Abstract
Fission yeast cells survive loss of the telomerase catalytic subunit Trt1 (TERT) through recombination-based telomere maintenance or through chromosome circularization. Although trt1Δ survivors with linear chromosomes can be obtained, they often spontaneously circularize their chromosomes. Therefore, it was difficult to establish genetic requirements for telomerase-independent telomere maintenance. In contrast, when the telomere-binding protein Taz1 is also deleted, taz1Δ trt1Δ cells are able to stably maintain telomeres. Thus, taz1Δ trt1Δ cells can serve as a valuable tool in understanding the regulation of telomerase-independent telomere maintenance. In this study, we show that the checkpoint kinase Tel1 (ATM) and the DNA repair complex Rad32-Rad50-Nbs1 (MRN) are required for telomere maintenance in taz1Δ trt1Δ cells. Surprisingly, Rap1 is also essential for telomere maintenance in taz1Δ trt1Δ cells, even though recruitment of Rap1 to telomeres depends on Taz1. Expression of catalytically inactive Trt1 can efficiently inhibit recombination-based telomere maintenance, but the inhibition requires both Est1 and Ku70. While Est1 is essential for recruitment of Trt1 to telomeres, Ku70 is dispensable. Thus, we conclude that Taz1, TERT-Est1, and Ku70-Ku80 prevent telomere recombination, whereas MRN-Tel1 and Rap1 promote recombination-based telomere maintenance. Evolutionarily conserved proteins in higher eukaryotic cells might similarly contribute to telomere recombination.
Telomeres are specialized DNA-protein complexes at the ends of linear chromosomes and function as protective caps to prevent normal chromosomal ends from end-to-end fusions and recombinational events (13). In most eukaryotic organisms, telomeric DNA is composed of short GT-rich repeat sequences. This DNA is synthesized by telomerase, a reverse transcriptase (RT) tightly associated with its own template RNA. The very end of telomeric DNA is a 3′ single-stranded GT-rich region, known as the G-tail. When cells replicate linear chromosomes without telomerase, telomeric DNA is gradually lost due to the inability of conventional DNA polymerases to fully replicate the ends of DNA molecules. Since GT-rich telomeric repeats provide binding sites for telomere-specific proteins that are important in creating protective caps at telomeres (14), shortening or loss of telomeric repeats leads to DNA damage checkpoint activation and “repair” (fusion and recombination) of telomeric DNA ends.
Given the telomere's ability to suppress DNA repair and DNA damage checkpoint responses, one might expect that functional telomeres would have the ability to prevent detection by DNA repair and DNA damage checkpoint proteins. However, DNA double-strand break (DSB) repair protein complexes, such as Ku70-Ku80 and Mre11-Rad50-Nbs1 (MRN), are bound to telomeres and necessary for normal telomere maintenance (63). Likewise, the checkpoint sensor complexes Rad1-Rad9-Hus1, Rad17-Rfc2-5, ATM, and ATR-ATRIP associate with telomeres and contribute to telomere maintenance (46, 56, 64).
The catalytic subunit of telomerase is known as telomerase RT (TERT) (49). In the fission yeast Schizosaccharomyces pombe, TERT is encoded by trt1+ gene (45), and trt1Δ cells progressively lose their telomeric DNA. The viability of trt1Δ cells drops to the lowest level around 120 divisions after deletion of trt1+, but cells eventually recover in growth and establish survivors. Most cells survive by circularizing all chromosomes to bypass the need for telomerase, while some cells survive by maintaining their telomeres, presumably through recombination (44). The telomerase regulatory subunit Est1 is also essential for telomerase-dependent telomere maintenance (4).
Taz1 is a telomeric double-stranded DNA-binding protein in S. pombe (12) thought to be the counterpart of two mammalian telomere proteins TRF1 and TRF2 (53). Taz1 is important for recruitment of Rap1 and Rif1 to telomeres (10, 30), and deletion of the taz1+ gene leads to massive telomere elongation by telomerase (12, 44). Taz1 also promotes replication of telomeric GT-rich repeats by DNA polymerases (39), and telomeres in taz1Δ cells are less protected against fusions by nonhomologous end-joining (NHEJ) mediated by Ku70-Ku80 and ligase IV (19). Interestingly, simultaneous deletion of taz1 and trt1 genes allows cells to robustly maintain telomeres rather than circularizing chromosomes (44). However, it remained unclear how telomeres are maintained in taz1Δ trt1Δ cells.
Studies in other organisms have established that telomerase-negative cells can survive loss of telomeric repeats by mechanisms that involve homologous recombination (HR) among telomeres (16, 47, 58). In Saccharomyces cerevisiae, these recombination-based survival mechanisms have been divided into two major types: type I survivors are characterized by amplification of subtelomeric Y′ elements but have relatively short telomeric GT-rich repeats, whereas type II survivors are characterized by long telomeric GT-repeat tracts. The HR protein Rad52 is required for generation of both types of survivors (58), whereas Rad51, Rad54, and Rad57 are essential for generation of type I survivors. The Mre11-Rad50-Xrs2 (MRX) complex and Sgs1 DNA helicase are important for generation of type II survivors (27, 57). Furthermore, Tel1 (ATM) and Mec1 (ATR) kinases play redundant roles in generating type II survivors (60).
In the present study, we identify Rad22 (an ortholog of S. cerevisiae Rad52), MRN, Tel1 (ATM), and Rap1 as contributing positively to recombination-based telomere maintenance in taz1Δ trt1Δ cells. Unexpectedly, efficient Taz1-dependent binding of Rap1 to telomeres is not required to promote recombination-based telomere maintenance. We also show that TERT-Est1, Taz1, and Ku70-Ku80 represent three redundant but interdependent mechanisms that operate at telomeres in preventing telomere recombination. Surprisingly, the ability of TERT to negatively regulate telomere recombination is not dependent on its RT activity and requires only the N-terminal half of the protein.
MATERIALS AND METHODS
Yeast strains.
The fission yeast strains used in the present study were constructed by standard techniques (1) and are listed in Table S1 in the supplemental material. Original sources for most mutations and epitope-tagged genes utilized in the present study were previously described (46), except for est1Δ (4), rap1Δ (30), and rap1-HA (30). For liquid growth curve experiments, appropriate heterozygous diploid cells were sporulated on ME plates, and the resulting tetrads were dissected on a YES plate (1). Genotypes of haploid strains were then determined based on growth on PMG minimal medium lacking selective amino acids. Triple mutant strains (taz1Δ trt1Δ plus additional deletion) used in Fig. 2 were generated by deleting the indicated gene from taz1Δ trt1Δ survivor cells by transformation of a deletion construct DNA or by genetic cross of a taz1Δ trt1Δ survivor strain with a strain carrying the desired deletion mutation. Triple mutant cells generated by transformation of taz1Δ trt1Δ survivor cells may not be epigenetically identical to triple mutant cells generated by genetic cross. However, for taz1Δ trt1Δ rad32Δ cells, strains produced by transformation and strains produced by genetic cross all generated survivor cells carrying circular chromosomes after repeated restreaking on agar plates. The remaining triple mutant strains (taz1Δ trt1Δ rad22Δ, taz1Δ trt1Δ rad50Δ, taz1Δ trt1Δ nbs1Δ, taz1Δ trt1Δ tel1Δ, and taz1Δ trt1Δ rad3Δ) were generated by genetic crosses, whereas taz1Δ trt1Δ pku70Δ and taz1Δ trt1Δ est1Δ strains were generated by transformation. taz1Δ trt1Δ pku70Δ tel1Δ strains were generated by transforming taz1Δ trt1Δ pku70Δ cells with a tel1Δ construct. taz1Δ est1Δ strains were generated by transforming taz1Δ cells with an est1Δ construct. taz1Δ pku70Δ est1Δ strains were generated by transforming taz1Δ pku70Δ cells with an est1Δ construct. rap1Δ trt1Δ cells were generated either by crossing trt1Δ cells carrying pWH5-trt1+ plasmid with rap1Δ cells or by transforming rap1Δ cells with a trt1Δ construct. taz1Δ trt1Δ rap1Δ strains were generated by transforming taz1Δ trt1Δ cells with a rap1Δ construct. Except for liquid growth curve experiments, cells were extensively restreaked on plates to ensure that cells reached their terminal phenotype before preparation of the chromosomal DNA plugs or genomic DNA.
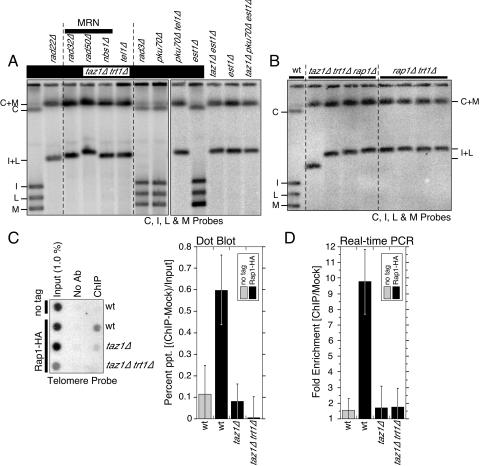
FIG. 2.
Determination of genes required for telomere maintenance in taz1Δ trt1Δ cells. (A) Rad22 and MRN-Tel1 are required for telomere maintenance in taz1Δ trt1Δ cells, whereas Rad3, Ku70, and Est1 are dispensable. Multiple independently derived clones were analyzed by PFGE after extensive restreaking on YES agar plates, but only one representative clone per genotype is shown. Chromosomal plugs were prepared from cells that were grown at least 150 generations after the strains were generated. (B) Rap1 is essential for telomere maintenance in taz1Δ trt1Δ or trt1Δ cells. Independently derived strains were restreaked extensively on YES agar plates before analysis by PFGE. (C and D) Rap1 association with telomeres requires Taz1 in both wild-type (trt1+) and trt1Δ cells. Telomere association of hemagglutinin-tagged Rap1 was determined by ChIP assay using dot blot hybridization with a telomeric probe (C) or quantitative PCR with primers against the TAS1 sequence (D). Error bars represent the standard deviation from at least three independent experiments.
Plasmids.
Plasmid pREP81x-taz1+ carries taz1+ gene behind the weakest version of nmt1 promoter, S. cerevisiae LEU2 gene, and S. pombe autonomous replication sequence (ars1+). Plasmid pKAN-trt1+ contains the ∼5.5-kb S. pombe genomic KpnI fragment bearing the trt1+ gene, kanMX4 marker, and S. pombe ars1+ (23). Plasmids pKAN-trt1-D590A, pKAN-trt1-D743A, pKAN-trt1-ΔNsi, pKAN-trt1-ΔPac, and pKAN-trt1-Δ[Nde-Xho] are essentially the same as pKAN-trt1+, except that they carry the indicated mutant alleles. Plasmids pKAN-trt1:Cmyc9, pKAN-trt1-D590A:Cmyc9, and pKAN-trt1-D743A:Cmyc9 express the indicated Trt1 alleles with the myc9 tag at the C terminus. Plasmid pWH5-trt1+ contains the ∼11-kb S. pombe genomic HindIII fragment bearing the trt1+ gene, S. cerevisiae LEU2 gene, and S. cerevisiae 2μ replication origin.
Cell growth analysis.
Liquid culture growth curve experiments were performed essentially as previously described (46). After cell cultures were used to inoculate 4 × 104 cells/ml in fresh YES liquid medium, the remaining cells were collected by centrifugation, washed once in SP1 buffer (1), and frozen at −80°C for later preparation of agarose plugs.
Pulsed-field gel electrophoresis (PFGE).
Chromosomal DNA samples were prepared in agarose plugs, and NotI-digested DNA samples were fractionated with the CHEF-DR III system (Bio-Rad) as previously described (46). The telomeric repeat probe and C, I, L, and M probes were prepared as previously described (44).
ChIP analysis.
Exponentially growing cells were processed for chromatin immunoprecipitation (ChIP) analysis as previously described (46, 48). Protein G-Sepharose beads (GE Amersham) were added to whole-cell extracts preincubated with monoclonal antibody 9B11 (anti-myc), 12CA5 (anti-HA), or no antibody (mock). After extensive washes, bead-bound DNA was recovered by using Chelex-100 resin (Bio-Rad) (48). Recovered DNA from ChIP was analyzed by SYBR green-based real-time PCR (Bio-Rad) using BAM136 and BAM137 primers (TAS1 region [46]). Fold enrichment values were calculated based on difference in Ct values between ChIP (antibody) and mock (no antibody) samples. For dot blot analysis, ChIP and input DNA samples were denatured by boiling at 100°C for 10 min in 0.4 M NaOH and 10 mM EDTA, snap chilled on ice, and blotted onto a Hybond XL membrane (GE Amersham Biosciences). Dot blots were then hybridized with a telomeric probe, and hybridization signals were quantified by using ImageQuant software. Percent precipitated DNA for ChIP samples were calculated as 100 × (ChIP - mock)/input.
RESULTS
taz1Δ trt1Δ cells show accelerated telomere fusions compared to trt1Δ cells.
Haploid taz1Δ trt1Δ cells generated by sporulation of heterozygous diploid taz1Δ/taz1+ trt1Δ/trt1+ cells reproducibly went through an initial low-viability phase soon after germination (44) (Fig. 1D; see also Fig. S1D in the supplemental material), although there was no discernible difference in the initial size of colonies or spore viability (see Fig. S1A and B in the supplemental material). In contrast, trt1Δ cells initially grew well, reached their slowest growth phase around day 10, and then recovered (Fig. 1B and C; see also Fig. S1C in the supplemental material). In both cases, reduction in cell growth rate was accompanied by an increase in appearance of highly elongated cells that can no longer divide and frequently contain fragmented DNA stuck between two dividing nuclei (44; data not shown).
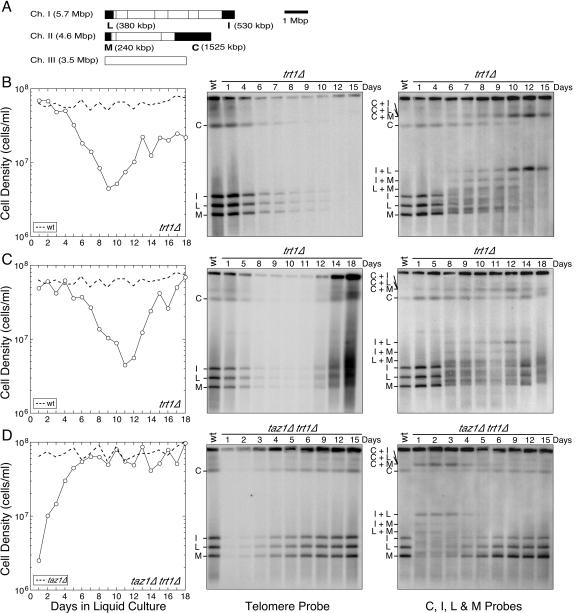
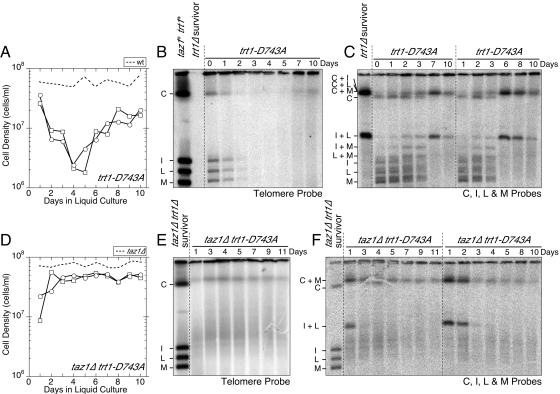
FIG. 1.
PFGE analysis for trt1Δ and taz1Δ trt1Δ cells. (A) NotI restriction map of S. pombe chromosomes. The telomeric fragments C, I, L, and M are filled in black. (B to D) trt1Δ/trt1+ or taz1Δ/taz1+ trt1Δ/trt1+ diploid strains were sporulated, and trt1Δ (B and C) or taz1Δ trt1Δ (D) cells were selected. For trt1Δ cells, two independent cultures, which produced survivors with an intermediate growth rate (B) or a wild-type-like growth rate (C), are shown. The left panels show the growth characteristics of cells after germination in liquid culture. For reference, results from growth curve experiments for wild-type (trt1+) or taz1Δ cells are also plotted (dotted lines). The middle panels show PFGE results for NotI-digested S. pombe chromosomal DNA hybridized with telomere GT-rich repeat specific probe. The right panels show PFGE results for NotI-digested S. pombe chromosomal DNA hybridized with probes for the C, I, L, and M bands.
To better understand the cause of the low viability exhibited by taz1Δ trt1Δ and trt1Δ cells, we analyzed changes in telomere stability by PFGE of NotI-digested chromosomal DNA during the course of liquid cell growth experiments (Fig. 1). When chromosomal DNA of trt1Δ cells was analyzed by Southern blotting with probes corresponding to telomeric fragments (C, I, L, and M probes, Fig. 1A), a heterogeneous distribution of slower-migrating bands gradually appeared as cells divided and became sick (Fig. 1B and C, right panels). Based on sequential hybridization with individual probes, we confirmed that some of these altered mobility bands represent inter- and intrachromosomal fusions (data not shown). These fusion bands no longer contain telomeric GT-rich repeats (Fig. 1B and C, middle panels), indicating that fusion events might occur after the total loss of telomeric repeats. Alternatively, the fusion events could be responsible for the loss of telomeric repeats. In any case, interchromosomal fusions, which create dicentric chromosomes, are detrimental to cell viability. Therefore, nondisjunction of fused chromosomes and/or fusion-breakage cycles, caused by uncapped chromosomal ends, are likely to be responsible for a loss of cell viability in trt1Δ cells. We also found that when our blots are hybridized to C, I, L, and M probes, additional diffused bands are detected just above the I, L, and M bands when the cells are in the low-viability phase (Fig. 1B and C). We are uncertain of the exact identities of these bands, but they do not seem to contain much telomeric GT-rich repeat sequences since they are not detected by a telomere probe. These diffused bands might be created by fusion-breakage cycles, or they might represent DNA repair intermediates, which could run abnormally on the pulsed-field gel due to their structures.
Although the decline in growth rate for independent trt1Δ liquid cultures was very reproducible, the extent of recovery was not always the same (Fig. 1B and C; see Fig. S1C in the supplemental material) (44, 46). For the majority of cultures tested, survivors initially grew at a reduced rate compared to trt1+ cells and had circular chromosomes (Fig. 1B). Fewer cultures generated survivors that grew similar to trt1+ cells and maintained linear chromosomes, although telomeric bands became much more heterogeneous in later generations (Fig. 1C). Even when cultures were able to establish fast-growing survivors, the growth rate was not always stable, and these cultures sometimes went through a second phase of low viability and slow growth (44; data not shown). Conversely, cultures containing predominantly circular chromosome survivors were often able to improve to a faster-growing state with very diffused weak telomeric bands after an extended period of selection beyond 2 to 3 weeks (data not shown). Such recovery is possible since extremely rare but faster-growing survivors within a population can eventually take over more common but slower-growing survivors in a competitive liquid culture growth environment (3). When telomere repeat length and telomere-associated sequence (TAS) pattern were analyzed by Southern blot hybridization of conventional agarose gels, serially diluted trt1Δ liquid cultures often displayed telomere elongation reminiscent of type II survivors in S. cerevisiae (see Fig. S4 and S5 in the supplemental material). However, as we have previously reported, these survivors are not always stable and cultures often went through repeated rounds of telomere attrition and low-viability phases (44; data not shown).
For taz1Δ trt1Δ cells, a heterogeneous distribution of slower-migrating NotI telomeric bands, characteristic of inter- and intrachromosome fusions, was observed at the very early low-viability phase (Fig. 1D, right panel, days 1 to 3), and telomeric repeats were lost from these fused telomeric fragments (Fig. 1D, middle panel). On the other hand, taz1Δ trt1Δ survivors that arose later (days 9 to 15) grew very well and were able to stably maintain linear chromosomes with sharp distinct bands for telomeric NotI fragments, which also contained stable telomeric repeats (Fig. 1D; see Fig. S1D in the supplemental material). Thus, the absence of Taz1 in trt1Δ cells resulted in a much earlier occurrence of telomere fusions, but taz1Δ trt1Δ cells were eventually able to reproducibly establish survivors with very stable telomeres. When these taz1Δ trt1Δ survivor cultures were analyzed for telomere length and the TAS region, they were found to contain heterogeneous telomeres and amplified TAS1 sequences (see Fig. S4 and S5 in the supplemental material). Since these cultures contain elongated telomeres, these survivors could be similar to type II survivors in budding yeast. On the other hand, amplified TAS is reminiscent of type I survivors in budding yeast. In any case, these observations indicated that Taz1 and TERT could have partially redundant roles in protecting telomeres from immediate fusions and in regulating recombination events at telomeres.
DNA repair and checkpoint proteins affect telomere maintenance in taz1Δ trt1Δ cells.
Although trt1Δ survivor cells with linear chromosomes can be obtained, they often spontaneously circularize their chromosomes. Therefore, it was difficult to establish genetic requirements for telomerase-independent telomere maintenance in trt1Δ cells. In contrast, taz1Δ trt1Δ survivor cells are able to stably maintain telomeres, and thus they can be utilized as a valuable tool in understanding the regulation of telomerase-independent telomere maintenance. In this setup, mutations that lead to the conversion of chromosome structures from linear to circular could identify genes involved in the promotion of telomerase-independent telomere maintenance and/or in protection of telomeres against telomere fusions. One should keep in mind, however, that the genetic requirement for telomere recombination in taz1Δ trt1Δ and trt1Δ cells may not be completely identical.
When the gene for the HR protein Rad22 (Rad52) was eliminated from taz1Δ trt1Δ cells, telomeric NotI bands (C, I, L, and M) were converted to C+M and I+L bands indicative of circular chromosomes (Fig. 2A). These fusion bands also lacked telomeric repeat sequences (data not shown). Therefore, Rad22-mediated HR is required for telomerase-independent telomere maintenance, much as in S. cerevisiae. In contrast, the NHEJ protein Ku70 was not required to maintain telomeres in taz1Δ trt1Δ cells since elimination of Ku70 did not lead to chromosome circularization (Fig. 2A). We can rule out the possibility that Ku70 is essential for chromosome end fusion, making it impossible for taz1Δ trt1Δ pku70Δ to circularize their chromosomes, based on observations that circular chromosomes can still be generated in trt1Δ pku70Δ cells (3), taz1Δ trt1Δ pku70Δ tel1Δ cells (Fig. 2A), taz1Δ pku70Δ est1Δ cells (Fig. 2A), or taz1Δ trt1Δ pku70Δ cells carrying a taz1+ plasmid (see Fig. 5C). Interestingly, after Ku is eliminated, the average telomere size appeared to increase compared to the parental taz1Δ trt1Δ cells (see Fig. S6 in the supplemental material). Such a phenotype may be an indication that recombination-based telomere maintenance of taz1Δ trt1Δ cells becomes even more efficient in the absence of Ku.
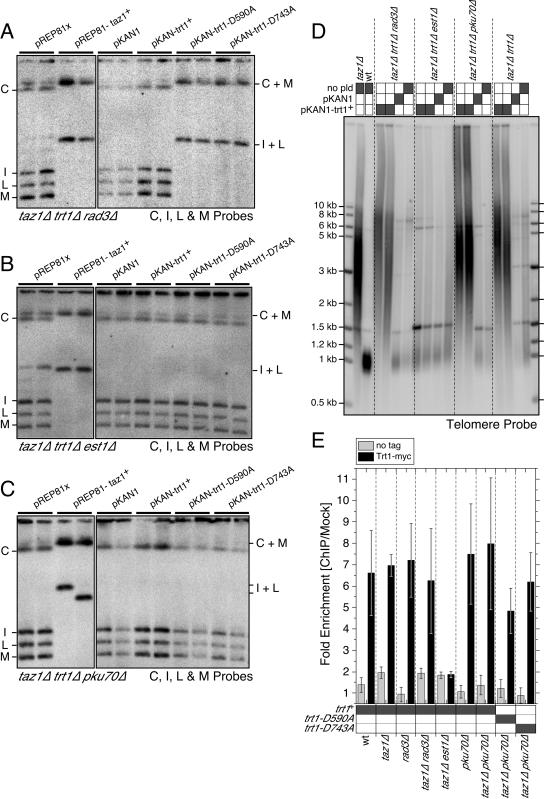
FIG. 5.
Est1 and Ku70 are required for catalytically inactive TERT to inhibit recombination-based telomere maintenance in taz1Δ trt1Δ cells. (A to C) PFGE analysis of taz1Δ trt1Δ rad3Δ, taz1Δ trt1Δ est1Δ, or taz1Δ trt1Δ pku70Δ cells transformed with indicated plasmids after extensive restreaking on appropriate selection plates. (D) Telomere length analysis for the indicated strains. After digestion with EcoRI, genomic DNA was fractionated on a 1% agarose gel and processed for Southern blot analysis with a telomere probe. (E) Trt1 association with telomeres requires Est1, but not Ku70 or Rad3. Telomere association of myc-tagged Trt1 was determined by ChIP assay using quantitative PCR against the TAS1 sequence. For taz1Δ pku70Δ cells, the recruitment of myc-tagged catalytically dead Trt1 (Trt1-D590A and Trt1-D743A) was also examined. Error bars represent the standard deviation from at least three independent experiments.
Next, we found that elimination of any of the MRN complex components (Rad32, Rad50, and Nbs1) or Tel1 leads to chromosome circularization and the complete loss of telomeric repeats in taz1Δ trt1Δ cells (Fig. 2A; data not shown). Therefore, the MRN complex and Tel1 contribute positively to telomerase-independent telomere maintenance. They may be important either for telomere-telomere recombination or the protection of telomeres from fusions. It was previously shown that the MRN complex and Tel1 work in the same telomere maintenance pathway in fission yeast (8, 46) and that Nbs1-Tel1 interaction recruits Tel1 to sites of DNA damage (67).
Because S. cerevisiae Tel1 has been shown to be important in the prevention of NHEJ-dependent telomere fusions in the absence of telomerase (9), we also examined whether the chromosome circularization phenotype we observed for taz1Δ trt1Δ tel1Δ cells depends on Ku by deleting tel1+ gene from taz1Δ trt1Δ pku70Δ survivor cells. Since taz1Δ trt1Δ pku70Δ tel1Δ cells still circularized their chromosomes (Fig. 2A), Tel1 is likely to prevent telomere fusions by promoting a mechanism(s) essential for telomere maintenance in taz1Δ trt1Δ cells rather than inhibiting Ku-dependent NHEJ at telomeres. On the other hand, a previous study has found that telomere-telomere fusions can be generated by a Ku-independent but ligase IV-dependent mechanism (37). Therefore, it is still possible that Tel1 may be involved in the inhibition of ligase IV-dependent fusions in taz1Δ trt1Δ cells.
In S. pombe, the MRN-Tel1 pathway and the Rad3-Rad26 (corresponds to S. cerevisiae Mec1-Ddc2 or mammalian ATR-ATRIP) pathway function redundantly to prevent telomere dysfunction and chromosome circularization (8, 42, 46). Accordingly, we tested whether the Rad3 checkpoint kinase might have a similar function as Tel1 in telomerase-independent telomere maintenance in taz1Δ trt1Δ cells. However, in contrast to Tel1, the elimination of Rad3 (ATR) checkpoint kinase did not lead to chromosome circularization in taz1Δ trt1Δ cells (Fig. 2A). Therefore, Rad3 does not appear to be essential for telomere maintenance when Taz1 and Trt1 are both missing from cells, although we found that taz1Δ trt1Δ rad3Δ cells showed a reduction in average telomere length compared to taz1Δ trt1Δ cells (see Fig. S6 in the supplemental material). We can rule out the possibility that Rad3 is essential for generating circular chromosomes since the reintroduction of the taz1+ plasmid into taz1Δ trt1Δ rad3Δ cells led to chromosome circularization (see Fig. 5A). A recent study has also found that taz1Δ trt1Δ rqh1Δ cells (Rqh1 is ortholog of S. cerevisiae Sgs1) cannot maintain linear chromosomes (32). Taken together, the recombination-based telomere maintenance mechanism observed in taz1Δ trt1Δ cells has genetic requirements very similar to those of S. cerevisiae type II survivors, which require Rad52, the MRX complex, Tel1, Mec1, and Sgs1 (27, 57, 58, 60).
Elimination of Est1 leads to chromosome circularization even in the absence of Taz1.
We also examined the effect of eliminating the telomerase regulatory subunit Est1 from taz1Δ or taz1Δ trt1Δ cells. In S. cerevisiae, Est1 is essential for telomerase function in vivo; it contributes to efficient telomere recruitment and activation of the telomerase catalytic subunit Est2 (TERT) (5, 18, 55). Similarly, S. pombe Est1 coimmunoprecipitates with Trt1, and est1Δ cells progressively lose telomeres (4). Therefore, we expected that deletion of est1+ would have similar effects on telomere maintenance as the deletion of trt1+.
As we have previously shown, when taz1Δ trt1Δ cells are generated by deleting trt1+ from taz1Δ cells, survivors with linear chromosomes are generated exclusively, since preexistence of the taz1Δ mutation strongly favors cell survival via recombination-based telomere maintenance over cell survival via chromosome circularization (44). Accordingly, we expected to see exclusive occurrences of survivors with linear chromosomes when we deleted est1+ from taz1Δ cells. Surprisingly, the deletion of est1+ from taz1Δ cells still led to chromosome circularization (Fig. 2A). In contrast, when we deleted est1+ from taz1Δ trt1Δ survivor cells, we found that all clones were able to stably maintain telomeres (Fig. 2A). In fact, the loss of Est1 appeared to lead to telomere lengthening in taz1Δ trt1Δ background (see Fig. S6 in the supplemental material). Thus, the loss of Est1 in the absence of Trt1 might further contribute to more enhanced telomere recombination. On the other hand, it appears that the presence of TERT, even when est1+ is deleted in taz1Δ cells, can inhibit the efficient use of recombination to maintain telomeres. Such a difference in the ability to generate recombination-based survivors between taz1Δ est1Δ and taz1Δ trt1Δ cells might provide an explanation for the previous observation that est1Δ/est1+ taz1Δ/taz1Δ diploid cells could not produce taz1Δ est1Δ haploid cells, whereas trt1Δ/trt1+ taz1Δ/taz1Δ diploid cells could efficiently generate taz1Δ trt1Δ haploid cells (4).
Taz1-independent role of Rap1 in the promotion of recombination-based telomere maintenance.
Next, we decided to test the effect of deleting rap1+ from taz1Δ trt1Δ cells on recombination-based telomere maintenance. Since previous studies have shown that recruitment of S. pombe Rap1 to telomeres depends on Taz1 (10, 30), we predicted that the resulting taz1Δ trt1Δ rap1Δ cells would be able to stably maintain linear chromosomes, much like taz1Δ trt1Δ cells.
Surprisingly, taz1Δ trt1Δ rap1Δ cells were unable to maintain telomeres and circularized their chromosomes (Fig. 2B), indicating that Rap1 is required for TERT-independent telomere maintenance. Furthermore, in agreement with a previous study (39), we found that rap1Δ trt1Δ cells cannot maintain telomeres even when rap1Δ trt1Δ cells were generated by deleting trt1+ from rap1Δ cells (Fig. 2B). Thus, Rap1 contributes to TERT-independent telomere maintenance in both taz1+ and taz1Δ backgrounds. We confirmed that efficient telomere recruitment of Rap1 to telomeres is indeed dependent on Taz1 (Fig. 2C and D). We also considered the possibility that Rap1 can be efficiently recruited to telomeres in the absence of Taz1 once Trt1 is also eliminated. However, we saw no evidence of Rap1 recruitment to telomeres in taz1Δ trt1Δ cells (Fig. 2C and D). Thus, Rap1 has a Taz1-independent function in promoting recombination-based telomere maintenance, even when the recruitment of Rap1 to telomeres is greatly reduced or abolished.
Catalytically inactive TERT affects the generation of recombination-based survivors.
Our data in Fig. 1D suggested that TERT and Taz1 redundantly provide protection against very rapid occurrence of telomere fusions. To test whether the predicted telomere protection function of TERT in the taz1Δ background could be separated from its telomere replication function, we monitored changes in cell growth and telomere structure for taz1Δ trt1-D743A cells, which were derived from taz1Δ/taz1+ trt1-D743A/trt1+ diploid cells. The trt1-D743A mutation (see Fig. 4A) has been shown to abolish catalytic activity without affecting protein expression level (23). If the catalytically inactive TERT could still function in telomere protection, we would expect to see that taz1Δ trt1-D743A cells exhibit a delayed loss of viability compared to taz1Δ trt1Δ cells.
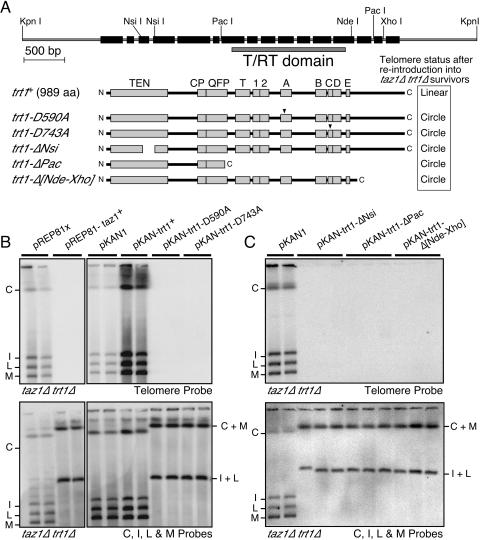
FIG. 4.
Taz1 and catalytically inactive TERT inhibit recombination-based telomere maintenance in taz1Δ trt1Δ cells. (A) The top diagram shows the S. pombe trt1+ gene structure. Exons are shown as black boxes, and the relevant restriction sites and the locations of conserved motifs and functional domains are indicated. Below are schematic representations of the various Trt1 constructs tested. (B and C) PFGE analyses of taz1Δ trt1Δ survivor cells transformed with the indicated plasmids. After transformation with plasmids, the colonies were extensively restreaked on agar plates appropriate for selection of plasmids before analysis by PFGE. The top panel shows hybridization with a telomere probe. The bottom panel shows hybridization with probes for the C, I, L, and M bands.
However, the loss of viability and telomere dysfunction in taz1Δ trt1-D743A cells occurred even earlier than in taz1Δ trt1Δ cells. (Fig. 3D; see also Fig. S2D in the supplemental material). While taz1Δ trt1-D743A cells were extremely sick immediately after the germination of spores, by the time cells were collected on day 1 of the liquid growth experiment, they had already started to recover in growth rate. PFGE analysis revealed the presence of mostly fused telomere bands (C+M and I+L) consistent with circular chromosomes on day 1 (Fig. 3F), but continued serial dilutions led to rapid disappearance of distinct telomeric NotI bands by day 3. The failure to detect distinct telomeric NotI fragments was not due to general degradation of chromosomal DNA, since we can detect expected bands for nontelomeric NotI fragments on ethidium bromide-stained agarose gels (see Fig. S3 in the supplemental material). On the other hand, we did reproducibly observe extremely broad diffused hybridization signals when blots for taz1Δ trt1-D743A samples were hybridized to probes specific to C, I, L, and M bands (Fig. 3F). Southern blot analysis of conventional agarose gels revealed that taz1Δ trt1-D743A survivor cultures from serial liquid culture dilution experiments often contained highly amplified TAS sequences but little telomere signal (see Fig. S4 and S5 in the supplemental material; also, data not shown). Such organization of subtelomeric regions is reminiscent of type I survivors from budding yeast. Thus, it appears that the presence of catalytically inactive TERT could prevent taz1Δ trt1-D743A cells from achieving a stable type II-like linear telomere maintenance mode normally observed in taz1Δ trt1Δ cells. Taken together, our results suggest that catalytically inactive Trt1-D743A protein can affect recombination efficiency at telomeres in a taz1Δ background.
FIG. 3.
PFGE analysis for trt1-D743A and taz1Δ trt1-D743A cells. A taz1Δ/taz1+ trt1-D743A/trt1+ diploid strain was sporulated, and trt1-D743A (A to C) or taz1Δ trt1-D743A (D to F) cells were selected. (A and D) Liquid growth characteristics of cells after germination. For references, results from growth curve experiments for wild-type and taz1Δ cells are also plotted (dotted lines). (B and E) PFGE of NotI-digested S. pombe chromosomal DNA hybridized with telomere probe. (C and F) PFGE of NotI-digested S. pombe chromosomal DNA hybridized with probes for the C, I, L, and M bands.
Liquid culture growth experiments also revealed that trt1-D743A cells reached their lowest viability 5 to 6 days earlier than trt1Δ cells (Fig. 3A; see also Fig. S2C in the supplemental material). This earlier loss of viability correlated with earlier telomere erosion and chromosome fusions (Fig. 3B and C). Although the parental heterozygous diploid cells had telomere length comparable to wt cells (data not shown), trt1-D743A cells appear to lose telomeres much faster than trt1Δ cells. Previously, trt1Δ cells that carry a Trt1-D743A expression plasmid were also shown to lose cell viability earlier than trt1Δ cells (23). Analysis of telomere length and the TAS region also revealed that trt1-D743A liquid culture survivors contain very little telomere repeat sequences and show much more prominent amplification of the TAS1 sequence than trt1Δ survivor cultures (see Fig. S4 and S5 in the supplemental material). Thus, the presence of inactive TERT appears to interfere with the generation of type II-like survivor cells in both taz1+ and taz1Δ cells even when survivor cells were selected in competitive liquid culture selection conditions.
Taz1 and catalytically inactive TERT inhibit recombination-based telomere maintenance.
The recombination-based telomere maintenance in taz1Δ trt1Δ cells is highly efficient, presumably because changes in the telomeric chromatin structure in the absence of Taz1 allow better access for recombination enzymes to telomeres (12). The taz1Δ trt1-D743A experiments also suggest that catalytically inactive TERT may be able to inhibit telomere recombination. To directly test whether Taz1 and TERT can inhibit recombination at telomeres, we reintroduced Taz1 or catalytically inactive TERT (Trt1-D590A or Trt1-D743A; Fig. 4A) (23) into taz1Δ trt1Δ survivor cells and then analyzed the telomere structure by PFGE after repeated restreaking on agar plates.
Reintroduction of Taz1, Trt1-D590A, or Trt1-D743A into taz1Δ trt1Δ cells interfered with recombination-based telomere maintenance, causing chromosome circularization (Fig. 4B). On the other hand, reintroduction of trt1+ into taz1Δ trt1Δ cells resulted in telomere elongation (Fig. 4B and 5D). Wild-type Trt1 is most likely as effective in inhibiting recombination at telomeres as the catalytically inactive Trt1, but since catalytically active Trt1 allows cells to stably maintain telomeric repeats without the help of recombination, chromosomes do not circularize after the reintroduction of wild-type Trt1. Taken together, these results indicate that TERT is as effective as Taz1 in preventing recombination-dependent telomere maintenance, and the inhibitory function of TERT on recombination can be separated from its RT activity.
Since the RT activity of TERT is not necessary to prevent telomere recombination, we next sought to determine whether the RT domain of TERT is necessary to inhibit telomere recombination by constructing a series of truncation mutants (Fig. 4A). The smallest tested TERT fragment that can efficiently inhibit telomere recombination was Trt1-ΔPac (Fig. 4C), which completely lacks the C-terminal RT domain. Thus, the N-terminal half of TERT, which includes the recently crystallized TEN (telomerase essential) domain, the CP motif, and most of the QFP motif (Fig. 4A), represents a functional subdomain that can efficiently inhibit recombination at telomeres even in the absence of Taz1 protein (28, 65).
Est1 and Ku70 are essential for catalytically inactive TERT to inhibit telomere recombination.
Next, we searched for mutations that would abrogate the ability of catalytically inactive TERT to inhibit recombination-based telomere maintenance in taz1Δ trt1Δ cells. Since catalytically inactive TERT is expected to inhibit telomere recombination only if it is recruited to telomeres, factors that are required for the recruitment of TERT to telomeres might be identified from such analyses. Alternatively, factors that are involved in suppressing telomere recombination might also be identified.
Rad3, Est1, and Ku70 are good candidates for proteins that might be involved in the recruitment of TERT to telomeres, since all three proteins have previously been shown to positively contribute to telomere length maintenance in taz1+ trt1+ cells (3, 4, 46). Moreover, we have already established that these proteins are dispensable for telomere maintenance in taz1Δ trt1Δ cells (Fig. 2A). Therefore, we reintroduced catalytically inactive TERT into triple-mutant cells (rad3Δ, est1Δ, or pku70Δ combined with taz1Δ trt1Δ) and examined their telomere structure by PFGE. As a positive control, we also reintroduced Taz1 into these cells. In all cases, reintroduction of Taz1 resulted in circular chromosomes (Fig. 5A to C), indicating that Rad3, Est1, and Ku70 are not necessary for chromosome circularization.
Reintroduction of Trt1-D590A or D743A into taz1Δ trt1Δ rad3Δ cells resulted in efficient chromosome circularization and the complete loss of telomeric repeats (Fig. 5A and data not shown). Conversely, reintroduction of wild-type Trt1 resulted in massive telomere elongation (Fig. 5D), and ChIP analysis confirmed the efficient recruitment of Trt1 to telomeres in both rad3Δ and taz1Δ rad3Δ cells (Fig. 5E). Thus, Rad3 is not essential for recruitment of Trt1 to telomeres.
Reintroduction of Trt1-D590A or D743A into taz1Δ trt1Δ est1Δ cells did not lead to chromosome circularization (Fig. 5B). Furthermore, reintroduction of wild-type Trt1 did not result in telomere elongation (Fig. 5D), and Trt1 was no longer detected at telomeres by ChIP analysis in taz1Δ est1Δ cells (Fig. 5E). Thus, Est1 is essential for recruitment of Trt1 to telomeres. This is the first direct demonstration outside of budding yeast that Est1 is involved in the recruitment of TERT to telomeres (5).
It was surprising that reintroduction of wild-type or catalytically inactive TERT into established taz1Δ trt1Δ est1Δ survivors did not result in chromosome circularization since we earlier showed that taz1Δ est1Δ cells, generated by deletion of est1+ from taz1Δ cells, carry circular chromosomes (Fig. 2A). Such a discrepancy might indicate that taz1Δ trt1Δ survivor cells establish an altered telomeric chromatin environment that no longer allows recruitment of TERT without Est1. In contrast, Est1 might be dispensable for the recruitment of TERT in taz1Δ cells, and inactive TERT (due to the lack of Est1) could interfere with the establishment of recombination-based survivors. Alternatively, Est1 might still be required for recruitment of TERT to telomeres even in taz1Δ cells, but a gradual decline in Est1 protein level after the deletion of est1+ could result in a situation where a limiting amount of Est1 is sufficient for Trt1 recruitment but not RT activation, thus mimicking the situation where catalytically inactive TERT interferes with the generation of recombination survivors. Further careful studies are necessary to distinguish these possibilities.
For taz1Δ trt1Δ pku70Δ, reintroduction of Trt1-D590A or D743A did not result in chromosome circularization (Fig. 5C). However, unlike in the case of taz1Δ trt1Δ est1Δ cells, reintroduction of wild-type Trt1 into taz1Δ trt1Δ pku70Δ cells resulted in massive telomere elongation (Fig. 5D). In addition, telomere recruitment for either wild-type or catalytically inactive Trt1 proteins was normal in taz1Δ pku70Δ cells based on ChIP analysis (Fig. 5E). Trt1 was also recruited efficiently in pku70Δ cells (Fig. 5E). Thus, elimination of pku70Δ appears to affect telomere recombination independent of TERT recruitment. Indeed, previous studies have shown that S. pombe Ku70 has a role in prevention of recombination at telomeres (3, 33). Thus, our results identify Ku70 as a third parallel and redundant factor besides Trt1-Est1 and Taz1 contributing to the prevention of recombination at telomeres.
DISCUSSION
While many proteins contribute to telomere maintenance in eukaryotic cells, it is not well understood how multitudes of proteins work collaboratively or antagonistically in either telomerase-dependent or -independent mechanisms of telomere maintenance. It is also not clear why telomerase-based telomere maintenance is the most prevalent mechanism in eukaryotic cells, since diverse non-telomerase-based mechanisms, such as telomere-telomere recombination and integration of retrotransposons, appear to be almost as effective in maintaining telomeres (7, 41). To understand these issues better, we investigated how the presence or absence of telomere-specific proteins, DNA repair proteins, and DNA damage checkpoint proteins contribute to the prevention or promotion of recombination and fusion events at telomeres in fission yeast.
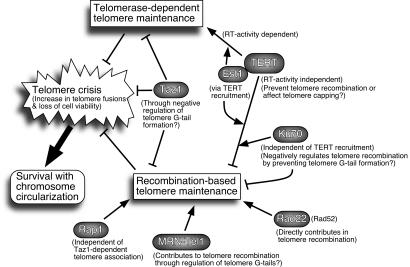
We first demonstrated that simultaneous loss of Taz1 and TERT results in accelerated telomere fusion compared to the loss of TERT alone (Fig. 1). Therefore, we envision Taz1 and TERT as two major factors that are essential for the prevention of telomere crisis, caused by increased fusion and recombination events at telomeres (Fig. 6). Since the elimination of either Taz1 or TERT alone does not result in immediate telomere crisis, their roles in the protection of telomeres are redundant. Because early telomere crisis cannot be averted by expression of catalytically inactive TERT (Fig. 3; see also Fig. S2 in the supplemental material), we suggest that the rapid loss of telomeric GT-rich repeats likely causes an immediate increase in telomere fusion events. This hypothesis is consistent with a recent study that showed that replication of telomeric repeats in taz1Δ cells is critically dependent on TERT (39). GT-rich telomeric repeats also represent the binding site for the telomere capping protein Pot1 (2), so if taz1Δ trt1Δ cells fail to maintain telomeric repeats, Pot1 might no longer be recruited to protect telomeres and thus lead to catastrophic telomere fusions.
FIG. 6.
Summary of genetic interactions found in the current study affecting telomerase-dependent and -independent telomere maintenance and chromosome circularization.
On the other hand, we found that simultaneous elimination of Taz1 and TERT also allows cells to very efficiently maintain telomeres by a Rad22 (Rad52)-dependent recombination mechanism (Fig. 1, 2, and 6). Since reintroduction of Taz1 into taz1Δ trt1Δ survivors causes chromosome circularization (Fig. 4), Taz1 functions as a potent inhibitor of telomere recombination. Rad22 is in fact essential for preventing NHEJ-dependent fusion of telomeres in taz1Δ cells, and taz1Δ cells also carry long telomeric G-tails, expected to be a favored substrate for HR (19, 59). However, it is difficult to know how quickly cells can establish long G-tails after the loss of Taz1. When Taz1 and TERT are simultaneously lost, as in the case of taz1Δ trt1Δ cells derived from taz1Δ/taz1+ trt1Δ/trt1+ diploid cells, the inability of taz1Δ cells to replicate through telomeres without telomerase (39) might initially dominate telomere dynamics and cause many taz1Δ trt1Δ cells to quickly lose telomeric repeats and fuse telomeres. Once a nuclease(s) acting at the telomeres (which are normally inhibited by Taz1) degrades the CA-rich strand of telomeres and establishes long G-tails, taz1Δ trt1Δ cells could then efficiently maintain telomeres through the Rad22-dependent HR mechanism. The fact that taz1Δ cells have already established long telomere tracts with extended G-tails can then explain why we observe only linear chromosome survivors when taz1Δ trt1Δ cells are generated by eliminating Trt1 from taz1Δ cells (44). In S. cerevisiae, combining a temperature-sensitive mutation of the G-tail binding protein Cdc13 (cdc13-1) and deletion of telomerase RNA TLC1 also caused cdc13-1 tlc1Δ cells to experience much earlier telomere dysfunction than tlc1Δ cells when the cells were grown at a semipermissive temperature (60). Moreover, these cells exclusively generate type II survivors (22, 60). It is worth noting that cdc13-1 cells grown at semipermissive temperature carry longer G-tails, much like taz1Δ cells.
Our analyses have also uncovered a surprising RT-independent role of TERT in the prevention of recombination at telomeres (Fig. 6). Reintroduction of catalytically inactive versions of TERT was as effective as reintroduction of Taz1 in causing chromosome circularization. The N-terminal 396- amino-acid portion of TERT (Trt1-ΔPac), which includes the recently crystallized TEN domain and the region implicated in telomerase RNA binding, was sufficient for the inhibition (28, 31). Although such results are also consistent with the interpretation that catalytically inactive TERT can disrupt the protective function of telomeres against fusions, we favor the idea that circularization is caused by inhibition of recombination-based survival in taz1Δ trt1Δ cells since Est1-dependent recruitment of TERT is required to cause circularization and removal of the Ku complex, which may further compromise telomere capping, can reverse the circularization phenotype (Fig. 5).
Our analysis also indicated that the Ku complex is essential for the TERT-dependent protection of telomeres from recombination in the absence of Taz1 (Fig. 5C and 6). While studies in S. cerevisiae have clearly demonstrated that the Ku complex plays a very important role in the recruitment of Est2 (TERT) to telomeres (20), we found no evidence that Ku is involved in the recruitment of TERT to telomeres in fission yeast (Fig. 5E). However, the S. cerevisiae Ku complex plays the most important role in the recruitment of Est2 in the G1 phase of the cell cycle (20). Given that exponentially growing S. pombe cell cultures contain very few cells in G1 phase, we cannot rule out the possibility that the recruitment of S. pombe TERT to telomeres in G1 phase might be affected by the loss of the Ku complex. On the other hand, previous studies have found that the Ku70-Ku80 heterodimer is required to prevent recruitment of the HR repair protein Rhp51 (Rad51) to telomeres in fission yeast (33) and is involved in the inhibition of telomere recombination in budding yeast and mammalian cells (6, 51). Therefore, we favor a model in which the Ku complex represents a third independent component involved in repressing recombination at telomeres, in addition to Taz1 and TERT-Est1 (Fig. 6).
Although previous studies have suggested that TERT could have telomere protection roles independent of its extension function (36, 54, 69), these putative “protective” functions of TERT generally still required TERT to be catalytically active. Therefore, even when the overall telomere length was not altered by the expression of TERT, it was difficult to rule out the possibility that TERT might selectively act on extremely short telomeres to improve chromosome stability or cell viability. In fact, the expression of catalytically inactive hTERT variants causes cell death or senescence, probably due to the increase in the loss of telomeric DNA (24, 25, 68). However, these studies also found that cells expressing catalytically inactive hTERT were not able to activate the recombination-based ALT (alternative lengthening of telomeres) survival mechanism to escape cell death, and thus the observations in these studies are consistent with the notion that catalytically inactive TERT can function as an effective protector against telomeric recombination. It was recently reported that catalytically inactive TERT (Est2) and telomerase RNA together could protect telomeres from excessive G-tail formation in Candida albicans (26). As mentioned earlier, long G-tails should serve as a good substrate for recombination, and thus the prevention of long G-tail formation may explain why catalytically inactive TERT can function as an inhibitor of telomeric recombination. It is worth noting that a human hTERTα splice variant, which lacks highly conserved critical amino acid residues within the RT domain required for telomerase activity, is expressed in development- and tissue-specific manners and has been shown to function as a dominant-negative inhibitor of telomerase activity (11, 66). A recent study has also shown that Arabidopsis POT1A can interact with the N-terminal splicing variant of TERT (52). Therefore, we suggest that the inhibition of telomeric recombination by the naturally occurring hTERTα splice variant could also have an important role in the regulation of telomere maintenance in human cells.
Our genetic analysis has uncovered that recombination-based telomere maintenance in taz1Δ trt1Δ cells requires the Tel1-MRN (ATM-MRN) complex (Fig. 2A and 6). Previously, type II survivors in S. cerevisiae and human ALT cells have been shown to also require the MRX/MRN complex (29, 57). Thus, MRN appears to be universally important for recombination-based telomere maintenance. Interestingly, ATM and MRN are also very important in the protection and/or maintenance of telomeres in Drosophila cells, where transpositions of retroelements are utilized in telomere maintenance (7). Moreover, studies in S. cerevisiae provide convincing evidence that Tel1-MRX plays a very important role(s) in the recruitment of telomerase components to telomeres (21, 61). Therefore, the ATM-MRN complex contributes positively to telomere maintenance involving telomerase, recombination, and retrotransposons. Since MRN is involved in the generation of G-tails at telomeres (35, 59) and the presence of G-tails is favorable for both efficient recruitment of telomerase and initiation of telomeric recombination (21, 62), ATM-MRN is one of the most important gatekeepers in regulating telomere accessibility to various modes of telomere maintenance. Our current finding that Taz1 and TERT can function as inhibitors of telomere recombination suggests that these telomere-specific factors can in turn regulate the ATM-MRN complex in ensuring that telomeres are protected against recombination and are maintained via telomerase-based extension.
In contrast to the Tel1-MRN complex, our analysis indicated that the Rad3-Rad26 (ATR-ATRIP) complex does not play a significant role in recombination-based telomere maintenance in the absence of Taz1 and TERT (Fig. 2 and 5). On the other hand, in the presence of wild-type Taz1 and Trt1, Rad3-Rad26 appears to play a more important role(s) in telomerase-dependent telomere maintenance than Tel1-MRN, since the deletion of Rad3-Rad26 causes telomeres to become much shorter than cells lacking Tel1 or MRN (8, 46). Our ChIP and chromosome circularization assays suggest that Rad3 is not essential for the recruitment of TERT to telomeres (Fig. 5), although we cannot rule out the possibility that Tel1-MRN and Rad3-Rad26 are redundantly required for the recruitment of TERT to telomeres, since tel1Δ rad3Δ cells are incapable of maintaining telomeres even in the presence of wild-type TERT (42). Thus, further studies are required to understand the exact contribution of the Rad3-Rad26 (ATR-ATRIP) complex in the telomerase-dependent telomere maintenance mechanism.
We were surprised by our observation that Rap1 is essential for the maintenance of telomeres in taz1Δ trt1Δ cells since previous studies established that the recruitment of Rap1 to telomeres largely depends on Taz1, although some residual Rap1 foci at telomeres were occasionally observed by fluorescence microscopy in taz1Δ cells undergoing meiosis (10, 30). Although it is still possible that a very small amount of Rap1 (undetectable by ChIP) is recruited to telomeres and contributes to the promotion of recombination or the prevention of fusions at telomeres, our data suggest that Rap1 can positively contribute to linear chromosome maintenance without being efficiently recruited to telomeres (Fig. 6). If there is very weak or transient residual binding of Rap1 undetectable by ChIP, such interaction might involve a Myb domain of Rap1 interacting with telomeric DNA or other telomere proteins. Therefore, it might be interesting to test whether Rap1 lacking a Myb domain might still be able to prevent chromosome circularization after elimination of TERT. A previous genetic study in S. pombe has found that the loss of Rap1 exacerbates the cold sensitivity of taz1Δ cells, which led the authors to conclude that Rap1 may have Taz1-independent telomere functions (38). However, a rap1Δ strain on its own is not cold sensitive, and only Taz1 (but not Rap1) has been found to promote replication of telomeric GT-rich repeats by DNA polymerases (38, 39). Thus, our current observation is the first report wherein rap1Δ cells show more severe defects in telomere stability than taz1Δ cells. Unlike S. cerevisiae cells, in which Rap1 directly binds to the telomeric DNA, recruitment of human Rap1 to the telomere is also dependent on TRF2 (an ortholog of S. pombe Taz1) (34). Therefore, it will be interesting to determine whether human Rap1 might also contribute to telomere maintenance independent from its TRF2-dependent recruitment to telomeres. In S. cerevisiae, Rap1 plays a well-established role in the transcriptional activation of various genes (40). It is possible that S. pombe Rap1 might also contribute to the transcriptional activation of genes responsible for the recombinational mode of telomere maintenance in taz1Δ trt1Δ cells. However, such a possibility awaits future investigations. It should also be noted that a recent study has provided evidence that Rap1 is necessary to prevent NHEJ in budding yeast cells (50).
Telomerase-based maintenance of telomeres is the most common method of telomere maintenance among eukaryotic cells. This can be partly explained by the fact that telomerase appears to have originated very early in the evolution of eukaryotic cells (43). However, a strong argument can be made that recombination-based or retrotransposon-based telomere maintenance might have even more ancient origins than modern day telomerase-based telomere maintenance (15). So, how could the telomerase-based mechanism be so effective in preventing underlying recombination- or transposon-based modes of telomere maintenance? Certainly, factors that specifically bind to telomeric GT-rich repeats, such as S. pombe Taz1, S. cerevisiae Rap1, and mammalian TRF1 and TRF2 proteins, play major roles in preventing excessive recombination at telomeres (53). In addition, our observations suggest that TERT is directly involved in preventing alternative telomere maintenance mechanisms. In fact, the N-terminal domain we found to be crucial for the prevention of telomeric recombination is not found in other RT proteins encoded by retrotransposons and viruses (17). It is worth noting that both TRF1/TRF2-type telomere-specific proteins and TERT are absent in Drosophila cells and that ALT cells generally lack functional telomerase. Thus, TERT's ability to efficiently compete and protect against other ancient DNA repair and damage response machineries is very important for understanding how telomere maintenance is regulated in eukaryotic cells.
Supplementary Material
Acknowledgments
We thank F. Ishikawa, J. P. Cooper, T. R. Cech, and P. Russell for strains and plasmids and J. P. Cooper, T. R. Cech, and E. Noguchi for helpful comments on the manuscript.
Initial portions of this work were carried out by T.M.N. at the University of Colorado and were supported by National Institutes of Health (NIH) grant GM28039 to T. R. Cech. Our work has been supported by UIC startup fund, the Sidney Kimmel Scholar Program, and NIH grant GM078253 to T.M.N. L.S. has been supported in part by a predoctoral fellowship from the American Heart Association.
Footnotes
Published ahead of print on 26 December 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alfa, C., P. Fantes, J. Hyams, M. McLoed, and E. Warbrick. 1993. Experiments with fission yeast. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 2.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 2921171-1175. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, P., and T. R. Cech. 2000. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell 113265-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beernink, H. T., K. Miller, A. Deshpande, P. Bucher, and J. P. Cooper. 2003. Telomere maintenance in fission yeast requires an Est1 ortholog. Curr. Biol. 13575-580. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi, A., S. Negrini, and D. Shore. 2004. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell 16139-146. [DOI] [PubMed] [Google Scholar]
- 6.Celli, G. B., E. L. Denchi, and T. de Lange. 2006. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 8885-890. [DOI] [PubMed] [Google Scholar]
- 7.Cenci, G., L. Ciapponi, and M. Gatti. 2005. The mechanism of telomere protection: a comparison between Drosophila and humans. Chromosoma 114135-145. [DOI] [PubMed] [Google Scholar]
- 8.Chahwan, C., T. M. Nakamura, S. Sivakumar, P. Russell, and N. Rhind. 2003. The fission yeast Rad32 (Mre11)-Rad50-Nbs1 complex is required for the S-phase DNA damage checkpoint. Mol. Cell. Biol. 236564-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, S. W., and E. H. Blackburn. 2003. Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol. Cell 111379-1387. [DOI] [PubMed] [Google Scholar]
- 10.Chikashige, Y., and Y. Hiraoka. 2001. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr. Biol. 111618-1623. [DOI] [PubMed] [Google Scholar]
- 11.Colgin, L. M., C. Wilkinson, A. Englezou, A. Kilian, M. O. Robinson, and R. R. Reddel. 2000. The hTERTα splice variant is a dominant-negative inhibitor of telomerase activity. Neoplasia 2426-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper, J. P., E. R. Nimmo, R. C. Allshire, and T. R. Cech. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385744-747. [DOI] [PubMed] [Google Scholar]
- 13.d'Adda di Fagagna, F., S. H. Teo, and S. P. Jackson. 2004. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 181781-1799. [DOI] [PubMed] [Google Scholar]
- 14.de Lange, T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 192100-2110. [DOI] [PubMed] [Google Scholar]
- 15.de Lange, T. 2004. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell. Biol. 5323-329. [DOI] [PubMed] [Google Scholar]
- 16.Dunham, M. A., A. A. Neumann, C. L. Fasching, and R. R. Reddel. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26447-450. [DOI] [PubMed] [Google Scholar]
- 17.Eickbush, T. H. 1994. Origin and evolutionary relationships of retroelements, p. 121-157. In S. S. Morse (ed.), The evolutionary biology of viruses. Raven Press, New York, NY.
- 18.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286117-120. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira, M. G., and J. P. Cooper. 2001. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell 755-63. [DOI] [PubMed] [Google Scholar]
- 20.Fisher, T. S., A. K. Taggart, and V. A. Zakian. 2004. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 111198-1205. [DOI] [PubMed] [Google Scholar]
- 21.Goudsouzian, L. K., C. T. Tuzon, and V. A. Zakian. 2006. Saccharomyces cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell 24603-610. [DOI] [PubMed] [Google Scholar]
- 22.Grandin, N., C. Damon, and M. Charbonneau. 2001. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 206127-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haering, C. H., T. M. Nakamura, P. Baumann, and T. R. Cech. 2000. Analysis of telomerase catalytic subunit mutants in vivo and in vitro in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 976367-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahn, W. C., S. A. Stewart, M. W. Brooks, S. G. York, E. Eaton, A. Kurachi, R. L. Beijersbergen, J. H. Knoll, M. Meyerson, and R. A. Weinberg. 1999. Inhibition of telomerase limits the growth of human cancer cells. Nat. Med. 51164-1170. [DOI] [PubMed] [Google Scholar]
- 25.Herbert, B., A. E. Pitts, S. I. Baker, S. E. Hamilton, W. E. Wright, J. W. Shay, and D. R. Corey. 1999. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc. Natl. Acad. Sci. USA 9614276-14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu, M., M. J. McEachern, A. T. Dandjinou, Y. Tzfati, E. Orr, E. H. Blackburn, and N. F. Lue. 2007. Telomerase core components protect Candida telomeres from aberrant overhang accumulation. Proc. Natl. Acad. Sci. USA 10411682-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, P., F. E. Pryde, D. Lester, R. L. Maddison, R. H. Borts, I. D. Hickson, and E. J. Louis. 2001. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 11125-129. [DOI] [PubMed] [Google Scholar]
- 28.Jacobs, S. A., E. R. Podell, and T. R. Cech. 2006. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 13218-225. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, W. Q., Z. H. Zhong, J. D. Henson, and R. R. Reddel. 2007. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene 264635-4647. [DOI] [PubMed] [Google Scholar]
- 30.Kanoh, J., and F. Ishikawa. 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 111624-1630. [DOI] [PubMed] [Google Scholar]
- 31.Kelleher, C., M. T. Teixeira, K. Forstemann, and J. Lingner. 2002. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 27572-579. [DOI] [PubMed] [Google Scholar]
- 32.Kibe, T., Y. Ono, K. Sato, and M. Ueno. 2007. Fission yeast Taz1 and RPA are synergistically required to prevent rapid telomere loss. Mol. Biol. Cell 182378-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kibe, T., K. Tomita, A. Matsuura, D. Izawa, T. Kodaira, T. Ushimaru, M. Uritani, and M. Ueno. 2003. Fission yeast Rhp51 is required for the maintenance of telomere structure in the absence of the Ku heterodimer. Nucleic Acids Res. 315054-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konig, P., L. Fairall, and D. Rhodes. 1998. Sequence-specific DNA recognition by the myb-like domain of the human telomere-binding protein TRF1: a model for the protein-DNA complex. Nucleic Acids Res. 261731-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larrivee, M., C. LeBel, and R. J. Wellinger. 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 181391-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masutomi, K., R. Possemato, J. M. Wong, J. L. Currier, Z. Tothova, J. B. Manola, S. Ganesan, P. M. Lansdorp, K. Collins, and W. C. Hahn. 2005. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl. Acad. Sci. USA 1028222-8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mieczkowski, P. A., J. O. Mieczkowska, M. Dominska, and T. D. Petes. 2003. Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 10010854-10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, K. M., M. G. Ferreira, and J. P. Cooper. 2005. Taz1, Rap1, and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 243128-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, K. M., O. Rog, and J. P. Cooper. 2006. Semi-conservative DNA replication through telomeres requires Taz1. Nature 440824-828. [DOI] [PubMed] [Google Scholar]
- 40.Morse, R. H. 2000. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 1651-53. [DOI] [PubMed] [Google Scholar]
- 41.Muntoni, A., and R. R. Reddel. 2005. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 14(Spec. No. 2)R191-R196. [DOI] [PubMed] [Google Scholar]
- 42.Naito, T., A. Matsuura, and F. Ishikawa. 1998. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat. Genet. 20203-206. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura, T. M., and T. R. Cech. 1998. Reversing time: origin of telomerase. Cell 92587-590. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura, T. M., J. P. Cooper, and T. R. Cech. 1998. Two modes of survival of fission yeast without telomerase. Science 282493-496. [DOI] [PubMed] [Google Scholar]
- 45.Nakamura, T. M., G. B. Morin, K. B. Chapman, S. L. Weinrich, W. H. Andrews, J. Lingner, C. B. Harley, and T. R. Cech. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277955-959. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura, T. M., B. A. Moser, and P. Russell. 2002. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 1611437-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Natarajan, S., and M. J. McEachern. 2002. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 224512-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nelson, J. D., O. Denisenko, and K. Bomsztyk. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1179-185. [DOI] [PubMed] [Google Scholar]
- 49.Nugent, C. I., and V. Lundblad. 1998. The telomerase reverse transcriptase: components and regulation. Genes Dev. 121073-1085. [DOI] [PubMed] [Google Scholar]
- 50.Pardo, B., and S. Marcand. 2005. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 243117-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polotnianka, R. M., J. Li, and A. J. Lustig. 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8831-834. [DOI] [PubMed] [Google Scholar]
- 52.Rossignol, P., S. Collier, M. Bush, P. Shaw, and J. H. Doonan. 2007. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J. Cell Sci. 1203678-3687. [DOI] [PubMed] [Google Scholar]
- 53.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73177-208. [DOI] [PubMed] [Google Scholar]
- 54.Stewart, S. A., W. C. Hahn, B. F. O'Connor, E. N. Banner, A. S. Lundberg, P. Modha, H. Mizuno, M. W. Brooks, M. Fleming, D. B. Zimonjic, N. C. Popescu, and R. A. Weinberg. 2002. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl. Acad. Sci. USA 9912606-12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taggart, A. K., S. C. Teng, and V. A. Zakian. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 2971023-1026. [DOI] [PubMed] [Google Scholar]
- 56.Takata, H., Y. Kanoh, N. Gunge, K. Shirahige, and A. Matsuura. 2004. Reciprocal association of the budding yeast ATM-related proteins Tel1 and Mec1 with telomeres in vivo. Mol. Cell 14515-522. [DOI] [PubMed] [Google Scholar]
- 57.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6947-952. [DOI] [PubMed] [Google Scholar]
- 58.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 198083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomita, K., A. Matsuura, T. Caspari, A. M. Carr, Y. Akamatsu, H. Iwasaki, K. Mizuno, K. Ohta, M. Uritani, T. Ushimaru, K. Yoshinaga, and M. Ueno. 2003. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol. Cell. Biol. 235186-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai, Y. L., S. F. Tseng, S. H. Chang, C. C. Lin, and S. C. Teng. 2002. Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol. Cell. Biol. 225679-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsukamoto, Y., A. K. Taggart, and V. A. Zakian. 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 111328-1335. [DOI] [PubMed] [Google Scholar]
- 62.Usui, T., H. Ogawa, and J. H. Petrini. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 71255-1266. [DOI] [PubMed] [Google Scholar]
- 63.Verdun, R. E., and J. Karlseder. 2007. Replication and protection of telomeres. Nature 447924-931. [DOI] [PubMed] [Google Scholar]
- 64.Verdun, R. E., and J. Karlseder. 2006. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127709-720. [DOI] [PubMed] [Google Scholar]
- 65.Xia, J., Y. Peng, I. S. Mian, and N. F. Lue. 2000. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 205196-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yi, X., D. M. White, D. L. Aisner, J. A. Baur, W. E. Wright, and J. W. Shay. 2000. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia 2433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.You, Z., C. Chahwan, J. Bailis, T. Hunter, and P. Russell. 2005. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 255363-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, X., V. Mar, W. Zhou, L. Harrington, and M. O. Robinson. 1999. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 132388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu, J., H. Wang, J. M. Bishop, and E. H. Blackburn. 1999. Telomerase extends the lifespan of virus-transformed human cells without net telomere lengthening. Proc. Natl. Acad. Sci. USA 963723-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.