Abstract
Several transcription factors are essential determinants of a cortical projection neuron identity, but their mode of action (instructive versus permissive) and downstream genetic cascades remain poorly defined. Here, we demonstrate that the proneural basic helix-loop-helix (bHLH) gene Ngn2 instructs a partial cortical identity when misexpressed in ventral telencephalic progenitors, inducing ectopic marker expression in a defined temporal sequence, including early (24 h; Nscl2), intermediate (48 h; BhlhB5), and late (72 h; NeuroD, NeuroD2, Math2, and Tbr1) target genes. Strikingly, cortical gene expression was much more rapidly induced by Ngn2 in the dorsal telencephalon (within 12 to 24 h). We identify the bHLH gene Math3 as a dorsally restricted Ngn2 transcriptional target and cofactor, which synergizes with Ngn2 to accelerate target gene transcription in the cortex. Using a novel in vivo luciferase assay, we show that Ngn2 generates only ∼60% of the transcriptional drive in ventral versus dorsal telencephalic domains, an activity that is augmented by Math3, providing a mechanistic basis for regional differences in Ngn2 function. Cortical bHLH genes thus cooperate to control transcriptional strength, thereby temporally coordinating downstream gene expression.
Advanced cognitive functioning is controlled by the cerebral cortex, which includes the six-layered neocortex, a brain region mainly comprised of excitatory, glutamatergic projection neurons and a smaller number of inhibitory, GABAergic (where GABA is γ-aminobutyric acid) interneurons. While all cortical projection neurons share a pyramidal morphology, dorsal telencephalic origin, and glutamatergic neurotransmitter phenotype, they also display laminar and region-specific differences in morphology, projection pattern, and gene expression (47, 57). The homeodomain transcription factors Lhx2, Pax6, and Emx2 are considered cortical selector genes as they are each required for cortical development (48, 49). Strikingly, in Pax6; Emx2 double mutants, the neocortex is converted to basal ganglia, a ventral telencephalic territory (49). A similar conversion of dorsal (cortical) to ventral regional identity also occurs in mice carrying mutations in β-catenin and Gli3, downstream transcriptional effectors in the Wnt and Shh signaling pathways, respectively (6, 72, 73). Moreover, ventral, GABAergic rather than dorsal, glutamatergic neurons differentiate in Ngn2 mutant cortices, a proneural gene encoding a basic-helix-loop-helix (bHLH) transcription factor (19, 62). Taken together, these studies demonstrate that the decision to differentiate into a glutamatergic versus GABAergic neuronal phenotype is a binary fate choice in the telencephalon, a phenomenon that is also observed in the thalamus, midbrain, and spinal cord (16, 50, 56, 62).
Gain-of-function studies can determine if genes act permissively or instructively to specify neuronal phenotypes. In the telencephalon, misexpression of Wnt pathway effectors has dorsalizing effects both in vitro and in vivo (6, 24, 42, 76). In addition, Emx1 and Emx2 are sufficient to convert medial telencephalic territories destined to form choroid plexus into cortex (75). Unexpectedly, overexpression of Pax6 inhibits rather than promotes neuronal differentiation, and while Pax6 can upregulate some cortex-specific genes when overexpressed in the cortex, it remains to be determined if it plays an instructive role in specifying cortical neuronal identities when misexpressed in ectopic sites (4, 10, 29). Likewise, it remains to be determined if Wnt pathway effectors and Emx genes only initiate the transcription of cortical progenitor genes or also induce markers of a mature projection neuron identity.
The proneural functions of the Ngn genes have been examined in various vertebrate species, revealing a role for these transcription factors in the induction of generic neuronal differentiation (14, 32, 33, 53, 55, 60). Specifically, the Ngn genes are thought to promote differentiation by inducing the expression of cascades of effector bHLH genes (66). Indeed, in the neocortex, as neuronal precursors differentiate, they initiate the expression of a number of such bHLH genes, including Ngn1/Ngn2, Math2, Math3, NeuroD, NeuroD2, Nscl1, Nscl2, and Bhlhb5. While mutations in most of these bHLH genes have been reported (with the exception of Bhlhb5), defects in the specification of cortical neurons have been shown only in Ngn1/Ngn2 mutants (19, 62). The lack of similar defects in other bHLH mutants may reflect either a later role for some of these factors or redundant functions between bHLH genes that share expression domains, as demonstrated in other systems (35, 74). Indeed, NeuroD2 is required postnatally for the survival of cortical neurons, while NeuroD2 and Math2 cooperate to regulate differentiation and survival of hippocampal granule neurons (34, 40, 51, 64, 65, 74).
In addition to specifying generic neuronal differentiation, the proneural genes have been implicated in the specification of neuronal subtype identities (63). However, while the Ngn genes act permissively to specify identities in some neural lineages, there are also examples whereby the Ngn genes are instructive for neuronal fate (31, 41, 53, 54, 60). To determine how Ngn2 specifies a glutamatergic projection neuron fate in the cortex, we misexpressed Ngn2 in the dorsal and ventral telencephalon via in utero electroporation (36, 59). Here, we show that Ngn2 is sufficient to induce a cascade of cortical gene expression in a temporally defined order in the ventral telencephalon, demonstrating that Ngn2 is partially instructive for a projection neuron identity. Furthermore, we implicate Math3 as a key cofactor in the Ngn2-regulated cortical differentiation cascade, demonstrating that Ngn2 and Math3 cooperate to temporally coordinate the onset of cortical gene transcription.
MATERIALS AND METHODS
Animals.
Embryos were staged using the morning of the vaginal plug as embryonic day 0.5 (E0.5). CD1 mice (Charles River) were used for in utero electroporation experiments. Ngn2 mutant lines in which a GFP cassette was knocked into the Ngn2 locus were maintained as heterozygotes on a CD1 background, and genotyping was performed as described previously (12).
In utero electroporation.
Previously described Ngn2NRAQ (38), NeuroD (37), and Math3 (70) cDNAs were subcloned into the pCIG2 expression vector (26) using standard procedures. Ngn2 cDNA was subcloned into pCIG2 by PCR amplification from an E13.5 murine cDNA library using the primers Ngn2F (GTGTGTGAATTCGTAGGATGTTCGTCAA) and Ngn2R (GTGTGTGAATTCCTCTAGATACAGTCC). Electroporations were performed at E12.5 as described previously (12, 36, 59) using column-purified endotoxin-free DNA (Qiagen) and platinum tweezer-style electrodes (5 mm; Protech) to apply seven 30-mS pulses at 50 V.
RNA in situ hybridization.
Electroporated brains were fixed overnight in 4% paraformaldehyde, dissolved in diethylpyrocarbonate-treated phosphate-buffered saline (PBS), serially cryosectioned at 10 μm, and collected on Superfrost Plus (Fisher) slides. Slides were processed for RNA in situ hybridization as described previously (2, 12). Probes were used for the following genes (gene aliases are given in parentheses): EGFP (Cairine Logan, University of Calgary), Ngn2 (Neucog2/Math4a) (22), Mash1 (ascl1) (23), Math2 (NeuroD6/Nex) (7), Math3 (NeuroD4/ath3/NeuroM) (70), NeuroD (NeuroD1) (37), Nscl1 (Nhlh1/Hen1/Tal2) (8), Nscl2 (Nhlh2/Hen2) (C. Glenn Begley, Amgen Inc.), Bhlhb5 (Beta3) (77), Id2 (Idb2) (25), Tbr1 (27), VGlut2 (Slc17a6) (20), GAD1 (GAD67) (9), Mef2c (43), Pax6 (68), FezL (FezF2/Zfp312) (28), Robo1 (Dutt1) (3), and RORβ (RZRβ/Nr1f2) (61). A NeuroD2 (Ndrf) probe was generated from an IMAGE Consortium (Lawrence Livermore National Laboratory) cDNA clone from Open Biosystems (Hunstville, AL) (IMAGE clone 6817440; GenBank accession no. BC058965).
Immunostaining and imaging.
Slides were processed for immunostaining as described previously (2, 12). Primary antibodies included mouse anti-NeuN (1/500; Chemicon, Temecula, CA), mouse anti-β-III-tubulin (1/500; Swant, Bellinzona, Switzerland), rabbit anti-green fluorescent protein (anti-GFP; 1/500; Chemicon), rabbit anti-ER81 (1/500; Tom Jessell and Susan Morton [5]), goat anti-NeuroD (1:100; Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-Ngn2 (1/4; David Anderson [41]), rabbit anti-Tbr1 (1:3,000; Chemicon), goat anti-Bhlhb5 (1:1,000; Santa Cruz), and rabbit anti-Otx1 (1/500; Flora Vaccarino [39]). Secondary antibodies were conjugated to Cy3, aminomethylcoumarin acetate (Jackson Immunoresearch, West Grove, PA) or Alexa488 (Molecular Probes) and diluted 1/500. Some sections were stained for 5 min with 4′,6-diamidino-2-phenylindole (DAPI; 1/10,000 dilution in 1× PBS; Santa Cruz), washed an additional three times with PBS, and mounted with AquaPolymount (Polysciences, Inc., Warrington PA). Bright-field and fluorescence microscopy was performed as described previously (12). The numbers of informative brains examined are shown in the figures (see Fig. 4 to 6, lower-right corners of panels; see also Fig. S5 in the supplemental material). Double in situ hybridization was performed as described previously (67). For cell counts of immunostained sections, the number of GFP-positive cells that coexpressed the marker of interest were enumerated from two to three sections from at least three independently transfected brains. For ventral transfections, the minimum number of GFP-positive cells counted per experimental group was 1,257 (range, 1,257 to 1,808), while in dorsal transfections the total number of cells counted was 1,767.
FIG. 4.
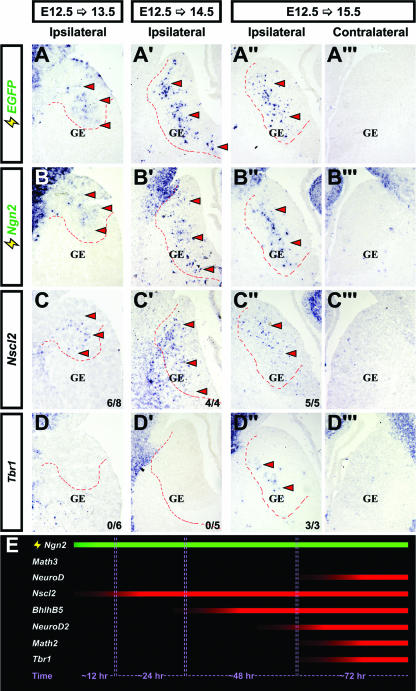
Kinetics of ventral telencephalic gene induction downstream of Ngn2. Frontal sections of ventral telencephalons were electroporated at E12.5 with pCIG2-Ngn2 and harvested at E13.5 (A to D), E14.5 (A′ to D′), or E15.5 (A" to D"). Untransfected E15.5 telencephalons (contralateral) are shown in panels A‴ to D‴. Expression of the EGFP, Ngn2, Nscl2, and Tbr1 genes (identified along the left side of the figure) is also shown. Dashed outlines indicate the approximate location of transfected cells. In panels C, C′, C", D, D′, and D", the numbers of times depicted results were observed are also indicated (lower-right corners). Arrowheads indicate ectopic transcripts. (E) Schematic illustrating the kinetics of gene induction downstream of Ngn2 in the ventral telencephalon (continuous expression; green bar), showing approximate time required for detectable levels of each transcript to accumulate.
FIG. 6.
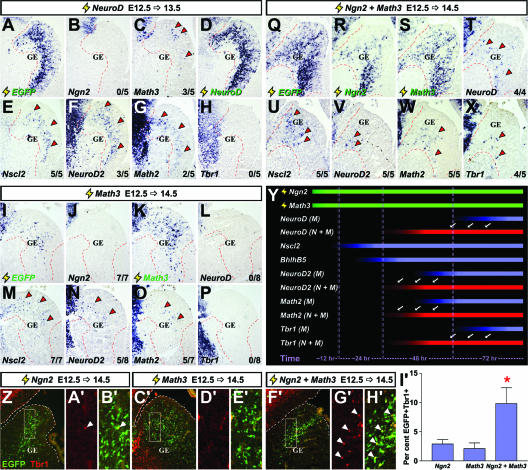
Comparing the kinetics of cortical gene induction downstream of NeuroD, Math3, and Ngn2 plus Math3 in the ventral telencephalon. (A to H) Frontal sections of brains electroporated at E12.5 with pCIG2-NeuroD (3 μg/μl) and harvested at E13.5. (I to P) Frontal sections of brains electroporated at E12.5 with pCIG2-Math3 (3 μg/μl) and harvested at E14.5. (Q to X) Frontal sections of brains electroporated at E12.5 with pCIG2-Ngn2 and pCIG2-Math3 (1.5 μg/μl each) and harvested at E14.5. Localizations of the transcripts are shown for the EGFP, Ngn2, Math3, NeuroD, Nscl2, NeuroD2, Math2, and Tbr1 genes (identified on the panels). Arrowheads in panels A to X indicate ectopic expression. (Y) Schematic depicting shifts in the kinetics of gene induction when Ngn2 and Math3 are expressed together (N+M) versus Math3 expressed alone (M). Green bars depict overexpressed genes, red bars depict genes with accelerated kinetics of induction when Ngn2 and Math3 are coexpressed (shift highlighted by arrows), and blue bars show gene expression induced by Math3 alone. (Z to H′) Frontal sections of brains electroporated at E12.5 with pCIG2-Ngn2 (3 μg/μl), pCIG2-Math3 (3 μg/μl), and pCIG2-Ngn2 plus pCIG2-Math3 (1.5 μg/μl each) harvested at E14.5 (genes are identified above the panels). White boxes in Z, C′, and F′ indicate the positions of higher magnification images displayed shown in panels A′, B′, D′, E′, G′, and H′). Sections were stained with Tbr1 (red; arrowheads indicate double-positive cells). (I′) The percentage of EGFP-positive cells that expressed Tbr1. *, significantly different versus Ngn2 (P < 0.05) and significantly different versus Math3 (P < 0.01).
Luciferase assays.
P19 cells (American Type Culture Collection) were transfected with a combination of the B1-1000 NeuroD luciferase plasmid (30) and the pβ-actin-lacZ plasmid at 0.5 μg/μl and 0.1 μg/μl, respectively, for P19 cell transfections and at an equivalent ratio of 1 μg/μl each for in vivo electroporations of the telencephalon. P19 cells were cultured in alpha minimal essential medium supplemented with l-glutamine and ribonucleosides (Invitrogen, Burlington, Ontario, Canada), to which sodium bicarbonate, penicillin/streptomycin, and 10% fetal bovine serum (Invitrogen) were added. For luciferase experiments, P19 cells were seeded into six-well plates at 100,000 cells per well and transfected with Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions. Luciferase assays were performed using Reporter Lysis Buffer and Luciferase Assay System (Promega, Madison WI) according to the manufacturer's protocol. For electroporated tissue, samples were placed in a microcentrifuge tube with 100 μl of Reporter Lysis Buffer and triturated with a 200-μl pipette tip before being subjected to freeze-thaw cycles. To detect β-galactosidase enzyme activity, fixed volumes (10 or 50 μl) of lysate were suspended in 500 μl of 5-bromo-4-chloro-3-indolyl-β′-d-galactopyranoside buffer (PBS, pH 7.0, with 2 mM MgCl2 and 50 mM β-mercaptoethanol), to which 400 μg (in 100 μl of double-distilled H2O) of o-nitrophenyl-β-d-galactopyranoside (Rockland, Gilbertsville, PA) was added. Samples were incubated for 1 to 16 h, and absorbance at 420 nm was analyzed in a Beckman DU 640 spectrophotometer. Data were normalized by dividing raw light units by the corresponding A420 values. To make specific comparisons between two sample sets in our in vivo luciferase studies, statistical analyses were performed using a Wilcoxon t test (Wilcoxon signed rank test). For luciferase assays in P19 cells, multiple comparisons were performed by applying a one-way analysis of variance and Tukey's multiple comparison test. Both tests were applied using Graphpad Prizm software (San Diego, CA).
GST pull-down assays.
To generate pGex2T-Ngn2, the Ngn2 open reading frame was excised from pCIG2-Ngn2 using EcoR1 and ligated into the EcoR1 site of pGex2T (Pharmacia). To subclone Math3 into EcoRI-linearized pBluescript II KS (Stratagene), the open reading frame was amplified by PCR from pCIG2-Math3 using the primers Math3S (GAGAGAATTCGATGGCAAAAATGTATATG) and Math3AS (GAGAGAATTCCTAATCAGAGAAGATCGTATTG), cut with EcoR1, and ligated with standard conditions. 35S-labeled Math3 and E47 proteins were generated using Express Protein Labeling Mix (Perkin Elmer) and a TNT Coupled Reticulocyte Lysate System (Promega) using the pBluescript-Math3 and pcDNA3-E47 plasmids as templates. Glutathione S-transferase (GST) pull-down assays were performed as described previously (22). Densitometry was performed on scanned images using Adobe Photoshop software.
RESULTS
Identification of candidate Ngn2 target genes in the cortex.
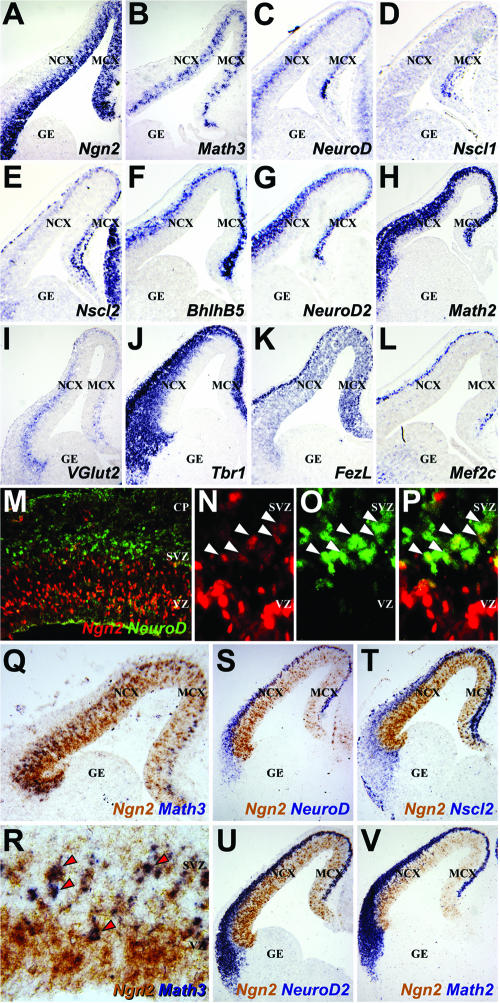
In Drosophila, proneural genes activate transcriptional cascades composed of bHLH and non-bHLH transcription factors (11), as do bHLH determination genes in vertebrate skeletal muscle (52). To characterize the cortical transcriptional cascade(s) executed downstream of Ngn2, we selected a panel of potential target genes using restricted or enriched expression in the dorsal telencephalon and deregulated expression in Ngn2 mutant cortices as criteria (19, 44, 62). At E13.5, Ngn2 was expressed in the ventricular zone (VZ) of the dorsal and not ventral telencephalon (Fig. 1A) (19, 21, 22, 26). Of the other cortical bHLH genes, Math3 expression was also confined to the VZ (Fig. 1B), while NeuroD was expressed in the subventricular zone (SVZ), a secondary population of cortical progenitors and newborn neurons (Fig. 1C). Nscl1 was weakly expressed in the medial-most preplate, a layer containing the first postmitotic neurons in the cortex (Fig. 1D), while the related gene Nscl2 was more robustly expressed throughout the preplate as well as at lower levels in the SVZ (Fig. 1E). BhlhB5, NeuroD2, and Math2 were primarily expressed in the developing cortical plate, which is populated by later-born neurons (Fig. 1F to H). Of the non-bHLH genes, VGlut2, a glutamatergic marker, was expressed in the E13.5 SVZ and preplate (Fig. 1I), as was the T-box gene Tbr1, which was also expressed in the cortical plate (Fig. 1J). The zinc finger gene FezL was expressed in the VZ and preplate (Fig. 1K) and was selected as a putative Ngn2 target gene as it is required to specify subplate and layer V neurons (15, 28, 46), as is Ngn2 (62). Finally, Mef2c (Fig. 1L) was analyzed as a nonregionalized, Ngn2-regulated, cortical gene (44), expressed dorsally in the preplate and cortical plate and ventrally in the ganglionic eminences (GE). With the exception of FezL, all of these genes have been previously reported to exhibit reduced expression in dorsomedial domains of Ngn2 mutant cortices, results that we have reproduced in E13.5 Ngn2 mutants for the purposes of clarity (see Fig. S1 in the supplemental material) (19, 44, 62). Importantly, Math3 expression was completely lost in E13.5 Ngn2 mutant cortices, at least at the level of detection afforded by RNA in situ hybridization (see Fig. S1B and B′ in the supplemental material), while other bHLH genes were less severely affected, due in part to the retained expression of Ngn1 (data not shown), a related gene that was previously reported to have overlapping functions with Ngn2 (19).
FIG. 1.
Expression profiles of putative Ngn2 target genes in the telencephalon. (A to L) RNA in situ hybridization of frontal sections of E13.5 brains with probes for Ngn2, Math3, NeuroD, Nscl1, Nscl2, BhlhB5, NeuroD2, Math2, VGlut2, Tbr1, FezL, and Mef2c (as labeled on panels). (M to P) Double immunolabeling of E13.5 cortex with anti-Ngn2 (red) and anti-NeuroD (green). Arrowheads mark double-positive cells at the VZ/SVZ border. (Q to V) Double in situ hybridization of frontal sections through the E12.5 telencephalon with probes for Ngn2 (brown) in combination with probes for (all in purple; identified on panels) Math3 (arrowheads mark double-positive cells, NeuroD, Nscl2, NeuroD2, or Math2. NCX, neocortex; MCX, medial cortex; CP, cortical plate.
Math3 and NeuroD are direct transcriptional targets of Ngn genes in Xenopus (71). We reasoned that if Math3 and/or NeuroD were direct Ngn2 targets in the cortex, double-positive cells should be evident. While the vast majority of E13.5 cortical cells expressed only Ngn2 or NeuroD protein, a layer of double-positive cells one to two cells thick was observed at the VZ/SVZ interface (Fig. 1M to P). Similarly, using two-color RNA in situ hybridization, Ngn2 and NeuroD transcripts colocalized at the E13.5 VZ/SVZ border but were exclusive elsewhere in the cortex (Fig. 1S). In striking contrast, most Math3-positive cells positioned in the upper VZ coexpressed Ngn2 (Fig. 1Q and R). Finally, while the dorsoventral expression limits of the remaining bHLH genes registered precisely with those of Ngn2 (Fig. 1Q to V), Ngn2 displayed limited coexpression with Nscl2, NeuroD2, and Math2, with double-positive cells concentrated at the VZ/SVZ interface (Fig. 1T to V), while Ngn2 and BhlhB5 were not detectably coexpressed (data not shown). Math3, NeuroD, Nscl2, NeuroD2, and/or Math2 could thus be direct transcriptional targets of Ngn2, albeit (with the exception of Math3) in a limited number of cells.
Ngn2 induces expression of a subset of dorsal regional markers in the ventral telencephalon.
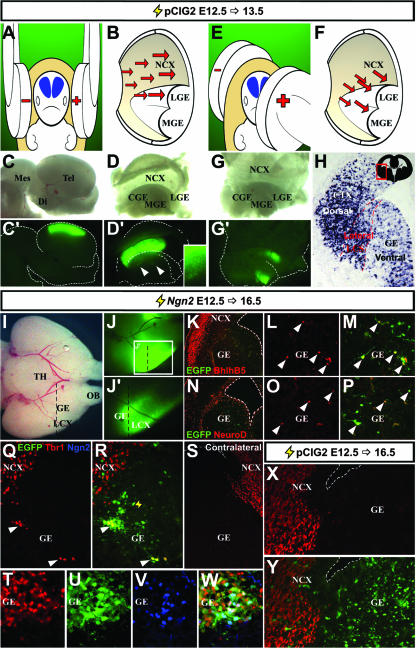
To understand how Ngn2 executes its neuronal specification functions in the cortex, we used a gain-of-function approach. Ngn2 was misexpressed in E12.5 telencephalic progenitors via in utero electroporation (36, 59) using a bicistronic expression vector for Ngn2 and an internal ribosome entry site 2-enhanced GFP (EGFP) cassette (pCIG2 [26]). At E12.5, the telencephalon, which is a bilateral structure located in the rostral-most region of the embryonic neural tube (Fig. 2A, blue structures), has a large fluid-filled ventricle where DNA constructs were introduced with micropipettes. To target the uptake of DNA expression constructs into the dorsolateral telencephalon, electrodes were placed parallel to the E12.5 head in utero (Fig. 2A and B) (36, 59). With this approach, dorsal telencephalic progenitors, which line the ventricular surface, were preferentially targeted, while only a small number of ventral progenitors in the lateral GE were transfected (Fig. 2C, C′, D, and D′). To increase the number of ventral progenitors targeted, the cathode was rotated ∼30° rostrally (Fig. 2E and F), resulting in reliable transfection of the E12.5 lateral GE and to a lesser extent the medial GE (83.2% electroporations with ventral cells targeted; n = 94/113) (Fig. 1G and G′). In sections taken through the telencephalon 24 h postelectroporation, typically, a continuum of dorsolateral (cortical) and ventral progenitors was electroporated (71.7% electroporations; n = 81/113) (Fig. 2H), while only 16.8% (19/113) and 11.5% (13/113) of the electroporations, respectively, exclusively targeted dorsal or ventral cells.
FIG. 2.
Ngn2 instructs a partial cortical, neuronal identity in the ventral telencephalon. (A and B) Schema of electroporation strategy to target dorsolateral cortex. (C and D) Example of E12.5 brain electroporated with pCIG2 at E12.5 and harvested 24 h later, showing whole-mount and opened views of dissected brain in bright-field (C and D) and fluorescence (C′ and D′) images, respectively. A small number of transfected GE cells are shown in the inset in panel D′. (E and F) Schema of electroporation strategy to target ventral telencephalon. (G and G′). Bright-field (G) and fluorescence (G′) images of dissected and opened brain electroporated at E12.5 and harvested 24 h later. (H) EGFP expression in electroporated E12.5 brain harvested after 24 h. Schematic in upper right corner shows a frontal section through the entire telencephalon, with the boxed area highlighting the location of the field in which EGFP expression is highlighted. (I to W) E12.5 brain electroporated with pCIG2-Ngn2 and harvested at E16.5. Ventral views of bright-field (I) and fluorescence (J and J′) images. Panel J′ is a higher magnification image of boxed area in panel J. Dashed lines indicate the approximate locations of the sections depicted in (K to Y). (K to W) Frontal sections through electroporated ventral telencephalon imaged for EGFP epifluorescence (green) and BhlhB5 (red; K to M), NeuroD (red; N to P), Tbr1 (red; Q to W), and Ngn2 (blue; V and W) immunostaining. Double-positive cells are marked by arrowheads (L, M, and O to R). An untransfected, contralateral hemisphere immunostained with Tbr1 (S) is also shown. (X and Y) Frontal section through an electroporated E12.5 ventral telencephalon transfected with an empty pCIG2 vector and imaged for EGFP epifluorescence (green; Y) and Tbr1 (red; X and Y) expression at E16.5. Mes, mesencephalon; Di, diencephalon; Tel, telencephalon; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; CGE, caudal ganglionic eminence; NCX, neocortex; LCX, lateral cortex; TH, thalamus; OB, olfactory bulb; LCS, lateral cortical stream.
To determine if Ngn2 could induce ectopic cortical gene expression ventrally, embryos electroporated at E12.5 were harvested after 96 h, and successful transfection of the ventral telencephalon was identified by EGFP epifluorescence (Fig. 2I, J, and J′). In sections, EGFP-positive cells were scattered throughout the GE (Fig. 2K, M, N, and P), although in well-transfected brains, EGFP-positive cells instead aggregated together to form distinct heterotopia (Fig. 2Q to W and 3). Strikingly, a subset of EGFP-positive cells in the GE ectopically expressed the cortical-specific markers BhlhB5 (Fig. 2K to M), NeuroD (Fig. 2N to P) and Tbr1 (Fig. 2Q, R, T, and W). Tbr1 (Fig. 2S) and other cortical markers (data not shown) were not expressed in the nontransfected, contralateral GE. Ngn2 protein was also detected in EGFP-positive cells (Fig. 2V and W), while ectopic Ngn2 transcripts were detectable up to 288 h posttransfection (longest time point analyzed) (see Fig. S2 in the supplemental material), indicating that Ngn2 expression was maintained. Ngn2 is thus sufficient to cell-autonomously instruct some aspects of cortical identity in the ventral telencephalon.
FIG. 3.
Identification of Ngn2 target genes in the ventral telencephalon. Frontal sections of E16.5 brain that was electroporated at E12.5 with pCIG2-Ngn2. Localizations are shown of transcripts for the EGFP, Ngn2, Nscl2, BhlhB5, NeuroD, NeuroD2, Math2, Tbr1, FezL, VGlut2, and GAD1 genes (identified along left sides of panels). The contralateral hemisphere was not transfected (A′ to K′). (L and M) False-color overlay between adjacent sections hybridized with probes for GAD1 (red) and EGFP (cyan; L) or VGlut2 (cyan; M). Arrowheads mark ectopic gene expression (A to H, J, and K). NCX, neocortex.
To determine if Ngn2 was sufficient to initiate the expression of all or only a subset of cortical genes, we expanded the number of markers analyzed by using RNA in situ hybridization. Riboprobes for EGFP (Fig. 3A) and Ngn2 (Fig. 3B) identified patches of Ngn2-transfected cells in the GE 96-h postelectroporation of the E12.5 telencephalon. In adjacent sections, genes ectopically induced by Ngn2 in the GE included Nscl2, BhlhB5, NeuroD, NeuroD2, Math2, and Tbr1 (Fig. 3C to H). In contrast, the dorsally restricted genes Math3 (data not shown) and FezL (Fig. 3I) and the nonregionalized gene Mef2c (data not shown) were not induced by Ngn2 in the GE (see Table S1 in the supplemental material). Verifying the specificity of gene induction by Ngn2, cells in the nontransfected, contralateral GE never expressed cortical genes (Fig. 3C′ to H′). Moreover, GE transfected with pCIG2, expressing EGFP alone (n = 15) (Fig. 2X and Y; see also Fig. S3 in the supplemental material) or Ngn2 with a mutated, nonfunctional DNA-binding domain (Ngn2NRAQ; n = 2) (see Fig. S4 in the supplemental material) failed to induce ectopic cortical markers. Thus, although Ngn2 can elicit some biological effects through DNA-binding-independent mechanisms (69), DNA binding is required to instruct a cortical identity.
Ngn2 is instructive for neurotransmitter but not laminar identity.
We next questioned if Ngn2 was sufficient to specify neurotransmitter and layer-specific phenotypes, which are perturbed in Ngn mutant cortices (19, 62). In brains electroporated at E12.5 and harvested 96 h later, Ngn2-expressing GE cells ectopically expressed VGlut2 (Fig. 3J), which was not expressed in the nontransfected, contralateral GE (Fig. 3J′) or in control (empty vector or Ngn2NRAQ) electroporated GE cells (see Fig. S3 and S4 in the supplemental material). Concomitantly, GAD1 expression was lost in Ngn2-transfected patches (Fig. 3K). Accordingly, the superimposition of adjacent sections showed that GE cells expressing EGFP (Fig. 3L), and hence Ngn2 (not shown) and VGlut2 (Fig. 3M), did not express GAD1 (Fig. 3L). These data support the notion that glutamatergic traits are acquired at the expense of GABAergic phenotypes.
Ngn2 is required to specify the identities of early-born, deep-layer neurons (62). To test if Ngn2 was sufficient to specify a deep-layer V/VI identity, we electroporated the dorsal telencephalon with Ngn2 or control (EGFP) constructs at E14.5, when neurons in upper layers II to IV are born (36). Three days postelectroporation, Ngn2-transfected cells, including neurons that had reached the cortical plate, did not ectopically express Otx1 or ER81, layer V/VI and V markers, respectively (see Fig. S2 in the supplemental material). Similarly, at postnatal day 7, when cortical migration is complete, neurons derived from both Ngn2- and control-transfected E14.5 progenitors predominantly localized to layer IV and expressed appropriate layer markers (RORβ, Bhlhb5, and low Tbr1), while deep-layer markers (FezL, Robo1, and ER81) were not ectopically expressed (see Fig. S2 in the supplemental material). Finally, in ventral GE electroporations, deep-layer markers, including Robo1, ER81, and Otx1, were not induced by Ngn2 (data not shown). Ngn2 is thus sufficient to specify aspects of a dorsal regional and glutamatergic neurotransmitter identity but cannot impart neocortical layer properties.
Ordering the Ngn2-dependent genetic cascade.
To examine the order of gene induction downstream of Ngn2, the stage of electroporation was kept constant (E12.5) while the time of analysis (6, 9, 12, 24, 48, and 72 h postelectroporation) was varied (see Table S1 and Fig. S5 in the supplemental material for a summary). Between 6 to 12 h posttransfection, no cortical genes were ectopically expressed in Ngn2-electroporated GE cells at levels detectable by RNA in situ hybridization. However, by 24 h, ectopic Nscl2 transcripts were detected in the GE (n = 6/8 embryos) (Fig. 4C and E). Nscl2 expression was maintained in Ngn2-transfected cells in the GE at 48 h postelectroporation in 4/4 embryos (Fig. 4C′), at which time, transcripts for BhlhB5 were also apparent (3/5 embryos) (see Fig. S5C′ in the supplemental material). By 72 h, in addition to Nscl2 (5/5 embryos) and Bhlhb5 (4/4), ectopic expression of NeuroD2 (3/3), Math2 (2/2), NeuroD (4/4), Tbr1 (3/3), and VGlut2 (2/2) was readily apparent in all Ngn2-electroporated GE (Fig. 4A″ to D″; see also Fig. S5C″ to G″ in the supplemental material). Notably, all of the cortical genes analyzed were not expressed in the nontransfected contralateral GE or in control transfections, indicating that ectopic gene expression is a consequence of Ngn2 misexpression (Fig. 4A‴ to D‴; see Fig. S3, S4, and S5C‴ to G‴ in the supplemental material). Ngn2 thus induces cortical gene expression in a temporally defined manner in the GE (Fig. 4E).
Ngn2 rapidly induces cortical gene expression in the neocortex.
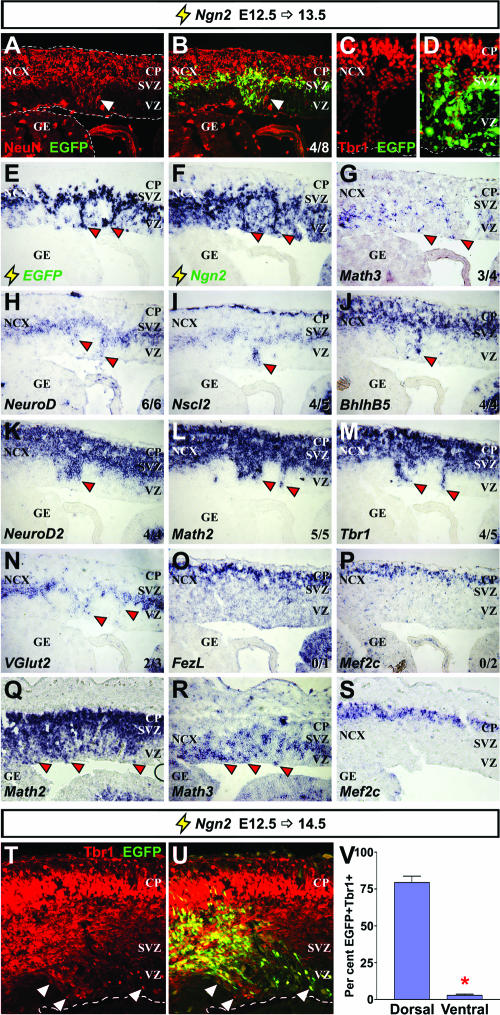
We next tested if Ngn2 could induce premature expression of cortical genes when misexpressed in the E12.5 dorsal telencephalon. At 12 h postelectroporation, Ngn2-misexpressing cells in the cortical VZ prematurely expressed Nscl2 (3/8 brains) and NeuroD (6/8), while transcripts for genes induced later in the GE, such as BhlhB5 (1/6) and Tbr1 (1/6), were induced less frequently (data not shown; see Table S1 in the supplemental material). At 24 h postelectroporation, in strongly transfected cortices, Ngn2 frequently induced the formation of columnar structures in the cortical VZ that were comprised of ectopic neurons positive for NeuN (columns formed in 5/10 brains; 5/5 columns were NeuN positive) (Fig. 5A and B), Tbr1 (Fig. 5C and D), and Tuj1 (data not shown). In Ngn2-induced cortical columns, the precocious expression of Math3 (3/4 columns), NeuroD (6/6), Nscl2 (4/5), Bhlhb5 (4/4), NeuroD2 (4/4), Math2 (5/5), Tbr1 (4/5), and VGlut2 (2/3) was observed after 24 h (Fig. 5G to N, Q, and R). However, even when massive VZ columns formed, FezL (0/1 column) and Mef2c (0/2) were not induced by Ngn2 (Fig. 5O, P, and S), similar to their lack of induction by Ngn2 ventrally. To further support the idea that Ngn2 more efficiently induces the expression of downstream genes in dorsal versus ventral telencephalic domains, we also showed that approximately 27.5-fold more Ngn2-electroporated cells (EGFP positive) coexpressed the cortical marker Tbr1 in dorsal versus ventral telencephalic domains 48 h postelectroporation (Fig. 5T to V).
FIG. 5.
Ngn2 more rapidly induces cortical gene expression in the dorsal telencephalon. Frontal sections of cortices were electroporated at E12.5 with pCIG2-Ngn2 and harvested at E13.5. (A to D) Visualization of EGFP epifluorescence (green; B and D) and immunolabeling for NeuN (red; A and B) or Tbr1 (red; C and D). (E to S) Localization of transcripts for EGFP, Ngn2, Math3, NeuroD, Nscl2, BhlhB5, NeuroD2, Math2, Tbr1, VGlut2, FezL, and Mef2c genes (identified on the panels). Panels Q to S show a more strongly transfected brain. The number of times each marker was ectopically expressed in cortical columns is indicated in panels B and G to P (lower-right corners). (T and U) Visualization of EGFP epifluorescence (green; U) and immunolabeling for Tbr1 (red; T and U) in the cortex. Arrowheads indicate ectopic transcript or protein expression. (V) Quantitation of EGFP/Tbr1 double-positive cells in dorsal and ventral transfections. The asterisk in panel V indicates a significant difference with a P value of <0.0001 (two-tailed t test). NCX, neocortex; CP, cortical plate.
We conclude that Ngn2 prematurely induces neurogenesis and the expression of a cortical transcriptional cascade in the dorsal telencephalon, acting at an accelerated rate compared to ventral domains, where many genes require 72 h for induction. We favor this interpretation over perturbed migration, as migration defects should result in Ngn2-induced cortical columns expressing all cortical markers, including Mef2c and FezL. Moreover, misexpression of Ngn2 repressed Pax6 expression in the cortical columns (see Fig. S6 in the supplemental material), as previously observed in the spinal cord (10), consistent with the idea that Ngn2 has specific effects on gene expression.
Positioning Math3 and NeuroD in the Ngn2-regulated genetic cascade.
Math3 and NeuroD are direct targets of Ngn genes in other systems (71). Consistent with similar genetic relationships existing in the cortex, both Math3 and NeuroD were coexpressed to some extent with Ngn2 (Fig. 1M to S) and are dependent on Ngn2 for their transcription (see Fig. S1 in the supplemental material) (62). To determine if Math3 and/or NeuroD are downstream effectors of Ngn2 in the cortex, we compared their abilities to induce ectopic dorsal gene expression in the GE (see Table S2 in the supplemental material). Within 24 h postelectroporation at E12.5, NeuroD induced the ectopic expression of Nscl2 (5/5 brains), NeuroD2 (3/5), and Math2 (2/5) in the GE (Fig. 6E to G), transcripts that remained detectable after 48 h, at which time Tbr1 (3/6; not shown) and VGlut2 (3/4; not shown) transcripts were also detected. Notably, NeuroD did not induce Ngn2 expression in the GE (0/5 after 24 h) (Fig. 6B). Instead, Ngn2 expression was suppressed by NeuroD in dorsal domains (see Fig. S6 in the supplemental material). However, in striking contrast to Ngn2, NeuroD induced ectopic Math3 expression in the E12.5 GE at 24 h postelectroporation (3/5) (Fig. 6C), albeit in a transient manner (0/6 brains after 48 h). NeuroD is thus sufficient to induce ectopic cortical gene expression in the GE, acting at an accelerated rate compared to Ngn2 and in a Ngn2-independent fashion, suggesting that NeuroD acts downstream of Ngn2 in the cortex. In contrast, in Math3-electroporated E12.5 GE analyzed after 24 h, ectopic expression of Nscl2 (2/2) but not Ngn2 (0/3) or NeuroD (0/3; not shown) was observed (see Table S3 in the supplemental material). After 48 h, Math3-electroporated GE cells continued to express Nscl2 (7/7) and additionally expressed BhlhB5 (5/8; not shown), NeuroD2 (5/8), and Math2 (5/7) but not NeuroD (0/8) or Tbr1 (0/8) (Fig. 6L to P), which were not detected until 72 h posttransfection. The rate of Math3-induced cortical gene transcription in the GE was thus intermediate between Ngn2 and NeuroD.
We were struck by several differences in Ngn2 function in dorsal versus ventral telencephalic domains. Firstly, Ngn2 induced cortical gene expression at an accelerated rate in dorsal compared to ventral domains. Secondly, while premature Math3 expression was induced by Ngn2 in the dorsal telencephalon, Math3 was not induced in the GE at any stage analyzed (6 to 96 h postelectroporation) (see Table S1 in the supplemental material). Finally, NeuroD was an early target of Ngn2 in dorsal transfections but was a relatively late target in the GE. In skeletal muscle, the bHLH gene MyoD induces the expression of late target genes through cooperative interactions with Myog, another bHLH gene that functions later in the differentiation process (13). We therefore hypothesized that Ngn2 could cooperate with Math3 to more efficiently induce the expression of later-onset cortical genes (e.g., NeuroD). In this scenario, the absence of Math3 induction by Ngn2 in ventral domains might explain the differences in the kinetics of Ngn2-mediated gene induction in the ventral versus dorsal telencephalon.
To test if Math3 cooperates with Ngn2, equivalent concentrations of each expression construct were coelectroporated in the E12.5 GE (1.5 μg/μl each, with a final concentration of 3 μg/μl, which is the same as the final concentration of DNA used for all single-construct transfections). While Nscl2 was the only gene ectopically induced by a coexpression of both Math3 and Ngn2 in the GE within 24 h (see Table S4 in the supplemental material), after 48 h, Math2 (5/5) and NeuroD2 (5/5) transcripts were readily detected in 100% of electroporated GE (Fig. 6V and W), greater than the 62.5% (5/8) and 20% (1/5) of Math3 and Ngn2 single electroporations, respectively, in which these cortical genes were induced (see Tables S1 and S3 in the supplemental material). Moreover, NeuroD (4/4) and Tbr1 (4/5) transcripts were induced within 48 h in GE transfected with Math3 plus Ngn2; these transcripts were never observed in this time frame when Math3 or Ngn2 was transfected individually (Fig. 6T and X). Accordingly, when transfected GE sections were stained for Tbr1 protein after 48 h (Fig. 6Z to H′), fewer than 3% of EGFP-positive cells expressed Tbr1 in individual Ngn2 or Math3 transfections. However, the percentage of EGFP-positive cells that coexpressed Tbr1 after coexpression of Ngn2 and Math3 was significantly increased to nearly 10% (Fig. 6I′). Ngn2 and Math3 thus cooperate to accelerate cortical gene induction in the GE (Fig. 6Y).
Cooperative transcriptional interactions between Math3 and Ngn2.
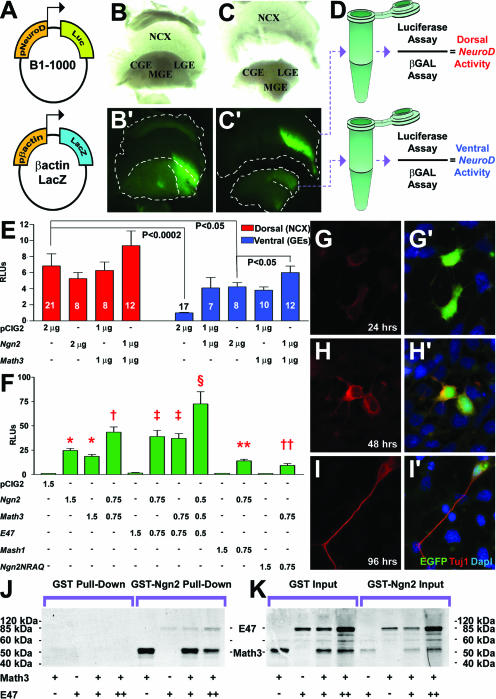
To quantitate transcriptional cooperativity between Ngn2 and Math3, we developed an in vivo luciferase assay using a Ngn-dependent NeuroD promoter to drive luciferase expression (30) and controlling for transfection efficiency with a β-actin-lacZ reporter (17) (Fig. 7A). Equivalent amounts of both promoter constructs were mixed with combinations of pCIG2, pCIG2-Ngn2, and/or pCIG2-Math3 (2 μg/μl total) and electroporated into the E12.5 telencephalon, which was divided into dorsal and ventral domains 24 h postelectroporation (Fig. 7B, B′, C, and C′). Luciferase and β-galactosidase activities were assayed in cortical and GE lysates, using β-galactosidase to normalize for transfection efficiency (Fig. 7D). Since NeuroD is not endogenously expressed in the ventral telencephalon, NeuroD promoter activity in the ventral telencephalon was arbitrarily assigned a value of 1, against which all other values were normalized. As expected, basal NeuroD promoter activity was higher in dorsal (6.85 ± 1.50; n = 21) than ventral (1.00 ± 0.07; n = 17) domains (P < 0.0001) (Fig. 7E). Cotransfection of Ngn2 did not further increase NeuroD promoter activity dorsally (5.28 ± 0.73; n = 8), ostensibly due to high levels of endogenous Ngn2 (Fig. 7E). In contrast, in the ventral telencephalon, Ngn2 increased NeuroD promoter activity approximately fourfold (1 μg/μl Ngn2, 4.11 ± 1.31 [n = 7; P = 0.0133]; 2 μg/μl Ngn2, 4.25 ± 0.53 [n = 8; P < 0.0001]), albeit only to ∼60% of the activity levels observed dorsally (P = 0.0076 for pCIG2 dorsal versus twice the amount of Ngn2). Math3 similarly had little effect on the NeuroD promoter in the dorsal telencephalon (1 μg/μl, 6.28 ± 0.73; n = 8) but efficiently induced NeuroD promoter activity in ventral domains (1 μg/μl, 3.84 ± 0.39; n = 10; P = 0.0002 versus pCIG2 ventral) (Fig. 7E). In contrast, coexpression of Ngn2 and Math3 increased promoter drive ∼1.4-fold in ventral (1 μg/μl each, 6.01 ± 0.82; n = 12; P = 0.014) domains, compared to equivalent amounts of Ngn2 alone. Importantly, comparisons were made between the luciferase values obtained following electroporations of 2 μg/μl of Ngn2 versus 1 μg/μl each of the coexpressed Ngn2 and Math3 (2 μg/μl together), such that the total amounts of bHLH DNA transfected were equivalent. We thus concluded that cotransfection of Ngn2 and Math3 synergizes on the NeuroD promoter in vivo, since additive interactions should have alternatively yielded equivalent activation levels in single and double electroporations.
FIG. 7.
Ngn2 and Math3 synergize on the NeuroD promoter. (A to E) In vivo luciferase assay. NeuroD-luciferase (A) and β-actin-lacZ (B) (1 μg/μl each) promoter constructs were combined with 2 μg/μl of pCIG2 combinations (pCIG2, Ngn2 and/or Math3; shown in E) and electroporated into E12.5 telencephalons. Bright-field (B and C) and dark-field (B′ and C′) images are shown of an electroporated telencephalic vesicle harvested after 24 h (B and B′) and dissected into dorsal and ventral halves (C and C′), which were then measured for luciferase and β-galactosidase activity (D). (E) Normalized luciferase activity levels, showing relative induction when NeuroD promoter was expressed with various pCIG2 constructs. (F) Luciferase experiment in P19 cells, showing normalized luciferase levels with a NeuroD promoter construct cotransfected with various expression vectors. *, significantly different versus pCIG2 (P < 0.01); †, significantly different versus pCIG2, Ngn2, and Math3 (P < 0.001); ‡, significantly different versus pCIG2 or E47 (P < 0.01) and not significantly different versus Ngn2 plus Math3; §, significantly different versus all other means (P < 0.001); **, significantly different versus pCIG2, Ngn2, and Mash1 (P < 0.001); ††, significantly different versus pCIG2, Ngn2, Math3, and Ngn2NRAQ (P < 0.05). (G to I and G′ to I′) P19 cells transfected with Ngn2 (green) expressed Tuj1 (red) 48 h posttransfection. Blue is DAPI counterstain. (J and K) GST pull-down experiments with GST or GST-tagged Ngn2 protein immobilized on glutathione beads and combined with 35S-labeled Math3 and/or E47, showing bound protein (J) and input protein (K). RLU, relative light units; NCX, neocortex; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; CGE, caudal ganglionic eminence; Luc, luciferase; β-Gal, β-galactosidase.
To investigate further the synergism of the combination of Ngn2 and Math3, we used P19 embryonal carcinoma cells, which differentiate into neurons in response to bHLH genes (18) (Fig. 7G to I and G′ to I′). P19 cells were transfected with equivalent amounts of the NeuroD and β-galactosidase promoter constructs and 1.5 μg/μl total of pCIG2 expression vectors (four independent experiments, with 12 replicates for each set of constructs) (Fig. 7F). While Ngn2 and Math3 transactivated the NeuroD promoter when transfected alone (pCIG2, 1.00 ± 0.01; Ngn2, 24.76 ± 2.24; Math3, 18.67 ± 1.95; P < 0.001 for Ngn2 and P < 0.01 for Math3, both versus pCIG2), cotransfection of Math3 and Ngn2 doubled NeuroD transactivation (43.52 ± 5.40; P < 0.001 versus all other means). Proneural bHLH proteins are thought to form obligate heterodimers with E2A bHLH proteins (e.g., E47) but are also capable of promiscuous heterodimerization (22). Accordingly, while E47 did not transactivate the NeuroD promoter by itself (1.67 ± 0.43), coexpression of E47 with Ngn2 or Math3 yielded about a twofold increase in promoter drive (Ngn2 plus E47, 39.05 ± 6.34; Math3 plus E47, 37.01 ± 5.08) (Fig. 7F). Strikingly, when the three bHLH genes were coexpressed in equal ratios (0.5 μg/μl each) and with the same total amount of bHLH DNA (1.5 μg/μl), there was a further 1.66-fold boost in promoter activity (72.55 ± 12.51; P < 0.001). The synergism between Ngn2 and Math3 was not generic to all bHLH genes as cotransfections of Ngn2 and the bHLH gene Mash1 (0.75 μg/μl each; 1.5 μg/μl total) resulted in activity levels that were roughly equivalent to transfecting a half-dosage of Ngn2 (14.12 ± 1.70; P < 0.001 versus 1.5 μg/μl Ngn2). Similarly, cotransfection of Math3 with Ngn2NRAQ, a DNA-binding domain mutant (0.75 μg/μl each) reduced transcriptional activity by half (9.35 ± 1.94) compared to double dosages (1.5 μg/μl) of Math3 (P < 0.01) or Ngn2 (P < 0.001). The DNA binding activity of Ngn2 is thus obligatory for cooperation with Math3.
To test if Ngn2 and Math3 proteins formed direct physical interactions, we performed GST pull-down assays. In vitro transcribed and translated, 35S-labeled Math3 efficiently bound immobilized GST-tagged Ngn2 but not GST alone (Fig. 7J). To test the effects of E47 on Ngn2-Math3 heterodimer formation, equivalent amounts of 35S-Math3 were mixed with increasing amounts of 35S-E47 (Fig. 7K, inputs) and then incubated with GST and GST-Ngn2-bound beads. Although E47 bound poorly to GST-Ngn2 relative to Math3, doubling the amount of E47 competitor reduced the amount of Math3 bound by approximately half (a 56% reduction in input-normalized optical density; normalized Math3 values were 255.7 with the initial amount of E47 and 113.2 with twice the amount of E47) (Fig. 7J). This strongly suggests that Math3 and E47 compete for the same binding site in Ngn2, likely the HLH domain.
DISCUSSION
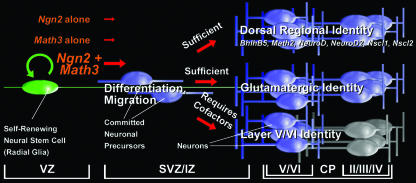
Neuronal differentiation occurs in a series of interdependent steps that require the strict temporal and spatial coordination of gene expression. In the cortex, several genes have been identified that participate in the specification of a glutamatergic projection neuron identity, yet few studies have examined how these genes are organized into genetic networks or conclusively determined if they are instructive or permissive determinants of neuronal identity. We report here that Ngn2 acts instructively to promote a cortical neuronal identity in the ventral telencephalon, inducing cortical gene expression in a defined temporal order. Surprisingly, however, for Ngn2 to efficiently and rapidly promote the expression of cortical genes, it must cooperate with a second bHLH gene, Math3, with which it forms physical interactions. Our study thus provides an important framework for understanding how bHLH genes cooperate to determine aspects of neuronal subtype identity in the cortex (Fig. 8).
FIG. 8.
Model of Ngn2 genetic cascades in the cortex. Ngn2, which is expressed in cycling cortical progenitors, initiates neuronal differentiation and specifies aspects of a neocortical projection neuron identity including neurotransmitter and regional identities. Ngn2 can induce Math3 expression in the dorsal telencephalon but requires context-specific cofactors which are not present in the ventral telencephalon to do so. The instructive power of Ngn2 does not extend to all cortical marker genes nor to lamina-specific genes, which must be activated by independent or codependent genetic pathways. CP, cortical plate; IZ, intermediate zone.
Ngn2 instructively induces the expression of a network of cortical genes.
The early expression of Ngn2 in VZ progenitors was consistent with its acting at the top of a cortical transcriptional cascade. Indeed, misexpression of Ngn2 in ventral telencephalic progenitors was sufficient to induce the ectopic expression of several cortical genes, including markers of a dorsal regional and glutamatergic neuronal identity. However, Ngn2 was not sufficient to induce expression of all cortical markers (e.g., FezL and Mef2c), including layer-specific genes, suggesting that it promotes a partial cortical identity. Nevertheless, this is one of the first examples in the central nervous system where a Ngn gene is instructive for neuronal subtype identity. For instance, in the spinal cord, Ngn2 is not sufficient to promote motoneuron or other ventral neuronal identities (45, 53, 55). Instead, Ngn2 mediates generic neuronal differentiation in motor neuron progenitors, an activity that is temporally coordinated with fate specification via interactions with the LIM homeodomain transcription factors Lhx3 and Islet1 (38). Ngn1 and Ngn2 also mediate generic neuronal differentiation and not cell fate specification in other contexts, such as the midbrain (32). In contrast, in the peripheral nervous system, Ngn1 and Ngn2 act instructively in neural crest cells to promote a sensory neuron identity, albeit in a context-dependent fashion (41, 54).
While at first glance our results conflict with a previous study in which Ngn2 was knocked into the Mash1 locus and was not sufficient to respecify ventral telencephalic progenitors (53), there are several key differences in the two studies. With the approach used herein, Ngn2 was expressed from a strong cytomegalovirus/β-actin enhancer/promoter that leads to high and protracted expression. This contrasts to the genetic approach, where Ngn2 was ectopically expressed in ventral telencephalic progenitors under the control of Mash1 regulatory sequences, leading to transient and more physiological expression levels (53). While the ability of Ngn2 to promote a cortical identity may be an artifact of the superphysiological levels of expression obtained by in utero electroporation, the ability of exogenous Ngn2 to transactivate the NeuroD promoter was considerably lower in the ventral telencephalon than the transactivation achieved with endogenous Ngn2 in cortical progenitors. Moreover, the initiation of cortical marker expression by Ngn2 took up to three times longer in the ventral telencephalon. We thus favor the interpretation that sustained expression (rather than overexpression) of Ngn2 is the critical difference in these two approaches. According to this model, Ngn2 would require a dorsally restricted cofactor(s) in order to achieve the rapid gene induction kinetics observed in the dorsal telencephalon. In its absence, Ngn2 would require more time to trigger gene expression (which does not occur in the knock-in model), either because its transactivation strength is subthreshold or because of additional mechanisms that repress target gene transcription in ventral domains (e.g., epigenetic modifications).
Synergistic interactions between Ngn2 and Math3.
Several observations suggested that Math3 is an essential cofactor that is required for Ngn2 to promote a cortical fate efficiently. First, Math3 is dorsally restricted and highly coexpressed with Ngn2 in cortical progenitors. Second, Math3 expression is lost (or sharply downregulated) in Ngn2 mutant cortices, suggesting that the loss of this gene may contribute to the downregulation of cortical gene expression observed in Ngn2 mutants. The future analysis of Math3 single mutants will be informative in this regard. Third, in ventral domains where Ngn2 activity is sharply attenuated, Math3 is not induced by ectopic Ngn2. This was surprising, as Math3 is a transcriptional target of Ngn in the spinal cord and Xenopus ectoderm (55, 71). However, in Ngn2 mutant retina, Math3 expression instead increases (1), suggesting that genetic interactions between Ngn2 and Math3 are context dependent. Fourth, Math3 activates the same transcriptional targets as Ngn2 in the ventral telencephalon, displaying similar, albeit slightly faster kinetics. Consistent with our model of cooperativity, coexpression of Math3 and Ngn2 in the ventral telencephalon boosted NeuroD promoter activity ∼1.4-fold, achieving transcriptional activation levels approaching those observed in the cortex in the absence of exogenous bHLH expression. Moreover, the observed boost in promoter drive correlated with the faster onset of target gene expression when Ngn2 and Math3 are coexpressed.
The mechanism(s) responsible for the observed cooperativity between Ngn2 and Math3 remains to be fully elucidated. One possibility is that Ngn2 and Math3, which, we showed, physically interact, might act as a heterodimer to activate target genes. Furthermore, our data suggest that while E47 can compete for the interaction domains of Ngn2 and Math3, it enhances functional cooperation between Ngn2 and Math3 rather than acting as a competitor. One possibility is that Ngn2, Math3, and E2A proteins generate a variety of heterodimer combinations. Ngn2/E47 and Math3/E47 heterodimers could act independently by binding to distinct promoter/enhancer elements or by recruiting different cofactors to modulate promoter architecture, as has been proposed for the myogenic bHLH genes MyoD and MyoG (13).
The nested expression of bHLH genes in progenitors and more differentiated cell types has been observed in neural and nonneural tissues. This has traditionally been interpreted to indicate that these genes act sequentially, first to determine and then to differentiate cells (11, 58). Although the bHLH genes Ngn2 and Math3 were thought to act in a linear sequence, we showed here that they cooperate to activate downstream gene expression. Yet we have also shown that Math3 or Ngn2 can also act independently to initiate gene expression, albeit with a temporal delay. Our data thus help explain why deleting a single bHLH gene often fails to yield a phenotype, except where very rapid and/or precisely timed differentiation events are required.
Supplementary Material
Acknowledgments
We thank David Anderson, Amparo Cano, Jay Cross, Masahiko Hibi, Tom Jessell, Ryoichiro Kageyama, Cairine Logan, Eric Olson, Sam Pfaff, Franck Polleux, Susan Morton, Ming-Jer Tsai, and Flora Vaccarino for generously providing reagents; Magdalena Götz for insightful comments; and Monica Vetter for critical reading of an earlier version of the manuscript.
C.S. is an Alberta Heritage Foundation for Medical Research (AHFMR) Senior Scholar. This work was supported by CIHR (MOP-44094) and Human Frontiers Science Program (RGY0019/2002) Operating Grants to C.S. L.M.L. and P.M. were supported by a CIHR training grant in Genetics, Child Development and Health; P.M. was also supported by a CIHR Canada Graduate Scholarship and AHFMR Studentship and is currently supported by a Heart and Stroke Foundation Studentship.
Footnotes
Published ahead of print on 26 December 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akagi, T., T. Inoue, G. Miyoshi, Y. Bessho, M. Takahashi, J. E. Lee, F. Guillemot, and R. Kageyama. 2004. Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J. Biol. Chem. 27928492-28498. [DOI] [PubMed] [Google Scholar]
- 2.Alam, S., D. Zinyk, L. Ma, and C. Schuurmans. 2005. Members of the Plag gene family are expressed in complementary and overlapping regions in the developing murine nervous system. Dev. Dyn. 234772-782. [DOI] [PubMed] [Google Scholar]
- 3.Anselmo, M. A., S. Dalvin, P. Prodhan, K. Komatsuzaki, J. T. Aidlen, J. J. Schnitzer, J. Y. Wu, and T. B. Kinane. 2003. Slit and robo: expression patterns in lung development. Gene Expr. Patterns 313-19. [DOI] [PubMed] [Google Scholar]
- 4.Arai, Y., N. Funatsu, K. Numayama-Tsuruta, T. Nomura, S. Nakamura, and N. Osumi. 2005. Role of Fabp7, a downstream gene of Pax6, in the maintenance of neuroepithelial cells during early embryonic development of the rat cortex. J. Neurosci. 259752-9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arber, S., D. R. Ladle, J. H. Lin, E. Frank, and T. M. Jessell. 2000. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell 101485-498. [DOI] [PubMed] [Google Scholar]
- 6.Backman, M., O. Machon, L. Mygland, C. J. van den Bout, W. Zhong, M. M. Taketo, and S. Krauss. 2005. Effects of canonical Wnt signaling on dorso-ventral specification of the mouse telencephalon. Dev. Biol. 279155-168. [DOI] [PubMed] [Google Scholar]
- 7.Bartholoma, A., and K. A. Nave. 1994. NEX-1: a novel brain-specific helix-loop-helix protein with autoregulation and sustained expression in mature cortical neurons. Mech. Dev. 48217-228. [DOI] [PubMed] [Google Scholar]
- 8.Begley, C. G., S. Lipkowitz, V. Gobel, K. A. Mahon, V. Bertness, A. R. Green, N. M. Gough, and I. R. Kirsch. 1992. Molecular characterization of NSCL, a gene encoding a helix-loop-helix protein expressed in the developing nervous system. Proc. Natl. Acad. Sci. USA 8938-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behar, T., W. Ma, L. Hudson, and J. L. Barker. 1994. Analysis of the anatomical distribution of GAD67 mRNA encoding truncated glutamic acid decarboxylase proteins in the embryonic rat brain. Brain Res. Dev. Brain Res. 7777-87. [DOI] [PubMed] [Google Scholar]
- 10.Bel-Vialar, S., F. Medevielle, and F. Pituello. 2007. The on/off of Pax6 controls the tempo of neuronal differentiation in the developing spinal cord. Dev. Biol. 305659-673. [DOI] [PubMed] [Google Scholar]
- 11.Bertrand, N., D. S. Castro, and F. Guillemot. 2002. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci 3517-530. [DOI] [PubMed] [Google Scholar]
- 12.Britz, O., P. Mattar, L. Nguyen, L. M. Langevin, C. Zimmer, S. Alam, F. Guillemot, and C. Schuurmans. 2006. A role for proneural genes in the maturation of cortical progenitor cells. Cereb. Cortex 16i138-i151. [DOI] [PubMed] [Google Scholar]
- 13.Cao, Y., R. M. Kumar, B. H. Penn, C. A. Berkes, C. Kooperberg, L. A. Boyer, R. A. Young, and S. J. Tapscott. 2006. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 25502-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cau, E., S. Casarosa, and F. Guillemot. 2002. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development 1291871-1880. [DOI] [PubMed] [Google Scholar]
- 15.Chen, B., L. R. Schaevitz, and S. K. McConnell. 2005. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc. Natl. Acad. Sci. USA 10217184-17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng, L., A. Arata, R. Mizuguchi, Y. Qian, A. Karunaratne, P. A. Gray, S. Arata, S. Shirasawa, M. Bouchard, P. Luo, C. L. Chen, M. Busslinger, M. Goulding, H. Onimaru, and Q. Ma. 2004. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat. Neurosci. 7510-517. [DOI] [PubMed] [Google Scholar]
- 17.Cross, J. C., M. L. Flannery, M. A. Blanar, E. Steingrimsson, N. A. Jenkins, N. G. Copeland, W. J. Rutler, and Z. Werb. 1995. Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development 1212513-2523. [DOI] [PubMed] [Google Scholar]
- 18.Farah, M. H., J. M. Olson, H. B. Sucic, R. I. Hume, S. J. Tapscott, and D. L. Turner. 2000. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development 127693-702. [DOI] [PubMed] [Google Scholar]
- 19.Fode, C., Q. Ma, S. Casarosa, S. L. Ang, D. J. Anderson, and F. Guillemot. 2000. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 1467-80. [PMC free article] [PubMed] [Google Scholar]
- 20.Fremeau, R. T., Jr., M. D. Troyer, I. Pahner, G. O. Nygaard, C. H. Tran, R. J. Reimer, E. E. Bellocchio, D. Fortin, J. Storm-Mathisen, and R. H. Edwards. 2001. The expression of vesicular glutamate transporters defines two classes of excitatory synapse. Neuron 31247-260. [DOI] [PubMed] [Google Scholar]
- 21.Ge, W., F. He, K. J. Kim, B. Blanchi, V. Coskun, L. Nguyen, X. Wu, J. Zhao, J. I. Heng, K. Martinowich, J. Tao, H. Wu, D. Castro, M. M. Sobeih, G. Corfas, J. G. Gleeson, M. E. Greenberg, F. Guillemot, and Y. E. Sun. 2006. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl. Acad. Sci. USA 1031319-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gradwohl, G., C. Fode, and F. Guillemot. 1996. Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev. Biol. 180227-241. [DOI] [PubMed] [Google Scholar]
- 23.Guillemot, F., and A. L. Joyner. 1993. Dynamic expression of the murine Achaete-Scute homologue Mash-1 in the developing nervous system. Mech. Dev. 42171-185. [DOI] [PubMed] [Google Scholar]
- 24.Gunhaga, L., M. Marklund, M. Sjodal, J. C. Hsieh, T. M. Jessell, and T. Edlund. 2003. Specification of dorsal telencephalic character by sequential Wnt and FGF signaling. Nat. Neurosci. 6701-707. [DOI] [PubMed] [Google Scholar]
- 25.Gunnersen, J. M., C. Augustine, V. Spirkoska, M. Kim, M. Brown, and S. S. Tan. 2002. Global analysis of gene expression patterns in developing mouse neocortex using serial analysis of gene expression. Mol. Cell Neurosci. 19560-573. [DOI] [PubMed] [Google Scholar]
- 26.Hand, R., D. Bortone, P. Mattar, L. Nguyen, J. I. Heng, S. Guerrier, E. Boutt, E. Peters, A. P. Barnes, C. Parras, C. Schuurmans, F. Guillemot, and F. Polleux. 2005. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron 4845-62. [DOI] [PubMed] [Google Scholar]
- 27.Hevner, R. F., L. Shi, N. Justice, Y. Hsueh, M. Sheng, S. Smiga, A. Bulfone, A. M. Goffinet, A. T. Campagnoni, and J. L. Rubenstein. 2001. Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29353-366. [DOI] [PubMed] [Google Scholar]
- 28.Hirata, T., Y. Suda, K. Nakao, M. Narimatsu, T. Hirano, and M. Hibi. 2004. Zinc finger gene fez-like functions in the formation of subplate neurons and thalamocortical axons. Dev. Dyn. 230546-556. [DOI] [PubMed] [Google Scholar]
- 29.Holm, P. C., M. T. Mader, N. Haubst, A. Wizenmann, M. Sigvardsson, and M. Gotz. 2007. Loss- and gain-of-function analyses reveal targets of Pax6 in the developing mouse telencephalon. Mol. Cell Neurosci. 3499-119. [DOI] [PubMed] [Google Scholar]
- 30.Huang, H. P., M. Liu, H. M. El-Hodiri, K. Chu, M. Jamrich, and M. J. Tsai. 2000. Regulation of the pancreatic islet-specific gene BETA2 (neuroD) by neurogenin 3. Mol. Cell. Biol. 203292-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeong, J. Y., Z. Einhorn, S. Mercurio, S. Lee, B. Lau, M. Mione, S. W. Wilson, and S. Guo. 2006. Neurogenin1 is a determinant of zebrafish basal forebrain dopaminergic neurons and is regulated by the conserved zinc finger protein Tof/Fezl. Proc. Natl. Acad. Sci. USA 1035143-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kele, J., N. Simplicio, A. L. Ferri, H. Mira, F. Guillemot, E. Arenas, and S. L. Ang. 2006. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development 133495-505. [DOI] [PubMed] [Google Scholar]
- 33.Kim, C. H., Y. K. Bae, Y. Yamanaka, S. Yamashita, T. Shimizu, R. Fujii, H. C. Park, S. Y. Yeo, T. L. Huh, M. Hibi, and T. Hirano. 1997. Overexpression of neurogenin induces ectopic expression of HuC in zebrafish. Neurosci. Lett. 239113-116. [DOI] [PubMed] [Google Scholar]
- 34.Kruger, M., and T. Braun. 2002. The neuronal basic helix-loop-helix transcription factor NSCL-1 is dispensable for normal neuronal development. Mol. Cell. Biol. 22792-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kruger, M., T. Schmid, S. Kruger, E. Bober, and T. Braun. 2006. Functional redundancy of NSCL-1 and NeuroD during development of the petrosal and vestibulocochlear ganglia. Eur. J. Neurosci. 241581-1590. [DOI] [PubMed] [Google Scholar]
- 36.Langevin, L. M., P. Mattar, R. Scardigli, M. Roussigne, C. Logan, P. Blader, and C. Schuurmans. 2007. Validating in utero electroporation for the rapid analysis of gene regulatory elements in the murine telencephalon. Dev. Dyn. 2361273-1286. [DOI] [PubMed] [Google Scholar]
- 37.Lee, J. E., S. M. Hollenberg, L. Snider, D. L. Turner, N. Lipnick, and H. Weintraub. 1995. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science 268836-844. [DOI] [PubMed] [Google Scholar]
- 38.Lee, S. K., and S. L. Pfaff. 2003. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron 38731-745. [DOI] [PubMed] [Google Scholar]
- 39.Lin, X., M. W. State, F. M. Vaccarino, J. Greally, M. Hass, and J. F. Leckman. 1999. Identification, chromosomal assignment, and expression analysis of the human homeodomain-containing gene Orthopedia (OTP). Genomics 6096-104. [DOI] [PubMed] [Google Scholar]
- 40.Liu, M., S. J. Pleasure, A. E. Collins, J. L. Noebels, F. J. Naya, M. J. Tsai, and D. H. Lowenstein. 2000. Loss of BETA2/NeuroD leads to malformation of the dentate gyrus and epilepsy. Proc. Natl. Acad. Sci. USA 97865-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo, L., E. Dormand, A. Greenwood, and D. J. Anderson. 2002. Comparison of the generic neuronal differentiation and neuron subtype specification functions of mammalian achaete-scute and atonal homologs in cultured neural progenitor cells. Development 1291553-1567. [DOI] [PubMed] [Google Scholar]
- 42.Machon, O., M. Backman, S. Krauss, and Z. Kozmik. 2005. The cellular fate of cortical progenitors is not maintained in neurosphere cultures. Mol. Cell Neurosci. 30388-397. [DOI] [PubMed] [Google Scholar]
- 43.Martin, J. F., J. J. Schwarz, and E. N. Olson. 1993. Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc. Natl. Acad. Sci. USA 905282-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattar, P., O. Britz, C. Johannes, M. Nieto, L. Ma, A. Rebeyka, N. Klenin, F. Polleux, F. Guillemot, and C. Schuurmans. 2004. A screen for downstream effectors of Neurogenin2 in the embryonic neocortex. Dev. Biol. 273373-389. [DOI] [PubMed] [Google Scholar]
- 45.Mizuguchi, R., M. Sugimori, H. Takebayashi, H. Kosako, M. Nagao, S. Yoshida, Y. Nabeshima, K. Shimamura, and M. Nakafuku. 2001. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron 31757-771. [DOI] [PubMed] [Google Scholar]
- 46.Molyneaux, B. J., P. Arlotta, T. Hirata, M. Hibi, and J. D. Macklis. 2005. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron 47817-831. [DOI] [PubMed] [Google Scholar]
- 47.Molyneaux, B. J., P. Arlotta, J. R. Menezes, and J. D. Macklis. 2007. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci 8427-437. [DOI] [PubMed] [Google Scholar]
- 48.Monuki, E. S., F. D. Porter, and C. A. Walsh. 2001. Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron 32591-604. [DOI] [PubMed] [Google Scholar]
- 49.Muzio, L., B. DiBenedetto, A. Stoykova, E. Boncinelli, P. Gruss, and A. Mallamaci. 2002. Conversion of cerebral cortex into basal ganglia in Emx2(−/−) Pax6(Sey/Sey) double-mutant mice. Nat. Neurosci. 5737-745. [DOI] [PubMed] [Google Scholar]
- 50.Nakatani, T., Y. Minaki, M. Kumai, and Y. Ono. 2007. Helt determines GABAergic over glutamatergic neuronal fate by repressing Ngn genes in the developing mesencephalon. Development 1342783-2793. [DOI] [PubMed] [Google Scholar]
- 51.Olson, J. M., A. Asakura, L. Snider, R. Hawkes, A. Strand, J. Stoeck, A. Hallahan, J. Pritchard, and S. J. Tapscott. 2001. NeuroD2 is necessary for development and survival of central nervous system neurons. Dev. Biol. 234174-187. [DOI] [PubMed] [Google Scholar]
- 52.Parker, M. H., P. Seale, and M. A. Rudnicki. 2003. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 4497-507. [DOI] [PubMed] [Google Scholar]
- 53.Parras, C. M., C. Schuurmans, R. Scardigli, J. Kim, D. J. Anderson, and F. Guillemot. 2002. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 16324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez, S. E., S. Rebelo, and D. J. Anderson. 1999. Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development 1261715-1728. [DOI] [PubMed] [Google Scholar]
- 55.Perron, M., K. Opdecamp, K. Butler, W. A. Harris, and E. J. Bellefroid. 1999. X-ngnr-1 and Xath3 promote ectopic expression of sensory neuron markers in the neurula ectoderm and have distinct inducing properties in the retina. Proc. Natl. Acad. Sci. USA 9614996-15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puelles, E., D. Acampora, R. Gogoi, F. Tuorto, A. Papalia, F. Guillemot, S. L. Ang, and A. Simeone. 2006. Otx2 controls identity and fate of glutamatergic progenitors of the thalamus by repressing GABAergic differentiation. J. Neurosci. 265955-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rash, B. G., and E. A. Grove. 2006. Area and layer patterning in the developing cerebral cortex. Curr. Opin. Neurobiol. 1625-34. [DOI] [PubMed] [Google Scholar]
- 58.Ross, S. E., M. E. Greenberg, and C. D. Stiles. 2003. Basic helix-loop-helix factors in cortical development. Neuron 3913-25. [DOI] [PubMed] [Google Scholar]
- 59.Saito, T., and N. Nakatsuji. 2001. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 240237-246. [DOI] [PubMed] [Google Scholar]
- 60.Scardigli, R., C. Schuurmans, G. Gradwohl, and F. Guillemot. 2001. Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron 31203-217. [DOI] [PubMed] [Google Scholar]
- 61.Schaeren-Wiemers, N., E. Andre, J. P. Kapfhammer, and M. Becker-Andre. 1997. The expression pattern of the orphan nuclear receptor RORbeta in the developing and adult rat nervous system suggests a role in the processing of sensory information and in circadian rhythm. Eur. J. Neurosci. 92687-2701. [DOI] [PubMed] [Google Scholar]
- 62.Schuurmans, C., O. Armant, M. Nieto, J. M. Stenman, O. Britz, N. Klenin, C. Brown, L. M. Langevin, J. Seibt, H. Tang, J. M. Cunningham, R. Dyck, C. Walsh, K. Campbell, F. Polleux, and F. Guillemot. 2004. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 232892-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schuurmans, C., and F. Guillemot. 2002. Molecular mechanisms underlying cell fate specification in the developing telencephalon. Curr. Opin. Neurobiol. 1226-34. [DOI] [PubMed] [Google Scholar]
- 64.Schwab, M. H., A. Bartholomae, B. Heimrich, D. Feldmeyer, S. Druffel-Augustin, S. Goebbels, F. J. Naya, S. Zhao, M. Frotscher, M. J. Tsai, and K. A. Nave. 2000. Neuronal basic helix-loop-helix proteins (NEX and BETA2/Neuro D) regulate terminal granule cell differentiation in the hippocampus. J. Neurosci. 203714-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwab, M. H., S. Druffel-Augustin, P. Gass, M. Jung, M. Klugmann, A. Bartholomae, M. J. Rossner, and K. A. Nave. 1998. Neuronal basic helix-loop-helix proteins (NEX, neuroD, NDRF): spatiotemporal expression and targeted disruption of the NEX gene in transgenic mice. J. Neurosci. 181408-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seo, S., J. W. Lim, D. Yellajoshyula, L. W. Chang, and K. L. Kroll. 2007. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 265093-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simmons, D. G., A. L. Fortier, and J. C. Cross. 2007. Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev. Biol. 304567-578. [DOI] [PubMed] [Google Scholar]
- 68.Stenman, J., R. T. Yu, R. M. Evans, and K. Campbell. 2003. Tlx and Pax6 co-operate genetically to establish the pallio-subpallial boundary in the embryonic mouse telencephalon. Development 1301113-1122. [DOI] [PubMed] [Google Scholar]
- 69.Sun, Y., M. Nadal-Vicens, S. Misono, M. Z. Lin, A. Zubiaga, X. Hua, G. Fan, and M. E. Greenberg. 2001. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 104365-376. [DOI] [PubMed] [Google Scholar]
- 70.Takebayashi, K., S. Takahashi, C. Yokota, H. Tsuda, S. Nakanishi, M. Asashima, and R. Kageyama. 1997. Conversion of ectoderm into a neural fate by ATH-3, a vertebrate basic helix-loop-helix gene homologous to Drosophila proneural gene atonal. EMBO J. 16384-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Talikka, M., S. E. Perez, and K. Zimmerman. 2002. Distinct patterns of downstream target activation are specified by the helix-loop-helix domain of proneural basic helix-loop-helix transcription factors. Dev. Biol. 247137-148. [DOI] [PubMed] [Google Scholar]
- 72.Theil, T., G. Alvarez-Bolado, A. Walter, and U. Ruther. 1999. Gli3 is required for Emx gene expression during dorsal telencephalon development. Development 1263561-3571. [DOI] [PubMed] [Google Scholar]
- 73.Tole, S., C. W. Ragsdale, and E. A. Grove. 2000. Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes(J). Dev. Biol. 217254-265. [DOI] [PubMed] [Google Scholar]
- 74.Tomita, K., K. Moriyoshi, S. Nakanishi, F. Guillemot, and R. Kageyama. 2000. Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 195460-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.von Frowein, J., A. Wizenmann, and M. Gotz. 2006. The transcription factors Emx1 and Emx2 suppress choroid plexus development and promote neuroepithelial cell fate. Dev. Biol. 296239-252. [DOI] [PubMed] [Google Scholar]
- 76.Watanabe, K., D. Kamiya, A. Nishiyama, T. Katayama, S. Nozaki, H. Kawasaki, Y. Watanabe, K. Mizuseki, and Y. Sasai. 2005. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat. Neurosci. 8288-296. [DOI] [PubMed] [Google Scholar]
- 77.Xu, Z. P., A. Dutra, C. M. Stellrecht, C. Wu, J. Piatigorsky, and G. F. Saunders. 2002. Functional and structural characterization of the human gene BHLHB5, encoding a basic helix-loop-helix transcription factor. Genomics 80311-318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.