Abstract
The Cdc42-like GTPase Wnt responsive Cdc42 homolog 1 (Wrch1) has several atypical features; it has an N-terminal proline-rich extension that confers binding to SH3 domains, and it harbors an extremely high intrinsic nucleotide exchange activity, which overrides the normal GTPase activity. As a result, Wrch1 resides mainly in the active, GTP-loaded conformation under normal cellular conditions. We have previously shown that ectopic expression of Wrch1 in fibroblasts resulted in an altered cell morphology visible as a formation of filopodia, a loss of stress fibers, and a reduction in focal adhesions. Here, we show that Wrch1 binds to the nonreceptor tyrosine kinase Pyk2. The interaction required Wrch1 to be in a GTP conformation and also required an intact N-terminal proline-rich extension as well as an intact effector loop. Wrch1 requires Pyk2 in imposing the cytoskeletal effects, seen as the formation of filopodia, since treatment of cells with a Pyk2-specific small interfering RNA abrogated this response. Interestingly, we found that the presence and activity of Src were needed for the formation of a Wrch1-Pyk2 complex as well as for the Wrch1-induced formation of filopodia. We propose a model in which Pyk2 and Src function to coordinate the Wrch1-dependent effects on cytoskeletal dynamics.
The Rho GTPases belong to the Ras superfamily of GTP-hydrolyzing enzymes and are known to be potent regulators of the actin filament system and thereby the morphogenic and migratory properties of vertebrate cells (8, 18). In addition, several recent studies have implicated that Rho signaling is overactive in cancer, in particular during metastasis of certain tumor types (13). The Rho GTPases consist of 20 family members in humans (1, 39, 40). A majority of the Rho GTPases follow similar schemes for activation and regulation, and they function as GTP-hydrolyzing enzymes (8, 18). The binding of GTP triggers a conformational switch, which results in the exposure of a structural element, the so-called effector loop, which mediates the binding to downstream targets of the Rho GTPases (8). GTP hydrolysis inactivates the Rho GTPases; however, the relatively low intrinsic activity requires the aid of GTPase-activating proteins (GAPs) (26). The hydrolyzed GDP can then be exchanged for a GTP, a process that requires the aid of guanine nucleotide exchange factors (GEFs) (25, 33). The C termini of the Rho GTPases contain the CAAX box, which undergoes posttranslational prenylations (in the case of the Rho GTPases commonly by a geranyl-geranyl moiety). This modification is critical for targeting of the Rho GTPases to the plasma membrane or to intracellular lipid bilayers. The Rho guanine nucleotide dissociation inhibitors (GDIs) bind the prenylated Rho GTPases and sequester them in the inactive conformation in the cytoplasm (10). Cell activation results in a disruption of the RhoGDI-Rho GTPase interaction, and the released Rho GTPases are translocated to the relevant cellular localization.
The Rho GTPase Wnt-1 responsive Cdc42 homolog (Wrch1) is another example of a Rho GTPase with atypical features (3, 34, 35, 36). This protein belongs to the Cdc42 subfamily of the Rho GTPases, which also comprises Cdc42, TC10, TCL, and Chp. The human Wrch1 protein shares 57% identity and 70% similarity with Cdc42. In contrast to Cdc42, it contains both N- and C-terminal extensions, with 46 and 21 amino acids, respectively, and the N-terminal extension contains several PXXP motifs (34, 36). Unlike Cdc42, Wrch1 does not seem to be prenylated in the CAAX box; instead, membrane targeting is achieved by C-terminal palmitoylation (3). Although Wrch1 harbors GTPase activity, its very high intrinsic guanine nucleotide exchange activity is likely to ensure that the protein is predominantly in the GTP-loaded conformation. PAK1, Nckβ, Grb2, and PLCγ have been identified as proteins interacting with Wrch1 (34, 35, 36). When expressed transiently in NIH 3T3 cells, Wrch1 reduced the stress fiber content about 90% and the cells had an up-rounded morphology (34). The expression of Wrch1 also induced extensive filopodium formation in these cells. Wrch1 was originally identified as a gene upregulated by Wnt-1 and was shown to induce Wnt-1-like cell transformation of mouse mammary epithelial cells (36). Constitutively active mutants of Wrch1 also induce anchorage-independent growth of NIH 3T3 fibroblasts (35).
We conducted a yeast two-hybrid screen using a constitutively active Wrch1 mutant, Wrch1(Q107L), as bait, employing an Epstein-Barr virus (EBV)-transformed B-cell library. One set of clones, representing potential Wrch1-binding partners, turned out to encode proline-rich tyrosine kinase 2 (Pyk2), also known as related adhesion focal tyrosine kinase, cell adhesion kinase β, or calcium-dependent protein tyrosine kinase (2). Pyk2 is a nonreceptor tyrosine kinase related to focal adhesion kinase (FAK), and both kinases have important functions in the regulation of cell adhesion and focal adhesion assembly (2, 41). We show that Wrch1 is likely to be a physiological binding partner of Pyk2 and that Wrch1 also binds FAK, albeit weakly. We provide evidence that Wrch1 requires Pyk2 and Src to impose the effects on cytoskeletal dynamics.
MATERIALS AND METHODS
Plasmids, inhibitors, and antibodies.
The pRK5FAK, pRK5Pyk2, and pRK5HA-Pyk2/KD (kinase-dead [KD]) plasmids were generous gifts from Ivan Dikic, Frankfurt, Germany. The construction of pRK5Myc, encoding the different mutants of Wrch1, has been described before (34). The Src kinase inhibitor SU6656 was purchased from Sigma. The following antibodies were used: rabbit polyclonal anti-Myc (Santa Cruz), mouse monoclonal antiphosphotyrosine (PY99; Santa Cruz), mouse monoclonal antivinculin (Sigma), tetramethyl rhodamine isothiocyanate (TRITC)-conjugated anti-mouse antibody (Jackson ImmunoResearch Laboratories), and aminomethylcoumarin acetate (AMCA)-conjugated anti-rabbit antibody (Jackson). Alexa Fluor 488-conjugated phalloidin (Molecular Probes) was used to visualize filamentous actin.
Yeast two-hybrid screen.
The Saccharomyces cerevisiae strain Y190 (genotype, MATa gal4-542 gal80-538 his3 trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL1-LacZ Lys2::GAL1-HIS3cyhr) was transformed with a cDNA encoding the human Wrch1 or Wrch1 mutants fused to the GAL4 DNA-binding domain (GAL4DB) in the pYTH6 vector. The Wrch1 constructs harbored cysteine-to-serine mutations in the CAAX box since we reasoned that this would facilitate nuclear translocation during the screening procedure. The GAL4DB-Wrch1(Q107L)-expressing yeast strain was used to screen a cDNA library from EBV-transformed human B cells fused to the GAL4 activation domain in the pACT vector.
Cell cultivation and immunocytochemistry.
Human embryonic kidney 293T (HEK293T) cells and mouse embryonic fibroblasts (MEFs) from wild-type mice as well as from FAK−/− (17) or SYF−/− (20) mice were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. Porcine aortic endothelial (PAE) cells stably transfected with mouse platelet-derived growth factor receptor β (PDGFRβ), referred to as PAE/PDGFRβ cells (clone β1:3), were cultured in Ham's F-12 medium supplemented with 10% FBS and penicillin-streptomycin. All cells were cultured at 37°C in an atmosphere of 5% CO2. For immunostaining purposes, cells were seeded on coverslips and transfected by Lipofectamine according to the procedure provided by the manufacturer. The cells were fixed 18 to 20 h after addition of the DNA-Lipofectamine mixture to the cells, and during the last 10 h, the cells were kept in serum-free medium. The cells were fixed in 2% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature. The cells were thereafter washed again and incubated in the presence of 10 mM glycine in PBS for 1 h. Primary and secondary antibodies were diluted in PBS containing 5% FBS. Cells were incubated with primary antibodies followed by secondary antibodies for intervals of 1 h. The coverslips were mounted on object slides in Fluoromount-G mounting medium (Southern Biotechnology Associates). The cells were photographed by a Hamamatsu ORCA charge-coupled-device digital camera employing QED imaging system software, using a Zeiss Axioplan 2 microscope.
For immunoprecipitations, HEK293T cells were transfected by calcium phosphate precipitation or by Lipofectamine. Transfected cells were harvested 24 h after transfection, washed with ice-cold PBS, and lysed on ice in Triton buffer (20 mM HEPES [pH 7.5], 0.1 M NaCl, 1% Triton X-100, 10% glycerol, 5 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, 1% aprotinin [Trasylol; Beyer], 1 mM pefablock [Roche]). Lysed cells were collected in Eppendorf tubes and centrifuged for 15 min. The resulting supernatants were subjected to immunoprecipitation experiments. For coimmunoprecipitations, supernatants were incubated together with antibodies as indicated in the figure legends for 1 h at 4°C, after which immunoprecipitates were collected on protein A-Sepharose (Immunosorb A; Medicago). The beads were washed three times with Triton buffer, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer was added to each sample. The immunoprecipitates as well as control cell lysates were subjected to SDS-PAGE and subsequent electrotransfer to Hybond C (GE Healthcare). Western blot analyses were performed with antibodies as specified in the figure legends followed by horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies (GE Healthcare). The Western blots were detected by Luminol immunoblotting reagent (Santa Cruz).
Knockdown of Pyk2 expression was triggered by transfection of MEFs with double-stranded Pyk2-specific small interfering RNA (siRNA) (Sigma) or a control siRNA employing the Silentfect transfection reagent (Bio-Rad). The cells were left overnight and then transfected with a plasmid encoding Myc-tagged Wrch1 by the Lipofectamine procedure described above, with the exception that the cells were not serum starved prior to fixation.
RESULTS
Wrch1 requires the proline-rich N-terminal domain to produce filopodia.
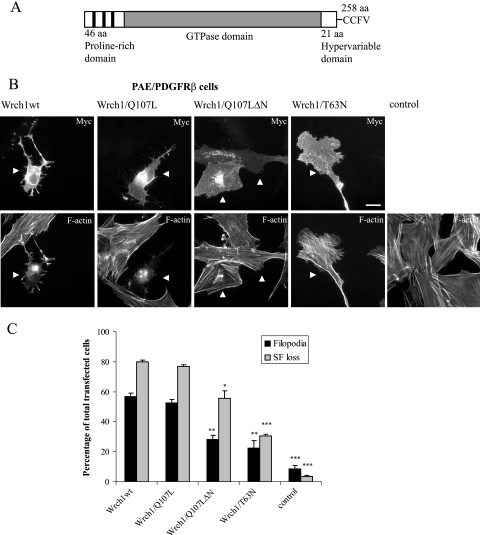
The human Wrch1 gene is located on chromosome 1 (1q42.11-q42.3) and encodes a polypeptide of 258 amino acid residues (Fig. 1A) (34, 36). We have shown previously that ectopic expression of Wrch1 in NIH 3T3 fibroblasts resulted in loss of focal adhesion, cell rounding, stress fiber dissolution, and the formation of filopodia (34). In the present study, we transfected PAE/PDGFRβ cells with Wrch1, Wrch1(Q107L), Wrch1(T63N), and a Q107L mutant with the N-terminal proline-rich domain deleted [Wrch1(Q107LΔN)] (Fig. 1B). Wild-type Wrch1 and Wrch1(Q107L) each induced filopodia and stress fiber dissolution to a similar extent (Fig. 1B and C). These responses required Wrch1 to be in the GTP-bound conformation, since the putatively GDP-bound Wrch1(T63N) mutant had a much reduced ability to induce dissolution of stress fibers and the formation of filopodia. Moreover, an intact N terminus was required for the formation of filopodia, since the Wrch1(Q107LΔN) mutant had a significantly reduced ability to induce filopodia (Fig. 1B and C).
FIG. 1.
Wrch1 is a Cdc42-like member of the Rho GTPases. (A) Schematic representation of the domains present in the human Wrch1: N-terminal proline-rich motifs, the Cdc42-like GTPase domain, and the C-terminal CAAX box. aa, amino acids. (B) Wrch1 (Wrch1wt) and Wrch1(Q107L) induce the formation of filopodia and dissolution of stress fibers. Wrch1 or Wrch1(Q107L) was transiently transfected into PAE/PDGFRβ cells. Myc-tagged Wrch1 and Wrch1(Q107L) were visualized with a rabbit anti-Myc antibody followed by an AMCA-conjugated anti-rabbit antibody. Filamentous actin was visualized by Alexa Fluor 488-conjugated phalloidin. The arrowheads mark transfected cells. (C) Quantification of the phenotypes observed in panel B was performed by microscopy analysis and scored as filopodium formation (filopodia) or stress fiber dissolution (SF loss). Stress fiber loss was scored when cells had less than one-fifth of the stress fiber content seen in the control cells. This is actually rather clear, since a majority of Wrch1-expressing cells retain only few and often disintegrated filamentous actin-containing structures. The values represent analyses of at least 100 transfectants in three independent experiments. The statistics analysis was carried out using Student's t test to compare the phenotypes induced by Wrch1wt to the phenotypes induced by Wrch1(Q107LΔN) and Wrch1(T63N). * represents P values of <0.05, ** represents P values of <0.01, and *** represents P values of <0.001.
Wrch1 binds the Pyk2 tyrosine kinase.
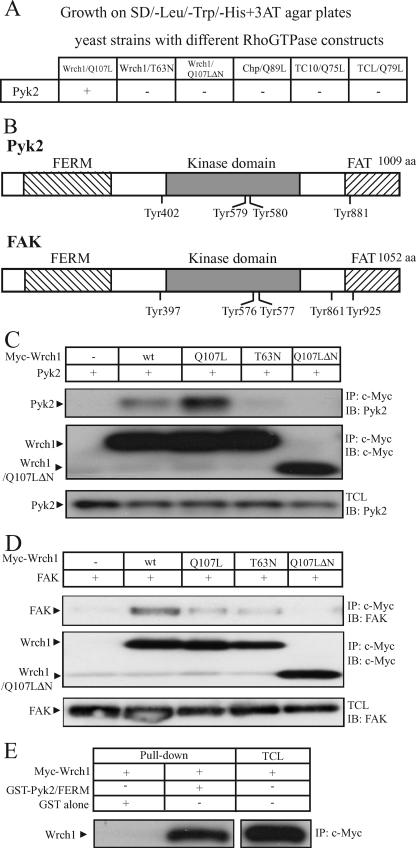
We wanted to learn more about the mechanisms underlying the Wrch1-dependent cellular effects. To this end, we performed a yeast two-hybrid screen in order to identify binding partners for the constitutively active mutant of Wrch1 [Wrch1(Q107L)]. S. cerevisiae cells expressing Wrch1(Q107L) were transformed with a cDNA library derived from EBV-transformed human B cells. Clones expressing potential Wrch1(Q107L)-interacting proteins were isolated, and the library plasmid DNA was recovered and subjected to sequence analysis. A total of 10 clones, encoding seven potential Wrch1-interacting proteins, were isolated. Three of the clones encoded Pyk2 (Fig. 2B). The insert lengths of the clones differed but all started in the N-terminal part of the FERM domain of Pyk2. We tested the potential binding to constitutively active members of the Cdc42 subfamily, but Pyk2 interacted only with Wrch1 (Fig. 2A). Moreover, the binding required Wrch1 to be in the GTP-bound conformation as well as to have an intact N-terminal domain, since the putatively GDP-bound mutant of Wrch1 [Wrch1(T63N)] and the N-terminal deletion mutant [Wrch1(Q107LΔN)] did not bind (Fig. 2A).
FIG. 2.
Wrch1 binds the Pyk2 and FAK nonreceptor tyrosine kinases. (A) Yeast two-hybrid assay for the interaction of the Cdc42 subfamily members with Pyk2. Interaction of GAL4 activation domain fusion proteins of Pyk2 with GAL4DB-Wrch1(Q107L), GAL4DB-Wrch1(T63N), GAL4DB-Wrch1(Q107LΔN), GAL4DB-ChpL89, GAL4DB-TC10L75, or GAL4DB-TCLL79 in the yeast two-hybrid system. Colonies growing on selective media as described in Materials and Methods were assayed for β-galactosidase activity. A positive interaction is scored as +. (B) Schematic representation of Pyk2 and FAK. aa, amino acids. (C) Interaction between Myc-tagged Wrch1 or Wrch1 mutants and Pyk2 was assessed by transient transfection into HEK293T cells. The presence of Pyk2 in Myc immunoprecipitates (IP) was determined by immunoblotting (IB). wt, wild type. (D) Interaction between Myc-tagged Wrch1 or Wrch1 mutants and FAK was assessed by transient transfections in HEK293T cells. The presence of FAK in Myc immunoprecipitates was determined by immunoblotting. (E) Interaction between the FERM domains of Pyk2 and Wrch1 was determined by incubating lysates from HEK293T cells transiently transfected with Myc-Wrch1 with Pyk2 fusion-proteins bound to glutathione-Sepharose beads (GST alone and GST-FERM). The presence of Myc-Wrch1 in the precipitate was detected by Western blotting employing a Flag-specific antibody.
We next tested whether Wrch1 could interact with Pyk2 in a coimmunoprecipitation experiment. We made a Wrch1-specific antiserum; however, it was not able to detect endogenous Wrch1 in cell lysates, possibly due to the low expression of the protein in cell lines. Instead, HEK293T cells with plasmids encoding different variants of Myc-tagged Wrch1 were transfected together with a plasmid encoding Pyk2. The cells were lysed, and Wrch1 was immunoprecipitated with a Myc-specific antibody. The presence of Pyk2 in the immunoprecipitates was examined by SDS-PAGE followed by immunoblotting. This way, it was found that Pyk2 bound to wild-type Wrch1 and to the constitutively active mutants of Wrch1. In agreement with the yeast two-hybrid results, we could not detect any binding of Pyk2 to the Wrch1(T63N) and Wrch1(Q107LΔN) mutants (Fig. 2C). Pyk2 is about 50% identical to the FAK (Fig. 2B) (2), and therefore, we tested the interaction between FAK and Wrch1. Interestingly, FAK also bound toWrch1; however, the interaction was much weaker than that between Wrch1 and Pyk2 (Fig. 2D). Finally, we used a glutathione S-transferase (GST) pulldown approach to test the interaction between Wrch1 and domains of Pyk2. This way, we found that Wrch1 binds strongly to the FERM domain of Pyk2 (Fig. 2E).
Wrch1 requires an intact Pyk2-binding ability for the formation of filopodia.
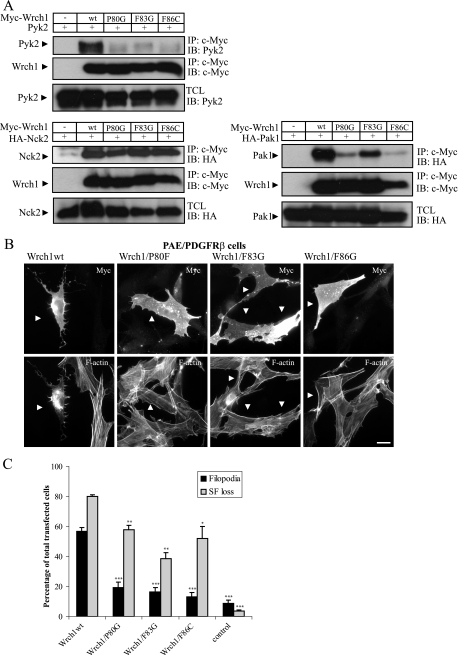
The Wrch1(Q107LΔN) mutant, which does not bind Pyk2, had a significantly reduced ability to trigger the formation of filopodia in PAE/PDGFRβ cells (Fig. 1B and C). To study this further, we produced three mutations in the effector loop of Wrch1 [Wrch1(P80G), Wrch1(F83G), and Wrch1(F86C)] and tested them for Pyk2 binding in a coimmunoprecipitation experiment. The interaction was abolished in all three cases (Fig. 3A). The Wrch1(F83G) and Wrch1(F86C) mutants are equivalent to the F37A and Y40C mutants in Rac1 and Cdc42 described by Lamarche et al. (22). In this study, it was found that Pak1 bound to Rac1(F37A) and not to Rac1(Y40C). We therefore tested whether Pak1 had a binding pattern similar to those of the Wrch1 effector loop mutants, and we could see that Pak1 bound to Wrch1(F83G) and only weakly to Wrch1(P80G) and Wrch1(F86C) (Fig. 3A). In contrast, Nck2, which requires the Wrch1 N-terminal domain for binding, bound with equal affinity to all three mutants (Fig. 3A). We next tested the abilities of the Wrch1 effector loop mutants to induce filopodia and stress fiber dissolution in PAE/PDGFRβ cells. All three mutants had significantly reduced abilities to induce filopodia, which was associated with reduced stress fiber dissolution. In particular, Wrch1(F83G) had a significantly reduced stress fiber breakdown (Fig. 3B and C). Taken together, the results suggest a correlation between the Pyk2-binding ability of Wrch1 and its ability to induce the cytoskeletal responses.
FIG. 3.
Pyk2 interaction requires an intact Wrch1 effector loop. (A) Interaction between Myc-tagged Wrch1 or Wrch1 mutants and Pyk2, Pak1, or Nck2 was assessed by transient transfection into HEK293T cells. The presence of Pyk2, Pak1, or Nck2 in Myc immunoprecipitates (IP) was determined by immunoblotting (IB). wt, wild type. (B) Wild-type Wrch1 and Wrch1 effector loop mutants [Wrch1(P80G), Wrch1(F83G), or Wrch1(F86C)] were transiently transfected into PAE/PDGFRβ cells. Myc-tagged Wrch1 was visualized with a rabbit anti-Myc antibody followed by an AMCA-conjugated anti-rabbit antibody. Filamentous actin was visualized Alexa Fluor 488-conjugated phalloidin. The arrowheads mark Wrch1-transfected cells. (C) Quantification of the phenotypes observed in panel B was performed by microscopy analysis and scored as filopodium formation (filopodia) or stress fiber dissolution (SF loss). The values represent analyses of at least 100 transfectants in three independent experiments. The statistics analysis was carried out using Student's t test to compare the phenotypes induced by Wrch1wt to the phenotypes induced by Wrch1 effector loop mutants. * represents P values of <0.05, ** represents P values of <0.01, and *** represents P values of <0.001.
Wrch1 affects the phosphorylation status of Pyk2.
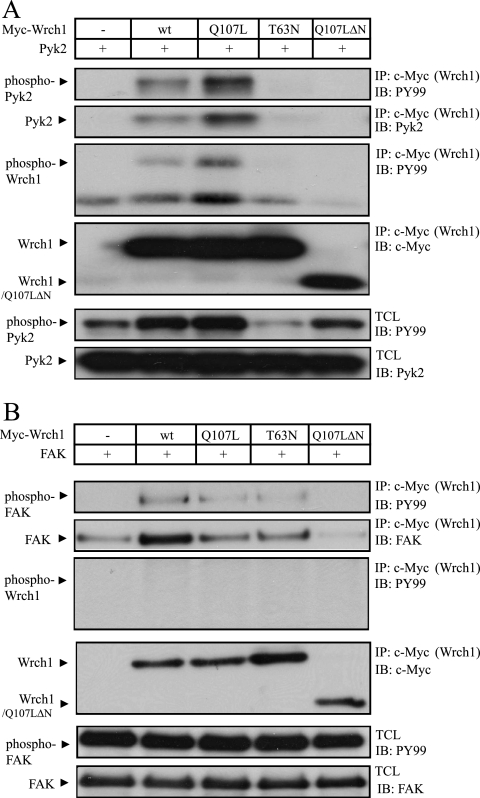
Pyk2 is a tyrosine kinase that needs to be phosphorylated to become active; therefore, we tested whether Wrch1 affected the phosphorylation of Pyk2. Again, wild-type Wrch1 and Wrch1 mutants were cotransfected with Pyk2 in HEK293T cells and Wrch1 was immunoprecipitated. The tyrosine phosphorylation of the precipitated Pyk2 and Wrch1 was assessed by a phosphotyrosine-specific antibody. We found that Pyk2 was tyrosine phosphorylated, and importantly, we also found that Wrch1 became tyrosine phosphorylated in the presence of Pyk2 (Fig. 4A). Interestingly, the phosphorylation of Pyk2 was markedly reduced in the presence of Wrch1(T63N), suggesting that the activity of Wrch1 has an impact on the activation of Pyk2 upstream of the kinase. FAK was also phosphorylated in the presence of Wrch1; however, in this case, Wrch1 was not tyrosine phosphorylated by FAK (Fig. 4B). Moreover, Wrch1(T63N) did not influence the phosphorylation status of FAK. We also attempted to identify the Wrch1-binding domains of Pyk2 and FAK. We performed coimmunoprecipitation experiments with Wrch1 together with isolated FERM, kinase, and focal adhesion targeting domains of Pyk2 or FAK. Despite several attempts, we were not able to determine the Wrch1-binding domains. This suggests that the Pyk2 and FAK tyrosine kinases will bind only in the context of the full-length protein. It is worth noticing that all of the Pyk2 clones isolated from the yeast two-hybrid system screen encoded almost the entire Pyk2 protein, only small parts in the N-terminal part of the FERM domains were missing.
FIG. 4.
Ectopic expression of Wrch1 affects the phosphorylation status of Pyk2 but not that of FAK. (A) The effect on Pyk2 phosphorylation by simultaneous expression of Wrch1 or Wrch1 mutants was assessed by immunoblotting (IB) using a phosphotyrosine-specific antibody. Note that Wrch1(T63N) induced a marked reduction of the Pyk2 phosphorylation. wt, wild type; IP, immunoprecipitation. (B) The effect on FAK phosphorylation by simultaneous expression of Wrch1 or Wrch1 mutants was assessed by immunoblotting using a phosphotyrosine-specific antibody.
Pyk2 is a potential Wrch1 effector.
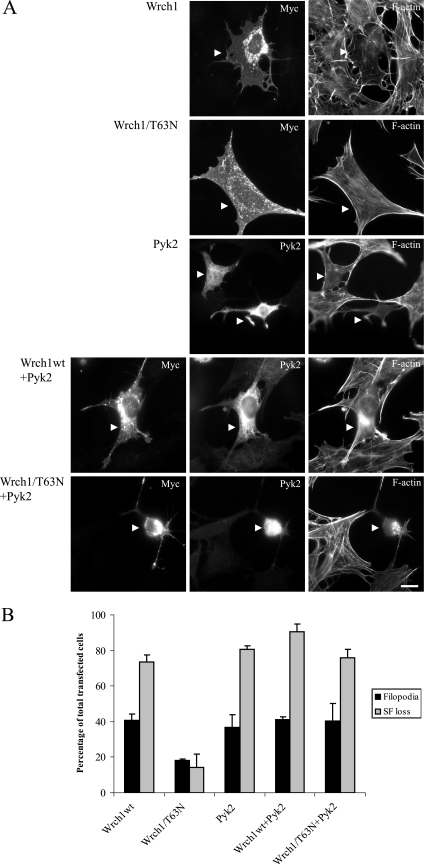
Having established that Pyk2 is a binding partner for Wrch1, we analyzed the effects of Wrch1 and Pyk2 expression on cell morphology in normal mouse fibroblasts. We found that Wrch1 was slightly less efficient in the ability to trigger filopodia in these cells than in PAE/PDGFRβ cells, but the effect on stress fiber dissolution was almost the same. Again, the dominant-negative Wrch1(T63N) mutant had a much reduced ability to induce filopodia and stress fiber breakdown (Fig. 5A and B). Interestingly, ectopic expression of Pyk2 induced a phenotype that to a large extent resembled the Wrch1 phenotype in filopodium formation and stress fiber dissolution; however, the Pyk2-expressing cells normally had fewer filopodia than the Wrch1-expressing cells. We reasoned that if Pyk2 could function as an effector of Wrch1, Pyk2 would bypass the dominant-negative phenotype associated with Wrch1(T63N)-expressing cells and it would give a phenotype which is more similar to Pyk2 or Wrch1 alone. Pyk2 transfected together with wild-type Wrch1 resulted in a phenotype which largely resembled the phenotype induced by each of the proteins alone (Fig. 5A and B). Importantly, the Pyk2 phenotype was dominant over the Wrch1(T63N) phenotype in cells expressing both proteins (Fig. 5A and B).
FIG. 5.
Pyk2 is potential effector to Wrch1. (A) Normal MEFs were transfected with Wrch1, Wrch1(T63N), Pyk2, Pyk2 plus Wrch1 (Wrch1wt+Pyk2), or Pyk2 plus Wrch1(T63N) (Wrch1/T63N+Pyk2). Myc-tagged Wrch1 was detected by a rabbit anti-Myc antibody followed by an AMCA-conjugated anti-rabbit antibody. Pyk2 was detected by a mouse anti-Pyk2 antibody followed by a TRITC-conjugated anti-mouse antibody. Filamentous actin was detected by Alexa Fluor 488-conjugated phalloidin. The arrowheads mark transfected cells. Bar, 20 μm. (B) Quantification of the phenotypes observed in panel A was performed by microscopy analysis and scored as filopodium formation (filopodia) or stress fiber dissolution (SF loss). The values represent analyses of at least 100 transfectants in three independent experiments. Wrch1wt, wild-type Wrch1.
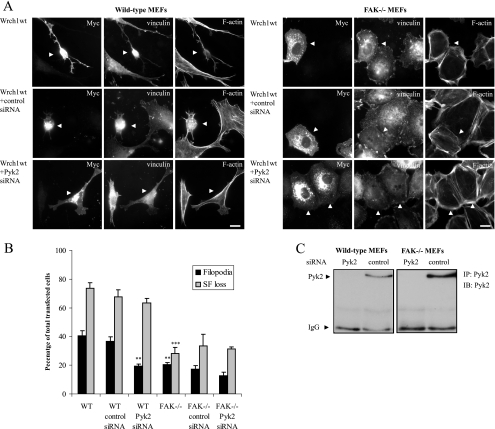
We next used an siRNA approach to knock down the expression of Pyk2 in normal mouse fibroblasts to see if the Wrch1-induced cytoskeletal reorganization required Pyk2. We designed Pyk2-specific siRNA and found it to be effective in reducing the Pyk2 expression (Fig. 6C). We next transfected Wrch1 in cells pretreated with the Pyk2-specific siRNA or a control siRNA. Wrch1 induced the formation of filopodia and stress fiber dissolution in cells transfected with Wrch1 alone or Wrch1 pretreated with a control siRNA (Fig. 6A and B). In contrast, the cells pretreated with the Pyk2-specific siRNA had a marked reduction in the filopodium response. Since Wrch1 also bound to FAK, we reasoned that this protein might rescue some of the defects associated with loss of Pyk2. Therefore, we repeated the experiment with MEFs derived from mice lacking FAK (17). Ectopic expression of Wrch1 in wild-type MEFs results in a clear reduction in filopodium formation, and this was associated with a reduced stress fiber breakdown (Fig. 6A and B). The filopodium formation was further reduced in the presence of Pyk2-specific siRNA, but the stress fiber dissolution was marginally further reduced (Fig. 6A and B). These data support a model suggesting that Pyk2 is able to function as a Wrch1 effector in the regulation of cytoskeletal dynamics.
FIG. 6.
Pyk2 and FAK are required for the Wrch1-induced filopodium formation or stress fiber dissolution. (A) Myc-tagged wild-type Wrch1 (Wrch1wt) was transfected into normal MEFs or MEFs isolated from FAK−/− mice in the presence or absence of Pyk2-specific siRNA or control siRNA. Myc-tagged Wrch1 was detected by a rabbit anti-Myc antibody followed by an AMCA-conjugated anti-rabbit antibody. Vinculin was detected by a mouse antivinculin antibody followed by a TRITC-conjugated anti-mouse antibody. Filamentous actin was detected by Alexa Fluor 488-conjugated phalloidin. The arrowheads mark transfected cells. Bar, 20 μm. (B) Quantification of the phenotypes observed in panel A was performed by microscopy analysis and scored as filopodium formation (filopodia) or stress fiber dissolution (SF loss). The values represent analyses of at least 100 transfectants in three independent experiments. The statistics analysis was carried out using Student's t test to compare the phenotypes induced by Wrch1 in the presence of control siRNA and Pyk2-specific siRNA or the Wrch1-induced phenotypes in FAK−/− and wild-type MEFs. * represents P values of <0.05, ** represents P values of <0.01, and *** represents P values of <0.001. (C) Transfection of siRNA specific for Pyk2 leads to a knockdown of Pyk2 expression in wild-type and FAK−/− MEFs. The expression of Pyk2 is rather low in these cell lines; therefore, Pyk2 was immunoprecipitated (IP) with a Pyk2-specific antibody and the presence of Pyk2 in the precipitates was detected by immunoblotting (IB) using a Pyk2-specific antibody. IgG, immunoglobulin G.
Src is necessary for Wrch1 binding to Pyk2.
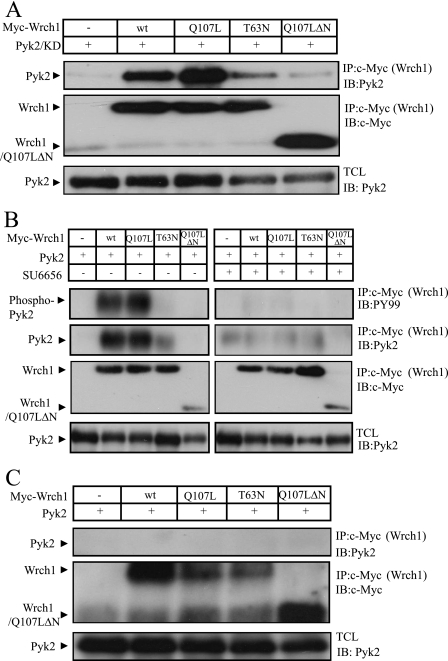
We tested whether kinase activity of Pyk2 was important for binding, and we found that KD Pyk2 bound with wild-type strength to Wrch1, indicating that kinase activity is not necessary for binding (Fig. 7A). It has previously been found that KD Pyk2 is probably functioning as an adapter protein and retains many of the functions of the wild-type Pyk2 protein (21). Binding of Src to Pyk2 is an important step in the activation of Pyk2 (2). Therefore, we treated the cells with a Src-specific low-molecular-weight inhibitor, SU6656. This treatment affected the tyrosine phosphorylation of Pyk2, but more importantly, it affected the binding between Pyk2 and Wrch1 (Fig. 7B). To confirm this result, we repeated the experiments with fibroblasts lacking the Src, Yes, and Fyn nonreceptor tyrosine kinases (SYF−/− MEFs) (20). Also in this case, no interaction between Pyk2 and Wrch1 was detected (Fig. 7C), strongly suggesting that the presence of an active Src kinase is needed for the formation of a Wrch1-Pyk2 complex. This interaction is not mediated by Src binding to Wrch1, since we could not detect any direct interaction between Wrch1 and Src (data not shown).
FIG. 7.
An active Src kinase is obligatory for the interaction between Wrch1 and Pyk2. (A) The interaction between Myc-tagged Wrch1 or Wrch1 mutants and KD Pyk2 (Pyk2/KD) was assessed by transient transfections into HEK293T cells. The presence of Pyk2/KD in Myc immunoprecipitates (IP) was determined by immunoblotting (IB). wt, wild type. (B) The necessity of the presence of an active Src kinase for the Wrch1 and Pyk2 interaction was assessed by transient transfections of Myc-tagged Wrch1 or Wrch1 mutants and Pyk2 into HEK293T cells. The cells were treated with the Src inhibitor SU6656 (2 μM) for 1 h prior to cell lyses. No Pyk2 was detected in Myc immunoprecipitates, as determined by immunoblotting. (C) The necessity of the presence of the Src family kinase for the Wrch1-and-Pyk2 interaction was further assessed by transient transfections of Myc-tagged Wrch1 or Wrch1 mutants and Pyk2 into SYF−/− MEFs. No Pyk2 could be detected in Myc immunoprecipitates, as determined by immunoblotting.
Src is necessary for Wrch1-induced formation of filopodia.
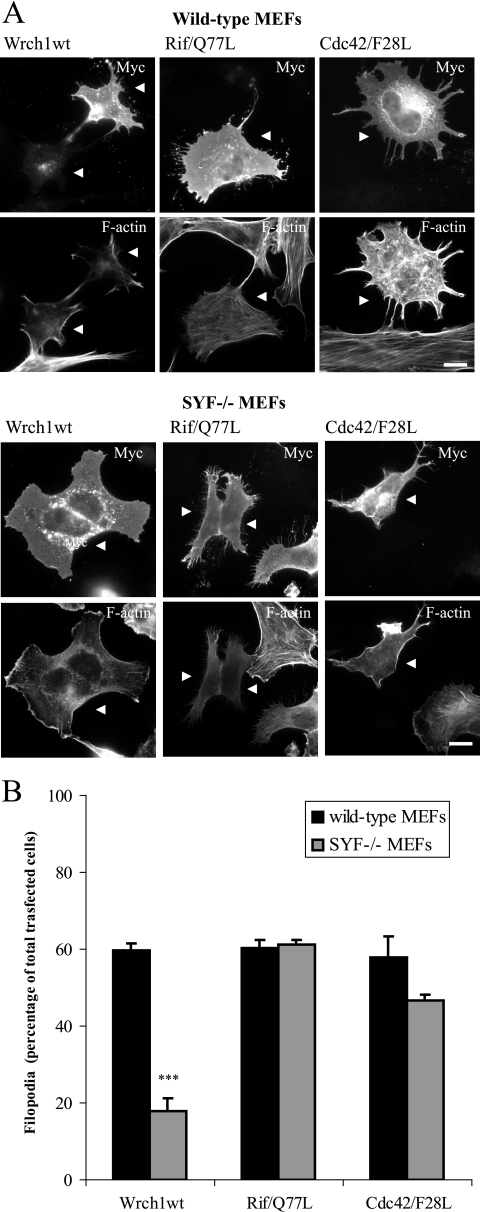
We next tested whether Src is necessary for the Wrch1 cellular phenotypes, and to this end, we analyzed the ability of Wrch1 to induce filopodium formation in the absence of Src. Wild-type and SYF−/− MEFs were transfected with wild-type Wrch1 and Wrch1(T63N). Interestingly, Wrch1 was much less effective in triggering filopodia in the SYF−/− MEFs, clearly showing that Src is necessary for the induction of filopodia by Wrch1 (Fig. 8A and B). In contrast, ectopic expression of Rif(Q77L) and Cdc42(F28L), which also induce the formation of a filopodium-like protrusion (1), formed these structures in wild-type and SYF−/− MEFs equally effectively, demonstrating that Src is specifically needed for the Wrch1-induced filopodia.
FIG. 8.
Src is required for the Wrch1-induced filopodia but not for the Rif- or Cdc42-induced filopodia. (A) Myc-tagged wild-type Wrch1 (Wrch1wt), Rif(Q77L), or Cdc42(F28L) was transfected into normal MEFs or MEFs isolated from SYF−/− mice. Myc-tagged Wrch1, Rif(Q77L), or Cdc42(F28L) was detected by a rabbit anti-Myc antibody followed by an AMCA-conjugated anti-rabbit antibody. Vinculin was detected by a mouse antivinculin antibody followed by a TRITC-conjugated anti-mouse antibody. Filamentous actin was detected by Alexa Fluor 488-conjugated phalloidin. The arrowheads mark transfected cells. Bar, 20 μm. (B) Quantification of the phenotypes observed in panel A was performed by microscopy analysis and scored as filopodium formation (filopodia) or stress fiber dissolution (SF loss). The values represent analyses of at least 100 transfectants in three independent experiments. The statistics analysis was carried out using Student's t test to compare the amounts of Wrch1-induced filopodia in wild-type and SYF−/− MEFs. * represents P values of <0.05, ** represents P values of <0.01, and *** represents P values of <0.001. The slightly reduced ability of Cdc42(F28L) to induce filopodia in SYF−/− MEFs is not statistically significant.
DISCUSSION
Wrch1 has a profound effect on cell morphology and cell adhesion (33). The finding that Wrch1 binds to the Pyk2 tyrosine kinase is of importance in this context and provides a plausible explanation for the decreased cell adhesion associated with ectopic expression of Wrch1. FAK is activated by integrin engagement, and FAK is needed for the dynamic turnover of focal adhesions (29, 41). Fibroblasts isolated from FAK−/− MEFs have enhanced focal adhesions and impaired migration. Pyk2 is activated by integrin engagement, but it is also activated by a number of other stimuli, such as Ca2+ signaling, protein kinase C, and G-protein-coupled receptors (2). Pyk2 and FAK have been associated with Rho signaling before, though in an indirect way (24, 29). For instance, FAK binds to the p190RhoGEF and the FAK-dependent tyrosine phosphorylation of p190RhoGEF is associated with an increased RhoA activation (42). FAK also binds to the Rho and Cdc42-specific GEF GRAF and can thereby influence the activated statuses of Cdc42 and Rho (16). Moreover, Pyk2 and FAK were shown to bind the Cdc42-specific RhoGAP PSGAP. Interestingly, Pyk2, but not FAK, had a negative influence on the catalytic activity of PSGAP, which resulted in an activation of Cdc42 (30). The direct interaction between Wrch1 and Pyk2 adds another level to the interconnection between signaling involving this family of tyrosine kinases and signaling employing Rho GTPases.
How is the activity of Wrch1 regulated? This is still an open question. The primary structure of Wrch1 is very similar to that of Cdc42, but it harbors several characteristics which make it unique among the Rho GTPases (34, 35, 36). The N-terminal proline-rich domain was found to bind SH3 domain-containing proteins, such as Nck-2, PLC-γ, and Grb2, and according to Shutes et al., the N terminus elicits a negative influence on the GTP-bound state of Wrch1, which is relieved by binding to SH3 domains (34, 35). Importantly, the intrinsic exchange activity of Wrch1 is much higher than the exchange activity of any other Rho GTPase. As a result, the protein is likely to reside more or less constantly in a GTP-bound state. This will clearly have a major impact on the function of the protein and suggests that the protein is regulated by a mechanism different from those of the classical Rho GTPases. So far, no GAPs or GEFs have been found for Wrch1, which does not necessarily imply that there are no such Wrch1-specific regulatory proteins. However, it is more likely that the activity of Wrch1 is regulated by mechanisms that affect the intracellular protein concentration of Wrch1. This has been seen for other atypical members of the Rho GTPases, such as the Rnd proteins (5). These proteins have amino acid substitutions in the amino acid residues critical for nucleotide binding, and as a result, the Rnd proteins are likely to always be in the active GTP-bound conformation. Rnd3/RhoE has been extensively studied, and its expression is induced by the Raf signaling pathway and by PDGF signaling (15, 31). In addition, stress-related stimuli, such as UV-B irradiation, have also been shown to upregulate the Rnd3/RhoE expression (28). Less is known about the downregulation of the Rnd proteins, but at least Rnd3/RhoE seems to be phosphorylated by a number of stimuli (5, 32). Phosphorylation seems to affect the stability for the protein, and the nonphosphorylated form seems to be degraded by the ubiquitin-proteasome pathway (31). We do not know what factors affect the transcription of Wrch1; however, Wrch1 was originally isolated as a gene upregulated in response to Wnt-1 (36). An increase in Wrch1 expression was also noticed in bone marrow macrophages during receptor activator of NF-κB ligand-induced osteoclastogenesis (4). Conversely, Wrch1 expression was found to be downregulated in response to β-estradiol in MCF-7 cells as well as in interstitial cells of Cajal in mice harboring mutations in the W locus (W/Wv mice) (9). However, despite these scattered observations, we know very little about what factors really define the activated status of Wrch1, and this is something that we hope will be sorted out in the near future.
Most members of the Rho GTPases are modified in their C-terminal CAAX boxes by a geranyl-geranyl isoprenoid; however, TC10, TCL, RhoD, and Rnd1-3 are instead farnesylated, whereas the Miro and RhoBTB GTPases (with the exception of RhoBTB3) lack this type of membrane-targeting motive (39). Surprisingly, Chp and Wrch1 do not seem to be prenylated at all; instead, a C-terminal palmitoylation seems to confer membrane targeting on the proteins (3, 6, 7). It is not known at the moment what this means in terms of the signaling abilities of the Chp and Wrch1 GTPases, but this constitutes yet another parameter that distinguishes these GTPases from the classical Rho GTPases. Membrane targeting of small GTPases and compartmentalization of the signaling pathways have received increased attention recently, in particular in the research on Ras signaling. Differences in posttranslational modification give unique signaling flavors to the different Ras isoenzymes (14, 27). This is a notion that will be of importance here also, in particular since Wrch1 shares binding partners with other members of the Cdc42 and Rac subfamilies. PAK1 serine/threonine kinase is a common binding partner for all Cdc42- and Rac-like Rho GTPases (34, 36). We have also seen that there is a weak, but detectable, interaction between Wrch1 and Par6C (1). The activation of a specific member of the Rho GTPases results in the specific subcellular localization of PAK1 or Par6C and, as a result, the induction of unique signaling cues. Sorting out how this is regulated will be a true challenge for future studies.
Interestingly, we found a clear requirement for Src for the Wrch1-Pyk2 interaction. Src does not seem to bind directly to Wrch1, but the SH2 domain of Src has a strong affinity for the phosphorylated tyrosine 402 of Pyk2 (2). A couple of previous studies have implicated Src in the activation of other members of the Rho GTPases. Cdc42, RhoA, and Rac1 were shown to be needed for the localization of Src to focal adhesions (37). Integrin activation of the Rho GTPases requires the Src-like kinase Fyn during morphological differentiation of oligodendrocytes, and Src is required for epidermal growth factor-stimulated activation of Cdc42 in fibroblast-like cells (23, 38). Moreover, Src is needed for the regulation of the Cdc42 GEF Cool-1/βPIX (11). Another study showed that the adherence junction protein nectin activates Cdc42 in a manner dependent on Src and Src-mediated activation of the Cdc42 GEF FRG (12). Src was shown to phosphorylate Cdc42 on tyrosine 64 in the switch II domain (38). This phosphorylation does not seem to affect the effector-binding capacity of Cdc42, but it increases the affinity of Cdc42 for RhoGDI. Pyk2, but not FAK, can phosphorylate Wrch1 on tyrosine residues, but this modification does not affect the interaction with Pyk2, and the impact of tyrosine phosphorylation of Wrch1 is currently not clear. We also noticed that the putatively GDP-bound mutant Wrch1(T63N) affected the tyrosine phosphorylation of Pyk2 but not that of FAK. This means that Wrch1(T63N) is interfering with the upstream signals responsible for Pyk2 phosphorylation. A similar situation has been described for FAK in neuronal cells where the dominant-negative Cdc42(T17N) mutant attenuated the muscarinic cholinergic receptor-mediated phosphorylation of FAK (2).
What is the physiological function of Wrch1? There seems to be an ability in Wrch1 to induce a transformed phenotype in different model systems, such as mouse mammary epithelial cells and NIH 3T3 fibroblasts (3, 35, 36). The ability to induce a transformed phenotype in fibroblasts is something Wrch1 shares with Chp (7). On the other hand, mRNA levels of Wrch1 were found to be lower in many tumor types than in normal tissue, but there are examples of the reverse: an increased expression of Wrch1 in certain tumors (19). In essence, most of the current data collectively suggest that the major role of Wrch1 is in the regulation of the morphological and adhesive processes that underlie cell migration, but future studies will hopefully find out more about the exact mechanisms for this regulation.
Acknowledgments
The research in the laboratory of the authors has been supported by funds from the Ludwig Institute for Cancer Research, The Swedish Cancer Society, and the Swedish Research Council.
We thank C.-H. Heldin for useful comments on the manuscript.
Footnotes
Published ahead of print on 17 December 2007.
REFERENCES
- 1.Aspenström, P., Å. Fransson, and J. Saras. 2004. Rho GTPases have diverse effects on the organisation of the actin filament system. Biochem. J. 377327-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avraham, H., S.-Y. Park, K. Schinkmann, and S. Avraham. 2000. RAFT/Pyk2-mediated cellular signalling. Cell. Signal. 12123-133. [DOI] [PubMed] [Google Scholar]
- 3.Berzat, A., J. E. Buss, E. J. Chenette, C. A. Weinbaum, A. Shutes, C. J. Der, A. Minden, and A. D. Cox. 2005. Transforming activity of the Rho family GTPase, Wrch-1, a Wnt-regulated Cdc42 homolog, is dependent on a novel carboxyl-terminal palmitoylation motif. J. Biol. Chem. 28033055-33065. [DOI] [PubMed] [Google Scholar]
- 4.Brazier, H., S. Stephens, S. Ory, P. Fort, N. Morrison, and A. Blangy. 2006. Expression profile of RhoGTPases and RhoGEFs during RANKL-stimulated osteoclastogenesis: identification of essential genes in osteoclasts. J. Bone Miner. Res. 211387-1398. [DOI] [PubMed] [Google Scholar]
- 5.Chardin, P. 2006. Function and regulation of Rnd proteins. Nat. Rev. Mol. Cell Biol. 754-62. [DOI] [PubMed] [Google Scholar]
- 6.Chenette, E. J., A. Abo, and C. J. Der. 2005. Critical and distinct roles of amino- and carboxyl-terminal sequences in regulation of the biological activity of the Chp atypical Rho GTPase. J. Biol. Chem. 28013784-13792. [DOI] [PubMed] [Google Scholar]
- 7.Chenette, E. J., N. Y. Mitin, and C. J. Der. 2006. Multiple sequence elements facilitate Chp Rho GTPase subcellular location, membrane association, and transforming activity. Mol. Biol. Cell 173108-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colicelli, J. 2004. Human RAS superfamily proteins and related GTPases. Sci. STKE 250RE13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daigo, Y., I. Takayama, B. A. J. Ponder, C. Caldas, S. M. Ward, K. M. Sanders, and M. Fujino. 2004. Novel human, mouse and xenopus genes encoding a member of the Ras superfamily of low-molecular-weight GTP-binding proteins and its downregulation in W/Wv jejunum. J. Gastroenterol. Hepatol. 19211-217. [DOI] [PubMed] [Google Scholar]
- 10.DerMardirossian, C., and G. M. Bokoch. 2005. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 15356-363. [DOI] [PubMed] [Google Scholar]
- 11.Feng, Q., D. Baird, X. Peng, J. Wang, J.-L. Guan, and R. A. Cerione. 2006. Cool-1 functions as an essential regulatory node for EGF receptor- and Src-mediated cell growth. Nat. Cell Biol. 8945-956. [DOI] [PubMed] [Google Scholar]
- 12.Fukuhara, T., K. Shimizu, T. Kawakatsu, T. Fukuyama, Y. Minami, T. Honda, T. Hoshino, T. Yamada, H. Ogita, M. Okada, and Y. Takai. 2004. Activation of Cdc42 by trans interactions of the cell adhesion molecules nectins through c-Src and Cdc42-GEF FRG. J. Cell Biol. 116393-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gómez del Pulgar, T., S. A. Benitah, P. F. Valerón, C. Espina, and J. C. Lacal. 2005. Rho GTPase expression in tumourigenesis: evidence for a significant link. Bioessays 27602-613. [DOI] [PubMed] [Google Scholar]
- 14.Hancock, J. F. 2003. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 4373-384. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, S. H., M. M. Zegers, M. Woodrow, P. Rodrigues-Viciana, P. Chardin, K. E. Mostov, and M. McMahon. 2000. Induced expression of Rnd3 is associated with transformation of polarized epithelial cell by the Raf-MEK-extracellular signal-regulated kinase pathway. Mol. Cell. Biol. 209364-9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hildebrand, J. D., J. M. Taylor, and J. T. Parsons. 1996. An SH3 domain-containing GTPase-activating protein for Rho and Cdc42 associates with focal adhesion kinase. Mol. Cell. Biol. 163169-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilic, D., Y. Furuta, S. Kanazawa, N. Takeda, K. Sobue, N. Nakatsuji, S. Nomura, J. Fujimoto, M. Okada, and T. Yamamoto. 1995. Reduced motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature 377539-544. [DOI] [PubMed] [Google Scholar]
- 18.Jaffe, A. B., and A. Hall. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21247-269. [DOI] [PubMed] [Google Scholar]
- 19.Kirikoshi, H., and M. Katoh. 2002. Expression of Wrch1 in human cancer and down-regulation of Wrch1 by beta-estradiol in MCF-7 cells. Int. J. Oncol. 20777-783. [PubMed] [Google Scholar]
- 20.Klinghoffer, R. A., C. Sachsenmaier, J. A. Cooper, and P. Soriano. 1999. Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J. 182459-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakkakorpi, P. T., A. J. Bett, L. Lipfert, G. A. Rodan, and L. T. Duong. 2003. PYK2 autophosphorylation, but not kinase activity, is necessary for adhesion-induced association with c-Src, osteoclast spreading, and bone resorption. J. Biol. Chem. 27811502-11512. [DOI] [PubMed] [Google Scholar]
- 22.Lamarche, N., N. Tapon, L. Stowers, P. D. Burbelo, P. Aspenström, T. Bridges, J. Chant, and A. Hall. 1996. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell 87519-529. [DOI] [PubMed] [Google Scholar]
- 23.Liang, X., N. A. Draghi, and M. D. Resh. 2004. Signaling from integrins to Fyn to Rho family GTPases regulates morphologic differentiation of oligodendrocytes. J. Neurosci. 247140-7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linseman, D. A., F. Hofmann, and S. K. Fisher. 2000. A role for the small molecular weight GTPases, Rho and Cdc42, in muscarinic receptor signalling to focal adhesion kinase. J. Neurochem. 742010-2020. [DOI] [PubMed] [Google Scholar]
- 25.Meller, N., S. Merlot, and C. Guda. 2005. CZH proteins: a new family of Rho-GEFs. J. Cell Sci. 1184937-4946. [DOI] [PubMed] [Google Scholar]
- 26.Moon, S. Y., and Y. Zheng. 2003. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 1313-22. [DOI] [PubMed] [Google Scholar]
- 27.Mor, A., and M. R. Philips. 2006. Compartmentalized Ras/MAPK signaling. Annu. Rev. Immunol. 24771-800. [DOI] [PubMed] [Google Scholar]
- 28.Murakami, T., M. Fujimoto, M. Ohtsuki, and H. Nakagawa. 2001. Expression profiling of cancer-related genes in human keratinocytes following non-lethal ultraviolet B irradiation. J. Dermacol. Sci. 27121-129. [DOI] [PubMed] [Google Scholar]
- 29.Parsons, J. T. 2003. Focal adhesion kinase: the first ten years. J. Cell Sci. 1161409-1416. [DOI] [PubMed] [Google Scholar]
- 30.Ren, X.-R., Q.-S. Du, Y.-Z. Huang, S. Z. Ao, L. Mei, and W. C. Xiong. 2001. Regulation of Cdc42 GTPase by proline-rich tyrosine kinase 2 interacting with PSGAP, a novel pleckstrin homology and Src homology 3 domain containing protein. J. Cell Biol. 152971-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riento, K., R. M. Guasch, R. Garg, B. Jin, and A. J. Ridley. 2003. RhoE binds to ROCK I and inhibits downstream signaling. Mol. Cell. Biol. 234219-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riento, K., N. Totty, P. Villalonga, R. Garg, R. Guasch, and A. J. Ridley. 2005. RhoE function is regulated by ROCK I-mediated phosphorylation. EMBO J. 241170-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossman, K. L., C. J. Der, and J. Sondek. 2005. GEF means go: turning on Rho GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6167-180. [DOI] [PubMed] [Google Scholar]
- 34.Saras, J., P. Wollberg, and P. Aspenström. 2004. Wrch1 is a GTPase-deficient Cdc42-like protein with unusual binding characteristics and cellular effect. Exp. Cell Res. 299356-369. [DOI] [PubMed] [Google Scholar]
- 35.Shutes, A., A. C. Berzat, A. D. Cox, and C. J. Der. 2004. Atypical mechanisms of regulation of the Wrch-1 Rho family small GTPase. Curr. Biol. 142052-2056. [DOI] [PubMed] [Google Scholar]
- 36.Tao, W., D. Pennica, L. Xu, R. F. Kalejata, and A. J. Levine. 2001. Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev. 151769-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timpson, P., G. E. Jones, M. C. Frame, and V. G. Brunton. 2001. Coordination of cell polarization and migration by the Rho family GTPases requires Src tyrosine kinase activity. Curr. Biol. 111836-1846. [DOI] [PubMed] [Google Scholar]
- 38.Tu, S., W. J. Wu, J. Wang, and R. A. Cerione. 2003. Epidermal growth factor-dependent regulation of Cdc42 is mediated by the Src tyrosine kinase. J. Biol. Chem. 27849293-49300. [DOI] [PubMed] [Google Scholar]
- 39.Wennerberg, K., and C. J. Der. 2004. Rho-family GTPases: it's not only Rac and Rho (and I like it). J. Cell Sci. 1171301-1312. [DOI] [PubMed] [Google Scholar]
- 40.Wherlock, M., and H. Mellor. 2002. The Rho GTPase family: a Racs to Wrchs story. J. Cell Sci. 115239-240. [DOI] [PubMed] [Google Scholar]
- 41.Wozniak, M. A., K. Modzelewska, L. Kwong, and P. J. Keely. 2004. Focal adhesion regulation of cell behaviour. Biochim. Biophys. Acta 1692103-119. [DOI] [PubMed] [Google Scholar]
- 42.Zhai, J., H. Lin, Z. Nie, J. Wu, R. Cañete-Soler, W. W. Schlaepfer, and D. D. Schlaepfer. 2003. Direct interaction of focal adhesion kinase with p190RhoGEF. J. Biol. Chem. 27824865-24873. [DOI] [PubMed] [Google Scholar]