Chem. 2791703-1712. [DOI] [PubMed] [Google Scholar]
- 12.Lehmann, A. R., M. Walicka, D. J. F. Griffiths, J. M. Murray, F. Z. Watts, S. McCready, and A. M. Carr. 1995. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol. Cell. Biol. 157067-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindroos, H. B., L. Strom, T. Itoh, Y. Katou, K. Shirahige, and C. Sjogren. 2006. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell 22755-767. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Sanchez, N., Z. Gonzalez-Fernandez, M. Niinobe, K. Yoshikawa, and J. M. Frade. 2007. A single Mage gene in the chicken genome encodes CMAGE, a protein with functional similarities to mammalian type II MAGE proteins. Physiol. Genomics 30156-171. [DOI] [PubMed] [Google Scholar]
- 15.Losada, A., and T. Hirano. 2005. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 191269-1287. [DOI] [PubMed] [Google Scholar]
- 16.McDonald, W. H., Y. Pavlova, J. R. Yates III, and M. N. Boddy. 2003. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J. Biol. Chem. 27845460-45467. [DOI] [PubMed] [Google Scholar]
- 17.Morikawa, H., T. Morishita, S. Kawane, H. Iwasaki, A. M. Carr, and H. Shinagawa. 2004. Rad62 protein functionally and physically associates with the Smc5/Smc6 protein complex and is required for chromosome integrity and recombination repair in fission yeast. Mol. Cell. Biol. 249401-9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasmyth, K., and C. H. Haering. 2005. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74595-648. [DOI] [PubMed] [Google Scholar]
- 19.Nishimura, I., S. Shimizu, J. Y. Sakoda, and K. Yoshikawa. 2007. Expression of Drosophila MAGE gene encoding a necdin homologous protein in postembryonic neurogenesis. Gene Expr. Patterns 7244-251. [DOI] [PubMed] [Google Scholar]
- 20.Palecek, J., S. Vidot, M. Feng, A. J. Doherty, and A. R. Lehmann. 2006. The SMC5-6 DNA repair complex: bridging of the SMC5-6 heads by the Kleisin, NSE4, and non-Kleisin subunits. J. Biol. Chem. 28136952-36959. [DOI] [PubMed] [Google Scholar]
- 21.Pebernard, S., W. H. McDonald, Y. Pavlova, J. R. Yates III, and M. N. Boddy. 2004. Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol. Biol. Cell 154866-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pebernard, S., J. Wohlschlegel, W. H. McDonald, J. R. Yates III, and M. N. Boddy. 2006. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell. Biol. 261617-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potts, P. R., M. H. Porteus, and H. Yu. 2006. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 253377-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potts, P. R., and H. Yu. 2005. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 257021-7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schleiffer, A., S. Kaitna, S. Maurer-Stroh, M. Glotzer, K. Nasmyth, and F. Eisenhaber. 2003. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol. Cell 11571-575. [DOI] [PubMed] [Google Scholar]
- 26.Sergeant, J., E. Taylor, J. Palecek, M. Fousteri, E. Andrews, S. Sweeney, H. Shinagawa, F. Z. Watts, and A. R. Lehmann. 2005. Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5-6) complex. Mol. Cell. Biol. 25172-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor, E. M., J. S. Moghraby, J. H. Lees, B. Smit, P. B. Moens, and A. R. Lehmann. 2001. Characterisation of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombe Rad18/Spr18 complex. Mol. Biol. Cell 121583-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torres-Rosell, J., F. Machin, S. Farmer, A. Jarmuz, T. Eydmann, J. Z. Dalgaard, and L. Aragon. 2005. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat. Cell Biol. 7412-419. [DOI] [PubMed] [Google Scholar]
- 29.Verkade, H. M., S. J. Bugg, H. D. Lindsay, A. M. Carr, and M. J. O'Connell. 1999. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell 102905-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao, X., and G. Blobel. 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 1024777-47782. [DOI] [PMC free article] [PubMed] [Google Scholar]