Abstract
Burkholderia is an important bacterial genus with a complex taxonomy that contains species of both ecological and pathogenic importance, including nine closely related species collectively termed the Burkholderia cepacia complex (BCC). In order to more thoroughly investigate the virulence of this bacterial complex of microorganisms, alternative infection models would be useful. To this end, we have adapted and developed the use of the Galleria mellonella wax moth larvae as a host for examining BCC infections. The experimental conditions affecting the BCC killing of the “wax worm” were optimized. BCC virulence levels were determined using 50% lethal doses, and differences were observed between both species and strains of the BCC. The BCC pathogenicity trends obtained compare favorably with results acquired using other published alternative infection models, as well as mammalian infection models. In addition, BCC killing activity was determined by directly measuring relative bacterial loads in three different BCC strains, thus demonstrating innate differences in BCC strain virulence. Finally, genetically mutated BCC strains were compared to a wild-type BCC strain in order to show concomitant reduction of BCC virulence and increased wax worm survival. For experimentation examining the virulent properties of the BCC, the wax worm has proven to be a useful alternative infection model.
Members of the Burkholderia cepacia complex (BCC) are closely related bacterial species that can act as opportunistic pathogens primarily affecting patients with cystic fibrosis (CF) (17, 33). While Pseudomonas aeruginosa may be considered the most prominent chronic respiratory pathogen for these patients (32), BCC infections have had a major impact on the quality of life and mortality of CF patients. Most troublesome is the fact that lung infections with BCC can culminate in a rapidly fatal condition in which patients develop a progressive, necrotizing pneumonia and sepsis commonly referred to as “cepacia syndrome” (23). More typically, CF patients infected with the BCC develop a chronic infection that leads to a slow decline in pulmonary function that is associated with increased mortality and morbidity (31). Because patient-to-patient spread of BCC bacteria is of concern to the CF community (30), social limitations have been placed upon infected individuals, thereby resulting in adverse isolation effects (42). Unfortunately, treatment of the BCC is problematic. Most of the BCC organisms are highly resistant to all major classes of antibiotics, including aminoglycosides (1, 29).
The BCC currently consists of a total of nine Burkholderia species: B. cepacia, B. multivorans, B. cenocepacia, B. stabilis, B. vietnamiensis, B. dolosa, B. ambifaria, B. anthina, and B. pyrrocinia (8, 9, 10, 44-47). The ability of the BCC to cause infections in the CF population is not species dependent, as members of all species have been recovered from infected individuals (42). However, the vast majority of clinical isolates in North America are B. cenocepacia and B. multivorans; in Canada, B. cenocepacia strains cause approximately 80% of BCC infections in CF patients, although this prevalence varies by region (42). Infections with B. cenocepacia are generally associated with the poorest clinical prognosis and the highest rates of transmissibility and mortality. Infections with B. multivorans, although quite common among CF patients, are associated with a better prognosis, having reportedly lower transmission and mortality rates (42). The differences in pathogenicity between all BCC species, however, are not well understood (3, 7, 18).
It is probable that opportunistic bacterial pathogens like the BCC use common virulence factors to infect different organisms. In order to better understand the factors that are important to disease causation by the BCC, we have developed an alternative infection model using the “wax worm.” Larvae of the Greater wax moth Galleria mellonella have been used previously as an infection model for the study of other bacterial human pathogens, including P. aeruginosa (19, 24, 38), Bacillus cereus (16), Proteus mirabilis (39) and, more recently, Francisella tularensis (2), as well as fungal pathogens Cryptococcus neoformans (40), Aspergillus spp. (41, 43), and Candida albicans (12). The innate immune systems of insects such as G. mellonella share a high degree of structural and functional homology to the innate immune systems of mammals (21). Although the immune systems of insects do not display memory or clonal selection mechanisms, they do offer powerful resistance to microbial infections (48). This defense against microorganisms involves both cellular and humoral defenses (21). The humoral immune response of insects consists of the processes of melanization, hemolymph clotting, and the production of a number of potent antimicrobial peptides. The cellular reactions include phagocytosis, nodulization, and large-scale encapsulation. Therefore, G. mellonella is an attractive alternative infection model for a number of reasons. Analysis of insect responses to pathogens can provide an accurate indication of the mammalian response to that pathogen (21, 25). Furthermore, substantial correlation between the virulence of certain microbes in mice and the G. mellonella model has been established (4, 24). While the use of higher animals, such as mice (7) and rats (3, 6), for the study of the BCC has provided invaluable information, alternative infection models that could provide comparable data but that are more cost-effective, less labor-intensive, and more ethically acceptable would be highly useful. Other alternative infection models have previously been tested with the BCC, including alfalfa seedlings (3), Caenorhabditis elegans (26, 35, 22), and Acanthamoeba species (27, 36). Unfortunately, all of these models exhibit at least one deficiency in reproducing the virulence observed with BCC in mice and rats. We have found that the use of G. mellonella as an alternative infection model for describing BCC virulence is in some cases more quantitative, more accurate, and more robust than other available alternative infection models.
MATERIALS AND METHODS
Strains and media.
The bacterial strains of the BCC used in this study were collected strains from the Burkholderia cepacia complex experimental strain panel (34) and the updated experimental strain panel (11). BCC strains were grown in one-half-strength Luria-Bertani (1/2 LB) medium at 30°C. ATCC 29424 mutants were grown in 1/2 LB supplemented with 100-μg/ml trimethoprim at 30°C. Overnight cultures were pelleted and resuspended in 10 mM MgSO4 supplemented with 1.2 mg/ml ampicillin. The presence of ampicillin in the inoculum was to prevent infection with bacteria naturally present on the surface of the larvae. Bacteria collected from the hemolymph of infected larvae were serially diluted in 10 mM MgSO4 and plated onto B. cepacia selective agar (20).
G. mellonella killing assays.
Larvae were stored in wood chips at 4°C. A 10-μl Hamilton syringe was used to inject 5-μl aliquots into G. mellonella via the hindmost left proleg. Following injection, larvae were placed in a static incubator in the dark at 30°C, the optimum temperature for insect growth and development (4). For 50% lethal dose (LD50) experiments, a series of 10-fold serial dilutions containing from 106 to 0 bacteria in 10 mM MgSO4 plus 1.2 mg/ml ampicillin were injected into G. mellonella larvae. Control larvae were injected with 5 μl of only 10 mM MgSO4 plus 1.2 mg/ml ampicillin in order to measure any potentially lethal effects of the physical injection process. Ten larvae were injected at each dilution, and larvae were scored as dead or alive 48 and 72 h postinfection (p.i.) at 30°C. Larvae were considered dead when they displayed no movement in response to shaking of the petri dish or touch with a pipette tip. For each strain, data from three independent experiments were combined, and LD50s were calculated using the Systat computer program as previously described (24). Briefly, Systat was used to fit a curve to the infection data of the following form: Y = [A + (1 − A)]/[1 + exp(B − G × lnX)], where Y is the fraction of larvae killed by the infection, A is the number of larvae killed by control injections, X is the number of bacteria injected, and B and G are Systat-generated variable parameters designed to best fit the curve to the data points. For linear relationships between X and Y, we used a linear regression model using the Systat computer program to determine the LD50.
For time-to-death experiments, live versus dead larvae were monitored every 24 h postinfection. Galleria mellonella larvae were injected with serially diluted bacteria (from 1 × 106 to 0 CFU) and monitored for their survival over a 72-h period. Three independent trials were conducted consisting of 10 worms per bacterial concentration for each specified BCC strain. No more than one control larva died in any given trial. In instances where greater than one control larva died, the data from infected larvae were not used. Results are shown for inoculum concentrations in which the differences between species could be most easily observed (1 × 106 or 1 × 103 CFU).
In order to monitor bacterial loads in larval hemolymph over time, larvae were injected with between 500 and 800 CFU. For the zero time point, larvae were infected and allowed to sit for 20 min before having their hemolymph collected. Equal volumes of hemolymph were collected from five living worms at each time point and combined into a microcentrifuge tube, serially diluted, and plated onto B. cepacia selective agar for quantification. Three groups of five worms were used for each time point in order to quantify bacterial loads.
BCC mutants in G. mellonella assays.
Mutations were isolated or constructed in B. vietnamiensis ATCC 29424 (49). Mutations were introduced with random TnMod-OTp′ plasposon mutagenesis, using a procedure described previously (14). In each example, the mutation was isolated by plasposon rescue, cloned in Escherichia coli, and identified by DNA sequence analysis. The resulting mutants were tested to ensure that none exhibited growth defects in 1/2 LB medium. Many different plasposon mutants were tested in the G. mellonella infection model, although only the mutants with the most significant virulence defects are shown. Alternatively, site-directed insertion mutagenesis was used to create double mutations in the genes for the transport/structural proteins ExbB1 and TolQ (CS1/BG1) or ExbB2 and TolQ (CH1/BG1). Briefly, genes were amplified using PCR primers designed to BCC database sequences for B. cenocepacia J2315 (http://www.sanger.ac.uk/Projects/B_cenocepacia/) and cloned into the E. coli plasmid pUC19, which is unable to replicate in BCC. The individual genes were interrupted with an appropriate selectable antibiotic resistance cassette, such as one of those encoding trimethoprim or tetracycline resistance (15), and the plasmids were electrotransformed into ATCC 29424 using standard protocols (13). Loss of the cloning vector and replacement of the wild-type gene by homologous recombination was confirmed by PCR and DNA sequence analysis. Prior to larval injection, all mutants and wild-type BCC ATCC 29424 cells were grown overnight at 30°C to similar optical density at 600 nm values. Larvae were injected with between 5 × 106 and 9 × 106 CFU of bacteria. Three trials were performed in which 10 larvae were injected for each bacterial strain.
RESULTS
LD50s of BCC strains.
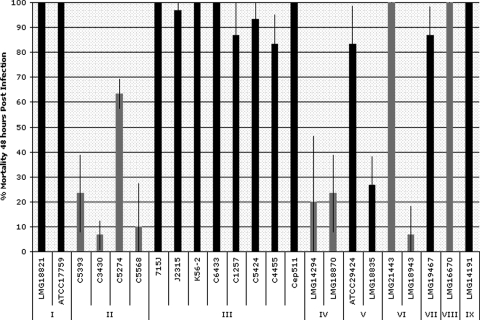
We determined the LD50s of 23 strains within the Burkholderia cepacia complex. As shown in Table 1, B. cepacia and B. cenocepacia strains had the lowest LD50s, while B. multivorans and B. stabilis strains had the highest LD50s. The most virulent strain tested, B. cepacia ATCC 17759, had an LD50 of one bacterium. Some strains within the complex did not exhibit enough larval killing to calculate an LD50, even at the highest dosage tested. Six strains within the complex also exhibited slower killing, and therefore LD50s for those strains could only be calculated after 72 h. Members of B. cenocepacia and B. cepacia were shown to have lower LD50s than B. stabilis and B. multivorans using an independent pairs t test (P < 0.00000001).
TABLE 1.
LD50s of BCC strains in G. mellonella larvae 48 h postinfection
| Species and strain | Alternative collection name | Strain sourcea | LD50 (CFU) in G. mellonella | Reference or strain source |
|---|---|---|---|---|
| B. cepacia strains | ||||
| Cep509 | LMG 18821 | CF, Australia | 30 | 34 |
| ATCC 17759 | LMG 2161 | Soil, Trinidad | 1 | 34 |
| B. multivorans strains | ||||
| C5393 | LMG 18822 | CF, Canada | >3,000,000b | 34 |
| C3430 | CF, Canada | >3,000,000b | CRRRd | |
| C5274 | CF, Canada | 1,000,000 | CRRRd | |
| C5568 | CF, Canada | >3,000,000b | CRRRd | |
| B. cenocepacia strains | ||||
| PC715j | CF, Canada | 4,000 | 37 | |
| J2315 | LMG 16656 | CF-e, United Kingdom | 100,000c | 34 |
| K56-2 | LMG 18863 | CF-e, Canada | 900 | 34 |
| C1257 | CF, Canada | 40,000c | CRRRd | |
| C4455 | CF, Canada | 100,000c | CRRRd | |
| C5424 | LMG 18827 | CF, Canada | 200,000 | 34 |
| C6433 | LMG 18828 | CF, Canada | 30,000 | 34 |
| Cep511 | LMG 18830 | CF, Austrailia | 80,000 | 34 |
| B. stabilis strains | ||||
| ATCC BA-67 | LMG 14294 | CF, Belgium | 2,000,000c | 34 |
| C7322 | LMG 18870 | CF, Canada | >2,000,000b | 34 |
| B. vietnamiensis strains | ||||
| DBO1 | ATCC 29424 | Soil, United States | 200,000 | 49 |
| PC259 | LMG 18835 | CF, United States | >3,000,000b | 34 |
| B. dolosa strains | ||||
| AU0645 | LMG 18943 | CF, United States | >4,000,000b | 8 |
| STM1441 | LMG 21443 | Rhizosphere (Senegal) | 40,000 | 11 |
| B. ambifaria strain | ||||
| Cep0996 | LMG 19467 | CF, Australia | 800,000 | 11 |
| B. anthina strain | ||||
| J2552 | LMG 16670 | Rhizosphere, United Kingdom | 300,000 | 11 |
| B. pyrrocinia strain | ||||
| ATCC 15958 | LMG 14191 | Soil, Japan | 300 | 11 |
Abbreviations: CF, cystic fibrosis infection; CF-e, strain that has spread epidemically among patients with CF.
Mortality never reached 50%, even at the highest dosage.
The LD50 was calculated after 72 h.
CRRR, from the Canadian Burkholderia cepacia complex Research and Referral Repository.
Although some strain variability was noted within BCC species, in general, the pathogenicity displayed toward G. mellonella as measured by LD50 values (Table 1) was similar between BCC strains of the same species. This similarity between the virulence of strains within a BCC species was particularly true for B. cepacia (very low LD50s), B. cenocepacia (moderate LD50s), and B. multivorans and B. stabilis (very high LD50s). Although fewer numbers of the more recently classified species were available for testing, members of B. vietnamiensis and B. dolosa showed greater variation in virulence between strains. In contrast, there were large differences in virulence toward G. mellonella between BCC species. In 48 h, the two B. cepacia strains tested killed on average 50% of G. mellonella larvae at bacterial numbers ranging from 1 to 30 CFU, while the eight B. cenocepacia strains tested killed on average 50% of G. mellonella larvae at numbers ranging from 900 to 200,000 CFU. In contrast, the four B. multivorans strains tested killed on average 50% of G. mellonella larvae under the same conditions, with numbers ranging from 1.0 × 106 CFU or higher, and the two B. stabilis strains tested were only lethal at 2.0 × 106 CFU or higher. A chart showing these trends is displayed in Fig. 1.
FIG. 1.
Differential virulence of Burkholderia cepacia complex strains in the Galleria mellonella infection model. Larvae were injected with approximately ∼106 bacteria and monitored for survival 48 h postinfection. Each bar is representative of three independent trials (n = 10), and standard deviations are shown. Groups according to BCC genomovar: I, B. cepacia; II, B. multivorans; III, B. cenocepacia; IV, B. stabilis; V, B. vietnamiensis; VI, B. dolosa; VII, B. ambifaria; VIII, B. anthina; IX, B. pyrrocinia.
Table 2 illustrates a comparison between BCC virulence in the G. mellonella model with the previously published alfalfa seedling and rat lung agar bead infection models (3). Although identical strains were not used for every species tested, good correlation was observed between the G. mellonella and the alfalfa seedling models. B. multivorans and B. stabilis strains were relatively avirulent in both models, producing disease symptoms in 0 to 13% of alfalfa seedlings, whereas B. cepacia and B. cenocepacia strains were much more virulent in both models, producing disease symptoms in 65 to 100% of the alfalfa seedlings (3). The rat lung agar bead model results similarly compared favorably with the results observed with the BCC in G. mellonella; the B. cepacia and B. cenocepacia strains that resulted in the lowest LD50s in G. mellonella (30 to 105) had the highest amounts of rat lung pathology after 7 days (22.8 to 43.0%). In comparison, B. multivorans, B. stabilis, and B. vietnamiensis strains, with LD50s of at least 106, exhibited the lowest amounts of rat lung pathology (13.3 to 19.7%). An exception was B. cenocepacia strain Cep511, which resulted in a moderate LD50 in G. mellonella (8 × 104), a very high percentage of alfalfa seedlings with disease symptoms (98%), and one of the lowest percentages of pathology observed in the rat lung agar bead model (12.7%) (3). In comparison to these models, the BCC-C. elegans infection model did not demonstrate a general virulence trend within species (5) and therefore did not correlate well with the general results observed for the G. mellonella model. Instead, the BCC-C. elegans infection model displayed significant strain-to-strain variation within species. However, similar to the results obtained for BCC in G. mellonella, B. multivorans was relatively avirulent, and the more recently classified BCC members, such as B. dolosa, B. ambifaria, B. anthina, and B pyrrocinia (11), exhibited significant toxicity in C. elegans.
TABLE 2.
Comparison of BCC virulence in different infection models
| Species and strain | LD50 in G. mellonella | % Alfalfa seedlings with symptomsa | Virulence in rats (% with pathology on day 7 p.i.)a | C. elegans pathogenicity scoreb |
|---|---|---|---|---|
| B. cepacia strains | ||||
| Cep509 | 30 | 85 ± 18c | 43.0 ± 2.0c | 0 |
| ATCC 17759 | 1 | 73 ± 13 | NDd | ND |
| B. multivorans strains | ||||
| C5393 | >3,000,000 | 0 | 13.3 ± 5.9 | 0 |
| C3430 | >3,000,000 | ND | ND | 0 |
| C5274 | 1,000,000 | ND | ND | 0 |
| C5568 | >3,000,000 | ND | ND | 1 |
| B. cenocepacia strains | ||||
| PC715j | 4,000 | 80 ± 18 | 22.8 ± 10.8 | ND |
| J2315 | 100,000 | 65 ± 10 | 38.0 ± 21.0 | 1 |
| K56-2 | 900 | 100 ± 0 | 40.5 ± 4.2 | 3 |
| C1257 | 40,000 | ND | ND | ND |
| C4455 | 100,000 | ND | ND | 1 |
| C5424 | 200,000 | ND | ND | 0 |
| C6433 | 30,000 | ND | ND | ND |
| Cep511 | 80,000 | 98 ± 3 | 12.7 ± 1.53 | 0 |
| B. stabilis strains | ||||
| ATCC BA-67 | 2,000,000 | 13 ± 12 | 16.0 ± 5.6 | 0 |
| C7322 | >2,000,000 | 10 ± 17 | ND | 0 |
| B. vietnamiensis strains | ||||
| DBO1 | 200,000 | ND | ND | ND |
| PC259 | >3,000,000 | 7 ± 6 | 19.7 ± 0.6 | 1 |
| B. dolosa strains | ||||
| AU0645 | >4,000,000 | 25 ± 0 | ND | 1 |
| STM1441 | 40,000 | ND | ND | 2 |
| B. ambifaria strain | ||||
| Cep0996 | 800,000 | 53 ± 16 | ND | 3 |
| B. anthina strain | ||||
| J2552 | 300,000 | ND | ND | 3 |
| B. pyrrocinia strain | ||||
| ATCC 15958 | 300 | ND | ND | 3 |
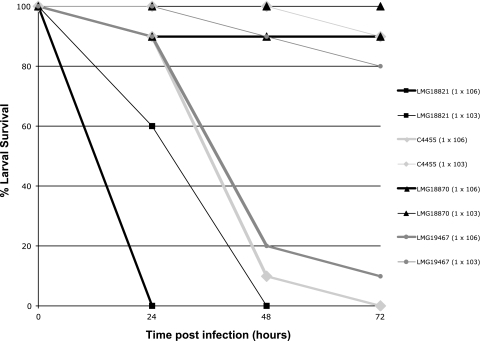
Representative time-to-death curves for four different BCC species at different bacterial doses are shown in Fig. 2. B. cepacia LMG 18821 kills all G. mellonella at 106 CFU within 24 h and 103 CFU within 48 h, faster than a 106 CFU dose of all other BCC species tested. B. cenocepacia C4455 and B. ambifaria LMG 19467 exhibit similar pathogenicities toward G. mellonella at both 106 CFU and 103 CFU over 72 h, intermediate to B. cepacia LMG 18821and B. stabilis LMG 18870. B. stabilis LMG 18870 was not toxic toward G. mellonella at a dose of 103 CFU and only able to kill 10% of the larvae at a dose of 106 CFU, although at higher doses more killing was observed (data not shown). These results were similar to those observed for B. multivorans, as well as some strains of B. vietnamiensis and B. dolosa, in which case LD50 values were difficult to calculate.
FIG. 2.
G. mellonella larvae survival over time when infected with strains of the Burkolderia cepacia complex. Galleria mellonella larvae were injected with bacteria (1 × 106 or 1 × 103) and monitored for their survival over a 72-h period. Each data set is representative of a single trial with the specified strain (n = 10). No more than one uninfected control larvae died in any given trial.
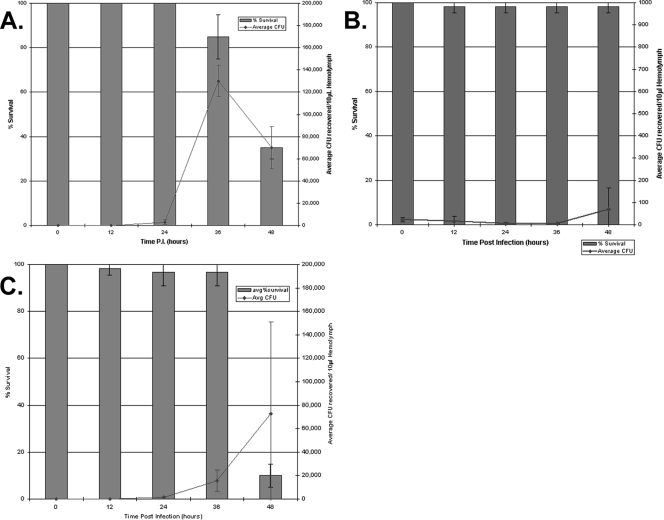
Survival and relative bacterial loads in infected G. mellonella larvae.
Larvae were monitored following infection with three different BCC strains (Fig. 3). The majority of larvae killed following infection with 500 to 800 CFU of B. cepacia LMG18821 or B. cenocepacia K56-2 died after 36 h p.i. As shown in Fig. 3A and C, the bacterial load for larvae infected with strain B. cepacia LMG18821 was approximately 10-fold less than for B. cenocepacia K56-2 at 36 h p.i. (1.6 × 104 compared to 1.3 × 105 CFU recovered/10 μl hemolymph), while infection with LMG 18821 resulted in more larval death at 48 h. Accordingly, the LD50 for B. cepacia LMG 18821 is 30 times lower than the LD50 of B. cenocepacia K56-2 (Table 1). Due to our ability to collect hemolymph only from living larvae, we anticipate that the relative bacterial loads taken at 48 h p.i. from B. cepacia LMG18821 and B. cenocepacia K56-2 are likely gross underestimates. B. multivorans C3430 was unable to establish significant bacterial loads in the hemolymph of infected wax worms at any of the time points following infection (Fig. 3B). These results indicate that this strain is relatively avirulent, and the results correspond with its relatively high LD50 value (>3 × 106 CFU/larvae).
FIG. 3.
Survival and relative bacterial load of G. mellonella infected with different BCC species over time. A. Larvae were infected with approximately 500 CFU B. cenocepacia K56-2 and maintained at 30°C. At the time points indicated, larvae were monitored for survival and bacterial load was quantified from living larvae by the collection of hemolymph as described in Materials and Methods. B. G. mellonella larvae were infected with approximately 700 CFU B. multivorans C3430 and monitored for survival and relative bacterial load over time. C. G. mellonella larvae were infected with B. cepacia LMG 18821 over time. Larvae were infected with approximately 800 CFU LMG 18821 and monitored for their survival and bacterial loads.
Pathogenicity of BCC mutants toward G. mellonella.
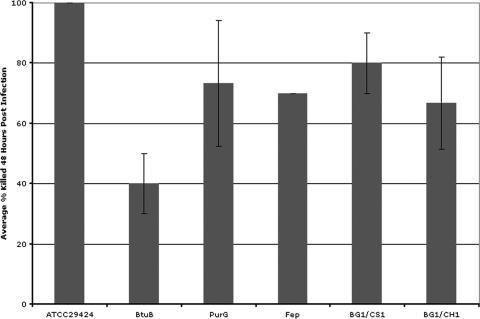
To determine whether the G. mellonella model would be useful for identifying BCC mutants with attenuated virulence, we examined the ability of mutants constructed in B. vietnamiensis strain ATCC 29424 to affect the survival of infected wax worm larvae. As shown in Fig. 4, five mutants constructed in different genes exhibited differing degrees of decreased virulence compared to the wild-type BCC strain. Plasposon mutants with decreased virulence in vivo contained mutations in the BCC orthologous genes encoding BtuB (the receptor for vitamin B12), Fep (a protein involved in ferri-siderophore uptake), or PurG (an enzyme involved in purine biosynthesis). Site-directed mutants with decreased virulence in vivo contained mutations in the genes encoding transport/structural proteins ExbB1 and TolQ (CS1/BG1) or ExbB2 and TolQ (CH1/BG1). The decreased virulence observed was not due to changes in growth or the presence of an antibiotic resistance cassette in the genome, since other similarly constructed mutants grown under identical conditions did not display decreased virulence (data not shown). The results obtained with these constructed mutants indicate that the G. mellonella model will be a valuable experimental system for determining the virulence properties of BCC genetic mutants.
FIG. 4.
Ability of B. vietnamiensis ATCC 29424 mutants to cause disease in the G. mellonella model. Larvae were injected with between 5 × 106 and 9 × 106 CFU. Three trials were performed in which 10 larvae were injected for each strain. Larvae were scored as dead or alive 48 h p.i. at 30°C.
DISCUSSION
In this study, we compare our results for a G. mellonella infection model with previously published BCC data from other infection models, taking care to use many of the identical BCC strains previously tested in order to ensure valid comparison. We observed that G. mellonella, as an infection model for the BCC, possessed several advantages over other infection models. Using G. mellonella, we obtained similar results to both mouse and other alternative infection models with identical BCC strains. In our experience, no other alternative infection models as tested were as sensitive, accurate (with respect to mice and rat lung model results), or as robust or simple to use as the G. mellonella model. This was despite the fact G. mellonella possesses a relatively complex innate immune response, which is lacking in plant infection models. Furthermore, due to ethical and regulatory issues, it is now difficult to perform LD50 experiments with mice. In addition, amoebae and C. elegans are somewhat more difficult to work with, and the BCC do not appear to give consistent results in C. elegans. All of these problems are eliminated or resolved by using the G. mellonella infection model. In a direct comparison of several BCC strains infecting G. mellonella, alfalfa seedlings, or C. elegans, we observed that the G. mellonella model was the most sensitive and consistent infection model with respect to the BCC, as well as being easy to use and relatively rapid (data not shown). We therefore proceeded to characterize this BCC infection model further.
BCC species have previously been characterized as having differing pathogenicity levels (3, 7). B. cenocepacia strains have been observed to cause a greater degree of illness than B. multivorans in a murine model of pulmonary infection (7). Similarly, Bernier et al. (3) was able to demonstrate that most BCC species exhibit similar levels of pathogenicity in an alfalfa infection model as well as a rat agar bead infection model and that B. multivorans strains did not produce severe symptoms of lung pathology in the rat agar bead model or disease symptoms in the alfalfa model. These findings are consistent with the results we obtained for the BCC with the G. mellonella infection model (Fig. 1; Tables 1 and 2). We have demonstrated that B. multivorans strains are relatively avirulent in G. mellonella. In contrast, the most virulent BCC strains tested in G. mellonella belonged to the BCC species B. cepacia and B. cenocepacia, and these were also the most virulent strains tested in the rat agar bead model and the alfalfa model (3). In the leukopenic mouse model (7), B. cenocepacia was more damaging to lung tissue but persisted less well than B. multivorans, which in general persisted longer but without toxicity. Two of the most virulent BCC strains tested in the rat agar bead model (B. cepacia Cep509 and B. cenocepacia K56-2) were also two of the most virulent BCC strains in the G. mellonella model, with LD50s of 30 and 900, respectively. B. stabilis strains tested in these models were found to be rapidly cleared or relatively avirulent (3, 7), which is consistent with what we have observed in the G. mellonella infection model. Therefore, in general, there is a good correlation between the pathogenicity of BCC species infecting G. mellonella and that observed in higher mammalian models.
In support of these findings suggesting G. mellonella is an ideal infection model for the BCC, a significant positive correlation was previously observed between the virulence of P. aeruginosa PA14 mutants in mice and G. mellonella (24). However, no significant correlation was observed between the virulence of these bacterial strains in mice compared to that observed in plants or nematodes, suggesting that G. mellonella may be a more predictive alternative model system for studying the infection process in mammals (24). Similarly, a good correlation was observed between the virulence of C. albicans mutants in the G. mellonella model and the virulence measured in a mouse model of infection (4).
In comparison to other BCC alternative infection models, G. mellonella may prove to be the most useful infection model currently available. For example, no special equipment is required for either the alfalfa or G. mellonella models; however, results can be obtained from the G. mellonella model within 72 h versus 9 days for the alfalfa model (3). Bacterial virulence in the alfalfa model is quantified based on the visual assessment of symptoms, such as yellow leaves, brown necrotic regions, and stunted roots, whereas the G. mellonella model uses larvae death as a finite assay end point. Furthermore, bacteria are exposed to and must overcome a relatively sophisticated innate immune system in the G. mellonella larvae versus that found in an alfalfa seedling.
With respect to nematode models of infection, BCC pathogenicity results are particularly inconsistent. For example, Markey et al. (35) reported that B. cenocepacia strain C5424 effectively kills C. elegans, while Cardona et al. (5) reported that the same strain was nonpathogenic toward C. elegans. Furthermore, Cardona et al. indicated that B. cenocepacia strains that are considered to be clonal exhibit considerably different pathogenicity phenotypes in C. elegans (5). These discrepancies suggest that results obtained with BCC using this infection host are not necessarily reliable. One potential reason for this inconsistency is that C. elegans can experience either fast or slow killing when exposed to bacterial pathogens (26). Furthermore, C. elegans is unable to survive temperatures similar to those observed in animal models, its optimal growth temperature being 20 to 23°C (28). Temperature-associated changes in bacterial virulence can be addressed in the G. mellonella infection model because wax worm larvae can be maintained at temperatures up to 37°C. In addition, both nematode age and the growth medium noticeably impact the ability of the BCC to kill C. elegans (26, 28), whereas G. mellonella larvae do not require feeding during the course of the infection. Finally, in the fast killing model, bacterial pathogenicity is based on subjective indices of C. elegans appearance, such as reduced locomotive capacity.
Free-living amoebae have been proposed as a reservoir for the acquisition and transmission of the BCC, and therefore the BCC have been investigated for their ability to cause infection in Acanthamoeba species (27, 36). Approximately one-third of B. cenocepacia strains tested in this model were found to infect Acanthamoeba, although most of the strains that tested positive for infection did so at the lower levels of scored infectivity, compared to P. aeruginosa PAO1 (36). Somewhat surprisingly, many strains of B. cepacia and B. vietnamiensis were able to infect Acanthamoeba species, while almost all B. multivorans and B. cenocepacia strains were noninfective (36). Therefore, this model may have limited use as a BCC infection model, given that the vast majority of clinical isolates are B. multivorans and B. cenocepacia (42). However, these results do support the idea that there are general differences in virulence between the BCC species, even though strain-to-strain variation does occur.
In order to examine the sensitivity of G. mellonella to the BCC in more detail, we observed the rate at which killing occurred in four different BCC species (Fig. 2). These results showed that in this model, as in other animal models, B. cepacia is exquisitely toxic, even at lower bacterial challenge concentrations. The nature of this toxicity is unknown; however, it is interesting that one strain of B. cepacia tested (ATCC 17759), originally isolated from soil, had an LD50 of 1 in G. mellonella. This is equivalent to the LD50 previously determined for P. aeruginosa in the G. mellonella infection model (LD50 of 1 to 10) (24). B. cenocepacia and B. ambifaria were less virulent but still able to kill 90% and 80% of the larvae, respectively, within 48 h at 106 CFU. However, this killing appeared to be dependent upon the bacterial concentration, since 103 CFU of either BCC strain did not result in more than 20% larval death, even at 72 h p.i. This suggests that either the mechanism of killing is significantly different in B. cenocepacia and B. ambifaria versus B. cepacia infections or that there is a similar mechanism of killing for all BCC strains but differences in virulence factor expression levels. Multiple mechanisms of killing have been reported for P. aeruginosa (19). In contrast, B. stabilis was avirulent in G. mellonella at 103 CFU and killed only 10% of the infected larvae at 106 CFU. This indicates that B. stabilis is relatively nonpathogenic in vivo, at least in this G. mellonella model, and this correlates well with its relative nonprevalence as a human pathogen (42).
In order to better understand what was occurring during BCC infection in vivo, we examined the bacterial loads of infected G. mellonella larvae over time, in three different BCC strains (Fig. 3). Prior to the majority of larval death at 36 h p.i., the level for B. cenocepacia K56-2 was significantly higher (P = <0.001) than the level for B. cepacia LMG 18821 in infected larvae. This suggests that the increased virulence of B. cepacia LMG 18821 compared to B. cenocepacia K56-2 (as also indicated by their LD50 values) is not mediated by high levels of bacterial growth within the hemolymph. Instead, these data suggest that B. cepacia LMG 18821 exerts its toxic effect on wax worm larvae by another means. In comparison, B. multivorans C3430 exhibited poor growth in vivo as determined by viable bacterial counts, even at 48 h p.i., and subsequently showed little toxicity toward the G. mellonella larvae. The bacterial loads at 48 h p.i. could not be determined as accurately in G. mellonella infected with the more virulent BCC strains, because bacteria could not be easily recovered from coagulated and melanized dead or nearly dead larvae. This unfortunately resulted in substantial amounts of error for bacterial numbers of B. cepacia LMG 18821 collected at 48 h p.i. (Fig. 3C).
As the pathogenic differences between BCC species are not well understood, the use of this G. mellonella model may provide a high-throughput, cost-effective screen with which to better elucidate the virulence mechanisms underlying these differences. As shown in Fig. 4, we tested several BCC genetic mutants in the G. mellonella model to demonstrate the usefulness of this approach. Compared to wild-type B. vietnamiensis ATCC 29424, all of the mutants displayed significantly less virulence toward G. mellonella larvae. We anticipate that some of these mutant strains are defective in a system important for in vivo growth. Proteins such as those involved in iron uptake (Fep and ExbB) have been previously been shown to be virulence factors, though not necessarily in the BCC. Under some growth conditions, purine pathway mutations will reduce physiological fitness, and this appears to be the case for the PurG mutant. However, we did not predict that a BCC mutation to btuB, the gene encoding the outer membrane receptor for vitamin B12, would produce such a significant reduction to BCC virulence. The results suggest that the BtuB mutation in B. vietnamiensis ATCC 29424 is significantly less virulent than the other mutants examined with virulence defects, and it is approximately 40% less virulent than the parental strain. Because this protein is as yet uncharacterized in BCC, it is impossible to know whether its function is similar to the BtuB protein in E. coli. However, this result clearly demonstrates the value of in vivo screening for virulence factors.
In conclusion, the differences in virulence toward G. mellonella observed between species of the BCC coincide with observations made using other BCC infection models. The G. mellonella infection model can be used to detect pathogenicity differences between both BCC species as well as different strains within a BCC species. It is likely that bacterial pathogens like the BCC use common virulence factors to infect different hosts. We have demonstrated that insect model systems can be useful for the identification and characterization of BCC virulence factors involved in causing disease in vivo. This model should provide a cost-effective, practical, ethically acceptable, user-friendly alternative for the study of the BCC which will further our understanding of this diverse group of opportunistic bacterial pathogens.
Acknowledgments
This study was funded by an operating grant (to J.J.D.) from the Canadian Cystic Fibrosis Foundation. K.D.S. was supported by a PGS-D award from the Natural Sciences and Engineering Research Council of Canada.
We thank D. Henry, P. Sokol, and G. Zylstra for strains used in this study. We thank M. R. Clark for assistance with statistical analysis, and B. Gee, C. Handford, and C. Shipp for technical assistance in the construction of bacterial mutants.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 14 January 2008.
REFERENCES
- 1.Aaron, S. D., W. Ferris, D. A. Henry, D. P. Speert, and N. E. Macdonald. 2000. Multiple combination bactericidal antibiotic testing for patients with cystic fibrosis infected with Burkholderia cepacia. Am. J. Respir. Crit. Care Med. 1611206-1212. [DOI] [PubMed] [Google Scholar]
- 2.Aperis, G., B. Burgwyn Fuchs, C. A. Anderson, J. E. Warner, S. B. Calderwood, and E. Mylonakis. 2007. Galleria mellonella as a model host to study infection by the Francisella tularensis live vaccine strain. Microbes Infect. 9729-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernier, S. P., L. Silo-Suh, D. E. Woods, D. E. Ohman, and P. A. Sokol. 2003. Comparative analysis of plant and animal models for characterization of Burkholderia cepacia virulence. Infect. Immun. 715306-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan, M., D. Y. Thomas, M. Whiteway, and K. Kavanagh. 2002. Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol. Med. Microbiol. 34153-157. [DOI] [PubMed] [Google Scholar]
- 5.Cardona, S. T., J. Wopperer, L. Eberl, and M. A. Valvano. 2005. Diverse pathogenicity of Burkholderia cepacia complex strains in the Caenorhabditis elegans host model. FEMS Microbiol. Lett. 25097-104. [DOI] [PubMed] [Google Scholar]
- 6.Cash, H. A., D. E. Woods, B. McCullough, W. G. Johanson, and J. A. Bass. 1979. A rat model of chronic respiratory infection with Pseudomonas aeruginosa. Am. Rev. Respir. Dis. 119453-459. [DOI] [PubMed] [Google Scholar]
- 7.Chu, K. K., D. J. Davidson, T. K. Halsey, J. W. Chung, and D. P. Speert. 2002. Differential persistence among genomovars of the Burkholderia cepacia complex in a murine model of pulmonary infection. Infect. Immun. 702715-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coenye, T., J. J. LiPuma, D. Henry, B. Hoste, K. Vandemeulebroecke, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 51271-279. [DOI] [PubMed] [Google Scholar]
- 9.Coenye, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 511481-1490. [DOI] [PubMed] [Google Scholar]
- 10.Coenye, T., P. Vandamme, J. R. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 393427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coenye, T., P. Vandamme, J. J. LiPuma, J. R. Govan, and E. Mahenthiralingam. 2003. Updated version of the Burkholderia cepacia complex experimental strain panel. J. Clin. Microbiol. 412797-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter, G., S. Doyle, and K. Kavanagh. 2000. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol. Med. Microbiol. 27163-169. [DOI] [PubMed] [Google Scholar]
- 13.Dennis, J. J., and P. A. Sokol. 1995. Electrotransformation of Pseudomonas. Methods Mol. Biol. 47125-133. [DOI] [PubMed] [Google Scholar]
- 14.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 642710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis, J. J., and G. J. Zylstra. 1998. Improved antibiotic-resistance cassettes through restriction site elimination using Pfu DNA polymerase PCR. BioTechniques 25772-776. [DOI] [PubMed] [Google Scholar]
- 16.Fedhila, S., N. Daou, D. Lereclus, and C. Nielsen-LeRoux. 2006. Identification of Bacillus cereus internalin and other candidate virulence genes specifically induced during oral infection in insects. Mol. Microbiol. 62339-355. [DOI] [PubMed] [Google Scholar]
- 17.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govan, J. R., J. E. Hughes, and P. Vandamme. 1996. Burkholderia cepacia: medical, taxonomic and ecological issues. J. Med. Microbiol. 45395-407. [DOI] [PubMed] [Google Scholar]
- 19.Hendrickson, E. L., J. Plotnikova, S. Mahajan-Miklos, L. G. Rahme, and F. M. Ausubel. 2001. Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects, and mice. J. Bacteriol. 1837126-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry, D. A., M. E. Campbell, J. J. LiPuma, and D. P. Speert. 1997. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J. Clin. Microbiol. 35614-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann, J. A. 1995. Innate immunity of insects. Curr. Opin. Immunol. 74-10. [DOI] [PubMed] [Google Scholar]
- 22.Huber, B., F. Feldmann, M. Kothe, P. Vandamme, J. Wopperer, K. Riedel, and L. Eberl. 2004. Identification of a novel virulence factor in Burkholderia cenocepacia H111 required for efficient slow killing of Caenorhabditis elegans. Infect. Immun. 727220-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104206-210. [DOI] [PubMed] [Google Scholar]
- 24.Jander, G., L. G. Rahme, and F. M. Ausubel. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 1823843-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kavanagh, K., and E. P. Reeves. 2004. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol. Rev. 28101-112. [DOI] [PubMed] [Google Scholar]
- 26.Kothe, M., M. Antl, B. Huber, K. Stoecker, D. Ebrecht, I. Steinmetz, and L. Eberl. 2003. Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell. Microbiol. 5343-351. [DOI] [PubMed] [Google Scholar]
- 27.Lamothe, J., S. Thyssen, and M. A. Valvano. 2004. Burkholderia cepacia complex isolates survive intracellularly without replication within acidic vacuoles of Acanthamoeba polyphaga. Cell. Microbiol. 61127-1138. [DOI] [PubMed] [Google Scholar]
- 28.Laws, T. R., S. V. Harding, M. P. Smith, T. P. Atkins, and R. W. Titball. 2004. Age influences resistance of Caenorhabditis elegans to killing by pathogenic bacteria. FEMS Microbiol. Lett. 234281-287. [DOI] [PubMed] [Google Scholar]
- 29.LiPuma, J. J. 1998. Burkholderia cepacia. Management issues and new insights. Clin. Chest Med. 19473-486. [DOI] [PubMed] [Google Scholar]
- 30.LiPuma, J. J., S. E. Dasen, D. W. Nielson, R. C. Stern, and T. L. Stull. 1990. Person-to-person transmission of Pseudomonas cepacia between patients with cystic fibrosis. Lancet 3361094-1096. [DOI] [PubMed] [Google Scholar]
- 31.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell III, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 16492-96. [DOI] [PubMed] [Google Scholar]
- 32.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahenthiralingam, E., A. Baldwin, and P. Vandamme. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51533-538. [DOI] [PubMed] [Google Scholar]
- 34.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Markey, K. M., K. J. Glendinning, J. A. Morgan, C. A. Hart, and C. Winstanley. 2006. Caenorhabditis elegans killing assay as an infection model to study the role of type III secretion in Burkholderia cenocepacia. J. Med. Microbiol. 55967-969. [DOI] [PubMed] [Google Scholar]
- 36.Marolda, C. L., B. Hauroder, M. A. John, R. Michel, and M. A. Valvano. 1999. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology 1451509-1517. [DOI] [PubMed] [Google Scholar]
- 37.McKevitt, A. I., S. Bajaksouzian, J. D. Klinger, and D. E. Woods. 1989. Purification and characterization of an extracellular protease from Pseudomonas cepacia. Infect. Immun. 57771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyata, S., M. Casey, D. W. Frank, F. M. Ausubel, and E. Drenkard. 2003. Use of the Galleria mellonella caterpillar as a model host to study the role of the type III secretion system in Pseudomonas aeruginosa pathogenesis. Infect. Immun. 712404-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morton, D. B., G. B. Dunphy, and J. S. Chadwick. 1987. Reactions of hemocytes of immune and non-immune Galleria mellonella larvae to Proteus mirabilis. Dev. Comp. Immunol. 1147-55. [DOI] [PubMed] [Google Scholar]
- 40.Mylonakis, E., R. Moreno, J. B. El Khoury, A. Idnurm, J. Heitman, S. B. Calderwood, F. M. Ausubel, and A. Diener. 2005. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 733842-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reeves, E. P., C. G. Messina, S. Doyle, and K. Kavanagh. 2004. Correlation between gliotoxin production and virulence of Aspergillus fumigatus in Galleria mellonella. Mycopathologia 15873-79. [DOI] [PubMed] [Google Scholar]
- 42.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.St. Leger, R. J., S. E. Screen, and B. Shams-Pirzadeh. 2000. Lack of host specialization in Aspergillus flavus. Appl. Environ. Microbiol. 66320-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. Govan. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res. Microbiol. 15491-96. [DOI] [PubMed] [Google Scholar]
- 45.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 471188-1200. [DOI] [PubMed] [Google Scholar]
- 46.Vandamme, P., E. Mahenthiralingam, B. Holmes, T. Coenye, B. Hoste, P. De Vos, D. Henry, and D. P. Speert. 2000. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 381042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vermis, K., T. Coenye, J. J. LiPuma, E. Mahenthiralingam, H. J. Nelis, and P. Vandamme. 2004. Proposal to accommodate Burkholderia cepacia genomovar VI as Burkholderia dolosa sp. nov. Int. J. Syst. Evol. Microbiol. 54689-691. [DOI] [PubMed] [Google Scholar]
- 48.Vilmos, P., and E. Kurucz. 1998. Insect immunity: Evolutionary roots of the mammalian innate immune system. Immunol. Lett. 6259-66. [DOI] [PubMed] [Google Scholar]
- 49.Walsh, T. A., and D. P. Ballou. 1983. Halogenated protocatechuates as substrates for protocatechuate dioxygenase from Pseudomonas cepacia. J. Biol. Chem. 25814413-14421. [PubMed] [Google Scholar]