Abstract
Eukaryotic initiation factor 2B (eIF2B) plays a key role in controlling the initiation of mRNA translation. eIF2B is heteropentamer whose catalytic (ɛ) subunit promotes GDP/GTP exchange on eIF2. We show here that depriving human cells of amino acids rapidly results in the inhibition of eIF2B, independently of changes in eIF2 phosphorylation. Although amino acid deprivation also inhibits signaling through the mammalian target of rapamycin complex 1 (mTORC1), the inhibition of eIF2B activity by amino acid starvation is independent of mTORC1. Instead, amino acids repress the phosphorylation of a novel site in eIF2Bɛ. We identify this site as Ser525, located adjacent to the known phosphoregulatory region in eIF2Bɛ. Mutation of Ser525 to Ala abolishes the regulation of eIF2B and protein synthesis by amino acids. This indicates that phosphorylation of this site is crucial for the control of eIF2B and protein synthesis by amino acids. These findings identify a new way in which amino acids regulate a key step in translation initiation and indicate that this involves a novel amino acid-sensitive signaling mechanism.
Amino acids positively regulate several components of the eukaryotic translational machinery, presumably to link their availability to their consumption by protein synthesis. Several are controlled through mammalian target of rapamycin complex 1 (mTORC1). These include the ribosomal protein S6 kinases (which also phosphorylate eukaryotic initiation factor 4B [eIF4B]) and eIF4E-binding protein 1 (4E-BP1). It is not yet clear how amino acids activate mTORC1 signaling, although amino acid starvation is reported to modulate the binding of mTORC1 to the small G protein, Rheb, which in its GTP-bound form, activates the kinase function of mTOR (29).
A second mechanism involves the protein kinase GCN2 (for general control of amino acid biosynthesis, nondepressing 2), which is activated by uncharged tRNA during amino acid starvation in yeast, the system in which Gcn2 was first identified (18). Gcn2 phosphorylates eIF2 at Ser51 in its α-subunit. Phosphorylated eIF2 competitively inhibits its guanine nucleotide exchange factor, eIF2B, thereby leading indirectly to the impairment of eIF2 function (9, 35) and, since eIF2 is required for all cytoplasmic translation initiation events, to inhibition of protein synthesis. Relatively little information is available on the importance of Gcn2 for the control of translation by amino acids in mammalian cells.
eIF2B plays a key role in translation initiation and in its regulation. eIF2B promotes the exchange of GDP for GTP on eIF2 to regenerate active eIF2.GTP, which binds the initiator methionyl-tRNA (Met-tRNAi) and recruits it to the 40S ribosomal subunit (reviewed in reference 18). eIF2B is composed of five different subunits, α to ɛ. The largest subunit, ɛ, contains the catalytic domain (see references 12 and 13 and references cited therein).
Many agents that stimulate protein synthesis activate eIF2B (11, 26, 49). One mechanism by which they do this involves the dephosphorylation of a conserved site in eIF2Bɛ (Ser540 in human eIF2Bɛ). Phosphorylation at this site inhibits eIF2B activity and is catalyzed by glycogen synthase kinase-3 (GSK3) (48, 50). GSK3 is inhibited by agents such as insulin, leading to the dephosphorylation and activation of eIF2B. However, dephosphorylation of the GSK3 site alone is insufficient to cause activation of eIF2B, indicating that there additional regulatory inputs into eIF2B activity (44). Further evidence for this is the fact that, in Chinese hamster ovary (CHO) cells, the activation of eIF2B by insulin requires that cells are supplied with amino acids (6), indicating an input from amino acids into the regulation of eIF2B.
Two possible mechanisms for the control of mammalian eIF2B by amino acids involve Gcn2 and mTORC1. However, in earlier work, eIF2α phosphorylation did not increase when CHO cells were deprived of amino acids, apparently ruling out this mechanism (6, 43). The rapamycin insensitivity of the activation of eIF2B by insulin suggests that mTOR is not involved. However, certain amino acid-regulated outputs from mTORC1 are insensitive to rapamycin, e.g., the phosphorylation of the N-terminal sites in 4E-BP1 (42), so that an input from mTORC1 cannot be ruled out on this basis.
In addition to the GSK3 site, several other phosphorylation sites have been identified in eIF2Bɛ. A site located four residues C-terminal to the GSK3 site “primes” eIF2B for phosphorylation by GSK3 (45). Such priming sites are found in several GSK3 substrates (8). Two other sites lie within the extreme C-terminal region that is required for binding to the substrate, eIF2 (45). Mutation of these sites (Ser717/718 in human eIF2Bɛ) to alanine prevents the interaction between eIF2Bɛ and eIF2. N-terminal to the catalytic domain lie two further sites of phosphorylation (Ser466/469), which are probably targets for casein kinase 1, but are of unknown significance (45).
We show here that amino acids positively regulate the activity of mammalian eIF2B, independently of changes in eIF2α phosphorylation. Based on several lines of evidence, this effect is also independent of signaling through mTOR. We demonstrate that amino acids modulate (repress) the phosphorylation of a novel site in eIF2Bɛ and identify it as Ser525, which lies close to the known regulatory GSK3 site. We demonstrate that Ser525 is required for the control of eIF2B activity and protein synthesis by amino acids. We also show that there is no apparent “cross talk” between the two known amino acid sensing and signaling pathways in mammalian cells, i.e., between Gcn2 and mTORC1. It is likely that the amino acid-regulated phosphorylation of eIF2Bɛ involves further unidentified amino acid-sensitive signaling events.
MATERIALS AND METHODS
Chemicals and biochemicals.
These were obtained from BDH-Merck or Sigma, unless indicated otherwise.
Plasmids.
The vectors encoding myc-tagged subunits of human eIF2B have been described previously (28). Additional mutants of eIF2Bɛ were created by using the QuikChange system (Stratagene). The vector encoding Rheb described earlier (42). The vector for the destabilized luciferase was a generous gift from Mark Coldwell and Anne Willis (Nottingham, United Kingdom).
Cell culture, transfection, treatment, and lysis.
Human embryonic kidney 293 (HEK293) cells were propagated and transfected as described earlier (14). Where indicated, at 30 h after transfection, the medium was changed into Dulbecco's phosphate-buffered saline containing 10 mM glucose (D-PBS; Gibco) containing or lacking amino acids for 2 h before lysis (unless otherwise indicated). To measure eIF2B activity, cells were harvested into buffer composed of 25 mM HEPES (pH 7.6), 25 mM β-glycerophosphate, 50 mM KCl, 0.5% Triton, 14 mM β-mercaptoethanol, 0.5 mM sodium orthovanadate, and proteinase inhibitors (1 mM benzamidine, 1 μg of pepstatin/ml, 1 μg of leupeptin/ml, 1 μg of antipain/ml, and 0.2 mM phenylmethylsulfonyl fluoride). For luciferase assays, cells were lysed with the 1× lysis buffer provided by the manufacturer (Promega). Otherwise, the cells were lysed in buffer containing: 25 mM HEPES (pH 7.6), 50 mM β-glycerophosphate, 50 mM KCl, 0.5 mM EGTA, 0.5 mM EDTA, 0.5% Triton, 14 mM β-mercaptoethanol, 0.5 mM sodium orthovanadate, and proteinase inhibitors (as described above). Gel electrophoresis and Western blotting were carried out as described previously (33). Jurkat and hepatoma G2 cells were grown as described earlier (5, 17). Gcn2+/+ and Gcn2−/− mouse embryonic fibroblasts (MEFs) were a generous gift from Ronald Wek (Indianapolis, IN) and were cultured as described earlier (20, 21). Affinity purification of eIF4E and associated proteins on m7GTP-Sepharose was performed as described previously (42).
Measurement of eIF2B activity.
The guanine nucleotide exchange activity of eIF2B was measured as described earlier by using purified human eIF2, complexed with [3H]GDP, as a substrate (28). To purify ectopically expressed eIF2B complexes from cell lysates prior to assay, we made use of the His6 tag on the eIF2Bɛ polypeptide to purify them on Ni-nitrilotriacetic acid (NTA)-agarose (Qiagen) (28). The data are shown as means ± the standard error of the mean.
Luciferase assay.
A total of 20 μl of 293 cell lysate (lysed with 1× manufacturer's lysis buffer) was applied to a well of a 96-well plate. The luciferase activity was assayed by using Firefly luciferase assay reagent (Promega), with a Labsystems Luminoskan Ascent luminometer. As a control for the contribution to the total luciferase activity of enzyme made prior to amino acid starvation, we also treated cells with the protein synthesis inhibitor cycloheximide for the same period of time as the amino acid starvation and subtracted the activity of observed from the other experimental values. The data are shown as means ± the standard error of the mean.
Radiolabeling of cells and peptide mapping.
HEK293 cells (10-cm plates) were transfected with the appropriate vector. After 30 h, the cells were washed three times with phosphate-free Krebs-Ringer bicarbonate buffer (20 mM HEPES, 107 mM NaCl, 5 mM KCl, 3 mM CaCl2, 1 mM MgSO4, 7 mM NaHCO3, 10 mM glucose) and then grown in this medium for 1 h before change to fresh buffer containing 2 mCi of 32Pi with or without amino acids for a further 2.5 h. Cells were lysed, and eIF2Bɛ was isolated by immunoprecipitation with anti-myc antibody. Subsequently, samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. The band corresponding to eIF2Bɛ was excised and digested with trypsin (and, in some cases, Asp-N) or CNBr (100 mg/ml in 70% [vol/vol] HCOOH at 37°C overnight in the dark), and the resulting peptides were resolved by two-dimensional mapping (45) or analyzed by 4 to 12% (wt/vol) acrylamide Novex gel electrophoresis, followed by autoradiography.
RESULTS
Amino acids can regulate the activity of eIF2B without altering the phosphorylation of eIF2α.
To investigate the regulation of eIF2B by amino acids, we used HEK293 cells since they can readily be transfected and are very sensitive to amino acid withdrawal, at least in terms of mTORC1 signaling (15, 42). This allows us to express tagged wild-type or mutant eIF2B, thereby facilitating the determination of eIF2B activity, which is otherwise hard to measure in HEK293 cell lysates.
Cells were transiently transfected with vectors encoding all five subunits of human eIF2B (α to ɛ), each equipped with a myc tag to facilitate detection (as shown in Fig. 1A) and isolation, respectively. eIF2Bɛ also had a His6 tag. The tags were N terminal, i.e., at the opposite end of eIF2Bɛ from the catalytic domain and the region that binds the substrate, eIF2 (1, 12). The expression levels of eIF2Bγ and ɛ were similar, presumably because they are components of the binary catalytic eIF2B subcomplex, but were not always identical to those of the other three: in the experiment shown in Fig. 1A, eIF2Bα, -β, and -δ were expressed at slightly higher levels. These subunits form a distinct trimeric regulatory subcomplex (32).
FIG. 1.
Expression of wild-type and catalytically inactive eIF2B complexes in HEK293 cells. HEK293 cells were transfected with vectors for myc-tagged human eIF2B (α to ɛ). The ɛ-subunit also has a His6 tag. In some cases, catalytically inactive eIF2Bɛ (E577A) was used. (A) Western blot (anti-myc) of crude lysate from HEK293 cells expressing wild-type or mutant (E577A) eIF2B complexes. The positions of the subunits of eIF2B are indicated. (B) eIF2B complexes were isolated by chromatography on nickel beads to isolate the His6-eIF2Bɛ and associated subunits. “n/s” denotes a band that is not an eIF2B subunit but is detected by anti-myc. (C) The guanine nucleotide exchange activity of eIF2B complexes isolated as in panel B was measured using eIF2.[3H]GDP as a substrate. The activity of wild-type eIF2B is set as unity (n = 5). (D) eIF2B complexes were isolated as in panel B but Western analysis was performed for eIF2Bɛ (lower section) or eIF2α (upper part). Vec, cells transfected with a similar amount of the corresponding empty vectors. The data are representative of three independent experiments. WT, wild type.
The ectopically expressed eIF2B was isolated by using the His6 tag, yielding purified complexes containing equimolar amounts for the five subunits, as judged by the equal strengths of the signals on anti-myc Western blots (Fig. 1B). The activity of the complexes can readily be determined (Fig. 1C) (see also reference 28). As a negative control, we also expressed a mutant in which Glu577 in the catalytic domain of eIF2Bɛ has been altered to alanine (E577A). This residue is predicted to be required for eIF2B activity on the basis of the three-dimensional structure of yeast eIF2Bɛ (4). Consistent with this, the E577A mutant displayed very low activity in vitro (Fig. 1C). Analysis of the amounts of eIF2α in His6 pull-downs of wild-type and mutant eIF2B showed that very little eIF2 copurified with the E577A mutant, the signal being similar to the “nonspecific” binding of eIF2 seen when lysates of cells transfected with empty vector were studied (Fig. 1D). Levels of expression of wild-type and mutant eIF2Bɛ were similar (Fig. 1D, lower section). Glu577 therefore appears to be required for the stable binding of eIF2B to eIF2.
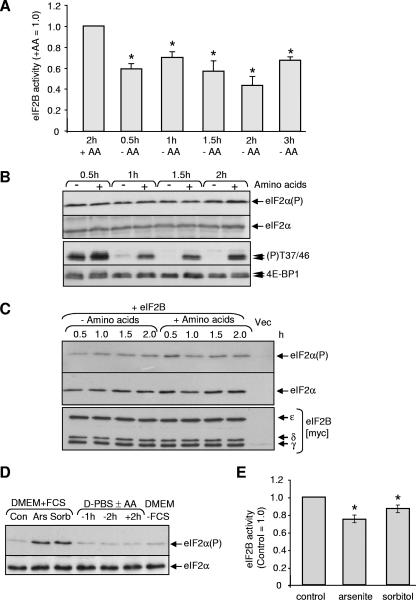
To test the effect of amino acid deprivation on the activity of eIF2B, cells that had been transfected with vectors for wild type eIF2B were transferred to D-PBS containing or lacking amino acids for time-periods up to 3 h. After the period of amino acid starvation, cells were lysed, eIF2B complexes were purified by virtue of the His6 tag on the ɛ-subunit, and the eIF2B activity was measured. Amino acid starvation caused a reduction in the activity of eIF2B measured in this assay (Fig. 2A). The effect of amino acid deprivation was rapid, being evident by 30 min and maximal by 2 h. We have previously noted in other cell types (3, 38) that (i) intracellular amino acids are not completely depleted after amino acid starvation, which may explain why the effects on eIF2B activity are partial, and (ii) amino acid starvation causes only a transient reduction in intracellular amino acid levels (they later increase, perhaps reflecting activation of autophagy, which is negatively controlled by mTOR) (27).
FIG. 2.
Effects of amino acid starvation on eIF2B activity and eIF2α phosphorylation in HEK293 cells. (A) HEK293 cells were transfected with vectors for all five subunits of human eIF2B (α to ɛ).(A) Cells were transferred to D-PBS containing (+AA) or lacking (−AA) for the times indicated prior to lysis. eIF2B complexes were isolated and the guanine nucleotide exchange activity of eIF2B complexes was measured. *, P < 0.05 versus the control (n = 3). (B) HEK293 cells were transferred to medium lacking or containing amino acids for the indicated times. Samples of lysates were analyzed by SDS-PAGE and immunoblotting for eIF2 or phosphorylated eIF2 (eIF2[αP]) or for 4E-BP1 using antibodies that recognize the amino acid sensitive sites Thr37/46 or total 4E-BP1. 4E-BP1 runs as two to three distinct species (which differ in their states of phosphorylation), as indicated by the small arrows. (C) Same as panel A, but isolated eIF2B complexes were analyzed by Western blotting for total or phosphorylated eIF2α (upper part) or for eIF2B subunits (anti-myc). “Vec” indicates cells were transfected with empty vectors. (D and E) Cells were maintained in Dulbecco modified Eagle medium (DMEM) (+ fetal calf serum [FCS]). Arsenite (0.5 mM) or sorbitol (0.4 M) was added for 25 min (or starved of amino acids as indicated). In panel D, samples of lysates were analyzed by SDS-PAGE and/or immunoblotting for eIF2 or phosphorylated eIF2 (eIF2[αP]). In panel E, samples were processed for measurement of eIF2B activity as in panel A. The activity of eIF2B in cells maintained in D-PBS plus amino acids is set as unity in panel A, while the activity in control cells is used as a reference in panel E. *, P < 0.05 versus the control (n = 4).
Starvation of HEK293 cells for amino acids did not affect the phosphorylation state of eIF2α (Fig. 2B) but did cause a profound drop in the phosphorylation of 4E-BP1, which is controlled by the amino acid-regulated mTORC1 pathway and serves as a “positive control” for the effects of amino acid starvation (see, e.g., reference 42).
One could argue that there might be a small, undetectable, change in eIF2α phosphorylation that nonetheless suffices to inhibit eIF2B. When phosphorylated, eIF2 binds more tightly to eIF2B (36). Amino acid starvation did not change the levels of bound eIF2(αP) or total eIF2α bound to isolated eIF2B complexes (Fig. 2C). It is thus very unlikely that the decreased eIF2B activity is due to increased binding of eIF2B to its competitive inhibitor, eIF2(αP).
Treatment of cells with arsenite or sorbitol markedly increased eIF2α phosphorylation (Fig. 2D), slightly increased the association of eIF2 with eIF2B complexes (data not shown), and also inhibited eIF2B activity (Fig. 2E). However, the change in eIF2B activity was considerably smaller than that seen in response to amino acid starvation (Fig. 2A). If the decrease in eIF2B activity caused by amino acid starvation were due to an undetectably small increase in eIF2α phosphorylation, then its effect on eIF2B activity should be less than that seen in response to arsenite or sorbitol, whereas the converse is true. Taken together, these data argue strongly that the decrease in eIF2B activity caused by amino acid starvation is not due to increased eIF2α phosphorylation.
Amino acid starvation does not alter eIF2α phosphorylation in certain other mammalian cell types.
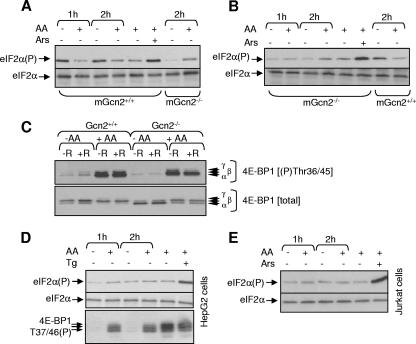
The observation that amino acid starvation does not increase eIF2α phosphorylation in HEK293 cells is similar to our earlier findings in CHO cells (6). Potential explanations for this include (i) that Gcn2 is not activated by amino acid starvation and (ii) that the effect of amino acid starvation on eIF2α phosphorylation differs according to the cell type. Starvation for amino acids did increase the phosphorylation of eIF2α in control (Gcn2+/+) MEFs (Fig. 3A) but not in MEFs in which the Gcn2 gene has been disrupted (Fig. 3B). If anything, a decrease was actually observed: we do not understand the reasons for this, although it was a quite consistent observation (see also Fig. 4C). Arsenite (a “positive control”) did increase eIF2α phosphorylation in the Gcn2−/− MEFs (Fig. 3B), ruling out the possibility that eIF2α phosphorylation is already maximal in these cells. It is not known how arsenite increases eIF2α phosphorylation: this is evidently independent of Gcn2. Thus, in MEFs, amino acid starvation does increase eIF2α phosphorylation, and this depends on Gcn2.
FIG. 3.
Effects of amino acids on eIF2α phosphorylation in various cell types. (A to E) The indicated cells were transferred to D-PBS containing or lacking amino acids (AA) for the indicated times (90 min in panel C). Cells were then placed in D-PBS with or without amino acids for 45 min and, where indicated (+R), rapamycin at 100 nM was added, and the incubation was continued for a further 45 min. In some cases, cells were kept in DMEM and treated with thapsigargin (panel D, Tg; 0.5 μM, 1 h) or sodium arsenite (panel E, Ars; 0.5 mM, 30 min). Samples were then analyzed by SDS-PAGE and/or Western blotting for phosphorylated or total eIF2α (A, B, D, and E), for 4E-BP1 phosphorylated at Thr37/46 (C to E), or “total” 4E-BP1 (C).
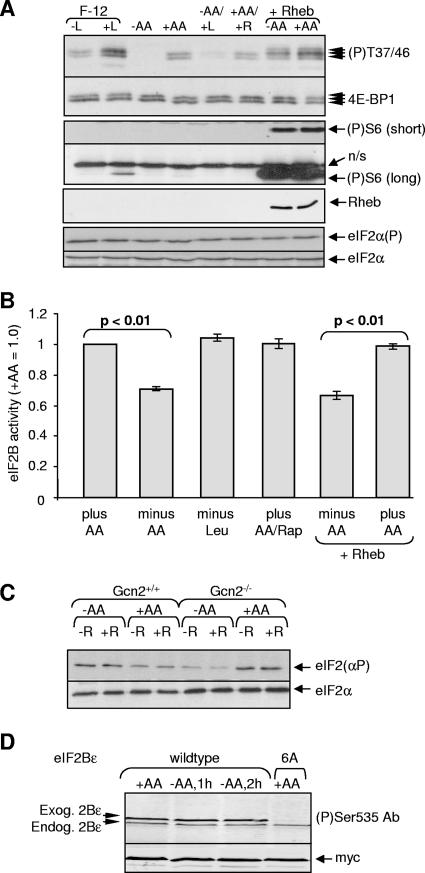
FIG. 4.
Investigation of the mechanism by which amino acids (AA) control eIF2B. (A) HEK293 cells were transferred to F-12 medium containing all amino acids (+L) or except leucine (−L), to D-PBS with or without amino acids (+AA or −AA), or to D-PBS containing only leucine (−AA/+L). Cells in D-PBS+AA were treated with rapamycin (100 nM, 45 min; +AA/+R). Where indicated (+Rheb), cells were transfected with a vector encoding Rheb and subsequently transferred to PBS containing or lacking amino acids. Cells were then lysed, and Western blots were performed for all 4E-BP1 or 4E-BP1 phosphorylated on Thr37/46, phosphorylated S6 (S235/236; long or short exposures shown), Rheb (Flag tagged), and total or phosphorylated eIF2α, as indicated. A total of four independent experiments was performed in each case. (B) Cells were transfected with vectors for eIF2B(α-ɛ) and 40 h later were transferred to D-PBS containing (+AA) or lacking amino acids (−AA) or only leucine for 2 h. Where indicated (Rap) cells were treated with rapamycin (100 nM, 45 min). In some cases, cells were also transfected with Rheb. eIF2B complexes were isolated and activity measured. The data are representative of six independent experiments. (C) Gcn2+/+ or Gcn2−/− cells were placed in D-PBS with or without amino acids for 45 min and, where indicated (+R), rapamycin (100 nM) was added, and the incubation was continued for a further 45 min. Samples of cell lysate were analyzed by SDS-PAGE and/or Western blotting for eIF2α or eIF2α phosphorylated on Ser51. (D) HEK293 cells expressing all five eIF2B subunits, including either wild-type or 6A mutant versions of eIF2Bɛ, were starved of amino acids as in panel A, but samples of isolated eIF2B complexes were analyzed by SDS-PAGE and/or immunoblotting using either a phospho-specific antibody for the GSK3 site in eIF2Bɛ or anti-myc (loading control). The 6A mutant of eIF2Bɛ lacks this site and five other previously identified ones. The positions of the overexpressed (exog.) and endogenous (endog.) eIF2Bɛ polypeptides are indicated.
When Gcn2−/− MEFs were starved for amino acids, 4E-BP1 underwent dephosphorylation and shifted to its less-phosphorylated α and β forms (Fig. 3C). Indeed, the extent of dephosphorylation of 4E-BP1 was consistently somewhat greater in the starved GCN2−/− cells than in the controls (e.g., the proportion of the hypophosphorylated α-species was markedly higher in the knockout cells; Fig. 3C). These data demonstrate that Gcn2 is not required for the inactivation of mTORC1 signaling in response to amino acid starvation. The larger effect of amino acid starvation in the knockout cells suggests that Gcn2 may even offer some “protection” against the inactivation of mTORC1 signaling, perhaps because in the Gcn2+/+ cells the phosphorylation of eIF2α normally leads to inhibition of protein synthesis, decreasing the utilization and consequent depletion of amino acids.
In contrast to the findings for Gcn2+/+ MEFs, when hepatoma G2 (Fig. 3D), Jurkat T (Fig. 3E), or HeLa (not shown) cells were deprived of amino acids for periods up to 2 h, no increase in the phosphorylation of eIF2α was observed. Sodium arsenite or thapsigargin, positive controls, did increase eIF2α phosphorylation (Fig. 3D and E), indicating that the absence of a change in eIF2α phosphorylation after amino acid starvation is not due to high basal phosphorylation. Amino acid starvation did decrease the phosphorylation of 4E-BP1 (a target for mTORC1 signaling that serves as a positive control) in hepatoma cells (46) (Fig. 3D). (Levels of 4E-BP1 were too low in Jurkat cells to allow us to assess this.) Thus, the absence of an effect of amino acid starvation on eIF2α phosphorylation is not unique to HEK293 cells, since it is also seen in several other cell lines (including also CHO cells) (6).
Control of eIF2B by amino acids appears not to be mediated through mTORC1.
What mechanisms might account for the control of eIF2B activity independently of changes in eIF2α phosphorylation? Initially, we focused on the possibility that signaling through mTORC1 controls eIF2B, as mTORC1 signaling is dependent upon amino acids and is important in the control of other translation factors (23, 46). As expected from earlier studies (see, for example, reference 42), starving HEK293 cells for amino acids impaired signaling through mTORC1, as indicated by the marked dephosphorylation of 4E-BP1. Starvation for leucine alone also impaired mTORC1 signaling (Fig. 4A). However, this did not affect eIF2B activity in cells kept in D-PBS (Fig. 4B) or in F-12 medium (data not shown). Starvation of cells for lysine or methionine (data not shown) also did not affect eIF2B activity.
In budding yeast, TOR signaling can regulate eIF2α phosphorylation by modulating the activity of GCN2 (7). However, treatment of HEK293 cells with rapamycin did not increase eIF2α phosphorylation (Fig. 4A), suggesting the absence of such a link in HEK293 cells. However, since amino acid starvation does not affect eIF2α phosphorylation in HEK293 cells, it was important to test the effects of rapamycin on eIF2α in cells where amino acid starvation does affect eIF2α phosphorylation and Gcn2 is presumably activated, such as the Gcn2+/+ MEFs. As shown in Fig. 4C, rapamycin was also without effect on eIF2α phosphorylation in these cells. This finding suggests that there is indeed no link between mTORC1 and the control of Gcn2 in mammalian cells. In this context, it is relevant to note that the TOR-regulated phosphorylation site in yeast GCN2 (Ser577) (7) is absent from its mammalian ortholog.
Treating amino acid-fed cells with rapamycin did not affect eIF2B activity either (Fig. 4B), again indicating eIF2B is not regulated by mTORC1 (which is consistent with our earlier studies) (51). However, some effects of mTORC1 are rather insensitive to rapamycin (42) (as typified by Thr37/46 in 4E-BP1, which corresponds to Thr36/45 in the mouse protein, see Fig. 3C). We therefore also studied the effects of overexpressing Rheb, a small G-protein that specifically activates mTORC1 signaling and increases the phosphorylation of the rapamycin-insensitive sites in 4E-BP1 (Thr37/46) (42). Overexpression of Rheb activated mTORC1 signaling in amino acid-starved HEK293 cells, as illustrated by the increased phosphorylation of 4E-BP1 and of S6 (Fig. 4A), in line with earlier data (39, 40). However, Rheb did not reverse the fall in eIF2B activity seen in amino acid-starved HEK293 cells (Fig. 4B), providing further evidence that eIF2B is not regulated by mTORC1.
Phosphorylation of eIF2Bɛ by GSK3 inhibits eIF2B activity (48). Although amino acids have not been shown to regulate GSK3, GSK3 can be phosphorylated and inhibited by S6 kinase, which is linked to the mTOR pathway (52): amino acid starvation might thus increase GSK3 activity and phosphorylation of eIF2B. However, amino acid starvation did not affect the phosphorylation of the GSK3 site in eIF2Bɛ, as assessed by using a phospho-specific antibody (Fig. 4D; the “6A” mutant, which lacks this site, serves as a “negative control”).
Importantly, these findings indicate that the control of eIF2B activity by amino acids must involve a novel mechanism that is distinct from mTORC1 signaling and independent of the phosphorylation of eIF2α.
Protein synthesis.
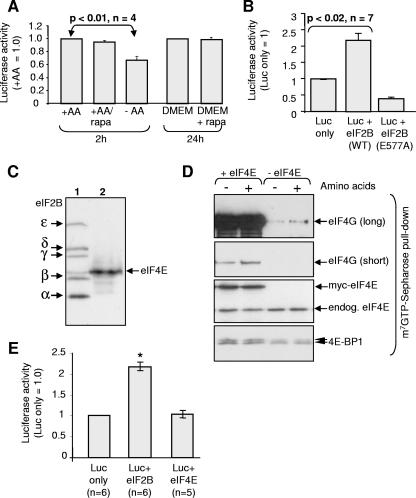
In order to assess the importance of controlling eIF2B activity for the overall control of mRNA translation, it was important to study the effect of amino acid starvation on protein synthesis. It is not appropriate to use the standard radiolabeling methods to study the effects of amino acid starvation on the rates of protein synthesis, since removing amino acids (including the cold methionine) from the medium greatly increases the specific radioactivity of the labeled amino acid within the cell (since “cold” amino acids no longer compete with the labeled one for import into the cells). As an alternative, we therefore transfected the cells with a reporter construct encoding a destabilized version of luciferase to allow us to assess changes in its rate of synthesis over relatively short time periods. Thirty hours later, we subjected the cells to amino acid starvation. We then measured the luciferase activity. Amino acid starvation for 2 h decreased luciferase expression (Fig. 5A). However, the fall in luciferase levels was a little smaller than that in eIF2B activity at 2 h (Fig. 2A; see also 4B), likely because the changes in eIF2B activity are only maximal by 2 h and are rather transient (Fig. 2A), whereas the levels of even the destabilized luciferase reflect its synthesis over a period of time. Treatment of cells with rapamycin for 2 h or even 24 h had no effect on luciferase levels (Fig. 5A). These data suggest that, although amino acid starvation does strongly impair mTORC1 signaling, this impairment is not responsible for the decrease in luciferase synthesis seen in amino acid starved cells. This interpretation is supported by the fact that overexpressing Rheb did not enhance luciferase activity (data not shown), although it does promote mTORC1 signaling (Fig. 4A).
FIG. 5.
Regulation of protein synthesis by eIF2B or amino acids in HEK293 cells. (A) Cells were transfected with a vector encoding destabilized luciferase (Luc). After transfer to D-PBS containing (+AA) or lacking (−AA) amino acids for 2 h, cells were then lysed, and the luciferase activity was determined (n = 4). In some cases, rapamycin (100 nM) was added to DMEM for 24 h prior to lysis. (B) Cells were cotransfected with the luciferase vector and, where indicated, vectors for eIF2Bα-δ and either wild-type (WT) or mutant (E577A) eIF2Bɛ. Cells were then lysed, and the luciferase activity was measured (n = 7). (C) Cells were transfected with a vector encoding myc-tagged versions of wild-type eIF2B or myc-eIF4E and then transferred to D-PBS lacking amino acids (as indicated) for 2 h. Western blots were performed for myc eIF2B and myc eIF4E. (D) Cells were transfected with a vector encoding eIF4E (or empty vector), and then transferred to D-PBS containing (+AA) or lacking (−AA) amino acids for 2 h. eIF4E/4G complexes were isolated on m7GTP Sepharose. The bound material was analyzed by SDS-PAGE and Western blotting with antibodies for eIF4G, eIF4E, and 4E-BP1. (E) Cells were cotransfected with the luciferase vector and either with wild-type eIF2B or eIF4E. Cells were transferred to D-PBS (without amino acids) for 2 h prior to lysis. Luciferase activity was determined. *, P < 0.05.
To assess the extent to which eIF2B activity contributes to the control of protein synthesis, we expressed the five-subunit eIF2B complex in HEK293 cells and then measured the levels of destabilized luciferase. A marked increase in luciferase expression was observed (Fig. 5B), suggesting that eIF2B activity is limiting for protein synthesis in HEK293 cells. As a control to establish that the effects really are due to eIF2B activity, we expressed the inactive E577A mutant of eIF2Bɛ together with wild-type eIF2Bα-δ. Expression of this mutant invariably inhibited luciferase synthesis, indicating that it has a dominant interfering phenotype (Fig. 5B). As further controls, we also tested the effects of ectopic expression of eIF2Bɛ alone or of another single subunit of eIF2B. Expressing eIF2Bɛ alone did stimulate luciferase production, a finding consistent with the observation that the activity of eIF2Bɛ does exhibit some activity (see, e.g., reference 10). In contrast, expressing other individual eIF2B subunits actually inhibited luciferase expression, a finding consistent with the fact that only eIF2Bɛ possesses exchange activity: the inhibition may reflect interference by the overexpressed single subunit with the proper formation of functional eIF2B holocomplexes. We also coexpressed all three eIF2 subunits with the luciferase reporter. Cells did not tolerate overexpression of eIF2 very well, and luciferase expression was actually inhibited. The reasons for this are unknown but may reflect sequestration by eIF2 of other factors to which it binds such as eIF5, ABC50, or maybe eIF2B. We have not pursued this further.
The initiation factor eIF4E is a well-studied target for control by mTORC1 and amino acids, its availability being regulated by the 4E-BPs (24, 44), and is widely considered to play a pivotal role in the control of translation initiation (30, 41). We therefore assessed the effects of overexpressing eIF4E on formation of initiation complexes containing eIF4G (Fig. 5C and D) and on luciferase expression (Fig. 5E). Since the eIF4E and eIF2B constructs are both singly myc tagged, it is evident from Fig. 5C that eIF4E expressed at a higher level than eIF2B. Overexpressing eIF4E greatly increased levels of eIF4G/eIF4E complexes (assessed after isolation of eIF4E by affinity chromatography, followed by Western blot analysis; Fig. 5D) under both amino acid-starved and amino acid-fed conditions. Slightly more 4E-BP1 was also recovered in m7GTP-pull downs from cells overexpressing eIF4E. (The apparent differential increases in binding of eIF4G and 4E-BP1 to eIF4E likely reflects both their relative levels in HEK293 cells and their relative affinities for eIF4E: 4E-BP1 presumably normally binds more tightly than eIF4G, since it outcompetes eIF4G for binding to eIF4E.) However, high-level expression of eIF4E had no detectable effect on luciferase levels in amino acid-starved cells, in contrast to the marked effect of expressing a lower level of eIF2B (Fig. 5E). As expected, overexpressing eIF2B had no effect on eIF4G binding to eIF4E (data not shown). These data indicate that eIF4E is less important than eIF2B for the control of protein synthesis in amino acid-starved HEK293 cells. Similar findings were made in amino acid fed cells, where eIF2B markedly stimulated luciferase levels, whereas eIF4E had very little effect (data not shown).
Amino acids regulate the phosphorylation of eIF2Bɛ at a novel site.
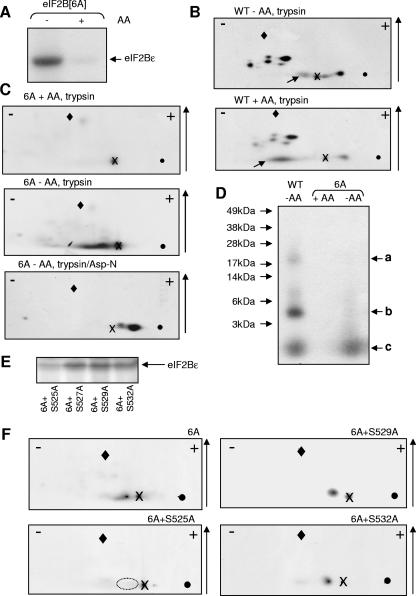
Given that the inhibition of eIF2B activity induced by amino acid starvation is apparently not due to eIF2α phosphorylation or mTORC1 signaling, we considered the possibility that eIF2B activity might be regulated by amino acid-sensitive phosphorylation of eIF2B itself. We have previously identified six phosphorylation sites in the ɛ-subunit of eIF2B (45). To determine whether eIF2Bɛ underwent amino acid-sensitive phosphorylation, we created a mutant in which these six sites were mutated to alanines (eIF2Bɛ[6A]) and expressed this, with the other four wild-type subunits, in HEK293 cells. The cells were subjected to metabolic labeling with [32P]orthophosphate. eIF2B complexes were then isolated and analyzed by SDS-PAGE and autoradiography. There was no detectable labeling of eIF2Bɛ[6A] in amino acid-fed cells (Fig. 6A) but, strikingly, it underwent marked phosphorylation in amino acid-starved cells. eIF2Bɛ therefore contains at least one additional (novel) phosphorylation site that is controlled by the amino acid status of the cells. The other subunits of eIF2B copurify with the eIF2Bɛ[6A] polypeptide: no difference in their radiolabeling was apparent when samples from amino acid-fed and from amino acid-starved cells were compared (data not shown).
FIG. 6.
Identification of the amino acid-regulated phosphorylation site in eIF2Bɛ. (A) HEK293 cells were transfected with vectors for all five subunits of eIF2B, including a mutant of eIF2Bɛ in which all six known sites of phosphorylation were converted to alanine (6A). After 30 h, cells were labeled with [32P]orthophosphate for 2.5 h. After lysis, eIF2B complexes were isolated by myc immunoprecipitation. Samples were analyzed by SDS-PAGE and autoradiography. (B) Same as in panel A, but using WT eIF2Bɛ. The eIF2Bɛ band was excised and digested with trypsin. The resulting peptides were resolved by two-dimensional mapping, where the polarity (+/−) and the direction of chromatography (↑) are shown. “X” marks the origin where the samples were applied, the diamond marks the position of the marker (DNP-lysine), and the solid circle shows where the marker was loaded. Diagonal arrows denote peptides discussed in the text. (C) Same as in panel A, and eIF2B complexes were isolated and resolved by SDS-PAGE as in panel B. Phosphopeptides from eIF2Bɛ were generated and displayed as in panel B, except that in one case they were further digested with Asp-N after the initial tryptic cleavage. (D) Same as in panel B, except that eIF2Bɛ was digested with CNBr, not trypsin, and peptides were resolved by SDS-PAGE (using 4 to 12% [wt/vol] acrylamide gels). The positions of the molecular weight markers and the three radiolabeled peptides a to c discussed in the text are indicated. (E) Same as in panel A, but cells were transfected with vectors for eIF2Bɛ[6A] in which the indicated additional serine residues had been mutated to alanine and in all cases cells were deprived of amino acids. (F) Same as in panel B, except that, in addition to the 6A mutant, the other indicated mutants were also analyzed. The dotted ellipse in the lower left panel shows where the peptide that is absent from this map, but present in the others, would be expected to migrate.
To study this further, the experiment was repeated using wild-type and 6A versions of eIF2Bɛ. The radiolabeled eIF2Bɛ bands were excised from the gel and digested with trypsin (followed in some cases by Asp-N). The resulting phosphopeptides were displayed by two-dimensional mapping, followed by autoradiography. Peptide maps from wild-type eIF2Bɛ contained about seven main phosphopeptides and some minor species (which may be partial digestion products; Fig. 6B), a finding consistent with the presence in eIF2Bɛ of at least six sites of phosphorylation (45). These maps differ from our earlier ones (45) because that study was performed using rat, not human, eIF2Bɛ. Although the pattern of peptides from wild-type eIF2Bɛ was almost identical for amino acid-starved (Fig. 6B, top) and amino acid-fed (Fig. 6B, bottom) cells, one striking difference was apparent: the position of one phosphopeptide shifted between the two conditions (spots arrowed in Fig. 6B). The peptide from eIF2B from amino acid-starved cells migrated further toward the anode, indicating a gain in negative charge. The peptide thus likely contains two sites of phosphorylation, only one of which is labeled under amino acid-fed conditions. The amino acid-regulated peptide scarcely migrated in chromatography and tended to run as a streak in electrophoresis, indicating a highly hydrophilic species (Fig. 6B).
Tryptic phosphopeptide maps from eIF2Bɛ(6A) isolated from amino acid-fed cells contained none of the peptides seen for the wild-type protein (Fig. 6C, top; cf. Fig. 6B). Thus, no sites eIF2Bɛ(6A) are phosphorylated under amino acid-fed conditions, implying that the second site in the peptide affected by amino acid starvation is a known one. Analysis of the eIF2Bɛ[6A] mutant from amino acid-starved cells yielded a one major phosphopeptide with net positive charge. The fact that this peptide was absent from maps derived from eIF2Bɛ from amino acid-fed cells (Fig. 6B) likely explains why it was not detected in our earlier experiments using amino acid-replete cells (45). Further digestion of this peptide with Asp-N yielded a single major peptide which migrated toward the anode (Fig. 6C, lower section), indicating that it represents the N-terminal part of the tryptic peptide discussed above (since the C-terminal section of would contain a Lys or Arg and have a net positive charge at this pH).
Careful inspection of the sequence of human eIF2Bɛ reveals 16 theoretical peptides defined by tryptic and Asp-N cleavage sites that contain serine and/or threonine residues and would have a net negative charge (if phosphorylated) at pH 1.9 (at which the electrophoresis step is run). However, this is insufficient to define the amino acid-regulated phosphorylation site(s) in eIF2Bɛ. To study this further, we used a third cleavage procedure, CNBr, which cleaves C-terminal to methionines. Since methionines are relatively uncommon (15 in human eIF2Bɛ), a smaller set of generally larger peptides is generated. Gel chips containing wild-type or 6A mutant eIF2Bɛ from radiolabeled cells that had been starved of amino acids were therefore digested with CNBr. As a control, the 6A mutant was also labeled in amino acid-fed cells. The resulting peptides were analyzed by 4 to 12% (wt/vol) acrylamide Novex gel electrophoresis (Fig. 6D). Wild-type eIF2Bɛ yielded three phosphopeptides with apparent Mrs of 20 kDa, 4 to 5 kDa, and ∼1.5 kDa, respectively (Fig. 6D, arrows a to c). The two larger species (arrows a and b in the figure) were absent from the 6A mutant. Based on their observed sizes, “a” presumably contains Ser466/469 identified earlier (predicted CNBr fragment = 20 kDa) whereas “b” likely contains the extreme C-terminal sites (Ser717/8). The GSK3 and priming sites are contained within a 14-residue fragment (likely species “c”). Consistent with this, no peptide is seen in this position for the 6A mutant from amino acid-fed cells. However, a fragment of this size is visible in the 6A sample from starved cells: thus, the amino acid-regulated site is located in a peptide similar in size to the one containing the GSK3 site, i.e., around 14 residues.
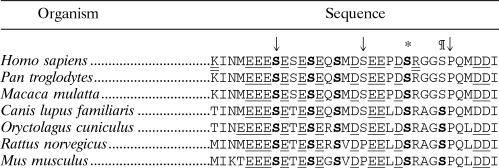
Identification of the amino acid-sensitive phosphorylation of eIF2Bɛ as Ser525.
Given these two sets of data (characteristics of tryptic and tryptic/Asp-N peptides; size of CNBr fragments), only one region of eIF2Bɛ emerges as a possible location for the novel phosphorylation site (Table 1). It contains five serines (525, 527, 529, 532, 535, and 540). However, Ser540 (marked with an asterisk in Table 1) is known to be the phosphorylation site for GSK3 (48) and is an alanine in the 6A mutant (so can be excluded). Ser527 is replaced by another phosphorylatable residue (threonine) in rodents (Table 1). Ser535 may be excluded because, after digestion with trypsin and Asp-N, it will be found in a positively charged peptide, whereas the peptide containing the amino acid-regulated site is negatively charged (Fig. 6C, lower section). Ser535 is also not conserved in other mammalian species (Table 1). Four residues thus remained as possible candidates. The tryptic peptide from wild-type eIF2Bɛ that contains these residues also contains Ser540, which, as already mentioned, is phosphorylated by GSK3. This concurs with the above interpretation (from data in Fig. 6B and from Fig. 6C, top part) that the tryptic phosphopeptide containing the amino acid-regulated site also contains a known site of phosphorylation.
TABLE 1.
Sequences of eIF2Bɛ from mammalian species showing the region containing the amino acid-regulated sitea
Conserved serine residues are in boldface (no threonines are conserved between these seven sequences). These include the known regulatory sites phosphorylated by GSK3 (*) and the requisite priming kinase (¶). Acidic residues are underlined, and tryptic cleavage sites in the human sequence are indicated by a double underline. The numbering is based upon the sequence of human eIF2Bɛ (used in the present study). Positions 525, 535, and 545 are indicated by the arrows from left to right, respectively.
One potential way to identify phosphorylation sites is by tandem mass spectrometry. We collaborated with three different laboratories that are expert in this technology, but none could identify the phosphorylation site. This likely reflects the fact that this peptide lacks net positive charge, while the instruments in these laboratories run in “positive-ion mode.” Although the tryptic fragment does contain a C-terminal arginine, the peptide is rich in acidic residues, and this negative charge is augmented both by the probable phosphorylation site and the propensity of methionine residues (two in this peptide) to undergo oxidation and gain negative charge.
As an alternative, we created further mutants of eIF2Bɛ in the “6A” background, replacing each of the serines 525, 527, 529, and 532 by alanine, to create four “7A” mutants. These mutants were expressed individually as his/myc tagged fusions in HEK293 cells (together with the other four eIF2B subunits α to δ). Cells were transferred to amino acid-free medium and labeled with [32P]orthophosphate. Samples were analyzed by SDS-PAGE and autoradiography (Fig. 6E), and then peptide maps were prepared from the labeled eIF2Bɛ as before (Fig. 6F). Labeling was observed for all of the mutants tested, but the incorporation into the “6A+S525A” mutant was strikingly lower than for the others (Fig. 6E and data not shown). The low residual level of phosphorylation reflects incorporation into the linker region (between the myc tag and the eIF2Bɛ sequence), as determined by mass spectrometric analysis of tryptic peptides (data not shown).
It is also clear that the peptide normally observed in maps from the 6A mutant is absent from that derived from the S525A mutant (indicated by dotted ellipse; Fig. 6F) but still present in maps from the S529A and S532A mutants (Fig. 6F) and in those from the mutant at the nonconserved serine (S527A; data not shown). Taking all of these data together, we deduce that Ser525 is the phosphorylation site in eIF2Bɛ that is regulated by amino acid availability.
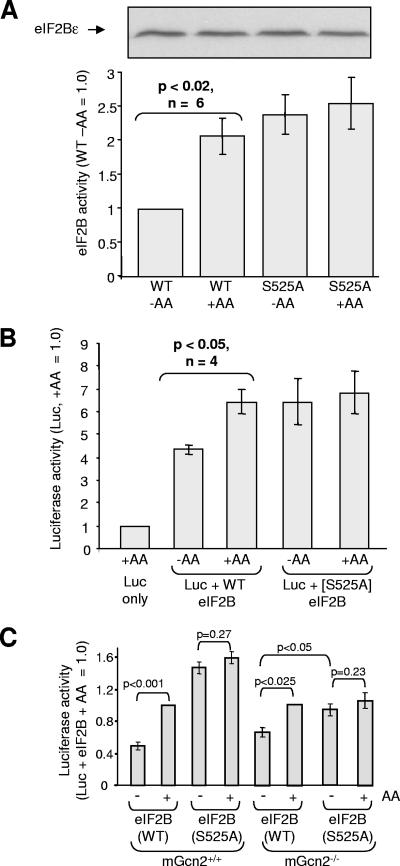
Mutating Ser525 renders eIF2B insensitive to amino acid starvation.
To test the role of Ser525 in regulating the activity of eIF2B in response to amino acids, we created a mutant in which this residue was replaced by a nonphosphorylatable one, alanine (eIF2B[S525A]). This mutant was expressed, with the other four wild-type eIF2B subunits, in HEK293 cells. Amino acid starvation had no significant effect on the activity of the S525A mutant, in contrast to the roughly twofold difference seen for wild-type eIF2B (Fig. 7A). Indeed, under both conditions the activity of the S525A mutant was at least as high as that of WT eIF2B from amino acid-fed cells. These data are consistent with the conclusion that the phosphorylation of wild-type eIF2Bɛ at Ser525 in the absence of amino acids serves to decrease its activity. The findings also indicate that Ser525 alone suffices to confer sensitivity to amino acids. Thus, no other regulatory mechanisms, intrinsic or extrinsic to eIF2B (e.g., other phosphorylation sites in the eIF2B complex or phosphorylation of eIF2α), are required for this effect.
FIG. 7.
Ser525 in eIF2Bɛ plays a key role in regulating eIF2B and protein synthesis. (A) HEK293 cells were transfected with vectors for all five subunits of eIF2B, including either wild-type eIF2Bɛ or the S525A mutant. Cells were subsequently transferred to D-PBS with or without amino acids (AA). Two hours later, cells were lysed, eIF2B complexes were isolated and their activities were determined (n = 6). The panel above the graph shows an immunoblot for myc-eIF2Bɛ. (B) As in in panel A, but cells were cotransfected with the vector for destabilized luciferase. The luciferase activity was measured (n = 4). A blot for myc-tagged eIF2Bɛ is shown above panel A. Other eIF2B subunits were also expressed at similar levels in all four samples (not shown). (C) MEFs (mGcn+/+ or mGcn2−/−, as indicated) were transfected with eIF2B and luciferase vectors as in panel B, but using GenePORTER (Genlantis, San Diego, CA). Cells were subsequently transferred to D-PBS with or without amino acids (AA) for 90 min (the time at which intracellular amino acid depletion is maximal) (3, 38). The luciferase activity was measured (n = 5 to 6). Again, immunoblot analysis revealed that wild-type and S525A eIF2Bɛ were expressed at similar levels (data not shown).
In some cases, glutamates or aspartates can mimic the effect of P-serine. Based on the above data, such mutations would be expected to reduce eIF2B activity under amino acid-fed conditions. However, when we expressed the relevant mutants of eIF2Bɛ in HEK293 cells, no differences between the activity of these eIF2B complexes and wild-type eIF2B complexes were seen (data not shown). The structures of Glu and Asp differ quite significantly from that of P-Ser, and it seems that in this case they do not mimic phosphorylation at Ser525.
Finally, we sought to determine whether the phosphorylation of Ser525 was important for the control of protein synthesis by amino acids by transfecting HEK293 cells with vectors for the luciferase reporter and vectors for eIF2B subunits, including either wild-type eIF2Bɛ or the S525A mutant. After 30 h, the cells were transferred to medium containing or lacking amino acids, and 2 h later the cells were lysed and the luciferase activity was measured. Under both conditions, expression of eIF2B again substantially enhanced luciferase levels (Fig. 7B). In cells expressing wild-type eIF2B, luciferase levels were significantly lower (P < 0.05) under amino acid-deprived conditions than under amino acid-fed ones. This is consistent with the ability of amino acids to promote the activity of the ectopically expressed wild-type eIF2B. In contrast, in cells expressing the S525A mutant, the luciferase levels were not affected by amino acid starvation and were similar to those seen in amino acid-fed cells expressing the wild-type protein. These data indicate (i) that eIF2B plays a key role in the control of protein production (synthesis) in response to amino acids and (ii) that the phosphorylation site at Ser525 is required for this.
mGcn2 is not required for the control of eIF2B by amino acids.
To test whether mGcn2 is needed for the control of eIF2B by amino acids through the phosphorylation of Ser525, we transfected wild-type MEFs and MEFs lacking mGcn2 with vectors for the luciferase reporter and either all five wild-type subunits of eIF2B or the eIF2Bɛ(S525A) mutant and wild-type versions of eIF2Bα-δ. In some cases, cells were starved of amino acids for 90 min (the time at which the effect of amino acid starvation on intracellular amino acid concentrations appears to be maximal, although it still only causes partial depletion at that time (3, 38). As shown in Fig. 7C, amino acid starvation impaired luciferase expression in wild-type MEFs, and this effect was essentially lost when the S525A mutant was used (but not with wild-type eIF2B). This suggests that the regulation of eIF2B via its phosphorylation at S525 in eIF2Bɛ also operates in cells where amino acid starvation does increase eIF2α phosphorylation. Given that eIF2(αP) acts as a competitive inhibitor of eIF2B (36), the fact that eIF2B is being overexpressed in these experiments may diminish the effects of eIF2α phosphorylation.
Amino acids still affected luciferase expression in mGcn2−/− MEFs. The effect was smaller than in the wild-type MEFs, presumably because in the wild-type cells the increased phosphorylation of eIF2α that is induced by amino acid withdrawal (Fig. 3A) also contributes to the impairment of luciferase expression. Expression of the S525A mutant again eliminated the difference in luciferase expression between amino acid-fed and amino acid-starved cells. Thus, it appears that mGcn2 is not required for the control of wild-type eIF2B by amino acids, implying that it is not the kinase that phosphorylates Ser525. This is consistent with data from our in vitro experiments using isolated recombinant mGcn2, where it was unable to phosphorylate purified eIF2B (data not shown).
DISCUSSION
Here, we report four main findings: (i) that amino acid starvation impairs the activity of human eIF2B, independently of changes in eIF2α phosphorylation; (ii) that the effect of amino acids on eIF2B activity is also apparently independent of mTOR, the major known amino acid regulated signaling pathway in mammalian cells; (iii) that amino acids regulate the phosphorylation of a novel conserved site in the catalytic subunit of eIF2B (ɛ), Ser525, which is adjacent to the known regulatory phosphorylation sites (Table 1); and (iv) that Ser525 is essential for the control of eIF2B activity by amino acids. Our data also indicate that that there is no “cross talk” between the mTORC1 and Gcn2 amino acid signaling mechanisms. An earlier report suggested that mTORC1 was inhibited by the accumulation of uncharged tRNA (19) and that this effect might be mediated through the activation of Gcn2. It is possible that uncharged tRNA does regulate mTORC1 under some conditions. Since the accumulation of uncharged tRNAs may prejudice the fidelity of translation, it may be advantageous to mammalian cells to possess mechanisms that can shut off translation independently of their build-up, such as the control of mTORC1 and probably the new mechanism for controlling eIF2B that we identify in this report.
In HEK293 cells, the control of eIF2B by amino acids is independent of the two previously identified ways in which translation factor activity can be controlled by amino acids. One of these involves mTOR, which couples control of several proteins involved in mRNA translation to amino acid availability (e.g., eIF4E [via the 4E-BPs]; S6 and eEF2 [46]). The control of eIF2B may be distinguished from the effects mediated by mTOR in several ways. First, it is not observed in response to starvation of cells only for leucine, a maneuver that causes profound inhibition of mTOR signaling in HEK293 cells (Fig. 4B) and, second, unlike the control of targets for mTOR, expression of the mTOR regulator Rheb cannot “rescue” the activity of eIF2B (Fig. 4B). In addition, most amino acid-dependent effects of mTOR are inhibited by rapamycin: however, pretreatment of cells with rapamycin had no effect on eIF2B activity (Fig. 4B) or its activation by insulin (51).
The second mechanism, best described from budding yeast, involves the phosphorylation of eIF2 by the protein kinase Gcn2p which leads to inhibition of eIF2B, since eIF2(αP) is a competitive inhibitor of eIF2B. Our data show that starvation of HEK293 cells for amino acids does not elicit increased phosphorylation of eIF2α and that there is no increase in the amount of eIF2(αP) bound to eIF2B in amino acid-starved cells.
The fact that the effect of amino acid starvation on eIF2B activity is partial may reflect one or more factors: e.g., amino acid depletion is likely not complete, the phosphorylation of Ser525 in eIF2Bɛ may also not be complete, and such phosphorylation may not totally inhibit eIF2B activity. Finally, it is possible that the in vitro assay conditions for eIF2B may not reveal the full extent of the change in eIF2B activity within the cell.
The amino acid-regulated phosphorylation site (Ser525) lies in a region adjacent to the other phosphorylation site that is known to control the activity of eIF2B, i.e., the site for GSK3 (45, 48). This region, which lies immediately N-terminal to the catalytic domain of eIF2Bɛ, thus appears to control eIF2B activity in response to amino acids and hormones or growth factors. The identity of the kinase and/or phosphatase that modulate eIF2B phosphorylation in response to amino acids remain to be established. Its identification is clearly an important goal but lies beyond the scope of the present study. It is unlikely to be Gcn2 firstly because amino acid starvation does not increase eIF2α phosphorylation in HEK293 cells (suggesting Gcn2 is not activated) and, secondly, because, when we tested whether Gcn2 could phosphorylate eIF2B in vitro, no phosphorylation of any subunit of eIF2B was observed (data not shown). Third, amino acid starvation still decreases luciferase expression in cells that lack mGcn2, and this effect is eliminated by expression the eIF2Bɛ(S525A) mutant, showing that mGcn2 is not required for the control of protein synthesis through the phosphorylation of eIF2B at this site. The S6 kinases are regulated by amino acids but are inactive in amino acid-starved cells (2) and can therefore be excluded as candidates for catalyzing the amino acid-suppressed phosphorylation of Ser525 in eIF2Bɛ.
In HEK293 cells, eIF2B appears to be a more important regulator of protein synthesis than proteins that are controlled by mTOR in a rapamycin-dependent manner, such as eIF4E, eIF4B, or S6. At least in these cells, the activity of eIF2B seems to play a greater role than the availability of eIF4E in determining the overall rate of protein synthesis, whether in the absence or in the presence of amino acids. This important role for eIF2B in controlling translation initiation is consistent with the presence in mammalian cells of no fewer than four distinct protein kinases that control eIF2B activity through the phosphorylation of its substrate, eIF2 (47) and with the operation of other regulatory mechanisms to modulate eIF2B activity (e.g., through GSK3 or mitogen-activated protein kinase signaling (25, 34, 48). Evidence that eIF2B plays a major role in regulating overall protein synthesis and cell growth come from studies by Sadoshima and coworkers, who showed that overexpression of eIF2Bɛ can drive the growth of neonatal cardiomyocytes (16). Overexpression of eIF2Bɛ also activates protein synthesis and promotes cell growth in adult rat cardiomyocytes (Y. Wang, B. Huang, and C. G. Proud, unpublished data). Our data are consistent with the finding that eIF2B, rather than eIF4F, plays a major role in the control of protein synthesis in response to amino acid starvation in L6 myoblasts (22). eIF2B is also implicated in the control of protein synthesis in response to amino acid imbalance (raised levels of leucine, glutamine, and tyrosine) in the perfused liver (22, 37).
Our data indicate that overexpression of eIF2B is much more effective than overexpression of eIF4E in increasing general protein synthesis, even though the levels of eIF4G/eIF4E complexes are greatly enhanced in the latter case. This is consistent with the idea that eIF4E promotes the translation of specific mRNAs (see, for example, reference 31), while eIF2B is needed for all initiation events, on account of its role in regenerating active eIF2.GTP.
In conclusion, these studies identify (i) a novel mechanism by which amino acid availability can modulate translation initiation and (ii) a new regulatory phosphorylation site in eIF2B which is controlled by amino acids. Furthermore, our findings point to a novel regulatory mechanism by which amino acids signal to the translational machinery, which is distinct from the two main known mechanisms in mammalian cells, the control of mTORC1 and of Gcn2. We also provide evidence that mTORC1 signaling does not regulate Gcn2 in mammalian cells (in contrast to the situation in yeast) (7) and that Gcn2 is not required for regulation of mTORC1 by amino acid starvation.
Acknowledgments
This study was supported by funding from the Wellcome Trust and the Faculty of Medicine, University of British Columbia.
We are very grateful to Ronald Wek (University of Indiana, Indianapolis) for providing Gcn2+/+ and Gcn2−/− MEFs and to Rui Liu for creating expression vectors of subunits of eIF2. We thank Nick Morrice (University of Dundee), Bryan Ballif (formerly Harvard University) and Jürgen Kast (University of British Columbia) for help with the mass spectrometric analyses.
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.Asano, K., T. Krishnamoorthy, L. Phan, G. D. Pavitt, and A. G. Hinnebusch. 1999. Conserved bipartite motifs in yeast eIF5 and eIF2Bepsilon, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 181673-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avruch, J., C. Belham, Q. Weng, K. Hara, and K. Yonezawa. 2001. The p70 S6 kinase integrates nutrient and growth signals to control translational capacity. Prog. Mol. Subcell. Biol. 26115-154. [DOI] [PubMed] [Google Scholar]
- 3.Beugnet, A., A. R. Tee, P. M. Taylor, and C. G. Proud. 2002. Evidence that intracellular amino acids regulate translation factor function in mammalian cells. Biochem. J. 372555-566. [Google Scholar]
- 4.Boesen, T., S. S. Mohammad, G. D. Pavitt, and G. R. Andersen. 2004. Structure of the catalytic fragment of translation initiation factor 2B and identification of a critically important catalytic residue. J. Biol. Chem. 27910584-10592. [DOI] [PubMed] [Google Scholar]
- 5.Buxade, M., J. L. Parra, S. Rousseau, N. Shpiro, R. Marquez, N. Morrice, J. Bain, E. Espel, and C. G. Proud. 2005. The Mnks are novel components in the control of TNFα biosynthesis and phosphorylate and regulate hnRNP A1. Immunity 23177-189. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, L. E., X. Wang, and C. G. Proud. 1999. Nutrients differentially modulate multiple translation factors and their control by insulin. Biochem. J. 344433-441. [PMC free article] [PubMed] [Google Scholar]
- 7.Cherkasova, V. A., and A. G. Hinnebusch. 2003. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev. 17859-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, P., and S. Frame. 2001. The renaissance of GSK3. Nat. Rev. Mol. Cell. Biol. 2769-776. [DOI] [PubMed] [Google Scholar]
- 9.Dever, T. E., A. C. Dar, and F. Sicheri. 2006. The eIF2α kinases, p. 319-344. In M. B. Mathews, N. Sonenberg, and J. W. B. Hershey (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 10.Fabian, J. R., S. R. Kimball, N. K. Heinzinger, and L. S. Jefferson. 1997. Subunit assembly and guanine nucleotide exchange factor activity of eukaryotic initiation factor eIF2B subunits expressed in Sf9 cells. J. Biol. Chem. 27212359-12365. [DOI] [PubMed] [Google Scholar]
- 11.Gilligan, M., G. I. Welsh, A. Flynn, I. Bujalska, C. G. Proud, and K. Docherty. 1996. Glucose stimulates the activity of the guanine nucleotide-exchange factor eIF-2B in isolated rat islets of Langerhans. J. Biol. Chem. 2712121-2125. [DOI] [PubMed] [Google Scholar]
- 12.Gomez, E., S. S. Mohammad, and G. D. Pavitt. 2002. Characterization of the minimal catalytic domain within eIF2B: the guanine-nucleotide exchange factor for translation initiation. EMBO J. 215292-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez, E., and G. D. Pavitt. 2000. Identification of domains and residues within translation initiation factor eIF2Bɛ required for guanine nucleotide-exchange reveals a novel activation function promoted by eIF2B complex formation. Mol. Cell. Biol. 203965-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall-Jackson, C. A., D. A. Cross, N. Morrice, and C. Smythe. 1999. ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene 186707-6713. [DOI] [PubMed] [Google Scholar]
- 15.Hara, K., K. Yonezawa, Q.-P. Weng, M. T. Kozlowski, C. Belham, and J. Avruch. 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF4E BP1 through a common effector mechanism. J. Biol. Chem. 27314484-14494. [DOI] [PubMed] [Google Scholar]
- 16.Hardt, S. E., H. Tomita, H. A. Katus, and J. Sadoshima. 2004. Phosphorylation of eukaryotic translation initiation factor 2Bɛ by glycogen synthase kinase-3beta regulates beta-adrenergic cardiac myocyte hypertrophy. Circ. Res. 94926-935. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi, A. A., and C. G. Proud. 2007. The rapid activation of protein synthesis by growth hormone requires signaling through the mammalian target of rapamycin, mTOR. Am. J. Physiol. Endocrinol. Metab. 292E1647-E1655. [DOI] [PubMed] [Google Scholar]
- 18.Hinnebusch, A. G. 2000. Mechanism and regulation of methionyl-tRNA binding to ribosomes, p. 185-243. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Iiboshi, Y., P. J. Papst, H. Kawasome, H. Hosoi, R. T. Abraham, P. J. Houghton, and N. Terada. 1999. Amino acid-dependent control of p70S6k: involvement of tRNA amino acylation in the regulation. J. Biol. Chem. 2741092-1099. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, H. Y., S. A. Wek, B. C. McGrath, D. Lu, T. Hai, H. P. Harding, X. Wang, D. Ron, D. R. Cavener, and R. C. Wek. 2004. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 241365-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, H. Y., S. A. Wek, B. C. McGrath, D. Scheuner, R. J. Kaufman, D. R. Cavener, and R. C. Wek. 2003. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-κB in response to diverse cellular stresses. Mol. Cell. Biol. 235651-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimball, S. R., R. L. Horetsky, and L. S. Jefferson. 1998. Implication of eIF2B rather than eIF4E in the regulation of global protein synthesis by amino acids in L6 myoblasts. J. Biol. Chem. 27330945-30953. [DOI] [PubMed] [Google Scholar]
- 23.Kimball, S. R., and L. S. Jefferson. 2006. Signaling pathways and molecular mechanisms through which branched-chain amino acids mediate translational control of protein synthesis. J. Nutr. 136227S-231S. [DOI] [PubMed] [Google Scholar]
- 24.Kimball, S. R., and L. S. Jefferson. 2002. Control of protein synthesis by amino acid availability. Curr. Opin. Clin. Nutr. Metab. Care. 563-67. [DOI] [PubMed] [Google Scholar]
- 25.Kleijn, M., and C. G. Proud. 2000. The activation of eukaryotic initiation factor (eIF)2B by growth factors in PC12 cells requires MEK/Erk signalling. FEBS Lett. 476262-265. [DOI] [PubMed] [Google Scholar]
- 26.Kleijn, M., G. I. Welsh, G. C. Scheper, H. O. Voorma, C. G. Proud, and A. A. M. Thomas. 1998. Nerve and epidermal growth factors induce protein synthesis and eIF2B activation in PC12 cells. J. Biol. Chem. 2735536-5541. [DOI] [PubMed] [Google Scholar]
- 27.Klionsky, D. J. 2005. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 1187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, W., X. Wang, M. S. Van der Knaap, and C. G. Proud. 2004. Mutations linked to leukoencephalopathy with vanishing white matter impair the function of the eukaryotic initiation factor 2B complex in diverse ways. Mol. Cell. Biol. 243295-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long, X., Y. Lin, S. Ortiz-Vega, K. Yonezawa, and J. Avruch. 2005. Rheb binds and regulates the mTOR kinase. Curr. Biol. 15702-713. [DOI] [PubMed] [Google Scholar]
- 30.Mamane, Y., E. Petroulakis, O. Lebacquer, and N. Sonenberg. 2006. mTOR, translation initiation and cancer. Oncogene 256416-6422. [DOI] [PubMed] [Google Scholar]
- 31.Mamane, Y., E. Petroulakis, Y. Martineau, T. A. Sato, O. Larsson, V. K. Rajasekhar, and N. Sonenberg. 2007. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS. ONE 2e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pavitt, G. D., K. V. A. Ramaiah, S. R. Kimball, and A. G. Hinnebusch. 1998. eIF2 independently binds two distinct eIF2B subcomplexes that catalyse and regulate guanine-nucleotide exchange. Genes Dev. 12514-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price, N. T., S. F. Nakielny, S. J. Clark, and C. G. Proud. 1989. The two forms of the beta-subunit of initiation factor-2 from reticulocyte lysates arise from proteolytic degradation. Biochim. Biophys. Acta 1008177-182. [DOI] [PubMed] [Google Scholar]
- 34.Quevedo, C., A. Alcazar, and M. Salinas. 2000. Two different signal transduction pathways are implicated in the regulation of initiation factor 2B activity in insulin-like growth factor-1-stimulated neuronal cells. J. Biol. Chem. 27519192-19197. [DOI] [PubMed] [Google Scholar]
- 35.Ron, D., and H. P. Harding. 2006. eIF2α phosphorylation in cellular stress responses and disease, p. 345-368. In M. B. Mathews, N. Sonenberg, and J. W. B. Hershey (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Rowlands, A. G., R. Panniers, and E. C. Henshaw. 1988. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J. Biol. Chem. 2635526-5533. [PubMed] [Google Scholar]
- 37.Shah, O. J., D. A. Antonetti, S. R. Kimball, and L. S. Jefferson. 1999. Leucine, glutamine and tyrosine reciprocally modulate the translation initiation factors eIF4F and eIF2B in perfused rat liver. J. Biol. Chem. 27436168-36175. [DOI] [PubMed] [Google Scholar]
- 38.Smith, E. M., S. G. Finn, A. R. Tee, G. J. Browne, and C. G. Proud. 2005. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J. Biol. Chem. 28018717-18727. [DOI] [PubMed] [Google Scholar]
- 39.Tee, A. R., J. Blenis, and C. G. Proud. 2005. Analysis of mTOR signaling by the small G-proteins, Rheb and RhebL1. FEBS Lett. 5794763-4768. [DOI] [PubMed] [Google Scholar]
- 40.Tee, A. R., B. D. Manning, P. P. Roux, L. C. Cantley, and J. Blenis. 2003. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 131259-1268. [DOI] [PubMed] [Google Scholar]
- 41.von der, H. T., J. D. Gross, G. Wagner, and J. E. McCarthy. 2004. The mRNA cap-binding protein eIF4E in posttranscriptional gene expression. Nat. Struct. Mol. Biol. 11503-511. [DOI] [PubMed] [Google Scholar]
- 42.Wang, X., A. Beugnet, M. Murakami, S. Yamanaka, and C. G. Proud. 2005. Distinct signaling events downstream of mTOR cooperate to mediate the effects of amino acids and insulin on initiation factor 4E-binding proteins. Mol. Cell. Biol. 252558-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, X., L. E. Campbell, C. M. Miller, and C. G. Proud. 1998. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem. J. 334261-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, X., M. Janmaat, A. Beugnet, F. E. M. Paulin, and C. G. Proud. 2002. Evidence that the dephosphorylation of Ser535 in the epsilon-subunit of eukaryotic initiation factor 2B is insufficient for the activation of eIF2B by insulin. Biochem. J. 367475-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, X., F. E. M. Paulin, L. E. Campbell, E. Gomez, K. O'Brien, N. Morrice, and C. G. Proud. 2001. Eukaryotic initiation factor 2B: identification of multiple phosphorylation sites in the epsilon subunit and their roles in vivo. EMBO J. 204349-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, X., and C. G. Proud. 2006. The mTOR pathway in the control of protein synthesis. Physiology 21362-369. [DOI] [PubMed] [Google Scholar]
- 47.Wek, R. C., H. Y. Jiang, and T. G. Anthony. 2006. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 347-11. [DOI] [PubMed] [Google Scholar]
- 48.Welsh, G. I., C. M. Miller, A. J. Loughlin, N. T. Price, and C. G. Proud. 1998. Regulation of eukaryotic initiation factor eIF2B: glycogen synthase kinase-3 phosphorylates a conserved serine which undergoes dephosphorylation in response to insulin. FEBS Lett. 421125-130. [DOI] [PubMed] [Google Scholar]
- 49.Welsh, G. I., and C. G. Proud. 1992. Regulation of protein synthesis in Swiss 3T3 fibroblasts: rapid activation of the guanine-nucleotide-exchange factor by insulin and growth factors. Biochem. J. 28419-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welsh, G. I., and C. G. Proud. 1993. Glycogen synthase kinase-3 is rapidly inactivated in response to insulin and phosphorylates eukaryotic initiation factor eIF-2B. Biochem. J. 294625-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welsh, G. I., C. M. Stokes, X. Wang, H. Sakaue, W. Ogawa, M. Kasuga, and C. G. Proud. 1997. Activation of translation initiation factor eIF2B by insulin requires phosphatidylinositol 3-kinase. FEBS Lett. 410418-422. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, H. H., A. I. Lipovsky, C. C. Dibble, M. Sahin, and B. D. Manning. 2006. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol. Cell 24185-197. [DOI] [PMC free article] [PubMed] [Google Scholar]