Abstract
During cardiac development, the T-box transcription factor Tbx5 displays dynamic changes in localization from strictly nuclear to both nuclear and cytoplasmic to exclusively cytoplasmic along the actin cytoskeleton in cells coexpressing its binding protein LMP4. Although nuclear localization signals (NLSs) have been described, the mechanism by which Tbx5 exits the nucleus remained elusive. Here, we describe for Tbx5 a nuclear export signal (NES) that is recognized by the CRM1 export protein. Site-directed mutagenesis of a critical amino acid(s) within this sequence determined the functionality of this NES. Confocal localization studies and luciferase transcriptional reporter assays with NES mutant Tbx5 forms demonstrated retention in the nucleus, regardless of the presence of LMP4. Coimmunoprecipitation and pharmacological interference studies demonstrated a direct interaction between Tbx5 and CRM1, revealing that Tbx5 is using the CRM1 pathway for nuclear export. In addition to Tbx5, we identified NESs in all T-box proteins and demonstrated interaction of the family members Tbx3 and Brachyury with the CRM1 exporter, suggesting general significance. This first demonstration of evolutionarily conserved NESs in all T-box proteins in conjunction with NLSs indicates a primordial function of T-box proteins to dynamically shuttle between nuclear and cytoplasmic compartments of the cell.
In vertebrates, the T-box proteins Tbx5 and Tbx4 have important functions in the specification, initiation, and outgrowth of the fore- and hindlimbs, respectively (37, 47). Similar to its role in the forelimb, Tbx5 appears to be the earliest determinant of vertebrate heart growth, and it has apparent functions in a variety of cardiac lineages and structures (10, 43). Loss-of-function experiments with mice (12) and mutant analyses of fish (20) have further established a dual developmental role for Tbx5 in the heart and limbs. Mutations in human TBX5 cause Holt-Oram syndrome (HOS), an autosomal dominant condition involving malformations of the arms, as well as cardiac atrial and ventricular septal defects (7, 11, 36, 41).
Guided by their protein structures, Tbx5 and related T-box family members are thought to function as transcription factors. Several Tbx genes are implicated to function upstream or downstream of fibroblast growth factor (FGF) signal transduction, suggesting that growth control by Tbx genes could be a more general conserved function in a variety of developing organs (43). Experimental evidence indicates that Tbx5 regulates the expression of Fgf10 and Wnt2b genes, thereby controlling cell proliferation during limb development (37). In addition, Tbx5 interacts with other proteins to form functional complexes. For instance, the DNA-binding domain of the Tbx5 protein can interact with those of the heart-specific transcription factors Nkx2.5 and Gata4, and the transcription factor complexes act in a cooperative manner to regulate target gene activities (10).
In contrast to the N-terminal DNA-binding domain, LMP4, a PDZ-LIM protein, has been identified by our laboratory as specifically interacting with the C-terminal transactivation domains of Tbx5 and Tbx4, suggesting a different functional role for these transcription factors (30). PDZ-LIM family proteins are proposed to mediate protein association with the cytoskeleton and with proteins involved in signal transduction cascades that regulate cell lineage specification and organ development (6, 16, 17, 29). In transfected cells, coexpression of the cytoplasmic LMP4 induces the relocalization of Tbx5 from the nucleus to the cytoplasm, where the Tbx5-LMP4 complex associates with the actin cytoskeleton (30). In the presence of LMP4, Tbx5 shuttles dynamically between the nucleus and the cytoplasm, thus allowing LMP4 to regulate the nuclear availability of Tbx5 and in turn modulate the transcriptional activity of the protein (13).
Work with chicken primary epicardial cells revealed that endogenous Tbx5 and LMP4 subcellular localization changes in response to differentiation stimuli (13). While the nature of the signal remains elusive, these in vitro differentiation studies provided the first indications that Tbx5 subcellular localization is not dependent solely on the presence of LMP4 within the cell but, rather, on a regulated event. In vivo studies of Tbx5 and LMP4 protein expression during chicken embryonic heart development confirmed this hypothesis (9). The two proteins were dynamically expressed both temporally and spatially in the developing heart. In coexpressing cells, Tbx5 localization was strictly nuclear, nuclear and cytoplasmic, or strictly cytoplasmic, depending on the developmental stage and individual cardiac cell lineage. Cytoplasmic localization of the Tbx5-LMP4 complex was also demonstrated for the developing chicken wing; however, the ratio of nuclear to cytoplasmic distribution of Tbx5 varied in different regions of the limb. These in vivo observations for multiple tissues would indicate a more general functional role for nuclear and cytoplasmic Tbx5 distribution. Furthermore, the dynamic localization of Tbx5 in these tissues during chicken development reveals a striking correlation with the tissues affected in humans diagnosed with HOS. Taken together, these results strongly emphasize the importance of appropriate subcellular localization of the protein during developmental processes.
Nuclear localization signals (NLSs) were initially identified for human TBX5. An analysis of selected fragments of the C-terminal region of TBX5 identified a NLS within the transactivation domain (NLS2) (51). Work with a series of N- and C-terminal deletion constructs and single-amino-acid substitutions revealed a second NLS in the DNA-binding domain (NLS1) (15). From these studies, it remained unclear, however, to what extent the critical residues within each given NLS are important for function and whether the two NLS motifs have a synergistic effect on nuclear localization.
The identification of NLS motifs within Tbx5 provided only a partial insight into how the protein shuttles across the nuclear membrane. In the present study, we describe the identification of a putative nuclear export signal (NES) in the DNA-binding domain of Tbx5, a core of hydrophobic amino acids that appear to be evolutionarily conserved in all T-box protein family members. Site-directed mutagenesis of this NES and pharmacological interference studies in combination with confocal imaging and coimmunoprecipitation demonstrated that Tbx5 uses this NES for binding to the export protein CRM1 in order to relocate from the nucleus to cytoplasmic sites. The current work provides a deeper understanding of the molecular machinery that enables dynamic nuclear/cytoplasmic shuttling of the Tbx5 transcription factor. Our findings suggest that Tbx5 is using a mechanism for protein relocalization that evolved early in the generation of the T-box protein family, thus setting a new paradigm for nuclear and cytoplasmic functions of potentially many T-box proteins during developmental processes.
MATERIALS AND METHODS
Plasmid constructs and mutagenesis.
Full-length chicken LMP4 was cloned into a pcDNA3.1 expression vector containing a C-terminal Myc tag. LMP4 was also recombined as an N-terminal fusion into a modified HcRed-C1 expression vector suitable for the Gateway recombination system (Invitrogen). Full-length chicken Tbx5, Tbx3, and Brachyury were cloned into pDONR/Zeo entry vectors suitable for subsequent Gateway recombination reactions as an N-terminal fusion into Gateway-modified enhanced green fluorescent protein (EGFP)-C1 or hemagglutinin (HA)-C1 expression vectors. A modification of a PCR-based site-directed mutagenesis method (1, 18) was used to generate the Tbx5 NES mutants. Briefly, nonoverlapping oligonucleotides incorporating the desired mutation(s) were designed to amplify the sequence outward, generating a double-stranded linear product using Pfu Turbo DNA polymerase (Stratagene). The ends of the PCR products were phosphorylated with T4 kinase (New England BioLabs) for 30 min at 37°C followed by heat inactivation for 20 min at 65°C. The parental methylated template plasmids were digested with DpnI at 37°C for 1 h. The products were purified using a QIAquick PCR purification kit (Qiagen), and an aliquot was used in a ligation reaction that was electroporated into DH10B cells. Mutant plasmids were extracted and verified by restriction digest and DNA sequencing. The resulting Tbx5 NES mutant pDONR/Zeo constructs were then recombined into the modified EGFP-C1 and HA-C1 expression vectors. The following primers were used to generate the Tbx5 NES point mutants: Tbx5 L152P, 5′-TCTGGAAGGAAACCGGCTGCCTC-3′; Tbx5 L152V, 5′-TCTGGAAGGAAACCACCTGCCTC-3′); Tbx5 L152A, 5′-TCTGGAAGGAAACCGCCTGCCTC-3′); and the multiple Tbx5 NES mutant F155A/L158A/L160A, 5′-CTGGGCGGAAACCAACTGCCTCATC-3′. Each of these Tbx5 NES mutant primers were paired with 5′-AGCTTAAGCTCACCAACAACCACCT-3′ for the respective mutagenesis reactions. To generate the Tbx5 NES L160A point mutant, the following primer pair was used: 5′-AGCTTAAGGCCACCAACAACCACCT-3′ and 5′-TCTGGAAGGAAACCAACTGCCTCAT-3′. To generate the Tbx5 mutNES mutant, the following primer pair was used: 5′-AAGGCCAAGGCCACCAACAACCACCTTGAC-3′ and 5′-CTGGGCGGAAACCGCCTGCCTCATACCGTG-3′. To generate the Tbx5 NLS mutants, the following primers were used: mutNLS1, 5′-ATGTTTCCCAGCTACAAAGTGGGGGTCACTGGAC-3′ and 5′-ACCCCCTCCAGCCCCTGTTATGATCATCTCCG-3′; mutNLS2, 5′-ACCACCGAGCATCCCTACGGTGGGCCCTACATGG-3′ and 5′-GGAACATTCCTCATCTTTTCCCCCGGTGCAGTG-3′. To generate the Tbx5 mutNLS1+2 mutant form, both of the individual NLS mutant plasmids were digested with BglII and EcoRV (New England BioLabs) in order to replace the NLS2 in Tbx5 mutNLS1 with the respective mutated form of Tbx5 mutNLS2. All three mutant Tbx5 NLS constructs were then used for subsequent recombination into the Gateway-modified EGFP-C1 expression vector.
Cell culture, transfection, and pharmacological inhibitors.
COS-7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For CRM1 inhibition studies, the cells were incubated in leptomycin B (LC Laboratories) overnight at the indicated concentrations followed by processing for confocal microscopy as described below.
Confocal microscopy.
Transfected COS-7 cells were fixed in 4% paraformaldehyde followed by a brief wash with phosphate-buffered saline (PBS). Confocal microscopy was performed using a 510 Meta system (Carl Zeiss MicroImaging, Inc.) equipped with a Plan Apochromat 63× objective/1.4 numerical aperture oil differential interference contrast lens. Images were processed by using Photoshop CS2 (Adobe).
Luciferase reporter assays.
For reporter assays, COS-7 cells were grown in 12-well culture dishes and transfected with 300 ng of atrial natriuretic factor (ANF)-luciferase reporter plasmid (12, 13), 10 ng of Renilla luciferase plasmid, 25 ng of Tbx5 expression plasmids, and 100 ng of LMP4 expression plasmid. The total amount of DNA transfected was held at 1 μg. Transfected cells were cultured for 36 h before lysis. Luciferase activity was measured by using a Dual-Luciferase assay system (Promega), and samples were read with a Lumat LB 9501 instrument (Bethold). All reporter assays were performed in triplicate, and the collected data from independent experiments were normalized to the Renilla luciferase activity. In order to compare values from independent experiments, the activation for wild-type Tbx5 alone was set at 100% and was used to standardize the experimental values from other data sets.
Immunoprecipitation and immunoblotting.
COS-7 cells were grown to 80 to 90% confluence in 60-mm culture dishes and transfected with the described plasmids. A modification of the coimmunoprecipitation protocol from Rastogi et al. (44) was used. Forty-eight hours posttransfection, the medium was aspirated and the cells were scraped using 1 ml of cold PBS, collected, and centrifuged at 3,000 rpm for 2 min. The cells were washed three more times with cold PBS, resuspended in 150 μl of M2 buffer (20 mM Tris [pH 7.6], 0.5% NP-40, 250 mM NaCl, 3 mM EGTA, 3 mM EDTA, and protease inhibitor cocktail [Sigma-Aldrich]) and incubated with rocking at 4°C for 30 min followed by centrifugation at 14,000 rpm for 15 min. The supernatants were transferred to new tubes, and after the addition of 5 μl of anti-CRM1 (H-300; Santa Cruz Biotechnology), the samples were incubated with rocking at 4°C for 1 h. To each tube, 100 μl of immunoprecipitation buffer (20 mM HEPES [pH 7.9], 40 mM KCl, 1 mM MgCl2, 0.1 mM EGTA, 0.1 mM EDTA, 0.1 mM dithiothreitol, 0.1 mM NaF, 0.1 mM Na3VO4, 0.5% NP-40, 3 mg/ml bovine serum albumin) and 15 μl of protein G Sepharose for Fast Flow (Amersham Bioscience) were added with further rocking at 4°C for 1 h. The reaction was transferred to a Zeba Spin column (Pierce) and centrifuged at 5,000 rpm for 2 min followed by five washes with the immunoprecipitation buffer. The protein was eluted from the Sepharose beads by adding 30 μl of sodium dodecyl sulfate (SDS) buffer, boiling for 5 min, and centrifuging at maximum speed for 2 min. The protein samples were resolved by SDS-polyacrylamide gel electrophoresis (PAGE), electrotransferred to nitrocellulose membranes, and immunoblotted with the indicated antibodies (anti-CRM1, H-300; Santa Cruz Biotechnology; anti-HA, HA-7; Sigma-Aldrich). Anti-mouse or anti-rabbit horseradish peroxidase-linked secondary antibodies were used with SuperSignal West Femto reagents (Pierce) to visualize the immunoreactive protein bands on film.
Three-dimensional cellular reconstruction.
COS-7 cells transfected with EGFP-Tbx5 and HcRed-LMP4 were fixed and imaged using confocal microscopy as described above, and optical sections were collected for the entire thickness of the cell. The three-dimensional rendering of the cell and QuickTime movie making was done using Velocity 4.0.1 software (Improvision; Image Processing & Vision Company, Ltd.).
Protein structural modeling.
Tbx5 three-dimensional protein structural modeling was done using NCBI's Molecular Modeling Database by utilizing the human TBX3 crystal structure (Protein Data Bank ID 1H6F) as a template. The resulting structure was developed using Cn3D, NCBI's three-dimensional structure viewing software, and the resulting image was processed with Photoshop CS2 (Adobe).
RESULTS
Role of NLS motifs in Tbx5 subcellular localization.
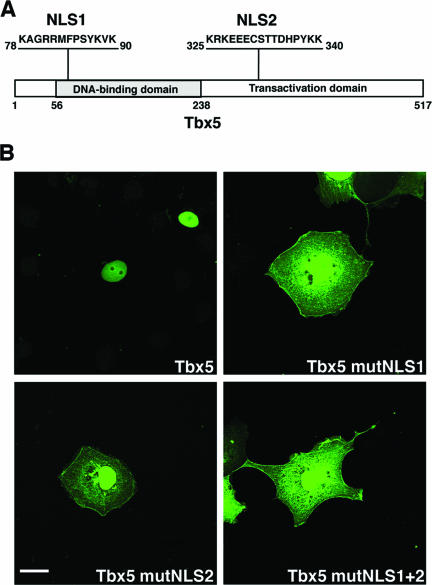
In agreement with its assumed role as transcription factor, Tbx5 had been detected in the nuclei of transfected cells, and this subcellular localization is a function of a bipartite pair of NLSs (15, 44). NLS1 is located in the N-terminal DNA-binding domain, whereas NLS2 is found in the C-terminal transactivation domain (Fig. 1A). The two NLS motifs in Tbx5 were identified through the generation of deletion constructs, with critical amino acids identified by generating point mutants and monitoring the subcellular localization of these mutated forms within cells as EGFP fusions (15, 51). Here, we extended these investigations by generating mutations of all of the identified critical amino acids within each NLS sequence (Tbx5 mutNLS1 or mutNLS2) and both NLS sequences in combination (Tbx5 mutNLS1+2) (see Fig. 3). Wild-type Tbx5 or NLS mutant Tbx5 constructs were expressed in COS-7 cells as EGFP fusion proteins, and subcellular localization was assessed. When expressed in COS-7 cells, wild-type Tbx5 displayed a strictly nuclear localization (Fig. 1B). In contrast, the individual NLS mutant constructs revealed significant cytoplasmic distribution of Tbx5. However, in addition to cytoplasmic Tbx5, some degree of nuclear localization persisted as well (Fig. 1B). In the double NLS mutant, Tbx5 protein localized to the cytoplasm and nucleus, a result comparable to that for the single mutants (Fig. 1B). Although each NLS alone can localize Tbx5 to the nucleus, both NLSs are required for efficient and complete localization of Tbx5 to the nucleus.
FIG. 1.
Tbx5 protein NLS sequences. (A) Schematic representation of the chicken Tbx5 protein (GenBank/EBI Data Bank number NP_989504) with the N-terminal DNA-binding domain and the C-terminal transactivation domain. The two NLS sequences are indicated and consist of amino acids 78 to 90 in the DNA-binding domain (NLS1) and amino acids 325 to 340 in the transactivation domain (NLS2). (B) Subcellular localization of Tbx5 NLS mutants. COS-7 cells were transfected with expression vectors encoding the indicated EGFP-Tbx5 fusion proteins. After transient expression, cells were fixed and imaged using a Zeiss 510 Meta scanning confocal microscope with a Plan Apochromat 63× objective/1.4 numerical aperture oil differential interference contrast lens. Tbx5 is detected in the nuclei of transfected cells, whereas all three of the Tbx5 NLS mutant forms reveal robust distribution of the protein in the cytoplasm as well as in the nuclei. White scale bar = 20 μm.
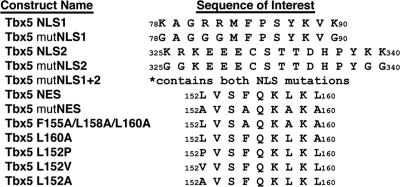
FIG. 3.
Wild-type and mutagenized Tbx5 expression constructs. Listed by name are the wild-type or mutagenized (mut) Tbx5 expression constructs used in these experiments. The positional amino acid sequence of interest for each is denoted by numbering the first and last amino acid within a given sequence.
Identification of an NES motif in the Tbx5 transcription factor.
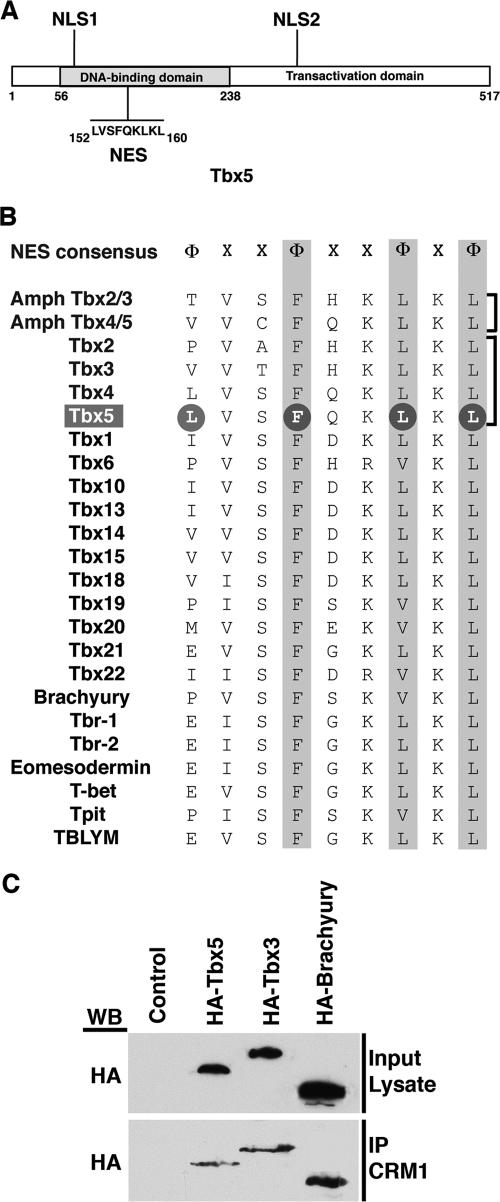
In contrast, our previous work demonstrated that Tbx5 and Tbx4 could assume a cytoplasmic actin-associated distribution in a complex with LMP4 (9, 13). The mechanism and the proteins involved in this nuclear/cytoplasmic shuttling, however, remained unknown. Many proteins that translocate out of the nucleus contain a leucine-rich NES with the consensus motif Φ-(X)2-3-Φ-(X)2-3-Φ-X-Φ, where Φ represents hydrophobic residues (L, I, F, V, M, P) and X represents any amino acid (22, 34). This conserved NES has been reported to be the target for binding by the nuclear exporting protein CRM1 (19, 42). We analyzed the Tbx5 protein for such a sequence motif and identified a putative NES (152LVSFQKLKL160) in the N-terminal DNA-binding domain between NLS1 and NLS2 (Fig. 2A). Having discovered the presence of an NES in Tbx5, we wondered whether this sequence motif was conserved among all members of the T-box protein family. An identical amino acid sequence was indeed present in Tbx4 (Fig. 2B), and in this context we note that Tbx4, similar to Tbx5, can also interact with LMP4 and colocalize in the cytoplasm at the actin cytoskeleton (30). Evolutionarily, the next closest pair of T-box proteins are Tbx2 and Tbx3, which together with the previous proteins define the Tbx2/3/4/5 subfamily. Tbx2 and Tbx3 share three of the four critical amino acids within the conserved NES motif. They diverge, however, in the initial position, and instead of a leucine, they have a proline and a valine, respectively (Fig. 2B). Going beyond the Tbx2/3/4/5 subfamily and including all T-box family members currently described for the mouse, we noticed the same theme: while the C-terminal three critical residues are well conserved, the N-terminal initial position is occupied by a range of amino acids, including L, P, V, I, M, E, and F. Thus, it appears that T-box proteins in general, in addition to NLS motifs, also contain a conserved NES motif.
FIG. 2.
T-box protein NES sequences. (A) Schematic representation of the chicken Tbx5 protein with the N-terminal DNA-binding domain and the C-terminal transactivation domain. The two NLS sequences are indicated, and the NES sequence indicated in the DNA-binding domain consists of amino acids 152 to 160. (B) Alignment of putative NES sequences in Tbx family members. The NES consensus sequence was derived from proteins that utilize the CRM1 nuclear export pathway, where Φ represents hydrophobic residues (L, I, F, V, M, P) and X represents any amino acid. The two primordial amphioxus family members Tbx2/3 and Tbx4/5 are followed by the derived cognate subfamily members Tbx2, -3, -4, and -5 in vertebrates. Other mouse Tbx family members are subsequently listed. Conserved residues are highlighted. Of note, the chicken Tbx5 used in this study has an NES amino acid sequence identical to that of the mouse. The GenBank/EBI Data Bank numbers for the sequences used are: Amph Tbx2/3, AF262563; Amph Tbx4/5, AF262564; Tbx1, P70323; Tbx2, NP_033350; Tbx3, NP_035665; Tbx4, NP_035666; Tbx5, NP_035667; Tbx6, P70327; Tbx10, Q810F8; Tbx13, AAC40116; Tbx14, AAC40115; Tbx15, O70306; Tbx18, Q9EPZ6; Tbx19, NP_114394; Tbx20, NP_919239; Tbx21, Q9JKD8; Tbx22, Q8K402; Brachyury, NP_033335; Tbr-1, NP_033348; Tbr-2, BAA83416; Eomesodermin, EDL09251; T-bet, AAF61242; Tpit, AAK27153; TBLYM, AAF00056. (C) COS-7 cells transfected with HA-Tbx5, HA-Tbx3, or HA-Brachyury expression constructs and protein lysates were generated for coimmunoprecipitation (IP) analysis. To verify expression of the proteins, an aliquot of lysate was used for Western blotting (WB) with antibodies against the HA tag. The remaining lysates were utilized for the coimmunoprecipitation reactions with antibodies against CRM1. All three T-box proteins do coimmunoprecipitate with CRM1.
As this core NES motif is present in all T-box protein family members, we wondered if they are capable of interacting with the CRM1 export protein. In addition to Tbx5, we selected a close family member, Tbx3, and a more distantly related T-box protein, Brachyury, for subsequent studies. To demonstrate physical interaction with CRM1, HA-tagged full-length T-box protein expression plasmids were transfected into COS-7 cells to determine if they would coimmunoprecipitate with CRM1 from cell lysates. Recombinant protein expression was verified by SDS-PAGE and Western blotting of the input lysate with an HA-tagged antibody (Fig. 2C). The remaining lysate was subjected to immunoprecipitation with the CRM1 antibody followed by Western blotting with the HA-tagged antibody. Tbx5, Tbx3, and Brachyury were all coimmunoprecipitated with CRM1 (Fig. 2C), indicating that T-box proteins in general can interact with the CRM1 exporter and suggesting that they are capable of nuclear/cytoplasmic shuttling.
Protein-protein interaction between Tbx5 and CRM1.
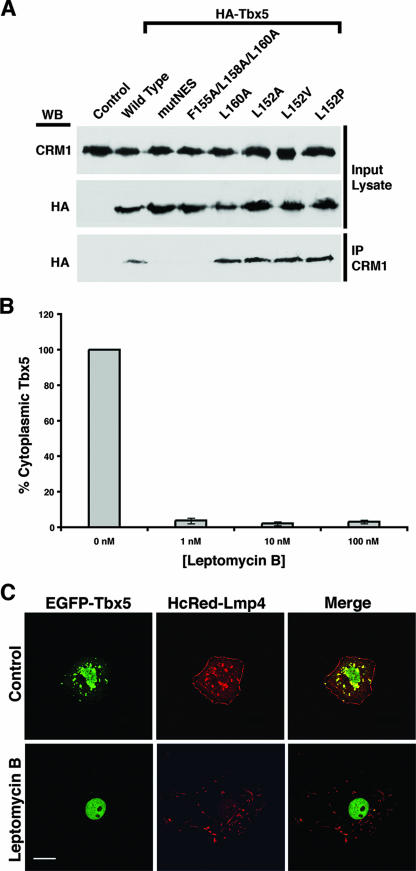
Having demonstrated that several members of the T-box family can bind to CRM1, we used Tbx5 as a model to investigate this interaction in more detail. To determine the core amino acids required for a physical interaction between Tbx5 and CRM1, COS-7 cells were transfected with expression plasmids containing HA-tagged wild-type and mutated forms to determine if they would coimmunoprecipitate with CRM1 from cell lysates. Equal protein levels before the immunoprecipitations were verified by SDS-PAGE and Western blots of the input cell lysate with antibodies against either CRM1 or the HA tag (see Fig. 4A). The remainder of the lysate was then subjected to immunoprecipitation with the CRM1 antibody. In order to detect coimmunoprecipitated Tbx5, a Western blot was performed using HA-tagged antibodies. The wild-type form of Tbx5 revealed a coimmunoprecipitate with CRM1, whereas the Tbx5 mutNES protein with all four critical NES residues exchanged with alanines did not bind to CRM1 (see Fig. 4A). Replacing the C-terminal NES residues with alanines (Fig. 3) similarly prevented the mutagenized protein from interacting with CRM1 (Fig. 4A). Surprisingly, single-amino-acid NES mutants in either the terminal or the initial position (Fig. 3) retained their ability to complex with CRM1 (Fig. 4A). In addition, an exchange of the initial leucine in the NES with either alanine or residues found in Tbx2, Tbx3, or Brachyury has no negative effect on binding the exporter protein. Thus, our work with wild-type and mutant forms of Tbx5 identifies a combination of three hydrophobic amino acids within the four-position NES to be essential for interaction with CRM1. Furthermore, the data suggest that apparently all T-box proteins contain a CRM1-interacting NES motif.
FIG. 4.
Interaction of Tbx5 and CRM1. (A) COS-7 cells were transfected with HA-Tbx5 wild-type or NES mutant expression constructs, and protein lysates were generated for coimmunoprecipitation (IP) analysis. To verify expression of the proteins, an aliquot of lysate was used for Western blotting (WB) with antibodies against CRM1 or the HA tag. The remaining lysates were utilized for the coimmunoprecipitation reactions with antibodies against CRM1. Wild-type Tbx5 and the Tbx5 NES single-amino-acid substitution mutants do coimmunoprecipitate with CRM1, whereas the Tbx5 mutNES and F155A/L158A/L160A proteins do not. (B) COS-7 cells were cotransfected with HcRed-LMP4 and wild-type EGFP-Tbx5 expression vectors and incubated overnight with or without the CRM1 inhibitor leptomycin B at the indicated concentrations. After transient expression, cells were fixed and imaged using a Zeiss 510 Meta scanning confocal microscope with a Plan Apochromat 63× objective/1.4 numerical aperture oil differential interference contrast lens. Cells coexpressing HcRed-LMP4 and EGFP-Tbx5 were counted, and the percentage with Tbx5 in the cytoplasm was determined. The graph represents two sets of independent experiments in which 85 to 95 cells were counted for each concentration of leptomycin B used. A >95% decrease in the percentage of cells with Tbx5 in the cytoplasm was observed after inhibitor treatment. Error bars represent the standard errors of the means for the given experiments. (C) COS-7 cells were cotransfected with HcRed-LMP4 and wild-type EGFP-Tbx5 expression vectors. Representative images of a cell receiving no leptomycin B treatment (Control) compared to a cell treated with leptomycin B (1 nM) are shown. The left column shows the localization of the EGFP-Tbx5 protein, the middle column shows the localization of the HcRed-LMP4 protein, and the right column shows both proteins together in a merged image. White scale bar = 20 μm.
The identification of a putative NES motif within Tbx5 was based on a consensus amino acid sequence found in proteins that utilize the CRM1 pathway for nuclear export (22, 34). The mutagenesis experiments demonstrated that these residues are important for Tbx5-CRM1 binding, but they did not conclusively indicate that Tbx5 is using the CRM1 export pathway. In order to directly demonstrate that Tbx5 utilizes the CRM1 pathway, we employed a specific inhibitor of CRM1-mediated nuclear export, leptomycin B (50). This inhibitor specifically interferes with the binding of the NES with CRM1 (31, 32). COS-7 cells were cotransfected with EGFP-Tbx5 and HcRed-LMP4 expression constructs and then treated with increasing concentrations (1 to 100 nM) of leptomycin B overnight. In order to obtain statistically significant values, cells (n = 85 to 95 cells per concentration used) from two independent experiments were counted and scored for the number of cells expressing cytoplasmic Tbx5 in the presence of LMP4. Even at the lowest concentration (1 nM) used, we observed an approximately 95% decrease in the number of cells with cytoplasmic Tbx5 (Fig. 4B). A representative set of imaged cells at 0 nM (control) and 1 nM leptomycin B are shown in Fig. 4C, demonstrating that in the presence of leptomycin B, Tbx5 protein cannot leave the nucleus (Fig. 4C). The ability of the pharmacological inhibitor leptomycin B to prevent nuclear export of Tbx5 in context with the NES mutagenesis data indicates that T-box transcription factors are utilizing the CRM1 pathway for nuclear export.
We wondered whether LMP4 would be part of the transport complex, temporarily entering the nucleus. Previous biochemical cell fractionation and coimmunoprecipitation experiments did not reveal any nuclear localization of LMP4 (13). However, here we confirmed an exclusive cytoplasmic distribution for LMP4 at the single-cell level by using a three-dimensional reconstruction of cells transfected with EGFP-Tbx5 and HcRed-LMP4 (see Movie S1 in the supplemental material).
Role of NES motif in Tbx5 subcellular localization.
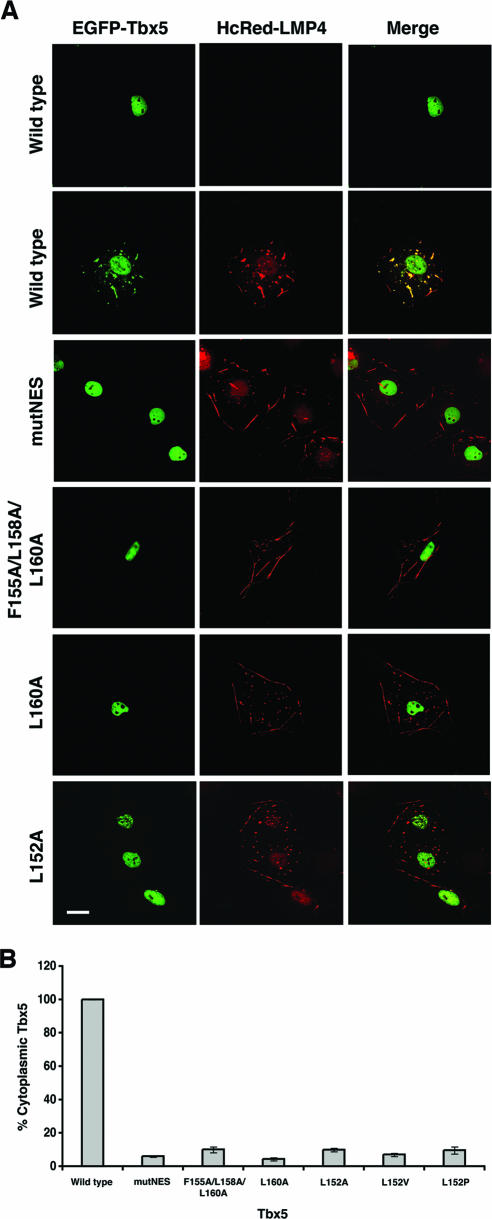
Consistent with our previous work (9, 13, 30), wild-type Tbx5 revealed strictly nuclear localization when expressed alone, but in the presence of LMP4, it was also distributed in the cytoplasm as a complex with LMP4 along the actin cytoskeleton (Fig. 5A). In order to investigate if the identified putative NES in Tbx5 was functional for export, the four critical amino acid residues in the conserved motif were replaced by alanines (Fig. 3). As with the NLS mutants, all of the NES mutant constructs were designed as EGFP fusions, but this time were coexpressed in COS-7 cells with an HcRed-LMP4 expression construct. Substitution of all the critical amino acids in the NES (mutNES) with alanines completely blocked the localization of Tbx5 to cytoplasmic sites (Fig. 5A). In support of the CRM1 coimmunoprecipitation results (Fig. 4A), the F155A/L158A/L160A mutant that did not bind CRM1 also did not localize outside of the nucleus (Fig. 5A). Interestingly, the apparent block of cytoplasmic localization of Tbx5 in the presence of LMP4 was recapitulated in each of the single point mutants (Fig. 5A) (see Fig. S1 in the supplemental material), even though they are capable of interaction with CRM1 (Fig. 4A). In order to verify that these observations were statistically significant, cells (n = 85 to 95 per construct) from two independent experiments were counted and scored for the number of cells expressing cytoplasmic Tbx5 in the presence of LMP4. The percentage of cotransfected cells with cytoplasmic Tbx5 dropped from 100% for wild-type Tbx5 to less than 10% for each of the NES mutants (Fig. 5B). In summary, these data suggest that Tbx5 contains two functional NLS motifs and, in addition, a novel NES motif that is responsible for the shuttling of the protein out of the nucleus.
FIG. 5.
Subcellular localization of Tbx5 NES mutants in the presence of LMP4. (A) COS-7 cells were cotransfected without or with HcRed-LMP4 and the indicated EGFP-Tbx5 expression vectors. After transient expression, cells were fixed and imaged using a Zeiss 510 Meta scanning confocal microscope with a Plan Apochromat 63× objective/1.4 numerical aperture oil differential interference contrast lens. The left column shows the localization of the EGFP-Tbx5 proteins, the middle column shows the localization of the HcRed-LMP4 protein, and the right column shows both proteins together in a merged image. When expressed alone, wild-type Tbx5 is detected only in the nuclei, while when coexpressed with LMP4, wild-type Tbx5 is detected in both the nuclei and colocalized with LMP4 in the cytoplasm. All of the Tbx5 NES mutants remain localized in the nucleus even in the presence of LMP4. White scale bar = 20 μm. (B) To quantify the localization observed in panel A, COS-7 cells expressing both HcRed-LMP4 and EGFP-Tbx5 were counted, and the percentage with Tbx5 in the cytoplasm was determined. The graph represents two sets of independent experiments in which 85 to 95 cells were counted for each Tbx5 construct used. The error bars represent the standard errors of the means for the given experiments.
Consequences of subcellular localization on Tbx5 transcriptional activity.
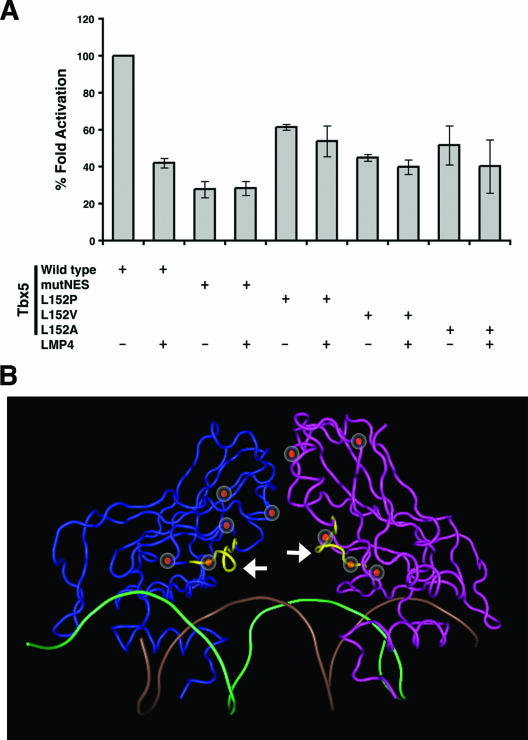
The function of Tbx5 as a transcription factor has been established in the developing limb and heart (12, 23). In the mouse, the heart-specific ANF and the limb-specific Fgf10 genes have been shown to be immediate downstream targets of Tbx5 (2, 12). Previous work from our group using ANF and Fgf10 promoter-luciferase reporter constructs demonstrated that Tbx5 transcriptional activity is modulated by LMP4. Altering the subcellular localization of Tbx5 out of the nucleus by LMP4 repressed the transcription factor's activity in a dose-dependent manner (13). We employed this sensitive luciferase reporter assay in combination with the Tbx5 NES mutants to independently determine if Tbx5 transcriptional activity would change in the presence of LMP4. COS-7 cells were transfected with the ANF-luciferase reporter and LMP4 and/or Tbx5 expression plasmids. In order to compare data sets from various experiments, the activation of luciferase activity for wild-type Tbx5 alone was set at 100% and was used to standardize the other experimental values. Consistent with previous findings (13), coexpression of Tbx5 and LMP4 resulted in a decrease in Tbx5 transcriptional activity by approximately 60% (Fig. 6A). Of note, all of the Tbx5 NES mutants tested exhibited reduced transcriptional activity in this reporter assay. However, there was no statistically significant change in activation between the mutant Tbx5-transfected and mutant Tbx5-LMP4-cotransfected samples (Fig. 6A). These findings are in agreement with the protein localization data in Fig. 5 and confirm that the Tbx5 NES mutants do not leave the nucleus.
FIG. 6.
Tbx5 subcellular localization and transcriptional activity. (A) Transcriptional activity of Tbx5 NES mutants. COS-7 cells were transfected with the ANF-luciferase reporter and 25 ng of EGFP-Tbx5 wild-type or NES mutant expression plasmids with or without 100 ng of the LMP4-Myc expression plasmid. Changes in luciferase reporter expression are indicated as activation normalized against Renilla signal. The activation for wild-type Tbx5 alone was set at 100% and was used to standardize the other experimental values. A 60% decrease in activation is observed for wild-type Tbx5 in the presence of LMP4, while the values for the Tbx5 NES mutants do not significantly change when LMP4 is present. Representative data from independent experiments performed in triplicate are shown; the error bars represent the standard errors of the means. (B) Three-dimensional representation of the Tbx5 DNA-binding domain based on human TBX3 crystal structure coordinates. The Tbx5 is modeled as a dimer (blue and magenta) positioned on DNA (brown and green helix). The identified NES motif is depicted in yellow (white arrows). Regions of Tbx5 important for protein-DNA interaction, protein-protein interaction, and HOS manifestation are indicated with the red circles.
The data from the transcriptional reporter assays revealed decreases in the overall activation of transcription to various degrees, depending on the individual NES mutant form used. All of the constructs were found to yield similar levels of translated protein in COS-7 cells (Fig. 4A and data not shown). In order to gain a better understanding of the cause of this activity drop, we determined the position of the mutagenized residues within the three-dimensional structure of the protein. Structural data on Tbx5 have not been published. However, given the high degree of conservation within the DNA-binding domain of T-box proteins, we used the human TBX3 crystal structure information (14) to model the conformation of the Tbx5 DNA-binding domain, which contains the NES motif. Three-dimensional modeling of the amino acid residues in the NES suggests that this site is positioned on the surface of the protein, making it available for interaction with other proteins, including those involved in nuclear export (Fig. 6B). Furthermore, the NES motif lies in close proximity to amino acid residues that are thought to be involved in DNA binding (K159, N162, and N163) (14, 39), homodimerization of the protein (P139 through T144) (14, 39), and HOS disease manifestation (G80, P139, and G169) (26). Therefore, the observed overall decrease in transcriptional activity of the Tbx5 NES mutants is most likely due to less-efficient DNA binding or protein dimerization.
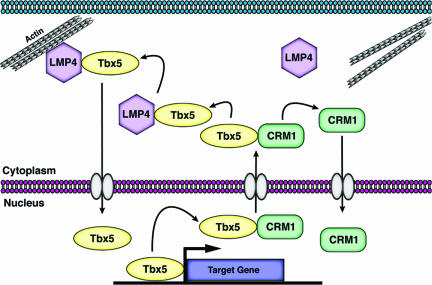
The data presented here provide evidence supporting the notion that Tbx5 cytoplasmic relocalization in the presence of LMP4 utilizes the CRM1 nuclear export pathway and that this process is dependent on the identified NES sequence present in the DNA-binding domain of the transcription factor. In addition to Tbx5, the data show that all T-box proteins contain an NES motif, are capable of interaction with CRM1, and potentially could shuttle between nuclear and cytoplasmic sites. In summary, new information presented in this study allowed an integration of previous and current findings into a new working model for Tbx5 nuclear-cytoplasmic shuttling as described in Fig. 7.
FIG. 7.
Model for Tbx5 relocalization between nuclear and cytoplasmic compartments. Tbx5 can interact with CRM1 to be exported across the nuclear membrane into the cytoplasm, where the proteins dissociate. CRM1 will return to the nucleus while Tbx5 is now free to interact with LMP4 and localize to the actin cytoskeleton. Tbx5 is also then able to return to the nucleus in order to regulate target gene activity, completing the dynamic shuttling between the two cellular compartments.
DISCUSSION
The interaction and complex formation of the Tbx5 transcription factor with the PDZ-LIM domain protein LMP4 lead to a change in Tbx5's subcellular localization from exclusively nuclear to a cytoplasmic distribution along the actin cytoskeleton (30). The exit of Tbx5 from the nucleus interferes with expression of downstream genes, demonstrating that LMP4 can modulate Tbx5 transcriptional activity (13). During heart and limb development in the chicken, Tbx5 is dynamically expressed in nuclear and/or cytoplasmic compartments (9). These findings indicate complex regulation of Tbx5 shuttling into and out of the nucleus and subsequent retention of the protein at specific subcellular locations.
In this study, we sought to gain a better understanding of the mechanisms facilitating Tbx5 transport across the nuclear envelope. Although two NLS motifs in Tbx5 had been recently identified, nuclear import was investigated with single-amino-acid point mutants and protein truncation constructs in one or the other NLS (15, 51). We confirmed and extended these studies demonstrating that NLS1 and NLS2 have a synergistic relationship and that both NLS motifs are essential for efficient nuclear localization. Surprisingly, but in agreement with the other studies, we could not achieve an exclusive cytoplasmic localization, even when both NLS1 and NLS2 were mutagenized. This result may suggest that additional, yet-unknown sites or mechanisms are involved in nuclear localization.
In order to explain Tbx5 nuclear export, we wondered if the CRM1-dependent nuclear export pathway utilized by other shuttling proteins is also employed by T-box proteins (38, 42). CRM1 is known to recognize and bind a conserved NES motif of four mostly large hydrophobic amino acids, thereby targeting the respective protein for nuclear export (38, 42). Our analysis of T-box amino acid sequences revealed that all known T-box protein family members contain the hydrophobic amino acid NES core motif. The CRM1 binding studies with Tbx5 and two additional protein family members experimentally confirmed that T-box proteins use the CRM1 pathway for nuclear export. The two closest family members, Tbx5 and Tbx4, share an identical NES motif within the DNA-binding domain. Interestingly, the evolutionarily more-divergent family members Tbx2 and Tbx3 (3) share the core NES but contain a proline and a valine, respectively, in place of the initial leucine. In this context, it was of interest to compare the data with those for the cephalochordate amphioxus, which has been used to track the gene structure changes of T-box genes in evolution (45). While in this species Tbx2/3 and Tbx4/5 gene clusters have formed from a primordial Tbx2/3/4/5 gene, this two-gene cluster had not been further extended to individual Tbx2, -3, -4, and -5 genes as seen for vertebrates. We identified an NES motif in amphioxus Tbx2/3 and Tbx4/5 with a threonine and a valine in the initial position, respectively, suggesting that all T-box proteins are shuttling proteins and that the particular NES sequence in vertebrate Tbx5 and Tbx4 was acquired later in evolution when new functional roles were adopted.
The other T-box protein family members use a diverse but limited range of hydrophobic residues in the initial NES position. Tbx1, -10, and -13 have an isoleucine, while Tbx2, -6, and Brachyury (T) have a proline in this position. Of note, the T-box protein Tbr-1 with a glutamic acid in the initial position in the cytoplasm of adult rodent brain cells has been described previously (24). In addition, recent work on Brachyury during mouse gastrulation described the protein in various embryonic and extraembryonic tissues as localizing predominantly to the cytoplasm (25), and based on experiments presented in this report, as also being capable of interacting with the CRM1 exporter. Thus, it is probable that T-box proteins with various NES motifs can utilize the CRM1 pathway for nuclear export; however, the mechanism of cytoplasmic retention for these proteins is still unknown.
We find in the T-box protein family a higher level of conserved amino acids with predominantly leucines or phenylalanines in the more C-terminal NES positions compared to the initial position that reveals a wider range of amino acid options. Using Tbx5 as a model, we investigated the functional role of the various NES core amino acids with regard to the extent to which they contribute to CRM1 binding. This was of interest since almost all previous studies mutagenized the four critical positions as a whole but did not evaluate individual point mutations. A set of systematic point mutations of the Tbx5 NES revealed the importance of particularly the three C-terminal positions for binding of CRM1. While we found the first position dispensable for interaction, the C-terminal positions of the NES appeared sufficient for binding to the export protein. This is in line with the observation that for NES activity in other shuttling proteins, the residues in the C-terminal position are more susceptible to mutation than those in the N-terminal position (33, 49). Replacing the initial leucine of Tbx5 with alanines or residues found in Tbx2, Tbx3, or Brachyury did not change the ability of the mutagenized protein to interact with CRM1. This result is consistent with our finding that full-length Tbx3 and Brachyury recombinant proteins can also bind to the CRM1 export protein. Interestingly, when we kept the N-terminal NES motif in Tbx5 as the wild type and mutagenized only the last position of the NES core, the ability to interact with CRM1 was retained. Thus, while four amino acids had been described to be critical for NES function, our experiments demonstrate that three of the hydrophobic core residues are sufficient for effective CRM1 binding. A robust protein interaction may be constrained by the three-dimensional surface features and therefore relies on a combinatorial and synergistic effect of the individual NES core residues with the target site on the exporter protein. Moreover, this interaction undoubtedly influences the formation of the export protein complex with the other proteins necessary for complete and efficient shuttling out of the nucleus. Besides sequence pattern, accessibility and flexibility appear to be additional important properties of NESs (35). In this context, it may be no coincidence that in the T-box transcription factors, the NES is integrated in the DNA-binding domain, a designated site for DNA and protein interactions.
Our studies demonstrated that the identified NES is responsible for CRM1-mediated nuclear export of Tbx5. This export activity is abolished by treatment with leptomycin B or site-directed mutagenesis of critical amino acids within the NES motif. Moreover, LMP4-mediated regulation of Tbx5 transcriptional activity is no longer functional when nuclear export is blocked. Replacement of just the initial leucine of the NES motif sequence does not interfere with CRM1 binding; however, it is sufficient to prevent cytoplasmic retention of Tbx5 as a complex with LMP4 and actin. Thus, the initial residue of the NES may be involved in the specific recognition of the cytoplasmic interacting protein LMP4. In this context, we recall that Tbx4 shares the identical NES sequence with Tbx5, interacts also with LMP4, and is retained in the cytoplasm. Brachyury, on the other hand, has a quite different NES, including a proline in the initial position but, as we have demonstrated, it does bind the CRM1 export protein and, as reported by others, it also localizes to cytoplasmic sites (25). In contrast to Tbx5 and Tbx4, for Brachyury and other T-box proteins the respective cytoplasmic interacting proteins are not yet known. It would not be surprising if there is a defined group of cytoplasmic proteins targeted for binding by individual T-box family members with unique NES sequences. Their identification will facilitate a deeper and more general understanding of T-box protein nuclear export and cytoplasmic retention and function.
In addition, specific residues in T-box proteins may be involved in posttranslational modifications and therefore contribute to the regulation of the subcellular localization of the protein. For example, phosphorylation has been shown to play a role in the nuclear export of several proteins that contain a CRM1-dependent NES sequence motif (27, 28, 40). It is therefore conceivable that regulated phosphorylation of T-box proteins may be involved in fine-tuning the protein's localization and its overall activity. Nuclear import and export sequences are important regulators of the subcellular localization of proteins. Thus, they provide new opportunities to manipulate the subcellular distribution of T-box proteins in vivo in order to investigate the resulting functional consequences on development, work that is ongoing in our laboratory.
The human disorder HOS is caused by mutations in TBX5, leading to skeletal and cardiac malformations. The majority of mutations critical for disease manifestation are considered to result in early protein terminations and haploinsufficiency (8). However, chromosome 12q2 duplications, which lead to increased TBX5 dosage, have also been reported to cause HOS (21, 48). These results suggest that a balanced level of Tbx5 protein in the cell is critical for its function, as under- and overexpression result in similar phenotypes. Tbx5 contains functional NLS and NES motifs, and the protein dynamically shuttles between nuclear and cytoplasmic sites (13). Our modeling studies revealed that amino acids linked with HOS manifestation are in close proximity to the NES, facing the inner surface of the protein dimer. It is therefore possible that conformational changes induced by these point mutations may also affect the NES interaction with proteins important for nuclear export. Thus, disease manifestation in HOS is potentially caused by compromised regulation of the subcellular location of TBX5, in addition to altered transcriptional activity, and therefore affects protein function in both nuclear and cytoplasmic cell compartments. We note a highly related NES motif in Tbx1, making this family member also a candidate for changing its localization between the nucleus and cytoplasm. In humans, TBX1 is linked to DiGeorge syndrome, a disease with phenotypes ranging from cardiac malformations, facial dismorphogenesis, and cleft palate to hypoplasia of the thymus and parathyroid glands (4, 5, 46), all attributed to disruption of TBX1 transcriptional activity. Based on sequence homology in the putative NES, it is possible that TBX1 may have additional functions through other, yet-unidentified binding partners, facilitating shuttling and distribution of the protein in the cytoplasm. As suggested for TBX5, alternative explanations for the observed disease phenotypes may rely on functions different from transcriptional regulation. The NES appears to be an evolutionarily highly conserved motif in T-box proteins. Consequently, we expect to see protein family members with distribution patterns and functions inside and outside the nucleus, calling for a paradigm shift of how we interpret T-box protein function. Our efforts to understand how this dynamic protein shuttling is regulated at the cell and tissue levels may also provide new insights into the signaling pathways that control the T-box protein subcellular localization and function.
Supplementary Material
Acknowledgments
We thank S. Mackem and M. Logan for sharing full-length Brachyury and Tbx3 cDNA clones, respectively; J. Topczewski for assistance with the mutagenesis protocols; T.-L. Chew for consultation regarding microscopy and three-dimensional cell reconstruction; S. Ahlgren and R. Dettman; Troy Camarata and Jennifer Krcmery for critical readings of various manuscript versions; and members of the Developmental Biology Core for stimulating discussions.
This work was supported by a grant from the National Institutes of Health (HL085834-01 to H.-G.S.).
Footnotes
Published ahead of print on 26 December 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adereth, Y., K. J. Champion, T. Hsu, and V. Dammai. 2005. Site-directed mutagenesis using Pfu DNA polymerase and T4 DNA ligase. BioTechniques 38864. 866-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal, P., J. N. Wylie, J. Galceran, O. Arkhitko, C. Li, C. Deng, R. Grosschedl, and B. G. Bruneau. 2003. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development 130623-633. [DOI] [PubMed] [Google Scholar]
- 3.Agulnik, S. I., N. Garvey, S. Hancock, I. Ruvinsky, D. L. Chapman, I. Agulnik, R. Bollag, V. Papaioannou, and L. M. Silver. 1996. Evolution of mouse T-box genes by tandem duplication and cluster dispersion. Genetics 144249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold, J. S., E. M. Braunstein, T. Ohyama, A. K. Groves, J. C. Adams, M. C. Brown, and B. E. Morrow. 2006. Tissue-specific roles of Tbx1 in the development of the outer, middle and inner ear, defective in 22q11DS patients. Hum. Mol. Genet. 151629-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold, J. S., U. Werling, E. M. Braunstein, J. Liao, S. Nowotschin, W. Edelmann, J. M. Hebert, and B. E. Morrow. 2006. Inactivation of Tbx1 in the pharyngeal endoderm results in 22q11DS malformations. Development 133977-987. [DOI] [PubMed] [Google Scholar]
- 6.Bach, I. 2000. The LIM domain: regulation by association. Mech. Dev. 915-17. [DOI] [PubMed] [Google Scholar]
- 7.Basson, C. T., D. R. Bachinsky, R. C. Lin, T. Levi, J. A. Elkins, J. Soults, D. Grayzel, E. Kroumpouzou, T. A. Traill, J. Leblanc-Straceski, B. Renault, R. Kucherlapati, J. G. Seidman, and C. E. Seidman. 1997. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat. Genet. 1530-35. [DOI] [PubMed] [Google Scholar]
- 8.Basson, C. T., T. Huang, R. C. Lin, D. R. Bachinsky, S. Weremowicz, A. Vaglio, R. Bruzzone, R. Quadrelli, M. Lerone, G. Romeo, M. Silengo, A. Pereira, J. Krieger, S. F. Mesquita, M. Kamisago, C. C. Morton, M. E. Pierpont, C. W. Muller, J. G. Seidman, and C. E. Seidman. 1999. Different TBX5 interactions in heart and limb defined by Holt-Oram syndrome mutations. Proc. Natl. Acad. Sci. USA 962919-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bimber, B., R. W. Dettman, and H. G. Simon. 2007. Differential regulation of Tbx5 protein expression and sub-cellular localization during heart development. Dev. Biol. 302230-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruneau, B. G. 2002. Transcriptional regulation of vertebrate cardiac morphogenesis. Circ. Res. 90509-519. [DOI] [PubMed] [Google Scholar]
- 11.Bruneau, B. G., M. Logan, N. Davis, T. Levi, C. J. Tabin, J. G. Seidman, and C. E. Seidman. 1999. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev. Biol. 211100-108. [DOI] [PubMed] [Google Scholar]
- 12.Bruneau, B. G., G. Nemer, J. P. Schmitt, F. Charron, L. Robitaille, S. Caron, D. A. Conner, M. Gessler, M. Nemer, C. E. Seidman, and J. G. Seidman. 2001. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106709-721. [DOI] [PubMed] [Google Scholar]
- 13.Camarata, T., B. Bimber, A. Kulisz, T. L. Chew, J. Yeung, and H. G. Simon. 2006. LMP4 regulates Tbx5 protein subcellular localization and activity. J. Cell Biol. 174339-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coll, M., J. G. Seidman, and C. W. Muller. 2002. Structure of the DNA-bound T-box domain of human TBX3, a transcription factor responsible for ulnar-mammary syndrome. Structure 10343-356. [DOI] [PubMed] [Google Scholar]
- 15.Collavoli, A., C. J. Hatcher, J. He, D. Okin, R. Deo, and C. T. Basson. 2003. TBX5 nuclear localization is mediated by dual cooperative intramolecular signals. J. Mol. Cell. Cardiol. 351191-1195. [DOI] [PubMed] [Google Scholar]
- 16.Dawid, I. B., J. J. Breen, and R. Toyama. 1998. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 14156-162. [DOI] [PubMed] [Google Scholar]
- 17.Fanning, A. S., and J. M. Anderson. 1999. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J. Clin. Investig. 103767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher, C. L., and G. K. Pei. 1997. Modification of a PCR-based site-directed mutagenesis method. BioTechniques 23570. 1:574. [DOI] [PubMed] [Google Scholar]
- 19.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 901051-1060. [DOI] [PubMed] [Google Scholar]
- 20.Garrity, D. M., S. Childs, and M. C. Fishman. 2002. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development 1294635-4645. [DOI] [PubMed] [Google Scholar]
- 21.Hatcher, C. J., M. S. Kim, C. S. Mah, M. M. Goldstein, B. Wong, T. Mikawa, and C. T. Basson. 2001. TBX5 transcription factor regulates cell proliferation during cardiogenesis. Dev. Biol. 230177-188. [DOI] [PubMed] [Google Scholar]
- 22.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256213-224. [DOI] [PubMed] [Google Scholar]
- 23.Hiroi, Y., S. Kudoh, K. Monzen, Y. Ikeda, Y. Yazaki, R. Nagai, and I. Komuro. 2001. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat. Genet. 28276-280. [DOI] [PubMed] [Google Scholar]
- 24.Hong, C. J., and Y. P. Hsueh. 2007. Cytoplasmic distribution of T-box transcription factor Tbr-1 in adult rodent brain. J. Chem. Neuroanat. 33124-130. [DOI] [PubMed] [Google Scholar]
- 25.Inman, K. E., and K. M. Downs. 2006. Localization of Brachyury (T) in embryonic and extraembryonic tissues during mouse gastrulation. Gene Expr. Patterns 6783-793. [DOI] [PubMed] [Google Scholar]
- 26.Isphording, D., A. M. Leylek, J. Yeung, A. Mischel, and H. G. Simon. 2004. T-box genes and congenital heart/limb malformations. Clin. Genet. 66253-264. [DOI] [PubMed] [Google Scholar]
- 27.Jain, A. K., and A. K. Jaiswal. 2006. Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. J. Biol. Chem. 28112132-12142. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, H., M. V. Olson, D. R. Medrano, O. H. Lee, J., Xu, Y. Piao, M. M. Alonso, C. Gomez-Manzano, M. C. Hung, W. K. Yung, and J. Fueyo. 2006. A novel CRM1-dependent nuclear export signal in adenoviral E1A protein regulated by phosphorylation. FASEB J. 202603-2605. [DOI] [PubMed] [Google Scholar]
- 29.Kadrmas, J. L., and M. C. Beckerle. 2004. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5920-931. [DOI] [PubMed] [Google Scholar]
- 30.Krause, A., W. Zacharias, T. Camarata, B. Linkhart, E. Law, A. Lischke, E. Miljan, and H. G. Simon. 2004. Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Dev. Biol. 273106-120. [DOI] [PubMed] [Google Scholar]
- 31.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 969112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242540-547. [DOI] [PubMed] [Google Scholar]
- 33.Kudo, N., H. Taoka, T. Toda, M. Yoshida, and S. Horinouchi. 1999. A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J. Biol. Chem. 27415151-15158. [DOI] [PubMed] [Google Scholar]
- 34.Kutay, U., and S. Guttinger. 2005. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 15121-124. [DOI] [PubMed] [Google Scholar]
- 35.la Cour, T., L. Kiemer, A. Molgaard, R. Gupta, K. Skriver, and S. Brunak. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 17527-536. [DOI] [PubMed] [Google Scholar]
- 36.Li, Q. Y., R. A. Newbury-Ecob, J. A. Terrett, D. I. Wilson, A. R. Curtis, C. H. Yi, T. Gebuhr, P. J. Bullen, S. C. Robson, T. Strachan, D. Bonnet, S. Lyonnet, I. D. Young, J. A. Raeburn, A. J. Buckler, D. J. Law, and J. D. Brook. 1997. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat. Genet. 1521-29. [DOI] [PubMed] [Google Scholar]
- 37.Logan, M. 2003. Finger or toe: the molecular basis of limb identity. Development 1306401-6410. [DOI] [PubMed] [Google Scholar]
- 38.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67265-306. [DOI] [PubMed] [Google Scholar]
- 39.Muller, C. W., and B. G. Herrmann. 1997. Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature 389884-888. [DOI] [PubMed] [Google Scholar]
- 40.New, L., Y. Jiang, and J. Han. 2003. Regulation of PRAK subcellular location by p38 MAP kinases. Mol. Biol. Cell 142603-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newbury-Ecob, R. A., R. Leanage, J. A. Raeburn, and I. D. Young. 1996. Holt-Oram syndrome: a clinical genetic study. J. Med. Genet. 33300-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ossareh-Nazari, B., C. Gwizdek, and C. Dargemont. 2001. Protein export from the nucleus. Traffic 2684-689. [DOI] [PubMed] [Google Scholar]
- 43.Plageman, T. F., Jr., and K. E. Yutzey. 2005. T-box genes and heart development: putting the “T” in heart. Dev. Dyn. 23211-20. [DOI] [PubMed] [Google Scholar]
- 44.Rastogi, S., B. Joshi, G. Fusaro, and S. Chellappan. 2006. Camptothecin induces nuclear export of prohibitin preferentially in transformed cells through a CRM-1-dependent mechanism. J. Biol. Chem. 2812951-2959. [DOI] [PubMed] [Google Scholar]
- 45.Ruvinsky, I., L. M. Silver, and J. J. Gibson-Brown. 2000. Phylogenetic analysis of T-box genes demonstrates the importance of amphioxus for understanding evolution of the vertebrate genome. Genetics 1561249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoller, J. Z., and J. A. Epstein. 2005. Identification of a novel nuclear localization signal in Tbx1 that is deleted in DiGeorge syndrome patients harboring the 1223delC mutation. Hum. Mol. Genet. 14885-892. [DOI] [PubMed] [Google Scholar]
- 47.Stopper, G. F., and G. P. Wagner. 2005. Of chicken wings and frog legs: a smorgasbord of evolutionary variation in mechanisms of tetrapod limb development. Dev. Biol. 28821-39. [DOI] [PubMed] [Google Scholar]
- 48.Vaughan, C. J., and C. T. Basson. 2000. Molecular determinants of atrial and ventricular septal defects and patent ductus arteriosus. Am. J. Med. Genet. 97304-309. [DOI] [PubMed] [Google Scholar]
- 49.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82463-473. [DOI] [PubMed] [Google Scholar]
- 50.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4139-147. [DOI] [PubMed] [Google Scholar]
- 51.Zaragoza, M. V., L. E. Lewis, G. Sun, E. Wang, L. Li, I. Said-Salman, L. Feucht, and T. Huang. 2004. Identification of the TBX5 transactivating domain and the nuclear localization signal. Gene 3309-18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.