Abstract
Interleukin 1 (IL-1) has been reported to stimulate the polyubiquitination and disappearance of IL-1 receptor-associated kinase 1 (IRAK1) within minutes. It has been thought that the polyubiquitin chains attached to IRAK1 are linked via Lys48 of ubiquitin, leading to its destruction by the proteasome and explaining the rapid IL-1-induced disappearance of IRAK1. In this paper, we demonstrate that IL-1 stimulates the formation of K63-pUb-IRAK1 and not K48-pUb-IRAK1 and that the IL-1-induced disappearance of IRAK1 is not blocked by inhibition of the proteasome. We also show that IL-1 triggers the interaction of K63-pUb-IRAK1 with NEMO, a regulatory subunit of the IκBα kinase (IKK) complex, but not with the NEMO[D311N] mutant that cannot bind K63-pUb chains. Moreover, unlike wild-type NEMO, the NEMO[D311N] mutant was unable to restore IL-1-stimulated NF-κB-dependent gene transcription to NEMO-deficient cells. Our data suggest a model in which the recruitment of the NEMO-IKK complex to K63-pUb-IRAK1 and the recruitment of the TAK1 complex to TRAF6 facilitate the TAK1-catalyzed activation of IKK by the TRAF6-IRAK1 complex.
Following infection by bacteria or viruses, components of these pathogens bind to Toll-like receptors (TLRs) on host cells, triggering the activation of signaling pathways that stimulate the production of inflammatory mediators such as proinflammatory cytokines, chemokines, and interferons. These substances are released into the circulation, where they mount responses to combat the invading pathogen. Apart from TLR3, the activation of all TLRs and the interleukin-1 receptor (IL-1R) recruits a signaling complex to the receptors that includes an adaptor protein, MyD88, and two protein kinases, termed IL-1R-associated kinase 1 (IRAK1) and IRAK4. IRAK4 activates IRAK1, leading to autophosphorylation of the latter, its dissociation from MyD88, and its interaction with tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6), with which it propagates the signal. IL-1 signaling is greatly impaired in cells that do not express either TRAF6 or IRAK1 (14, 20, 30).
A key step in the activation of “downstream” signaling pathways by bacterial lipopolysaccharide (LPS) acting via TLR4 or IL-1 acting via the IL-1R is thought to be the formation of polyubiquitinated TRAF6 in which ubiquitin molecules are linked to one another by isopeptide bonds formed between lysine 63 of ubiquitin and the C-terminal carboxyl moiety of the preceding ubiquitin in the chain. The formation of Lys63-linked polyubiquitin (K63-pUb) chains attached to TRAF6 appears to enable the recruitment and activation of transforming growth factor-activated kinase 1 (TAK1) (31) because the TAB2 and TAB3 regulatory subunits of the TAK1 complex contain C-terminal nuclear zinc finger (NZF) motifs that interact with K63-pUb chains (15). TAK1 is then thought to phosphorylate and activate the IκBα kinase (IKK) complex because cells that lack TAK1 (27) or express a truncated inactive form of this protein (24) display greatly reduced activation of NF-κB in response to IL-1. Once activated, IKK phosphorylates its substrates, which include the inhibitory IκBα subunit of the NF-κB transcription factor and, in the case of LPS and TNF-α signaling, the p105 (also called NF-κB1) regulatory subunit of the protein kinase Tpl2 (tumor progression locus 2, also called COT [cancer Osaka thyroid oncogene]). The phosphorylation of these proteins induces their Lys48-linked polyubiquitination and destruction by the proteasome, leading to activation of NF-κB (reviewed in reference 4) and Tpl2 (2, 32). The role of Tpl2 is to activate mitogen-activated protein kinase kinases 1 and 2 and hence extracellular signal-regulated kinase 1 (ERK1) and ERK2 (9).
In contrast to signaling via TLRs and the IL-1R, signaling by TNF-α does not induce the formation of a TRAF6-IRAK1 complex but instead induces the formation of a complex between TRAF2 or TRAF5 and receptor-interacting protein 1 (RIP1). This is followed by the formation of K63-pUb-RIP1, which can then recruit the IKK complex, because NF-κB essential modifier (NEMO), a regulatory subunit of the IKK complex, binds to K63-pUb chains rather specifically (11, 34). A mutation in the K63-pUb-binding domain of NEMO (NEMO[D311N]), which causes an immunoinsufficiency disease (8), prevents binding to K63-pUb chains in vitro and to K63-pUb-RIP1 in cells (11, 34). Moreover, unlike wild-type NEMO, the NEMO[D311N] mutant cannot restore TNF signaling to NF-κB in NEMO-deficient cells (11, 34). K63-pUb-RIP1 also recruits and activates TAK1 (11), and the colocalization of TAK1 and IKK on a K63-pUb-RIP1 scaffold is thought to facilitate the TAK1-catalyzed activation of IKK. Embryonic fibroblasts that do not express the TAK1 catalytic subunit (27) or express an inactive TAK1 protein (24) display reduced activation of NF-κB in response to TNF.
LPS and IL-1 are reported to induce the polyubiquitination of IRAK1, which is followed by the disappearance of IRAK1 (12, 18, 35). It was thought that the pUb chains attached to IRAK1 are linked via Lys48 and that, by analogy with IκBα, the formation of K48-pUb-IRAK1 leads to its destruction by the proteasome, explaining the rapid IL-1/LPS-stimulated disappearance of IRAK1. Indeed, proteasome inhibitors were reported to slightly delay the disappearance of IRAK1 in MRC-5 cells (35). Here, we demonstrate that this is not the case in IL-1R-expressing HEK 293 cells. In these cells, IL-1-stimulates the formation of K63-pUb-IRAK1 and not K48-pUb-IRAK1 and the IL-1-induced disappearance of IRAK1 is not blocked by inhibition of the proteasome. We also show that IL-1 induces the interaction of K63-pUb-IRAK1 with NEMO but not with the NEMO[D311N] mutant form, which cannot bind K63-pUb chains. Moreover, unlike wild-type NEMO, the NEMO[D311N] mutant form is unable to restore IL-1-stimulated NF-κB-dependent gene transcription to NEMO-deficient cells. Our data suggest a model in which the recruitment of the NEMO-IKK complex to K63-pUb-IRAK1 and TAK1 to TRAF6 facilitates the activation of IKK by the TRAF6-IRAK1 complex.
MATERIALS AND METHODS
Antibodies and other proteins.
Antibodies that recognize IRAK1, IκBα, NEMO, TRAF6, Tpl2, and TAK1 were from Santa Cruz; rabbit polyclonal antiubiquitin was from DakoCytomation; monoclonal mouse anti-rabbit glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was from Research Diagnostics Inc. (RDI); and antihemagglutinin (anti-HA) was from Sigma. Phospho-specific antibodies that recognize p105 phosphorylated at Ser933 and the activated forms of IKKα/β and ERK1/ERK2 were from Cell Signaling Technologies, as were antibodies that recognize all forms of IKKα or all forms of IKKβ. Anti-GST, anti-TAB1, and anti-TAB2 antibodies (5); an antibody that immunoprecipitates Tpl2; and an antibody that recognizes TAK1 phosphorylated at Thr187 (raised against the peptide IQTHMT*NNKGS, where T* is phosphothreonine) were generated in sheep by the Division of Signal Transduction Therapy, University of Dundee. The phosphospecific TAK1 antibody was affinity purified on a phosphopeptide antigen-agarose column and used for immunoblotting in the presence of 10 μg/ml of the unphosphorylated form of the peptide immunogen. Rabbit-, mouse-, and sheep-specific secondary antibodies conjugated to horseradish peroxidase were from Pierce. Protein G coupled to horseradish peroxidase was from Bio-Rad, and murine IL-1α and human IL-1β were obtained from Sigma.
DNA constructs.
NEMO (AAH00299) was amplified from IMAGE 6062527 and cloned into pGEX6P-1 and EBG6P using BamHI and NotI sites. The mutant form of NEMO (NEMO[D311N]) was made by using standard protocols. DNAs encoding ubiquitin, ubiquitin[K48R], and ubiquitin[K63R] (a gift from David Lane, University of Dundee, Scotland) were cloned into pCMV5 for expression as HA-tagged proteins in mammalian cells.
Cell culture, transfection, cell lysis, and NF-κB reporter assay.
Human embryonic kidney (HEK) 293 cells overexpressing the IL-1 receptor (IL-1R cells), provided by Tularik Inc., CA, and mouse embryonic fibroblasts (MEFs) lacking NEMO (a gift from Manolis Pasparakis, University of Cologne, Germany) or expressing a truncated version of TAK1 (a gift from Shizuo Akira, Osaka University, Japan) were cultured in 15-cm dishes in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal calf serum and extracted with lysis buffer (50 mM Tris-HCl [pH 7.5], 1 mM EGTA, 1 mM EDTA, 1% [wt/wt] Triton X-100, 1 mM Na3VO4, 50 mM NaF, 5 mM sodium pyrophosphate, 10 mM sodium glycerophosphate, 0.27 M sucrose, 50 mM iodoacetamide, 1 mM benzamidine, 0.2 mM phenylmethylsulfonyl fluoride). EDTA inhibits protein phosphorylation and protein ubiquitination by chelating Mg2+, while dephosphorylation is inhibited by fluoride, pyrophosphate, vanadate, and glycerophosphate, and deubiquitination is inhibited by iodoacetamide, which inhibits ubiquitin-specific proteases. Phosphorylation and ubiquitination are therefore frozen in the state they were in at the moment of extraction. After centrifugation of cell lysates for 15 min at 18,000 × g, the supernatant (termed cell extract) was decanted. Lysis in sodium dodecyl sulfate (SDS) was performed with 2% SDS in 50 mM Tris-HCl (pH 7.5)-1 mM EDTA-1 mM EGTA-50 mM iodoacetamide-1 mM benzamidine-0.2 mM phenylmethylsulfonyl fluoride. Lysates were sonicated to shear the DNA, and after centrifugation for 15 min at 18,000 × g, the supernatant was decanted. Cells were transfected with polyethyleneimine as previously described (10).
For measurement of NF-κB-dependent luciferase gene expression, cells were lysed in passive lysis buffer (Promega) and activity measured using a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase activity.
RESULTS
IL-1 stimulates the interaction of NEMO with polyubiquitinated IRAK1.
The IL-1-stimulated formation of K63-pUb-TRAF6 induces the interaction of TAK1 complexes with TRAF6, leading to the oligomerization, autophosphorylation, and activation of TAK1 (see the introduction). Since the NEMO regulatory subunit of the IKK complex also binds to K63-pUb chains specifically, we initially investigated whether IL-1 induces the interaction of K63-pUb-TRAF6 with NEMO. A vector expressing GST-NEMO was therefore transfected into HEK 293 cells that stably express the IL-1R (termed IL-1R cells), and after purification on glutathione-Sepharose, we examined whether endogenous TRAF6 and TAK1 were associated with this protein. These experiments demonstrated an IL-1-dependent but transient interaction of NEMO with both TRAF6 and TAK1 that could be detected after 2 min and was maintained for 5 to 10 min but disappeared by 30 min (Fig. 1A, lanes 1 to 5). The form of TRAF6 associated with NEMO appeared to be mainly unmodified TRAF6, the higher-molecular-mass species corresponding to polyubiquitinated TRAF6 being barely visible.
FIG. 1.
Interaction of NEMO with polyubiquitinated IRAK1 in IL-1-stimulated IL-1R cells. (A) IL-1R cells were transfected with a vector expressing either wild-type GST-NEMO or the GST-NEMO[D311N] mutant form. After 24 h, the cells were starved for 16 h, stimulated with 5 ng/ml IL-1β for the times indicated, and then extracted with lysis buffer. To precipitate proteins bound to GST-NEMO, an aliquot of cell extract (0.5 mg protein) was added to 20 μl of glutathione-Sepharose beads and after 1 h of incubation at 4°C, the beads were collected by centrifugation and washed three times with 1 ml of lysis buffer and once with 1 ml 10 mM Tris-HCl (pH 8). The bound proteins were released by denaturation in 1% SDS, subjected to SDS-polyacrylamide gel electrophoresis, and immunoblotted with a variety of antibodies to detect the proteins indicated. In the third panel from the top, the band corresponding to unmodified TRAF6 is marked by an arrow and a band that cross-reacts nonspecifically with the anti-TRAF6 antibody and corresponds to the position of GST-NEMO is marked by an asterisk. In addition, the cell extracts (20 μg protein) were analyzed by SDS-polyacrylamide gel electrophoresis, followed by immunoblotting with the antibodies indicated (bottom two panels). The small difference in IRAK1 expression in extracts from cells expressing GST-NEMO and GST-NEMO[D311N] was not observed in other experiments and is therefore not significant. (B) IL-1R cells were deprived of serum for 16 h, stimulated with 5 ng/ml IL-1β for the times indicated, and then extracted with lysis buffer. A 0.5-mg sample of cell extract was incubated for 45 min with an anti-IRAK1 antibody or control immunoglobulin G (IgG). Twenty microliters of protein G-Sepharose beads was added, and after another incubation of 45 min, the beads were collected by centrifugation and washed three times with 1 ml lysis buffer and once with 1 ml 10 mM Tris-HCl (pH 8). The bound proteins were released by denaturation in 1% SDS, subjected to SDS-polyacrylamide gel electrophoresis, and immunoblotted with antibodies recognizing NEMO and IRAK1. To detect NEMO, horseradish peroxidase-coupled protein G was used instead of a secondary antibody because this protein comigrates with the IgG heavy chain; this explains why a faint signal is still detected at the positions of these proteins in the control IgG lane and at time zero. (C) IL-1R cells were deprived of serum for 16 h, stimulated with 5 ng/ml IL-1β for the times indicated, and then extracted with lysis buffer. IRAK1 (top three panels) was immunoprecipitated from 3 mg of cell extract protein as in panel B. The bound proteins were released by denaturation in 1% SDS, subjected to SDS-polyacrylamide gel electrophoresis, and immunoblotted with antibodies recognizing IKKα, IKKβ, and IRAK1. In addition, the cell extracts (20 μg protein) were analyzed by SDS-polyacrylamide gel electrophoresis, followed by immunoblotting with antibodies recognizing phosphorylated IKKα/IKKβ and GAPDH as a loading control (bottom two panels). Abbreviations: IP, immunoprecipitation; p-IRAK1, phosphorylated IRAK1.
Since TRAF6 interacts with IRAK1, we next investigated whether IRAK1 was also present in the GST-NEMO “pull downs.” These experiments showed that IRAK1 did indeed interact with NEMO in an IL-1-dependent manner (Fig. 1A, lanes 1 to 5, top panel). This interaction was detectable after 2 min but, in contrast to the binding of TRAF6 and TAK1, became much stronger after 5 to 10 min and only decreased moderately after 30 min. Moreover, strikingly and in contrast to the major species of IRAK1 detected in the cell lysates (Fig. 1A, second lowest panel), the IRAK1 associated with NEMO was present not as the unmodified protein but as a smear of much higher-molecular-mass species characteristic of polyubiquitinated species (Fig. 1A, lanes 1 to 5, top panel). Immunoblotting with an antiubiquitin antibody showed that IL-1 had indeed induced a parallel association of polyubiquitin chains with NEMO (Fig. 1A, lanes 1 to 5, second panel from the top). The ubiquitinated material and the modified forms of IRAK1 both associated with NEMO at the same time and had identical electrophoretic mobilities, suggesting that the polyubiquitin was attached to IRAK1.
IL-1 did not induce the association of IRAK1, polyubiquitin chains, TRAF6, or TAK1 with the NEMO[D311N] mutant that is unable to bind to K63-pUb chains (Fig. 1A, lanes 6 to 10), indicating that NEMO was interacting with a polyubiquitinated component(s) of the IRAK1-TRAF6 complex. The results shown in Fig. 1A suggest that NEMO can bind to K63-pUb-IRAK1 because K63-pUb-IRAK1 is still associated with NEMO 30 min after IL-1 stimulation, a time at which TRAF6 and TAK1 are no longer bound to NEMO. It could be argued that TRAF6 is still present but not visible after 30 min because it has been converted to a large number of different polyubiquitinated species. However, this is not the case because, as shown later (see Fig. 5), the polyubiquitination of TRAF6 is transient, peaking after 10 min and almost returning to basal levels after 20 to 30 min. Moreover, since TRAF6 forms a rather stable complex with TAK1 and TAK1 also dissociates from NEMO after 30 min, this implies that TRAF6 has also left.
FIG. 5.
IL-1 induces the recruitment of TAB2-TAK1 to TRAF6 and IRAK1. IL-1R cell extracts from Fig. 1B (0.5 mg protein) were used to immunoprecipitate TAB2 (A) or TAB1 (B) as described in the legend to Fig. 1, with 2 μg of anti-TAB2 or anti-TAB1 antibodies, respectively. Immunoprecipitated (IP) proteins were analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting with anti-TAK1 and with an antibody that recognizes TAK1 phosphorylated at Thr187. (C) Further aliquots of the cell extract (20 μg protein) were immunoblotted with antibodies that recognize TRAF6, IκBα, and the active phosphorylated forms of ERK1/2 (p-ERK1/2).
The experiments presented in Fig. 1A demonstrated that overexpressed NEMO could interact with endogenous IRAK1, but it was important to know whether endogenous NEMO was also capable of binding to IRAK1. We therefore stimulated IL-1R cells with IL-1 and immunoprecipitated IRAK1 from the cell extracts. These experiments demonstrated that IL-1 induced the association of endogenous NEMO with endogenous polyubiquitinated IRAK1 (Fig. 1B). Consistent with this finding, the IKKα and IKKβ components of the IKK complex could also be detected in IRAK1 immunoprecipitates (Fig. 1C, upper panels). The recruitment of IKKα and IKKβ to IRAK1 correlated with the activation of these protein kinases (Fig. 1C, lower panels).
The NEMO[D311N] mutant does not restore IL-1-dependent NF-κB gene transcription to NEMO-deficient cells.
It has been shown previously that the NEMO[D311N] mutant, which is unable to bind to K63-linked polyubiquitin chains, cannot reconstitute TNF-α signaling to NF-κB in NEMO-deficient cells (11, 34). To address the question of whether the NEMO[D311N] mutant is able to reconstitute IL-1-induced NF-κB activation, we transfected MEFs that do not express NEMO (NEMO−/−) with constructs expressing wild-type NEMO or NEMO[D311N] and analyzed the NF-κB-dependent transcription of a luciferase reporter gene (Fig. 2). Wild-type NEMO was able to reconstitute basal and IL-1-stimulated NF-κB activity in NEMO−/− MEFs to 60% of the level observed in wild-type MEFs (NEMO+/+). In contrast, NEMO[D311N] was unable to reconstitute NF-κB-dependent gene transcription, suggesting that the binding of NEMO to K63-linked polyubiquitin chains is required for activation of NF-κB by IL-1.
FIG. 2.
The NEMO[D311N] mutant form does not reconstitute IL-1-stimulated NF-κB-dependent gene transcription in NEMO-deficient cells. Wild-type NEMO (+/+) or NEMO-deficient (−/−) MEFs were cotransfected with 0.5 μg of DNA encoding an NF-κB luciferase reporter construct, 0.5 μg pTK-RL plasmid DNA (encoding Renilla luciferase), and 5 μg of empty pCMV5 vector (empty vector) or a plasmid encoding N-myc-tagged wild-type NEMO (NEMO[WT]) or NEMO[D311N] by using the Amaxa Nucleofection MEF2 kit according to the manufacturer's instructions. Subsequently, the MEFs were cultivated in Dulbecco modified Eagle medium containing 10% fetal calf serum and either left unstimulated (open bars) or stimulated (black bars) with 5 ng/ml murine IL-1α. Sixteen hours later, the cells were lysed in passive lysis buffer (Promega) and luciferase activity was measured with a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase activity. The error bars show the standard deviations of duplicate experiments. To analyze protein expression, aliquots of the cell extract (10 μg protein) were immunoblotted with antibodies that recognize NEMO or GAPDH as a loading control. The positions of transfected myc-tagged NEMO and endogenous wild-type NEMO are indicated on the right.
IRAK1 undergoes Lys63-linked polyubiquitination in IL-1R cells.
The results presented in Fig. 1 indicated that IL-1 induced the binding of polyubiquitinated IRAK1 to NEMO. Since NEMO binds K63-pUb chains specifically (11, 34), these observations suggested that the polyubiquitin chains attached to IRAK1 were also linked via Lys63. To investigate whether this was the case, we transfected DNA encoding either wild-type HA-tagged ubiquitin or an HA-tagged ubiquitin mutant form in which Lys63 or Lys48 was mutated to Arg. After stimulating the cells with IL-1, we immunoprecipitated the endogenous IRAK1 from the cell lysates and assessed the incorporation of HA-ubiquitin into polyubiquitinated IRAK1 by immunoblotting with an anti-HA antibody. To exclude the possibility that the polyubiquitin chains associated with immunoprecipitated IRAK1 were not attached covalently to IRAK1 but attached to an IRAK1-binding protein, such as TRAF6 or NEMO, the IRAK1 immunoprecipitates were resuspended in 1% sodium dodecyl sulfate (SDS) without any reducing agent, followed by incubation for 5 min at 70°C to disrupt, by denaturation, the interaction of IRAK1 with other proteins. After dilution to 0.1% SDS, IRAK1 was again immunoprecipitated and immunoblotting experiments established that this treatment had completely freed IRAK1 from TRAF6 and NEMO (results not shown).
These experiments established that IL-1 induced the covalent attachment of HA-ubiquitin molecules to endogenous IRAK1 when the cells were transfected with HA-WT-ubiquitin (Fig. 3A, lanes 1 to 4) or HA-ubiquitin[K48R] (Fig. 3B, lanes 5 to 8). Polyubiquitination was much weaker when HA-ubiquitin[K63R] replaced HA-WT-ubiquitin (Fig. 3A, lanes 5 to 8), indicating that the HA-ubiquitin was largely attached to IRAK1 via Lys63. The HA-ubiquitin[K48R] mutant form was expressed at higher levels than HA-WT-ubiquitin or HA-ubiquitin[K63R] (Fig. 3, third panel from the bottom), which may explain why the HA-ubiquitin[K48R] attached to IRAK1 appeared to be greater than with HA-WT-ubiquitin (Fig. 3B, top panel). The expression of HA-ubiquitin in IL-1R cells did not affect IL-1 signaling, as judged by the phosphorylation of ERK1/ERK2 (Fig. 3, second panel from the bottom). It was important in these experiments to limit the amount of DNA transfected to 2.5 μg/10-cm-diameter dish, because if more DNA was transfected, polyubiquitination of IRAK1 was observed even without IL-1 stimulation.
FIG. 3.
IRAK1 undergoes Lys63-linked polyubiquitination. Dishes (10-cm diameter) of IL-1R cells were transfected with 7.5 μg empty vector DNA (control) or 5 μg empty vector DNA plus 2.5 μg of vector DNA expressing wild-type HA-tagged ubiquitin (HA-Ub WT) or an HA-tagged mutant form of ubiquitin with either Lys63 (HA-Ub [K63R]) or Lys48 (HA-Ub [K48R]) mutated to Arg. After 24 h, the cells were deprived of serum for 16 h, stimulated for 5 to 30 min with or without IL-1β (5 ng/ml), and then lysed. IRAK1 was immunoprecipitated (IP) from 0.5 mg of cell extract protein (see the legend to Fig. 1) and washed once with lysis buffer, twice with lysis buffer plus 500 mM NaCl, and once with 10 mM Tris-HCl (pH 8). The immunoprecipitated IRAK1 protein was resuspended in 1% SDS-50 mM Tris-HCl (pH 8) and incubated for 5 min at 70°C. After dilution to 0.1% SDS with lysis buffer, the IRAK1 protein was reimmunoprecipitated, released from the beads by denaturation in 1% SDS, subjected to SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and immunoblotted (IB) with anti-HA, antiubiquitin (Ub), or anti-IRAK1 antibody. A further aliquot of the cell extract (20 μg of protein) was subjected to SDS-polyacrylamide gel electrophoresis, transferred to PVDF membranes, and immunoblotted with antibodies recognizing the HA tag of transfected ubiquitin, phosphorylated ERK1/2 (p-ERK1/2), and GAPDH (bottom three panels).
IL-1 decreases the level of IRAK1 by a proteasome-independent mechanism.
The IL-1-stimulated polyubiquitination of IRAK1 has been reported previously, but the ubiquitin was thought to be linked via Lys48, rather than Lys63, and to be responsible for targeting IRAK1 to the proteasome for degradation, explaining the rapid disappearance of IRAK1 from the cell extracts (Fig. 1A, second panel from the bottom; see also the introduction) (18, 35). However, the present finding that IRAK1 is predominantly polyubiquitinated via Lys63 of ubiquitin, which is not known to trigger degradation, raised the question of the mechanism underlying the disappearance of IRAK1 from IL-1-stimulated cells. We therefore tested the effect of MG-132, a specific inhibitor of the proteasome. As expected, MG-132 prevented the IL-1-stimulated disappearance of IκBα from the cells, which is triggered by the Lys48-linked polyubiquitination of this protein and allowed the accumulation of polyubiquitinated IκBα (Fig. 4A, second panel from the top). Similarly, MG-132 also prevented the IL-1-induced disappearance of the protein kinase Tpl2 (Fig. 4A, third panel from the top), which is also mediated by the proteasome (2, 17), and increased the overall level of polyubiquitinated proteins in cell extracts (Fig. 4A, second panel from the bottom), presumably by preventing the proteasomal destruction of K48-pUb proteins. In contrast, MG-132 had no effect on the IL-1-stimulated disappearance of IRAK1 from the same cells (Fig. 4A, uppermost panel), nor did it enhance the formation of polyubiquitinated IRAK1 (Fig. 4B, upper panel).
FIG. 4.
IRAK1 degradation is independent of the proteasome. (A) IL-1R cells were deprived of serum for 16 h, treated for 1 h with 25 μM MG-132 (Calbiochem) or DMSO as a control, stimulated with 5 ng/ml IL-1β for the times indicated, and then extracted with lysis buffer. An aliquot of the cell extract (20 μg of protein) was subjected to SDS-polyacrylamide gel electrophoresis and immunoblotted (IB) with antibodies recognizing IRAK1, IκBα, Tpl2, ubiquitin, and GAPDH. (B) IRAK1 was immunoprecipitated (IP) from 0.5 mg of the cell extract protein used in panel A as described in the legend to Fig. 1. Beads were washed once with lysis buffer, twice with lysis buffer plus 500 mM NaCl, and once with 10 mM Tris-HCl (pH 8). Bound proteins were released from the beads by denaturation in 1% SDS, subjected to SDS-polyacrylamide gel electrophoresis, and immunoblotted with antibodies recognizing ubiquitin and IRAK1. (C) IL-1R cells were deprived of serum for 16 h, stimulated with 5 ng/ml IL-1β for the times indicated, and extracted with lysis buffer containing either 1% Triton X-100 or 2% SDS. An aliquot of the cell extract (20 μg of protein) was subjected to SDS-polyacrylamide gel electrophoresis and immunoblotted with antibodies recognizing IRAK1 and GAPDH.
Taken together, these results are consistent with our finding that IRAK1 undergoes mainly Lys63-linked polyubiquitination in response to IL-1 and indicates that IL-1 induces the disappearance of IRAK1 from cell extracts by a proteasome-independent mechanism. The disappearance of IRAK1 from cell extracts is not explained by its translocation to a cellular compartment that is not disrupted by the standard cell lysis buffer containing 1% Triton X-100 (Fig. 4C, lanes 1 to 4), because a similar disappearance of IRAK1 was observed when the cells were lysed in 2% SDS to solubilize all cellular proteins from membranes and the cytoskeleton (Fig. 4C, lanes 5 to 8).
IL-1 recruits endogenous TAB2-TAK1 to endogenous TRAF6 and IRAK1.
It has been reported that the IL-1-induced activation of TAK1 is triggered by the binding of K63-pUb-TRAF6 to the C-terminal NZF motif of the TAB2/TAB3 regulatory subunits of the TAK1 complex (15). In the present study, we found that IL-1 induced the association of endogenous TAB2 with both endogenous TRAF6 (Fig. 5A, top panel) and IRAK1 (Fig. 5A, second panel from the top) within 2 min (Fig. 5A). After 2 min of IL-1 stimulation, the most prominent species of IRAK1 and TRAF6 associated with TAB2 were the unmodified forms of these proteins (Fig. 5A). The polyubiquitination of TRAF6 increased with time, became maximal between 5 and 10 min, and correlated with the activation of TAK1, as judged by the phosphorylation of its activation loop at Thr187 (Fig. 5B, top panel). The polyubiquitination of IRAK1 was maximal between 10 and 30 min and correlated with the activation of IKK and its downstream target Tpl2, as judged by the degradation of IκBα and the phosphorylation of ERK1/2, respectively (Fig. 5C).
The TAK1 protein is required for the IL-1-stimulated activation of IKK and the phosphorylation of its targets.
It has been reported that the IL-1-stimulated phosphorylation (activation) of IKKα/β, the degradation of IκBα, and the induction of NF-κB DNA binding are greatly decreased, although not eliminated, in IL-1-stimulated MEFs that do not express the TAK1 catalytic subunit (27) or express a truncated, inactive form of TAK1 lacking the ATP-binding domain (24) (Fig. 6A, bottom panel). However, these studies did not show that TAK1 was required for the IL-1-induced, IKK-catalyzed phosphorylation of p105, which is known to be required for the LPS- or TNF-α-induced activation of Tpl2 and hence the activation of ERK1/ERK2 (2, 32). Here we confirmed that IL-1 does not induce the phosphorylation of IKKα/β or degradation of IκBα in immortalized MEFs expressing the truncated, inactive mutant form of TAK1 (Fig. 6A, top two panels). In addition, we demonstrated that the phosphorylation of p105 and the activation of ERK1/ERK2 were also impaired (Fig. 6A, third and fourth panels from the top). Thus, an intact TAK1 protein is required for the IL-1-stimulated phosphorylation of p105 and the activation of ERK1/ERK2 in MEFs. The IL-1-stimulated activation of ERK1/ERK2 is also impaired in Tpl2−/− MEFs (7), consistent with the phosphorylation of p105 inducing the activation of Tpl2.
FIG. 6.
TAK1 is required for the IL-1-induced activation of IKKα/β and subsequent degradation of IκBα and Tpl2-dependent ERK1/2 activation. (A) MEFs expressing wild-type TAK1 (TAK1+/+) or a truncated TAK1 form lacking kinase activity (TAK1−/−) were deprived of serum for 16 h, stimulated with 5 ng/ml IL-1α for the times indicated, and extracted with lysis buffer. An aliquot of the cell extract (20 μg protein) was subjected to SDS-polyacrylamide gel electrophoresis and immunoblotted with the indicated antibodies. TAK1 was immunoprecipitated (IP) with an anti-TAK1 antibody from 0.5 mg of cell extract protein as described in the legend to Fig. 3, subjected to SDS-polyacrylamide gel electrophoresis, and immunoblotted (IB) with the same anti-TAK1 antibody. (B) IL-1R cells were deprived of serum for 16 h, stimulated with 5 ng/ml IL-1β for the times indicated, and extracted with lysis buffer. An aliquot of the cell extract (20 μg of protein) was subjected to SDS-polyacrylamide gel electrophoresis and immunoblotted with the indicated antibodies. (C) IL-1R cells were deprived of serum for 16 h and stimulated for 15 min with 5 ng/ml IL-1β. Following cell lysis, 0.25 mg of extract protein was incubated for 2 h at 4°C with 5 μg of immunoprecipitating anti-Tpl2 antibody. Protein G-Sepharose (15 μl packed beads) was then added, and after mixing for 1 h, the beads were collected, washed, and assayed for Tpl2 activity as described previously for Raf (1).
We have also shown that, as in MEFs, IL-1 stimulates the phosphorylation of p105, the activation of ERK1/ERK2 (Fig. 6B), and the activation of Tpl2 (Fig. 6C) in the IL-1R cells used in this study. In LPS- or TNF-α-stimulated macrophages or TNF-α-stimulated MEFs, the activation of Tpl2 is followed by the rapid proteolytic degradation of the 58-kDa form of this protein kinase, but not the 52-kDa form (33), and this was also observed in IL-1-stimulated IL-1R cells (Fig. 6B).
Taken together, the experiments described above demonstrate that IL-1 stimulates the activation of ERK1/ERK2 by activating Tpl2 in both MEFs and IL-1R cells in a manner similar to that shown previously for LPS and TNF-α. The IL-1-stimulated activation of Tpl2 has also been demonstrated in HeLa cells (23).
DISCUSSION
Several lines of evidence presented in this paper establish that IL-1 stimulates the formation of K63-pUb-IRAK1 predominantly. Firstly, the ubiquitin[K63R] mutant form was not incorporated efficiently into the polyubiquitin chains attached to IRAK1 (Fig. 3). Secondly, polyubiquitinated IRAK1 was able to bind to wild-type NEMO but not to the NEMO[D3111N] mutant form that does not bind to K63-pUb chains (Fig. 1). Thirdly, the proteasome inhibitor MG-132, which prevents the degradation of K48-pUb proteins, such as IκB, and therefore enhances their level in cells (Fig. 4A), does not increase the polyubiquitination of IRAK1 (Fig. 4B). Moreover, MG132 did not affect the IL-1-induced disappearance of IRAK1, indicating that this occurs by a mechanism that is independent of the proteasome (Fig. 4).
These findings raise the question of how K63-pUb-IRAK1 is formed in cells, and recent work has suggested a mechanism by which this may take place. It has been known for several years that IRAK1 interacts with Pellino isoforms (reviewed in reference 26) and more recently that the cotransfection of IRAK1 with these proteins induces the polyubiquitination of IRAK1 (25, 28). These observations, and the presence of a RING-like domain at the C terminus of Pellino isoforms, suggested that they were E3 ubiquitin ligases (25). We (22) and others (3) recently established by assays performed in vitro that this was indeed the case, and we also found that the E3 ligase function of Pellino 1 and Pellino 3b was enormously enhanced by phosphorylation catalyzed by IRAK1 or IRAK4 in vitro (22). Moreover, in the presence of the E2 ubiquitin-conjugating complex Ubc13-Uev1a, Pellino 1 was able to induce the formation of K63-pUb chains specifically in vitro (3, 22). Importantly, the cotransfection of wild-type Pellino isoforms with wild-type IRAK1 in HEK 293 cells induced the formation of polyubiquitinated IRAK1, but cotransfection of a catalytically inactive mutant form of IRAK1 with wild-type Pellino or of wild-type IRAK1 with an E3 ligase-deficient mutant form of Pellino did not (22, 25). This is consistent with IRAK1 catalyzing the activation of Pellino, which then mediates the polyubiquitination of IRAK1 in cells. Crucially, the cotransfection of Pellino 2 and IRAK1 was found to induce the formation of K63-pUb-IRAK1 and its interaction with NEMO, but not NEMO[D311N]. Only traces of K48-pUb-IRAK1 were detectable in these experiments (22). Taken together, these findings suggest that one or more Pellino isoforms may be the E3 ligases that mediate the formation of K63-pUb-IRAK1 in response to IL-1. An important role for Pellino 1 in IL-1 signaling is supported by the finding that small interfering RNA-mediated knockdown of this protein has been reported to impair the activation of NFκB (13).
It is currently unknown whether it is IRAK1 and/or IRAK4 that activates Pellino in vivo. However, the observation that a catalytically inactive mutant form of IRAK1 (18), or fragments of IRAK1 that lack the catalytic domain (19, 36), can restore IL-1-stimulated NF-κB signaling to IRAK1-deficient cells could be explained by the IRAK4-catalyzed activation of Pellino, followed by the Lys63-linked polyubiquitination of catalytically inactive IRAK1 under these conditions. Importantly, all of the fragments of IRAK1 that were able to restore IL-1-stimulated, TAK1-dependent signaling to IRAK1-deficient cells underwent polyubiquitination in response to IL-1 (36).
The polyubiquitination of the IRAK1 fragments mentioned above was reported to be prevented by the mutation of Lys134 to Arg, and this mutation also suppressed the IL-1-stimulated degradation of IκBα (36), implying that the activation of IKK was impaired. However, when we used IRAK1-deficient cells (18) to generate stable cell lines expressing wild-type IRAK1 or IRAK1[K134R], we found that the IL-1-stimulated polyubiquitination of IRAK1[K134R] and that of wild-type IRAK1 were similar. Moreover, IL-1 stimulated a similar activation of ERK1/2 (which is dependent on IKK activation) in cells expressing wild-type IRAK1 or IRAK1[K134R] (Mark Windheim and Philip Cohen, results not shown). These findings indicate that wild-type IRAK1 is polyubiquitinated at additional sites and that the polyubiquitination of Lys134 is not essential for IL-1-signaling.
The finding that IL-1 stimulates the formation of K63-pUb-IRAK1 raises the question of the role of this modification in signaling. It has been reported previously that IRAK1 can be detected in NEMO immunoprecipitates when HeLa cells are stimulated with IL-1 (6). Here we provide evidence that the mechanism involves the interaction of K63-pUb-IRAK1 with NEMO, thereby recruiting the IKK complex to IRAK1 (Fig. 1).
We also show that the formation of K63-pUb-IRAK1 is maximal at 10 min, the time at which IKK is activated, as judged by the proteolytic destruction and disappearance of IκBα, the phosphorylation of the p105 subunit of the Tpl2 complex, and the activation of ERK1/ERK2 (Fig. 5 and 6). In contrast, the IL-1-induced interaction of TRAF6 with the TAK1 complex and the activation of TAK1 precede the polyubiquitination of IRAK1 and the activation of IKK (Fig. 5A), suggesting that TAK1 may activate IKK when the NEMO-IKK complex has been recruited to K63-pUb-IRAK1. Consistent with a role for TAK1 in activating IKK, the IL-1-stimulated activation of IKK did not occur in MEFs expressing a truncated, inactive form of TAK1 (Fig. 6A).
TRAF6-deficient cells do not respond to IL-1, demonstrating a critical role for TRAF6 in signaling downstream of the IL-1R (16, 20). Since TRAF6 is an E3 ligase, it is thought that the formation of K63-pUb-TRAF6 is an autoubiquitination event that occurs in the presence of E2 ubiquitin-conjugating complexes that specify the formation of K63-pUb chains. Indeed, it has been reported that the TAK1 complex can be activated in vitro if it is incubated with TRAF6, Ubc13-Uev1a (E2), E1, ubiquitin, and MgATP (31). However, whether the E3 ligase activity of TRAF6 is required for IL-1 signaling in vivo is unclear because IL-1 responsiveness is reported to be restored to TRAF6-deficient cells by transfection with a truncated form of TRAF6 in which the RING domain that carries the E3 ligase activity has been deleted (16). It is also unlikely that TRAF6 is the E3 ligase that mediates the formation of K63-pUb-IRAK1 because the IL-1-stimulated polyubiquitination and disappearance of IRAK1 are not impaired in IL-1-stimulated TRAF6−/− fibroblasts (M. Windheim, unpublished experiments; see also the Discussion in reference 36).
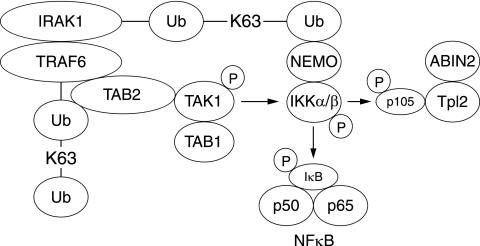
Taken together, our results suggest the following model for the IL-1-stimulated activation of IKK by TAK1 (Fig. 7). IL-1 initially stimulates the interaction of IRAK1 with TRAF6. The TAB2/TAB3-TAK1-TAB1 complexes then bind to the TRAF6 component of the IRAK1-TRAF6 complex, interaction being mediated by the TAB2/3 regulatory subunits. TAB2 has been shown to bind to TRAF6, but not to IRAK1, in a yeast “two-hybrid” analysis (29), and this interaction may be enhanced by the binding of the K63-pUb chains of TRAF6 to the NZF motifs of TAB2/TAB3 (15). The formation of the IRAK1-TRAF6-TAK1 complex in IL-1R cells is followed by the formation of K63-pUb-TRAF6, which correlates with the time at which phosphorylation/activation of TAK1 is maximal, and this is followed by the formation of K63-pUb-IRAK1, the binding recruitment and activation of the IKK complex, and the phosphorylation of its substrates. The proposed association of NEMO-IKK with K63-pUb-IRAK1 and TAB2/3-TAK1-TAB1 with K63-pUb-TRAF6 may colocalize these two protein kinases to the IRAK1-TRAF6 complex, facilitating the activation of IKK by TAK1. Such an arrangement may also facilitate the termination of signaling when the TRAF6-IRAK1 complex dissociates. In order to establish whether this hypothesis is correct, it is necessary to find ways of inhibiting the polyubiquitination of IRAK1 specifically without affecting the polyubiquitination of TRAF6. It should also be emphasized that TAK1 may not be the only protein kinase that activates the IKK complex in IL-1-stimulated cells, because although the activation of IKK by IL-1 or TNF-α does not occur in TAK1−/− MEFs, the activation of IKK is not impaired in TAB1−/− MEFs that also lack TAK1 activity (21). Therefore, a protein kinase distinct from TAK1 seems to activate IKK in TAB1-deficient cells.
FIG. 7.
Suggested model for how TAK1 and IKKα/β colocalize with the TRAF6-IRAK1 complex. The results presented in this paper, together with data from other laboratories, suggest that IL-1 induces the binding of the TAB2-TAK1-TAB1 complex to K63-pUb-TRAF6, while NEMO-IKK binds to K63-pUb-IRAK1 in the TRAF6-IRAK1 complex. These interactions may colocalize TAK1 and IKKα/β to this complex, which may facilitate the phosphorylation of IKKα/β by TAK1. Once activated, IKK phosphorylates the IκBα component of the NF-κB complex and the p105 regulatory subunit of Tpl2. This triggers the proteasomal destruction of these proteins, leading to the activation of NF-κB and Tpl2.
In summary, TRAF6 is not the only component of the TRAF6-IRAK1 complex that undergoes Lys63-linked polyubiquitination in response to IL-1. IRAK1 also undergoes this modification, adding a new level of complexity to this signaling system that has still to be fully worked out. Nevertheless, the tethering of protein kinases to K63-pUb-protein scaffolds is clearly critical for the activation of the innate immune system, since the NEMO[D311N] mutant form, which does not bind to K63-pUb chains, is the cause of a severe human immunodeficiency disease (8).
Acknowledgments
We thank David Lane (University of Dundee, Scotland) for providing vectors for the expression of tagged ubiquitin and ubiquitin mutant forms, Shizuo Akira (Osaka University, Japan) for TAK1−/− cells, Manolis Pasparakis (University of Cologne, Germany) for NEMO−/− cells, and Tak Mak (University of Toronto, Canada) for TRAF6−/− cells.
Mark Windheim acknowledges a postdoctoral position from the EU Research Training Framework 5 Programme. This work was supported by the UK Medical Research Council, The European Union Framework 5 program, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck and Co., Merck KGaA, and Pfizer.
Footnotes
Published ahead of print on 7 January 2008.
REFERENCES
- 1.Alessi, D. R., P. Cohen, A. Ashworth, S. Cowley, S. J. Leevers, and C. J. Marshall. 1995. Assay and expression of mitogen-activated protein kinase, MAP kinase kinase, and Raf. Methods Enzymol. 255279-290. [DOI] [PubMed] [Google Scholar]
- 2.Beinke, S., M. J. Robinson, M. Hugunin, and S. C. Ley. 2004. Lipopolysaccharide activation of the TPL-2/MEK/extracellular signal-regulated kinase mitogen-activated protein kinase cascade is regulated by IκB kinase-induced proteolysis of NF-κB1 p105. Mol. Cell. Biol. 249658-9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler, M. P., J. A. Hanly, and P. N. Moynagh. 2007. Kinase-active interleukin-1 receptor-associated kinases promote polyubiquitination and degradation of the Pellino family: direct evidence for PELLINO proteins being ubiquitin-protein isopeptide ligases. J. Biol. Chem. 28229729-29737. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Z. J. 2005. Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 7758-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung, P. C., D. G. Campbell, A. R. Nebreda, and P. Cohen. 2003. Feedback control of the protein kinase TAK1 by SAPK2a/p38α. EMBO J. 225793-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooke, E. L., I. J. Uings, C. L. Xia, P. Woo, and K. P. Ray. 2001. Functional analysis of the interleukin-1-receptor-associated kinase (IRAK-1) in interleukin-1β-stimulated nuclear factor κB (NF-κB) pathway activation: IRAK-1 associates with the NF-κB essential modulator (NEMO) upon receptor stimulation. Biochem. J. 359403-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das, S., J. Cho, I. Lambertz, M. A. Kelliher, A. G. Eliopoulos, K. Du, and P. N. Tsichlis. 2005. Tpl2/Cot signals activate ERK, JNK, and NF-κB in a cell-type and stimulus-specific manner. J. Biol. Chem. 28023748-23757. [DOI] [PubMed] [Google Scholar]
- 8.Döffinger, R., A. Smahi, C. Bessia, F. Geissmann, J. Feinberg, A. Durandy, C. Bodemer, S. Kenwrick, S. Dupuis-Girod, S. Blanche, P. Wood, S. H. Rabia, D. J. Headon, P. A. Overbeek, F. Le Deist, S. M. Holland, K. Belani, D. S. Kumararatne, A. Fischer, R. Shapiro, M. E. Conley, E. Reimund, H. Kalhoff, M. Abinun, A. Munnich, A. Israel, G. Courtois, and J.-L. Casanova. 2001. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-κB signaling. Nat. Genet. 27277-285. [DOI] [PubMed] [Google Scholar]
- 9.Dumitru, C. D., J. D. Ceci, C. Tsatsanis, D. Kontoyiannis, K. Stamatakis, J. H. Lin, C. Patriotis, N. A. Jenkins, N. G. Copeland, G. Kollias, and P. N. Tsichlis. 2000. TNF-α induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell 1031071-1083. [DOI] [PubMed] [Google Scholar]
- 10.Durocher, Y., S. Perret, and A. Kamen. 2002. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ea, C. K., L. Deng, Z. P. Xia, G. Pineda, and Z. J. Chen. 2006. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22245-257. [DOI] [PubMed] [Google Scholar]
- 12.Hu, J., R. Jacinto, C. McCall, and L. Li. 2002. Regulation of IL-1 receptor-associated kinases by lipopolysaccharide. J. Immunol. 1683910-3914. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, Z., H. J. Johnson, H. Nie, J. Qin, T. A. Bird, and X. Li. 2003. Pellino 1 is required for interleukin-1 (IL-1)-mediated signaling through its interaction with the IL-1 receptor-associated kinase 4 (IRAK4)-IRAK-tumor necrosis factor receptor-associated factor 6 (TRAF6) complex. J. Biol. Chem. 27810952-10956. [DOI] [PubMed] [Google Scholar]
- 14.Kanakaraj, P., P. H. Schafer, D. E. Cavender, Y. Wu, K. Ngo, P. F. Grealish, S. A. Wadsworth, P. A. Peterson, J. J. Siekierka, C. A. Harris, and W. P. Fung-Leung. 1998. Interleukin (IL)-1 receptor-associated kinase (IRAK) requirement for optimal induction of multiple IL-1 signaling pathways and IL-6 production. J. Exp. Med. 1872073-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanayama, A., R. B. Seth, L. Sun, C. K. Ea, M. Hong, A. Shaito, Y. H. Chiu, L. Deng, and Z. J. Chen. 2004. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol. Cell 15535-548. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi, N., Y. Kadono, A. Naito, K. Matsumoto, T. Yamamoto, S. Tanaka, and J. Inoue. 2001. Segregation of TRAF6-mediated signaling pathways clarifies its role in osteoclastogenesis. EMBO J. 201271-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang, V., A. Symons, S. J. Watton, J. Janzen, Y. Soneji, S. Beinke, S. Howell, and S. C. Ley. 2004. ABIN-2 forms a ternary complex with TPL-2 and NF-κ B1 p105 and is essential for TPL-2 protein stability. Mol. Cell. Biol. 245235-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, X., M. Commane, C. Burns, K. Vithalani, Z. Cao, and G. R. Stark. 1999. Mutant cells that do not respond to interleukin-1 (IL-1) reveal a novel role for IL-1 receptor-associated kinase. Mol. Cell. Biol. 194643-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, X., M. Commane, Z. Jiang, and G. R. Stark. 2001. IL-1-induced NFκB and c-Jun N-terminal kinase (JNK) activation diverge at IL-1 receptor-associated kinase (IRAK). Proc. Natl. Acad. Sci. USA 984461-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomaga, M. A., W. C. Yeh, I. Sarosi, G. S. Duncan, C. Furlonger, A. Ho, S. Morony, C. Capparelli, G. Van, S. Kaufman, A. van der Heiden, A. Itie, A. Wakeham, W. Khoo, T. Sasaki, Z. Cao, J. M. Penninger, C. J. Paige, D. L. Lacey, C. R. Dunstan, W. J. Boyle, D. V. Goeddel, and T. W. Mak. 1999. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 131015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendoza, H., D. G. Campbell, K. Burness, J. Hastie, N. Ronkina, J. H. Shim, J. S. Arthur, R. J. Davis, M. Gaestel, G. L. Johnson, S. Ghosh, and P. Cohen. 2008. Roles for TAB1 in regulating the IL-1-dependent phosphorylation of the TAB3 regulatory subunit and activity of the TAK1 complex. Biochem. J. 409711-722. [DOI] [PubMed] [Google Scholar]
- 22.Ordureau, A., H. Smith, M. Windheim, M. Peggie, E. Carrick, N. Morrice, and P. Cohen. 2008. The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochem. J. 40943-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodríguez, C., M. Pozo, E. Nieto, M. Fernandez, and S. Alemany. 2006. TRAF6 and Src kinase activity regulates Cot activation by IL-1. Cell. Signal. 181376-1385. [DOI] [PubMed] [Google Scholar]
- 24.Sato, S., H. Sanjo, K. Takeda, J. Ninomiya-Tsuji, M. Yamamoto, T. Kawai, K. Matsumoto, O. Takeuchi, and S. Akira. 2005. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 61087-1095. [DOI] [PubMed] [Google Scholar]
- 25.Schauvliege, R., S. Janssens, and R. Beyaert. 2006. Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: a role as novel RING E3-ubiquitin-ligases. FEBS Lett. 5804697-4702. [DOI] [PubMed] [Google Scholar]
- 26.Schauvliege, R., S. Janssens, and R. Beyaert. 2007. Pellino proteins: novel players in TLR and IL-1R signalling. J. Cell. Mol. Med. 11453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shim, J. H., C. Xiao, A. E. Paschal, S. T. Bailey, P. Rao, M. S. Hayden, K. Y. Lee, C. Bussey, M. Steckel, N. Tanaka, G. Yamada, S. Akira, K. Matsumoto, and S. Ghosh. 2005. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 192668-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strelow, A., C. Kollewe, and H. Wesche. 2003. Characterization of Pellino2, a substrate of IRAK1 and IRAK4. FEBS Lett. 547157-161. [DOI] [PubMed] [Google Scholar]
- 29.Takaesu, G., J. Ninomiya-Tsuji, S. Kishida, X. Li, G. R. Stark, and K. Matsumoto. 2001. Interleukin-1 (IL-1) receptor-associated kinase leads to activation of TAK1 by inducing TAB2 translocation in the IL-1 signaling pathway. Mol. Cell. Biol. 212475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas, J. A., J. L. Allen, M. Tsen, T. Dubnicoff, J. Danao, X. C. Liao, Z. Cao, and S. A. Wasserman. 1999. Impaired cytokine signaling in mice lacking the IL-1 receptor-associated kinase. J. Immunol. 163978-984. [PubMed] [Google Scholar]
- 31.Wang, C., L. Deng, M. Hong, G. R. Akkaraju, J. Inoue, and Z. J. Chen. 2001. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412346-351. [DOI] [PubMed] [Google Scholar]
- 32.Waterfield, M., W. Jin, W. Reiley, M. Zhang, and S. C. Sun. 2004. IκB kinase is an essential component of the Tpl2 signaling pathway. Mol. Cell. Biol. 246040-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterfield, M. R., M. Zhang, L. P. Norman, and S. C. Sun. 2003. NF-κB1/p105 regulates lipopolysaccharide-stimulated MAP kinase signaling by governing the stability and function of the Tpl2 kinase. Mol. Cell 11685-694. [DOI] [PubMed] [Google Scholar]
- 34.Wu, C. J., D. B. Conze, T. Li, S. M. Srinivasula, and J. D. Ashwell. 2006. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation. Nat. Cell Biol. 8398-406. [DOI] [PubMed] [Google Scholar]
- 35.Yamin, T. T., and D. K. Miller. 1997. The interleukin-1 receptor-associated kinase is degraded by proteasomes following its phosphorylation. J. Biol. Chem. 27221540-21547. [DOI] [PubMed] [Google Scholar]
- 36.Yao, J., T. W. Kim, J. Qin, Z. Jiang, Y. Qian, H. Xiao, Y. Lu, W. Qian, M. F. Gulen, N. Sizemore, J. DiDonato, S. Sato, S. Akira, B. Su, and X. Li. 2007. Interleukin-1 (IL-1)-induced TAK1-dependent versus MEKK3-dependent NFκB activation pathways bifurcate at IL-1 receptor-associated kinase modification. J. Biol. Chem. 2826075-6089. [DOI] [PubMed] [Google Scholar]