Abstract
The recently discovered nucleotide binding domain-leucine rich repeat (NLR) gene family is conserved from plants to mammals, and several members are associated with human autoinflammatory or immunodeficiency disorders. This family is defined by a central nucleotide binding domain that contains the highly conserved Walker A and Walker B motifs. Although the nucleotide binding domain is a defining feature of this family, it has not been extensively studied in its purified form. In this report, we show that purified Monarch-1/NLRP12, an NLR protein that negatively regulates NF-κB signaling, specifically binds ATP and exhibits ATP hydrolysis activity. Intact Walker A/B motifs are required for this activity. These motifs are also required for Monarch-1 to undergo self-oligomerization, Toll-like receptor- or CD40L-activated association with NF-κB-inducing kinase (NIK) and interleukin-1 receptor-associated kinase 1 (IRAK-1), degradation of NIK, and inhibition of IRAK-1 phosphorylation. The stable expression of a Walker A/B mutant in THP-1 monocytes results in increased production of proinflammatory cytokines and chemokines to an extent comparable to that in cells in which Monarch-1 is silenced via short hairpin RNA. The results of this study are consistent with a model wherein ATP binding regulates the anti-inflammatory activity of Monarch-1.
Nucleotide binding domain-leucine rich repeat (NLR) proteins share strong structural homology to the largest subgroup of plant disease resistance (R) proteins. These proteins share a trimeric domain architecture consisting of an N-terminal effector domain, a central nucleotide binding domain (NBD), and C-terminal leucine rich repeats (LRR). Mounting evidence suggests that NLR genes are important for the host response to pathogens and the regulation of inflammation. Interest in these genes has been further propelled by the realization that mutations in certain NLR genes are linked to human autoinflammatory and immunodeficiency diseases. For example, mutations in CIITA, the major histocompatability complex (MHC) class II transactivator, lead to a severe immunodeficiency disease, bare lymphocyte syndrome (22). Mutations in NOD2/CARD15 are associated with Crohn's disease and Blau syndrome, two human disorders with hyperinflammatory manifestations (3, 7, 18, 20). Finally, mutations in the cold-induced autoinflammatory syndrome-1 gene (CIAS1, also NALP3) are associated with a spectrum of autoinflammatory disorders which likely represent similar diseases with various levels of severity: familial cold-induced autoinflammatory syndrome, Muckle-Wells syndrome, and neonatal-onset multisystem inflammatory disease/chronic infantile neurologic, cutaneous, articular syndrome (1, 5, 8, 16, 19). Most notably, the majority of known disease-associated mutations within these NLR genes reside within the NBD domain. However, the influence of these mutations on the nucleotide binding activity remains poorly understood.
Several recent studies have provided greater detail regarding the mechanism and biological significance of nucleotide binding by NLR and NLR-related proteins. Apoptotic protease activating factor-1 (APAF-1) is a protein that contains a central NBD and C-terminal WD-40 repeats and thus is closely related to the NLR family. Under apoptotic conditions, APAF-1 binds cytochrome c. This interaction stimulates APAF-1 binding to dATP, leading to the formation of an APAF-1 heptamer that activates caspase-9. Thus, dATP binding by APAF-1 is a key regulatory step in the apoptotic process (6, 10). Similar to APAF-1, the NLR protein NLRP3 (previously known as cryopyrin, CIAS1, or NALP3) also requires nucleotide binding for its activity. Our group has recently demonstrated that, in the absence of a functional NBD, NLRP3 cannot form an active inflammasome. This results in reduced interleukin-1β (IL-1β) processing and decreased cell death (4). Most importantly, inactivation of the NBD of NLRP3 abolishes the hyperreactive phenotype of naturally occurring disease-associated mutations of this NLR protein. This demonstrates that nucleotide binding is required for the inflammatory phenotype of NLRP3-linked diseases.
In light of these findings, we examined the contribution of nucleotide binding to the functional role of an NLR protein, Monarch-1/NLRP12. Monarch-1 is expressed predominantly in cells of the myeloid lineage, including monocytes and granulocytes (28). Furthermore, single nucleotide polymorphism analysis has recently demonstrated a genetic link between Monarch-1 and atopic dermatitis (15). However, in contrast to most NLR proteins that promote inflammation, Monarch-1 functions as an attenuator of inflammatory responses. Monarch-1 inhibits Toll-like receptor (TLR)-mediated hyperphosphorylation of IL-1 receptor-associated kinase 1 (IRAK-1), a necessary step in IRAK-1 signaling pathways (27). In addition, Monarch-1 binds to and destabilizes the mitogen-activated protein kinase kinase kinase NF-κB-inducing kinase (NIK) and blocks NIK-mediated processing of NF-κB2/p100 to p52 (12). The p52 subunit is an important downstream mediator of signaling by TLRs as well as tumor necrosis factor family receptors, including CD40 and lymphotoxin-beta receptor. The introduction of short hairpin RNA (shRNA) specific for Monarch-1 greatly enhances NF-κB activation and the transcription of NIK-dependent genes induced by TLR and/or tumor necrosis factor family receptor activation (12). These earlier studies suggest that Monarch-1 performs an important anti-inflammatory role as an inhibitory molecule of innate immune activation.
In this report, we explore the role of nucleotide binding in the anti-inflammatory activity of Monarch-1. Herein, we demonstrate that Monarch-1, purified to homogeneity, specifically binds ATP. Nucleotide binding is indispensable for the biological function of Monarch-1, as an NBD mutant form of Monarch-1 does not inhibit IRAK-1 hyperphosphorylation nor does it inhibit NIK-dependent p52 production. Moreover, THP-1 monocytes stably expressing the NBD mutant form of Monarch-1 secrete elevated levels of proinflammatory cytokines and chemokines. These results open the door for the characterization of the nucleotide binding properties of other NLR members and will facilitate the design of pharmacological agents that modulate the functions of this family of proteins.
MATERIALS AND METHODS
Reagents.
The TLR2 agonist, the synthetic lipoprotein S-[2,3-bis(palmitoyloxy)-2(2-RS)-propyl]-N-palmitoyl-(R)-Cys-(S)-Ser-Lys-4-OH trihydrochloride (Pam3Cys4) was obtained from InvivoGen and used at a final concentration of 200 ng/ml. CD40 ligand (CD40L) was obtained from PeproTech and used at a final concentration of 250 ng/ml. Anti-six-His-horseradish peroxidase conjugates were obtained from Santa Cruz Biotechnology (Santa Cruz, CA), and anti-Flag M2-horseradish peroxidase was obtained from Sigma. THP-1 cell lines stably expressing empty vector (THP-EV), hemagglutinin (HA)-tagged wild-type (WT) Monarch-1 (THP-WT) or shRNA targeting Monarch-1 (THP-shMon) have been described previously (12, 27). The THP-1 cell line stably expressing HA-tagged Monarch-1 containing the Walker A/B mutation (THP-mutA/B) was generated by the same procedure.
Expression and purification of bacterial MBP-Monarch-1-NBD fusion protein.
The cDNA sequence encoding amino acids 188 to 448 of Monarch-1 and including the Walker A/B motifs was amplified by PCR using PFU Turbo polymerase (Stratagene). Restriction enzyme sites for HindIII and BamHI were incorporated into the 5′ and 3′ ends of the PCR product, respectively. A DNA sequence encoding a six-His tag was also introduced in the 3′ reverse primer. The amplified product was digested by HindIII and BamHI (New England BioLabs) and cloned into the C terminus of the maltose binding protein (MBP) in the vector pMAL-c2E (New England BioLabs). Mutations in both Walker A and Walker B were generated by site-directed mutagenesis (Stratagene). All constructs were confirmed by DNA sequencing. The MBP-Monarch-1-NBD fusion plasmids were transformed into the Escherichia coli strain Rosetta-Origami B (EMD Biosciences). One liter of LB with 100 μg/ml ampicillin was inoculated with 5 ml of an overnight bacterial culture. The culture was grown at 37°C to an A600 of 0.8, and then isopropylthio-β-d-galactoside (IPTG) was added to a final concentration of 0.3 mM to induce the expression of the MBP fusion proteins. After 3 h of induction at 25°C, the cells were harvested by centrifugation at 6,000 rpm. Cell pellets were washed once with cold phosphate-buffered saline and resuspended in ice-cold lysis buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, and Roche protease inhibitor cocktail). Resuspension was facilitated by sonication for 2 min. Bacteria were then lysed with recombinant lysozyme (EMD Biosciences) and then treated with benzonase (EMD Biosciences) to degrade bacterial DNA and RNA. Bacterial lysates were clarified by centrifugation twice at 15,000 × g for 30 min. The supernatant was filtered through a 0.2-μm-pore-size low-protein binding filter. Amylose resin (New England BioLabs) was washed twice with column buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol) and added directly to the bacterial lysate. The lysate-resin mix was rotated at 4°C for 1 h and then transferred into an empty column. The resin was washed with 10 volumes of column buffer and eluted with 5 volumes of column buffer containing 10 mM maltose. The eluate containing MBP fusion protein was concentrated with an Amicon centrifugal filter device. The partially purified MBP-Monarch-1-NBD fusion proteins were further purified over a fast protein liquid chromatography size exclusion column (Bio-Silect 400; Bio-Rad). Each fraction was tested for nucleotide binding activity. The fractions with high nucleotide binding activity were pooled and subsequently purified on a cobalt-based metal affinity column (Sigma) and eluted with 300 mM imidazole.
Expression and purification of mammalian cell-derived Monarch-1ΔLRR.
cDNA encoding Monarch-1 amino acids 1 to 686, which correspond to the pyrin and NBD domains, was PCR amplified and cloned into the pCEP4 vector (Invitrogen) by standard molecular cloning procedures. This expression construct was introduced into the HEK293EBNA cell line (ATCC CRL10852) by use of polyethyleneimine (Polyscience). The transfected HEK293EBNA cells were then harvested and lysed in hypotonic lysis buffer (25 mM HEPES-KOH [pH 7.5], 10 mM KCl, 5 mM MgCl2, 0.1 mM phenylmethylsulfonyl fluoride, and Roche protease inhibitor cocktail) for 15 min on ice followed by a brief sonication for 40 seconds. Lysates were cleared by centrifugation at 20,000 rpm for 30 min and filtered through a 0.45-μm-pore-size filter. The lysate was then subjected to cobalt metal affinity resin purification (Clontech). The eluate was further purified over an anti-Flag affinity matrix and eluted with excess Flag peptide (Sigma). These eluates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were visualized with Coomassie blue stain.
Nucleotide binding assay.
A rapid filter binding assay was developed to measure nucleotide binding to Monarch-1 fusion proteins. [γ-35S]ATP (1,250 Ci/mmol) (Perkin Elmer Life and Analytical Sciences) was mixed with the indicated amount of recombinant Monarch-1 in a final volume of 100 μl of binding buffer (50 mM Tris-HCl, 150 mM NaCl, 20 mM MgCl2, 2 mM dithiothreitol, 5% glycerol, pH 7.5) and was incubated at 30°C for 1 h. After this incubation, the samples were filtered through a 96-well nitrocellulose plate (Millipore) and immediately washed twice with 200 μl of ice-cold binding buffer by vacuum filtration (Millipore). The filter plate was then air dried, and radioactivity was measured using a scintillation counter. For homologous competition assays, Monarch-1-NBD fusion protein (2 μg) or Monarch-1ΔLRR (450 ng) was incubated with 90 nM [γ-35S]ATP and increasing concentrations of the indicated unlabeled nucleotide. Homologous competitive binding curves represent nonlinear regression fit to the single-site competition model [Y = Bottom + (Top − Bottom)/(1 + 10X − logIC50), where IC50 is the 50% inhibitory concentration] using GraphPad Prism 5 software. In homologous competitive binding assays, Kd (dissociation constant) = IC50 − [radioligand] (4).
ATPase assay.
ATP hydrolysis was measured by visualizing the conversion of [α-32P]ATP to [α-32P]ADP using thin-layer chromatography (TLC). A total of 5 μg of purified Monarch-1ΔLRR was incubated with 10 μM ATP and 0.1 μM [α-32P]ATP (3,000 Ci/mmol; Perkin Elmer Life and Analytical Science) in a total volume of 40 μl reaction buffer (25 mM Tris-HCl, pH 7.5; 150 mM NaCl; 10 mM MgCl2; 1 mM dithiothreitol; 1 mM EDTA; 0.1 mM phenylmethylsulfonyl fluoride) for 2 h. The reaction was quenched by adding an equal volume of TLC development solvent (1 M formic acid, 0.5 M LiCl). A total of 2 μl of the reaction mixture was spotted on a polyethyleneimine cellulose TLC plate and developed with 1 M formic acid with 0.5 M LiCl in a TLC chamber. The TLC plate was then exposed to X-ray film.
Detection of cytokines and chemokines.
The indicated THP-1-derived cells lines were stimulated with 200 ng/ml Pam3Cys4 for 18 h and then 250 ng/ml CD40L for an additional 5 h. The supernatant was applied to a RayBiotech Human G series 2000 glass slide array and processed according to the manufacturer's protocol. The signal strength for each cytokine was normalized to the signal strength of a spiked, internal control for each slide. Normalized data for each treatment group were then represented as a change compared to unstimulated samples. For the data set from the array, see Tables S1 and S2 and Fig. S1 in the supplemental material. The array was performed for screening purposes, and caution should be used when interpreting data not verified by enzyme-linked immunosorbent assay (ELISA). For the ELISA studies, THP-1-derived cell lines were stimulated as described above. Cytokine and chemokine levels in cell supernatants were analyzed by sandwich ELISA as recommended (R&D Systems).
Immunoprecipitation and Western blot analysis.
HEK293T and THP-1 cell lines, stimulated with the conditions indicated, were lysed in buffer containing 1% Triton X-100, 150 mM NaCl, 50 mM Tris-HCl (pH 8), 50 mM NaF, 2 mM EDTA, plus a protease inhibitor cocktail (Roche). Immunoprecipitates were washed four times in lysis buffer and eluted by boiling in reducing sample buffer. Samples were fractionated by SDS-PAGE and transferred to nitrocellulose. Western blots were probed with the indicated antibodies and then visualized by enhanced chemiluminescence (Pierce) and exposure to photographic film (Genesee Scientific). The films were scanned into Adobe Photoshop, and whole images were adjusted for brightness. The images were cropped and formatted in Adobe Illustrator. Nuclear/cytoplasmic fractions were prepared using the Pierce NE-PER nuclear and cytoplasmic extraction reagent kit. The following antibodies were used: anti-HA 12CA5 (Roche), anti-IRAK-1 (C-20), anti-NIK (H-248), and anti-CagA (b-300, isotype control; Santa Cruz Biotechnology).
RESULTS
Enrichment and characterization of recombinant Monarch-1.
Initial attempts to express full-length Monarch-1 protein using Escherichia coli and baculovirus expression systems failed to yield a sufficient quantity of soluble protein for biochemical analysis despite extensive testing and optimization of expression conditions and parameters. This is a common problem encountered in the study of this family of proteins (14). Therefore, to improve solubility, we employed an MBP expression system, in which MBP was fused to the NBD of Monarch-1.
The NBD can be divided into a NACHT domain (NAIP, CIITA, HET-E, and TP1) (11) and NACHT-associated domains (NAD) (2). The NACHT domain of NLR proteins contains well-conserved nucleotide binding structures, including the ATP/GTP-specific phosphate binding loop called Walker A and an Mg2+ coordination site called Walker B (21, 25, 26). We generated a fusion protein containing MBP fused to amino acids 188 to 448 of Monarch-1 (Fig. 1A). In addition to the N-terminal MBP moiety, a six-His tag was added to the C terminus of the Monarch-1-NBD fusion proteins to facilitate purification by dual-affinity chromatography. The region of amino acids 188 to 448 of Monarch-1 comprises the NACHT domain and NAD1. We predicted this region to have nucleotide binding properties based upon the following: (i) the crystal structure of the related dATP binding protein Apaf-1 and (ii) a recent molecular modeling study in which the previously broadly defined NBD domain was subdivided into the NACHT domain and three subsequent NAD sequences (2).
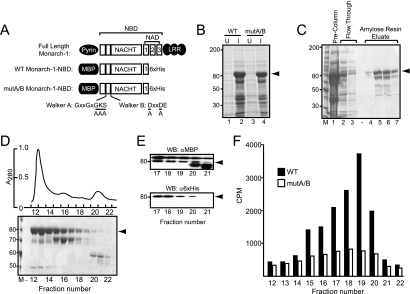
FIG. 1.
The generation and purification of Monarch-1-NBD fusion proteins. (A) Schematic diagram of domain structure of full-length Monarch-1 and WT and mutA/B Monarch-1-NBD fusion proteins. The positions of the Walker A and Walker B motifs are depicted as solid bars within the NACHT domain. The conserved amino acids within both motifs are shown, and the underlined residues were substituted with alanine by site-directed mutagenesis. (B) WT Monarch-1-NBD and mut A/B Monarch-1-NBD were transformed into E. coli, and soluble bacterial extracts were analyzed by SDS-PAGE and Coomassie blue staining. Arrowhead, Monarch-1-NBD fusion protein; U, uninduced; I, IPTG-induced. The sizes of molecular weight markers (in thousands) are indicated on the left. (C) Bacterial lysates were passed over an amylose resin column targeting MBP. Eluted proteins were resolved by SDS-PAGE and visualized with Coomassie blue stain. (D) Eluates from the amylose resin column were further purified by size exclusion chromatography. The absorbance profile of size exclusion purification is depicted (upper panel), and the size exclusion fractions were analyzed by SDS-PAGE and visualized by Coomassie blue staining (lower panel). (E) Western blots from size exclusion fractions 17 to 21 were probed with an anti-MBP or anti-His antibody to verify the intactness of the N terminus and C terminus, respectively, of the Monarch-1-NBD fusion protein. (F) Size exclusion fractions from WT or mutA/B Monarch-1-NBD were tested for ATP binding activity using [γ-35S]ATP, and specific binding was normalized to the protein concentration of each fraction.
NACHT domains consist of six α-helices and a β-sheet core structure, which contains the highly conserved Walker A and Walker B motifs responsible for nucleotide binding. The Walker A motif contains the consensus sequence GXXGXGK[T/S], wherein the conserved lysine residue is responsible for the coordination of the β- and γ-phosphate moieties of ATP. The Walker B motif, DXXDE, contains two well-conserved aspartic acid residues which are involved in the coordination of Mg2+ and ATP hydrolysis. To ensure the complete disruption of nucleotide binding, mutations were introduced within both the Walker A and Walker B motifs and will be referred to hereafter as mutA/B.
Plasmids encoding the WT or mutA/B Monarch-1-NBD fusion proteins were transfected into E. coli and were predominantly expressed as soluble proteins with a molecular weight of approximately 80,000 (Fig. 1B). An affinity-based amylose resin targeting MBP was used to enrich the fusion proteins and yielded a partially purified product (Fig. 1C). Although MBP increases the solubility of its cargo protein, a large portion of the fusion protein may remain in a misfolded, high-molecular-weight soluble aggregate that is biochemically inactive. To separate functionally active protein from these soluble aggregates and further purify the protein, we applied the amylose resin-enriched product to a size exclusion column. Figure 1D depicts the fast protein liquid chromatography elution profile of WT Monarch-1-NBD and the Coomassie blue-stained SDS-PAGE gel containing the size exclusion fractions. The mutA/B Monarch-1-NBD fusion protein exhibited expression and chromatographic profiles identical to those of the WT protein (data not shown). A large percentage of the protein (70 to 80%) eluted in fractions 12 to 14. These fractions consisted of the excluded volume of the column (>700,000) and represent proteins existing in a high-molecular-weight aggregation state. Fractions 15 to 19 contained the Monarch-1-NBD fusion protein existing in lower-molecular-weight states (∼80,000 to 300,000), which likely represented monomeric, dimeric, and trimeric complexes. Fractions 20 to 22 contained degradative products (Fig. 1D). The presence of intact Monarch-1-NBD was confirmed by Western blot analysis of fractions 17 to 21 using antibodies against N-terminal MBP and C-terminal six-His (Fig. 1E). The MBP antibody detected full-length Monarch-1-NBD protein as well as lower-molecular-weight moieties. The anti-six-His antibody detected only full-length Monarch-1-NBD proteins. This suggests that the smaller moieties detected by the MBP antibody in fractions 20 to 21 were likely MBP fusion proteins missing an intact C terminus.
To determine if the gel filtration fractions contained biochemically active Monarch-1-NBD, an ATP binding assay was performed. In this assay, recombinant proteins from each fraction were incubated with radiolabeled [γ-35S]ATP, a nonhydrolysable ATP analog, and then loaded onto a nitrocellulose filter plate. Protein-bound nucleotides were trapped on the membrane, while free nucleotides passed through the membrane by repeated buffer washes. Fractions 12 to 14, corresponding to the high-molecular-weight aggregates, exhibited poor ATP binding activity (Fig. 1F). This suggests that these high-molecular-weight soluble aggregates of Monarch-1-NBD likely remained in a misfolded, inactive state. In contrast, ATP binding activity significantly increased in the lower-molecular-weight fractions, with fractions 18 and 19 exhibiting the highest ATP binding activity. To assess if the Walker A and B motifs are required for nucleotide binding, we performed this ATP binding assay using size exclusion chromatography fractions containing mutA/B Monarch-1-NBD. Mutations within the Walker A/B sequences dramatically reduced the nucleotide binding activity of the protein (Fig. 1F), demonstrating their importance for Monarch-1 nucleotide binding. The greatest difference was observed in fraction 19, where the ATP binding activity of WT Monarch-1-NBD was fivefold higher than that of the corresponding mutA/B form of the protein. Together, these results indicate that Monarch-1 binds ATP and that the Walker A/B motifs are required for nucleotide binding.
Monarch-1 specifically binds ATP.
To more accurately determine both the affinity and specificity of ATP binding, we further purified WT Monarch-1-NBD by taking advantage of the C-terminal six-His tag. Fractions 18 and 19, which demonstrated the highest level of nucleotide binding (Fig. 1F), were pooled and applied to a cobalt-based metal affinity purification column. The resulting product was highly enriched and migrated as a single band at ∼80,000 as examined by Coomassie blue staining (Fig. 2A). To determine the affinity of ATP binding, highly enriched WT Monarch-1-NBD was incubated with radiolabeled [γ-35S]ATP along with increasing concentrations of unlabeled [γ-S]ATP. The dissociation constant (Kd) of [γ-35S]ATP binding was determined to be 100 nM (Fig. 2B). Next we tested the nucleotide binding preference of WT Monarch-1-NBD. The fusion protein was preincubated with radiolabeled [γ-35S]ATP for 2 h, and then unlabeled [γ-S]ATP, ATP, GTP, or CTP was added to compete for WT Monarch-1-NBD binding. Unlabeled [γ-S]ATP and ATP successfully competed with radiolabeled [γ-35S]ATP (Fig. 2C). However, even at this high nucleotide concentration, GTP and CTP displayed only a slight ability to compete with [γ-35S]ATP. This strongly suggests that WT Monarch-1-NBD specifically binds ATP.
FIG. 2.
Purified Monarch-1-NBD fusion protein specifically binds ATP. (A) Gel filtration fractions 18 to 19 (depicted in Fig. 1) were pooled and further purified by cobalt metal affinity chromatography. The purity of the final purification product was assessed by SDS-PAGE and Coomassie blue staining. Arrowhead, Monarch-1-NBD fusion protein. The sizes of molecular weight markers (in thousands) are indicated on the left. (B) Homologous competition assays were performed to assess the ATP binding affinity of Monarch-1-NBD. (C) Specificity of ATP binding was determined by incubating Monarch-1-NBD fusion protein with [γ-35S]ATP and 10 μM of the indicated unlabeled competitor nucleotide. Significance of the difference from control samples was estimated by Student's t test. “None” indicates no nucleotide. A double asterisk signifies a P value of <0.000003. A single asterisk signifies a P value of 0.011 for GTP or 0.0085 for CTP.
Recombinant Monarch-1 derived from mammalian cells binds ATP.
The experiments described above employed WT Monarch-1-NBD fusion proteins derived from bacteria to demonstrate ATP binding activity. However, these fusion proteins lacked the N-terminal pyrin domain and C-terminal LRR domain found in the full-length protein. The crystal structure of Apaf-1 indicates that its N-terminal effector domain folds back on the ATP binding pocket within the NBD (9). Although this does not affect dATP binding by Apaf-1, this observation led us to question if the pyrin domain or the LRR domain of Monarch-1 affects nucleotide binding.
To determine the influence of these domains on ATP binding activity, we purified full-length Monarch-1 using a mammalian expression system. In order to enhance protein expression, we employed an Epstein-Barr virus-based episomal replication system. Sequences encoding the WT and mutA/B Monarch-1 were cloned into Epstein-Barr virus expression vectors, and an N-terminal 10-His tag and a C-terminal Flag tag were added to facilitate dual-affinity purification. Again, we were unable to obtain sufficient quantities of full-length Monarch-1. However, soluble Monarch-1 protein was successfully obtained after deleting the LRR domain (Monarch-1ΔLRR) (Fig. 3A). The WT and mutA/B Monarch-1ΔLRR were first partially purified by cobalt metal affinity. The recombinant proteins in this eluate were then further purified by anti-Flag affinity chromatography. Coomassie blue staining along with anti-His and anti-Flag Western blotting revealed a single band that corresponded to the purified WT and mutA/B Monarch-1ΔLRR (Fig. 3B and C).
FIG. 3.
Mammalian cell-derived recombinant Monarch-1ΔLRR binds and hydrolyzes ATP. (A) Diagram depicting full-length Monarch-1 and the WT and mutA/B Monarch-1ΔLRR proteins produced in HEK293EBNA cells. (B) Soluble extracts of HEK293EBNA cells expressing WT and mutA/B Monarch-1ΔLRR were enriched for Monarch-1ΔLRR using a cobalt metal affinity column (lanes 1 and 2) and further purified over an anti-Flag affinity matrix (lanes 3 and 4). The purity was evaluated by Coomassie blue staining. The sizes of molecular weight markers (in thousands) are indicated on the left. (C) The double-purified Monarch-1ΔLRR proteins were analyzed by Western blotting using anti-His and anti-Flag antibodies. (D) ATP binding activity of purified WT and mutA/B Monarch-1ΔLRR was determined by incubating 500 ng purified protein with [γ-35S]ATP. Error bars represent the standard deviations of ATP binding measurements in triplicate. (E) The ATP binding affinity of WT Monarch-1ΔLRR was determined by homologous competition binding assays. (F) The nucleotide binding preference of WT Monarch-1ΔLRR was determined by incubating WT Monarch-1ΔLRR with [γ-35S]ATP and increasing concentrations of unlabeled nucleotide. Significance of the differences from control samples (nucleotide concentrations of 10−9 M) was estimated by Student's t test. A double asterisk signifies a P value of <0.0005. A single asterisk signifies a P value of <0.02. (G) The ATPase activity of purified WT or mutA/B Monarch-1ΔLRR was measured by visualizing the conversion of [α-32P]ATP to [α-32P]ADP by TLC. HEK293EBNA lysate and purified bovine serum albumin (BSA) were used as positive and negative controls, respectively. Arrowhead, Monarch-1-NBD fusion protein.
To determine if Monarch-1ΔLRR exhibited nucleotide binding activity, an ATP binding assay was performed. As seen in the bacterial expression system, purified WT Monarch-lΔLRR exhibited strong ATP binding activity, while this binding activity was dramatically reduced in the mutA/B form of the protein (Fig. 3D). Nucleotide competition assays determined the binding affinity to be 84 nM (Fig. 3E). This was comparable to the binding affinity observed for the WT Monarch-1-NBD fusion protein derived from bacterial expression. In addition, similar to the bacterium-derived WT Monarch-1-NBD fusion protein, unlabeled ATP successfully competed for [γ-35S]ATP binding (Fig. 3F). Unlabeled CTP and GTP, however, demonstrated only a slight ability to compete for [γ-35S]ATP at 100-fold higher concentrations of nucleotide. Together, these results demonstrate that Monarch-1 specifically binds ATP.
Recombinant Monarch-1 derived from mammalian cells possesses ATPase activity.
To determine if WT Monarch-1ΔLRR possessed ATP hydrolysis activity, the purified protein was incubated with radiolabeled [α-32P]ATP and the reaction mixture was analyzed by TLC. Whole-cell lysate was used as a positive control to show the conversion of radiolabeled ATP to ADP, while bovine serum albumin, a non-ATPase, was used as a negative control. Purified WT Monarch-1ΔLRR exhibited ATPase activity as shown by the conversion of ATP to ADP (Fig. 3G). In contrast, the mutA/B Monarch-1ΔLRR construct which bears the Walker A/B mutations did not exhibit ATPase activity. Together, these results clearly demonstrate that Monarch-1 binds ATP and possesses ATPase activity; however, we have not been able to separate these two activities at this point.
Nucleotide binding regulates Monarch-1 self association.
The oligomerization of NLR proteins has been shown to be important for their activity (6, 13). To determine if nucleotide binding is required for Monarch-1 self association, Flag-tagged WT Monarch-1 was cotransfected into HEK293T cells along with HA-tagged WT Monarch-1 or HA-tagged mutA/B Monarch-1. Protein complexes were immunoprecipitated with anti-Flag antibody, and Western blots were probed with anti-HA to detect self-associating, homomeric Monarch-1 complexes. As expected, WT Monarch-1 exhibited self association, as HA-tagged WT Monarch-1 coprecipitated with Flag-tagged WT Monarch-1 (Fig. 4, lane 3). In addition, WT Monarch-1 also associated with mutA/B Monarch-1, as HA-tagged forms of the NBD mutant were detected in WT Monarch-1 precipitates (Fig. 4, lane 4). Interestingly, however, when Flag-tagged and HA-tagged forms of mutA/B Monarch-1 were coexpressed, they failed to form homomeric complexes, as HA-tagged mutA/B Monarch-1 was not detected in anti-Flag immunoprecipitates (Fig. 4, lane 5). Together, these results suggest that complex formation among Monarch-1 molecules requires nucleotide binding but that not every member of the complex must bind nucleotides. This phenomenon was also observed for the NLR protein, CIITA, suggesting that it may be a common feature of NLR proteins (13).
FIG. 4.
Nucleotide binding is required for Monarch-1 self association. HEK293T cells were transfected with full-length forms of HA- or Flag-tagged WT or mutA/B Monarch-1. Flag-tagged proteins were immunoprecipitated (IP) with an anti-Flag antibody, and Western blots (IB) were probed with anti-HA antibody to detect self-associated Monarch-1.
Nucleotide binding regulates the ability of Monarch-1 to inhibit NIK activity.
Recently, we demonstrated that Monarch-1 suppresses the activation of noncanonical NF-κB by associating with NIK. To determine the role of nucleotide binding in the formation of this molecular complex, HEK293 cells were cotransfected with NIK and full-length forms of HA-tagged WT or mutA/B Monarch-1. Cell lysates were immunoprecipitated with anti-NIK antibodies, and Western blots were probed with anti-HA antibody to detect Monarch-1. As previously reported, WT Monarch-1 coprecipitated with NIK (Fig. 5A). Similarly, mutA/B Monarch-1 also associated with NIK, suggesting that nucleotide binding is not a requirement for complex formation in this overexpression model. To further assess the role of nucleotide binding in Monarch-NIK complex formation, THP-1 monocytes stably expressing HA-tagged WT or mutA/B Monarch-1 were treated with CD40L to induce the activation of endogenous NIK. Cell extracts were immunoprecipitated with anti-NIK antibodies, and Western blot analyses were performed to detect coprecipitating Monarch-1. As previously described, CD40L treatment enhanced the association of WT Monarch-1 with endogenous NIK (Fig. 5B). In contrast, the association between mutA/B Monarch-1 and NIK failed to increase upon activation. Thus, while nucleotide binding is not an absolute requirement for NIK binding, it is required for activation-induced complex formation.
FIG. 5.
Nucleotide binding is required for Monarch-1 to suppress NIK-mediated p100 processing. (A) HEK293T cells were cotransfected with NIK and HA-tagged WT or mutA/B Monarch-1. Cell lysates were immunoprecipitated (IP) with anti-NIK antibodies, and Western blots (IB) were probed with anti-HA antibodies to detect Monarch-1. ctrl lg, isotype control antibody. (B) THP-WT or THP-mutA/B cells were stimulated with CD40L for indicated times. Endogenous NIK was immunoprecipitated, and Western blots were probed with anti-HA to detect coprecipitating Monarch-1. Control immunoprecipitations were performed with an isotype control antibody to monitor specificity. Control Western blottings were performed to monitor the expression of Monarch-1 and NIK in cellular lysates. (C) THP-EV, THP-WT, or THP-mutA/B cells were stimulated with Pam3Cys4 for 18 h to induce p100 expression. The cells were then treated with CD40L for an additional 5 h to induce p100 cleavage to p52. Cells were fractionated into nuclear and cytoplasmic fractions, and proteins from each fraction were separated by SDS-PAGE. Western blots were probed with anti-p100 to detect p100 and its cleaved form, p52. Anti-HA was used to monitor Monarch-1 expression. These results are representative of at least five separate experiments.
NIK activates noncanonical NF-κB by inducing proteolytic processing of NF-κB2/p100 to p52. This smaller active form of NF-κB2 rapidly translocates to the nucleus to regulate the transcription of inflammatory genes. In contrast, the unprocessed form, p100, functions as an inhibitor of NF-κB activity (17). We recently demonstrated that the association of Monarch-1 with NIK results in the suppression of p100 processing. To determine the role of nucleotide binding in this function, THP-1 monocytes expressing an empty vector, WT Monarch-1, or mutA/B Monarch-1 were stimulated via TLR2 to induce p100 expression. The cells were then treated with CD40L to induce NIK-dependent p100 processing. As previously reported, WT Monarch-1 suppressed p100 processing, as demonstrated by a sharp reduction in nuclear p52 in THP-WT Monarch-1 cells (Fig. 5C, lane 4). Moreover, this reduction in nuclear p52 occurred in the presence of elevated levels of cytoplasmic p100 in these cells. Finally, the accumulation of p100 in the nucleus was consistently detected in THP-WT Monarch-1 cells, further emphasizing the role of Monarch-1 in inhibiting p100 processing. In contrast to WT Monarch-1, mutA/B Monarch-1 did not affect CD40L-induced activation of noncanonical NF-κB, as nuclear p52 levels were comparable to those seen in control THP-EV cells. Thus, consistent with the inability of mutA/B Monarch-1 to bind NIK following activation, the NBD mutant did not inhibit p100 processing. These results demonstrate that nucleotide binding is required for Monarch-1 to suppress noncanonical NF-κB.
Nucleotide binding regulates the ability of Monarch-1 to inhibit IRAK-1 activation.
In addition to NIK, we have also shown that upon TLR stimulation, Monarch-1 binds IRAK-1 and inhibits its hyperphosphorylation (27). To determine if nucleotide binding is required for the association of Monarch-1 with IRAK-1, THP-1 cells expressing WT or mutA/B Monarch-1 were stimulated with the TLR2 agonist, Pam3Cys4. Endogenous IRAK-1 complexes were captured by immunoprecipitation, and Western blot analyses were performed to detect Monarch-1. In agreement with our previous findings, the complex formation between WT Monarch-1 and IRAK-1 strengthened upon TLR stimulation (Fig. 6A). However, similar to NIK, mutA/B Monarch-1 associated more strongly with IRAK-1 in resting cells, and activation-induced association was abrogated by Walker A/B mutations. Accordingly, TLR2 stimulation resulted in the hyperphosphorylation of IRAK-1 in THP-mutA/B Monarch-1 at levels comparable to those of control THP-EV cells (Fig. 6B). In agreement with our previous findings, WT Monarch-1 suppressed the accumulation of these hyperphosphorylated forms of IRAK-1 (Fig. 6B) (12). These data confirm the requirement for nucleotide binding in Monarch-1-mediated suppression of inflammatory signaling.
FIG. 6.
Nucleotide binding by Monarch-1 is required for the suppression of IRAK-1 hyperphosphorylation. (A) THP-WT or THP-mutA/B cells were stimulated for the indicated times with Pam3Cys4. Endogenous IRAK-1 was immunoprecipitated (IP), and Western blots (IB) were probed with anti-HA to detect Monarch-1. Control samples were immunoprecipitated with an isotype-matched antibody. Control Western blottings were performed on cellular lysates to monitor the levels of Monarch-1 and IRAK-1. (B) THP-EV, THP-WT, and THP-mutA/B cells were stimulated with Pam3Cys4 for the indicated times. Lysates were separated by SDS-PAGE, and Western blots were probed with anti-IRAK-1 antibodies. Control Western blottings were performed and probed with anti-HA to monitor Monarch-1 expression. These results are representative of at least five separate experiments.
Nucleotide binding regulates the anti-inflammatory activity of Monarch-1.
Previously, we demonstrated that Monarch-1 suppresses the production of proinflammatory cytokines and chemokines in stimulated monocytic cells (12, 27). To determine the role of nucleotide binding in this anti-inflammatory activity, THP-1 cells stably expressing full-length WT or mutA/B Monarch-1 were stimulated, and supernatants were applied to a cytokine/chemokine antibody array. THP-1 cells stably transfected with empty vector (THP-EV) were used as controls. In agreement with our previous reports, THP-1 cells expressing WT Monarch-1 produced lower levels of inflammatory cytokines and chemokines than those of the THP-EV control samples. These included IL-6 and CXCL13, which have been previously shown to be suppressed by WT Monarch-1 (see Fig. S1 in the supplemental material) (12, 27). In contrast, THP-1 monocytes expressing mutA/B Monarch-1 produced increased levels (more than twofold) of these proteins compared to control samples (see Fig. S1 in the supplemental material). A large number of cytokines and chemokines were not substantially induced by Pam3Cys4/CD40L stimulation (see Table S1 in the supplemental material). Another set of proteins was increased less than twofold in THP-1 cells expressing mutA/B compared to control cells and was not further investigated (see Table S2 in the supplemental material).
Of the 79 cytokines and chemokines examined, we found the greatest differences in IL-6, CXCL6, and CXCL13 (Fig. 7). To confirm these results from the array, we performed ELISAs. For these experiments, THP-1 cells expressing shRNA targeting endogenous Monarch-1 (THP-shMon) were included. As expected, the expression of WT Monarch-1 resulted in the decreased production of IL-6, CXCL13, and CXCL6 compared to THP-EV cells (Fig. 7). In contrast, elevated levels of these cytokines/chemokines were detected in supernatants from THP-shMon cells, confirming our earlier reports demonstrating that silencing endogenous Monarch-1 results in a hyperinflammatory response (12, 27). Similar to THP-shMon cells, THP-mutA/B Monarch-1 cells also produced increased levels of these inflammatory mediators. These results demonstrate that nucleotide binding is required for the anti-inflammatory activity of Monarch-1. In addition, these results suggest that the presence of a nucleotide binding-deficient form of Monarch-1 can block the activity of endogenous Monarch-1.
FIG. 7.
Nucleotide binding is required for Monarch-1-mediated suppression of proinflammatory cytokine and chemokine production. THP-EV, THP-WT, THP-mutA/B, and THP-shMon were stimulated with Pam3Cys4 for 18 h and then CD40L for an additional 5 h. Cell culture supernatants were harvested, and cytokine/chemokine levels determined by ELISA.
DISCUSSION
Despite the general perception that members of the NLR family function as nucleotide binding proteins, this property has not been extensively studied. One group has shown data derived from plant R proteins that demonstrate the nucleotide binding properties of these proteins (23). Furthermore, despite this lack of convincing data, the assumption has been that nucleotide binding is required for the function of these proteins. This concept is largely modeled after the analysis of Apaf-1 (9). Thus, a key finding in this report is that highly enriched Monarch-1 specifically binds ATP, and this is required for its anti-inflammatory activity. Monarch-1 is also unique in that it functions as a brake on innate immune activation, as opposed to several of the other NLR proteins.
The requirement of nucleotide binding for Monarch-1 activity supports a recent report by our group that focused on the NLR protein cryopyrin/NLRP3. In that report, we demonstrated that NLRP3 binds ATP and that this binding activity is required for NLRP3-mediated IL-1β processing (4). Prior to these studies, only two other reports suggested nucleotide binding as a regulatory step in NLR function. An earlier report from our group suggested that CIITA binds GTP and that mutations within the Walker A/B motifs block CIITA-mediated transcriptional activation. In that study, however, nucleotide binding assays were performed by analyzing CIITA proteins that had been immunoprecipitated from transfected cells. Since CIITA forms large protein complexes, the nucleotide binding activities of coprecipitating proteins could not be ruled out. More recently, glutathione S-transferase fusion proteins containing the NBD of the NLR protein IPAF were shown to bind ATP (14). Mutations with the Walker A motif of IPAF result in reduced caspase-1 activity. However, similar to the CIITA study, protein purity remains an issue in this study.
In this report, we produced highly enriched Monarch-1 fusion proteins derived from both prokaryotic and eukaryotic expression systems to conclusively demonstrate the nucleotide binding activity of Monarch-1. The importance of Monarch-1 nucleotide binding in suppressing NF-κB activation and inflammatory signaling has been clearly demonstrated in this report. Expression of a nucleotide binding-deficient form of Monarch-1 (mutA/B Monarch-1) in monocytes resulted in dramatically increased production of proinflammatory mediators. These levels were comparable to those observed in THP-shMon cells in which endogenous Monarch-1 expression was silenced. This hyperinflammatory phenotype of cells expressing mutA/B Monarch-1 correlates well with the inability of this mutant to suppress NF-κB2/p100 processing and the inability of this mutant to efficiently bind NIK and IRAK-1 following stimulation.
Interestingly, an association was detected between these kinases and mutA/B Monarch-1 in resting cells. This suggests that the mutations within the Walker A/B sequences are subtle enough not to disrupt complex formation in resting cells but render Monarch-1 unable to bind these kinases upon stimulation. This loss of activation-induced NIK and IRAK-1 binding by mutA/B Monarch-1 may be due to its inability to form homomeric structures, a property required for the activity of CIITA, APAF-1, and NALP3. A second possibility relates to the functional dissection of ATP binding and ATP hydrolysis. For instance, it has been reported that ATP binding is required for the activity of the plant R protein I-2, yet the hydrolysis of this bound ATP moiety suppresses I-2 function (23, 24). Thus, the binding and hydrolysis of ATP may represent an on-off switch of NLR protein function. Future studies will be conducted to fully characterize the ATP hydrolysis cycle of Monarch-1 and to determine its contribution to the anti-inflammatory role of Monarch-1.
Despite exhaustive attempts to optimize conditions, we were not able to produce sufficient quantities of soluble, full-length purified protein. Instead, we generated recombinant Monarch-1 in mammalian cells by deleting the LRR domain. Thus, while it is clear that Monarch-1 binds ATP, it remains uncertain whether or not the LRR domain regulates this activity. In our recent report describing ATP binding by cryopyrin, purified proteins contained intact LRR sequences and these domains did not affect ATP binding. Therefore, due to the sequence similarities between Monarch-1 and NALP3, it is unlikely that the LRR domain disrupts nucleotide binding.
The nucleotide binding property of NLRs lends these proteins to pharmacologic intervention, as there are abundant nucleotide and nucleoside analog libraries available for drug screening. Thus, there now emerges the possibility of screening for nucleotide analogs that can bind to Monarch-1, cause sustained blockage of its function, and potentiate innate immune responses. In summary, our work demonstrates that Monarch-1 is an ATP binding protein and that this ATP binding activity is essential for Monarch-1 to perform its inhibitory role in innate immune signaling.
Supplementary Material
Acknowledgments
We thank Tsan Xiao for providing the reagents to establish the mammalian expression system. We also thank Janelle Arthur for critically reading the manuscript.
This work was supported by National Institutes of Health grants R01AI057157, R01AI063031, R01DE16326, and SERCEB A1-02-031 (J.P.-Y.T.); The American Cancer Society Postdoctoral Fellowship (J.D.L.); the Amgen/FOCIS Fellowship Award (K.L.W.); the Juvenile Diabetes Research Foundation Postdoctoral Fellowship (C.B.M.); the Pfizer Fellowship in Infectious Diseases (J.A.D.), and the National Institutes of Health Career Development Award K12RR023248 (J.A.D.). J.P.-Y.T. is a recipient of a Senior Investigator award of the Sandler Program in Asthma Research.
Footnotes
Published ahead of print on 26 December 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aksentijevich, I., M. Nowak, M. Mallah, J. J. Chae, W. T. Watford, S. R. Hofmann, L. Stein, R. Russo, D. Goldsmith, P. Dent, H. F. Rosenberg, F. Austin, E. F. Remmers, J. E. Balow, Jr., S. Rosenzweig, H. Komarow, N. G. Shoham, G. Wood, J. Jones, N. Mangra, H. Carrero, B. S. Adams, T. L. Moore, K. Schikler, H. Hoffman, D. J. Lovell, R. Lipnick, K. Barron, J. J. O'Shea, D. L. Kastner, and R. Goldbach-Mansky. 2002. De novo CIAS1 mutations, cytokine activation, and evidence for genetic heterogeneity in patients with neonatal-onset multisystem inflammatory disease (NOMID): a new member of the expanding family of pyrin-associated autoinflammatory diseases. Arthritis Rheum. 463340-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, M., and F. L. Takken. 2006. Update on the domain architectures of NLRs and R proteins. Biochem. Biophys. Res. Commun. 339459-462. [DOI] [PubMed] [Google Scholar]
- 3.Beutler, B. 2001. Autoimmunity and apoptosis: the Crohn's connection. Immunity 155-14. [DOI] [PubMed] [Google Scholar]
- 4.Duncan, J. A., D. T. Bergstralh, Y. Wang, S. B. Willingham, Z. Ye, A. G. Zimmermann, and J. P. Ting. 2007. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc. Natl. Acad. Sci. USA 1048041-8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman, H. M., J. L. Mueller, D. H. Broide, A. A. Wanderer, and R. D. Kolodner. 2001. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat. Genet. 29301-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu, Y., M. A. Benedict, L. Ding, and G. Nunez. 1999. Role of cytochrome c and dATP/ATP hydrolysis in Apaf-1-mediated caspase-9 activation and apoptosis. EMBO J. 183586-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hugot, J. P., M. Chamaillard, H. Zouali, S. Lesage, J. P. Cezard, J. Belaiche, S. Almer, C. Tysk, C. A. O'Morain, M. Gassull, V. Binder, Y. Finkel, A. Cortot, R. Modigliani, P. Laurent-Puig, C. Gower-Rousseau, J. Macry, J. F. Colombel, M. Sahbatou, and G. Thomas. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411599-603. [DOI] [PubMed] [Google Scholar]
- 8.Hull, K. M., N. Shoham, J. J. Chae, I. Aksentijevich, and D. L. Kastner. 2003. The expanding spectrum of systemic autoinflammatory disorders and their rheumatic manifestations. Curr. Opin. Rheumatol. 1561-69. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, X., and X. Wang. 2000. Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to Apaf-1. J. Biol. Chem. 27531199-31203. [DOI] [PubMed] [Google Scholar]
- 10.Kim, H. E., F. Du, M. Fang, and X. Wang. 2005. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl. Acad. Sci. USA 10217545-17550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koonin, E. V., and L. Aravind. 2000. The NACHT family—a new group of predicted NTPases implicated in apoptosis and MHC transcription activation. Trends Biochem. Sci. 25223-224. [DOI] [PubMed] [Google Scholar]
- 12.Lich, J. D., K. L. Williams, C. B. Moore, J. C. Arthur, B. K. Davis, D. J. Taxman, and J. P. Ting. 2007. Cutting edge: Monarch-1 suppresses non-canonical NF-κB activation and p52-dependent chemokine expression in monocytes. J. Immunol. 1781256-1260. [DOI] [PubMed] [Google Scholar]
- 13.Linhoff, M. W., J. A. Harton, D. E. Cressman, B. K. Martin, and J. P. Ting. 2001. Two distinct domains within CIITA mediate self-association: involvement of the GTP-binding and leucine-rich repeat domains. Mol. Cell. Biol. 213001-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu, C., A. Wang, L. Wang, M. Dorsch, T. D. Ocain, and Y. Xu. 2005. Nucleotide binding to CARD12 and its role in CARD12-mediated caspase-1 activation. Biochem. Biophys. Res. Commun. 3311114-1119. [DOI] [PubMed] [Google Scholar]
- 15.Macaluso, F., M. Nothnagel, Q. Parwez, E. Petrasch-Parwez, F. G. Bechara, J. T. Epplen, and S. Hoffjan. 2007. Polymorphisms in NACHT-LRR (NLR) genes in atopic dermatitis. Exp. Dermatol. 16692-698. [DOI] [PubMed] [Google Scholar]
- 16.McDermott, M. F., and I. Aksentijevich. 2002. The autoinflammatory syndromes. Curr. Opin. Allergy Clin. Immunol. 2511-516. [DOI] [PubMed] [Google Scholar]
- 17.Mercurio, F., J. A. DiDonato, C. Rosette, and M. Karin. 1993. p105 and p98 precursor proteins play an active role in NF-kappa B-mediated signal transduction. Genes Dev. 7705-718. [DOI] [PubMed] [Google Scholar]
- 18.Miceli-Richard, C., S. Lesage, M. Rybojad, A. M. Prieur, S. Manouvrier-Hanu, R. Hafner, M. Chamaillard, H. Zouali, G. Thomas, and J. P. Hugot. 2001. CARD15 mutations in Blau syndrome. Nat. Genet. 2919-20. [DOI] [PubMed] [Google Scholar]
- 19.Neven, B., I. Callebaut, A. M. Prieur, J. Feldmann, C. Bodemer, L. Lepore, B. Derfalvi, S. Benjaponpitak, R. Vesely, M. J. Sauvain, S. Oertle, R. Allen, G. Morgan, A. Borkhardt, C. Hill, J. Gardner-Medwin, A. Fischer, and G. de Saint Basile. 2004. Molecular basis of the spectral expression of CIAS1 mutations associated with phagocytic cell-mediated autoinflammatory disorders CINCA/NOMID, MWS, and FCU. Blood 1032809-2815. [DOI] [PubMed] [Google Scholar]
- 20.Ogura, Y., D. K. Bonen, N. Inohara, D. L. Nicolae, F. F. Chen, R. Ramos, H. Britton, T. Moran, R. Karaliuskas, R. H. Duerr, J. P. Achkar, S. R. Brant, T. M. Bayless, B. S. Kirschner, S. B. Hanauer, G. Nunez, and J. H. Cho. 2001. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411603-606. [DOI] [PubMed] [Google Scholar]
- 21.Saraste, M., P. R. Sibbald, and A. Wittinghofer. 1990. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15430-434. [DOI] [PubMed] [Google Scholar]
- 22.Steimle, V., L. A. Otten, M. Zufferey, and B. Mach. 1993. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell 75135-146. [PubMed] [Google Scholar]
- 23.Tameling, W. I., S. D. Elzinga, P. S. Darmin, J. H. Vossen, F. L. Takken, M. A. Haring, and B. J. Cornelissen. 2002. The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 142929-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tameling, W. I., J. H. Vossen, M. Albrecht, T. Lengauer, J. A. Berden, M. A. Haring, B. J. Cornelissen, and F. L. Takken. 2006. Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 1401233-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traut, T. W. 1994. The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide-binding sites. Eur. J. Biochem. 2229-19. [DOI] [PubMed] [Google Scholar]
- 26.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams, K. L., J. D. Lich, J. A. Duncan, W. Reed, P. Rallabhandi, C. Moore, S. Kurtz, V. M. Coffield, M. A. Accavitti-Loper, L. Su, S. N. Vogel, M. Braunstein, and J. P. Ting. 2005. The CATERPILLER protein Monarch-1 is an antagonist of Toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J. Biol. Chem. 28039914-39924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams, K. L., D. J. Taxman, M. W. Linhoff, W. Reed, and J. P. Ting. 2003. Cutting edge: Monarch-1: a pyrin/nucleotide-binding domain/leucine-rich repeat protein that controls classical and nonclassical MHC class I genes. J. Immunol. 1705354-5358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.