Abstract
Rad23 is required for efficient protein degradation and performs an important role in nucleotide excision repair. Saccharomyces cerevisiae Rad23, and its human counterpart (hHR23), are present in a complex containing the DNA repair factor Rad4 (termed XPC, for xeroderma pigmentosum group C, in humans). XPC/hHR23 was also reported to bind centrin-2, a member of the superfamily of calcium-binding EF-hand proteins. We report here that yeast centrin, which is encoded by CDC31, is similarly present in a complex with Rad4/Rad23 (called NEF2). The interaction between Cdc31 and Rad23/Rad4 varied by growth phase and reflected oscillations in Cdc31 levels. Strikingly, a cdc31 mutant that formed a weaker interaction with Rad4 showed sensitivity to UV light. Based on the dual function of Rad23, in both DNA repair and protein degradation, we questioned if Cdc31 also participated in protein degradation. We report here that Cdc31 binds the proteasome and multiubiquitinated proteins through its carboxy-terminal EF-hand motifs. Moreover, cdc31 mutants were highly sensitive to drugs that cause protein damage, failed to efficiently degrade proteolytic substrates, and formed altered interactions with the proteasome. These findings reveal for the first time a new role for centrin/Cdc31 in protein degradation.
The microtubule organizing center regulates cytoskeletal integrity, and is termed the centrosome in higher eukaryotes, and the spindle pole-body (SPB) in yeast. The duplication of the SPB/centrosome is a key event that initiates entry into the cell cycle. The SPB is a proteinaceous structure that is embedded in the nuclear envelope, and its duplication marks exit from the G1 phase and the onset of chromosome duplication. The composition of the SPB has been defined, and the steps that lead to its duplication have been studied extensively using biochemical, genetic, and microscopic methods (3, 5, 6, 18, 24, 28, 30, 43, 44). An important component of the SPB is Cdc31 (3, 13), which in yeast is encoded by the essential CDC31 gene. CDC31 was isolated as a suppressor of kar1, a mutant that is unable to duplicate the SPB at the nonpermissive temperature (6). As with kar1, defects in CDC31 (cdc31) also cause G1 phase arrest, due to a failure in SPB duplication (48). Intriguingly, the kar1 defect was suppressed by Dsk2 (5), which has functional and structural resemblance to Rad23 (15, 29, 34, 40).
The three centrin proteins that are expressed in human cells (30, 31) display differential tissue-specific levels (27, 36, 49). However, all three isoforms share the key structural features that are present in EF-hand proteins (32). Human CEN3 is closely related to Cdc31 and can interfere with SPB duplication when overexpressed in yeast (30). An N-terminal regulatory domain is proposed to regulate the activity of the C-terminal domain, possibly through Ca2+-mediated signaling (42). The C terminus binds various cellular proteins (1, 7, 11, 12, 16, 19-23, 28, 37, 46, 50), although the effect of these interactions in regulating growth control is not well understood. We found that the C-terminal domain of Cdc31 binds Rad4, and this interaction does not require Rad23. However, our findings indicate that Cdc31 forms regulated interactions with a preexisting complex comprising Rad23 and Rad4 that varies under different growth conditions. A mutant form of Cdc31, which formed a weak interaction with Rad4, showed reduced tolerance to UV-induced DNA damage.
We report for the first time that centrin/Cdc31 plays an important role in protein degradation by the ubiquitin/proteasome system. Specifically, we discovered that Cdc31 binds the proteasome and can be purified with multiubiquitinated proteins. Moreover, mutant Cdc31 proteins had altered interactions with both proteasomes and ubiquitinated substrates and conferred proteolytic defects. A role for Cdc31 in protein degradation was implied by its interactions with Dsk2 (5), a protein that functions in the ubiquitin/proteasome pathway (15, 39, 40). We showed that a related protein, Rad23, is a shuttle factor that can translocate ubiquitinated proteins to the proteasome (8). The loss of both shuttle factors (rad23Δ dsk2Δ) causes a temperature-sensitive growth defect resulting from a failure to duplicate the SPB (5). Strikingly, high-copy-number expression of Cdc31 could overcome the SPB duplication defect of rad23Δ dsk2Δ (23) and predicted a role for protein degradation in regulating entry into the cell cycle.
The presence of a single essential gene in Saccharomyces cerevisiae that encodes a centrin protein provides a genetically tractable system for characterizing the link between the cellular responses to stress and damage and the signaling pathways that control growth progression. For instance, DNA damage-induced growth arrest could be facilitated by blocking SPB duplication (1) in a mechanism involving the protein degradation function of Cdc31.
MATERIALS AND METHODS
Plasmids and strains.
Yeast genes were amplified using PCRs and cloned into plasmid YEplac181 with an amino-terminal FLAG epitope. The PCR products were cloned using 5′ EcoRI and 3′ KpnI DNA restriction sites and expressed from the copper-inducible PCUP1 promoter. Additionally, fragments containing 5′ BamHI and 3′ EcoRI DNA restriction sites were cloned into pGEX2TK vector, and the expression of glutathione S-transferase (GST)-fusion proteins in Escherichia coli was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to the growth medium. DNA oligonucleotide primers were used to isolate full-length CDC31 as well as the mutant genes using chromosomal DNA prepared from the relevant strains. Following cloning of the wild-type gene, the amino- and carboxy-terminal fragments were also generated by PCR. These constructs expressed the amino-terminal EF hands (amino acid residues 1 to 99) or the carboxy-terminal EF hands (residues 85 to 161). All PCR-amplified DNAs were examined by DNA sequencing, and the sequences of mutant genes were confirmed. The cdc31-1 and cdc31-2 mutant strains as well as a cdc31Δ strain were provided by B. Davis and M. Rose (Princeton University). A plasmid for expressing Pre2-hemagglutinin (HA) from its endogenous chromosomal locus was obtained from J. Dohmen (University of Cologne). DNA plasmids encoding His-tagged derivatives of Rpt1 to Rpt6 were a generous gift from D. Skowyra (St. Louis University).
Antibodies.
Antibodies against ubiquitin (polyclonal) and FLAG (FLAG-M2-agarose and monoclonal anti-FLAG-horseradish peroxidase) were purchased from Sigma Chemical Co. (St. Louis, MO). Monoclonal antibodies against HA as well as anti-HA affinity matrix were obtained from Roche. Antibodies against the His tag were purchased from BD Biosciences (San Jose, CA). Polyclonal antibodies against yeast Cdc31, Rpt1, and Rad23 were generated by Pocono Laboratories, using recombinant Cdc31, GST-Rpt1, and GST-Rad23, respectively. Polyclonal anti-Rpn12 was generously provided by D. Skowyra (St. Louis University). Monoclonal antibodies against β-galactosidase were purchased from Promega (Madison, WI).
Preparation of protein extracts and immunological methods.
Yeast strains expressing plasmids were grown in synthetic medium and pelleted in late-logarithmic phase (A600 of ∼4). Yeast cell pellets were suspended in 500 μl of lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM EDTA, and 1% Triton X-100) containing a protease inhibitor cocktail (Roche). In studies where we characterized the ubiquitination of Cdc31, we added 40 mM N-ethylmaleimide to the lysis buffer. The suspension was combined with 3 g of 0.45 mM glass beads and lysed by agitation in a Fast Prep PF120 disruptor. The supernatant was collected following 2 min of centrifugation at 12,000 × g at 4°C. Protein concentration was determined using the Bradford assay (Bio-Rad). Equal amounts of protein extract were applied to anti-FLAG-M2 agarose to immunoprecipitate FLAG-tagged proteins or applied to anti-HA affinity matrix to immunoprecipitate proteasome subunit Pre2-HA. Similarly, equal amounts of protein extract were combined with glutathione-Sepharose (GE Healthcare) to purify GST-tagged proteins from E. coli BL21 cells. His-tagged Rpt1 to Rpt6 proteins were also expressed in strain BL21 in the presence of 1 mM IPTG, and cells were lysed by sonication. Equal amounts of total protein extract were combined with recombinant GST-Cdc31 for 2 h to examine interaction. The beads were washed three times with 1 ml of lysis buffer containing an additional 1% Triton X-100, and the immobilized proteins were released by boiling in sodium dodecyl sulfate (SDS)-containing electrophoresis sample buffer. The proteins were resolved in 12% SDS-tricine polyacrylamide gels, transferred to nitrocellulose, and characterized by immunoblotting.
Protein stability and drug sensitivity measurements.
We measured protein stability by using [35S]methionine pulse-chase methods, as described previously (35). To determine sensitivity of cdc31 mutants to protein synthesis and translation inhibitors, yeast cells were grown to logarithmic phase, serial 10-fold dilutions were prepared in sterile water, and 3-μl aliquots were spotted on agar medium containing 0.5 μg/ml cycloheximide or 0.2 mM hygromycin B. The plates were incubated at 30°C for 3 to 5 days.
Two-dimensional (2D) electrophoresis.
FLAG-Cdc31 was immunoprecipitated (from 500 μg of total yeast protein) using anti-FLAG-agarose beads and washed three times with lysis buffer and two times with 20 mM Tris, pH 7.0. Similarly, GST-UBA1 and GST-UBA2 were purified from yeast extracts on glutathione-Sepharose beads. FLAG-agarose and glutathione-Sepharose beads were solubilized for 9 h in 200 μl of rehydration buffer [8 M urea, 0.5% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate), 0. 2% dithiothreitol (DTT)], 0.002% bromophenol blue) containing 2% immobilized polyacrylamide gel buffer (ampholine; pH 3 to 10; GE Healthcare, NJ). The beads were removed following brief centrifugation, and the supernatants were transferred to a DryStrip Re-swelling Tray (GE Healthcare) containing an 11-cm Immobiline DryStrip gel (pH 3 to 10, linear; GE Healthcare). The protein solutions were allowed to soak into the DryStrip for 16 h, and the gels were resolved in the first dimension for 5 h using Multiphor II (at 15 kV/h).
Proteins were resolved by isoelectric focusing, and the gel strips were rinsed in 50 ml of equilibration buffer 1 (2% SDS, 50 mM Tris, pH 8.8, 6 M urea, 30% glycerol, 0.002% bromophenol blue, 1% DTT) for 15 min, followed by a second wash in 50 ml of equilibration buffer 2 (equilibration buffer 1 lacking DTT and containing 4% iodoacetamide) for an additional 15 min. The gels were then layered onto an SDS-polyacrylamide gel and separated in the second dimension. The resolved proteins were transferred to nitrocellulose and examined by immunoblotting using antibodies against Cdc31 and ubiquitin.
RESULTS
Cdc31 binds Rad4.
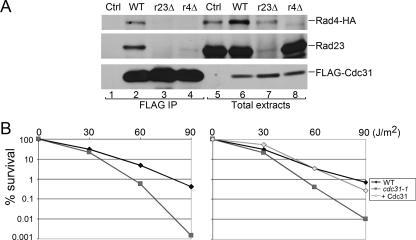
FLAG-Cdc31 was expressed from a plasmid and immunoprecipitated from yeast extracts. The interaction with Rad4 was determined by measuring the copurification of an epitope-tagged derivative (Rad4-HA) that was expressed at physiological levels from the chromosome (35). The expression of FLAG-Cdc31 was similar in total extracts prepared from wild type (Fig. 1A, lane 6), rad23Δ (lane 7), and rad4Δ (lane 8). We also examined a control yeast strain that did not express FLAG-Cdc31 (lane 5). Protein extracts were incubated with FLAG-agarose, and the immunoprecipitated proteins were characterized by immunoblotting (Fig. 1A, lanes 1 to 4). We found that FLAG-Cdc31 could copurify Rad4-HA in a wild-type strain (lane 2), consistent with the reported interaction between purified human xeroderma pigmentosum group C (XPC) and centrin-2 (1). The immunoblot was subsequently incubated with antibodies against Rad23, and this nucleotide excision repair (NER) factor was also coprecipitated (lane 2), confirming the formation of a trimeric complex containing Rad23/Rad4 (NEF2) and Cdc31. To determine if Cdc31/NEF2 interaction required Rad23, we measured FLAG-Cdc31 interactions with Rad4-HA and Rad23 in rad23Δ and rad4Δ strains, respectively. Following immunoprecipitation of FLAG-Cdc31, we detected low levels of Rad4-HA in rad23Δ cells, demonstrating that Cdc31 can bind Rad4 directly (lane 3). The significantly lower abundance of Rad4-HA in rad23Δ is due to its rapid degradation by the ubiquitin/proteasome system (35). In contrast, immunoprecipitation of FLAG-Cdc31 from rad4Δ did not copurify Rad23, demonstrating that the trimeric complex involves a specific interaction between Cdc31 and Rad4 and a separate interaction between Rad4 and Rad23. Rad23 expression was unaffected in rad4Δ (Fig. 1A, lane 8). (A weak reaction seen in lanes 4 and 7 of Fig. 1A is a nonspecific interaction with anti-Rad23 antibodies against an unrelated protein.)
FIG. 1.
Cdc31 interacts with the NEF2 (Rad4/Rad23) DNA repair complex. (A) FLAG-Cdc31 was expressed in wild-type, rad23Δ (r23Δ), and rad4Δ (r4Δ) strains. A strain lacking a FLAG-tagged protein was also examined (lanes 1 and 5). Total protein extracts were examined (lanes 5 to 8). Equal amounts of protein extract were incubated with FLAG-agarose, and the bound proteins were examined by immunoblotting. The expression of FLAG-Cdc31 was similar in all the strains. Similarly, the abundance of Rad23 was unaffected by the loss of Rad4. However, Rad4-HA levels were decreased in the rad23Δ strain, as noted previously. Following immunoprecipitation of FLAG-Cdc31, both Rad23 and Rad4-HA were copurified from a wild-type strain (lane 2). Low levels of Rad4-HA were also detected in rad23Δ (lane 3), although a significantly lower amount of Rad4 accumulated in this mutant (compare lanes 6 and 7). None of the proteins was nonspecifically bound to the beads (lane 1). (B) The UV survival of centrin mutant cdc31-1 was compared to the wild-type strain. Yeast cells were grown to exponential phase and then transferred to 37°C for 1 h. The cells were pelleted and plated on synthetic complete agar medium and irradiated at 1.5 J/m2 for 0 to 90 s. Cell survival was determined in two independent experiments, and the mean values are shown. cdc31-1 consistently showed moderate UV sensitivity at higher UV doses. However, this sensitivity was completely overcome when cdc31-1 was transformed with a plasmid expressing wild-type Cdc31 (right panel). WT, wild type; IP, immunoprecipitation; Ctrl, control.
cdc31-1 mutant is sensitive to UV light.
The role of Rad23/Rad4 in DNA repair is well characterized (33). Based on the formation of a Rad23/Rad4/Cdc31 complex, we investigated if cdc31 mutants were sensitive to UV-induced DNA damage. Because CDC31 is essential for viability, we characterized the UV sensitivity of a temperature-sensitive mutant (cdc31-1, a gift from M. Rose, Princeton University) (48). Yeast cultures were grown at the permissive temperature (23°C) for 15 h and then transferred to fresh medium that was preequilibrated to 37°C. After a 1-h incubation the cells were pelleted, suspended in phosphate-buffered saline, plated, and exposed to 254-nm UV light. The plates were stored in the dark at 23°C for 4 days. We found that the cdc31-1 strain displayed a dose-dependent loss of viability following DNA damage (Fig. 1B, left graph). A potential role in DNA repair is consistent with a previous report, which showed that centrin-2 could stimulate the DNA incision activity of XPC (33). Expression of wild-type Cdc31 (FLAG-Cdc31) in cdc31-1 fully restored UV resistance to normal levels (Fig. 1B, right graph). Although the cdc31-1 strain is not as sensitive to UV-induced DNA damage as most NER mutants (38), we note that the brief incubation at the nonpermissive temperature can only provide an approximation, since the cells resume repair of DNA lesions following their return to the permissive temperature. The characterization of cdc31-2 (48) was inconclusive due to the poor growth of this temperature-sensitive mutant.
Growth-specific interactions between Cdc31 and NEF2.
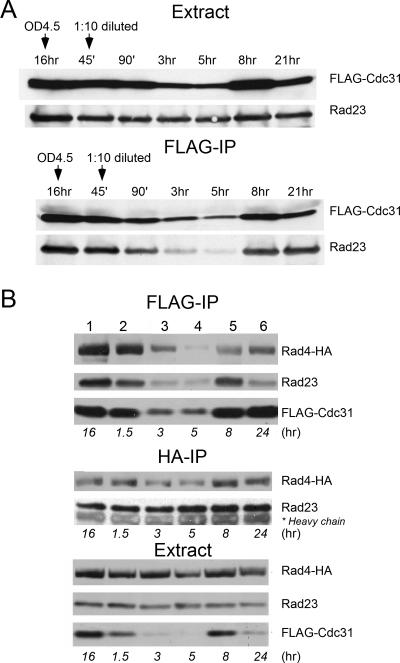
Because of the important role of Cdc31 in regulating entry into the cell cycle, we examined its expression during cell cycle progression and its interaction with the NEF2 DNA repair complex. In preliminary studies, we treated yeast cells with alpha-factor mating pheromone, hydroxyurea, or nocodozole to arrest growth at different cell cycle stages. We examined the interaction between FLAG-Cdc31 and Rad23/Rad4 at different cell cycle stages and found that Cdc31 levels and the interaction of Cdc31 with Rad23/Rad4 were constant during the cell cycle (data not shown). However, these exploratory studies revealed significant changes in the abundance of soluble FLAG-Cdc31 in different growth phases. To further explore this finding, we diluted early-stationary-phase cells (A600 of ∼4) into fresh medium and periodically withdrew aliquots from the culture. Protein extracts were prepared and examined by immunoblotting. We initially measured the level of the NEF2 complex that was purified with FLAG-Cdc31 (Fig. 2A). We examined the copurification of Rad23 with FLAG-Cdc31 since all cellular Rad4 is bound to Rad23 (1, 17). Rad23 levels were similar in exponential and stationary phases of growth (Fig. 2A, Extract), while FLAG-Cdc31 levels decreased markedly in actively growing cells (Fig. 2A, Extract). Consequently, the level of Rad23 that was purified with FLAG-Cdc31 was noticeably reduced in actively growing cells (Fig. 2A, FLAG-IP). FLAG-Cdc31 levels increased in stationary phase (Fig. 2A, at 8 and 21 h of growth), and correspondingly higher amounts of Rad23 were copurified.
FIG. 2.
Growth phase oscillations in Cdc31 levels. (A) A wild-type yeast strain expressing FLAG-Cdc31 was grown to late logarithmic phase and then diluted 10-fold into fresh medium. Aliquots were withdrawn at the times indicated. Total protein extract was prepared and examined by immunoblotting. The abundance of Rad23 (that was expressed at physiological levels) was unaffected in different growth stages in total extract (upper panel). In contrast, a noticeable decrease in FLAG-Cdc31 levels is evident by 3 h following transfer to fresh medium. An equal amount of extract was also precipitated on FLAG-agarose, and the copurification of Rad23 was tested. Because Cdc31 binds Rad4 (which is in a complex with Rad23), this assay measured the association of Cdc31 with both proteins. Decreased levels of FLAG-Cdc31 in actively growing cells are reflected by a proportionate reduction in the amount of Rad23 that was copurified (lower panel). (B) To directly test the interaction with Rad4-HA that was expressed at physiological levels, we transformed FLAG-Cdc31 into a wild-type strain containing integrated RAD4-HA. As described in panel A, Cdc31 was purified on FLAG-agarose, and the copurification of both Rad4-HA and Rad23 was determined (FLAG-IP). Lower expression of FLAG-Cdc31 (Extract) significantly reduced the amounts of Rad4-HA and Rad23 that were copurified (FLAG-IP, lanes 3 and 4). The same extracts were also incubated with HA-agarose to measure the interaction between Rad23 and Rad4. We found that the formation of the NEF2 complex was unaffected by growth phase (middle panel), suggesting that Cdc31 interacts with a preassembled Rad4/Rad23 complex. IP, immunoprecipitation; OD, optical density.
To examine Cdc31 interaction with Rad4-HA, we generated an independent set of strains that expressed Rad4-HA from the chromosome (35). Yeast cells were grown to stationary phase and then diluted into fresh medium. In agreement with earlier results (Fig. 2A), we found that FLAG-Cdc31 levels decreased in exponential-phase cells (Fig. 2B, Extract, lanes 3 and 4) but recovered in stationary phase. Consequently, immunoprecipitation of FLAG-Cdc31 yielded dramatically reduced amounts of both NEF2 components (Fig. 2B, FLAG-IP, Rad23 and Rad4-HA) during active growth (lanes 3 and 4). However, immunoprecipitation with anti-HA antibodies revealed constant amounts of Rad23 and Rad4-HA complex formation (Fig. 2B, HA-IP), suggesting that Cdc31 has regulated interactions with a preexisting complex of Rad23/Rad4.
Cdc31 can bind the 26S proteasome and multiubiquitinated proteins.
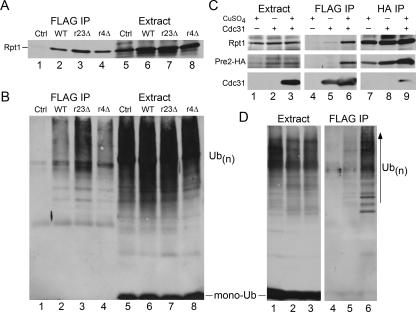
Rad23 and Dsk2 are substrate shuttle factors that perform significant roles in protein degradation (8, 9, 26, 39, 40). Because Cdc31 can genetically and biochemically interact with both shuttle factors, we investigated if Cdc31 might similarly contribute to proteolysis. We examined the immunoblots shown in Fig. 1A and found that the proteasome subunit Rpt1 was copurified with FLAG-Cdc31 (Fig. 3A, lane 2). Extracts prepared from a wild-type strain lacking FLAG-Cdc31 were also applied to FLAG-agarose, and Rpt1 was not detected (lane 1). Purification of FLAG-Cdc31 from rad23Δ (lane 3) and rad4Δ (lane 4) yielded similar amounts of Rpt1 compared to wild-type extracts (lane 2), demonstrating that Cdc31 interaction with the proteasome does not require Rad23. The same immunoblots were subsequently incubated with antibodies against ubiquitin. Despite the similar overall levels of high-molecular-weight multiubiquitinated proteins in wild-type and rad23Δ strains (Fig. 3B, lanes 6 and 7), noticeably higher amounts were purified with FLAG-Cdc31 from rad23Δ (compare lanes 2 and 3).
FIG. 3.
Cdc31 binds the proteasome and multiubiquitinated proteins. (A) FLAG-Cdc31 was immunoprecipitated, and an immunoblot was incubated with antibodies against proteasome subunit Rpt1. FLAG-Cdc31 coprecipitated the proteasome in all strains examined (lanes 2 to 4). The expression of Rpt1 was similar in all the strains, and nonspecific interaction with the matrix was not observed (lane 1). (B) The same filters were subsequently incubated with antibodies against ubiquitin, and high-molecular-weight ubiquitinated species were detected in total extracts (lanes 5 to 8). However, higher levels were recovered with FLAG-Cdc31 from rad23Δ cells (r23Δ; lane 3). (C) To confirm that Cdc31 could bind the intact proteasome, we also examine the levels of a subunit in the 20S proteasome catalytic particle. The expression of FLAG-Cdc31 was strongly induced by the addition of 100 μM CuSO4 (+) to the medium (Extract, lane 3). Immunoprecipitation of FLAG-Cdc31 yielded low levels of Rpt1 and Pre2-HA in uninduced cells (lane 5) and significantly higher levels following copper induction (lane 6). In a reciprocal assay, we immunoprecipitated the proteasome using antibodies against Pre2-HA and detected FLAG-Cdc31 (lane 9). Purification of Pre2-HA yielded the 19S proteasome subunit Rpt1 under all conditions examined (lanes 7 to 9). (D) The concentration-dependent interaction between Cdc31 and multiubiquitinated proteins was confirmed following copper induction of FLAG-Cdc31. The level of total multiubiquitinated proteins was similar in a strain lacking FLAG-Cdc31 and when FLAG-Cdc31 was expressed at different levels. However, following incubation with FLAG-agarose, significantly higher amounts of ubiquitinated species were isolated with FLAG-Cdc31. WT, wild type; IP, immunoprecipitation; Ctrl, control; Ub, ubiquitin; Ub(n), multiubiquitin; r4Δ, rad4Δ.
To verify the proteasome and ubiquitin binding properties of Cdc31 we conducted a reciprocal immunoprecipitation using an integrated derivative of the proteasome subunit Pre2-HA (a gift from J. Dohmen, University of Cologne). We also controlled the expression of FLAG-Cdc31 by using the copper-inducible PCUP1 promoter. Yeast cultures were grown in the presence or absence of copper sulfate, and protein extracts were incubated with FLAG- or HA-agarose to purify FLAG-Cdc31 and Pre2-HA, respectively. The expression of FLAG-Cdc31 was strongly increased by the addition of 100 μM CuSO4 to the growth medium (Fig. 3C, lane 3). The expression of proteasome subunits Rpt1 (in the 19S regulatory particle) and Pre2-HA (in the 20S regulatory particle) were unaffected by high-level expression of FLAG-Cdc31 (Fig. 3C, Extract). Low levels of Rpt1 and Pre2-HA were coimmunoprecipitated with FLAG-Cdc31 from uninduced cells (Fig. 3C, lane 5). Significantly higher levels of these proteasome subunits were purified when the expression of FLAG-Cdc31 levels was increased (lane 6). In the reciprocal study, Pre2-HA was expressed at physiological levels and precipitated, and the copurification of FLAG-Cdc31 was confirmed in extracts containing FLAG-Cdc31 (Fig. 3C, lane 9).
The dose-dependent interaction between FLAG-Cdc31 and multiubiquitinated proteins was also tested by immunoblotting. In the absence of copper sulfate, only low levels of multiubiquitinated proteins were copurified with FLAG-Cdc31 (Fig. 3D, lane 5). In contrast, significantly higher levels were copurified when FLAG-Cdc31 levels were induced in the presence of copper sulfate (lane 6). Overall levels of multiubiquitinated proteins were not affected by high expression of Cdc31 (Fig. 3D, Extract). These findings do not establish whether Cdc31 binds multiubiquitinated proteins directly or whether the multiubiquitinated proteins are associated with proteasomes that are efficiently bound by Cdc31.
Significant proteolytic defects in cdc31 mutant strains.
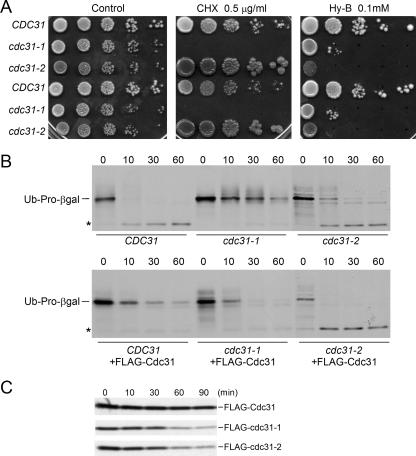
To examine the proteolytic defects of cdc31 mutants, we measured the sensitivity of the cdc31-1 and cdc31-2 strains to drugs that increase the levels of misfolded and truncated proteins. We determined that the cdc31-1 mutant is unable to grow in medium containing cycloheximide (Fig. 4A). In contrast, cdc31-2 displayed strong resistance that is caused by a secondary mutation in the CYH2 gene (B. Davis and M. Rose, personal communication), which confers resistance to cycloheximide. To verify the sensitivity of cdc31 mutants to translation inhibitors, we tested growth in medium containing hygromycin B. Both mutant strains showed strong sensitivity to this drug (Fig. 4A). Transformation of a plasmid expressing Cdc31 restored resistance to hygromycin B in both cdc31-1 and cdc31-2, demonstrating that the sensitivity to translation inhibitors is related to cdc31 (data not shown). Because mutations in the ubiquitin/proteasome pathway can cause sensitivity to drugs that generate damaged protein (10, 41), these findings support the hypothesis that Cdc31 functions in protein degradation.
FIG. 4.
A protein degradation defect in Cdc31 mutants. (A) The sensitivity of Cdc31 mutants to translation inhibitors is shown. Tenfold dilutions were spotted (in duplicate sets) on medium containing either 0.5 μg/ml cycloheximide (CHX) or 0.2 mM hygromycin B (Hy-B). (B) A plasmid encoding the proteasome substrate ubiquitin (Ub)-Pro-β-Gal was transformed into CDC31, cdc31-1, and cdc31-2 strains. Cultures were grown in 2% galactose and labeled with [35S]methionine-cysteine for 5 min. Aliquots were withdrawn at the times indicated during the chase. Equal amounts of radiolabeled protein were immunoprecipitated by anti-β-galactosidase antibody, resolved by SDS-PAGE, and examined by autoradiography. An asterisk signifies a stable degradation product. The upper panel shows stabilization of ubiquitin-Pro-β-Gal in cdc31 mutants. The lower panel shows that degradation of this substrate is restored after the wild-type Cdc31 was expressed in the three strains. (C) Pulse-chase measurements were conducted to determine the stability of wild-type and mutant cdc31 proteins. These proteins were expressed from a plasmid in a wild-type strain to avoid the growth defects of cdc31-1 and cdc31-2.
To further test this model, we measured the in vivo stability of a test protein of the ubiquitin/proteasome system. CDC31, cdc31-1, and cdc31-2 strains were transformed with a plasmid that expressed ubiquitin-Pro-β-galactosidase (ubiquitin-Pro-β-Gal), a well-studied reporter substrate (2). Protein stability was measured by 35S pulse-chase methods (8). Ubiquitin-Pro-β-Gal was rapidly degraded in CDC31, with a calculated half-life of <3 min during the initial 10 min of the chase (Fig. 4B). In contrast, ubiquitin-Pro-β-Gal was strongly stabilized in the cdc31-1 mutant, and the accumulation of multiubiquitinated intermediates was noticeable in the 10- and 30-min chase time-points. Ubiquitin-Pro-β-Gal was stabilized to a lesser degree in cdc31-2, although higher levels were detected in the first two time points in (0 and 10 min of chase). The degradation of ubiquitin-Pro-β-Gal in cdc31 mutants was restored following reexpression of Cdc31 from a plasmid (Fig. 4B, lower panel), confirming its requirement for efficient protein degradation. Surprisingly, overexpression of Cdc31 in CDC31 caused modest stabilization of ubiquitin-Pro-β-Gal, suggesting that both depletion and overexpression of Cdc31 can affect protein degradation. There is precedence for this observation. For instance, both depletion and overexpression of Ndc1 and Dsk2 proteins can disrupt SPB duplication (5). Additionally, both high and low levels of Kar1 interfere with Cdc31 localization to the SPB (6).
In an effort to understand the proteolytic defects of the cdc31 mutants, we examined the in vivo stabilities of FLAG-cdc31-1 and FLAG-cdc31-2 proteins. We found that wild-type Cdc31 was stable, in agreement with a previous report (5). In contrast, both mutant proteins were more unstable than the wild-type protein (Fig. 4C), suggesting that the depletion of cdc31 mutant proteins could cause the defects in the mutant strains. Centrin has been reported to oligomerize, although it is not known if the cdc31-1 and cdc31-2 mutant proteins are more prone to this effect and whether this could alter their stabilities. Because we conducted our studies in a wild-type background (to avoid the growth defects of cdc31-1 and cdc31-2), it is conceivable that an association with native Cdc31 could affect the stability of mutant Cdc31 proteins.
Domain-structure analysis of Cdc31 and the defect of mutants.
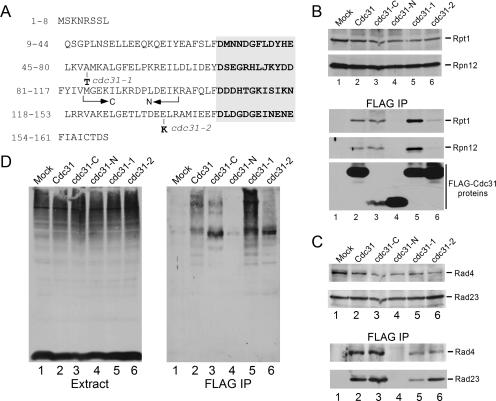
Allele-specific functional differences in cdc31 mutants may be due to the location of mutations in distinct regions of this multidomain protein. Numerous studies have shown that the carboxy terminus of centrin plays a key role in binding cellular partners, while the N terminus performs a regulatory role. The unique defects of cdc31 alleles are evidenced by the ability of Pkc1 to suppress only cdc31-2 but not cdc31-1 or cdc31-5 (23). We confirmed that the amino acid change in cdc31-1 is located between EF-1 and EF-2 in the amino-terminal regulatory domain (A48→T48), and the mutation in cdc31-2 is present between EF-3 and EF-4 in the carboxy-terminal Ca2+-binding fold (E133→K133) (48) (Fig. 5A).
FIG. 5.
Domain-structure analysis reveals strong interaction between the carboxy terminus of Cdc31 and the ubiquitin/proteasome system. (A) The amino acid sequence of the Cdc31 protein is shown. The conserved EF hands are aligned on the right in bold. The residues that are mutated in cdc31-1 and cdc31-2 are also indicated. The arrows indicate the regions that were cloned to generate the amino- and carboxy-terminal fragments. EF-1 and EF-2 hands are present in the amino-terminal domain (cdc31-N), while EF-3 and EF-4 domains are present in the carboxy-terminal domain (cdc31-C). (B) The expression of proteasome subunits (Rpt1 and Rpn12) was unaffected by expression of the Cdc31 proteins and truncated polypeptides (upper panel). However, expression of cdc31-C was reduced 10- to 20-fold compared to the full-length proteins or cdc31-N (see lower panel for expression levels of FLAG-Cdc31 proteins). Following immunoprecipitation, cdc31-C showed similar binding to the proteasome (FLAG-IP, lane 3) compared to the wild-type protein (lane 2). Interestingly, the cdc31-1 mutant protein interacted more strongly than the wild-type protein with the proteasome (lane 5), while cdc31-2 had a much weaker interaction (lane 6) than the wild-type protein. Lane 1 shows the absence of a nonspecific interaction with the matrix in a strain lacking a FLAG-tagged protein. (C) The same Cdc31 constructs were expressed in yeast containing Rad4-HA. The expression of Rad4-HA and Rad23 was similar in a wild-type strain that expressed the various Cdc31 derivatives (upper panel). Following immunoprecipitation on FLAG-agarose, we detected a strong interaction with cdc31-C (FLAG-IP, lane 3) and no interaction with the amino terminus (cdc31-N, lane 4). In contrast to proteasome binding, FLAG-cdc31-1 and FLAG-cdc31-2 showed weaker interactions with both Rad4 and Rad23 (lower panel). (D) An immunoblot containing total protein was incubated with antibodies against ubiquitin. The abundance of high-molecular-weight ubiquitinated proteins was similar in all strains (left panel). However, following immunoprecipitation, FLAG-cdc31-1 showed a significant increase in ubiquitinated proteins. Similarly, cdc31-C, but not cdc31-N, copurification of purified multiubiquitinated proteins (right panel). IP, immunoprecipitation.
The functionally distinct pairs of EF hands in centrin/Cdc31 proteins are separated by a flexible linker (4, 16, 32, 42). Previous studies showed that the carboxy-terminal domain of centrin proteins (containing EF-3 and EF-4) is required for Ca2+ binding and interaction with XPC (37). We generated a set of Cdc31 derivatives that expressed full-length Cdc31 and the amino (cdc31-N) and carboxy (cdc31-C) domains. We also expressed the cdc31-1 and cdc31-2 mutant proteins (Fig. 5A). These proteins contained the FLAG epitope and were expressed in wild-type yeast. The expression of cdc31-C was ∼10- to 20-fold lower than the wild-type protein (Fig. 5B, bottom panel, lane 3). An equal amount of protein extract was incubated with FLAG-agarose, and the bound proteins were examined by immunoblotting. The filter was reacted sequentially with antibodies against proteasome subunits Rpt1 and Rpn12. We found that the expression of proteasome subunits was not affected by the Cdc31 derivatives (Fig. 5B, upper panels). However, the Cdc31 derivatives formed markedly different interactions with the proteasome. Although cdc31-C was expressed at much lower levels than full-length Cdc31 (Fig. 5B, FLAG IP; compare lanes 2 and 3), it formed a very efficient interaction with the proteasome. In contrast, the amino terminal domain (cdc31-N) failed to bind the proteasome (Fig. 5B, FLAG-IP, lane 4), despite high levels of expression. We purified mutant cdc31 proteins and found that FLAG-cdc31-1 (bearing a mutation in the amino terminus) formed a stronger interaction with the proteasome (lane 5) than wild-type Cdc31, while FLAG-cdc31-2 (that contains a mutation in the carboxy terminus) had reduced binding (lane 6).
With the availability of the above-described Cdc31 derivatives, we examined interaction with DNA repair factor NEF2 (Rad23 and Rad4). The FLAG-Cdc31 proteins were purified from a yeast strain that expressed physiological levels of both native Rad23 and Rad4-HA from the chromosome. Protein extracts were incubated with FLAG-agarose, and the interaction between the Cdc31 derivatives and NEF2 subunits was determined by immunoblotting. Consistent with the results shown in Fig. 5B, we determined that cdc31-C mediated the interaction with Rad4-HA (Fig. 5C). However, in contrast to their interaction with the proteasome (Fig. 5A), both cdc31-1 and cdc31-2 had reduced interactions with Rad23 and Rad4 (Fig. 5C, lanes 5 and 6) compared to the full-length protein (lane 2).
The same immunoblot was stripped and incubated with anti-ubiquitin antibodies to test for interaction with multiubiquitinated proteins. Although the overall levels of multiubiquitinated proteins were unchanged (Fig. 5D, Extract), observable differences in binding efficiency were evident. Both wild-type Cdc31 and cdc31-C interacted with multiubiquitinated proteins (Fig. 5D, FLAG-IP, lanes 2 and 3). In contrast, cdc31-N did not bind multiubiquitinated proteins (lane 4) or the proteasome. The cdc31-1 mutant interacted strongly with multiubiquitinated proteins, while cdc31-2 demonstrated a very weak interaction, despite a high level of expression (Fig. 5B). Because the mutation in cdc31-2 is located between EF-3 and EF-4, these results are consistent with the model that the carboxy terminus of centrin mediates interactions with cellular partners. Collectively, these results suggest that Cdc31 interaction with the proteasome may be linked to its binding to multiubiquitinated proteins. Because the carboxy terminus mediated all the interactions that we identified, we investigated if it was sufficient for restoring viability in a cdc31 null mutant. We expressed the truncated derivatives of Cdc31 in the cdc31Δ strain that was supported by CDC31 expressed from a URA3-based plasmid. The ability of these mutant proteins to replace the wild-type protein was tested by plating the cultures on medium containing 5-fluoroorotic acid to identify yeast that could tolerate the loss of the plasmid-encoded CDC31 gene. Expression of the amino or carboxy terminus of Cdc31 failed to complement the cdc31Δ mutant (data not shown). These findings demonstrate the essential requirement of both domains, and their failure to complement is consistent with the view that the N terminus contains regulatory sequences that can influence the activities of the carboxy-terminal EF hands.
Copurification of Cdc31 with ubiquitin-associated domains.
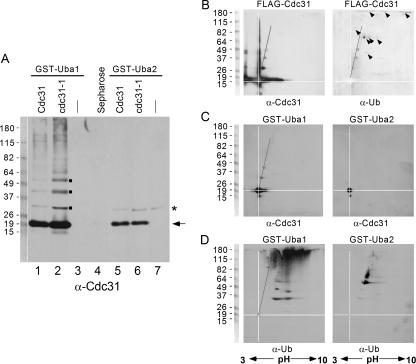
The interaction between Cdc31 and the proteasome could be caused by its ligation to a multiubiquitin chain. Although we considered this unlikely since FLAG-Cdc31 is a stable protein (Fig. 4C), it was conceivable that a multiubiquitin chain could promote interaction with the proteasome without affecting Cdc31 stability. To address this question, we determined if FLAG-Cdc31 could be copurified with ubiquitin-associated domains, which bind multiubiquitin chains. Specifically, if unmodified Cdc31 were purified with a ubiquitin-associated domain protein, it would support our hypothesis that Cdc31 can interact with multiubiquitin chains. In contrast, we would not expect unmodified Cdc31 to be purified with a ubiquitin-associated domain protein if the ubiquitin cross-reacting bands that were purified with FLAG-Cdc31 represented conjugates exclusively on Cdc31 (Fig. 3D). We purified FLAG-Cdc31 and FLAG-cdc31-1 from a wild-type strain that also expressed GST-UBA1 or GST-UBA2 (that were derived from yeast Rad23). Protein extracts were incubated with glutathione-Sepharose, and the bound proteins were examined by immunoblotting (Fig. 6A). Incubation with Cdc31 antibodies showed that predominantly unmodified FLAG-Cdc31 and FLAG-cdc31-1 were purified with both UBA1 and UBA2. We note that although the Cdc31 antibodies formed a strong interaction with unconjugated FLAG-Cdc31/cdc31-1, a low level of higher-molecular-weight bands was also detected (Fig. 6A, lanes 1 and 2). Remarkably, FLAG-Cdc31/cdc31-1 interactions with UBA2 showed no evidence for higher-molecular-weight forms of either FLAG-Cdc31 or cdc31-1. A minor high-molecular-weight band that was detected with GST-UBA2 (Fig. 6A, asterisk) was also present in the control lane (lane 7) that lacked FLAG-Cdc31. In Fig. 6A, lane 3 contains FLAG-Cdc31 but lacks GST-UBA1, and lane 4 contains GST-UBA1 but lacks FLAG-Cdc31.
FIG. 6.
Cdc31 can be purified with UBA domains. (A) GST-UBA1 and GST-UBA2 domains from yeast Rad23 protein can bind multiubiquitin chains. Both fusion proteins were expressed in wild-type yeast that contained either FLAG-Cdc31 or FLAG-cdc31-1. Equal amounts of protein extract were incubated with glutathione-Sepharose, and the bound proteins were resolved by SDS-PAGE, transferred to nitrocellulose, and incubated with anti-Cdc31 antibodies. A predominant interaction against unmodified Cdc31/cdc31-1 was detected. (Excess N-ethylmaleimide was present in the lysis buffer to prevent deubiquitination). The higher-molecular-weight species seen in the immunoblot (lanes 1 and 2) may be caused by either oligomerization of Cdc31, which is a property of centrin proteins, or by its ligation to ubiquitin. GST-UBA2 also formed a strong interaction with unmodified Cdc31/cdc31-1 (lanes 5 and 6). Lanes 3 and 7 contain GST-UBA1 and GST-UBA2, respectively, but the added extracts did not contain FLAG-Cdc31. In lane 4 an extract containing FLAG-Cdc31 was added to glutathione-Sepharose that did not contain an immobilized GST-UBA domain. These controls show that the interactions observed are specific. (B) FLAG-Cdc31 was immunoprecipitated, resolved in 2D gels, and examined by immunoblotting. The major fraction of FLAG-Cdc31 was distributed across a pH range of ∼3, suggesting extensive modification. FLAG-Cdc31 was also present in an intense vertical smear, which could be caused by oligomerization. The pI of the entire smear was identical to that of monomeric Cdc31 (pH 4.3). We also detected three spots that extended diagonally from the monomeric species (indicated by a line). The filter was stripped and incubated with anti-ubiquitin antibodies (right panel), and the three spots (as well as a number of other spots, indicated by arrowheads) were detected. These findings demonstrate that a small fraction of FLAG-Cdc31 can be ligated to a few ubiquitins. Because a number of additional ubiquitin-cross-reacting species did not react to the anti-Cdc31 antibody, we propose that FLAG-Cdc31 can also bind other ubiquitinated proteins. (C) Because FLAG-Cdc31 was copurified with UBA domains, as described in panel A, we separated proteins bound to GST-UBA in 2D gels. Monomeric FLAG-Cdc31 was efficiently purified with both GST-UBA1 and GST-UBA2. Because UBA domains bind multiubiquitin chains, the copurification of unmodified FLAG-Cdc31 suggests that it can also be purified with multiubiquitinated proteins. (D) We incubated the same filters with anti-ubiquitin antibodies and confirmed that high-molecular-weight ubiquitinated proteins were isolated with UBA1. In agreement with our earlier findings, UBA2 formed a weaker interaction with ubiquitin. Ub, ubiquitin; α, anti.
The pattern of some of the bands seen in lanes 1 and 2 of Fig. 6A (indicated by small squares) suggested the conjugation of one, two, and three ubiquitins to Cdc31. We characterized this further by 2D gel electrophoresis. FLAG-Cdc31 was immunoprecipitated, and following 2D analysis, an immunoblot was incubated with antibodies against Cdc31 (Fig. 6B, left panel). We found that a major fraction of FLAG-Cdc31 was apparently modified, as suggested by its distribution over a broad pH range. We also observed an intense vertical streak of anti-Cdc31-cross-reacting material extending from ∼20 kDa to ∼110 kDa. Because human centrin can self-assemble, this streak might represent the oligomerization of FLAG-Cdc31. This idea is further supported by the observation that the pI values of all these higher-molecular-weight species were identical to Cdc31. Intriguingly, we also detected three minor spots (corresponding in size to the bands identified in Fig. 6A, lanes 1 and 2) that migrated along a diagonal (Fig. 6B, left panel). This array is consistent with the conjugation of ubiquitins to Cdc31, since the distinct pI of ubiquitin will progressively shift the position of each conjugate on Cdc31 during isoelectric focusing (pI of Cdc31p is 4.3; pI of ubiquitin is 7.9). To verify this possibility, the filter was stripped and reprobed with antibodies against ubiquitin (Fig. 6B, right panel). The three spots described above (and corresponding to the squares in Fig. 6A) reacted with the anti-ubiquitin antibody, demonstrating that Cdc31 is conjugated to a few ubiquitins. It might be significant that oligomeric forms of ubiquitinated Cdc31 (Fig. 6B) were not detected, as this would be revealed by a vertical smear extending above each ubiquitin-Cdc31 spot. The white lines are aligned to intersect at the position of FLAG-Cdc31, while a solid diagonal line tracks the positions of three ubiquitin conjugates on FLAG-Cdc31 (Fig. 6B). In addition to these three spots, a number of additional ubiquitin cross-reacting spots were detected (Fig. 6B, arrowheads). However, these species represent other ubiquitinated cellular proteins since their positions deviated from the diagonal line, and they were not detected with anti-Cdc31 antibodies. Collectively, these findings show that Cdc31 can bind other multiubiquitinated proteins although a small fraction is also conjugated to ubiquitin.
To extend these findings we purified GST-UBA1 and GST-UBA2 from a wild-type strain that expressed FLAG-Cdc31. The purified GST-UBA proteins were resolved by 2D electrophoresis, and the filter was incubated with anti-Cdc31 antibodies (Fig. 6C). High levels of unmodified FLAG-Cdc31 (pI, ∼4.3) were copurified with both ubiquitin-associated domains. In contrast, very low levels of the ubiquitinated forms of FLAG-Cdc31 were detected with UBA1 (left panel), and the overwhelming fraction was unconjugated. Moreover, none of ubiquitinated forms of FLAG-Cdc31 were purified with UBA2 (right panel). The highly abundant oligomeric forms of FLAG-Cdc31 (Fig. 6B) were not isolated with either UBA domain. The filters used for the experiment shown in Fig. 6C were incubated with antibodies against ubiquitin, and, as expected, a significant amount of ubiquitinated proteins was purified with GST-UBA1 (Fig. 6D, left panel), while a smaller amount was isolated with GST-UBA2 (right panel). Note the intense antibody reaction at the interface of the stacking and resolving gels in the second dimension ranging from pH 4 to 10 (Fig. 6D, left panel). Similarly, purified FLAG-Cdc31 also showed a faint signal at the top of the gel (Fig. 6B, right panel, indicated by three arrowheads), suggesting that a fraction of higher-molecular-weight ubiquitinated proteins does not enter the SDS-polyacrylamide gel.
Cdc31 forms a direct interaction with AAA class ATPases in the proteasome.
In an effort to independently establish that Cdc31 can bind multiubiquitin chains and the proteasome, we characterized the binding using recombinant GST-Cdc31 and HA-Cdc31. Increasing amounts of total yeast protein extract were incubated with a fixed amount of GST-Cdc31, and the level of bound multiubiquitinated proteins was determined by immunoblotting (Fig. 7A). Not unexpectedly, higher levels of ubiquitin-cross-reacting material were purified with GST-Cdc31 when increasing amounts of extract were added (Fig. 7A, lanes 3 to 5). The presence of ubiquitinated species smaller than GST-Cdc31 (∼50 kDa) is consistent with an interaction between Cdc31 and other ubiquitinated cellular proteins (including some that are smaller than GST-Cdc31). However, compared to the level of total ubiquitinated proteins in cell extract (lane 1), it is evident that their interaction with Cdc31 is weak. One interpretation of this result is that the copurification of multiubiquitinated proteins with Cdc31 is not direct but reflects its interaction with the proteasome, which is bound to ubiquitinated proteins. This idea is supported by our results that cdc31-1 interacted strongly with both multiubiquitinated proteins and the proteasome, while cdc31-2 had significantly weaker interactions with both multiubiquitinated proteins and the proteasome (Fig. 5).
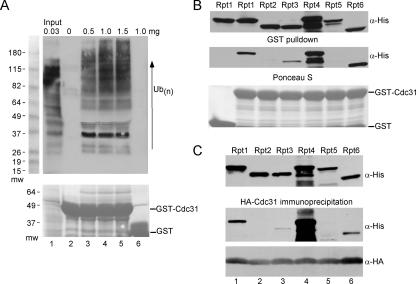
FIG. 7.
Cdc31 can bind specific AAA class ATPases in the proteasome. We further tested the Cdc31 interaction with multiubiquitinated proteins and the proteasome by examining interactions in vitro. (A) Total protein extract was prepared from a wild-type strain, and various amounts were applied to immobilized GST-Cdc31. Following a 2-h incubation, the beads were washed, and the level of bound multiubiquitin-cross-reacting material was measured by immunoblotting. Increasing amounts of extract resulted in progressively higher amounts of multiubiquitinated proteins in association with GST-Cdc31. Multiubiquitinated proteins were not recovered when the extract was combined with glutathione-Sepharose that was bound to only GST (lane 6). (B) Recombinant His-tagged Rpt1 to Rpt6 proteins were expressed in E. coli BL21 cells. The E. coli protein lysates were incubated with purified GST-Cdc31 that was bound to glutathione-Sepharose beads. Following a 2-h incubation, the amount of Rpt proteins that was bound to GST-Cdc31 was determined. Immunoblots were probed with anti-His antibodies, and strong signals were detected against Rpt1, Rpt4, and Rpt6. (C) HA-Cdc31 was purified from E. coli BL21 and combined with the Rpt-containing bacterial extracts, and the interaction was examined by immunoblotting using anti-HA antibodies. Taken together, these two studies strongly support our view that HA-Cdc31 interacts with the proteasome directly, without a need for prior conjugation to a multiubiquitin chain.
We also examined the Cdc31 interaction with proteasome subunits in vitro. We initially surveyed a number of proteasome subunits for their ability to bind GST-Cdc31 (data not shown). Based on these preliminary studies, we focused our attention on the ATPase subunits. Plasmids that expressed a collection of His epitope-tagged ATPase subunits (Rpt1 to Rpt6) in E. coli were generously provided by D. Skowyra (St. Louis University). GST-Cdc31 was purified from E. coli and immobilized on glutathione-Sepharose. E. coli lysates, containing nearly equivalent amounts of each Rpt protein (Fig. 7B), were incubated with GST-Cdc31 bound to glutathione-Sepharose. The purified proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), and an immunoblot was incubated with antibodies against the His epitope. We found that Cdc31 formed a significant interaction with Rpt1, Rpt4, and Rpt6, which reside in the 19S regulatory particle (Fig. 7B, upper panel). In an alternate method we combined E. coli protein extracts containing each of the six Rpt proteins with a similar E. coli extract containing HA-tagged Cdc31. Consistent with the earlier results (Fig. 7B), Rpt1, Rpt4, and Rpt6 were coimmunoprecipitated with HA-Cdc31 (Fig. 7C). The direct interaction with several AAA class subunits, using two different epitopes, provides compelling evidence that Cdc31 can bind the proteasome without prior conjugation to ubiquitin.
DISCUSSION
Cdc31 and Dsk2 were isolated as suppressors of a kar1 mutant, which is defective in SPB duplication. Dsk2 resembles Rad23, since both proteins contain proteasome-binding (UBL) and ubiquitin-binding (UBA) domains. Rad23 functions as a shuttle factor that can deliver multiubiquitinated substrates to the proteasome, and a similar function is predicted for Dsk2, which can also bind multiubiquitinated proteins and the proteasome. Furthermore, Dsk2 has partially overlapping roles with Rad23 (39, 40), and both proteins have biochemical and genetic interactions with Cdc31. The association of Cdc31 with two related proteolytic factors anticipated a role for protein degradation in SPB duplication and cell cycle control. It is significant in this regard that loss of both Dsk2 and Rad23 (dsk2Δ rad23Δ) causes a failure in SPB duplication that could be suppressed by Cdc31. A role for Rad23 in cell cycle control was suggested by the defective SPB duplication in a rad23Δ dsk2Δ mutant (5) and by a transient G2 phase growth delay in a rad23Δ rpn10Δ mutant (26). Taken together, these diverse genetic and biochemical findings suggest that Cdc31, Rad23, and Dsk2 function in a common pathway that regulates cell cycle progression.
The regulation of Cdc31 function is unclear although in metazoans Ca2+ is likely to be a critical signaling mediator. Because yeast Cdc31 interacts very weakly with Ca2+ (37), it is uncertain if its interactions with multiubiquitinated proteins and proteasomes are regulated by this or by other divalent metals. In addition to Ca2+-binding, a poorly described covalent modification of human centrin has been described (36). Our characterization of FLAG-Cdc31 by 2D gel electrophoresis also provides evidence for extensive modification. However, further study will be required to determine if the posttranslational modification of Cdc31 affects its interaction with cellular proteins. For instance, it would be interesting to determine if Cdc31 forms exclusive interactions with Rad4 and the proteasome and if these interactions are regulated following DNA damage.
Centrins are conserved EF-hand proteins bearing strong structural similarity to the calmodulin family of regulatory proteins (4, 16). The yeast Cdc31 protein contains two pairs of EF hands that are separated by a linker sequence (Fig. 5A). Each pair operates as a functional unit, although only the carboxyl pair of EF hands in yeast Cdc31 has been reported to bind calcium (37). Most, if not all, Cdc31/centrin interactions are mediated by the carboxy-terminal domain. However, the EF hands in the amino-terminal domain may influence the function of the C terminus. Cdc31 has been reported to bind SPB components (5, 21, 24, 44), regulators of mRNA export (14, 25), DNA repair factors (1), and signal transduction proteins (23, 45). The Kar1 and Dsk2 proteins play an important role in targeting Cdc31 to the SPB. Cdc31 binds Sfi1, a central component in the SPB (28). An intriguing question is how the centrin proteins bind such a diverse collection of proteins through a single domain. An important clue to the nature of this interaction is offered by the characterization of calmodulin, which binds an amphiphilic α-helix in its target proteins. Similar structures have been identified in Kar1 and XPC/Rad4 (7, 22, 37, 46, 50). The interactions between Cdc31 and diverse cellular proteins were revealed in genetic studies (23) and are consistent with multiple roles. Moreover, a significant fraction of Cdc31 is not present in the SPB (36). In addition to SPB duplication defects, cdc31 mutants have also shown cell morphology and integrity defects (45).
Human CEN2 was discovered in a complex with the DNA repair factors XPC and hHR23 (1) and was reported to stimulate XPC-mediated DNA incision (33). The related DNA repair proteins in yeast are encoded by the RAD4 and RAD23 genes, and both proteins are present in the NEF2 repair complex (17). The interaction between Cdc31 and NEF2 could provide a way to convey the cellular response to DNA damage to the pathways that arrest cell cycle progression. To explore these ideas we characterized the single centrin-encoding gene in yeast (CDC31). We show here that yeast NEF2 contains Cdc31. Although this interaction is mediated by Rad4 and does not require Rad23, we note that Cdc31 forms regulated interactions with a preassembled Rad23/Rad4 complex. Significantly, a cdc31 mutant that formed reduced binding to Rad4 showed sensitivity to UV-induced DNA damage, suggesting that Cdc31 might promote the cellular response to DNA damage by regulating cell cycle progression.
We describe significant new interactions that define a novel role for centrin/Cdc31 in the ubiquitin/proteasome system. Cdc31 can bind the 26S proteasome. Cdc31 can also be copurified with multiubiquitinated proteins, which could reflect its interaction with the proteasome. Additionally, Cdc31 is conjugated to one to three ubiquitins, although it is a stable protein. Since the presence of one to three ubiquitins does not promote a stable interaction with the proteasome, we speculate that this modification might affect Cdc31 function. We note, for instance, that oligomeric forms of ubiquitinated Cdc31 were not detected (Fig. 6C), suggesting that ubiquitination might prevent self-assembly and aggregation.
We found that Cdc31 could bind several AAA class ATPases that are present in the 19S regulatory particle. Although the effect of this interaction is currently not known, we stress that this binding with purified proteins strongly validates our hypothesis that Cdc31 can interact with the proteasome. Furthermore, these in vitro results demonstrate that the Cdc31-proteasome interaction does not require prior attachment to a multiubiquitin chain.
We propose that Cdc31 interactions with the ubiquitin/proteasome system are biologically significant because cdc31 mutants are highly sensitive to drugs that generate protein damage and are unable to degrade proteolytic substrates efficiently. Moreover, the cdc31-2 protein interacted at dramatically reduced levels with both proteasomes and multiubiquitinated proteins.
The binding of Cdc31 to its various cellular partners (including XPC/Rad4, Kar1, Sfi1, proteasome, and multiubiquitinated substrates) is mediated by its carboxy-terminal domain. Removal of the amino terminus resulted in ∼20-fold decreased abundance of the carboxy-terminal domain (cdc31-C), although a strong interaction with its cellular partners was retained. In contrast, the amino-terminal domain (cdc31-N) was stable but did not bind any of the aforementioned proteins. These results are consistent with the view that the amino-terminal EF hand exerts a regulatory effect. Moreover, both domains are essential for function since neither cdc31-N nor cdc31-C could suppress the inviability of cdc31Δ.
The regulation of Cdc31 activities is not well understood. Although Cdc31 belongs to a family of conserved Ca2+-binding proteins, it does not bind Ca2+ with high affinity. Cdc31 interactions with Kic1 and Kar1 (6, 45), as well as its binding to proteasomes and multiubiquitinated proteins, are apparently Ca2+ independent. Cdc31 interactions with the proteasome and multiubiquitinated proteins were similarly unaffected by EGTA or excess Ca2+ (data not shown). In contrast, studies using purified human CEN2 and the carboxy-terminal EF-hand domain showed Ca2+-dependent interaction with a peptide derived from XPC (37). Subtle changes in calcium levels may affect Cdc31 structure and influence its interaction with the proteasome and multiubiquitinated proteins.
CDC31 encodes the essential Cdc31 protein in S. cerevisiae.
The availability of genetic mutants permitted our preliminary characterization of Cdc31 and our discovery that it plays an important role in protein degradation. Cdc31 mutants harboring defects in various cellular functions have been identified and characterized. Because of Cdc31's critical role in controlling SPB duplication, Cdc31 might be ideally positioned to regulate cell cycle arrest in response to environmental stresses. Therefore, we speculate that an important role for Cdc31 might involve integrating a DNA damage signal to execute checkpoint growth arrest. Delaying SPB duplication is expected to facilitate efficient NER. This model is consistent with the UV sensitivity of the cdc31-1 mutant, the previously described interaction between centrin and XPC, and our finding that Cdc31 binds NEF2. However, it remains to be determined if the proteolytic functions of Cdc31 are required for a putative checkpoint function. We note that a number of genetic and biochemical interactions with components of the ubiquitin/proteasome system (including Dsk2 and Rad23) have been described, and it is significant that double mutants involving Rad23 (rad23Δ rpn10Δ or rad23Δ dsk2Δ) cause defects in cell cycle control.
Acknowledgments
This work was supported by Public Health Service grants (CA83875 and GM83321) from the National Institutes of Health to K.M.
We thank J. Dohmen and M. Rose for plasmids and strains. We are especially grateful to D. Skowyra for generously sharing reagents before their description in the literature.
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.Araki, M., C. Masutani, M. Takemura, A. Uchida, K. Sugasawa, J. Kondoh, Y. Ohkuma, and F. Hanaoka. 2001. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 27618665-18672. [DOI] [PubMed] [Google Scholar]
- 2.Bachmair, A., D. Finley, and A. Varshavsky. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234179-186. [DOI] [PubMed] [Google Scholar]
- 3.Baum, P., C. Furlong, and B. Byers. 1986. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc. Natl. Acad. Sci. USA 835512-5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharya, D., J. Steinkotter, and M. Melkonian. 1993. Molecular cloning and evolutionary analysis of the calcium-modulated contractile protein, centrin, in green algae and land plants. Plant Mol. Biol. 231243-1254. [DOI] [PubMed] [Google Scholar]
- 5.Biggins, S., I. Ivanovska, and M. D. Rose. 1996. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J. Cell Biol. 1331331-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biggins, S., and M. D. Rose. 1994. Direct interaction between yeast spindle pole body components: Kar1p is required for Cdc31p localization to the spindle pole body. J. Cell Biol. 125843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charbonnier, J. B., P. Christova, A. Shosheva, E. Stura, M. H. Le Du, Y. Blouquit, P. Duchambon, S. Miron, and C. T. Craescu. 2006. Crystallization and preliminary X-ray diffraction data of the complex between human centrin 2 and a peptide from the protein XPC. Acta Crystallogr. Sect. F 62649-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, L., and K. Madura. 2002. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 224902-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, L., U. Shinde, T. G. Ortolan, and K. Madura. 2001. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuang, S. M., L. Chen, D. Lambertson, M. Anand, T. G. Kinzy, and K. Madura. 2005. Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol. Cell. Biol. 25403-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox, J. A., F. Tirone, I. Durussel, C. Firanescu, Y. Blouquit, P. Duchambon, and C. T. Craescu. 2005. Calcium and magnesium binding to human centrin 3 and interaction with target peptides. Biochemistry 44840-850. [DOI] [PubMed] [Google Scholar]
- 12.Craig, T. A., L. M. Benson, H. R. Bergen 3rd, S. Y. Venyaminov, J. L. Salisbury, Z. C. Ryan, J. R. Thompson, J. Sperry, M. L. Gross, and R. Kumar. 2006. Metal-binding properties of human centrin-2 determined by micro-electrospray ionization mass spectrometry and UV spectroscopy. J. Am. Soc. Mass Spectrom. 171158-1171. [DOI] [PubMed] [Google Scholar]
- 13.Errabolu, R., M. A. Sanders, and J. L. Salisbury. 1994. Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J. Cell Sci. 1079-16. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, T., S. Rodriguez-Navarro, G. Pereira, A. Racz, E. Schiebel, and E. Hurt. 2004. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat. Cell Biol. 6840-848. [DOI] [PubMed] [Google Scholar]
- 15.Funakoshi, M., T. Sasaki, T. Nishimoto, and H. Kobayashi. 2002. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. USA 99745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geier, B. M., H. Wiech, and E. Schiebel. 1996. Binding of centrins and yeast calmodulin to synthetic peptides corresponding to binding sites in the spindle pole body components Kar1p and Spc110p. J. Biol. Chem. 27128366-28374. [DOI] [PubMed] [Google Scholar]
- 17.Guzder, S. N., P. Sung, L. Prakash, and S. Prakash. 1998. Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. J. Biol. Chem. 27331541-31546. [DOI] [PubMed] [Google Scholar]
- 18.Helfant, A. H. 2002. Composition of the spindle pole body of Saccharomyces cerevisiae and the proteins involved in its duplication. Curr. Genet. 40291-310. [DOI] [PubMed] [Google Scholar]
- 19.Hu, H., and W. J. Chazin. 2003. Unique features in the C-terminal domain provide caltractin with target specificity. J. Mol. Biol. 330473-484. [DOI] [PubMed] [Google Scholar]
- 20.Hu, H., J. H. Sheehan, and W. J. Chazin. 2004. The mode of action of centrin. Binding of Ca2 + and a peptide fragment of Kar1p to the C-terminal domain. J. Biol. Chem. 27950895-50903. [DOI] [PubMed] [Google Scholar]
- 21.Jaspersen, S. L., T. H. Giddings, Jr., and M. Winey. 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 159945-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kateb, F., D. Abergel, Y. Blouquit, P. Duchambon, C. T. Craescu, and G. Bodenhausen. 2006. Slow backbone dynamics of the C-terminal fragment of human centrin 2 in complex with a target peptide probed by cross-correlated relaxation in multiple-quantum NMR spectroscopy. Biochemistry 4515011-15019. [DOI] [PubMed] [Google Scholar]
- 23.Khalfan, W., I. Ivanovska, and M. D. Rose. 2000. Functional interaction between the PKC1 pathway and CDC31 network of SPB duplication genes. Genetics 1551543-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilmartin, J. V. 2003. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 1621211-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohler, A., P. Pascual-Garcia, A. Llopis, M. Zapater, F. Posas, E. Hurt, and S. Rodriguez-Navarro. 2006. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol. Biol. Cell 174228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambertson, D., L. Chen, and K. Madura. 1999. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics 15369-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laoukili, J., E. Perret, S. Middendorp, O. Houcine, C. Guennou, F. Marano, M. Bornens, and F. Tournier. 2000. Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J. Cell Sci. 1131355-1364. [DOI] [PubMed] [Google Scholar]
- 28.Li, S., A. M. Sandercock, P. Conduit, C. V. Robinson, R. L. Williams, and J. V. Kilmartin. 2006. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 173867-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medicherla, B., Z. Kostova, A. Schaefer, and D. H. Wolf. 2004. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 5692-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Middendorp, S., T. Kuntziger, Y. Abraham, S. Holmes, N. Bordes, M. Paintrand, A. Paoletti, and M. Bornens. 2000. A role for centrin 3 in centrosome reproduction. J. Cell Biol. 148405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Middendorp, S., A. Paoletti, E. Schiebel, and M. Bornens. 1997. Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc. Natl. Acad. Sci. USA 949141-9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakayama, S., N. D. Moncrief, and R. H. Kretsinger. 1992. Evolution of EF-hand calcium-modulated proteins. II. Domains of several subfamilies have diverse evolutionary histories. J. Mol. Evol. 34416-448. [DOI] [PubMed] [Google Scholar]
- 33.Nishi, R., Y. Okuda, E. Watanabe, T. Mori, S. Iwai, C. Masutani, K. Sugasawa, and F. Hanaoka. 2005. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol. Cell. Biol. 255664-5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohno, A., J. Jee, K. Fujiwara, T. Tenno, N. Goda, H. Tochio, H. Kobayashi, H. Hiroaki, and M. Shirakawa. 2005. Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition. Structure 13521-532. [DOI] [PubMed] [Google Scholar]
- 35.Ortolan, T. G., L. Chen, P. Tongaonkar, and K. Madura. 2004. Rad23 stabilizes Rad4 from degradation by the Ub/proteasome pathway. Nucleic Acids Res. 326490-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paoletti, A., M. Mohammed, M. Paintrand, J. L. Salisbury, and M. Bornens. 1996. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 1093089-3102. [DOI] [PubMed] [Google Scholar]
- 37.Popescu, A., S. Miron, Y. Blouquit, P. Duchambon, P. Christova, and C. T. Craescu. 2003. Xeroderma pigmentosum group C protein possesses a high affinity binding site to human centrin 2 and calmodulin. J. Biol. Chem. 27840252-40261. [DOI] [PubMed] [Google Scholar]
- 38.Prakash, S., and L. Prakash. 2000. Nucleotide excision repair in yeast. Mutat. Res. 45113-24. [DOI] [PubMed] [Google Scholar]
- 39.Rao, H., and A. Sastry. 2002. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J. Biol. Chem. 27711691-11695. [DOI] [PubMed] [Google Scholar]
- 40.Saeki, Y., A. Saitoh, A. Toh-e, and H. Yokosawa. 2002. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem. Biophys. Res. Commun. 293986-992. [DOI] [PubMed] [Google Scholar]
- 41.Seufert, W., and S. Jentsch. 1990. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 9543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheehan, J. H., C. G. Bunick, H. Hu, P. A. Fagan, S. M. Meyn, and W. J. Chazin. 2006. Structure of the N-terminal calcium sensor domain of centrin reveals the biochemical basis for domain-specific function. J. Biol. Chem. 2812876-2881. [DOI] [PubMed] [Google Scholar]
- 43.Spang, A., I. Courtney, U. Fackler, M. Matzner, and E. Schiebel. 1993. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J. Cell Biol. 123405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spang, A., I. Courtney, K. Grein, M. Matzner, and E. Schiebel. 1995. The Cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J. Cell Biol. 128863-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sullivan, D. S., S. Biggins, and M. D. Rose. 1998. The yeast centrin, cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J. Cell Biol. 143751-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, J. R., Z. C. Ryan, J. L. Salisbury, and R. Kumar. 2006. The structure of the human centrin 2-xeroderma pigmentosum group C protein complex. J. Biol. Chem. 28118746-18752. [DOI] [PubMed] [Google Scholar]
- 47.Reference deleted.
- 48.Vallen, E. A., W. Ho, M. Winey, and M. D. Rose. 1994. Genetic interactions between CDC31 and KAR1, two genes required for duplication of the microtubule organizing center in Saccharomyces cerevisiae. Genetics 137407-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfrum, U., and J. L. Salisbury. 1998. Expression of centrin isoforms in the mammalian retina. Exp. Cell Res. 24210-17. [DOI] [PubMed] [Google Scholar]
- 50.Yang, A., S. Miron, L. Mouawad, P. Duchambon, Y. Blouquit, and C. T. Craescu. 2006. Flexibility and plasticity of human centrin 2 binding to the xeroderma pigmentosum group C protein (XPC) from nuclear excision repair. Biochemistry 453653-3663. [DOI] [PubMed] [Google Scholar]