Abstract
The positive elongation factor P-TEFb appears to function as a crucial C-terminal-domain (CTD) kinase for RNA polymerase II (Pol II) transcribing immediate early genes (IEGs) in neuroendocrine GH4C1 cells. Chromatin immunoprecipitation indicated that in resting cells Pol II occupied the promoter-proximal regions of the c-fos and junB genes, together with the negative elongation factors DSIF and NELF. Thyrotropin-releasing hormone (TRH)-induced recruitment of positive transcription elongation factor b (P-TEFb) abolished the pausing of Pol II and enhanced phosphorylation of CTD serine 2, resulting in transcription elongation. In addition, P-TEFb was essential for splicing and 3′-end processing of IEG transcripts. Importantly, the MEK1-extracellular signal-regulated kinase (ERK) signaling pathway activated by TRH up-regulated nuclear CDK9 and CDK9/cyclinT1 dimers (i.e., P-TEFb), facilitating the recruitment of P-TEFb to c-fos and other IEGs. Thus, in addition to established gene transcription control via promoter response elements, the MEK1-ERK signaling pathway controls transcription elongation by Pol II via the up-regulation of nuclear CDK9 integrated into P-TEFb.
Gene transcription by RNA polymerase II (Pol II) proceeds through multiple steps: preinitiation, initiation, elongation, and termination (45). Historically, preinitiation and initiation have been considered the rate-limiting steps. Consequently, most studies on transcription control mechanisms have focused on the cis- and trans-acting elements in promoters. However, it has become increasingly evident in recent years that transcription elongation of many inducible genes is controlled by external stimuli (36, 51). These include the immediate early genes (IEGs) c-fos, c-myc, and MKP-1 (mitogen-activated protein kinase [MAPK] phosphatase 1), for which, based upon in vitro nuclear run-on experiments, a block to elongation has been postulated. Pol II transcription initiation of such genes is constitutive even under cellular resting conditions; however, transcripts are not elongated unless extracellular stimuli trigger intracellular signals, which permit transcription elongation to produce full-length transcripts (11, 13, 30, 41, 42, 54).
Progress through the transcription steps is tightly linked to the phosphorylation state of the C-terminal domain (CTD) in a large subunit of Pol II (37). The CTD consists of repeats of the YSPTSPS motif—52 repeats in mammalian cells. Various CTD kinases, including cyclin-dependent kinases (CDKs), selectively phosphorylate the serine residues at positions 2 and 5 (Ser-2 and Ser-5, respectively). The phosphorylation pattern of the CTD is changed in a dynamic fashion during the activation and attenuation of transcription. Pol II, which has initiated transcription, is phosphorylated on Ser-5, elongating Pol II in addition on Ser-2 (6, 10, 29, 37, 43). We have recently shown that the rate of c-fos transcription in vivo is continuously regulated at the level of elongation and that this regulation is reflected by the dynamic changes of Pol II CTD phosphorylation along the c-fos gene (41). Positive transcription elongation factor b (P-TEFb), a complex of cyclin T1 with CDK9, a kinase preferentially phosphorylating Ser-2 of the CTD, is upon activation recruited massively to the transcription unit on the whole c-fos gene. It thus appears that a Pol II complex that can overcome a block to elongation necessarily includes P-TEFb, presumably in order to keep Pol II CTD Ser-2 phosphorylated.
A number of P-TEFs and negative transcription elongation factors (N-TEFs) have been identified as regulators to accelerate or attenuate transcription elongation by Pol II (12, 36, 44). Transcription elongation control mechanisms involving P-TEFs and N-TEFs have been studied extensively in vitro (33, 52, 53, 59-61). 5,6-Dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) sensitivity-inducing factor (DSIF) and negative elongation factor (NELF) are recruited in the promoter-proximal region of a gene, causing Pol II to pause. Once P-TEFb is recruited to the gene, it will phosphorylate the CTD of Pol II and the C-terminal repeats (CTR) of Spt5, a subunit of DSIF. As a consequence, paused Pol II will resume transcription elongation of nascent transcripts. In addition to the in vitro reports, some in vivo observations, especially in Drosophila cells, have shown that NELF is present in the promoter-proximal regions of heat shock genes in resting cells, and P-TEFb is recruited on these genes after heat shock to induce their transcription (3, 6, 26, 56, 57). DSIF associates with Drosophila heat shock genes, not only in resting cells, but also during active transcription (3, 26, 56). Spt5 and its phosphorylation by P-TEFb are required for epidermal growth factor-induced transcription elongation on the c-fos gene in HeLa cells (58). We have recently described gene-specific recruitment of DSIF before and during stimulated transcription of the MKP-1 gene in neuroendocrine cells (18). Thus, although DSIF was discovered as an N-TEF, these reports suggest that it plays a dual role, functioning as an N-TEF and a P-TEF during resting and active transcription, respectively. Note that DSIF is still referred to as a “negative” elongation factor in reference to the background of its discovery.
P-TEFb plays a general role in transcription elongation and primary transcript processing, not only for induced genes, but also for genes that are expressed constitutively (9). The question thus arises of whether and how activation of intracellular signaling would lead to enhanced activity of P-TEFb. To address this, we studied the induction of IEGs by thyrotropin-releasing hormone (TRH) in pituitary GH4C1 cells. We first examined in detail how P-TEFb-regulated transcription elongation of the c-fos and junB genes was induced by TRH via CTD phosphorylation and modulation of the negative elongation factors NELF and DSIF. Next, we examined the relationship of P-TEFb with TRH-induced signaling cascades. A major intracellular signaling pathway stimulated by TRH is the MEK1-extracellular signal-regulated kinase (ERK) cascade. Several mechanisms by which this pathway regulates transcription have been well studied. They include especially the phosphorylation of CREB and of TCF to address the response elements CRE and SRE in the c-fos promoter (49), as well as the phosphoryation of histones (48, 64). To study the potential role in which this pathway contributes to transcription elongation regulation, we progressed with the working hypothesis that the MEK1-ERK pathway would affect directly or indirectly the function of P-TEFb. We show that the MEK1-ERK pathway is decisive for TRH-induced recruitment of P-TEFb to IEGs and subsequent transcript elongation and processing. Evidence is presented that the activation of the MEK1-ERK signaling pathway enhances the nuclear availability of CDK9 and thus of P-TEFb. Such a process could contribute to induced transcription elongation of IEGs, for which P-TEFb may be rate limiting.
MATERIALS AND METHODS
Cell culture and stimulation.
Pituitary GH4C1 cells were usually grown at 37°C in Ham's F-10 medium (Invitrogen, Carlsbad, CA) containing 2.5% (vol/vol) fetal bovine serum and 15% (vol/vol) horse serum in a humidified atmosphere of 5% CO2. GH4C1 cells incubated in Ham's F-10 serum-free medium for 24 h were incubated with 30 μM DRB (Sigma, St. Louis, MO) for 2 h, 10 μM U0126 (Promega, Madison, WI) for 1 h, or 10 μM ERK activation inhibitor peptide I (Calbiochem, San Diego, CA) for 2 h and then stimulated with 100 nM TRH (Roche, Indianapolis, IN).
ChIP assay.
An anti-cyclin T1 polyclonal antibody (H-245; Santa Cruz Biotechnology, Santa Cruz, CA), an anti-Spt5 monoclonal antibody (BD Bioscience, Lexington, KY), an anti-NELF-A polyclonal antibody (A-20; Santa Cruz Biotechnology), an anti-Pol II polyclonal antibody (N-20; Santa Cruz Biotechnology), and anti-Pol II monoclonal antibodies (8WG16, H2, and H5; Convance, Princeton, NJ) were used. Chromatin immunoprecipitation (ChIP) assays were performed as described previously (18, 41). DNAs precipitated with the antibodies were used as templates for quantitative real-time PCR with Sybr green PCR master mix (Applied Biosystems, Foster City, CA) or Universal PCR master mix (Applied Biosystems). Primers and TaqMan probes used in this experiment were as follows: c-fos promoter-proximal region (−62 to +14), forward primer, 5′-CTCATGACGTAGTAAGCCATTCAAG-3′, and reverse primer, 5′-GCAATCGCGGTTGGAGTAGT-3′; c-fos exon 1 (+123 to +187), forward primer, 5′-CCCTCGCCGAGCTTTGC-3′, and reverse primer, 5′-GCCTCGTAGTCCGCGTTGA-3′; TaqMan probe, 5′-6-carboxyfluorescein (FAM)-CAAACCACGACCATGATGTTCTCGGGT-6-carboxytetramethylrhodamine (TAMRA)-3′; c-fos exon 1 and intron 1 (+244 to +316), forward primer, 5′-ATTCCCCAGCCGACTCCTT-3′, and reverse primer, 5′-CGACTGCACAAAGCCAAACTC-3′; c-fos intron 1 (+502 to +595), forward primer, 5′-CGCGGCAGGTTTACTCTGA-3′, and reverse primer, 5′-AGCGAGTCTTTGCTAGAGACTTGTT-3′; c-fos exon 3 (+1746 to +1818), forward primer, 5′-AAGGGAAAGGAATAAGATGGCTG-3′, and reverse primer, 5′-CGCTTGGAGCGTATCTGTCA-3′; TaqMan probe, 5′-FAM-CCTCCGATTCCGGCACTTGGCT-TAMRA-3′; c-fos exon 4 (+2423 to +2495), forward primer, 5′-TCCCAGCTGCACTACCTATACG-3′, and reverse primer, 5′-TGCGCAGCTAGGGAAGGA-3′; TaqMan probe, 5′-FAM-CTTCCTTTGTCTTCACCTACCCCGAGGC-TAMRA-3′; junB 5′ (+63 to +133), forward primer, 5′-GCACCACGGAGGGAGAGA-3′, and reverse primer, 5′-CAGCGGCCAGTTGGTAGCT-3′; junB 5′b (+277 to +341), forward primer, 5′-ACGAAAATGGAACAGCCTTTCTA-3′, and reverse primer, 5′-GGGCTCCGACCGTATCCT-3′; junB 3′ (+1147 to +1237), forward primer, 5′-CTGGAGGACAAGGTGAAGACACT-3′, and reverse primer, 5′-GCTTGAGCTGCGCCACTT-3′; MKP-1 exon 1 (+21 to +94), forward primer, 5′-GGGACGCGCGGTGAAG-3′, and reverse primer, 5′-GATCTTGTGCGGTTTTTTGTGG-3′; TaqMan probe, 5′-FAM-CCTAAGTCCTCAAGTGCTCGCTGATCCTAATCT-TAMRA-3′; MKP-1 exon 1b (+227 to +287), forward primer, 5′-CTTCTGGATTGTCGCTCCTTCT-3′, and reverse primer, 5′-CGTTCACTGAGCCCACGAT-3′; MKP-1 exon 4 (+2043 to +2109), forward primer, 5′-CCCTGTTCACCCCACGAA-3′, and reverse primer, 5′-GCAGCTCGGAGAGGTTGTG-3′; TaqMan probe, 5′-FAM-TGCCCTGAACTACCTTCAAAGCCCCA-TAMRA-3′; nongenetic region, forward primer, 5′-GATTCCCCAGAACCCACATG-3′, and reverse primer, 5′-GATCCCTGGGTCTGGCATTA-3′.
RNA preparation and quantitative reverse transcriptase (RT) PCR.
Total RNA was extracted from GH4C1 cells with an acid phenol-guanidinium reagent (TRI-Reagent; Molecular Research Center, Cincinnati, OH) according to the manufacturer's instructions. Quantifications of c-fos, junB, and MKP-1 RNAs was performed with the total RNA by using real-time PCR with Sybr Green PCR master mix (Applied Biosystems) or Universal PCR master mix (Applied Biosystems) as described previously (42). The primers and TaqMan probes used in this experiment were as follows; c-fos spliced mRNA, forward primer, 5′-GACAGCCTTTCCTACTACCATTCC-3′, and reverse primer, 5′-AAAGTTGGCACTAGAGACGGACAG-3′; c-fos total RNA, forward primer, 5′-CTTCAGCGTCCATGTTCATTGT-3′, and reverse primer, 5′-ACGTTTTCATGGAAAACTGTTAATGTC-3′; c-fos 3′-end noncleaved RNA, forward primer, 5′-TGCGACCACCTCGTTGCT-3′, and reverse primer, 5′-CCCATCTTGACAAACTGGTCTCT-3′; junB total RNA, forward primer, 5′-GGAGCAGGAGGGCTTTGC-3′, and reverse primer, 5′-CGTCACGTGGTTCATCTTGTG-3′; junB 3′-end noncleaved RNA, forward primer, 5′-TAATTTCTGTGGTCTCTTTCTTCCAA-3′, and reverse primer, 5′-CCCGAAAGGAGAGAACTACAAACT-3′; MKP-1 spliced mRNA, forward primer, 5′-CGCGCTCCACTCAAGTCTTC-3′, and reverse primer, 5′-GGTGGACTGTTTGCTGCACA-3′; TaqMan probe, 5′-FAM-AGCCGAAAACGCTTCATATCCTCCTTGG-TAMRA-3′; MKP-1 total RNA, forward primer, 5′-CCCTGTTCACCCCACGAA-3′, and reverse primer, 5′-GCAGCTCGGAGAGGTTGTG-3′; TaqMan probe, 5′-FAM-TGCCCTGAACTACCTTCAAAGCCCCA-TAMRA-3′; MKP-1 3′-end noncleaved RNA, forward primer, 5′-TATTGAGTTCTCGGTGCTTTGTTTC-3′, and reverse primer, 5′-GGTTCAGTGCCCCATACTATGC-3′. For normalization, amplification of 18S rRNA was performed under standard conditions by using 18S rRNA predeveloped assay reagent (Applied Biosystems).
Preparation of cytosolic and nuclear extracts.
GH4C1 cells (about 2 × 107 cells) stimulated with TRH for 0 to 48 min were lysed in 1 ml of lysis buffer (10 mM Tris-HCl, pH 8.0, 60 mM KCl, 1 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 50 mM NaF, 1 mM Na3VO4, 1 μg/ml leupeptin, 1 μg/ml aprotinin) for 15 min on ice and centrifuged (1,500 × g) at 4°C for 4 min. The supernatant and precipitate were collected as cytosolic extract and debris including the nucleus, respectively. After being washed with the lysis buffer without NP-40, the debris was incubated with nuclear extraction buffer (20 mM Tris-HCl, pH 8.0, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM PMSF, 1 mM Na3VO4, 50 mM NaF, 1 μg/ml leupeptin, 1 μg/ml aprotinin) for 30 min at 4°C. After centrifugation (16,000 × g) at 4°C for 10 min, the supernatant was collected as the nuclear extract.
Immunoprecipitation and in vitro phosphorylation assay.
Nuclear fraction (90 μg) in immunoprecipitation buffer (20 mM Tris-HCl, pH 8.0, 25% glycerol, 150 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% NP-40, 1 mM PMSF, 1 mM Na3VO4, 50 mM NaF, 1 μg/ml leupeptin, 1 μg/ml aprotinin) was incubated with 1 μg of an anti-cyclin T1 antibody (H-245; Santa Cruz Biotechnology) at 4°C for 2 h and then mixed with 30 μl of protein A-Sepharose beads (50% [vol/vol] slurry; Amersham Pharmacia) at 4°C for 2 h. The beads were washed three times with immunoprecipitation buffer and then boiled with sample buffer (125 mM Tris-HCl, pH 6.8, 4% sodium dodecyl sulfate, 0.2% bromophenol blue, 10% 2-mercaptoethanol). For in vitro phosphorylation assays, the beads washed with immunoprecipitation buffer were washed another two times with CTD kinase buffer (20 mM Tris-HCl, pH 7.4, 5 mM MgCl2). The immunoprecipitated protein complex was incubated with 1 μg of glutathione S-transferase (GST)-CTD protein (prepared as described by Wada et al. [53]), 50 μM ATP, and 370 kBq [γ-32P]ATP in CTD kinase buffer at 30°C for 25 min. The reaction was stopped by adding sample buffer and boiling the mixture. The GST-CTD proteins were separated in sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then subjected to autoradiography.
Western blotting.
The immunoprecipitated protein complex denatured in sample buffer was subjected to Western blotting with an anti-cyclin T1 polyclonal antibody (H-245; Santa Cruz Biotechnology) and an anti-CDK9 monoclonal antibody (D-7; Santa Cruz Biotechnology). The cytosolic and nuclear extracts denatured in sample buffer were subjected to Western blotting using an anti-phospho-p44/42 MAPK (Thr202/Tyr204; Cell Signaling Technology, Danvers, MA), an anti-ERK1 (BD Bioscience), an anti-CDK9 (C-20; Santa Cruz Biotechnology), an anti-cyclin T1 (H-245; Santa Cruz Biotechnology), an anti-TFIIB (C-18; Santa Cruz Biotechnology), and an antiactin (Sigma) polyclonal antibody. The protein bands were visualized by using a horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody (BD Bioscience) or a horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G antibody (BD Bioscience) with LumiGlo reagent and peroxide (Cell Signaling Technology). The intensities of the visualized bands were analyzed using Scion Imaging Software (Scion Corporation, Frederick, MD).
RESULTS
Occupancy of the c-fos and junB genes by P-TEFb, NELF, and DSIF.
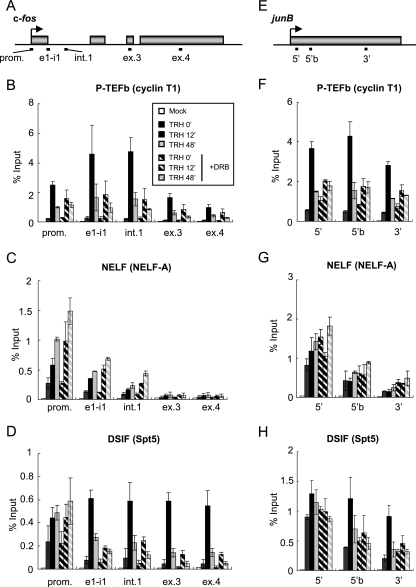
We first assessed quantitatively in vivo the association of P-TEFb, NELF, and DSIF with the IEG c-fos. To this end, ChIP was combined with quantitative real-time PCR. We reported previously that c-fos transcription in GH4C1 cells is immediately induced by TRH stimulation and reaches maximal rates at around 12 min thereafter (41). Beyond this time point, the transcription rate is attenuated but remains significantly elevated over starting levels for 48 to 96 min. In this study, we quantified the association of the three elongation factors at three time points, representing (i) the basal state prior to induction (0 min), (ii) the most active state of transcription (12 min after TRH stimulation), and (iii) the attenuated but sustained steady state of induced transcription (48 min after TRH stimulation). In consideration of the postulated block to elongation in the promoter-proximal region (40) and/or in intron 1 (11, 30) on the c-fos gene, we designed three real-time PCR primer sets probing the promoter, including the promoter-proximal region, the exon 1-intron 1 junction, and intron 1. Two further probes monitored exon 3 and exon 4 (Fig. 1A).
FIG. 1.
Association of P-TEFb, NELF, and DSIF with the c-fos and junB genes. (A) Rat c-fos (GeneID, 314322) genomic locus; the boxes represent exons. The bars below the genes show the positions of the primer sets used in ChIP assays: promoter-proximal region (prom.), exon 1/intron 1 junction (e1-i1), intron 1 (int.1), exon 3 (ex.3), and exon 4 (ex.4). (B to D) Distribution of P-TEFb and N-TEFs on the c-fos gene. ChIP assays were performed with an anti-cyclin T1 antibody (B), an anti-NELF-A antibody (C), and an anti-Spt5 antibody (D) with chromatin at various time points after TRH stimulation (0 [basal condition], 12, and 48 min) in the absence (filled bars) or presence (hatched bars) of DRB. The density on the c-fos gene was quantified by using real-time PCR and is presented as a percentage of the input (the mean of two experiments is shown; the error bars represent the range). (E) Rat junB (GeneID, 24517) genomic locus; the box represents the single exon. The bars below the genes show the positions of the primer sets used in ChIP assays: the 5′ region (5′), 5′ region b (5′b), and 3′ region (3′). (F to H) Distribution of P-TEFb and N-TEFs on the junB gene (as in panels B to D).
The localization of P-TEFb, which consists of cyclin T1 and CDK9, was examined by ChIP with an anti-cyclin T1 antibody (Fig. 1B). Prior to TRH stimulation, P-TEFb was present only in the 5′ region of the c-fos gene. Although very modest, the ChIP signals obtained with probes for the promoter-proximal, exon 1-intron 1 junction, and intron 1 regions were clearly higher than the background ChIP signal obtained for a nongenetic region (see Fig. S1 in the supplemental material), showing that some P-TEFb is present in the 5′ regions of the gene even under cellular resting conditions. At 12 min after TRH stimulation, P-TEFb was recruited to the whole gene. While exon 1 and intron 1 were most prominently occupied by P-TEFb, the elongation factor also significantly occupied the downstream parts of the c-fos gene. At 48 min after TRH stimulation, P-TEFb associations with all parts of the c-fos gene were reduced in parallel to between 20% and 40% of the maxima but remained significantly elevated compared to resting cells (TRH, 0 min). We have shown previously that CDK9 activity is essential for the recruitment of P-TEFb and DSIF to the transcribed parts of the MKP-1 gene (18). Treatment of GH4C1 cells with DRB, a specific inhibitor of CDK9 kinase, markedly reduced the induced occupancy by P-TEFb of the transcribed part of the c-fos gene, especially in the promoter-proximal, exon 1-intron 1 junction, and intron 1 regions. Assuming that P-TEFb occupancy is linked to Pol II occupancy (Fig. 2), this would be consistent with the need for CDK9 activity phosphorylating Pol II to overcome the block to elongation (11, 30, 40).
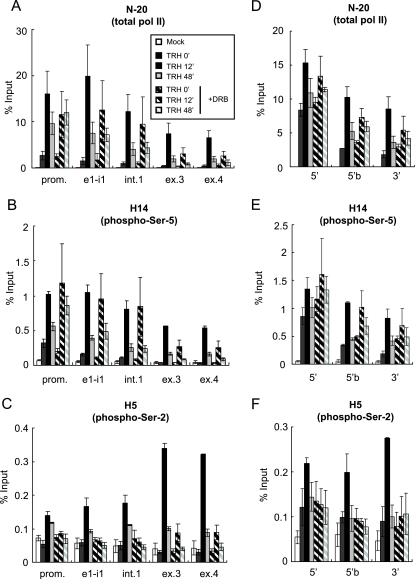
FIG. 2.
Ser-2 CTD phosphorylation by P-TEFb is essential for transcription elongation on the c-fos and junB genes. Shown are the distributions of Pol II on the c-fos (A to C) and junB (D to F) genes. ChIP assays were performed with specific anti-Pol II antibodies: N-20, which is used for immunoprecipitation of total Pol II (A and D); H14, directed against phospho-Ser-5 CTD (B and E); and H5, directed against phospho-Ser-2 CTD, which also reacts well with doubly Ser-2/Ser-5-phosphorylated CTD (C and F). The densities on the c-fos and junB genes at various time points after TRH stimulation (0 [basal condition], 12, and 48 min) in the absence (filled bars) or presence (hatched bars) of DRB were quantified by real-time PCR and are presented as percentages of the input (the mean of two experiments is shown; the error bars represent the range). The positions of primer sets used in real-time PCR are shown in Fig. 1A and E.
Next, we examined distribution patterns on the c-fos gene of NELF, which is composed of four subunits (A, B, C or D, and E). ChIP with an anti-NELF-A antibody revealed the presence of NELF confined to the promoter-proximal region (Fig. 1C). While more and more NELF was associated with the c-fos gene after TRH stimulation, all NELF remained confined to the 5′ part of the gene. NELF occupancy was maximal 48 min after TRH stimulation, when Pol II and P-TEFb on the c-fos gene were already reduced and the transcription rate was attenuated. Interestingly, DRB treatment potentiated the recruitment of NELF at the prompter-proximal region at both 12 and 48 min after TRH stimulation. It has been reported that NELF-E is phosphorylated by P-TEFb in vitro and that the phosphorylation induces dissociation of NELF from the Pol II complex (17). Increased association of NELF with the promoter-proximal region after DRB treatment may be due to the fact that unphosphorylated NELF would not detach from the Pol II complex in vivo.
The distribution pattern of DSIF, which consists of Spt4 and Spt5, was in part similar to that of NELF (Fig. 1D). ChIP with an anti-Spt5 antibody showed a limited association of DSIF with the promoter-proximal region in resting cells that was remarkably similar to that of NELF. In contrast to NELF, DSIF was also largely recruited to the whole transcribed part of the c-fos gene following TRH. It was particularly striking that at the time of maximal transcription rate (12 min), abundant DSIF was found on exons 3 and 4. Such TRH-induced association with the 3′ region suggests that following induction of transcription elongation, DSIF progresses together with Pol II (Fig. 2A). At 48 min, DSIF occupancy was very markedly diminished on the transcribed part of the c-fos gene (most markedly at 3′) but not at all on the promoter-proximal region. While DRB treatment did not affect the occupancy by DSIF of the promoter-proximal region, the recruitment of DSIF to the transcribed part of the c-fos gene was quite sensitive to CDK9 inhibition. In particular, the TRH-induced occupancy by DSIF of the 3′ regions was nearly abolished by DRB treatment. Thus, the recruitment of DSIF in the promoter-proximal region is independent of CDK9 activity, but progress along the gene is strongly dependent upon CDK9 activity. Taken together, these observations suggest that under basal conditions DSIF and NELF were present, possibly as a complex, in the promoter-proximal region, together with Pol II (Fig. 2A).
There have been a limited number of reports demonstrating in vivo recruitment of P-TEFb, NELF, and DSIF on IEGs in mammalian cells. We therefore wondered whether the stimulus-induced recruitment of those transcription regulation factors would be common to IEGs and whether P-TEFb would always be a key regulator for the recruitment machinery. Therefore, we extended our observations obtained for c-fos and previously for the MKP-1 gene (18) to yet another IEG, junB, for which Pol II pausing at the promoter-proximal region was previously reported (2). The junB gene, in contrast to c-fos and MKP-1, is intronless. Three primer sets probing the promoter-proximal (5′) region, the region downstream of the 5′ region (5′b), and the 3′ region of junB gene were used (Fig. 1E). ChIP experiments with an anti-cyclin T1 antibody showed the TRH-induced association of P-TEFb, which was remarkably sensitive to DRB (Fig. 1F). As on the c-fos gene, DSIF and NELF were confined to the promoter-proximal region under resting conditions. After TRH stimulation, DSIF also occupied the 3′ region of junB (Fig. 1G and H). DRB did not affect the presence of DSIF in the 5′ region but abolished it in the 3′ region (Fig. 1H). Thus, the transcriptional elongation machinery shown in Fig. 1, including P-TEFb, NELF, and DSIF, appears to be common to IEGs.
Transcription elongation of the c-fos and junB genes is tightly linked to phosphorylation by P-TEFb of Ser-2 in the Pol II CTD.
In order to define the role of Pol II CTD phosphorylation by P-TEFb, we next observed the distribution of CTD-phosphorylated Pol II on the c-fos gene in the presence or absence of DRB. We used three different anti-Pol II antibodies: N-20, which recognizes the N-terminal region of a large subunit of Pol II and is used for immunoprecipitation of total Pol II irrespective of CTD phosphorylation, and to differentiate between the different phosphorylation states of the CTD of Pol II, the commonly used antibodies H14, recognizing phospho-Ser-5 CTD, and H5, directed against phospho-Ser-2 CTD, distinguished between phospho-Ser-5 CTD and doubly (Ser-2/Ser-5) phosphorylated CTD plus phospho-Ser-2 CTD (25; see also the comment on antibody specificities in the supplemental data in reference 41).
As is shown in Fig. 2A, prior to TRH stimulation, significant occupancy by Pol II was detected mainly in the promoter-proximal region on the gene and less on exons 3 and 4. At 12 min after TRH stimulation, the occupancy by Pol II of the entire c-fos gene was increased, more significantly on exons 3 and 4 (15- to 20-fold over the basal condition) than in the promoter-proximal region (5-fold increased). These results indicate that in order to reach maximal transcription rates, TRH stimulated transcription elongation by Pol II, which had already initiated transcription in addition to further initiating Pol II. At 48 min after TRH stimulation, Pol II occupancy of the c-fos gene was reduced but remained significantly elevated over the basal rate. DRB treatment strongly suppressed TRH-induced Pol II occupancy of the 3′ part of the c-fos gene, whereas it slightly affected Pol II occupancy of its promoter-proximal region under basal and induced conditions. TRH-induced recruitment of Pol II to the promoter-proximal region was reduced by DRB to 75% at 12 min after TRH stimulation but was increased by DRB after 48 min, possibly due to stacking of initiated Pol II in the attenuated steady state of TRH-induced transcription. DRB effects on Pol II occupancy of 5′ transcribed parts of the gene (the exon 1-intron 1 junction and intron 1) were similar to those seen on the promoter-proximal region. Taken together, these data suggested that transcription elongation is much more dependent upon CDK9 activity than transcription initiation. ChIP experiments with the H14 antibody yielded patterns of basal and TRH-induced occupancy and DRB effects on them that were very similar to those seen for total Pol II with the N-20 antibody (Fig. 2B). This illustrated that the phospho-Ser-5 CTD distribution is similar to that of total Pol II. In contrast, the distribution patterns seen with the H5 antibody were very different from those of total Pol II. The ChIP data in Fig. 2C indicated that occupancy of the c-fos gene by Pol II Ser-2 phosphorylated on its CTD was not detectable anywhere prior to stimulation but was very highly induced by TRH at 12 min on exons 3 and 4, significantly more than on the 5′ part of the transcribed region. At the attenuated steady state (48 min), occupancy of the c-fos gene by Pol II Ser-2 phosphorylated on its CTD was much reduced. DRB treatment completely prevented such TRH-induced occupancy throughout the gene. DRB would also affect the kinase activity of CDK9 that had been recruited by TRH stimulation to the c-fos gene, thus largely preventing the phosphorylation of Pol II CTD Ser-2. Taken together, the data in Fig. 1B and 2 are consistent with the notion, mainly derived in vitro, that CDK9 in P-TEFb is responsible for the phosphorylation of CTD Ser-2 but not of Ser-5 (1, 34). TRH-induced phosphorylation by CDK9 in P-TEFb of Pol II CTD Ser-2 was required for transcription elongation, rather than initiation of the c-fos gene.
We also investigated the distribution patterns of total Pol II, phospho-Ser-5 CTD, and phospho-Ser-2 CTD on the junB gene. The distributions of total Pol II and phospho-Ser-5 CTD Pol II were quite similar, with higher occupancy in the promoter-proximal region (Fig. 2D and E). In addition, the effect of DRB treatment on the recruitment of phospho-Ser-5 CTD was small (Fig. 2E). In contrast, Pol II Ser-2 phosphorylated on its CTD, which 12 min after TRH was manifest on the whole junB gene, was fairly sensitive to DRB treatment. These results are analogous to the observations of the c-fos (Fig. 2B-D) and MKP-1 (18) genes. Taken together, these data highlight the fact that inducible transcription of IEGs appears to be regulated by a common mechanism involving a change in Ser-2 CTD phosphorylation induced by P-TEFb.
P-TEFb regulates IEG transcript processing.
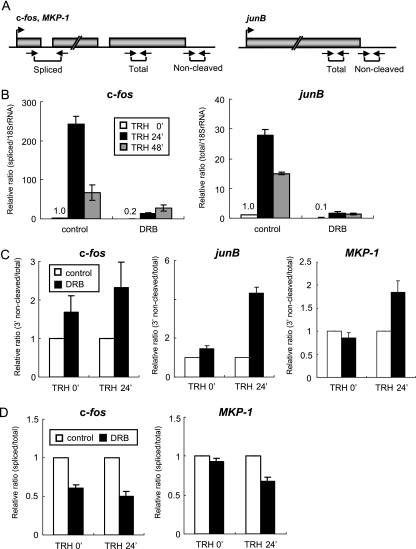
Transcription and primary transcript processing are coordinated to ensure efficient production of mature mRNA, with a key role played by CTD phosphorylation (22, 36). For example, capping of mRNA coincides with Ser-5 CTD phosphorylation (23, 29, 43). Splicing and 3′-end processing (cleavage and polyadenylation) are regulated by Ser-2 CTD phosphorylation (1, 5, 34). To investigate the role of P-TEFb in these processes, we first evaluated the effects of DRB on the production of mature c-fos mRNA, using RT-PCR with a primer set spanning the end of exon 1 and the start of exon 2 and detecting only spliced c-fos mRNA (Fig. 3A). As shown in Fig. 3B and reported previously (41), c-fos mRNA levels rose dramatically after TRH stimulation (24 min) and were later reduced to levels still markedly enhanced over basal levels (48 min). Under DRB treatment, under both basal and TRH-stimulated conditions, c-fos mRNA production was much reduced (Fig. 3B). The same kinetic pattern seen for c-fos mRNA was observed for junB RNA when junB transcripts were quantified with a primer set amplifying a sequence in the middle of the only exon (Fig. 3A and B). Like c-fos mRNA, junB RNA levels were very sensitive to DRB treatment regardless of TRH stimulation. As DRB at the concentration and exposure used here did not completely shut off transcription of both genes, it was possible to see whether transcription and primary transcript processing were still well coordinated or whether DRB inhibition of Ser-2 CTD phosphorylation might cause defects of splicing and 3′-end processing. To establish the relative 3′-end processing ratio of the IEG transcripts, we contrasted the 3′ primary transcript normally removed upon polyadenylation with the total transcripts (Fig. 3A), monitoring the 3′-end cleavage ratio. DRB inhibition of CDK9 reduced 3′-end cleavage of c-fos primary transcripts accumulated over 24 min of TRH stimulation; c-fos transcripts without the cleavage in total c-fos RNA were up to four times more abundant than without DRB (Fig. 3C). Thus, 3′-end cleavage was dependent upon CDK9 activity in P-TEFb, especially at times of most active transcription. We also examined 3′-end cleavage efficiency on junB and another IEG MKP-1 gene, whose transcription was also suppressed strongly but not completely by DRB at 30 μM concentration (18). After 24 min of TRH stimulation, DRB treatment obviously increased the transcripts without the cleavage of both genes (Fig. 3C). As for splicing efficiency, we contrasted spliced transcripts with the total transcripts. DRB treatment resulted in significantly reduced splicing of c-fos transcripts under both basal and TRH-stimulated conditions (24 min) (Fig. 3D). Reduced splicing was also observed in MKP-1 transcripts at times of most active transcription (Fig. 3D). Taking the data together, P-TEFb is related to the efficiency of 3′-end processing and splicing of TRH-induced IEG transcripts.
FIG. 3.
DRB suppresses IEG transcription and transcript processing. (A) Schematic diagrams, including exons and introns, with the locations of the primer sets used in the experiments. The primer sets for amplification of objective transcripts were designed based on the genetic information (c-fos, GenBank X06769; junB, GenBank NM_021836; MKP-1, GenBank NM_053769). (B) Relative amounts of c-fos and junB transcripts quantified by real-time RT-PCR (normalized to 18S RNA). (C) PCR-based analysis of the 3′-end cleavage ratio. Transcripts of the c-fos, junB, and MKP-1 genes were amplified with primer sets for detection of 3′-uncleaved and total RNAs. The values of 3′-uncleaved RNAs normalized to total RNAs are shown as relative 3′-end noncleavage ratios. (D) PCR-based analysis of the splicing ratio. c-fos and MKP-1 transcripts were amplified with primer sets for detection of spliced versus total RNA. The value of spliced RNA normalized to total RNA are shown as relative splicing ratios. (B to D) Each experiment was performed three times, and the relative ratio is shown as the mean ± standard error of the mean.
Activation of the MEK-ERK signaling pathway is decisive for TRH induction of IEG transcription and P-TEFb recruitment to IEGs.
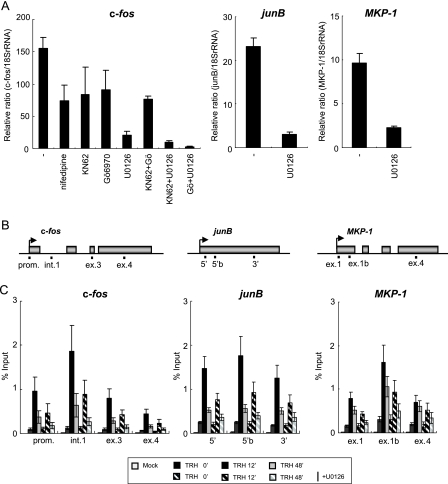
TRH activates multiple intracellular signaling pathways. To assess their relative importance for induction of the IEGs (c-fos, junB, and MKP-1), we used specific pharmacological inhibitors that target different signaling pathways (Fig. 4A). To inhibit the MEK-ERK signaling pathway, we used the specific MEK1/2 inhibitor U0126, which is generally accepted as specific and hardly affecting other protein kinases (protein kinase C [PKC], Abl, Raf, MEKK, ERK, JNK, MKK-3, MKK-4/SEK, MKK-6, Cdk2, Cdk4, etc.) (16). U0126 most strongly attenuated TRH-induced c-fos transcription. Blocking of other pathways, including Ca2+ influx via voltage-activated Ca2+ channels (nifedipine), PKC (Gö6970), or CaMKs (KN62) alone or even in combination, was less effective. Although MEK1/2 may be activated directly by several upstream kinases (Raf-1, MOS, A-RAf, B-Raf, etc.), no substrates for MEK1/2 other than ERK1/2 have been identified Therefore, induction of c-fos transcription by TRH should depend primarily upon the activation of the MEK-ERK signaling pathway, which was rapid and sustained (Fig. 5B). In addition, being strongly inhibited by U0126, TRH-induced junB and MKP-1 transcription also appeared to be strongly dependent upon this same signaling pathway (Fig. 4A).
FIG. 4.
TRH-induced P-TEFb recruitment on IEGs is dependent upon the MEK-ERK signaling pathway. (A) Induction by TRH involves multiple signaling pathways, most importantly the MEK-ERK pathway. Cells were incubated for 1 h with the inhibitors for L-type Ca2+ channels (nifedipine, 1 μM), CaMKs (KN62, 10 μM), PKCs (Gö6970, 1 μM), and MEK (U0126, 10 μM) and then stimulated with TRH (100 nM) for 30 min. c-fos, junB, and MKP-1 transcripts were quantified by using real-time RT-PCR and normalized to 18S RNA (mean ± standard error of the mean [SEM]; n = 3). (B) Rat c-fos (GeneID, 314322), junB (GeneID, 24517), and MKP-1 (GeneID, 114856) genomic locus with the primer positions (prom., promoter-proximal region; e1-i1, exon 1 and intron 1 junction; int.1, intron 1; ex.3, exon 3; ex.4, exon 4; 5′, 5′ region; 5′b, 5′ region b; 3′, 3′ region; ex.1b, exon 1b). (C) U0126 reduced occupancy of P-TEFb on the c-fos, junB, and MKP-1 genes. ChIP assays were performed with an anti-cyclin T1 antibody and chromatin at various time points of TRH stimulation (0, 12, and 48 min) in the absence (filled bars) or presence (hatched bars) of U0126. The density of P-TEFb was quantified by real-time PCR and is presented as a percentage of the input (mean ± SEM; n = 5).
FIG. 5.
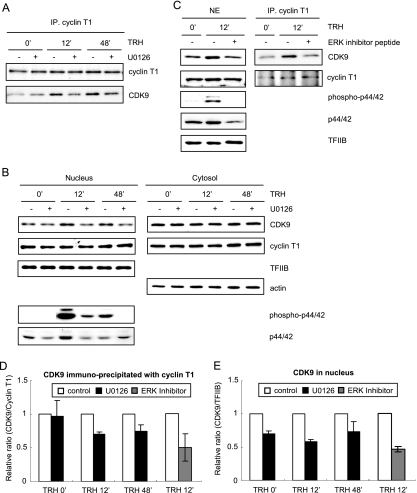
Activated MEK1-ERK signaling up-regulates nuclear CDK9 and enhances its association with cyclin T1 to form P-TEFb. (A) Cyclin T1, free or incorporated into P-TEFb, was immunoprecipitated from nuclear extracts of GH4C1 cells prepared at various time points of TRH stimulation (0 [basal condition], 12, and 48 min) in the presence or absence of U0126 (10 μM) using an anti-cyclin T1 antibody. Cyclin T1 and CDK9 were analyzed by Western blotting with an anti-cyclin T1 and an anti-CDK9 antibody. Shown is a typical experiment repeated three times. (B) Nuclear and cytosolic extracts prepared at various time points after TRH stimulation (0, 12, and 48 min) in the presence or absence of U0126 were subjected to Western blotting with an anti-CDK9, an anti-cyclin T1, an anti-TFIIB (nuclear extracts), and an anti-actin (cytosolic extracts) antibody. Nuclear extracts were separately analyzed for phospho-ERKs and ERKs by Western blotting with an anti-phospho-ERK and an anti-ERK antibody. Shown are typical experiments repeated three times. (C) Nuclear extract prepared with or without TRH stimulation or ERK activation inhibitor peptide I treatment (2 μM) were subjected to immunoprecipitation using an anti-cyclin T1 antibody. The nuclear extract and the immunoprecipitated proteins were subjected to Western blotting with an anti-CDK9, an anti-cyclin T1, an anti-TFIIB, an anti-phospho-ERK, and an anti-ERK antibody. Shown are typical experiments repeated two times. (D and E) The reduction of CDK9/cyclin T1 dimers (i.e., P-TEFb) (D) or that of CDK9 (E) in the nucleus was calculated by quantification of the bands derived from experiments shown in panels A to C and is shown as a graph. The relative ratio is shown as the mean ± standard error of the mean (n = 3; U0126) or the mean of two experiments with the range (ERK inhibitor).
The simplest hypothesis linking ERKs to Pol II would be that active phospho-ERKs directly phosphorylate the CTD of Pol II on the IEGs, thus favoring elongation. Indeed phospho-ERKs can phosphorylate the Pol II CTD in vitro (4, 7, 14, 50). However, ChIP failed to provide any evidence for recruitment of phospho-ERKs to the c-fos gene either prior to or after TRH stimulation (data not shown), suggesting that the phospho-ERKs did not directly phosphorylate Pol II CTD in vivo. Note that it is possible that the anti-ERK antibody we used does not work sufficiently for ChIP assays.
To evaluate the role of the MEK-ERK signaling pathway on TRH-induced recruitment of P-TEFb on the IEGs c-fos, junB, and MKP-1, we next performed a ChIP assay with an anti-cyclin T1 antibody under U0126 treatment. As shown in Fig. 4B and C, U0126 treatment inhibited the association of P-TEFb by up to 50% on the whole IEGs at 12 and 48 min after TRH treatment. This inhibition was observed equally even when we performed ChIP with an anti-CDK9 antibody (data not shown). These results suggest that the implication of the MEK-ERK signaling pathway during TRH-induced recruitment of P-TEFb is common to the three IEGs. It thus appeared likely that ERK somehow modulated P-TEFb occupancy of IEGs.
The MEK1-ERK signaling pathway does not regulate the catalytic activity of CDK9 but up-regulates constitution of P-TEFb.
We next examined in detail how the MEK-ERK signaling pathway modulated P-TEFb. We postulated two possibilities: the signaling pathway regulated the amount of P-TEFb in the nucleus or CDK9 activity, which is necessary for recruitment of P-TEFb on IEGs (Fig. 1B and F). P-TEFb was therefore immunoprecipitated from GH4C1 cells stimulated by TRH in the presence or absence of U0126 and analyzed by Western blotting (Fig. 5A). Whereas the amount of cyclin T1 immunoprecipitated with an anti-cyclin T1 antibody was constant irrespective of the conditions, CDK9 precipitated together with cyclin T1 varied with TRH stimulation and MEK inhibition: TRH increased the amount of CDK9 interacting with cyclin T1, whereas U0126 treatment abolished the augmented interaction. A similar pattern was observed even with complete nuclear extracts (Fig. 5B). In contrast, neither CDK9 nor cyclin T1 in the cytosolic extracts was affected by either TRH or U0126. To confirm the specific implication of the MEK-ERK signaling pathway for nuclear up-regulation of CDK9, we used as an inhibitor a 13-amino-acid peptide corresponding to the N terminus of MEK1 (ERK activation inhibitor peptide I) (27, 46) instead of U0126. ERK activation inhibitor peptide I interacts specifically with ERK1/2 without any effect on other, closely related MAPKs, c-Jun amino-terminal kinase, or p38 protein kinase; thus, the ERK activation inhibitor peptide competitively inhibits interaction of MEK1 with ERK1/2 (27), thus preventing ERK1/2 activation. The possibility that a yet unidentified substrate of MEK would also be inhibited by ERK activation inhibitor peptide cannot be excluded. As shown in Fig. 5C, like U0126, the peptide completely suppressed TRH-induced phosphorylation of ERK1/2 and augmentation of CDK9 in the nucleus. Consistently, TRH-induced interaction of CDK9 with cyclin T1 was also clearly reduced by the peptide. We also calculated the reduction of CDK9/cyclinT1 dimers or that of CDK9 in the nucleus by quantification of the bands in Western blotting (Fig. 5D and E). Compared to the control, at 12 min after TRH stimulation, 40 to 50% of the CDK9/cyclinT1 dimers or half of the CDK9 was abolished in the nucleus by inhibition of the MEK1-ERK signaling pathway. Taken together, the data in Fig. 5 illustrate that TRH increases the number of CDK9 and cyclin T1/CDK9 dimers, i.e., the assembly of P-TEFb in the nucleus via the MEK1-ERK signaling pathway.
We next addressed whether the catalytic activity of CDK9 was also regulated by the MEK1-ERK pathway. To this end, the protein kinase activity of P-TEFb immunoprecipitated from GH4C1 cells stimulated by TRH in the presence or absence of U0126 was determined by an in vitro phosphorylation assay with GST-CTD as a substrate (53). As shown in Fig. S2 in the supplemental material, in vitro phosphorylation activities normalized with the amount of immunoprecipitated CDK9 (the ratio of phospho-CTD versus CDK9) were not significantly changed by U0126 treatment. Thus, the in vitro phosphorylation assay did not provide any evidence for a change in the catalytic activity of CDK9.
In summary, these data would be consistent with the hypothesis that the MEK1-ERK signaling pathway increased nuclear CDK9, leading to enhanced dimerization with cyclin T1 to form abundant P-TEFb, thus facilitating recruitment of P-TEFb to the c-fos gene and other induced genes. In this manner, MEK1-ERK would contribute to P-TEFb-driven Pol II elongation.
Up-regulation of P-TEFb by the MEK1-ERK signaling pathway induces transcription elongation of c-fos.
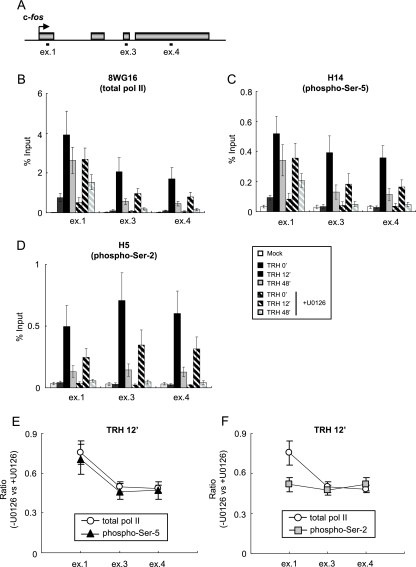
If activation of the MEK1-ERK signaling pathway facilitates P-TEFb recruitment to the IEGs, inhibition of the pathway would cause reduction of transcription elongation involving P-TEFb-mediated Pol II CTD Ser-2 phosphorylation. We thus performed ChIP assays in order to assess the distribution pattern of Pol II transcribing the c-fos gene under U0126 treatment. Occupancy by total and CTD-phosphorylated Pol II was monitored at exons 1, 3, and 4 (Fig. 6A). We first examined the distribution of total Pol II with an anti-CTD antibody (8WG16) (47) that recognizes CTD, irrespective of its phosphorylation, with efficiency similar to that of the N-20 antibody (41) (Fig. 6B). U0126 treatment reduced TRH-induced Pol II occupancy in all three regions, with more pronounced reduction in the downstream region (exons 3 and 4) than in the promoter-proximal region (exon 1), where 70% of Pol II recruitment resisted U0126 treatment. These results suggest that inhibition of the MEK1-ERK signaling pathway mainly suppressed transcription elongation. The distribution pattern of the phospho-Ser-5 CTD examined with the H14 antibody was very similar to that of total Pol II (Fig. 6C). On the other hand, ChIP with the H5 antibody revealed that U0126 treatment reduced TRH-induced occupancy by the phospho-Ser-2 CTD more severely on exon 1 than total Pol II and phospho-Ser-5 CTD (Fig. 6D). The selective effect of U0126 on Ser-2 CTD phosphorylation became more apparent when the ChIP data were plotted 12 min after TRH stimulation (Fig. 6E and F). U0126 treatment reduced exon 1 occupancy by total or Ser-5 CTD-phosphorylated Pol II only to 70% of the control (Fig. 6E); in contrast, occupancy by Ser-2 CTD-phosphorylated Pol II was reduced to 50% (Fig. 6F). Similar data were obtained 48 min after TRH treatment (data not shown). Note that DRB, a strong and selective suppressor of CDK9 phosphorylation of the CTD (9), not only markedly reduced TRH-induced recruitment of P-TEFb on the c-fos gene (Fig. 1B), but also nearly abolished phosphorylation of CTD Ser-2 for Pol II on the whole gene (Fig. 2C). U0126 inhibited half of the recruitment of P-TEFb on the c-fos gene (Fig. 4C) but did not affect CDK9 activity (see Fig. S2 in the supplemental material), which may explain why U0126 reduced Ser-2 CTD phosphorylation of Pol II in the 3′ region by only 50%. Thus, these data illustrate a role for the MEK1-ERK signaling pathway in transcription elongation, most probably via the phosphorylation of CTD Ser-2.
FIG. 6.
The MEK1-ERK signaling cascade is required for Pol II elongation via phosphorylation of CTD Ser-2 on the c-fos gene. (A) Rat c-fos genomic locus with the primer positions (ex.1, exon 1; ex.3, exon 3; ex.4, exon 4). (B to D) ChIP assays were performed with specific anti-Pol II antibodies: 8WG16, which is used for immunoprecipitation of total Pol II (B); H14, directed against phospho-Ser-5 CTD (C); and H5, directed against phospho-Ser-2 CTD, which also reacts well with doubly Ser-2/Ser-5-phosphorylated CTD (D). The Pol II densities on the c-fos gene at various time points of TRH stimulation (0 [basal condition], 12, and 48 min) in the absence (filled bars) or presence (hatched bars) of U0126 were quantified by real-time PCR and are presented as percentages of the input (mean ± standard error of the mean [SEM]; n = 4). (E and F) The reduction of phospho-Ser-5 CTD (E) or phospho-Ser-2 CTD (F) on exons 1, 3, and 4. The relative ratio derived from experiments in B and C (E) or B and D (F) is shown as the mean ± SEM (n = 4).
DISCUSSION
The c-fos transcription rate is regulated mainly via the control of transcription elongation by sophisticated mechanisms that involve P-TEFs and N-TEFs. Yamada et al. reported that transcription elongation of the c-fos gene is regulated by the TEFs DSIF and P-TEFb in HeLa cells (58). More recently, we showed that dynamic recruitment of P-TEFb and CTD phosphorylation on the c-fos gene is induced by TRH stimulation in the neuroendocrine GH4C1 cells (41). The present study consolidates the regulation by P-TEFb and DSIF of Pol II transcribing the c-fos gene and shows TRH-induced recruitment of NELF to the proximity of the c-fos promoter. CDK9 activity would be required to permit transcription elongation manifested through the presence of Pol II phosphorylated on Ser-2 of its CTD. P-TEFb, as well as DSIF, occupied the 3′ part of the c-fos gene in parallel with Pol II. Importantly, we showed that the transcriptional elongation machinery recruited to the c-fos gene upon induction of transcription is common to the other IEGs, junB and MKP-1. The MEK1-ERK signaling pathway, essential for the induction of IEG transcription, up-regulated nuclear CDK9 and P-TEFb. This novel and comprehensive mechanism contributes to the regulation of transcription elongation of IEGs in GH4C1 cells in vivo.
P-TEFb, NELF, and DSIF control transcription elongation on IEGs.
Prior to TRH stimulation, DSIF, NELF, and Pol II are similarly confined to the 5′ part of the c-fos and junB genes (Fig. 1 and 2), consistent with in vitro findings that showed that DSIF and NELF collaboratively stall Pol II promoter proximally (33, 52, 53, 59-61). NELF and DSIF occupancy of the c-fos gene in resting cells in vivo consolidated the notion of frequent promoter-proximal transcription pause (40), but not a further block to elongation in intron 1 (11, 41, 42, 54). Interestingly, the association of DSIF and NELF in the promoter-proximal region increased gradually after TRH stimulation (Fig. 1C, D, G, and H), in parallel with the increase of Pol II density in the promoter-proximal region after TRH stimulation (Fig. 2A and D). Indeed, the pausing of Pol II by DSIF and NELF may be indispensable for appropriate modification of pausing Pol II and/or for maintenance and modification of its nascent transcripts, e.g., for rendering Pol II elongation competent or to inhibit cleavage (38) and ensure capping of the nascent transcript (31).
What triggers the release from the pause? Ivanov et al. reported that phosphorylation of Spt5 by P-TEFb invalidates the role of DSIF as a negative regulator in vitro (24). Yamada et al. observed that the phosphorylated form of Spt5 seems to progress together with Pol II as an active regulator for Pol II elongation in vivo (58). The observation that progress of DSIF, together with Pol II, along the c-fos and junB genes occurs concomitantly with massive recruitment of P-TEFb on the genes in vivo (Fig. 1) supports the idea that P-TEFb modifies Spt5, causing the functional change of DSIF. Thus, P-TEFb would be required to release Pol II from the pause caused by DSIF and NELF and for the functional change of DSIF. Indeed, inhibition by DRB of CDK9 enhances NELF occupancy of the 5′ parts of those IEGs (Fig. 1), consistent with the proposal that phosphorylation by P-TEFb of NELF-E would trigger NELF detachment from the complex (17).
CTD phosphorylation by P-TEFb is essential for well-controlled gene transcription. There remains some controversy as to the sites in CTD that are phosphorylated by P-TEFb (37, 65). This study shows that P-TEFb is mainly responsible for Ser-2 CTD phosphorylation during induced transcription elongation and that CDK9 is not essential for induced Pol II occupancy of the 5′ parts of those IEGs (Fig. 2). P-TEFb is also necessary for maturation of IEG transcripts (c-fos, junB, and MKP-1) (Fig. 3). Being associated with all of the genes, P-TEFb may continually function to facilitate cotranscriptional splicing and 3′-end processing. Currently, it is assumed that RNA modification factors recognize specialized phosphorylation patterns on the CTD to associate with Pol II during transcription elongation (22, 36). Such variable phosphorylation patterns, called CTD code, are updated in a time- and position-dependent manner during transcription (8), with the participation of P-TEFb in the complex elongating IEG transcripts. Thus, the phosphorylation by P-TEFb may provide the optimal scaffold for RNA modification factors.
General versus gene-specific functions of P-TEFb and N-TEFs.
P-TEFb is considered to be of general importance for gene transcription, and indeed, DRB can block almost any gene expression in mammalian cells (9). In this report, we demonstrated that not only c-fos, but also another IEG, junB, is regulated by P-TEFb, NELF, and DSIF during Pol II elongation in GH4C1 cells. Moreover, the transcription elongation on the MKP-1 gene is also regulated by those elongation factors (18). The distribution patterns of P-TEFb, NELF, and DSIF on the junB and MKP-1 genes are similar to those on the c-fos gene, suggesting that the transcription elongation machinery including those three factors is common to c-fos, junB, MKP-1, and possibly other rapidly inducible IEGs. However, when the induction of stress-activated p53-dependent genes was analyzed, some exceptions were found, notably the p21 gene, which can be transcribed in spite of DRB inhibition of CDK9 (20, 55). Furthermore, our ChIP assays suggest that a housekeeping gene, such as the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, also appears to be occupied by P-TEFb, NELF, and DSIF (unpublished data). It thus remains puzzling that inducible and constant gene transcription should be tuned by common factors in an apparently gene- and inducer-specific manner.
The MEK1-ERK signaling pathway regulates Pol II elongation on IEGs through up-regulation of P-TEFb.
TEFs are regulated in multiple ways. Some processes favor or repress their recruitment to transcription complexes; others affect their nuclear availability (39). Sequestering P-TEFb in inactive heteropolymers, 7SK snRNA and HEXIM1/2, controls the availability of active P-TEFb (15, 62, 63). Cyclin T1 has a potential role in the translocation of P-TEFb into the nucleus (32). In this study, we found that activation of the MEK1-ERK signaling pathway favors assembly of cyclin T1/CDK9 heterodimers, which form the active P-TEFb. Thus, in addition to well-established action to phosphorylate several transcription factors, such as TCF proteins, which activate c-fos transcription via the SRE element (21, 49), the MEK1-ERK signaling pathway modulates the nuclear availability of active P-TEFb complex (Fig. 5). Furthermore, total CDK9, but not cyclin T1, is up-regulated in the nucleus by the MEK1-ERK signaling pathway. The causal relationship between these two observations remains open. In fact, the latter could be simply the consequence of reduced CDK9 degradation; free CDK9 is much less stable than CDK9 in a heterodimer with cyclin T1. CDK9 degradation is initiated by its ubiquitination via a SCFSKP2-dependent system (28). Cyclin T1, which is itself not ubiquitinated, functions as a scaffold connecting ubiquitinated CDK9 and the proteasome system. Garriga et al. have argued that this specific CDK9 degradation mechanism applies only to overexpressed CDK9 in some specific cell types (19). It has been proposed that the constitution of active P-TEFb involves a specific chaperone pathway including the heat shock proteins HSP70 and HSP90 (35). The TRH-induced MEK1-ERK signaling pathway may stabilize the nuclear CDK9 by facilitation of its interaction with HSP70 and HSP90. Alternatively, increased nuclear availability of P-TEFb may result from translocation of CDK9 from the cytoplasm to the nucleus favored by the MEK1-ERK signaling pathway, as CDK9 has the intrinsic property of shuttling between the nucleus and cytoplasm (32). We are currently investigating these potential mechanisms.
Taken together, our results clearly indicate that activation of the MEK1-ERK signaling pathway up-regulates nuclear CDK9 and P-TEFb, facilitating recruitment of P-TEFb to actively transcribed genes. Under cellular resting conditions, the number of CDK9/cyclin T1 dimers (i.e., P-TEFb) constantly present in the nucleus is likely sufficient to ensure elongation and transcript processing of the limited number of constantly active genes. However, after strong cellular stimulation (e.g., by TRH), initiation of transcription of a large number of IEGs may exhaust nuclear P-TEFb, as this factor remains associated with transcribing Pol II. Thus, in this situation, in which P-TEFb is rate limiting, the MEK1-ERK signaling pathway may contribute to regulating elongation by controlling the nuclear availability of P-TEFb. Therefore, the MEK1-ERK signaling pathway would eventually activate not only transcription initiation via a traditional pathway, but also elongation and transcript processing.
Conclusions.
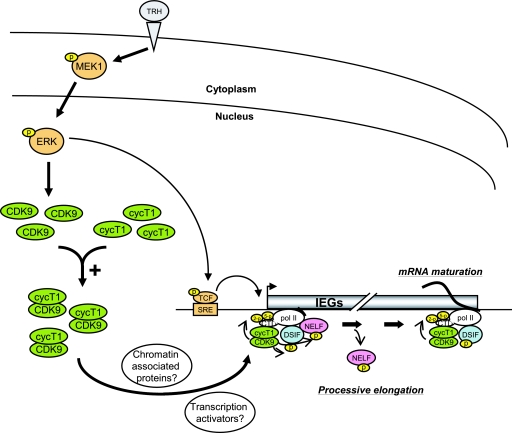
The dual action of the MEK1-ERK signaling pathway on the transcription elongation machinery, which includes P-TEFb, NELF, and DSIF and controls Pol II transcription of IEGs, is illustrated in the scheme in Fig. 7. TRH stimulation activates, among other signaling pathways, the MEK1-ERK cascade. On one hand, this causes transcription factors to bind to the SRE and thus promotes transcription initiation and subsequently favors the recruitment of P-TEFb to the transcription complex pausing in the promoter-proximal region. On the other hand, active ERK by a yet unknown mechanism enhances the number of cyclinT1/CDK9 dimers, i.e., active P-TEFb, thus facilitating its recruitment to c-fos and other induced genes. As recently reviewed (39), P-TEFb interacts with transcription activators (e.g., CIITA, NF-κB, c-Myc, and MyoD) or chromatin-associated proteins (e.g., Brd). Such interactions would favor gene- and stimulus-specific recruitment of up-regulated P-TEFb to induced genes. CDK9 phosphorylation of CTR of Spt5, NELF-E, and the Ser-2 CTD of Pol II triggers the functional change of DSIF and the detachment of NELF, allowing Pol II to resume elongation. P-TEFb and DSIF progress together with Pol II, maintaining the phosphorylation status of the Pol II CTD in a position-dependent manner and allowing the association of splicing and 3′-end processing complexes with the transcript. The MEK1-ERK-linked up-regulation of active P-TEFb would largely affect transcription of IEGs in general. This novel mechanism may thus facilitate the massive induction of gene transcription occurring upon cell cycle and/or cell fate transitions, in which the MEK1-ERK pathway plays a predominant role.
FIG. 7.
Transcription elongation controlled by P-TEFb, NELF, and DSIF through the MEK1-ERK signaling pathway. TRH stimulation activates the MEK1-ERK signaling pathway, resulting in up-regulation of nuclear CDK9 and cyclin T1/CDK9 complex (i.e., P-TEFb). This up-regulation may be caused by stabilization or translocation of CDK9. The up-regulated P-TEFb is subsequently recruited on IEGs (c-fos, junB, MKP-1, etc.), probably through transcription activators or chromatin-associated proteins. On IEGs, P-TEFb phosphorylates CTR of Spt5, NELF, and CTD Ser-2 of Pol II, favoring transcription elongation and maturation of mRNA by the Pol II complex associated with P-TEFb and DSIF.
Supplementary Material
Acknowledgments
We are very grateful to T. Wada and R. Young for providing us the GST-CTD construct.
This investigation was supported in part by the Swiss National Science Foundation grant 3100A0-102147 and by the Fondation pour Recherches Médicales.
Footnotes
Published ahead of print on 17 December 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 1367-76. [DOI] [PubMed] [Google Scholar]
- 2.Aida, M., Y. Chen, K. Nakajima, Y. Yamaguchi, T. Wada, and H. Handa. 2006. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol. Cell. Biol. 266094-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 142635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellier, S., M. F. Dubois, E. Nishida, G. Almouzni, and O. Bensaude. 1997. Phosphorylation of the RNA polymerase II largest subunit during Xenopus laevis oocyte maturation. Mol. Cell. Biol. 171434-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird, G., D. A. Zorio, and D. L. Bentley. 2004. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol. Cell. Biol. 248963-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boehm, A. K., A. Saunders, J. Werner, and J. T. Lis. 2003. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 237628-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnet, F., M. Vigneron, O. Bensaude, and M. F. Dubois. 1999. Transcription-independent phosphorylation of the RNA polymerase II C-terminal domain (CTD) involves ERK kinases (MEK1/2). Nucleic Acids Res. 274399-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buratowski, S. 2003. The CTD code. Nat. Struct. Biol. 10679-680. [DOI] [PubMed] [Google Scholar]
- 9.Chao, S. H., and D. H. Price. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 27631793-31799. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, C., and P. A. Sharp. 2003. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and gamma-actin genes. Mol. Cell. Biol. 231961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collart, M. A., N. Tourkine, D. Belin, P. Vassalli, P. Jeanteur, and J. M. Blanchard. 1991. c-fos gene transcription in murine macrophages is modulated by a calcium-dependent block to elongation in intron 1. Mol. Cell. Biol. 112826-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25375-380. [DOI] [PubMed] [Google Scholar]
- 13.Coulon, V., J. L. Veyrune, N. Tourkine, A. Vie, R. A. Hipskind, and J. M. Blanchard. 1999. A novel calcium signaling pathway targets the c-fos intragenic transcriptional pausing site. J. Biol. Chem. 27430439-30446. [DOI] [PubMed] [Google Scholar]
- 14.Dubois, M. F., V. T. Nguyen, M. E. Dahmus, G. Pages, J. Pouyssegur, and O. Bensaude. 1994. Enhanced phosphorylation of the C-terminal domain of RNA polymerase II upon serum stimulation of quiescent cells: possible involvement of MAP kinases. EMBO J. 134787-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dulac, C., A. A. Michels, A. Fraldi, F. Bonnet, V. T. Nguyen, G. Napolitano, L. Lania, and O. Bensaude. 2005. Transcription-dependent association of multiple positive transcription elongation factor units to a HEXIM multimer. J. Biol. Chem. 28030619-30629. [DOI] [PubMed] [Google Scholar]
- 16.Favata, M. F., K. Y. Horiuchi, E. J. Manos, A. J. Daulerio, D. A. Stradley, W. S. Feeser, D. E. Van Dyk, W. J. Pitts, R. A. Earl, F. Hobbs, R. A. Copeland, R. L. Magolda, P. A. Scherle, and J. M. Trzaskos. 1998. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 27318623-18632. [DOI] [PubMed] [Google Scholar]
- 17.Fujinaga, K., D. Irwin, Y. Huang, R. Taube, T. Kurosu, and B. M. Peterlin. 2004. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 24787-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita, T., S. Ryser, S. Tortola, I. Piuz, and W. Schlegel. 2007. Gene-specific recruitment of positive and negative elongation factors during stimulated transcription of the MKP-1 gene in neuroendocrine cells. Nucleic Acids Res. 351007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garriga, J., S. Bhattacharya, J. Calbo, R. M. Marshall, M. Truongcao, D. S. Haines, and X. Grana. 2003. CDK9 is constitutively expressed throughout the cell cycle, and its steady-state expression is independent of SKP2. Mol. Cell. Biol. 235165-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomes, N. P., G. Bjerke, B. Llorente, S. A. Szostek, B. M. Emerson, and J. M. Espinosa. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, C. S., and R. Treisman. 1995. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell 80199-211. [DOI] [PubMed] [Google Scholar]
- 22.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 141415-1429. [PubMed] [Google Scholar]
- 23.Ho, C. K., and S. Shuman. 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3405-411. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 202970-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, J. C., H. P. Phatnani, T. A. Haystead, J. A. MacDonald, S. M. Alam, and A. L. Greenleaf. 2004. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J. Biol. Chem. 27924957-24964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan, C. D., J. R. Morris, C. Wu, and F. Winston. 2000. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 142623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelemen B. R., K. Hsiao, and S. A. Goueli. 2002. Selective in vivo inhibition of mitogen-activated protein kinase activation using cell-permeable peptides. J. Biol. Chem. 2778741-8748. [DOI] [PubMed] [Google Scholar]
- 28.Kiernan, R. E., S. Emiliani, K. Nakayama, A. Castro, J. C. Labbe, T. Lorca, K. Nakayama, and M. Benkirane. 2001. Interaction between cyclin T1 and SCF(SKP2) targets CDK9 for ubiquitination and degradation by the proteasome. Mol. Cell. Biol. 217956-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 142452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krumm, A., T. Meulia, and M. Groudine. 1993. Common mechanisms for the control of eukaryotic transcriptional elongation. Bioessays 15659-665. [DOI] [PubMed] [Google Scholar]
- 31.Mandal, S. S., C. Chu, T. Wada, H. Handa, A. J. Shatkin, and D. Reinberg. 2004. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl. Acad. Sci. USA 1017572-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Napolitano, G., P. Licciardo, R. Carbone, B. Majello, and L. Lania. 2002. CDK9 has the intrinsic property to shuttle between nucleus and cytoplasm, and enhanced expression of cyclin T1 promotes its nuclear localization. J. Cell Physiol. 192209-215. [DOI] [PubMed] [Google Scholar]
- 33.Narita, T., Y. Yamaguchi, K. Yano, S. Sugimoto, S. Chanarat, T. Wada, D. K. Kim, J. Hasegawa, M. Omori, N. Inukai, M. Endoh, T. Yamada, and H. Handa. 2003. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 231863-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni, Z., B. E. Schwartz, J. Werner, J. R. Suarez, and J. T. Lis. 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 1655-65. [DOI] [PubMed] [Google Scholar]
- 35.O'Keeffe, B., Y. Fong, D. Chen, S. Zhou, and Q. Zhou. 2000. Requirement for a kinase-specific chaperone pathway in the production of a Cdk9/cyclin T1 heterodimer responsible for P-TEFb-mediated tat stimulation of HIV-1 transcription. J. Biol. Chem. 275279-287. [DOI] [PubMed] [Google Scholar]
- 36.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108439-451. [DOI] [PubMed] [Google Scholar]
- 37.Palancade, B., and O. Bensaude. 2003. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 2703859-3870. [DOI] [PubMed] [Google Scholar]
- 38.Palangat, M., D. B. Renner, D. H. Price, and R. Landick. 2005. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc. Natl. Acad. Sci. USA 10215036-15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterlin, B. M., and D. H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23297-305. [DOI] [PubMed] [Google Scholar]
- 40.Pinaud, S., and J. Mirkovitch. 1998. Regulation of c-fos expression by RNA polymerase elongation competence. J. Mol. Biol. 280785-798. [DOI] [PubMed] [Google Scholar]
- 41.Ryser, S., T. Fujita, S. Tortola, I. Piuz, and W. Schlegel. 2007. The rate of c-fos transcription in vivo is continuously regulated at the level of elongation by dynamic stimulus-coupled recruitment of positive transcription elongation factor b. J. Biol. Chem. 2825075-5084. [DOI] [PubMed] [Google Scholar]
- 42.Ryser, S., S. Tortola, G. van Haasteren, M. Muda, S. Li, and W. Schlegel. 2001. MAP kinase phosphatase-1 gene transcription in rat neuroendocrine cells is modulated by a calcium-sensitive block to elongation in the first exon. J. Biol. Chem. 27633319-33327. [DOI] [PubMed] [Google Scholar]
- 43.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 142435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sims, R. J. III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 152437-2468. [DOI] [PubMed] [Google Scholar]
- 45.Svejstrup, J. Q. 2004. The RNA polymerase II transcription cycle: cycling through chromatin. Biochim. Biophys. Acta 167764-73. [DOI] [PubMed] [Google Scholar]
- 46.Tanoue, T., M. Adachi, T. Moriguchi, and E. Nishida. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2110-116. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, N. E., T. H. Steinberg, D. B. Aronson, and R. R. Burgess. 1989. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J. Biol. Chem. 26411511-11520. [PubMed] [Google Scholar]
- 48.Thomson, S., A. L. Clayton, and L. C. Mahadevan. 2001. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol. Cell 81231-1241. [DOI] [PubMed] [Google Scholar]
- 49.Treisman, R. 1994. Ternary complex factors: growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev. 496-101. [DOI] [PubMed] [Google Scholar]
- 50.Trigon, S., H. Serizawa, J. W. Conaway, R. C. Conaway, S. P. Jackson, and M. Morange. 1998. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J. Biol. Chem. 2736769-6775. [DOI] [PubMed] [Google Scholar]
- 51.Uptain, S. M., C. M. Kane, and M. J. Chamberlin. 1997. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 66117-172. [DOI] [PubMed] [Google Scholar]
- 52.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 177395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Werlen, G., D. Belin, B. Conne, E. Roche, D. P. Lew, and M. Prentki. 1993. Intracellular Ca2+ and the regulation of early response gene expression in HL-60 myeloid leukemia cells. J. Biol. Chem. 26816596-16601. [PubMed] [Google Scholar]
- 55.Wood, A., and A. Shilatifard. 2006. Transcriptional blackjack with p21. Genes Dev. 20643-647. [DOI] [PubMed] [Google Scholar]
- 56.Wu, C. H., Y. Yamaguchi, L. R. Benjamin, M. Horvat-Gordon, J. Washinsky, E. Enerly, J. Larsson, A. Lambertsson, H. Handa, and D. Gilmour. 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 171402-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, C. H., C. Lee, R. Fan, M. J. Smith, Y. Yamaguchi, H. Handa, and D. S. Gilmour. 2005. Molecular characterization of Drosophila NELF. Nucleic Acids Res. 331269-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamada, T., Y. Yamaguchi, N. Inukai, S. Okamoto, T. Mura, and H. Handa. 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 20227-237. [DOI] [PubMed] [Google Scholar]
- 59.Yamaguchi, Y., T. Wada, and H. Handa. 1998. Interplay between positive and negative elongation factors: drawing a new view of DRB. Genes Cells 39-15. [DOI] [PubMed] [Google Scholar]
- 60.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 9741-51. [DOI] [PubMed] [Google Scholar]
- 61.Yamaguchi, Y., N. Inukai, T. Narita, T. Wada, and H. Handa. 2002. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 222918-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414317-322. [DOI] [PubMed] [Google Scholar]
- 63.Yik, J. H., R. Chen, R. Nishimura, J. L. Jennings, A. J. Link, and Q. Zhou. 2003. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12971-982. [DOI] [PubMed] [Google Scholar]
- 64.Zhong, S. P., W. Y. Ma, and Z. Dong. 2000. ERKs and p38 kinases mediate ultraviolet B-induced phosphorylation of histone H3 at serine 10. J. Biol. Chem. 27520980-20984. [DOI] [PubMed] [Google Scholar]
- 65.Zhou, M., M. A. Halanski, M. F. Radonovich, F. Kashanchi, J. Peng, D. H. Price, and J. N. Brady. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 205077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.