Abstract
The progesterone receptor (PR) plays a critical role during ovulation. Mice lacking the PR gene are anovulatory due to a failure in the rupture of the preovulatory follicles. The pathways that operate downstream of PR to control ovulation are poorly understood. Using gene expression profiling, we identified peroxisome proliferator-activated receptor γ (PPARγ) as a target of regulation by PR in the granulosa cells of the preovulatory follicles during the ovulatory process. To investigate the function of PPARγ during ovulation, we created a conditional knockout mouse in which this gene was deleted via Cre-Lox-mediated excision in granulosa cells. When these mutant mice were subjected to gonadotropin-induced superovulation, the preovulatory follicles failed to rupture and the number of eggs released from the mutant ovaries declined drastically. Gene expression analysis identified endothelin-2, interleukin-6, and cyclic GMP-dependent protein kinase II as novel targets of regulation by PPARγ in the ovary. Our studies also suggested that cycloxygenase 2-derived metabolites of long-chain fatty acids function as endogenous activating ligands of PPARγ in the preovulatory follicles. Collectively, these studies revealed that PPARγ is a key mediator of the biological actions of PR in the granulosa cells and activation of its downstream pathways critically controls ovulation.
Ovulation is a key female reproductive event which involves the release of a fertilizable oocyte from a mature ovarian follicle (28). In mammals, it is a highly coordinated process regulated by endocrine/paracrine factors produced by the hypothalamic-pituitary-ovarian axis. During each reproductive cycle in the rodent, follicle-stimulating hormone, synthesized by the pituitary gonadotrophs, promotes the growth and development of a selected pool of preantral follicles to the preovulatory stage. The action of follicle-stimulating hormone is followed by a timely surge of another pituitary gonadotropin, luteinizing hormone (LH), which triggers ovulation. The preovulatory surge of LH sets in motion a complex cascade of gene expression events in the follicular cells that culminates in the rupture of the preovulatory follicle (27). Genetic analyses of the mouse established that the genes encoding progesterone (P) receptor (PR) (26), cyclooxygenase 2 (COX-2) (7, 21), and CCAAT/enhancer binding protein β (36), which are induced in the granulosa cells following LH action, play obligatory roles during ovulation.
The steroid hormone P, acting via its cellular receptor, plays an essential role during ovulation (22, 23). Previous studies have shown that the PR is rapidly induced in the mural granulosa cells of the preovulatory follicles following the LH surge (26, 29). Development of a mouse model lacking PR by Lydon et al. revealed an indispensable role for this hormone receptor during ovulation (23). In PR-null mice, follicular development to the antral stage proceeds unaffected. These mutant mice, however, fail to ovulate due to a block in follicular rupture, even when the animals are subjected to stimulation with exogenous gonadotropins (23). Due to these distinctive ovarian phenotypes, the PR-null mouse has emerged as an attractive model to explore the PR-regulated pathways that underlie the ovulation process. It is postulated that PR, a ligand-inducible transcription factor, controls ovulation by regulating the expression of a unique network of downstream genes. Consistent with this hypothesis, Richards and coworkers identified three novel genes, a disintegrin and metalloprotease with thromboxan motif 1 (ADAMTS-1) (29), cyclic GMP-dependent protein kinase II (cGKII) (34), and synaptosome-associated protein 25 (Snap25) (32), as potential mediators of PR function in the ovary. Our laboratory has recently reported that endothelin-2 (ET-2), which is induced in the preovulatory follicles in a PR-dependent manner, is a critical regulator of follicular rupture (25). Despite these significant advances, the identities of downstream pathways that convey PR action during the ovulatory process are largely unknown and the precise molecular mechanisms by which these pathways are controlled during ovulation remain unclear.
Recently, we employed a global gene expression profiling strategy to identify the molecular targets of PR during ovulation (J. Kim and M. K. Bagchi, unpublished results). Using this approach, we have discovered that the gene for peroxisome proliferator-activated receptor γ (PPARγ), a well-studied member of the nuclear receptor superfamily, is a candidate PR-regulated gene during the ovulatory process. PPARγ has been implicated in numerous biological processes, including adipocyte differentiation (4, 38), regulation of lipid and glucose homeostasis (18, 20), and control of inflammatory responses (24). Little, however, is known regarding its role in reproduction (15). Ovarian expression of PPARγ has been reported previously, although no distinct function has been ascribed to it (16).
To analyze the ovulatory function of PPARγ, we have generated mice in which the gene encoding this transcription factor is excised from the genome of mural granulosa cells of the preovulatory follicles by the Cre-LoxP strategy. We found that conditional deletion of the PPARγ gene in these ovarian cells severely impairs follicular rupture. Our study also indicated that COX-derived metabolites serve as activating ligands of PPARγ during the ovulatory process, thereby establishing a critical functional link between multiple key pathways that regulate ovulation. We also demonstrated that PPARγ controls a unique network of downstream genes, including those encoding ET-2, interleukin-6 (IL-6), and cGKII, which mediate, at least in part, the critical effects of PR during ovulation.
MATERIALS AND METHODS
Hormone treatment and sample collection.
Ovulation was experimentally induced in mice by the administration of exogenous gonadotropins, namely, pregnant mare serum gonadotropin (PMSG; Sigma Inc., St. Louis, MO) and human chorionic gonadotropin (hCG or pregnyl; Organon Inc., West Orange, NJ), as described previously (29). Briefly, mice were primed with 10 IU of PMSG for 48 h to promote follicular growth prior to the administration of 5 IU of hCG to trigger ovulation, resulting in the release of fertilizable eggs. Typically, ovulation takes place approximately 12 to 14 h after administration of hCG. All injections were given intraperitoneally.
The ovaries were collected at 5 h after administration of hCG from wild-type (WT) (129Sv) or PR-null mice (n = 4) primed with PMSG for 48 h. The tissues were pooled, immediately frozen in liquid nitrogen, and kept at −80°C until RNA isolation. Alternatively, these tissues were fixed in paraformaldehyde for in situ hybridization or immunohistochemistry.
Generation of conditional PPARγ-null mice.
Conditional PPARγ-null mice (PPARγflox/flox PRcre/+) were generated by crossing PPARγ-floxed mice (1) with PR-Cre knock-in mice (33). Briefly, female homozygous PPARγ-floxed mice (PPARγflox/flox) were mated with male homozygous PR-Cre mice (PRcre/cre) to produce offspring (PPARγflox/+ PRcre/+) that are heterozygous for both mutations. These heterozygous mice (PPARγflox/+ PRcre/+) were then bred with each other or with homozygous PPARγ-floxed mice (PPARγflox/flox) to produce conditional mutant mice with the genotype PPARγflox/flox PRcre/+. In these mice, Cre-mediated excision of floxed PPARγ led to null mutation of this gene in PR-expressing tissues. Genomic DNA isolated from tail biopsy samples was used for PCR to verify the genotypes of mice as described previously (1, 33). The PR-Cre-mediated excision of floxed PPARγ alleles in PPARγflox/flox PRcre/+ mice was assessed by PCR analysis of genomic DNA isolated from the ovaries of mice treated with PMSG for 48 h and subsequently with hCG for 5 h.
Assessment of ovulation.
Ovulated eggs were counted at 18 to 19 h after administration of hCG from PPARγflox/flox or PPARγflox/flox PRcre/+ mice primed with PMSG for 48 h. Briefly, oviducts were removed from both ovaries of a mouse and placed into a fresh dish (35 mm). Subsequently, 20 μl of 0.1% hyaluronidase (prepared in M2 medium) was directly added to the collected oviducts. The ampulla portion of each oviduct was then opened with a fine forceps under a dissecting microscope to release the cumulus-oocyte complexes. The eggs were subsequently counted under an inverted microscope. The ovaries of these animals were also collected, fixed in 10% neutrally buffered formalin at room temperature for 24 h, paraffin embedded, and examined histologically following hematoxylin and eosin staining.
Mouse primary granulosa cell cultures.
Primary granulosa cells were isolated by needle puncture (30-gauge needle) of the preovulatory follicles obtained from the ovaries of either prepubertal WT or PR-null mice primed with PMSG for 48 h. The primary cells were then washed three times in 1% fetal bovine serum-Dulbecco modified Eagle medium (DMEM)-F12 supplemented with penicillin (100 IU) and streptomycin (100 μg/ml) by centrifugation at 400 × g for 5 min at 4°C. These cells were subsequently plated in 35-mm culture dishes and cultured overnight at 37°C in the presence of 5% CO2. To mimic LH signaling, the primary cells were treated in serum-free DMEM-F12 with forskolin (10 μM; Calbiochem, San Diego, CA) and phorbol 12-myristate 13-acetate (PMA; 20 nM; Sigma, St. Louis, MO) for 4, 8, 12, and 16 h in the presence or absence of RU486 (100 nM). In other experiments, instead of RU486, indomethacin (20, 50, 100, or 200 μM), NS-398 (10 μM), or GW9662 (Cayman Chemical Co., Ann Arbor, MI) was added to cultures for 12 h. After the cells were washed in diethyl pyrocarbonate-phosphate-buffered saline (PBS), TRIzol (Invitrogen, Carlsbad, CA) was added to extract total RNA, which was then converted to cDNA to be analyzed by real-time quantitative PCR.
Real-time PCR analysis.
Total RNA was isolated from ovarian tissue and converted to cDNA by standard protocols. The cDNA was amplified by real-time PCR to quantify gene expression using gene-specific primers and Sybr (Bio-Rad Laboratories, Hercules, CA). For each gene, the threshold cycle (Ct) was determined at an amplification threshold of 150 for control (0 h hCG or 0 h forskolin and PMA) and test treatments (hCG or forskolin and PMA for different times). The expression level of the gene for 36B4, a gene encoding a ribosomal protein, was used as a loading control. For a given treatment, the mean Ct and standard deviation were calculated from individual Ct values obtained from three or four replicates of a sample. The normalized mean Ct was computed as ΔCt by subtracting the mean Ct of 36B4 from the Ct of a target gene for each treatment. ΔΔCt was then calculated as the difference between the ΔCt values of a control and each treatment group. The n-fold change in gene expression relative to a control (0 h hCG WT or 0 h forskolin and PMA) was computed as 2−ΔΔCt. The error bars indicate 2−ΔΔCt ± the standard deviation.
The PCR primers used were as follows: PPARγ, CGTGAAGCCCATCGAGGACATC (forward) and TGGAGCACCTTGGCGAACAG (reverse); PPARγ2, GGGTGAAACTCTGGGAGATTC (forward) and TCTTGTGAAGTGCTCATAGGC (reverse); COX-2, TCACGAAGGAACTCAGCACTG (forward) and GGAAGAGCATCGCAGAGGTG (reverse); ADAMTS-1, TGCTCCAAGACATGCGGCTCAG (forward) and TGGTACTGGCTGGCTTCACTTCC (reverse); cGKII, GCCCGATTCTCCTCAACCTCCC (forward) and TCCACTCTTCCGAACCCACCAAC (reverse); ET-2, CCTGTGCTACCTTCTGCCATC (forward) and CCCTCAGCAGTCCACATCTTG (reverse); IL-6, CCGCTATGAAGTTCCTCTCTGC (forward) and AGGGAAGGCCGTGGTTGTC (reverse); 36B4, CGACATCACAGAGCAGGC (forward) and CACCGAGGCAACAGTTGG (reverse).
In situ hybridization.
Ovaries were fixed in fresh 4% paraformaldehyde for 24 h at 4°C. The fixed tissues were cryoprotected by using increasing concentrations of sucrose solution as follows: 15 min in 10% sucrose at room temperature (RT), 1 h in 20% sucrose at RT, and overnight in 30% sucrose at 4°C. For cryosectioning, the frozen ovary was embedded in optimum cutting temperature compound, sectioned at 8 μm, and placed onto microscope slides (Superfrost Plus; Fisher Scientific, Pittsburgh, PA), which were stored at −80°C until further use. For in situ hybridization, frozen tissue sections were dried at 42°C for 1 h, fixed in 4% paraformaldehyde for 15 min, and subsequently treated with proteinase K solution (1 μg/ml) for 15 min, followed by 0.2 N HCl and triethanolamine (0.1 M)-0.25% (vol/vol) acetic anhydride for 20 min each. The sections were then dehydrated in increasing concentrations of ethanol (EtOH; 50%, 70%, 80%, 95%, and 100%) for 3 min each. Dehydrated slides were then prehybridized in hybridization buffer (20 mM Tris-HCl [pH 8], 0.3 M NaCl, 10% dextran sulfate, 50% formamide, single-stranded salmon sperm DNA [200 μg/ml], 1× Denhardt's solution) for 2 h at 55°C prior to hybridization with digoxigenin DIG-labeled RNA probes in a humid chamber overnight at 55°C. Sense or antisense DIG-labeled RNA probes were synthesized from a 510-bp PPARγ cDNA with a DIG RNA labeling kit (Roche Molecular Biochemicals, Indianapolis, IN). The PPARγ cDNA (+1008 to +1517) was subcloned into pPCR-script Amp SK(+) vector (Stratagene, La Jolla, CA) and served as a template for in vitro transcription of RNA probes using T3, T7, or SP6 RNA polymerase and DIG-labeled nucleotides. On the next day, slides were first washed in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min at 50°C, followed by two high-stringency washes (50% formamide, 2× SSC) for 15 min each at 50°C. Slides were then treated with RNase A (1 μg/ml; Sigma, St. Louis, MO) in a buffer (0.5 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA) for 15 min at 37°C. After being washed twice in maleic acid buffer (0.1 M maleic acid, 0.15 M NaCl, pH 7.5), sections were incubated with sheep polyclonal anti-DIG alkaline phosphatase-conjugated antibody in a humid chamber overnight at 4°C. After removal of excess antibodies by washing twice in detection buffer (0.1 M Tris-HCl, 50 mM MgCl2, 0.1 M NaCl, pH 9.5) for 15 min each, the substrates 5-bromo-4-chloro-3-indolylphosphate (BCIP) and nitroblue tetrazolium salt were added for color development, which was carried out for 8 to 24 h in the dark until optimal staining was achieved. Staining was then terminated in tap water. Sections were mounted using an aqueous mounting medium (Crystal/Mount; Biomeda Corp., Foster City, CA) prior to visualization under a light microscope (BX51; Olympus, Melville, NY).
Immunohistochemistry.
Ovaries were collected from WT or PR-null mice treated with PMSG for 48 h, followed by hCG for 5 h. The tissues were fixed in 10% formalin, freshly prepared in sterile PBS (pH 7.4), for 2 h at RT with gentle rocking. The ovaries were then washed twice in sterile PBS, dehydrated in increasing concentrations of EtOH (25%, 50%, 70%, 83%, 95%, 95%, 100%, and 100%, 30 min each), permeabilized in methyl salicylate, and embedded in paraffin. The paraffin blocks were sectioned at 4 to 5 μm and mounted onto microscope slides (Superfrost Plus; Fisher Scientific, Pittsburgh, PA). For immunostaining, paraffin sections were deparaffinized in xylene (twice, 5 min each), rehydrated in decreasing concentrations of EtOH (100%, 100%, and 95%, 3 min each), and rinsed in PBS for 5 min. Antigen retrieval was subsequently performed by boiling the sections in 0.01 M sodium citrate (pH 6) for 10 min, followed by incubation at RT for 20 min. To quench endogenous peroxidase, these sections were treated in 1.5% H2O2 (prepared in PBS) for 20 min at RT and rinsed three times with PBS for 3 min each. The sections were then incubated with 1% goat nonimmune serum in blocking solution (3% bovine serum albumin, 0.1% Triton X-100, sterile PBS) for 10 min at RT in a humid chamber. Further incubation was carried out using a rabbit polyclonal antibody against PPARγ (catalog no. 2492, diluted 1:50 in blocking solution; Cell Signaling Technology Inc., Danvers, MA) in a humid chamber overnight at 4°C. After washing three times with PBS, the sections were incubated with goat anti-rabbit biotinylated immunoglobulin G (Histostain Kit; Zymed Laboratories Inc., San Francisco, CA) for 30 min at RT, followed by three additional PBS washes. Finally, horseradish peroxidase-conjugated streptavidin was added and incubation was performed for 30 min at RT. After three washes with PBS, the sections were stained in AEC Solution (Zymed Laboratories Inc., San Francisco, CA) for 1 to 3 min until optimal signals were obtained. Staining was subsequently terminated by washing with tap water. The slides were mounted using aqueous mounting medium (20% glycerol, 96 mM Tris [pH 8.5], 8% [vol/vol] polyvinyl alcohol, sterile water) and examined under a light microscope.
RESULTS
PR regulates the expression of PPARγ mRNA in preovulatory follicles.
To identify the PR-regulated pathways that control ovulation in mice, we performed gene expression profiling using the ovarian tissues of prepubertal WT (129Sv) and PR-null (similar genetic background) female mice under superovulation conditions. It is well established that maximal expression of PR mRNA in the preovulatory follicles occurs at ∼3 to 4 h following the LH surge prior to ovulation (26). To identify the primary targets of PR regulation, we analyzed RNA samples isolated from PMSG-primed WT and PR-null mice that were treated with hCG for 5 h. The microarray analysis, using an Affymetrix mouse chip (GeneChip Mouse Genome 430A 2.0 array), interrogated more than 14,000 well-characterized mouse genes and uncovered several genes whose expression was up- or down-regulated in the PR-null ovaries compared to the WT ovaries (J. Kim and M. K. Bagchi, unpublished results). We noted with interest that the gene for PPARγ, a well-known member of the nuclear receptor superfamily of ligand-activated transcription factors, is one of the genes whose expression was significantly down-regulated in the PR-null ovaries.
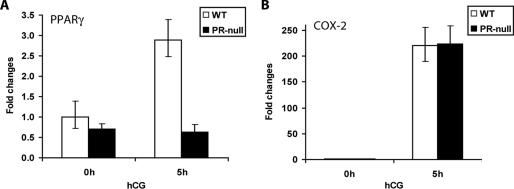
To confirm the results of our microarray analysis, we carried out a real-time PCR analysis using mRNA isolated from the WT and PR-null ovaries of mice subjected to superovulation. As shown in Fig. 1A, we observed a marked enhancement of the level of PPARγ mRNA in the ovary when PMSG-primed WT mice were treated with hCG for 5 h. This induction was absent in the ovaries of PR-null mice. In contrast, the expression level of COX-2, which is robustly induced in the ovaries of WT mice following hCG treatment, remained unaltered in the ovaries of PR-null mice under similar treatment conditions (Fig. 1B). These results clearly indicated that PPARγ is a specific downstream target of PR regulation in the ovarian tissue during the ovulatory process.
FIG. 1.
PR-mediated induction of PPARγ in the ovary during ovulation. Age-matched prepubertal WT (+/+) and PR-null (−/−) mice were treated with PMSG for 48 h prior to stimulation of ovulation by hCG. The ovaries were collected from these mice at 0 h (48 h PMSG) and 5 h after administration of hCG. Total RNA was isolated and converted by reverse transcription to cDNA. The cDNA was analyzed by real-time quantitative PCR using primer sets specific for PPARγ (A) and COX-2 (B), respectively. The expression of 36B4 mRNA, encoding a ribosomal protein, was used to normalize the variability of mRNA amounts in the samples.
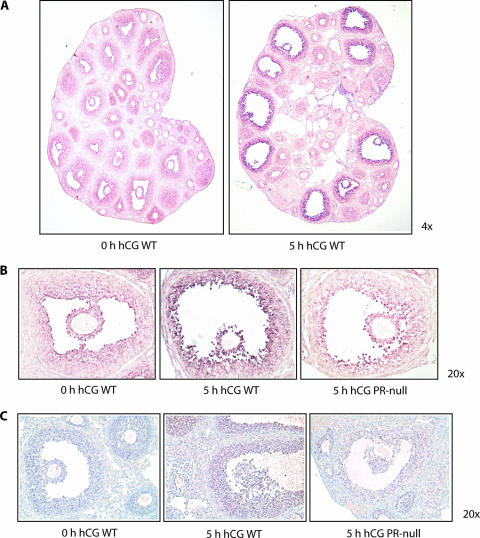
We next examined the spatial pattern of hCG-induced expression of PPARγ mRNA and protein in the ovary using in situ hybridization and immunohistochemistry, respectively. We noted that PPARγ mRNA was expressed at low levels in the mural and cumulus granulosa cells of all types of follicles, ranging from the primary to the preovulatory, in PMSG-treated WT ovaries (Fig. 2A). Strikingly, upon administration of hCG, the level of PPARγ mRNA was enhanced exclusively in the granulosa cells of the preovulatory follicles (Fig. 2A, right half, and B, left and middle parts). This induction of PPARγ was absent in the preovulatory follicles of the PR-null ovary (Fig. 2B, right part). Using an antibody raised against PPARγ, we detected specific immunostaining in both mural and cumulus granulosa cells of the preovulatory follicles of WT ovaries (Fig. 2C, middle part). This immunostaining was not observed in the ovarian cells of PR-null mice (Fig. 2C, right part). Taken together, these results are consistent with our view that the enhanced expression of PPARγ mRNA and protein in the granulosa cells during the ovulatory period is driven by hCG-induced expression of PR in these cells.
FIG. 2.
Localization of PPARγ in the ovary and its regulation by PR. (A) Spatial expression of PPARγ mRNA during ovulation. In situ hybridization was performed by using sense and antisense RNA probes corresponding to PPARγ cDNA to examine its expression in the ovaries of prepubertal WT (+/+) and PR-null (−/−) mice at 0 and 5 h after administration of hCG following a 48-h treatment with PMSG. Signals corresponding to the antisense RNA probe are shown. No signal for PPARγ mRNA was seen when the sense RNA probe was used. (B) Examination of PR regulation of PPARγ mRNA in the ovary during ovulation. Ovaries were collected from prepubertal WT (+/+) and PR-null (−/−) mice at 0 and 5 h after administration of hCG following a 48-h treatment with PMSG. The tissues were processed and subjected to in situ hybridization as described in Materials and Methods. (C) Analysis of PPARγ protein expression in the ovary. Ovaries from WT (0 and 5 h after administration of hCG) and PR-null (5 h after administration of hCG) mice were processed and subjected to immunohistochemical analysis using a rabbit polyclonal anti-PPARγ antibody as described in Materials and Methods.
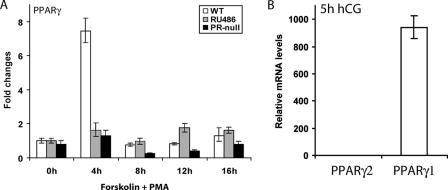
The PR regulation of PPARγ expression was further analyzed in vitro in primary cultures of granulosa cells obtained from the ovaries of WT and PR-null mice. Granulosa cells isolated from the preovulatory follicles of PMSG-primed WT or PR-null mice were treated with forskolin and PMA, which activate the protein kinase A and C pathways, respectively, thereby mimicking LH signaling (6). It was shown previously that expression of PR is maximally induced in the granulosa cells at 4 h following treatment with these reagents (35). Interestingly, we observed a robust induction of PPARγ mRNA expression at 4 h following treatment of primary granulosa cells with PMA and forskolin (Fig. 3A). In contrast, this induction was greatly reduced in granulosa cells isolated from PR-null ovaries. We also observed that treatment of WT granulosa cells with the PR antagonist RU486 prevented the induction of PPARγ expression by PMA and forskolin (Fig. 3A). These results further strengthened our hypothesis that PR is a critical regulator of PPARγ expression in granulosa cells during ovulation.
FIG. 3.
Expression of PPARγ in primary mouse granulosa cell cultures. (A) PR-dependent expression. Primary granulosa cells were extracted from ovaries of prepubertal (21 to 23 days old) WT (+/+) and PR-null (−/−) mice primed with PMSG for 48 h. To stimulate LH signaling, these cells were treated with a combination of forskolin (10 μM), an adenylyl cyclase activator that stimulates the protein kinase A pathway, and PMA (10 nM), a diacylglycerol analog that activates the protein kinase C pathway. RU486, a PR antagonist, was added to the WT primary cells along with forskolin and PMA to block P action. (B) Estimation of the relative levels of PPARγ1 and PPARγ2 transcripts in gonadotropin-stimulated granulosa cells. Granulosa cells were extracted from the ovaries of WT mice treated with PMSG for 48 h, followed by hCG for 5 h. Total RNA isolated from these cells was subjected to real-time PCR employing two distinct sets of primers, one specifically detecting PPARγ2 and the other detecting both isoforms. Since PPARγ2 transcripts were virtually undetectable, we infer that the PPARγ mRNAs represent transcripts corresponding to the γ1 isoform. The n-fold induction indicates the gene expression level at 5 h of hCG treatment relative to 0 h of hCG treatment as described in Materials and Methods.
Two isoforms of PPARγ, PPARγ1 and PPARγ2, have been described (5, 38). The mouse PPARγ2 isoform, which is mainly expressed in adipocytes, contains an additional 30 amino acids in the amino-terminal end compared to the PPARγ1 isoform (38). To determine the identity of the PPARγ isoform that is expressed in mouse ovarian granulosa cells during ovulation, we employed quantitative reverse transcription-PCR using two distinct sets of primers, one set detecting specifically PPARγ2 transcripts and the other set measuring transcripts encoding both the PPARγ1 and PPARγ2 isoforms. We found that the PPARγ2 transcripts were virtually undetectable in the gonadotropin-treated granulosa cells, whereas abundant expression of PPARγ transcripts, presumably representing the PPARγ1 isoform, was observed in these cells (Fig. 3B). By our estimate, at 5 h following hCG stimulation, the PPARγ1 mRNAs are expressed at a level that is at least 1,000-fold greater than that of PPARγ2 mRNAs.
Generation of mice with granulosa cell-specific excision of the PPARγ gene.
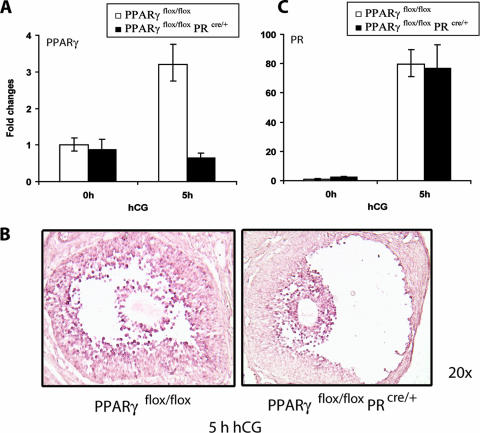
Mice carrying a PPARγ-null mutation die in utero due to a defect in placental development (2). To address the functional role of PPARγ during ovulation in adult mice, we therefore employed the Cre-LoxP strategy to generate a conditional mutant lacking the expression of this transcription factor in the ovary. We achieved granulosa cell-specific deletion of PPARγ by crossing female mice harboring a “floxed” PPARγ gene (1) with PR-Cre (33) male mice in which the gene encoding Cre is inserted at the PR gene locus. When the resulting PPARγflox/flox PRcre/+ female progenies were subjected to gonadotropin-induced superovulation, the expression of Cre, which is controlled by the regulatory regions of the endogenous PR gene, was induced in the granulosa cells. This led to the Cre-mediated excision of the “floxed” PPARγ gene in these cells, as evidenced by the complete loss of hCG-induced enhancement of PPARγ mRNA expression in the ovaries of the conditional mutant mice during the ovulatory period (Fig. 4A). In situ hybridization analysis confirmed that this Cre-mediated excision is restricted to the mural granulosa cells of the preovulatory follicles, consistent with the fact that PR expression occurs exclusively in these cells during ovulation (Fig. 4B). In contrast, no significant excision of the “floxed” PPARγ gene was detected in the cumulus granulosa cells, where PR is not expressed, indicating the cell-specific nature of the conditional mutation. We also noted that upon gonadotropin stimulation, the ovarian expression level of PR mRNA is not significantly different between the PPARγflox/flox mice containing both PR alleles and the PPARγflox/flox PRcre/+ mice containing only one PR allele (Fig. 4C). These results ruled out the possibility that the greatly reduced level of PPARγ in the ovaries of PPARγflox/flox PRcre/+ mice is a consequence of the deletion of one allele of the PR gene due to the targeted insertion of Cre at this locus.
FIG. 4.
PR-Cre-mediated excision of PPARγ in the preovulatory follicles of conditional PPARγ mutant mice. (A) PR-Cre-mediated ablation of the PPARγ floxed gene in the ovary. Cre-mediated excision of the floxed PPARγ gene was assessed by measuring the expression levels of PPARγ mRNA in ovaries of PPARγflox/flox and PPARγKO (PPARγflox/flox PRcre/+) mice following gonadotropin-induced ovulation. Ovaries were collected from PPARγflox/flox and PPARγKO (PPARγflox/flox PRcre/+) mice at 0 h (48 h PMSG) and 5 h after administration of hCG following 48 h of PMSG treatment. Total RNA was extracted and converted to cDNA by reverse transcription. The level of PPARγ expression was analyzed by real-time quantitative PCR using 36B4 as a normalizing control. (B) Cell type-specific loss of PPARγ expression in the ovary. PPARγ mRNA expression was examined by in situ hybridization in ovaries collected from PPARγflox/flox) and PPARγKO (PPARγflox/flox PRcre/+) mice at 5 h hCG following 48 h of PMSG treatment by using digoxigenin-labeled sense and antisense RNA probes specific for PPARγ. (C) Expression of PR mRNA in PPARγ conditional mutant ovaries. Ovaries were collected from PPARγflox/flox and PPARγflox/flox PRcre/+ mice at 0 and 5 h after administration of hCG following 48 h of PMSG treatment. Total RNA was isolated and converted to cDNA, and the levels of PR mRNA expression were analyzed by real-time quantitative PCR using 36B4 as a normalizing control. Changes were calculated corresponding to the PR mRNA level in 0-h PPARγflox/flox ovaries.
Loss of PPARγ expression in the preovulatory follicles impairs ovulation.
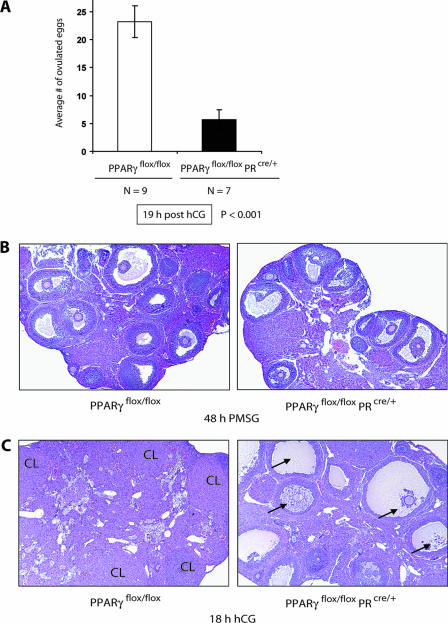
To assess the functional impact of granulosa cell-specific deletion of PPARγ in the preovulatory follicles, age-matched PPARγ mutant (PPARγflox/flox PRcre/+) and control (PPARγflox/flox) females were subjected to a superovulation protocol using PMSG for 48 h, followed by hCG treatment (Fig. 5A). The ovulated eggs were collected and counted at 19 h after administration of hCG. The number of eggs released from the ovaries of the conditional knockout mice (5.7 ± 2.8, n = 7) was markedly lower (∼75%, P < 0.001) than that released from the ovaries of the control mice (23.2 ± 1.7, n = 9). These results revealed that the loss of PPARγ expression in the preovulatory follicles results in significant impairment of ovulation. We also performed an extensive 6-month-long breeding study crossing PPARγflox/flox or PPARγflox/flox PRcre/+ females with PPARγflox/flox males under normal mating conditions. The results obtained from 16 breeding pairs of each type indicated that an average of 4.2 ± 2.2 pups were born to the conditional mutant mice, compared to the 9.0 ± 1.9 pups born to the control mice. Although the majority of mutant females delivered a significantly reduced number of pups (ranging from 60 to 80% fewer) compared to the control, a small number of the mutant mice showed only a modest reduction in litter size, which we speculate could be due to incomplete excision of the floxed PPARγ alleles in the granulosa cells during ovulation in these mice. Nevertheless, the overall marked reduction in litter size in the mutant mice was consistent with impaired ovulation and indicated that these mice are indeed subfertile.
FIG. 5.
Phenotypic effects of PPARγ gene deletion during ovulation. (A) Ovulatory effects of a PPARγ conditional mutation in the preovulatory follicles. Age-matched PPARγflox/flox and PPARγflox/flox PRcre/+ (PPARγ conditional null) mice were superovulated with PMSG for 48 h, followed by hCG for 18 h, and the ovulated eggs were counted. The average ± the standard error of the mean was estimated, and statistical significance was assessed at a critical P value (α) of 0.05. (B) Follicular development in the conditional mutant ovary. The effect of PPARγ deletion on follicle development was assessed by examining the ovarian histology of PPARγflox/flox and PPARγflox/flox PRcre/+ mice following stimulation of follicular development by treatment with PMSG for 48 h. The ovaries were fixed in 10% formalin. Paraffin-embedded sections were stained with hematoxylin and eosin and examined by light microscopy (magnification, ×5). (C) Impaired rupture of preovulatory follicles deficient in PPARγ. The ovaries of PPARγflox/flox and PPARγflox/flox PRcre/+ mice were collected at 18 h after administration of hCG following the superovulation experiment as described for panel A and examined histologically to determine the state of follicular rupture. The arrows (right) refer to unruptured preovulatory follicles. Although the oocyte was not visible in some of the unruptured follicles, its presence was confirmed in adjacent paraffin sections obtained from the same tissue. A small number of corpora lutea were also seen in the mutant ovaries.
Conditional loss of the PPARγ gene in granulosa cells does not affect ovarian follicular development but prevents follicular rupture.
We next examined whether the loss of ovulatory function in the mutant mice harboring a granulosa cell-specific deletion of the PPARγ gene is a consequence of a defect in follicular development or follicular rupture. Age-matched control (PPARγflox/flox) and PPARγ mutant (PPARγflox/flox PRcre/+) mice were treated with PMSG for 48 h to stimulate follicular growth and development. The ovarian sections obtained from these mice exhibited similar numbers and patterns of distribution of the primary, secondary, and preovulatory follicles (Fig. 5B), indicating that Cre-mediated excision of PPARγ did not affect follicular development in these mutant mice.
To determine whether the compromised ovulation observed in the conditional PPARγ-null mice arises from a lack of follicular rupture, we conducted a histological examination of the ovaries of PPARγ-null and control mice that were subjected to PMSG-hCG-induced superovulation (Fig. 5C). In this protocol, follicular rupture typically takes place between 11 and 12 h after administration of hCG. At 19 h after administration of hCG, the control (PPARγflox/flox) ovaries were composed of numerous corpora lutea, indicating that the follicles had ruptured and luteinzation ensued. In contrast, the ovaries of PPARγflox/flox PRcre/+ mice were found to contain very few corpora lutea but harbored many unruptured preovulatory follicles with trapped oocytes at 19 h after administration of hCG (Fig. 5C, right half). These results clearly indicated that PPARγ plays a vital role during follicular rupture.
PPARγ regulates a subset of PR target genes in granulosa cells.
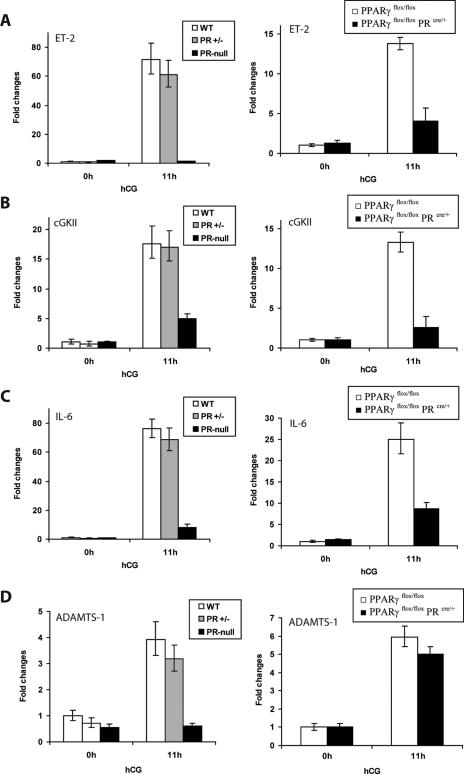
Our findings that PPARγ is a downstream target of regulation by PR in the ovary and the observation that the functional loss of each of these transcription factors results in impaired follicular rupture strongly suggested that these molecules are components of a linear signaling cascade during ovulation. To consider the possibility that PPARγ is a downstream mediator of PR-dependent control of gene expression during ovulation, we investigated whether it regulates known PR target genes. Previous studies by Richards and coworkers identified ADAMTS-1 and cGKII as PR-regulated genes in the ovary (29, 34). We have recently employed gene expression profiling to identify the genes for ET-2 and IL-6 as additional PR target genes (25). We therefore examined the expression of ADAMTS-1, ET-2, cGKII, and IL-6 in the ovaries of mutant mice lacking either PR or PPARγ expression.
Consistent with previous reports, administration of hCG to PMSG-primed WT or heterozygous (PR+/−) mice led to robust induction of the expression of the ET-2, cGKII, IL-6, and ADAMTS-1 mRNAs in the ovaries of these animals (Fig. 6A, B, C, and D, left halves). In contrast, similar treatment of homozygous (PR−/−) mice failed to induce the expression of these genes in the ovaries of these mutant animals, thereby confirming the PR regulation of these genes. Interestingly, we noted that the hCG-mediated enhancement of the ET-2, cGKII, and IL-6 mRNAs was markedly diminished upon Cre-mediated deletion of the PPARγ gene in the ovaries of PPARγflox/flox PRcre/+ mice compared to that seen in the ovaries of their control (PPARγflox/flox) littermates (Fig. 6A, B, and C, right halves). These results indicated that ET-2, cGKII, and IL-6 are under dual regulation by PR and PPARγ. In contrast, there was no significant difference in the levels of ADAMTS-1 mRNA in PPARγflox/flox and PPARγflox/flox PRcre/+ ovaries at 11 h after administration of hCG. These results indicated that ADAMTS-1 expression is regulated by PR, but not by PPARγ, during ovulation. Collectively, these results indicated that PPARγ controls the ovarian expression of a specific subset of PR-regulated genes, including those for ET-2, cGKII, and IL-6, and that it is a likely mediator of the PR-controlled functional events during ovulation.
FIG. 6.
PPARγ regulation of target genes during the ovulatory period. Age-matched WT (PR+/+), heterozygous (PR+/−), and PR-null (PR−/−) females were treated with hCG for 11 h following PMSG priming for 48 h. Ovaries were collected from these mice; total RNA was isolated and converted to cDNA; and ET-2 (A), cGKII (B), IL-6 (C), and ADAMTS-1 (D) mRNA levels were assessed by real-time quantitative reverse transcription-PCR analysis (left side). Similar expression analyses were also performed with ovaries collected from PPARγflox/flox and PPARγflox/flox PRcre/+ (PPARγ conditional null) mice to determine the PPARγ regulation of these genes (right side).
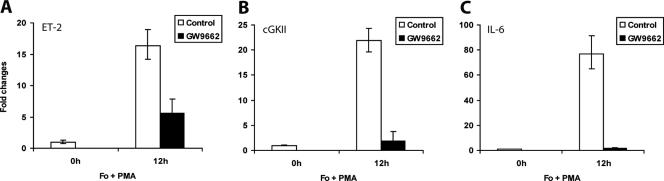
To further ascertain these in vivo findings, we next examined PPARγ regulation of cGKII, ET-2, and IL-6 in primary granulosa cell cultures by using a selective antagonist of this nuclear receptor. Treatment of primary granulosa cells isolated from PMSG-primed mice with PMA and forskolin dramatically enhanced the expression of the ET-2, cGKII, and IL-6 mRNAs within 12 h of treatment (Fig. 7A, B, and C). When these cells were treated with the PPARγ-specific antagonist GW9662 (3, 11) along with forskolin and PMA, the gene induction was sharply reduced in each case. The administration of GW9662 did not alter the expression level of PR, PPARγ, or COX-2 (data not shown), indicating that the inhibitory effects of this antagonist are gene specific. These results confirmed that PPARγ regulates the expression of cGKII, ET-2, and IL-6 in the granulosa cells of the preovulatory follicles.
FIG. 7.
A PPARγ-selective antagonist inhibits the expression of several PR target genes in primary granulosa cells. Primary granulosa cells were extracted from the ovaries of prepubertal mice primed with PMSG for 48 h and were cultured overnight in 1% fetal bovine serum-DMEM-F12. The cells were then treated with forskolin (Fo; 10 μM) and PMA (20 nM) to activate LH signaling in the presence or absence of GW9662 (1 μM), a PPARγ-selective antagonist. Total RNA was isolated from these cells, and the expression of mRNAs corresponding to ET-2 (A), cGKII (B), and IL-6 (C) was then measured by real-time quantitative reverse transcription-PCR using gene-specific primers as described in Materials and Methods.
COX-derived metabolites of endogenous long-chain fatty acids are potential activating ligands of PPARγ.
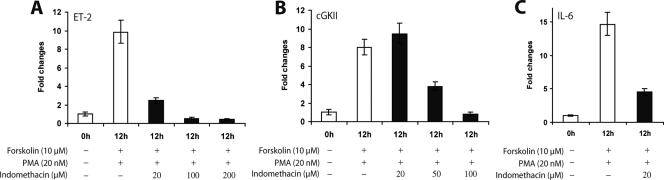
Although PPARγ was initially identified as an orphan nuclear receptor (5, 38), it is now known to be activated by an array of metabolites derived from unsaturated long-chain fatty acids such as arachidonic acid and linoleic acid. The cyclopentanoic prostaglandin derivative 15d-PGJ2, a COX-derived metabolite of arachidonic acid, was first reported to be a natural ligand for PPARγ (10, 13). It is also well established that COX-2, an LH/hCG-induced COX enzyme that catalyzes the conversion of arachidonic acid and related fatty acids into prostaglandins (12), is essential for ovulation (7, 21). Taken together, these facts raised the possibility that the COX enzymes may contribute to the ovulatory process by providing an endogenous activating ligand(s) for PPARγ. To test this hypothesis, we employed indomethacin, a nonsteroidal antiinflammatory drug that inhibits the activity of the COX enzymes and also blocks ovulation (37). We examined the effects of this inhibitor on the expression of the PPARγ target genes: the ET-2, cGKII, and IL-6 genes. Primary granulosa cells extracted from PMSG-primed mouse ovaries were treated with forskolin and PMA in the presence of increasing doses of indomethacin. As shown in Fig. 8, the expression of all three genes was markedly reduced in the granulosa cells treated with indomethacin compared to that in untreated cells. A similar suppression of the expression of these genes was seen when the primary granulosa cell cultures were treated with NS-398 (9), a selective COX-2 inhibitor (Fig. 9A, B, and C, lanes 1 to 3). These results provided strong support to our hypothesis that COX-derived metabolites of arachidonic acid or related fatty acids serve as endogenous activators of PPARγ signaling during ovulation. Most importantly, this finding provided, for the first time, an insight into the mechanisms by which two apparently unrelated regulatory routes downstream of LH/hCG signaling, namely, the PR-PPARγ pathway and the COX-2 pathway, may functionally converge to regulate the events leading to ovulation.
FIG. 8.
Inhibition of COX enzymes represses PPARγ-mediated gene expression. Primary granulosa cells were isolated from the ovaries of prepubertal mice primed with PMSG for 48 h. The cells were stimulated with forskolin (10 μM) and PMA (20 nM) in the presence or absence of increasing concentrations of indomethacin (20, 50, 100, and/or 200 μM), an inhibitor of COX enzymes. The expression of mRNAs corresponding to ET-2 (A), cGKII (B), and IL-6 (C) was then measured by real-time quantitative reverse transcription-PCR using gene-specific primers as described in Materials and Methods. Note that in the case of IL-6, only 20 μM indomethacin was used.
FIG. 9.
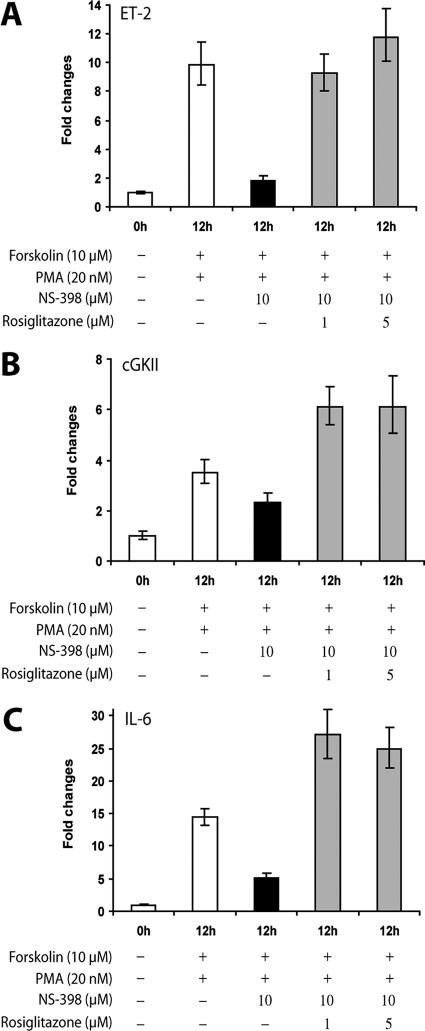
Repression of PPARγ target genes by a COX-2-selective inhibitor is reversed by rosiglitazone. Primary granulosa cells were isolated from the ovaries of prepubertal mice primed with PMSG for 48 h. The cells were stimulated with forskolin (10 μM) and PMA (20 nM) in the presence or absence of NS-398 (10 μM), a COX-2-specific inhibitor. Rosiglitazone (1 or 5 μM), a PPARγ-specific agonist ligand, was added as indicated. Gene expression was analyzed by real-time quantitative PCR using gene-specific primers for ET-2 (A), cGKII (B), and IL-6 (C), respectively.
If one or more COX-derived metabolites control the gene-regulatory function of PPARγ in granulosa cells, one would predict that the administration of an exogenous agonist ligand of this nuclear receptor would overcome indomethacin- or NS-398-mediated inhibition of ET-2, cGKII, and IL-6 gene expression in these cells. Indeed, as shown in Fig. 9A, B, and C, addition of rosiglitazone (19), a potent synthetic agonist of PPARγ, to granulosa cell cultures completely reversed NS-398-mediated inhibition of ET-2, cGKII, and IL-6 gene expression, providing further evidence in favor of our hypothesis that activation of PPARγ by COX-derived ligands generated in granulosa cells is a critical step in regulating downstream gene networks that participate in the ovulatory process.
DISCUSSION
In this study, we have identified the transcription factor PPARγ as a critical regulator of ovulation. An extensive literature links PPARγ to numerous biological and pathological processes (4, 17, 18, 24, 30, 39). It is recognized as a key regulator of adipocyte differentiation, lipid and glucose homeostasis, insulin sensitization, inflammation, and atherosclerosis. The evidence for a role for PPARγ in female reproduction is, however, limited. In the rat ovary, PPARγ is expressed in preantral and antral follicles, although the functional significance of this expression remains unexplored. Since a null mutation of the PPARγ gene caused embryonic lethality in mice (2), we generated a conditional mutant mouse that is deficient in PPARγ expression in the granulosa cells of the preovulatory ovarian follicles. Using this mutant mouse model, we demonstrated that granulosa cell-specific deletion of PPARγ results in marked impairment of ovulation due to defective follicular rupture and thereby uncovered a critical ovarian function of this factor.
Our study provided strong evidence linking PR function and PPARγ expression during ovulation. Gonadotropin-stimulated ovaries of WT mice at 5 h after administration of hCG exhibited markedly higher PPARγ mRNA expression compared to PR-null ovaries under similar treatment conditions (Fig. 1). Additionally, when primary granulosa cells isolated from PMSG-primed ovaries of WT and PR-null mice were cultured and treated with forskolin and PMA to mimic LH signaling, the expression of PPARγ mRNA was drastically reduced in cells from PR-null ovaries relative to that in cells from WT ovaries. Consistent with these results, PPARγ induction in granulosa cells obtained from WT ovaries was suppressed in the presence of RU486 (22), a potent PR antagonist. Collectively, these findings established that PPARγ is a downstream target of PR regulation in granulosa cells of the preovulatory follicle during the ovulatory process. Considering that the PR expression in the granulosa cells of preovulatory follicles temporally overlaps that of PPARγ, it is conceivable that PR regulates PPARγ expression during ovulation by directly interacting with a DNA sequence containing a palindromic P response element (PRE) in the regulatory regions of the PPARγ gene. However, an in silico analysis revealed no readily identifiable PRE in the 5′-flanking region of the PPARγ gene, although multiple imperfect motifs containing PRE half sites were located. It is known that PR can regulate its target genes via tethering to other promoter-binding transcription factors such as Sp1 and AP1 (8). Further investigations are necessary to determine whether PR regulates PPARγ via direct or indirect mechanisms.
The generation of a PPARγ conditional null model using PR-Cre mice (33) not only avoided embryonic lethality of the PPARγ null mutation but also ensured that within the ovary the excision of the floxed PPARγ gene would occur only in those follicular cells which naturally express PR during ovulation. This unique strategy restricted the excision of PPARγ to the mural granulosa cells of the preovulatory follicles where PR is exclusively expressed during the ovulatory process. No excision of PPARγ was seen in the smaller developing follicles or cumulus cells surrounding the oocyte, the sites where PR is not expressed (Fig. 4B). Gonadotropin-stimulated follicular development was indistinguishable between the ovaries of control mice (PPARγflox/flox) and those of conditional PPARγ-null mice (PPARγflox/flox PRcre/+), thus ruling out any defect in folliculogenesis in the mutant mice (Fig. 5B). In contrast, Cre-mediated ablation of this gene in the mural granulosa cells of the preovulatory follicles results in the release of a drastically reduced number of eggs than in control animals (PPARγflox/flox) during ovulation (Fig. 5A). Although PR-driven Cre expression in the gonadotrophs of the anterior pituitary is expected to delete the PPARγ gene in these cells, the administration of exogenous gonadotropins in the superovulation experiments bypasses the requirement for endogenous synthesis of these hormones and therefore rules out the possibility that the observed ovulatory phenotype originates from impaired gonadotropin production by the pituitary. Examination of the ovaries of conditional PPARγ-null mice beyond the time of ovulation revealed the presence of trapped oocytes, indicating that failure of follicular rupture accounts for the dramatic decline in egg release (Fig. 5C). Since a similar defect in follicular rupture is also observed in PR-null mice, our results are consistent with the view that PPARγ is a critical mediator of the ovulatory effects of PR.
To explore the molecular pathways controlled by PPARγ during ovulation, we considered the possibility that it is a downstream mediator of certain of the gene regulatory effects of PR during ovulation. Recent ovarian gene expression profiling experiments in our laboratory and elsewhere have indicated that several genes, such as those for ET-2, cGKII, and IL-6, are regulated by PR during the ovulatory process, although the precise mode of this regulation remains unclear. It is of interest that PR mRNA expression in the preovulatory follicles is maximal at 4 to 5 h following LH/hCG stimulation but sharply declines thereafter and becomes undetectable near the time of ovulation. In contrast, the mRNAs encoding ET-2, cGKII, and IL-6 are induced between 10 and 11 h following LH/hCG treatment, raising the possibility that PR regulates the expression of these genes indirectly via a primary target gene product such as PPARγ. Consistent with this prediction, the expression of ET-2, cGKII, and IL-6 was markedly reduced in conditional PPARγ mutant mouse ovaries relative to control (PPARγflox/flox) ovaries. Interestingly, the expression of ADAMTS-1, another known PR target gene, remained unaffected in PPARγ-null ovaries, indicating that PR regulation of this gene occurs via a signaling pathway independent of PPARγ. Therefore, PPARγ is likely to regulate the expression of a specific subset of PR-regulated genes during ovulation. The existence of distinct signaling pathways, PPARγ dependent and PPARγ independent, channeling the ovulatory effects of PR may also help explain why the ovulatory defect seen in PPARγ-null mice is somewhat less severe compared to the anovulatory phenotype displayed by PR-null mice.
We also investigated the possible mode of activation of PPARγ during the ovulatory process. Previous studies indicated that a variety of metabolites of long-chain unsaturated fatty acids, such as arachidonic and linoleic acids, are able to function as endogenous activating ligands for PPARγ and regulate its transcriptional activity (17, 31). These ligands include the COX-derived metabolites, such as the prostaglandin derivative 15d-PGJ2, and the lipoxygenase-derived metabolites, such as 12- and 15-hydroxyeicosatetraenoic acids and 9- and 13-hydroxyoctadecadienoic acids (10, 13, 20, 24). It is well documented that COX-2 and its metabolic products play critical roles in ovulation (7). Indomethacin, a nonsteroidal anti-inflammatory drug and a broad-spectrum COX inhibitor, is an efficient blocker of ovulation (37). It is conceivable that COX-2-derived metabolites function in ovulation by serving as activating ligands of PPARγ. In support of this concept, we found that administration of indomethacin or the selective COX-2 inhibitor NS-398 strongly suppressed PPARγ-dependent gene expression in ovarian granulosa cells. This finding provided a novel functional link between the pathways downstream of PR and COX-2, two key molecules that are induced during ovulation and are essential for this process. We forward the hypothesis that metabolism of long-chain unsaturated fatty acids by COX-2 in mural granulosa cells may provide the endogenous ligand(s) that activates PPARγ in the ovarian follicles and regulates the expression of its downstream target genes to control ovulation. Interestingly, the expression of PPARγ is also seen in the cumulus cells during the ovulatory period, although the functional significance of this expression remains unclear. It is possible that COX-2, which is also expressed in the cumulus cells, furnishes the modulatory ligands, which in turn control the function of PPARγ in these cells.
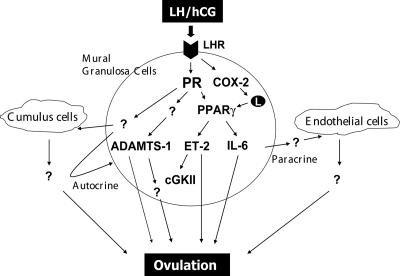
The biological events and the precise mechanisms by which PPARγ and its downstream targets, ET-2, cGKII, and IL-6, regulate follicular rupture and ovulation are not yet fully understood. Interestingly, recent studies in our laboratory and elsewhere have identified ET-2 as a critical regulator of ovulation in the mouse (14, 25). Blockade of ET-2 action using an antagonist of its cellular receptor during gonadotropin-induced superovulation resulted in severe impairment of follicular rupture. We have also identified cGKII and IL-6 as downstream targets of regulation by ET-2 (25). It is likely that PPARγ mediates its ovulatory effects in part by regulating the transcription of ET-2, cGKII, and IL-6. Our results, therefore, unveiled a novel signaling network comprising PR, PPARγ, ET-2, cGKII, IL-6, and possibly other downstream molecules that control ovulation (Fig. 10). Further studies using the conditional PPARγ-null mouse model will clarify the precise molecular interplay that conveys the signals that direct the ovulatory program leading to follicular rupture.
FIG. 10.
A network of PR- and PPARγ-regulated genes controls ovulation. Ovulatory action is initiated by LH, which acts via its receptor on mural granulosa cells of the preovulatory follicles to induce expression of PR in these cells. PR, in turn, induces the expression of PPARγ, a ligand-inducible transcription factor, which is activated by fatty acid metabolites generated by LH-induced COX-2. Activated PPARγ mediates PR function by controlling the expression of a subset of PR target genes, including ET-2, cGKII, and IL-6. It is likely that additional PR-dependent pathways, which are not regulated by PPARγ, such as ADAMTS-1, also contribute to the process of follicular rupture. The products of certain of the PPARγ target genes, such as ET-2 and IL-6, are thought to be released from mural granulosa cells of the preovulatory follicles and act on other ovarian cell types, such as endothelial cells of blood vessels and cumulus cells, via a paracrine mode. They may also act on mural granulosa cells via autocrine mechanisms. We envision that the concerted action of these autocrine and paracrine effectors ultimately brings about ovulation.
Acknowledgments
We thank Frank Gonzalez of the National Cancer Institute for providing PPARγ-floxed mice. We also thank David Roh for help on genotyping and Lori Raetzman and her graduate student Pamela Monahan for sharing their insight on immunohistochemistry.
This work is supported by NIH grants R01 HD044611 (to M.K.B.), U54 HD299901-12 (to I.C.B. and Regine L. Sitruk-Ware), and CA 77530 (to J.P.L.).
Footnotes
Published ahead of print on 2 January 2008.
REFERENCES
- 1.Akiyama, T. E., S. Sakai, G. Lambert, C. J. Nicol, K. Matsusue, S. Pimprale, Y.-H. Lee, M. Ricote, C. K. Glass, H. B. Brewer, Jr., and F. J. Gonzalez. 2002. Conditional disruption of the peroxisome proliferator-activated receptor γ gene in mice results in lowered expression of ABCA1, ABCG1, and apoE in macrophages and reduced cholesterol efflux. Mol. Cell. Biol. 222607-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barak, Y., M. C. Nelson, E. S. Ong, Y. Z. Jones, P. Ruiz-Lozano, K. R. Chien, A. Koder, and R. M. Evans. 1999. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell 4585-595. [DOI] [PubMed] [Google Scholar]
- 3.Bendixen, A. C., N. K. Shevde, K. M. Dienger, T. M. Willson, C. D. Funk, and J. W. Pike. 2001. IL-4 inhibits osteoclast formation through a direct action on osteoclast precursors via peroxisome proliferator-activated receptor γ1. Proc. Natl. Acad. Sci. USA 982443-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chawla, A., E. J. Schwarz, D. D. Dimaculangan, and M. A. Lazar. 1994. Peroxisome proliferator-activated receptor (PPAR) γ: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology 135798-800. [DOI] [PubMed] [Google Scholar]
- 5.Chen, F., S. W. Law, and B. W. O'Malley. 1993. Identification of two mPPAR related receptors and evidence for the existence of five subfamily members. Biochem. Biophys. Res. Commun. 196671-677. [DOI] [PubMed] [Google Scholar]
- 6.Clemens, J. W., R. L. Robker, W. L. Kraus, B. S. Katzenellenbogen, and J. S. Richards. 1998. Hormone induction of progesterone receptor (PR) messenger ribonucleic acid and activation of PR promoter regions in ovarian granulosa cells: evidence for a role of cyclic adenosine 3′,5′-monophosphate but not estradiol. Mol. Endocrinol. 121201-1214. [DOI] [PubMed] [Google Scholar]
- 7.Davis, B. J., D. E. Lennard, C. A. Lee, H. F. Tiano, S. G. Morham, W. C. Wetsel, and R. Langenbach. 1999. Anovulation in cyclooxygenase-2-deficient mice is restored by prostaglandin E2 and interleukin-1β. Endocrinology 1402685-2695. [DOI] [PubMed] [Google Scholar]
- 8.Doyle, K. M., D. L. Russell, V. Sriraman, and J. S. Richards. 2004. Coordinate transcription of the ADAMTS-1 gene by luteinizing hormone and progesterone receptor. Mol. Endocrinol. 182463-2478. [DOI] [PubMed] [Google Scholar]
- 9.Elvin, J. A., C. Yan, and M. M. Matzuk. 2000. Growth differentiation factor-9 stimulates progesterone synthesis in granulosa cells via a prostaglandin E2/EP2 receptor pathway. Proc. Natl. Acad. Sci. USA 9710288-10293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forman, B. M., P. Tontonoz, J. Chen, R. P. Brun, B. M. Spiegelman, and R. M. Evans. 1995. 15-Deoxy-δ12,14-prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell 83803-812. [DOI] [PubMed] [Google Scholar]
- 11.Huang, J. T., J. S. Welch, M. Ricote, C. J. Binder, T. M. Willson, C. Kelly, J. L. Witztum, C. D. Funk, D. Conrad, and C. K. Glass. 1999. Interleukin-4-dependent production of PPAR-γ ligands in macrophages by 12/15-lipoxygenase. Nature 400378-382. [DOI] [PubMed] [Google Scholar]
- 12.Joyce, I. M., F. L. Pendola, M. O'Brien, and J. J. Eppig. 2001. Regulation of prostaglandin-endoperoxide synthase 2 messenger ribonucleic acid expression in mouse granulosa cells during ovulation. Endocrinology 1423187-3197. [DOI] [PubMed] [Google Scholar]
- 13.Kliewer, S. A., J. M. Lenhard, T. M. Willson, I. Patel, D. C. Morris, and J. M. Lehmann. 1995. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor γ and promotes adipocyte differentiation. Cell 83813-819. [DOI] [PubMed] [Google Scholar]
- 14.Ko, C., M. C. Gieske, L. Al-Alem, Y. Hahn, W. Su, M. C. Gong, M. Iglarz, and Y. Koo. 2006. Endothelin-2 in ovarian follicle rupture. Endocrinology 1471770-1779. [DOI] [PubMed] [Google Scholar]
- 15.Komar, C. M. 2005. Peroxisome proliferator-activated receptors (PPARs) and ovarian function—implications for regulating steroidogenesis, differentiation, and tissue remodeling. Reprod. Biol. Endocrinol. 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komar, C. M., and T. E. Curry, Jr. 2003. Inverse relationship between the expression of messenger ribonucleic acid for peroxisome proliferator-activated receptor γ and P450 side chain cleavage in the rat ovary. Biol. Reprod. 69549-555. [DOI] [PubMed] [Google Scholar]
- 17.Lazar, M. A. 2005. PPARγ, 10 years later. Biochimie 879-13. [DOI] [PubMed] [Google Scholar]
- 18.Lee, C.-H., and R. M. Evans. 2002. Peroxisome proliferator-activated receptor-γ in macrophage lipid homeostasis. Trends Endocrinol. Metab. 13331-335. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann, J. M., L. B. Moore, T. A. Smith-Oliver, W. O. Wilkison, T. M. Willson, and S. A. Kliewer. 1995. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ (PPARγ). J. Biol. Chem. 27012953-12956. [DOI] [PubMed] [Google Scholar]
- 20.Li, Q., Y.-P. Cheon, A. Kannan, S. Shanker, I. C. Bagchi, and M. K. Bagchi. 2004. A novel pathway involving progesterone receptor, 12/15-lipoxygenase-derived eicosanoids, and peroxisome proliferator-activated receptor γ regulates implantation in mice. J. Biol. Chem. 27911570-11581. [DOI] [PubMed] [Google Scholar]
- 21.Lim, H., B. C. Paria, S. K. Das, J. E. Dinchuk, R. Langenbach, J. M. Trzaskos, and S. K. Dey. 1997. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell 91197-208. [DOI] [PubMed] [Google Scholar]
- 22.Loutradis, D., R. Bletsa, L. Aravantinos, K. Kallianidis, S. Michalas, and A. Psychoyos. 1991. Preovulatory effects of the progesterone antagonist mifepristone (RU486) in mice. Hum. Reprod. 61238-1240. [DOI] [PubMed] [Google Scholar]
- 23.Lydon, J. P., F. J. DeMayo, C. R. Funk, S. K. Mani, A. R. Hughes, C. A. Montgomery, Jr., G. Shyamala, O. M. Conneely, and B. W. O'Malley. 1995. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 92266-2278. [DOI] [PubMed] [Google Scholar]
- 24.Nagy, L., P. Tontonoz, J. G. Alvarez, H. Chen, and R. M. Evans. 1998. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARγ. Cell 93229-240. [DOI] [PubMed] [Google Scholar]
- 25.Palanisamy, G. S., Y. P. Cheon, J. Kim, A. Kannan, Q. Li, M. Sato, S. R. Mantena, R. L. Sitruk-Ware, M. K. Bagchi, and I. C. Bagchi. 2006. A novel pathway involving progesterone receptor, endothelin-2, and endothelin receptor B controls ovulation in mice. Mol. Endocrinol. 202784-2795. [DOI] [PubMed] [Google Scholar]
- 26.Park, O. K., and K. E. Mayo. 1991. Transient expression of progesterone receptor messenger RNA in ovarian granulosa cells after the preovulatory luteinizing hormone surge. Mol. Endocrinol. 5967-978. [DOI] [PubMed] [Google Scholar]
- 27.Richards, J. S. 1994. Hormonal control of gene expression in the ovary. Endocr. Rev. 15725-751. [DOI] [PubMed] [Google Scholar]
- 28.Richards, J. S., D. L. Russell, S. Ochsner, and L. L. Espey. 2002. Ovulation: new dimensions and new regulators of the inflammatory-like response. Annu. Rev. Physiol. 6469-92. [DOI] [PubMed] [Google Scholar]
- 29.Robker, R. L., D. L. Russell, L. L. Espey, J. P. Lydon, B. W. O'Malley, and J. S. Richards. 2000. Progesterone-regulated genes in the ovulation process: ADAMTS-1 and cathepsin L proteases. Proc. Natl. Acad. Sci. USA 974689-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosen, E. D., and B. M. Spiegelman. 2001. PPARγ: a nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 27637731-37734. [DOI] [PubMed] [Google Scholar]
- 31.Schopfer, F. J., Y. Lin, P. R. S. Baker, T. Cui, M. Garcia-Barrio, J. Zhang, K. Chen, Y. E. Chen, and B. A. Freeman. 2005. Nitrolinoleic acid: an endogenous peroxisome proliferator-activated receptor γ ligand. Proc. Natl. Acad. Sci. USA 1022340-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimada, M., Y. Yanai, T. Okazaki, Y. Yamashita, V. Sriraman, M. C. Wilson, and J. S. Richards. 2007. Synaptosomal-associated protein 25 (Snap25) gene expression is hormonally regulated during ovulation and is involved in cytokine/chemokine exocytosis from granulosa cells. Mol. Endocrinol. 212487-2502. [DOI] [PubMed] [Google Scholar]
- 33.Soyal, S. M., A. Mukherjee, K. Y. Lee, J. Li, H. Li, F. J. DeMayo, and J. P. Lydon. 2005. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis 4158-66. [DOI] [PubMed] [Google Scholar]
- 34.Sriraman, V., M. D. Rudd, S. M. Lohmann, S. M. Mulders, and J. S. Richards. 2006. Cyclic 5′-monophosphate-dependent protein kinase II is induced by luteinizing hormone and progesterone receptor-dependent mechanisms in granulosa cells and cumulus oocyte complexes of ovulating follicles. Mol. Endocrinol. 20348-361. [DOI] [PubMed] [Google Scholar]
- 35.Sriraman, V., S. C. Sharma, and J. S. Richards. 2003. Transactivation of the progesterone receptor gene in granulosa cells: evidence that Sp1/Sp3 binding sites in the proximal promoter play a key role in luteinizing hormone inducibility. Mol. Endocrinol. 17436-449. [DOI] [PubMed] [Google Scholar]
- 36.Sterneck, E., L. Tessarollo, and P. F. Johnson. 1997. An essential role for C/EBPβ in female reproduction. Genes Dev. 112153-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka, N., L. L. Espey, T. Kawano, and H. Okamura. 1991. Comparison of inhibitory actions of indomethacin and epostane on ovulation in rats. Am. J. Physiol. 260E170-E174. [DOI] [PubMed] [Google Scholar]
- 38.Tontonoz, P., E. Hu, R. A. Graves, A. I. Budavari, and B. M. Spiegelman. 1994. mPPARγ2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 81224-1234. [DOI] [PubMed] [Google Scholar]
- 39.Tontonoz, P., L. Nagy, J. G. Alvarez, V. A. Thomazy, and R. M. Evans. 1998. PPARγ promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell 93241-252. [DOI] [PubMed] [Google Scholar]