Abstract
IQGAPs are multidomain scaffolding proteins that integrate Rho GTPase and Ca2+/calmodulin signals with cell adhesive and cytoskeletal reorganizational events. Targeted disruption of the murine Iqgap2 gene resulted in the age-dependent development of apoptosis and hepatocellular carcinoma (HCC), characterized by the overexpression of IQGAP1, the loss of membrane E-cadherin expression, the cytoplasmic translocation (and activation) of β-catenin, and the overexpression of a nuclear target of β-catenin, cyclin D1. In normal hepatocytes, IQGAP2 was found to exist as one component of a multifunctional scaffolding complex comprising IQGAP1, β-catenin, and E-cadherin, with no evidence for direct IQGAP1-IQGAP2 interactions. Interbreeding of Iqgap2−/− mice into the Iqgap1−/− background resulted in the phenotypic correction of the preexisting hepatopathy, decreases in the incidence and sizes of HCC tumors, and the normalization of overall survival rates compared to those of Iqgap2−/− mice, suggesting that maximal penetrance of the Iqgap2−/− HCC phenotype requires the coordinate expression of IQGAP1. These results identify Iqgap2 as a novel tumor suppressor gene specifically linked to the development of HCC and the activation of the Wnt/β-catenin signaling pathway, while also suggesting that IQGAP1 and IQGAP2 retain functionally divergent roles in hepatocellular carcinogenesis.
Hepatocellular carcinoma (HCC) accounts for over 80% of all human liver cancer (16) and is responsible for between 500,000 and 1 million deaths worldwide annually (29). Predisposing etiologies for HCC include chronic hepatitis B and C virus infections, exposure to aflatoxin B1, chronic alcohol consumption, and any hepatic disease having associated cirrhosis. While the molecular mechanisms leading to HCC may differ by etiology, HCC generally evolves through a multistep process involving hepatocyte destruction, proliferation, and regeneration. At the molecular level, both genetic and epigenetic alterations that result in the abnormal expression of genes involved in cell cycle control, cell growth and proliferation, apoptosis, and cell-cell interactions have been observed (26, 47). The known genomic heterogeneity of human HCCs may be attributed to different causative agents. Recurrent allelic losses or gains on 14 chromosome arms have been detected in more than 30% of all HCCs analyzed (47). The deregulation of the canonical Wnt/Frizzled signaling cascade has been shown to occur in 33 to 67% of human HCCs (29), and HCC is among the most common malignancies with mutations in the Wnt pathway, typically involving the gene encoding β-catenin (CTNNb1) (15).
β-Catenin is a convergence point for Wnt and cellular adhesion pathways, functioning as a structural adaptor protein linking type I cadherins to the actin cytoskeleton and as a transcriptional cofactor in the Wnt pathway when in complex with T-cell factor/lymphoid enhancer factor (39). Normally, the majority of β-catenin is associated with E-cadherin at cell-cell junctions, and β-catenin is maintained at low cytoplasmic levels. In the absence of Wnt signaling, cytoplasmic β-catenin is bound to its destruction complex of Axin, adenomatosis polyposis coli, and glycogen synthase 3β kinase (GSK-3β), where it is targeted for ubiquitination and degradation in the 26S proteosome. Upon the activation of Wnt/β-catenin pathways, the β-catenin destruction complex dissociates and β-catenin escapes phosphorylation by GSK-3β and translocates to the nucleus, where it activates an array of target genes and cell cycle regulators such as c-myc and cyclin D1 (5, 29). β-catenin deficiency in mice causes lethality at embryonic day 7 (20), while liver-specific disruption of murine Apc (encoding adenomatosis polyposis coli) results in β-catenin signaling activation and the development of HCC (12). In contrast, hepatocyte-restricted overexpression of an oncogenic β-catenin transgene causes hepatomegaly without HCC formation (9). Similarly, the adenovirus-mediated expression of a dominant stable β-catenin mutant failed to cause neoplastic liver foci, suggesting that in contrast to adenomatosis polyposis coli, the activation of the Wnt signaling pathway by stabilized β-catenin is insufficient for HCC development, further suggesting that additional genetic or epigenetic changes are involved (21).
We now provide evidence that IQGAP2 functions as a critical component of a broader IQGAP1-IQGAP2-β-catenin-E-cadherin scaffold and that the loss of IQGAP2 expression by the targeted disruption of the murine Iqgap2 gene leads to the development of HCC. Of equal importance, mice deficient in both Iqgap1 and Iqgap2 genes (Iqgap1−/− Iqgap2−/−) displayed relative protection against HCC and corrected long-term survival rates relative to those of Iqgap2−/− mice, suggesting that IQGAP1 and IQGAP2 retain functionally divergent roles in hepatocellular carcinogenesis. IQGAPs are conserved homologues of an extended family of proteins found in yeast (Saccharomyces cerevisiae) and amoebae (Dictyostelium discoideum), studied in mammalian systems largely as scaffolding proteins with the capacity of integrating intracellular signals with cytoskeletal membrane events (1, 7, 32, 42). In mammals, two homologous IQGAPs (IQGAP1 and IQGAP2) have been characterized previously (8, 22, 27, 34), although a third (IQGAP3) was recently identified on the basis of homology search and implicated in axonal elongation in PC12 cells and rat hippocampal neurons (50). IQGAPs bind F-actin through the calponin homology domain (10) and interact with multiple calmodulin molecules in a predominantly Ca2+-independent fashion through repetitive IQ motifs (IQXXXRGXXR) (8) and with Rho GTPases cdc42 and rac1 by means of a C-terminal RasGAP-related domain (GRD) (8). While IQGAP1 is known to capture microtubules through its association with cytoplasmic linker protein 170 (CLIP-170) (17) and to modulate cadherin-based adhesion by acting as a competitive inhibitor of β-catenin-cadherin complex formation (6, 28), the physiological significance of these biochemical interactions remains unknown. These data establish the first identified pathological consequence of IQGAP2 deficiency and provide a novel function for Iqgap2 as a tumor suppressor gene coupled to the activation of the Wnt/β-catenin signaling pathway.
MATERIALS AND METHODS
Generation and characterization of Iqgap2-deficient and Iqgap1- and Iqgap2-deficient mice.
An ∼150-kb bacterial artificial chromosome clone derived from murine chromosome 13 and encompassing exons 7 to 34 of the Iqgap2 gene was isolated as previously described (14). The targeting vector was constructed by the insertion of an 8.5-kb genomic fragment containing Iqgap2 exons 15 to 17 into the BamHI site of plasmid pMJK-KO (available from Thomas Rosenquist, State University of New York [SUNY]—Stony Brook), upstream from the phosphoglycerate kinase promoter-neomycin (PGK-Neo) reporter cassette. An 8-kb Iqgap2 genomic fragment encompassing exon 31 was cloned into the XhoI site downstream of the PGK-Neo cassette. The targeting vector also contained a thymidine kinase gene (PGK-tk) as a second selection marker (see Fig. 1A). The linearized targeting vector was introduced into 129J1 embryonic stem (ES) cells by electroporation, and the Iqgap2 null allele was generated by homologous recombination using dual selection (33). Genomic DNA from individual ES clones was screened by Southern analysis using ApaI-digested DNA and a 32P-radiolabeled, PCR-generated 500-bp fragment as a probe (forward, 5′-GCTGGCAGTGGGGAGCACAGTGCA-3′; reverse, 5′-AGGGCAGGAAAGGCAGCAGCACTT-3′). ES clone no. 57 was used for microinjections into C57BL/6 blastocysts to generate chimeric mice (33) and bred into 129J1 and C57BL/6 backgrounds for at least eight generations prior to further studies. All functional studies were completed using 129J1 mice. F1 progeny of the chimeric mice were initially genotyped by Southern blot analysis (as outlined above) using tail DNA. Genotyping of subsequent progeny was completed by PCR. Oligonucleotide pair P3/P4 (P3, 5′-AAGGCATGATTCATTCACCTGAGA-3′; P4, 5′-AGGGCAGGAAAGGCAGCAGCACTT-3′) was used for the detection of the wild-type allele, and N2/N3 (N2, 5′-GTCAAGAAGGCGATAGAAGG-3′; N3, 5′-TTGAACAAGATGGATTGCACGCA-3′) was used for the null allele. The 35-cycle PCR included a denaturation step at 94°C for 1 min and 15 s, a 2-min annealing step at 55°C, and a 3-min primer extension step at 72°C.
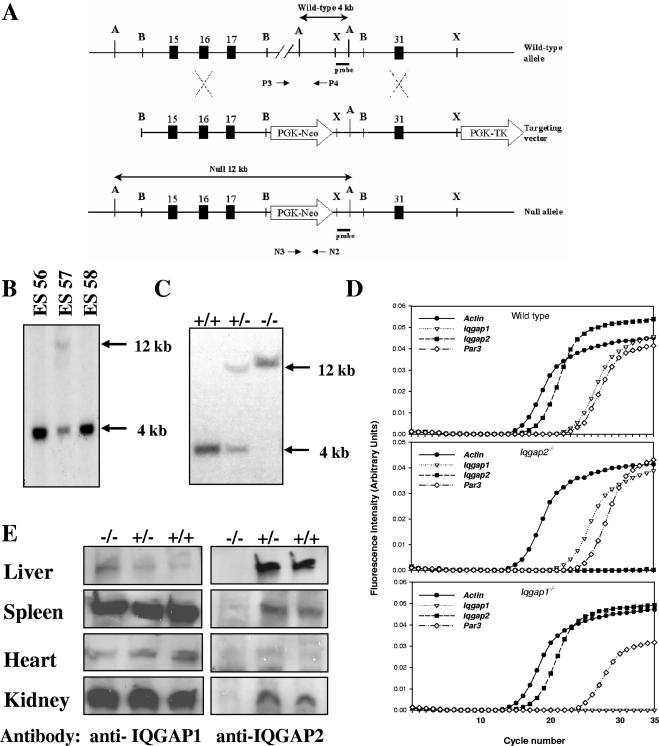
FIG. 1.
Generation and characterization of Iqgap2−/− mice. (A) Schema depicting the targeting construct and the Iqgap2 wild-type and null alleles, resulting in the replacement of an ∼36-kb genomic fragment (corresponding to amino acid residues Ala705 to Lys1306) with the PGK-Neo cassette. The deleted fragment encompasses a portion of the IQ2 (Gln696-to-Gly725), IQ3 (Asn726-to-Asp755), and IQ4 (His756-to-Pro785) domains and the GRD (Gln891 to Val1189) (8). Selected numbered exons are depicted by rectangles, and the relative positions of oligonucleotides and the 32P-radiolabeled probe used for allele identification are shown. A, ApaI; B, BamHI; X, XbaI. (B and C) Southern blot analysis was performed using 10 μg of ApaI-digested genomic DNA isolated from various ES clones (B) or Iqgap2−/− (−/−), Iqgap2+/− (+/−), or wild-type (+/+) mice (C) specifically generated from ES clone no. 57, and DNA was probed with the 32P-radiolabeled 500-bp genomic fragment depicted in panel A. The 4-kb wild-type and 12-kb mutant alleles are evident. (D) qRT-PCR was completed using total RNA isolated from livers of 2-month-old wild-type, Iqgap1−/−, or Iqgap2−/− mice and primers specific for the β-actin gene, Iqgap1, Iqgap2, and Par3 (known to be encompassed within the Iqgap2 gene [14]). (E) Total protein lysates (10 μg per lane) isolated from organs of 2-month-old Iqgap2−/− (−/−) or Iqgap2+/− (+/−) mice or wild-type (+/+) littermate controls were size fractionated by 4 to 15% SDS-PAGE, and IQGAP1 and IQGAP2 expression was assessed by immunoblotting. Note the absence of IQGAP2 in all Iqgap2−/− organs, with a modest reciprocal increase in IQGAP1 in Iqgap2−/− livers only.
Mice with targeted disruption of the Iqgap1 gene were maintained on a 129J1 background (kindly provided by Andre Bernards, Harvard Medical School). Iqgap1−/− Iqgap2−/− mice were generated by pairwise interbreeding of Iqgap1−/− and Iqgap2−/− mice with the 129J1 background. Offspring were born with predicted Mendelian frequencies, establishing that the Iqgap1−/− Iqgap2−/− genotype did not result in embryonic lethality; more detailed characterization of these mice will be given elsewhere. All mice were handled according to guidelines for the humane care and use of experimental animals, and procedures were approved by the SUNY—Stony Brook Institutional Animal Care and Use Committee.
Molecular genetic analyses.
The integrity of liver mitochondrial DNA from Iqgap2−/− mice was assessed by Southern blotting using a 2-kb EcoRI fragment of a mouse mitochondrial genome as a probe (43). Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA). RNA integrity was established using an Agilent 2100 bioanalyzer.
Quantitative reverse transcription-PCR (qRT-PCR) was performed using fluorescence-based real-time PCR technology (19); in brief, 5 μg of total RNA from murine livers was used for first-strand cDNA synthesis with random hexamer primers (Invitrogen, Carlsbad, CA). Total RNA in 7 μl of H2O was mixed with 5 μl of hexamer primers (100 ng/μl), and the mixture was heated to 85°C for 3 min and placed on ice. The reaction mix was prepared separately according to the Invitrogen protocol and contained 4 μl of 5× first-strand buffer, 2 μl of 0.1 M dithiothreitol, and 1 μl of a mix of deoxynucleoside triphosphates (15 mM each). RNA with primers was then added to the reaction mix, the mixture was incubated at 42°C for 3 min, and the reverse transcription was started by the addition of 1 μl of SuperScript II reverse transcriptase (200 U/μl; Invitrogen [catalog no. 18064]). The reverse transcription reaction was carried out at 42°C for 2 h and stopped by incubation of the reaction mixture at 85°C for 10 min.
The following PCR step was performed in a 96-well plate using an Opticon-I PCR machine (Bio-Rad). The reverse transcription reaction mixture was diluted up to 1:15 and equally divided among primer pairs for each target gene (Table 1). Oligonucleotide primer pairs for each target gene were generated using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and were designed to amplify PCR products of 200 ± 1 bp at the same annealing temperatures. The plate was heated at 95°C for 15 min and subjected to a 40-cycle PCR (94°C for 30 s, 55°C for 30 s, and 72°C for 1 min and fluorescence reading for 10 s). Melting curves were measured for every well of the plate at temperatures from 65 to 95°C, with a reading interval of 0.2°C and 5 s of holding between the readings. mRNA levels were quantified by monitoring the real-time fluorimetric intensity of SYBR green I (Invitrogen). Known amounts of DNA were used as standards to generate a calibration curve covering a range from 1 to 10−6 ng of a target per well. The relative mRNA abundance was determined from triplicate assays performed in parallel for each primer pair and calculated using the comparative threshold cycle number (23), and results were normalized to the amount for cellular β-actin mRNA, all as previously described (18).
TABLE 1.
Oligonucleotide primers for qRT-PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Iqgap1 | 5′-TGCAGCTGACACTTTTACGG-3′ | 5′-TTCCCTTGGATTTCAGCTTG-3′ |
| Iqgap2 | 5′-TCAAGATTGGACTGCTGGTG-3′ | 5′-AGGTTTGGTCTGGAGGAGGT-3′ |
| Par3 | 5′-TCAATGGCAACAACTGGGTA-3′ | 5′-ATGACCCACACTATGCCACA-3′ |
| β-Actin gene | 5′-TACCACAGGCATTGTGATGG-3′ | 5′-TTTGATGTCACGCACGATTT-3′ |
| 16S rRNA | 5′-GGGACTAGCATGAACGGCTA-3′ | 5′-CCCCAACCGAAATTTCAAAC-3′ |
| COXa gene | 5′-CTGAGCGGGAATAGTGGGTA-3′ | 5′-TGGGGCTCCGATTATTAGTG-3′ |
| Cytochrome b gene | 5′-ACGTCCTTCCATGAGGACAA-3′ | 5′-GAGGTGAACGATTGCTAGGG-3′ |
COX, cytochrome c oxidase subunit 1.
Murine phenotypic and electron microscopy analyses.
Freshly isolated mouse organs were immediately snap-frozen in liquid nitrogen or paraffin embedded by submersion in 10% neutral buffered formalin for 16 h at 4°C prior to being sectioned and stained using hematoxylin and eosin. For electron microscopy, freshly isolated mouse liver and skeletal muscle samples were fixed by immersion in a solution of 2% paraformaldehyde and 2.5% electron microscopy-grade glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). After fixation, samples were placed in 2% osmium tetroxide in 0.1 M sodium cacodylate buffer (pH 7.4), dehydrated in a graded series of ethyl alcohol preparations, and embedded in Durcupan resin (Electron Microscopy Sciences, Fort Washington, PA). Ultrathin sections of 80 nm were generated with a Reichert-Jung UltracutE ultramicrotome (Leica Microsystems, Inc., Bunnockburn, IL) and placed on Formvar-coated slot copper grids. Sections were then counterstained with uranyl acetate and lead citrate and viewed with a Tecnai12 BioTwinG2 electron microscope (FEI Company, Hillsboro, OR). Digital images were acquired using an AMT XR-60 charge-coupled device digital camera system (Advanced Microscopy Techniques Co., Danvers, MA). All microscopic and ultramicroscopic analyses were performed blindly to the genotype. The presence of HCC was established by the examination of the gross morphological features and a microscopic review of at least two sections per liver; for all specimens, microscopy was carried out in a blinded fashion and complete concordance was observed between the results of the examination of gross morphology and microscopic HCC detection. Histologic confirmation of HCC was established on paraffin sections using Gordon and Sweet's silver reticulin stain.
Biochemical determinations (for hepatic transferases, bilirubin, and alkaline phosphatase) were completed using sera isolated from retroorbital blood samples and quantified using a MODULAR PP serum work area analyzer (Roche Diagnostics, Indianapolis, IN). Automated blood counts were obtained on a Coulter counter specifically gated to detect murine blood cells, and blood smears were subjected to Wright-Giemsa stain.
Hepatocyte TUNEL and mitochondrial assays.
The quantification of apoptotic hepatocytes was completed using terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining (31). Briefly, liver sections were deparaffinized in xylene, rehydrated in decreasing alcohol concentrations (from 100 to 0%), and digested with 20 μg/ml of proteinase K for 10 min at 25°C, and the digestion was subsequently quenched by incubation with 3% hydrogen peroxide in phosphate-buffered saline for 10 min at 25°C. Washed sections were then incubated for 60 min at 37°C with terminal deoxynucleotidyltransferase enzyme for dUTP-biotin nick end labeling (Chemicon, Temecula, CA), after which they were incubated for 1 h at 25°C with the digoxigenin-conjugated anti-dUTP antibody. Detection was completed using 3,3′-diaminobenzidine as the substrate. Sections were counterstained with 0.5% methyl green. Apoptotic cells (among 1,000 cells per section) were scored and quantified in a blinded fashion by two investigators, and the results were expressed as the mean ± the standard error of the mean (SEM) for each age group studied.
Mitochondria were isolated from individual livers after isotonic homogenization in an ice-cold buffer containing 0.25 M sucrose, 10 mM HEPES buffer, pH 7.4, and 1 mM EGTA. The mitochondrial fraction was obtained by differential centrifugation at 4°C (48), and the protein content was measured by a bicinchoninic acid assay (Pierce, Rockford, IL). The integrity and functional capacity of mitochondria were assessed by tetramethylrhodamine ethyl ester loading (50 mM for 10 min at 25°C) and FACScan analysis using logarithmic-gain settings for light scattering and fluorescence (Becton Dickenson, Mountain View, CA). For all studies, 10,000 gated events were acquired.
Immunodetection and immunoprecipitation studies.
Immunohistochemistry analysis of paraffin-embedded liver sections was performed essentially as described previously (25) using mouse monoclonal antibodies specific for β-catenin (1:50; clone 14, BD Biosciences, San Jose, CA), E-cadherin (1:50; BD Biosciences), IQGAP1 (1:15; BD Biosciences), and IQGAP2 (1:50; Upstate Biotechnology/Millipore, Billerica, MA). The detection of primary antibody was carried out using a biotinylated mouse-specific secondary antibody (1:100), streptavidin peroxidase (Rockland Immunochemicals for Research, Gilbertsville, PA), and 3,3′-diaminobenzidine as the substrate.
Protein lysates from various organs were prepared as previously described (42). Lysates were centrifuged at 600 × g for 10 min at 4°C, and protein supernatants were quantified by a bicinchoninic acid assay prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 4 to 15% gradient gels (Bio-Rad Laboratories, Hercules, CA). Immunoblot analysis was completed as previously described (2). Primary antibodies, all mouse monoclonal antibodies, were as follows: IQGAP1 (1:1,000; BD Biosciences), IQGAP2 (1:1,000; Upstate Biotechnology/Millipore), actin (1:1,000; Chemicon, Temecula, CA), glyceraldehyde-3-phosphate dehydrogenase (1:1,000; Upstate Biotechnology/Millipore), cytochrome c (1:1,000; Cell Signaling Technology, Danvers, MA), complex I and II (both 1:200; Molecular Probes, Eugene, OR), prohibitin (1:500; Lab Vision Co., Fremont, CA), β-catenin (1:1,000; clone 14; BD Biosciences), active β-catenin (anti-ABC; 1:1,000; clone 8E7; Upstate Biotechnology/Millipore), E-cadherin (1:1,000; BD Biosciences), and cyclin D1 (1:400; Upstate Biotechnology/Millipore). Relative protein abundance was determined by densitometric analysis using Gel-Pro Analyzer software (version 3.0; Media Cybergenetics, Silver Spring, MD).
Immunoprecipitation experiments were completed using freshly isolated murine livers essentially as described previously (42). In brief, intact livers were homogenized in 25 mM HEPES-0.15 M NaCl buffer, pH 7.45, supplemented with 0.1% Triton X-100 and standard protease inhibitors. After centrifugation at 10,000 × g for 15 min at 4°C, 5 to 20 mg of whole-cell lysate was precleared in a 1-ml volume by using 50 μl of protein G agarose (25-μl bed volume; Roche Diagnostics GmbH, Mannheim, Germany) for up to 4 h at 4°C. After centrifugation at 12,000 × g for 20 s, supernatants were incubated with 5 μg of protein G agarose-conjugated antibodies overnight at 4°C. Pellets were then collected by centrifugation (12,000 × g for 20 s) and washed three times with lysis buffer prior to immunoblotting as outlined above.
Alternatively, immunoprecipitation experiments were carried out with COS1 lysates. COS1 cells were grown in Dulbecco minimal essential medium supplemented with 10% fetal bovine serum, 100 U/ml of penicillin, and 100 μg/ml of streptomycin sulfate. Transient transfections with full-length human IQGAP1 cDNA (kindly provided by David Sacks, Harvard Medical School) (24) and IQGAP2 cDNA (a gift from Andre Bernards, Harvard Medical School) (8) were completed using Fugene reagent (Roche Diagnostics). Transfection with EGFP-C1 plasmid (Clontech Laboratories, Mountain View, CA) served as a control; 24 h posttransfection, cells were washed and solubilized in a buffer containing 25 mM HEPES-0.15 M NaCl buffer, pH 7.45, supplemented with 0.1% Triton X-100 and standard protease inhibitors. Lysates were subsequently used for immunoprecipitation studies as outlined above.
Statistical analyses.
Kaplan-Meier survival curves and age-dependent HCC incidence data were generated using SPSS software (Statistical Package for the Social Sciences, version 14.0). For all analyses, differences in means between groups were analyzed by a χ2 test or two-tailed Student's t test using a P value of <0.05 as a measure of statistical significance.
RESULTS
Generation of Iqgap2-deficient mice.
To inactivate the Iqgap2 gene, a 36,241-bp genomic fragment spanning exons 18 to 30 (corresponding to amino acids Ala705 to Lys1306, encompassing a portion of the IQ2 [Gln696 to Gly725] motif, the entire IQ3 [Asn726 to Asp755] and IQ4 [His756 to Pro785] motifs, and the GRD [Gln891 to Val1189]) was replaced with a PGK-neomycin resistance cassette via site-specific homologous recombination (Fig. 1A to C). Interbreeding of heterozygous (Iqgap2+/−) founder mice on both 129J1 and C57BL/6 backgrounds demonstrated that progeny were born at normal Mendelian ratios, eliminating the possibility that Iqgap2 expression is required for normal embryonic development. Iqgap2−/− mice were fertile and clinically normal at up to 12 months of age. To exclude the possibility that Iqgap2 loss caused the reciprocal induction of the homologous Iqgap1 gene, qRT-PCR was completed using hepatocytes from Iqgap1−/− and Iqgap2−/− mice (Fig. 1D). In parallel, transcript expression of the murine protease-activated receptor 3 (Par3) gene was assessed to exclude inadvertent disruption. Murine Par3 is known to be encompassed within Iqgap2 intron 13 (14), and unaltered expression established that the thrombin-PAR3 receptor system (38, 41) was uninvolved in subsequent phenotypic defects. Transcript analysis confirmed that Iqgap2 was the predominant isoform expressed in murine livers, with no clear change in Iqgap1 gene expression. Immunoblot analysis using tissues known to express Iqgap2 (14) demonstrated a clear loss of protein, with the exception of a modest increase in hepatic IQGAP1 content (Fig. 1E), as assessed by immunoblot densitometry (data not shown). Thus, comparable to the Iqgap1−/− mouse (30), the Iqgap2−/− genotype did not result in intrauterine or immediate postnatal death, nor was there any evidence that compensatory induction of Iqgap1 masked the lack of an essential Iqgap2 function in adult tissues.
Iqgap2−/− mice display age-dependent hepatocellular apoptosis.
The initial survey of organs from 6-week-old Iqgap2−/− mice demonstrated no histological defects in any of the tissues studied; similarly, animals were fertile and maintained normal body weights. More-detailed hematological and biochemical studies of mice from distinct age groups (1, 4, and 8 months old) revealed normal hemograms, although Iqgap2−/− mice demonstrated an age-dependent increase in levels of distinct hepatocyte-derived aminotransferases in sera. Total bilirubin levels remained normal in mice up to 8 months of age, and enzymatic abnormality at this age was restricted to elevations of aspartate aminotransferase (AST) (Fig. 2A), to the exclusion of a rise in alanine aminotransferase (data not shown). A nonstatistically significant rise in AST was evident at 4 months, with clear differences in 8-month-old Iqgap2−/− mice (P = 0.04) compared to wild-type littermates. The AST was most likely hepatic (nonmuscular) in origin, since corresponding serum creatine phosphokinase levels remained normal and values for the two groups were statistically indistinguishable (53.6 ± 6.3 U/liter in wild-type mice versus 72.4 ± 20.59 U/liter in Iqgap2−/− mice). Similarly, the extensive organ survey failed to identify histologic defects in any nonhepatic sources. Although this pattern evident in Iqgap2−/− mice is typical of human alcoholic liver disease (i.e., disproportional elevations of AST compared to alanine aminotransferase), hepatic nonesterified fatty acid and triglyceride levels were normal (data not shown).
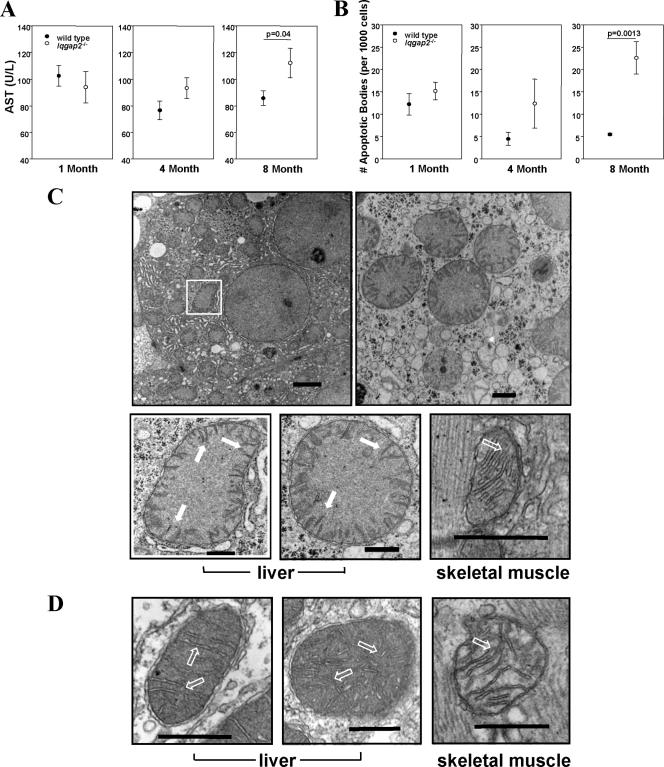
FIG. 2.
Age-dependent hepatopathy and mitochondrial damage in Iqgap2−/− mice. (A) Sera from mice in three age groups (1, 4, and 8 months old; n = 10 mice per age group) were analyzed for the quantification of AST. (B) Liver sections (n = 3 per age group) were analyzed for apoptotic cells by TUNEL staining. Results shown in panels A and B are means ± SEM; statistically significant P values are shown. (C and D) Transmission electron micrographs from livers and skeletal muscles of 12-month-old Iqgap2−/− mice (low and high magnifications) (C) or wild-type mice (high magnification) (D) demonstrate an edematous mitochondrial matrix with collapsed cristae (solid arrows) restricted to Iqgap2−/− mitochondria; hollow arrows indicate normal cristae. Note the normal architecture of mitochondria from skeletal muscles of both genotypes. Structural defects are representative of those seen in 8- and 12-month-old mice (n = 3 per group), with comparable but less-extensive defects seen in 4-month-old Iqgap2−/− mice. Size bars represent 2 μm (magnification, ×4,800 [C, top left panel]), 500 nm (magnification, ×18,500 [C, top right panel]), and 200 nm (magnification, ×49,000 [C, bottom panels, and D]). The white box denotes an Iqgap2−/− mitochondrion shown magnified at ×49,000 in the image below.
TUNEL staining using livers from wild-type and Iqgap2−/− littermates specifically demonstrated no differences in the numbers of apoptotic cells in 1-month-old mice; however, gradual and progressive evidence for spontaneous apoptosis in 4- and 8-month-old Iqgap2−/− mice was observed (Fig. 2B). Enhanced hepatocellular loss was evident (but not statistically significant) by 4 months, and that in 8-month-old mice was highly significant (P = 0.0013). Thus, the biochemical evidence for liver damage paralleled that for an associated age-dependent accumulation of apoptotic hepatocytes.
Structural mitochondrial abnormalities in Iqgap2−/− hepatocytes.
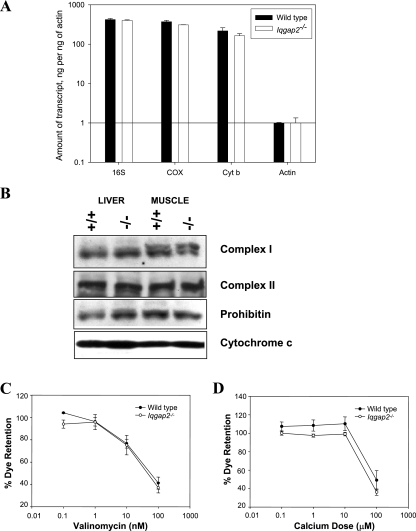
The age-related onset of the apoptotic phenotype evident in Iqgap2−/− hepatocytes is characteristic of many mitochondrial diseases and is postulated to result from the senescent decline in oxidative phosphorylation seen in postmitotic tissues such as liver (52). An ultrastructural review of hepatocytes from Iqgap2−/− mice (ages 1, 4, 8, and 12 months; n = 3 per age group) displayed distinct age-dependent abnormalities restricted to mitochondria (Fig. 2C and D). These defects were not present in any age-matched wild-type mice examined (n = 8), were more pronounced with age, and were restricted to the liver (to the exclusion of skeletal muscle). Furthermore, there was no evidence for associated abnormalities of mitochondrial number, as assessed by quantitative PCR and genomic blotting of mitochondrial DNA (data not shown), qRT-PCR analysis of mitochondrial RNA transcripts, or immunoblot analysis of key mitochondrial proteins (Fig. 3A and B) (35). Finally, the functional competence of the mitochondrial permeability transition pore (mtPTP; the complex regulating cytochrome c release as a primary apoptotic signal [13]) was found to be normal, as assessed by comparable dose-response sensitivities to calcium (the fundamental mtPTP activator) and valinomycin (a potassium-specific inducer of the mtPTP [45]) (Fig. 3C and D). Collectively, these data suggested that IQGAP2 deficiency caused hepatocellular apoptosis through a mitochondrial pathway unrelated to a concomitant intrinsic defect of mitochondrial function.
FIG. 3.
Mitochondrial characterization of Iqgap2−/− mice. (A) qRT-PCR was completed using total liver RNA from 12-month-old Iqgap2−/− mice or age-matched wild-type controls. 16S, 16S subunit rRNA; COX, cytochrome c oxidase subunit 1; Cyt b, cytochrome b. The bar graphs show means ± standard deviations of results from triplicate wells, normalized using β-actin; the results shown represent one complete set of experiments, repeated on two occasions. (B) Immunoblot analysis of liver and skeletal muscle lysates (10 μg/lane) from wild-type (+/+) and Iqgap2−/− (−/−) mice using antibodies specific to complex I, complex II, prohibitin, and cytochrome c. (C and D) Flow cytometry analysis of mitochondria (equivalent to 300 μg of protein/sample) isolated from hepatocytes from 8-month-old Iqgap2−/− or age-matched wild-type mice, loaded with tetramethylrhodamine ethyl ester, and incubated with the specified concentrations of valinomycin (C) or CaCl2 (D). For all experiments, the percentage of dye retention was determined from the geometric mean for 10,000 events, with 100% representing the baseline fluorescence of intact mitochondria. Results are the means ± SEM from two distinct experiments.
The development of HCC in Iqgap2−/− mice is associated with the reciprocal induction of IQGAP1 expression and the activation of the Wnt/β-catenin pathway.
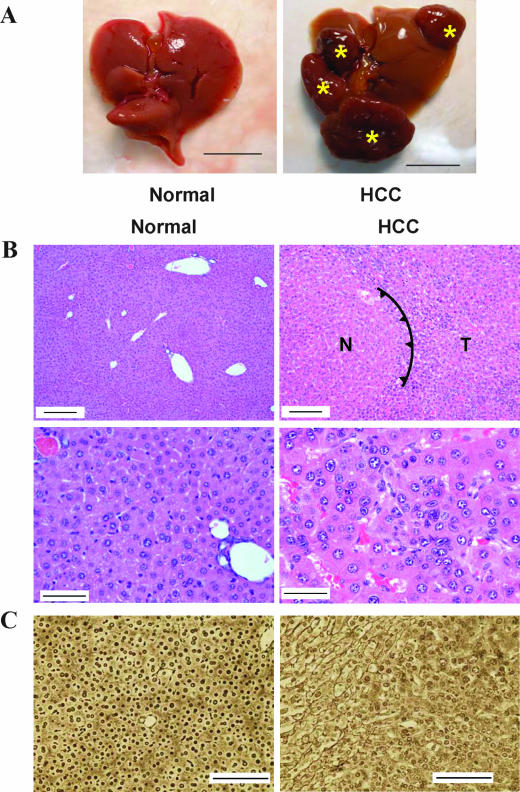
Since apoptosis is a hallmark of many hepatocellular disorders, mice were monitored for up to 2 years after birth. Detailed necroscopy of 18- to 24-month-old mice demonstrated a high incidence of HCC in Iqgap2−/− mice (18 of 21; 86%) compared to that in wild-type controls (0 of 15) (P < 0.0001) (see below). Both sexes were equally affected. In all situations, there was complete concordance between the presence of grossly abnormal livers and microscopic features of HCC (i.e., there were no instances of microscopic HCC in visibly normal livers) (Fig. 4A); histologically, the neoplasms displayed well-differentiated features with trabecular and pseudoglandular patterns (Fig. 4B). More detailed histological analysis using reticulin staining confirmed the presence of distorted architecture of the hepatic cords in Iqgap2−/− liver tumors, findings typically identified in HCC (Fig. 4C). Except for the occasional presence of uterine hemangiomas, no other tumors or abnormalities in Iqgap2−/− mice were detected. Similarly, no steatosis or other hepatic abnormalities were evident.
FIG. 4.
Development of HCC in Iqgap2−/− mice. (A) Gross morphological evidence of HCC in the liver of a 2-year-old Iqgap2−/− mouse compared to the liver of an age-matched wild-type control; asterisks denote multiple large, vascularized nodules. The scale bars correspond to 1 cm. (B) Representative hematoxylin- and eosin-stained sections from wild-type or Iqgap2−/− livers demonstrating HCC in Iqgap2−/− mice. The spiked line (right upper panel) delineates the border between normal (N) and tumor (T) tissue. Scale bars correspond to 150 μm (top panels) and 50 μm (bottom panels). (C) Representative reticulin-stained livers from wild-type and Iqgap2−/− mice showing HCC. Note the discontinuous patterns of reticulin fibers in the HCC liver tissue. Scale bars correspond to 200 μm.
Since the Wnt/β-catenin pathway is frequently involved in the development of human HCC (29), hepatocellular expression patterns for β-catenin and E-cadherin were studied; IQGAP1 (a known β-catenin binding protein) (28) was studied in parallel. Immunohistochemistry analysis of normal wild-type hepatocytes demonstrated membrane-associated expression of E-cadherin, β-catenin, and IQGAP1 (Fig. 5A), with both submembrane and more diffuse cytoplasmic patterns of IQGAP2 expression (data not shown). In contrast, a loss of IQGAP1 expression in plasma membranes and enhanced cytoplasmic expression of IQGAP1 in Iqgap2−/− HCC hepatocytes were evident, and this pattern occurred in parallel with the cytoplasmic translocation and accumulation of β-catenin; loss of E-cadherin expression in plasma membranes was also evident. Although β-catenin expression in Iqgap2−/− hepatocytes of younger mice without HCC was minimally affected, these mice displayed altered IQGAP1 expression, characterized primarily by diffused cytoplasmic staining (Fig. 5A).
FIG. 5.
Molecular analysis of HCC in Iqgap2−/− mice. (A) Immunohistochemical detection of IQGAP1, β-catenin, and E-cadherin in livers from 4-month-old wild-type or Iqgap2−/− mice without HCC or 2-year-old Iqgap2−/− mice with HCC. Note the loss of E-cadherin from cell membranes, along with the enhanced cytoplasmic expression and translocation of both IQGAP1 and β-catenin, occurring only with HCC. Scale bars correspond to 50 μm. (B) Immunoblot analysis was completed using an anti-β-catenin monoclonal antibody and liver lysates (10 μg/lane) from mice (2 years old) with HCC or age-matched controls; the arrow indicates the 92-kDa β-catenin band, with clear evidence for truncated β-catenin mutant forms in the case of HCC. (C) Immunoblot (10 μg of lysate/lane) demonstrating the presence of active (dephosphorylated) β-catenin mutant forms in HCC liver tissue. Results for livers from two 4-month-old wild-type (WT) and Iqgap2−/− (−/−) mice are also shown. Cell lysate from the A431 cell line (Upstate Biotechnology/Millipore) was used as a positive control. (D) Immunoblot analysis (10 μg of lysate/lane) to assess E-cadherin, cyclin D1, and IQGAP1 expression in Iqgap2−/− liver lobes without HCC or affected by HCC; paired normal (N) and tumor (T) samples were collected from the same animals. (E) Immunoblot densitometric analysis of E-cadherin, cyclin D1, and IQGAP1 expression was conducted using liver lysates (10 μg/lane) from lobes affected by HCC (n = 4), those of 4- and 12-month-old Iqgap2−/− mice without HCC (n = 3), and those of 4-month-old wild-type mice (n = 2). Data are presented as mean ratios ± standard deviations of the expression of individual proteins relative to that of proteins in the wild-type samples. (F) IQGAP1 expression was assessed by immunoblotting of liver lysates (10 μg/lane) from wild-type (WT) and Iqgap2−/− (−/−) mice of various ages (6 weeks and 4, 12, and 24 months). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific antibody served as a control for the loading of equal quantities. A representative immunoblot is shown. (G) Immunoblot densitometric analysis of hepatic IQGAP1 expression in wild-type and Iqgap2−/− mice of various ages. Data are presented as means ± SEM of the integrated optical densities of bands. n = 3 for each age group.
In an immunoblot analysis, β-catenin bands with a lower-than-normal (92-kDa) molecular mass were detected exclusively in HCC liver lysates, confirming the presence of previously described mutated forms of β-catenin known to contain somatic activating mutations in mouse and human HCC (15) (Fig. 5B). The presence of these mutated forms was confirmed by RT-PCR using RNA isolated from HCC livers and primers spanning the β-catenin gene (data not shown). These mutations prevent β-catenin from being phosphorylated, leading to its cellular redistribution and accumulation in the cytoplasm and/or nuclei (β-catenin activation) (15). To further investigate the age-dependent activation of the Wnt/β-catenin pathway in Iqgap2−/− livers, liver lysates from 4-month-old wild-type and Iqgap2−/− mice, as well as those from 2-year-old Iqgap2−/− mice affected with HCC, were probed with an antibody specifically recognizing the active form of β-catenin, dephoshorylated on Ser37 or Thr41 (Upstate Biotechnology/Millipore). As shown in Fig. 5C, the dephosphorylated (active) β-catenin was specifically detected in HCC livers and not in samples from younger wild-type or Iqgap2−/− mice (without HCC). Immunoblot densitometry confirmed that the cytoplasmic accumulation of β-catenin, another indicator of Wnt/β-catenin pathway activation, was 53% greater in HCC livers (n = 5) than in those from wild-type control mice (n = 8; P = 0.06) (data not shown). Furthermore, this effect was associated with an approximately ninefold increase in cytoplasmic IQGAP1 expression and an approximately eightfold increase in levels of cyclin D1 (Fig. 5D and E), the cell cycle regulator known to be a downstream target of β-catenin (46). The expression of E-cadherin in HCC livers was ∼12% of that in wild-type livers, consistent with immunohistochemistry data. Noteworthily, more-extensive studies demonstrated enhanced (∼2.5-fold) cyclin D1 and IQGAP1 expression in the livers of younger Iqgap2−/− mice without HCC, establishing that IQGAP1 upregulation and cyclin D1 activation predated histological evidence for HCC development (Fig. 5E). To assess the timing of IQGAP1 upregulation in greater detail, liver lysates from wild-type and Iqgap2−/− mice ranging in age from 1.5 to 24 months were analyzed for IQGAP1 protein expression. As shown in Fig. 5F and G, IQGAP1 was overexpressed in all Iqgap2−/− samples compared to expression in wild-type samples, irrespective of the age of the mice, and this overexpression thus clearly predated the onset of HCC development. Therefore, while IQGAP1 is evidently upregulated in Iqgap2−/− mice with HCC, the reciprocal induction appears to be present early on (by 16 weeks of age) and is sustained with age. Remarkably, no change in IQGAP1 expression was observed in other organs known to express IQGAP1, including hearts, lungs, kidneys, and spleens, from Iqgap2−/− 4- and 12-month-old mice (data not shown).
A multiprotein β-catenin-E-cadherin-IQGAP1-IQGAP2 scaffold exists in hepatocytes.
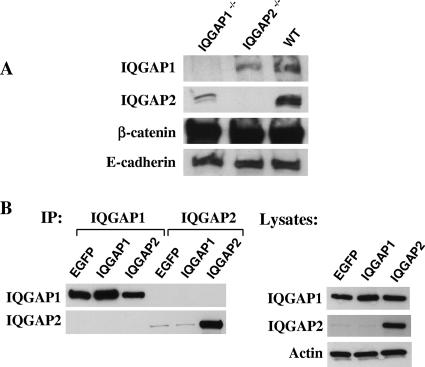
To further probe the relationships between IQGAP1, IQGAP2, β-catenin, and E-cadherin, a series of immunoprecipitation experiments were conducted using whole-liver lysates from Iqgap1−/−, Iqgap2−/−, and wild-type mice. As shown in Fig. 6A, all these proteins are clearly associated as components of a multiprotein scaffolding complex present in normal hepatocytes, as established using a β-catenin pull-down assay. In contrast, we saw no evidence for specific interaction between E-cadherin and IQGAP2 in either E-cadherin- or IQGAP2-specific immunoprecipitations (data not shown). Interestingly, in the β-catenin pull-down assays of Iqgap1−/− and Iqgap2−/− livers, β-catenin was associated with less IQGAP2 and IQGAP1, respectively, than in assays of wild-type livers, suggesting that IQGAP1 and IQGAP2 were functionally cooperative within this complex. Using normal hepatocytes, however, neither IQGAP1- nor IQGAP2-specific pull-down assays coprecipitated the corresponding homologous protein (data not shown). Transient transfections of COS1 cells (which endogenously express IQGAP1) were subsequently used as an in vitro model system to specifically address the existence of putative IQGAP1-IQGAP2 heterodimers. As evident in Fig. 6B, we were unable to pull down IQGAP1 or IQGAP2 by using reciprocal immunoprecipitations. Thus, while IQGAP1 and IQGAP2 are normally found as components of a broader quarternary scaffolding complex in hepatocytes, there appears to be no evidence that their presence within this scaffold occurs via direct (heterodimeric) interactions.
FIG. 6.
Molecular interactions involving IQGAPs, β-catenin, and E-cadherin. (A) Immunoprecipitations were completed using liver lysates (20 mg of total protein per sample) from 12-month-old wild-type (WT), Iqgap1−/−, or Iqgap2−/− mice and a protein G agarose-conjugated anti-β-catenin antibody (5 μg per sample). Complexes were fractionated by 4 to 15% gradient SDS-PAGE and immunodetected using antibodies specific to IQGAP1, IQGAP2, β-catenin, and E-cadherin. Results represent one complete set of experiments reproduced on two different occasions. (B) Immunoprecipitation (IP) experiments using COS1 lysates (20 mg of total protein per sample) transiently expressing enhanced green fluorescent protein (EGFP) as a control or human IQGAP1 or IQGAP2 (24 h posttransfection) were completed using protein G agarose-conjugated antibodies (5 μg per sample) directed against IQGAP1 or IQGAP2. Immunodetection was completed using the same antibodies. An immunoblot of corresponding lysates is shown to document protein expression. Note the high level of endogenous IQGAP1 expression in COS1 cells transfected with enhanced green fluorescent protein, with no evidence of IQGAP1-IQGAP2 complexes. Results represent one complete set of experiments reproduced on three different occasions.
IQGAP1 expression is necessary for complete penetrance of the HCC phenotype.
While the data cited above suggest that both IQGAPs associate with the β-catenin-E-cadherin complex, it remained unclear if HCC development results because of IQGAP2 deficiency per se or unattended downstream effects of upregulated IQGAP1 occurring in conjunction with the destabilization of the E-cadherin-β-catenin axis. To specifically address this issue, we postulated that if IQGAP1 upregulation is indeed causally implicated in HCC development in Iqgap2−/− mice, Iqgap1 Iqgap2 double-null mice would be protected against HCC; furthermore, such a result would suggest that the molecular mechanism for HCC formation in Iqgap2−/− mice is due to unattended IQGAP1 function occurring in conjunction with β-catenin activation.
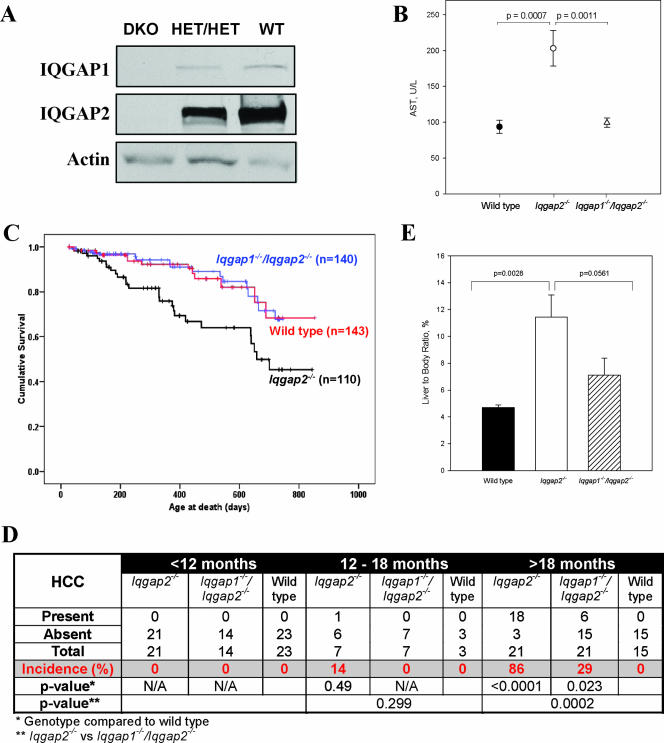
Accordingly, we generated Iqgap1−/− Iqgap2−/− mice for more-detailed analyses, assessing the development of HCC as the primary end point (more-detailed characterization of these mice will be presented elsewhere). A lack of hepatic IQGAP1 and IQGAP2 expression was clearly evident by immunoblot analysis, with residual IQGAP1 and IQGAP2 expression in Iqgap1+/− Iqgap2+/− double heterozygotes (Fig. 7A). Progressive, age-dependent increases in hepatic AST levels in 18- to 24-month-old Iqgap2−/− mice were demonstrated (compare results in Fig. 7A to Fig. 2A results for 4- and 8-month-old mice), with normal levels found in Iqgap1−/− Iqgap2−/− mice of this age group (Fig. 7B). Kaplan-Meier survival curves for mice up to 24 months of age confirmed the progressive increase in the death rate of Iqgap2−/− mice compared to that of wild-type control mice (Fig. 7C). Statistically significant (P < 0.05) separation of these survival curves was first evident by 200 days after birth, with progressive divergence over time. Entirely consistent with the normalization of the hepatic enzyme defect, Iqgap1−/− Iqgap2−/− mice had survival durations that were essentially identical to those of wild-type controls and were clearly improved compared to those of Iqgap2−/− mice.
FIG. 7.
Results from comparative studies of Iqgap2−/− and Iqgap1−/− Iqgap2−/− mice. (A) Immunoblots of liver lysates (30 μg/lane) from age-matched (4-month-old) Iqgap1−/− Iqgap2−/− (double-knockout [DKO]), Iqgap1+/− Iqgap2+/− double-heterozygote (HET/HET), and wild-type (WT) mice were obtained using either anti-IQGAP1 or anti-IQGAP2 antibodies. Antibody against actin was used to document the loading of equal amounts of protein. (B) AST levels in sera from 18- to 24-month-old wild-type (n = 12), Iqgap2−/− (n = 14), and Iqgap1−/− Iqgap2−/− (n = 23) mice were determined; data are presented as the means ± SEM, and P values are indicated. (C) Aggregate Kaplan-Meier survival curves for wild-type, Iqgap2−/−, and Iqgap1−/− Iqgap2−/− mice are displayed, with cohort numbers in parentheses; intergroup Mantel-Cox analyses demonstrated highly significant survival-rate differences between Iqgap2−/− and either wild-type or Iqgap1−/− Iqgap2−/− mice (P = 0.002), with no difference in survival rates between wild-type and Iqgap1−/− Iqgap2−/− mice. (D) Incidence of HCC among wild-type, Iqgap2−/−, and Iqgap1−/−/Iqgap2−/− mice of different ages as established by liver histology (P values are shown). N/A, not applicable. (E) Liver weights as percentages of total body weights of wild-type mice (n = 18) and Iqgap2−/− (n = 10) and Iqgap1−/− Iqgap2−/− (n = 6) mice affected with HCC. The bar graph shows means ± SEM, and P values are displayed.
Random necroscopy of Iqgap2−/− mice established that HCC was not found in mice younger than 12 months of age (n = 21). To more precisely delineate the incidence and characteristics of HCC in Iqgap2−/− and Iqgap1−/− Iqgap2−/− mice, livers from mice aged 18 to 24 months were isolated. The improved overall survival of Iqgap1−/− Iqgap2−/− mice compared with that of Iqgap2−/− mice occurred in parallel with lower HCC incidence in Iqgap1−/− Iqgap2−/− mice than in Iqgap2−/− mice (as assessed by both macroscopic and microscopic analyses), although HCC still occurred at a frequency greater than that seen in wild-type controls (Fig. 7D). As determined by liver-to-body-weight ratios used as a quantitative parameter for HCC mass, the mean weight of Iqgap2−/− mouse livers affected with HCC was significantly increased compared to that of livers from wild-type controls. Masses of HCCs from Iqgap1−/− Iqgap2−/− mice (when HCCs were identified) were smaller than those of HCCs found in Iqgap2−/− mice (Fig. 7E), yet the histological characteristics of the HCCs were similar to those of HCCs from Iqgap2−/− mice (data not shown). These data suggest that the inactivation of IQGAP1 in mouse liver impairs tumorigenesis caused by IQGAP2 deficiency.
DISCUSSION
Iqgap2 deficiency in mice is associated with a high incidence of age-dependent HCC development, establishing a novel function for IQGAP2 as a tumor suppressor causally implicated in a distinct cancer subtype linked to the Wnt/β-catenin signaling pathway. HCC in Iqgap2−/− mice is characterized by the loss of E-cadherin from plasma membranes, an increase in β-catenin expression, the translocation of β-catenin to the cytoplasm, and the overexpression of the β-catenin nuclear target cyclin D1. The evolution of HCC in Iqgap2-deficient mice recapitulates that of the human counterpart, proceeding through stepwise phases of hepatocellular apoptosis associated with mitochondrial damage to proliferation with polyploidization and culminating in HCC development (16). Furthermore, the presence of elevated cyclin D1 in younger mice without HCC suggests the development of an early preneoplastic state in Iqgap2−/− livers. Although Iqgap2 mRNA was initially described as liver specific (8), more recent data have demonstrated that it is generally expressed late in development (after embryonic day 15), with postnatal expression found not only in liver tissue (its predominant organ site), but also in kidney, prostate, and thyroid tissue and salivary glands (14). No pathological defects in any of these other tissues from Iqgap2−/− mice were identified. Thus, while various HCC-related transgenic mouse models have been described previously (see the Mouse Tumor Biology Database [http://tumor.informatics.jax.org/mtbwi/]), the restricted phenotype makes the Iqgap2−/− mouse model a valuable conventional knockout model of HCC, unassociated with preexisting steatosis or viral or other toxic exposures.
What is the relationship between IQGAP1 and IQGAP2 in HCC development? In normal hepatocytes, IQGAP2 is the predominant one of the two homologues, although IQGAP1 is reciprocally induced in Iqgap2−/− livers. This induction was readily evident by 16 weeks of age (preceding the onset of HCC) and continued throughout the lifetimes of the Iqgap2−/− mice. As importantly, our data on HCC development in Iqgap1−/− Iqgap2−/− mice suggest that maximal phenotypic penetrance is IQGAP1 dependent. Thus, although HCC did develop in Iqgap1−/− Iqgap2−/− mice, the incidence was considerably lower (29%) than the 86% incidence in Iqgap2−/− mice in the age group of 18 months and older. Furthermore, the overall aggressiveness of hepatic cancer in Iqgap1−/− Iqgap2−/− mice was diminished compared to that in Iqgap2−/− mice, as evidenced by smaller HCC tumors than those in Iqgap2−/− mice, coupled with cumulative survival rates that were indistinguishable from those of wild-type controls. Thus, despite the homology of IQGAP1 and IQGAP2, these collective observations suggest that these proteins retain functionally divergent (and nonredundant) roles in hepatocellular carcinogenesis. These data are consistent with results in previous reports demonstrating IQGAP1 overexpression in colon cancer (37), gastric cancer (44), and metastatic melanoma (11), although alterations of IQGAP2 expression were not reported previously. Notably, the knockout of murine IQGAP1 does not result in altered levels of IQGAP2 (30), although Iqgap1−/− mice displayed late-onset gastric hyperplasia. Progression to cancer was not evident, again reinforcing the functional divergence of these homologues.
Although IQGAP1 and IQGAP2 were collectively identified within the broader scaffolding complex comprising E-cadherin and β-catenin, there was no evidence that IQGAP1's modulatory effect on HCC development was mediated by direct interaction with IQGAP2. Immunoprecipitation experiments conducted with wild-type and null (Iqgap1−/− and Iqgap2−/−) hepatocytes confirmed the direct association of IQGAP2 with the β-catenin-E-cadherin complex and demonstrated this association to be unrelated to the presence of IQGAP1 (although there was a suggestion of IQGAP1-dependent functional binding cooperativity). Nonetheless, results from both hepatocyte and in vitro studies with COS1 cells gave no evidence for direct association between IQGAP1 and IQGAP2. Thus, despite evidence for the presence of IQGAP1 homodimers that may regulate protein function (3, 40), our data do not support the presence of direct IQGAP1-IQGAP2 interactions (i.e., heterodimers) that could modulate function via dominant negative effects. While these conclusions are based on results from studies completed with fasting animals, we cannot exclude the possibility that functionally dynamic changes within the scaffolding complex occur during times of altered hepatic metabolic states.
The overexpression of IQGAP1 was associated with acquired mutations in β-catenin, and the accumulation of dephosphorylated (active) β-catenin was specifically detected in HCC livers and not in livers from younger wild-type or Iqgap2−/− mice (without HCC). These data are consistent with previous observations that 26% of human HCCs and 50% of mouse HCCs have β-catenin-activating mutations, located predominantly in the potential GSK-3β phosphorylation site (15). Interestingly, IQGAP1 is known to display distinct effects on β-catenin function by inhibiting the ability of β-catenin to promote cell-cell adhesion (28). Furthermore, IQGAP1 overexpression in SW480 colon carcinoma cells stimulates the translocation of β-catenin to the nucleus, with a concomitant increase in transcriptional activity (6). Nonetheless, β-catenin overexpression appears to be insufficient for the development of HCC (9), suggesting that the coordinated overexpression and translocation of IQGAP1 and β-catenin may act in concert to promote carcinogenesis, primarily in the setting of IQGAP2 deficiency. Furthermore, it is intriguing that IQGAP1 has recently been identified as an RNA binding protein with the capacity to modulate gene expression (51), although whether isolated IQGAP1 overexpression is sufficient for HCC development remains unestablished.
Iqgap2−/− hepatocytes display an interesting structural defect in mitochondrial cristae occurring in concert with apoptosis and hepatic enzyme release. Neither the enzyme release nor the mitochondrial defects were evident in Iqgap1−/− Iqgap2−/− mice. Although cristae in Iqgap2−/− mice appeared to be shortened compared to those in wild-type mice (with vast portions of matrix devoid of cristae altogether), these structural defects were unassociated with mitochondrial genome integrity and mtPTP function (Fig. 2 and 3). Based on retained functional mitochondrial integrity, we hypothesize that the structural defects occur as phenomena secondary to an apoptotic stimulus in Iqgap2−/− hepatocytes. There is no evidence that IQGAP2 may be a structural component of hepatocyte mitochondria (4). Moreover, normal β-oxidation levels in Iqgap2−/− livers (our unpublished observations) indirectly confirm the absence of excessive reactive oxygen species formation (49), since the induction of the β-oxidation pathway is a significant source of reactive oxygen species. Thus, the structural defect in Iqgap2−/− mitochondria is unlikely to be causally implicated in HCC development but rather to be a secondary event. Of note, identical mitochondrial defects linked to apoptotic triggers have been observed in livers of mice transgenic for the hepatitis C virus core gene, with the subsequent development of HCC by 16 months of age (36).
Therapeutic options for the control of the Wnt/β-catenin signaling pathway are currently limited. One of the promising approaches is the employment of small molecule antagonists that reduce β-catenin interaction with its binding partners (further reviewed in references 5 and 29). Our studies using a mouse model offer two novel (but functionally opposing) candidate genes for targeted therapy of HCC. If our results are confirmed for humans, means of abolishing IQGAP1 or enhancing IQGAP2 function and/or critical interactions would represent logical approaches for targeted therapy of human HCC.
Acknowledgments
We thank Thomas Rosenquist (Transgenic Mouse Facility, SUNY—Stony Brook) for assistance in generating the Iqgap2−/− mice, Bernard Lane for expert interpretation of electron microscopy images, Susan Van Horn (Central Microscopy Imaging Center, SUNY—Stony Brook) and Gregory Rudomen for electron microscopy technical assistance, John Chen and Emily Huang for statistical analysis, and Lesley Scudder and Jean Wainer for excellent technical support.
This work was supported by NIH grants DK62040 (V.A.S.) and HL49141 (W.F.B.).
Footnotes
Published ahead of print on 7 January 2008.
REFERENCES
- 1.Adachi, H., Y. Takahashi, T. Hasebe, M. Shirouzu, S. Yokoyama, and K. Sutoh. 1997. Dictyostelium IQGAP-related protein specifically involved in the completion of cytokinesis. J. Cell Biol. 137891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahou, W. F., L. Scudder, D. Rubenstein, and J. Jesty. 2004. A shear-restricted pathway of platelet procoagulant activity is regulated by IQGAP1. J. Biol. Chem. 27922571-22577. [DOI] [PubMed] [Google Scholar]
- 3.Bashour, A., A. Fullerton, M. J. Hart, and G. S. Bloom. 1997. IQGAP1, a Rac- and Cdc42-binding protein, directly binds and cross-links actin. J. Cell Biol. 1371555-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu, S., E. Bremer, C. Zhou, and D. F. Bogenhagen. 2006. MiGenes: a searchable interspecies database of mitochondrial proteins curated using gene ontology annotation. Bioinformatics 22485-492. [DOI] [PubMed] [Google Scholar]
- 5.Breuhahn, K., T. Longerich, and P. Schirmacher. 2006. Dysregulation of growth factor signaling in human hepatocellular carcinoma. Oncogene 253787-3800. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, M. W., Z. Li, and D. B. Sacks. 2002. IQGAP1-mediated stimulation of transcriptional co-activation by beta-catenin is modulated by calmodulin. J. Biol. Chem. 2777453-7465. [DOI] [PubMed] [Google Scholar]
- 7.Briggs, M. W., and D. B. Sacks. 2003. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 4571-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brill, S., S. Li, C. Lyman, D. Church, J. Wasmuth, L. Weissbach, A. Bernards, and A. Snijders. 1996. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol. Cell. Biol. 164869-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cadoret, A., C. Ovejero, S. Saadi-Kheddouci, E. Souil, M. Fabre, B. Romagnolo, A. Kahn, and C. Perret. 2001. Hepatomegaly in transgenic mice expressing an oncogenic form of beta-catenin. Cancer Res. 613245-3249. [PubMed] [Google Scholar]
- 10.Carugo, K. D., S. Banuelos, and M. Saraste. 1997. Crystal structure of a calponin homology domain. Nat. Struct. Biol. 4175-179. [DOI] [PubMed] [Google Scholar]
- 11.Clark, E. A., T. R. Golub, E. S. Lander, and R. O. Hynes. 2000. Genomic analysis of metastasis reveals an essential role for RhoC. Nature 406532-535. [DOI] [PubMed] [Google Scholar]
- 12.Colnot, S., T. Decaens, M. Niwa-Kawakita, C. Godard, G. Hamard, A. Kahn, M. Giovannini, and C. Perret. 2004. Liver-targeted disruption of Apc in mice activates beta-catenin signaling and leads to hepatocellular carcinomas. Proc. Natl. Acad. Sci. USA 10117216-17221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crompton, M. 1999. The mitochondrial permeability transition pore and its role in cell death. Biochem. J. 341233-249. [PMC free article] [PubMed] [Google Scholar]
- 14.Cupit, L. D., V. A. Schmidt, F. Miller, and W. F. Bahou. 2004. Distinct PAR/IQGAP expression patterns during murine development: implications for thrombin-associated cytoskeletal reorganization. Mamm. Genome 15618-629. [DOI] [PubMed] [Google Scholar]
- 15.de La Coste, A., B. Romagnolo, P. Billuart, C. A. Renard, M. A. Buendia, O. Soubrane, M. Fabre, J. Chelly, C. Beldjord, A. Kahn, and C. Perret. 1998. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc. Natl. Acad. Sci. USA 958847-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farazi, P. A., and R. A. DePinho. 2006. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer 6674-687. [DOI] [PubMed] [Google Scholar]
- 17.Fukata, M., T. Watanabe, J. Noritake, M. Nakagawa, M. Yamaga, S. Kuroda, Y. Matsuura, A. Iwamatsu, F. Perez, and K. Kaibuchi. 2002. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell 109873-885. [DOI] [PubMed] [Google Scholar]
- 18.Gnatenko, D. V., L. D. Cupit, E. C. Huang, A. Dhundale, P. L. Perrotta, and W. F. Bahou. 2005. Platelets express steroidogenic 17beta-hydroxysteroid dehydrogenases. Distinct profiles predict the essential thrombocythemic phenotype. Thromb. Haemost. 94412-421. [DOI] [PubMed] [Google Scholar]
- 19.Gnatenko, D. V., J. J. Dunn, S. R. McCorkle, D. Weissmann, P. L. Perrotta, and W. F. Bahou. 2003. Transcript profiling of human platelets using microarray and serial analysis of gene expression. Blood 1012285-2293. [DOI] [PubMed] [Google Scholar]
- 20.Haegel, H., L. Larue, M. Ohsugi, L. Fedorov, K. Herrenknecht, and R. Kemler. 1995. Lack of beta-catenin affects mouse development at gastrulation. Development 1213529-3537. [DOI] [PubMed] [Google Scholar]
- 21.Harada, N., H. Miyoshi, N. Murai, H. Oshima, Y. Tamai, M. Oshima, and M. M. Taketo. 2002. Lack of tumorigenesis in the mouse liver after adenovirus-mediated expression of a dominant stable mutant of beta-catenin. Cancer Res. 621971-1977. [PubMed] [Google Scholar]
- 22.Hart, M. J., M. G. Callow, B. Souza, and P. Polakis. 1996. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 152997-3005. [PMC free article] [PubMed] [Google Scholar]
- 23.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6986-994. [DOI] [PubMed] [Google Scholar]
- 24.Ho, Y. D., J. L. Joyal, Z. Li, and D. B. Sacks. 1999. IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J. Biol. Chem. 274464-470. [DOI] [PubMed] [Google Scholar]
- 25.Javois, L. C. (ed.). 1994. Methods in molecular biology, vol. 34. Immunocytochemical methods and protocols. Humana Press, Totowa, NJ.
- 26.Kim, Y., R. C. Sills, and C. D. Houle. 2005. Overview of the molecular biology of hepatocellular neoplasms and hepatoblastomas of the mouse liver. Toxicol. Pathol. 33175-180. [DOI] [PubMed] [Google Scholar]
- 27.Kuroda, S., M. Fukata, K. Kobayashi, M. Nakafuku, N. Nomura, A. Iwamatsu, and K. Kaibuchi. 1996. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J. Biol. Chem. 27123363-23367. [DOI] [PubMed] [Google Scholar]
- 28.Kuroda, S., M. Fukata, M. Nakagawa, K. Fujii, T. Nakamura, T. Ookubo, I. Izawa, T. Nagase, N. Nomura, H. Tani, I. Shoji, Y. Matsuura, S. Yonehara, and K. Kaibuchi. 1998. Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin-mediated cell-cell adhesion. Science 281832-835. [DOI] [PubMed] [Google Scholar]
- 29.Lee, H. C., M. Kim, and J. R. Wands. 2006. Wnt/Frizzled signaling in hepatocellular carcinoma. Front. Biosci. 111901-1915. [DOI] [PubMed] [Google Scholar]
- 30.Li, S., Q. Wang, A. Chakladar, R. Bronson, and A. Bernards. 2000. Gastric hyperplasia in mice lacking the putative Cdc42 effector IQGAP1. Mol. Cell. Biol. 20697-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, X., F. Traganos, M. R. Melamed, and Z. Darzynkiewicz. 1995. Single-step procedure for labeling DNA strand breaks with fluorescein- or BODIPY-conjugated deoxynucleotides: detection of apoptosis and bromodeoxyuridine incorporation. Cytometry 20172-180. [DOI] [PubMed] [Google Scholar]
- 32.Lippincott, J., and R. Li. 1998. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140355-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansour, S. L., K. R. Thomas, and M. R. Capecchi. 1988. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature 336348-352. [DOI] [PubMed] [Google Scholar]
- 34.McCallum, S., W. Wu, and R. Cerione. 1996. Identification of a putative effector for Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J. Biol. Chem. 27121732-21737. [DOI] [PubMed] [Google Scholar]
- 35.Mootha, V. K., J. Bunkenborg, J. V. Olsen, M. Hjerrild, J. R. Wisniewski, E. Stahl, M. S. Bolouri, H. N. Ray, S. Sihag, M. Kamal, N. Patterson, E. S. Lander, and M. Mann. 2003. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115629-640. [DOI] [PubMed] [Google Scholar]
- 36.Moriya, K., H. Fujie, Y. Shintani, H. Yotsuyanagi, T. Tsutsumi, K. Ishibashi, Y. Matsuura, S. Kimura, T. Miyamura, and K. Koike. 1998. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 41065-1067. [DOI] [PubMed] [Google Scholar]
- 37.Nabeshima, K., Y. Shimao, T. Inoue, and M. Koono. 2002. Immunohistochemical analysis of IQGAP1 expression in human colorectal carcinomas: its overexpression in carcinomas and association with invasion fronts. Cancer Lett. 176101-109. [DOI] [PubMed] [Google Scholar]
- 38.Nakanishi-Matsui, M., Y. W. Zheng, D. Sulciner, E. Weiss, M. Ludeman, and S. Coughlin. 2000. PAR3 is a cofactor for PAR4 activation by thrombin. Nature 404609-613. [DOI] [PubMed] [Google Scholar]
- 39.Nelson, W. J., and R. Nusse. 2004. Convergence of Wnt, beta-catenin, and cadherin pathways. Science 3031483-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren, J. G., Z. Li, D. L. Crimmins, and D. B. Sacks. 2005. Self-association of IQGAP1: characterization and functional sequelae. J. Biol. Chem. 28034548-34557. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt, V. A., W. C. Nierman, D. R. Maglott, L. D. Cupit, K. A. Moskowitz, J. A. Wainer, and W. F. Bahou. 1998. The human proteinase-activated receptor-3 (PAR-3) gene. Identification within a PAR gene cluster and characterization in vascular endothelial cells and platelets. J. Biol. Chem. 27315061-15068. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt, V. A., L. Scudder, C. E. Devoe, A. Bernards, L. D. Cupit, and W. F. Bahou. 2003. IQGAP2 functions as a GTP-dependent effector protein in thrombin-induced platelet cytoskeletal reorganization. Blood 1013021-3028. [DOI] [PubMed] [Google Scholar]
- 43.Suomalainen, A., and J. Kaukonen. 2001. Diseases caused by nuclear genes affecting mtDNA stability. Am. J. Med. Genet. 10653-61. [DOI] [PubMed] [Google Scholar]
- 44.Takemoto, H., Y. Doki, H. Shiozaki, H. Imamura, T. Utsunomiya, H. Miyata, M. Yano, M. Inoue, Y. Fujiwara, and M. Monden. 2001. Localization of IQGAP1 is inversely correlated with intercellular adhesion mediated by e-cadherin in gastric cancers. Int. J. Cancer 91783-788. [DOI] [PubMed] [Google Scholar]
- 45.Tedeschi, H. 2005. Old and new data, new issues: the mitochondrial Δψ. Biochim. Biophys. Acta 1709195-202. [DOI] [PubMed] [Google Scholar]
- 46.Tetsu, O., and F. McCormick. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398422-426. [DOI] [PubMed] [Google Scholar]
- 47.Thorgeirsson, S. S., and J. W. Grisham. 2002. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 31339-346. [DOI] [PubMed] [Google Scholar]
- 48.von Ahsen, O., C. Renken, G. Perkins, R. M. Kluck, E. Bossy-Wetzel, and D. D. Newmeyer. 2000. Preservation of mitochondrial structure and function after Bid- or Bax-mediated cytochrome c release. J. Cell Biol. 1501027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallace, D. C. 2001. Mouse models for mitochondrial disease. Am. J. Med. Genet. 10671-93. [DOI] [PubMed] [Google Scholar]
- 50.Wang, S., T. Watanabe, J. Noritake, M. Fukata, T. Yoshimura, N. Itoh, T. Harada, M. Nakagawa, Y. Matsuura, N. Arimura, and K. Kaibuchi. 2007. IQGAP3, a novel effector of Rac1 and Cdc42, regulates neurite outgrowth. J. Cell Sci. 120567-577. [DOI] [PubMed] [Google Scholar]
- 51.Willingham, A. T., A. P. Orth, S. Batalov, E. C. Peters, B. G. Wen, P. Aza-Blanc, J. B. Hogenesch, and P. G. Schultz. 2005. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 3091570-1573. [DOI] [PubMed] [Google Scholar]
- 52.Yen, T. C., Y. S. Chen, K. L. King, S. H. Yeh, and Y. H. Wei. 1989. Liver mitochondrial respiratory functions decline with age. Biochem. Biophys. Res. Commun. 165944-1003. [DOI] [PubMed] [Google Scholar]