Abstract
Campylobacter jejuni is a leading cause of bacterial gastroenteritis in humans throughout the world, but infection of animals, especially poultry, results in a commensal colonization of the intestines. We previously found that a mutant lacking docA, which encodes a putative cytochrome c peroxidase (CCP), demonstrates up to a 105-fold reduction in colonization of the chick cecum compared to wild-type C. jejuni strain 81-176. Predictions from genomic sequences identified CJJ0382 as a second locus in C. jejuni encoding a CCP, making the bacterium unusual in having two putative CCPs. To understand what advantages are imparted by having two putative CCPs, we compared the colonization requirements of C. jejuni mutants lacking DocA or Cjj0382. Unlike the ΔdocA mutant, a ΔCJJ0382 mutant demonstrates a maximal 50-fold colonization defect that is dependent on the inoculum dose. The colonization differences of mutants lacking DocA or Cjj0382 suggest that the two predicted CCPs are unlikely to perform redundant functions during in vivo growth. In the characterizations of DocA and Cjj0382, we found that they are stable periplasmic proteins with an apparent heme-dependent peroxidase activity, which are characteristics of bacterial CCPs. However, the peroxidase activities of the proteins do not appear to contribute to resistance to hydrogen peroxide. Instead, we found that resistance to hydrogen peroxide in C. jejuni is mostly attributed to the cytoplasmic catalase KatA. Our data suggest that DocA and Cjj0382 have characteristics of CCPs but likely perform different physiological functions for the bacterium in colonization that are not related to resisting oxidative stress.
The gram-negative bacterium Campylobacter jejuni is a frequent cause of bacterial gastroenteritis in humans in the United States and throughout the world (4). Infection of humans can result in a mild to severe inflammatory gastroenteritis leading to a bloody diarrheal syndrome (30). Whereas infection of humans is usually self-limiting, secondary sequelae associated with having a previous C. jejuni infection can occur. One such complication is Guillain-Barré syndrome, which is an autoimmune disorder that results in an acute paralysis of the peripheral nervous system and in rare cases can lead to death (13, 24).
Sporadic cases of C. jejuni gastroenteritis in humans are most often associated with the consumption or handling of poultry meats (9). In contrast to human infections, C. jejuni colonization of many wild and agriculturally important animals, including avian species, is an asymptomatic intestinal infection that results in a commensal relationship between the bacterium and the host. Whereas infection of these animals is harmless to the hosts, it creates large reservoirs of C. jejuni in the environment that can contaminate the human food or water supply.
To understand the requirements of C. jejuni for colonization of poultry that result in commensalism, we previously employed a genetic selection procedure using signature-tagged transposon mutants to identify genes of the bacterium involved in growth within the avian cecum (17). Among the 22 different genes we identified were ones required for flagellar motility, protein glycosylation, and a putative cytochrome c peroxidase (CCP) that we annotated as docA (for determinant of chick colonization). docA is designated cjj81176_0047c and cj0020c in the C. jejuni strain 81-176 and strain NCTC11168 genomic sequences, respectively (8, 25). Deletion of docA from C. jejuni strain 81-176 results in 10- to 105-fold-lower bacterial loads in the ceca of chicks at 7 days postinfection compared to those of chicks infected with wild-type bacteria (17).
Bacterial CCPs are periplasmic proteins that reduce potentially toxic hydrogen peroxide compounds to water (1). Two heme molecules that are bound by each CCP protein receive electrons from cytochrome c to reduce hydrogen peroxide to water without generating other reactive oxygen intermediates. Despite having a peroxidase activity in vitro, the physiological roles of this activity and, more globally, the contribution of the CCPs to biological functions in bacteria are not well understood (1). The CCP of Neisseria gonorrhoeae may have a part in providing resistance to hydrogen peroxide, since a mutant lacking the CCP is slightly more sensitive to the compound than wild-type bacteria (32). However, the level of resistance to hydrogen peroxide provided by the cytoplasmic catalase of N. gonorrhoeae is much greater, suggesting that the primary role of the CCP in this bacterium may not be protection from oxidative stress. Considering that many gram-negative bacteria do not have a CCP and these proteins are absent from gram-positive organisms, CCPs are not believed to have a universal role in bacteria in promoting resistance to oxidative stress (1).
Bioinformatic analysis of the C. jejuni genome has predicted that the gene CJJ0382 (annotated as cjj81176_0382 in the genome of C. jejuni 81-176 [8] and cj0358 in the genome of C. jejuni NCTC11168 [25], but herein designated CJJ0382) encodes another putative CCP. In order to understand the importance of docA and CJJ0382 in colonization, we compared the ability of mutants lacking each gene to promote commensal colonization of 1-day-old chicks. We found that the docA mutant has up to 1,000-fold-greater cecal colonization defects in chickens than the CJJ0382 mutant. These data suggest that DocA and Cjj0382 may not perform redundant functions for C. jejuni during colonization of poultry. We also found that even though these proteins are located in the periplasm and have heme-dependent peroxidase activity, which are characteristics of typical CCPs, they do not appear to have a physiological role in promoting resistance to hydrogen peroxide. Instead, hydrogen peroxide resistance is largely attributed to katA, the cytoplasmic catalase. Further analysis revealed that a ΔkatA mutant of C. jejuni demonstrates a less severe colonization deficiency than a mutant lacking docA. This study provides insight into the colonization determinants that are required for optimal in vivo growth of C. jejuni in poultry. Furthermore, DocA and Cjj0382 have characteristics of CCPs but likely perform different functions for C. jejuni during colonization. Further studies will be required to elucidate the specific physiological roles of DocA and Cjj0382 in C. jejuni.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Campylobacter jejuni strain 81-176 is a clinical isolate that has been shown to promote gastroenteritis in humans as well as commensal colonization of the chick gastrointestinal tract (3, 15, 17, 21). C. jejuni was typically grown on Mueller-Hinton (MH) agar containing 10 μg/ml trimethoprim (TMP) under microaerobic conditions (85% nitrogen, 10% carbon dioxide, 5% oxygen) at 37°C. Antibiotics for C. jejuni were used at the following concentrations: chloramphenicol, 15 μg/ml; streptomycin, 100 μg/ml or 0.5, 1, 2, or 5 mg/ml; cefoperazone, 30 μg/ml. All C. jejuni strains were stored in 85% MH broth-15% glycerol solution at −80°C. Escherichia coli DH5α was grown in Luria-Bertani (LB) agar or broth, and antibiotics were used at the following concentrations as necessary: ampicillin, 100 μg/ml; chloramphenicol, 15 μg/ml; tetracycline, 12.5 μg/ml. All E. coli strains were stored in 80% LB broth-20% glycerol solution at −80°C.
Construction of point mutations and deletion mutants.
DRH212 (81-176 Smr) and DRH1169 (81-176 Smr ΔdocA) were constructed as described previously (16, 17). DRH1169 contains a deletion of codons 2 to 293 of docA from the chromosome of C. jejuni. To construct a deletion mutant of CJJ0382, we first amplified a 2.8-kb fragment that includes approximately 800 nucleotides upstream and downstream of the CJJ0382 coding sequence from the chromosome of C. jejuni 81-176 by PCR using primers with 5′ BamHI restriction sites. This fragment was cloned into BamHI-digested pUC19 to create pDRH308. In-frame, chromosomal deletion mutants were constructed as previously described (16). To interrupt CJJ0382, cat-rpsL (obtained from SmaI-digested pDRH265 [16]) was ligated into SwaI-digested pDRH308 to create pDRH1573. PCR-mediated mutagenesis was used to create an in-frame deletion of CJJ0382 (22). pDRH308 was used as the template with primers designed to delete the entire coding sequence of CJJ0382 to create pDRH1748. DRH212 was transformed by electroporation with pDRH1573, and transformants were recovered on MH agar with chloramphenicol to obtain LKB140 (81-176 Smr CJJ0382::cat-rpsL). This insertional mutant was then transformed by electroporation with pDRH1748, and MH agar with streptomycin was used to select for transformants that had replaced CJJ0382::cat-rpsL with the in-frame deletion construct on the chromosome of C. jejuni. This approach allowed the recovery of LKB151 (81-176 Smr ΔCJJ0382).
An in-frame deletion mutant of katA was generated by first amplifying a 3.1-kb fragment with approximately 800 nucleotides upstream and downstream of the katA coding sequence from the chromosome of C. jejuni 81-176 using primers with 5′ BamHI restriction sites. This fragment was cloned into BamHI-digested pUC19 to create pLKB106. An MscI site was generated by PCR-mediated mutagenesis by making a C-to-G mutation at nucleotide 181 of the katA coding sequence to create pLKB141. This plasmid was then digested with MscI for insertion of the SmaI-digested cat-rpsL cassette into the coding sequence of katA to create pLKB147. pLKB106 was also used for PCR-mediated mutagenesis to make an in-frame fusion removing the entire coding sequence of katA to create pLKB131. DRH212 was transformed by electroporation with pLKB147 to create LKB236, in which katA is interrupted with cat-rpsL. This insertional mutant was then transformed by electroporation with pLKB131, and transformants were recovered on MH agar with streptomycin to obtain LKB246 (81-176 Smr ΔkatA).
For construction of 81-176 Smr ΔdocA ΔCJJ0382, LKB151 (81-176 Smr ΔCJJ0382) was transformed by electroporation with pDRH1161 (pUC19 containing docA::cat-rpsL) (17) to create LKB165 (81-176 Smr ΔCJJ0382 docA::cat-rpsL). This strain was then transformed by electroporation with pDRH1147 (pUC19 containing ΔdocA) (17), resulting in LKB177 (81-176 Smr ΔdocA ΔCJJ0382). To create 81-176 Smr ΔdocA ΔkatA, DRH1169 (81-176 Smr ΔdocA) was transformed by electroporation with pLKB147 to create LKB170 (81-176 Smr ΔdocA katA::cat-rpsL). This strain was then transformed by electroporation with pLKB131, resulting in LKB181 (81-176 Smr ΔdocA ΔkatA). To create 81-176 Smr ΔCJJ0382 ΔkatA, LKB151 was transformed by electroporation with pLKB147 to create LKB164 (81-176 Smr ΔCJJ0382 katA::cat-rpsL). This strain was then transformed by electroporation with pLKB131, resulting in LKB231 (81-176 Smr ΔCJJ0382 ΔkatA). Lastly, to generate 81-176 Smr ΔdocA ΔCJJ0382 ΔkatA, LKB181 (81-176 Smr ΔdocA ΔkatA) was transformed by electroporation with pDRH1573 to create LKB213 (81-176 Smr ΔdocA ΔkatA CJJ0382::cat-rpsL). This strain was then transformed by electroporation with pDRH1748, resulting in LKB226 (81-176 Smr ΔdocA ΔCJJ0382 ΔkatA).
Heme-binding site (HBS) mutants were constructed by creating point mutations in the first cysteine and last histidine residue of the CXXCH motif of each binding site in docA and CJJ0382, generating Cys-to-Ser and His-to-Ala mutants. All point mutants were generated by PCR-mediated mutagenesis using pDRH765 (17) for docA HBS mutants or pLKB366 (pUC19 containing CJJ0382 with myc epitope at the C terminus) for CJJ0382 HBS mutants. For construction of pLKB366, pUC19::CJJ0382 was used for PCR-mediated mutagenesis to add the myc epitope to the C-terminal end of CJJ0382. For construction of docAC56S, a G-to-C point mutation at nucleotide 167 in the docA coding sequence was generated, creating pLKB477. For construction of docAH60A, C-to-G and A-to-C point mutations at nucleotides 178 and 179 in the docA coding sequence were generated, creating pSMS176. For construction of docAH203A, C-to-G and A-to-C point mutations at nucleotides 607 and 608 in the docA coding sequence were generated, creating pSMS201. DRH1168 (81-176 Smr docA::cat-rpsL) (17) was transformed by electroporation with pLKB477, pSMS176, and pSMS201 to create LKB506 (81-176 Smr docAC56S), LKB122 (81-176 Smr docAH60A), and LKB101 (81-176 Smr docAH203A), respectively. For construction of CJJ0382C80S, a G-to-C point mutation at nucleotide 239 in the CJJ0382 coding sequence was generated, creating pLKB428 (pUC19 containing CJJ0382C80S-myc). For construction of CJJ0382H84A, C-to-G and A-to-C point mutations at nucleotides 250 and 251 of the CJJ0382 coding sequence were generated, creating pLKB429. For construction of CJJ0382C225S, a G-to-C point mutation at nucleotide 674 in the CJJ0382 coding sequence was generated, creating pLKB430. For construction of CJJ0382H229A, C-to-G and A-to-C point mutations at nucleotides 685 and 686 in the CJJ0382 coding sequence were generated, creating pLKB431. LKB140 (81-176 Smr CJJ0382::cat-rpsL) was transformed by electroporation with pLKB428, pLKB429, pLKB430, and pLKB431 to create LKB469 (81-176 Smr CJJ0382C80S), LKB455 (81-176 Smr CJJ0382H84A), LKB447 (81-176 Smr CJJ0382C225S), and LKB450 (81-176 Smr CJJ0382H229A), respectively.
The 81-176 Smr ΔdocA and ΔCJJ0382 mutants were complemented in trans with pRY112 derivatives containing docA or CJJ0382 (33). Primers were designed with 5′ BamHI restriction sites to amplify DNA fragments containing docA and CJJ0382 with their putative promoter elements. Each fragment was digested with BamHI and ligated into BamHI-digested pRY112 to create pLKB260 and pLKB268 (containing docA or CJJ0382, respectively). These plasmids or pRY112 were then transferred to DH5α/pRK212.1 for subsequent conjugation into the 81-176 Smr ΔdocA or ΔCJJ0382 mutants (7, 12).
Protein homology and domain analyses.
Homology searches were performed with BLASTP and PSI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST). Analyses of N-terminal signal sequences were performed with the SignalP 3.0 server (http://cbs.dtu.dk/services/SignalP/).
Chick colonization assays.
One-day-old White Leghorn strain Δ chicks were orally infected with C. jejuni strains to assess the cecal commensal colonization capacity of wild-type and mutant strains of C. jejuni 81-176 Smr as previously described (17). Briefly, fertilized chicken eggs (SPAFAS) were incubated for 21 days at 37.8°C under appropriate humidity conditions and rotation of the eggs in a Sportsman II model 1502 incubator (Georgia Quail Farms Manufacturing Company). Approximately 12 to 36 h after hatch, chicks were orally infected with 100 μl of phosphate-buffered saline (PBS) containing approximately 102, 104, and 106 CFU of wild-type and mutant strains of C. jejuni. To prepare strains for infection, bacteria were grown from frozen stocks for 48 h on MH agar with TMP at 37°C under microaerobic conditions. Strains were then streaked heavily on MH agar with TMP and grown for 16 h. Bacteria were resuspended from the plates and diluted to the appropriate inoculum in PBS. Dilutions of the inocula were plated to determine the number of bacteria in the inoculation dose. Seven days postinfection, chicks were sacrificed, the cecal contents were recovered and resuspended in PBS, and dilutions were plated on MH agar containing 10 μg/ml TMP and 30 μg/ml cefoperazone to determine the number of C. jejuni per gram of cecal contents. Statistical analysis on results from colonization experiments was performed with the Mann-Whitney U test.
Biphasic culture system.
Bacteria were grown from frozen stocks for 48 h on MH agar with TMP at 37°C under microaerobic conditions. Strains were then streaked heavily on MH agar with TMP and grown for 16 h. Bacteria were resuspended from the plates and diluted to an optical density at 600 nm (OD600) of 0.1 with MH broth. Ten milliliters of culture was added to T25 flasks containing 5 ml solidified MH agar and incubated under microaerobic conditions at 37°C for 24 h. Growth rates were analyzed by monitoring the OD600 of each strain.
Real-time RT-PCR analysis.
One-day-old chicks were infected with 104 CFU wild-type C. jejuni 81-176 Smr (DRH212) as described above, and total RNA was extracted with Trizol reagent (Invitrogen) from the cecal contents of chicks 7 days postinfection and then treated with DNase prior to analysis. A final concentration of 50 ng/μl of RNA was used in a Sybr green PCR master mix. Real-time reverse transcription-PCR (RT-PCR) was performed using a 7500 real-time PCR system (Applied Biosystems). Detection of mRNA for gyrA, encoding DNA gyrase, served as an endogenous control, and the in vivo transcript levels of docA and CJJ0382 were compared to each other. The following primer pairs were used for real-time RT-PCR analysis: docA F, 5′-AACTTTATGGAGAAGTTACGGTAGAAAAC-3′, and docA R, 5′-ATAGCGATCAAAAGGAGAATTTGG-3′; CJJ0382 F, 5′-TTGCTGAAACTGCTCCATATTTTC-3′, and CJJ0382 R, 5′-GCCAAGTTGCACACTACCCATT-3′; gyrA F, 5′-CGACTTACACGGCCGATTTC-3′, and gyrA R, 5′-ATGCTCTTTGCAGTAACCAAAAAA-3′.
Fractionation of C. jejuni strains.
Fractionation of C. jejuni into subcellular compartments for analysis of localization of proteins was performed as previously described, with slight modifications (31). Briefly, C. jejuni strains were grown from frozen stocks on MH agar with either TMP or chloramphenicol at 37°C under microaerobic conditions for 48 h. Strains were then streaked heavily on MH agar and grown for 16 h. After growth, each strain was resuspended in MH broth and diluted to an OD600 of 0.8. This method of preparing each strain was used for all fractionation procedures described below. For whole-cell lysates (WCL), 1 ml of bacterial culture was pelleted, washed once in PBS, and resuspended in 50 μl 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. To generate periplasmic and cytoplasmic fractions, 20 ml of bacterial culture was washed twice with 2 ml of PBS containing 0.1% gelatin (PBSG) and then resuspended in 2 ml of PBSG containing 20 mg/ml polymyxin B sulfate (Sigma) to compromise the outer membrane and release the periplasmic contents. After centrifugation with a Sorvall Biofuge Pico microcentrifuge at 16,000 × g for 30 min, the supernatant was saved, and the recovered pellets consisted of whole spheroplasts. The supernatants were centrifuged again for 30 min at 16,000 × g to ensure removal of contaminating spheroplasts from the periplasmic fraction. The pellets consisting of whole spheroplasts were resuspended in 1 ml PBSG and sonicated with a Branson Sonifier 450 set at an amplitude of 4.5 with a constant duty cycle. Samples were kept on ice between bursts, and each sample was sonicated four times. Bursts were approximately 8 seconds long with a 2-minute resting period between each burst. After centrifugation at 16,000 × g for 30 min to remove insoluble membranes, the supernatants represented soluble cytoplasmic proteins. To obtain inner and outer membrane fractions, 5-ml aliquots of bacteria were pelleted and washed once with 1 ml of 10 mM HEPES (pH 7.4). Bacteria were then resuspended in 1 ml of 10 mM HEPES for sonication as described above, and total membranes were recovered after centrifugation for 30 min at 16,000 × g. Membranes were resuspended in 10 mM HEPES containing 1% N-lauroylsarcosine sodium salt to solubilize the inner membrane fraction. The soluble inner membrane proteins were separated from the insoluble outer membrane proteins by centrifugation for 30 min at 16,000 × g.
Preparation of antisera.
Primers were designed to amplify the coding sequences of docA from codon 33 to the stop codon, CJJ0382 from codon 50 to the stop codon, and atpF, encoding the b subunit of ATP synthase, from codon 54 to the stop codon. In-frame BamHI restriction sites were added to the 5′ ends of the primers. After amplification from the chromosome of C. jejuni strain 81-176, the gene fragments were digested with BamHI and ligated into BamHI-digested pGEX-4T-2 (GE Healthcare). Transformation into E. coli BL21(DE3) allowed for recovery of plasmids designated pDRH2302 (containing a portion of atpF), pDRH2556 (containing a portion of docA), and pDRH2557 (containing a portion of CJJ0382). For purification of glutathione-S-transferase fusion proteins, 500-ml cultures were grown in LB at 37°C and induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 4 h. Bacteria were lysed with an EmusliFlex-C5 cell disrupter (Avesin) at 15,000 to 20,000 lb/in2. Proteins were purified from the soluble fraction with glutathione-Sepharose 4B according to the manufacturer's instructions (GE Healthcare). Each protein was then injected into five different mice for production of polyclonal antisera.
Immunoblot analyses.
Proteins were separated by SDS-PAGE using 10% acrylamide gels. For WCL, protein samples were loaded to represent the proteins recovered from 200-μl aliquots of bacterial cultures which had been equilibrated to the same density as described above. For the fractions representing outer membrane, periplasmic, inner membrane, and cytoplasmic proteins, the amounts loaded represented protein samples obtained from 400 μl of bacterial culture. To detect DocA or Cjj0382, a 1:1,500 dilution of murine anti-DocA M10 antiserum or a 1:2,000 dilution of murine anti-Cjj0382 M17 antiserum was used. For control immunoblots detecting known proteins contained within specific fractions, the following antibodies were used: anti-RpoA M1 at a dilution of 1:1,500 was used to detect RpoA, a cytoplasmic protein (31); anti-AtpF M3 at a dilution of 1:1,000 was used to detect AtpF, an inner membrane protein; anti-FlgP M1 at a dilution of 1:1,500 was used to detect FlgP, an outer membrane protein (31). Horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G antiserum was used as the secondary antibody (Bio-Rad).
Arylsulfatase assays.
To determine the purity of the protein preparation derived from the periplasmic fractionation procedure, the amount of sample derived from an equivalent of 400 μl of bacterial culture described above was tested for arylsulfatase activity in assays similar to ones previously described (14, 18, 34). For comparison, we also analyzed the cytoplasmic fractions from an equivalent amount of bacterial culture. Samples were incubated in a final volume of 400 μl of 10 mM nitrophenylsulfate and 1 mM tyramine for 1 h at 37°C. Reactions were stopped, and the amount of nitrophenol released in each sample was determined by a spectrophotometric reading at 410 nm. The obtained readings were compared to a standard curve of OD410 readings from known concentrations of nitrophenol to determine the number of arylsulfatase units produced by each sample. Each sample was tested in triplicate.
Heme stain assay.
The method of Feissner et al. was used to detect heme bound to DocA and Cjj0382 in soluble periplasmic fractions (6). Approximately 25 μg of proteins isolated from the periplasmic fraction, as described above, was mixed with SDS-PAGE loading buffer that lacked β-mercaptoethanol. Samples were then loaded onto 10% SDS-polyacrylamide gels without boiling the samples. Preparing the samples in this manner is considered a nondenaturing condition that is required for this assay so that heme compounds remain covalently bound to proteins. After the proteins were transferred to a nitrocellulose membrane, the membrane was washed three times with PBS and exposed to SuperSignal West Femto chemiluminescent substrate (Pierce Biotechnology) for 5 min. The peroxidase activity of the bound heme reacts with the chemiluminescent substrate to generate light, which is detected upon exposure to film.
Peroxide resistance assays.
Wild-type and mutants strains of C. jejuni were grown from frozen stocks on MH agar with TMP at 37°C under microaerobic conditions. Bacteria then were streaked heavily on MH agar and grown for 16 h, resuspended in MH broth, and diluted to an OD600 of 0.4. One-milliliter samples of bacteria were treated with 0.5 mM hydrogen peroxide, cumene hydroperoxide, or tert-butyl hydroperoxide for 30 min at 37°C under microaerobic conditions. After incubation, serial dilutions were plated on MH agar with TMP and the number of surviving bacteria relative to the number of viable bacteria at the beginning of the assay was determined. Each assay was performed three times in triplicate.
RESULTS
ΔdocA and ΔCJJ0382 mutants display different colonization capacities.
Through a genetic selection procedure using signature-tagged transposon mutants that identified genes of C. jejuni 81-176 involved in commensal colonization of the chick cecum, we discovered that a docA mutant of C. jejuni is severely attenuated for colonization (17). Bioinformatic analysis suggests that DocA, which is predicted to function as a CCP, is similar to another putative CCP encoded by CJJ0382 in C. jejuni 81-176 (8). DocA and Cjj0382 share 62% similarity and 44% identity, including two different putative heme-binding motifs in each protein. These heme-binding sites are predicted to be essential for binding heme for the peroxidase activity that reduces hydrogen peroxide to water (1). The largest region of nonhomology occurs in the N-terminal regions of the proteins, which contain predicted N-terminal signal sequences for transport out of the bacterial cytoplasm. The presence of two CCPs in an individual bacterium appears to be uncommon (1). Phylogenetic analyses have predicted that Cjj0382 clusters closely to other classical CCPs that are typically found in various gram-negative bacteria (Cjj0382 is the same protein as Cj0358 encoded in the C. jejuni NCTC 11168 genome, which was used in the phylogenetic analysis listed in reference 1). However, DocA is not closely related phylogenetically to any predicted bacterial CCP and is currently the sole member of a separate clade.
Since a previous analysis by our laboratory revealed that a ΔdocA mutant (which was constructed by fusing the start codon to the last 12 codons, deleting the intervening 292 codons) was attenuated for colonization of the chick cecum (17), we analyzed whether Cjj0382 is required by C. jejuni for efficient colonization of chicks. We created a mutant of CJJ0382 in 81-176 Smr (DRH212) (16) by deleting the entire coding sequence of the gene from the chromosome.
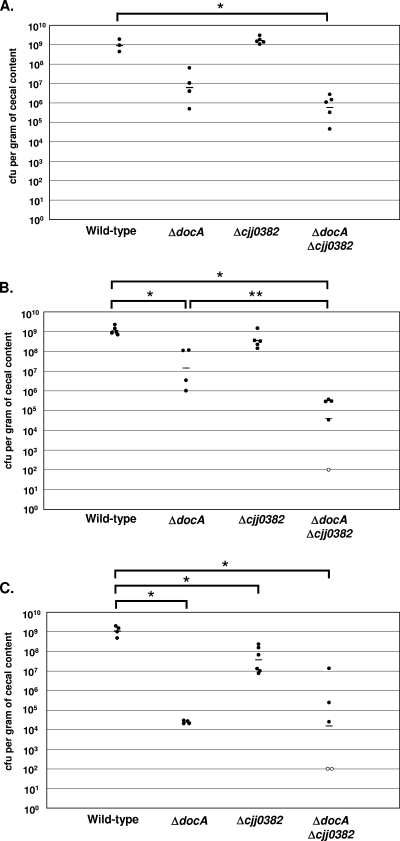
The ability of the ΔCJJ0382 mutant to colonize the chick cecum was compared to wild-type 81-176 Smr and 81-176 Smr ΔdocA by orally infecting 1-day-old chicks with inocula of approximately 106, 104, or 102 organisms. Wild-type bacteria consistently colonizes the chick cecum at approximately 109 CFU per gram of cecal content at day 7 postinfection, regardless of the inoculum size (Fig. 1A to C). At inocula of approximately 106 and 104, the ΔdocA mutant colonizes at levels 10- to 1,000-fold lower than wild-type bacteria, with a further reduction up to 10,000-fold at an inoculum of 102 bacteria (Fig. 1A to C). When we analyzed the ΔCJJ0382 mutant, we discovered that this strain displays a less severe colonization defect than the ΔdocA mutant. At the higher inocula (106 and 104 bacteria), the ΔCJJ0382 mutant demonstrates levels of colonization similar to wild type with only a very slight colonization defect of approximately twofold at the 104 inoculum (Fig. 1A and B). However, at the lowest inoculum (approximately 102 bacteria), we did observe a statistically significant 10- to 50-fold reduction in colonization for this mutant (Fig. 1C). The defects in colonization of the mutants are specific for in vivo growth, as we could not detect any differences in growth between the wild-type and mutant strains during in vitro growth in laboratory media (data not shown).
FIG. 1.
The ΔCJJ0382 mutant of C. jejuni displays a less severe colonization defect than the ΔdocA mutant. One-day-old chicks were orally infected with 100 μl of C. jejuni 81-176 Smr (DRH212), 81-176 Smr ΔdocA (DRH1169), 81-176 Smr ΔCJJ0382 (LKB151), or 81-176 Smr ΔdocA ΔCJJ0382 (LKB177) with inocula of approximately 106 (A), 104 (B), or 102 (C) bacteria. Chicks were sacrificed 7 days postinfection to determine the cecal colonization capacity of each C. jejuni strain. Each filled symbol represents the amount of C. jejuni recovered from the cecum of a single chick, reported as the number of CFU per gram of cecal content. Each open symbol represents chicks in which the level of C. jejuni colonization was below the limit of detection (<100 CFU per gram of cecal content). The geometric mean of the bacterial loads from each set of chicks is denoted by a short horizontal line. Statistical analysis was performed using the Mann-Whitney U test (P < 0.05). *, significant difference between wild-type and mutant strains; **, significant difference between the 81-176 Smr ΔdocA mutant and the 81-176 Smr ΔdocA ΔCJJ0382 mutant. The actual inoculum doses ranged from 5 × 105 to 1.7 × 106 CFU (A), 4.4 × 103 to 2.19 × 104 CFU (B), and 58 to 208 CFU (C).
To more thoroughly characterize the requirements for DocA and Cjj0382 for colonization of the chick cecum, we made a mutant that lacked both docA and CJJ0382 (81-176 Smr ΔdocA ΔCJJ0382). At inocula of approximately 106 and 104, the colonization defect of the ΔdocA ΔCJJ0382 mutant was greater than mutants lacking only docA or CJJ0382, with the differences in colonization with the latter inoculum being statistically significant (Fig. 1A and B). In addition, with the 104 inoculum, one chick contained levels of C. jejuni that were below the limit of detection. Even though two chicks contained C. jejuni at levels below the limit of detection with an inoculum of 102, we could not detect a significant difference between the overall colonization capacities of the ΔdocA and the ΔdocA ΔCJJ0382 mutant even though both mutants colonized at significantly lower levels than the wild-type strain (Fig. 1C). Together these data suggest that both proteins are involved in promoting efficient commensal colonization of the chick cecum, yet a greater dependency on DocA for colonization compared to Cjj0382 is evident.
Either a lack of expression of CJJ0382 or an increased expression of docA relative to CJJ0382 during in vivo growth could explain why the ΔCJJ0382 mutant is only slightly attenuated for in vivo growth in chicks compared to the ΔdocA mutant. To determine if either of these hypotheses were valid, we compared the relative levels of docA and CJJ0382 transcripts by real-time RT-PCR from wild-type C. jejuni directly isolated from the chick ceca 7 days postinfection. Contrary to our hypotheses, we discovered that both genes are expressed in vivo and that the levels of expression of CJJ0382 were approximately 15-fold greater than docA (data not shown). Therefore, expression of CJJ0382 is not a mitigating factor for the difference in colonization capacities of the ΔdocA and ΔCJJ0382 mutants. However, we cannot rule out translational control mechanisms that may affect the different levels of production of DocA or Cjj0382 proteins in vivo to explain the less-attenuated colonization phenotype of the ΔCJJ0382 mutant.
DocA and Cjj0382 are localized to the periplasm of C. jejuni.
Since C. jejuni mutants lacking either DocA or Cjj0382 display dramatically different colonization phenotypes, we hypothesized that these two proteins may perform different physiological roles in the bacterium necessary for optimal colonization of poultry. Because both proteins are predicted to function as bacterial CCPs, we determined if these proteins have features common to this class of proteins.
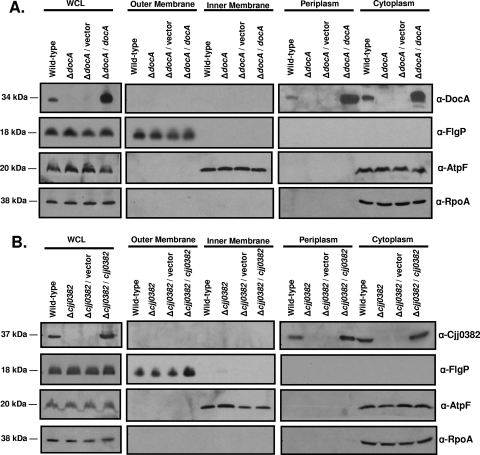
Since bacterial CCPs are periplasmic proteins, we first determined the location of DocA and Cjj0382 in C. jejuni by separating wild-type and mutant strains into outer membrane, inner membrane, periplasmic, and cytoplasmic fractions and performing immunoblot analyses on each fraction with antisera against DocA or Cjj0382. In whole-cell lysates, DocA and Cjj0382 appear as 34-kDa and 37-kDa proteins, respectively, in wild-type C. jejuni (Fig. 2A and B). Analysis of wild-type and ΔdocA mutants revealed DocA to be a soluble protein in the periplasmic and cytoplasmic fractions (Fig. 2A). Similarly, Cjj0382 was found in the periplasm and cytoplasm (Fig. 2B). Whereas localization to the periplasm is characteristic of a typical bacterial CCP, we suspect that the cytoplasmic forms of these proteins are likely nascently translated proteins that have yet to be secreted to the periplasm. As a control for a periplasmic protein, we performed arylsulfatase assays to detect AstA, an enzymatic protein with a predicted signal sequence for localization to the periplasm. In fractionation samples representing periplasmic and cytoplasmic proteins, arylsulfatase activity was present in both fractions (data not shown). Thus, proteins localized to the periplasm in C. jejuni appear in the cytoplasm as well. However, we do not believe that the presence of DocA or Cjj0382 in the periplasm is due to contamination of this fraction with cytoplasmic proteins, as RpoA, a component of RNA polymerase, was only detected in cytoplasmic fractions (Fig. 2A and B). Neither DocA nor Cjj0382 was found in the outer or inner membrane fractions. Verification of the purity of outer and inner membrane fractions was confirmed by detecting the FlgP flagellar protein in the outer membrane (31) and AtpF, a component of ATP synthase, in the inner membrane (Fig. 2A and B). In addition, the levels of the control proteins (RpoA, FlgP, and AtpF) were similar among the wild-type and mutant strains analyzed, ensuring that equal amounts of proteins were analyzed for each fraction from the various strains.
FIG. 2.
DocA and Cjj0382 are periplasmic proteins in C. jejuni. Proteins were separated by 10% SDS-PAGE. Anti-DocA and anti-Cjj0382 antisera were used for detection of the respective proteins. (A) Analysis of DocA localization. Strains used include C. jejuni 81-176 Smr (DRH212), ΔdocA (DRH1169), ΔdocA/pRY112 (LKB307), and ΔdocA/pRY112::docA (LKB313). (B) Analysis of Cjj0382 localization. Strains used include C. jejuni 81-176 Smr (DRH212), ΔCJJ0382 (LKB151), ΔCJJ0382 /pRY112 (LKB310), and ΔCJJ0382/pRY112::CJJ0382 (LKB277). For both panels A and B, fractions analyzed included those of the WCL, outer membrane, inner membrane, periplasm, and cytoplasm. Anti-FlgP, anti-AtpF, and anti-RpoA were used to verify fractionation procedures for the outer membrane, inner membrane, and cytoplasm, respectively. Proteins from WCL representing 200 μl of bacterial culture and proteins from the other fractions representing 400 μl of bacterial culture were used for immunoblotting.
Complementation of the ΔdocA or ΔCJJ0382 mutants in trans with a plasmid expressing each respective gene from their predicted native promoters restored the presence of the proteins in the periplasm and the cytoplasm, indicating that our antisera are specific for only DocA or Cjj0382. Our analysis suggests that, like typical bacterial CCPs, both proteins are localized to the periplasm. However, these findings do not provide insight into the differing physiological roles that the proteins may have during commensal colonization.
DocA and Cjj0382 are heme-bound proteins.
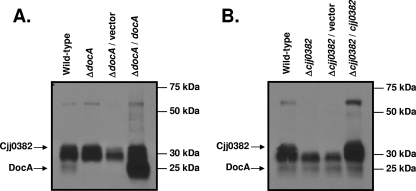
Most CCPs bind two heme compounds via different CXXCH heme-binding site motifs. DocA and Cjj0382 each contain two CXXCH motifs at residues 56 to 60 and 199 to 203 in DocA and residues 80 to 84 and 225 to 229 in Cjj0382. To determine if DocA and Cjj0382 bind heme, we performed a technique described previously by Feissner et al. that specifically detects heme-bound proteins (6). A very similar but less sensitive method has been used to detect bound heme in the CCP of N. gonorrhoeae (32). As shown in Fig. 3A and B, we could detect three separate heme-bound proteins with peroxidase activity from the periplasmic fraction of C. jejuni that migrated closely to each other. The lowest protein band appears to represent DocA, while the uppermost band appears to be Cjj0382. The faint band representing DocA is absent in ΔdocA mutants and appears as an abundant protein band when docA is overexpressed in trans from a plasmid (Fig. 3A). Similarly, the uppermost band representing Cjj0382 is absent in ΔCJJ0382 mutants but restored upon expression of the gene in trans (Fig. 3B). The middle band in all blots likely represents another heme-bound periplasmic protein of C. jejuni whose identity currently remains unknown. This assay not only provides evidence that these proteins are likely to bind heme but also reveals that the hemes, and consequently DocA and Cjj0382, have the characteristic peroxidase activity associated with CCPs.
FIG. 3.
DocA and Cjj0382 have heme-associated peroxidase activities. A heme stain assay was performed on proteins isolated from the periplasm. (A) Analysis of DocA heme-binding ability. Strains used included C. jejuni 81-176 Smr (DRH212), ΔdocA (DRH1169), ΔdocA/pRY112 (LKB307), and ΔdocA/pRY112::docA (LKB313). (B) Analysis of Cjj0382 heme-binding ability. Strains used include C. jejuni 81-176 Smr (DRH212), ΔCJJ0382 (LKB151), ΔCJJ0382/pRY112 (LKB310), and ΔCJJ0382 /pRY112::CJJ0382 (LKB277). The upper band represents Cjj0382, the faint lower band is DocA, and the middle band is an uncharacterized heme-bound protein of C. jejuni.
To further explore the ability of DocA and Cjj0382 to bind heme, we created point mutations in the CXXCH heme-binding site motifs of each protein by mutating cysteine residues to serine or histidine residues to alanine. However, mutation of the heme-binding sites in docA resulted in unstable mutant proteins that could not be detected in whole-cell lysates via immunoblotting (data not shown). Similar mutations in CJJ0382 resulted in unstable mutant proteins or proteins that were detected at lower levels in whole-cell lysates, but none of these proteins could be detected in the periplasm (data not shown). Thus, we are unable to confirm that the heme-binding motifs of DocA and Cjj0382 are required for attachment of heme.
Neither DocA nor Cjj0382 contributes substantially to resistance to hydrogen peroxide in vitro.
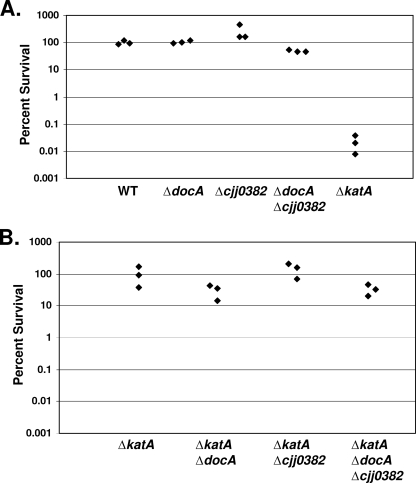
The physiological role of many previously studied bacterial CCPs remains uncertain (1). A role in hydrogen peroxide resistance has been demonstrated for the CCP of N. gonorrhoeae, in which a mutant lacking its CCP was slightly more sensitive to hydrogen peroxide (32). Considering that the heme-binding detection assay suggested that DocA and Cjj0382 have peroxidase activity, we tested the ability of DocA and Cjj0382 to promote survival of C. jejuni upon exposure to hydrogen peroxide. The wild type and mutants of C. jejuni lacking docA or CJJ0382 were exposed to 0.5 mM hydrogen peroxide for 30 min, and the number of bacteria surviving treatment was determined. By performing this analysis, we found that the ΔdocA, ΔCJJ0382, and ΔdocA ΔCJJ0382 mutants demonstrate a wild-type level of resistance to hydrogen peroxide (Fig. 4A).
FIG. 4.
DocA and Cjj0382 do not promote significant resistance to hydrogen peroxide in vitro. C. jejuni strains were treated with 0.5 mM hydrogen peroxide for 30 min at 37°C under microaerophilic conditions. (A) The percent survival of wild-type bacteria was set at 100%, and all other strains were normalized to this value. Strains used include C. jejuni 81-176 Smr (DRH212), ΔdocA (DRH1169), ΔCJJ0382 (LKB151), ΔdocA ΔCJJ0382 (LKB177), and ΔkatA (LKB246). (B) The percent survival of the ΔkatA mutant was set at 100%, and all other strains were normalized to this value. Strains used include C. jejuni Smr ΔkatA (LKB246), ΔkatA ΔdocA (LKB181), ΔkatA ΔCJJ0382 (LKB231), and ΔkatA ΔdocA ΔCJJ0382 (LKB226). For both panels A and B, each diamond represents the percent survival of a bacterial sample after exposure to hydrogen peroxide.
C. jejuni produces catalase, encoded by katA, that has been shown to promote resistance to hydrogen peroxide (5, 11). We reasoned that the presence of cytoplasmic catalase in the ΔdocA or ΔCJJ0382 mutants may be masking any resistance to hydrogen peroxide promoted by DocA or Cjj0382. We generated a ΔkatA mutant in C. jejuni 81-176 Smr and observed that this mutant was up to 10,000-fold more sensitive to hydrogen peroxide than wild-type bacteria (Fig. 4A). We then constructed mutants lacking docA, CJJ0382, or both genes in the ΔkatA background and determined if we could observe any increased sensitivity to hydrogen peroxide that could be attributed to the lack of DocA or Cjj0382. In this assay, the level of survival of the ΔkatA mutant to 0.5 mM hydrogen peroxide was set at 100%; the percent survival of all other mutant strains was calculated relative to that of the ΔkatA mutant. Removal of CJJ0382 in the ΔkatA mutant did not affect the survival of C. jejuni. Deletion of docA from the ΔkatA or ΔkatA ΔCJJ0382 mutants resulted in no more than a twofold increase in sensitivity to hydrogen peroxide. These results when combined largely indicate that neither DocA nor Cjj0382 contributes significantly to survival upon exposure to hydrogen peroxide in vitro. Rather, this phenotype is largely attributed to catalase. We also analyzed the sensitivity of C. jejuni to other peroxides, such as cumene hydroperoxide and tert-butyl hydroperoxide, but we found no evidence that Cjj0382 or DocA contribute to survival upon exposure to these compounds either (data not shown). Taken together, these data suggest that DocA and Cjj0382 have peroxidase activity, but this activity does not contribute to an overall resistance to peroxide stress.
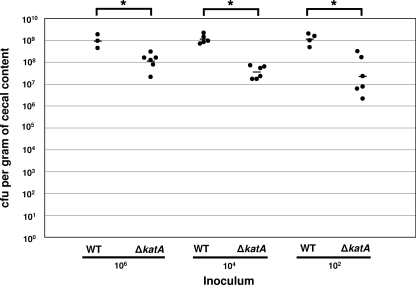
We explored if resistance to peroxide stress is a major determinant for C. jejuni in promoting commensal colonization of chicks by analyzing the ability of the ΔkatA mutant, which is hypersensitive to hydrogen peroxide, to colonize the ceca of chicks. Previous studies in Campylobacter coli found that a catalase-deficient mutant had a colonization capacity similar to wild type (27). In our colonization experiments, a ΔkatA mutant showed 10- to 50-fold-reduced bacterial loads in the ceca of chicks 7 days postinfection regardless of the inoculum size (Fig. 5). In comparison to the ΔdocA mutant, the ΔkatA mutant is much less attenuated for colonization (compare Fig. 1A to C with 5). Considering that the katA mutant is hypersensitive to hydrogen peroxide yet does not demonstrate a large defect in colonization, we suspect that peroxide stress is not a large factor for C. jejuni to overcome during commensal colonization.
FIG. 5.
A ΔkatA mutant of C. jejuni has a moderate defect for cecal colonization in chicks. One-day-old chicks were orally infected with 100 μl of C. jejuni 81-176 Smr (DRH212) or 81-176 Smr ΔkatA (LKB246) with inocula of approximately 106, 104, or 102 bacteria. Chicks were sacrificed 7 days postinfection to determine the cecal colonization capacity of each C. jejuni strain. Each filled symbol represents the amount of C. jejuni recovered from the cecum of a single chick, reported as the number of CFU per gram of cecal content. The geometric mean of the bacterial loads from each set of chicks is denoted by a short horizontal line. Statistical analysis was performed using the Mann-Whitney U test (P < 0.05). *, significant difference between wild-type and mutant strains. The actual inoculum doses ranged from 1.36 × 106 to 2.38 × 106 CFU (left), 5.9 × 103 to 8.2 × 103 CFU (middle), and 58 to 59 CFU (right). The colonization assay of wild-type C. jejuni shown in this figure is from the same experiments shown in Fig. 1A to C.
DISCUSSION
In this study, we found that CJJ0382 of C. jejuni encodes a protein with significant homology to DocA, a known determinant important for the bacterium to promote commensal colonization of poultry (17). A ΔdocA mutant displays up to a 10,000-fold colonization defect, whereas the ΔCJJ0382 mutant shows a maximal 50-fold defect only at a low inoculum. These data suggest that DocA and Cjj0382 may not have redundant functions during in vivo growth even though both proteins are similar to each other and predicted to serve as CCPs. Additional analysis confirmed that these two proteins have certain characteristics common to CCPs: (i) they are located in the periplasm, and (ii) they bind heme for an inherent peroxidase activity that can be detected in vitro. However, neither DocA nor Cjj0382 provides any significant level of resistance to hydrogen peroxide in vitro. Further analysis suggested that the major resistance to hydrogen peroxide stress is mediated by the cytoplasmic catalase. Since the docA mutant is 10- to 100-fold more attenuated for colonization than the catalase mutant, which is hypersensitive to hydrogen peroxide, we suspect that DocA may perform a physiological function in vivo other than promoting resistance to oxidative stress.
The observation that DocA and Cjj0382 do not play a major role in survival from exogenous hydrogen peroxide may not be surprising. In fact, only the CCP of N. gonorrhoeae has been shown to aid in survival to exposure to hydrogen peroxide (32). However, catalase provides the majority of the resistance to hydrogen peroxide in this bacterium. Instead of promoting resistance to hydrogen peroxide, the CCP in Bacteroides fragilis functions in survival of the bacterium to exposure to organic peroxides, such as cumene hydroperoxide and tert-butyl hydroperoxide (19). However, we did not find that DocA or Cjj0382 protected C. jejuni from either of these organic peroxides. It is possible that DocA and Cjj0382 do provide resistance to hydrogen peroxide in vivo, but our in vitro assays are not sensitive enough to detect a difference in viability of the bacterium upon exposure to reactive oxygen species. Secondly, there may be additional factors or environmental conditions during in vivo growth that contribute to DocA and Cjj0382 mediating resistance to peroxides that we are unable to reproduce in our in vitro assays. Most likely, the majority of hydrogen peroxide protection in C. jejuni is due to catalase, which we could observe in our in vitro assays and others have shown previously for Campylobacter species (5, 11).
One of the most common hypotheses for the physiological function of CCPs in various bacteria is that they may play a role during respiration or metabolism. For instance, when formate is used as an electron donor in Campylobacter mucosalis, hydrogen peroxide is generated as a by-product and may be toxic to the bacterium or harmful to other physiological activities (10). Evidence suggests that C. jejuni produces a formate dehydrogenase, but it is unknown if this complex is required by the bacterium for colonization of chickens or if it generates periplasmic hydrogen peroxide (20). In Thiosphaera pantotropha and Rhodobacter capsulatus, hydrogen peroxide inhibits reduction of nitrate, an important activity for respiration (28). Thus, limiting hydrogen peroxide concentrations in the periplasm is essential for nitrate respiration. C. jejuni has nitrate reductase activity, but this activity has not been tested for a critical function during C. jejuni colonization of chickens (26, 29). Multiple electron transport chains have been proposed for C. jejuni, and some of these pathways result in transfer of electrons to cytochrome c (23, 29). Since DocA and Cjj0382 may be like most other CCPs in receiving electrons from cytochrome c, any metabolic pathways that pass down electrons to cytochrome c could be impacted in mutants lacking DocA or Cjj0382. Further experimentation is necessary to determine if these metabolic pathways are important for C. jejuni in promoting colonization of poultry and if DocA or Cjj0382 influences these aspects of metabolism.
Infection of poultry with C. jejuni results in a commensal colonization of the cecum characterized by the lack of a significant inflammatory response from the host. Therefore, it may be hypothesized that the bacterium is not exposed to large amounts of hydrogen peroxide generated by an inflammatory response. Very few inflammatory cells, such as heterophils (the avian equivalent to human neutrophils) or macrophages, migrate to the cecal epithelium or lamina propria during colonization, limiting the interactions of C. jejuni with these cellular components of innate immunity and their associated damaging oxidative bursts (2). Thus, surviving oxidative stress produced by an inflammatory response would be predicted to not be a major hurdle for C. jejuni to overcome for colonizing poultry. This hypothesis is supported by the colonization phenotype of the ΔkatA mutant, which is hypersensitive to hydrogen peroxide but is only moderately attenuated for in vivo growth.
These findings with DocA and Cjj0382 have created interesting questions regarding the composition and biological activities of C. jejuni. First, possessing two different proteins that may function in some form as CCPs is somewhat unusual for a single bacterium. Most gram-negative bacteria have only one CCP, if they have one at all, and these proteins are absent from gram-positive organisms (1). Secondly, our results indicate that DocA and Cjj0382 likely perform nonredundant biological activities for C. jejuni during in vivo growth. Despite their homology, phylogenetic analysis of various bacterial CCPs suggests that Cjj0382 is grouped in a clade of classical CCPs. However, DocA, despite its similarity to Cjj0382, is the sole member of its own clade (1). This observation suggests that DocA may be truly unique among CCPs, with a potentially uncommon or unique function. Further exploration is required to uncover the specific physiological functions of DocA and Cjj0382 in vivo that are critical for the ability of the bacterium to promote commensalism in poultry.
Acknowledgments
We thank Bob Kranz for helpful discussions regarding assays detecting heme-bound proteins. In addition, we thank Shawn Sommerlad for construction of plasmids pSMS176 and pSMS201.
This work was supported by the U.S. Department of Agriculture National Research Initiative grant no. 2006-35201-17382 from the USDA Cooperative State Research, Education, and Extension Service Food Safety (32.0) program and the National Institutes of Health grant R01 AI065539 to D.R.H.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 17 December 2007.
REFERENCES
- 1.Atack, J. M., and D. J. Kelly. 2007. Structure, mechanism and physiological roles of bacterial cytochrome c peroxidases. Adv. Microb. Physiol. 5273-106. [DOI] [PubMed] [Google Scholar]
- 2.Beery, J. T., M. B. Hugdahl, and M. P. Doyle. 1988. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol. 542365-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157472-479. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2006. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, United States, 2005. Morb. Mortal. Wkly. Rep. 55392-395. [PubMed] [Google Scholar]
- 5.Day, W. A., Jr., J. L. Sajecki, T. M. Pitts, and L. A. Joens. 2000. Role of catalase in Campylobacter jejuni intracellular survival. Infect. Immun. 686337-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feissner, R., Y. Xiang, and R. G. Kranz. 2003. Chemiluminescent-based methods to detect subpicomole levels of c-type cytochromes. Anal. Biochem. 31590-94. [DOI] [PubMed] [Google Scholar]
- 7.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 761648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman, C. R., R. M. Hoekstra, M. Samuel, R. Marcus, J. Bender, B. Shiferaw, S. Reddy, S. D. Ahuja, D. L. Helfrick, F. Hardnett, M. Carter, B. Anderson, and R. V. Tauxe. 2004. Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis. 38(Suppl. 3)S285-S296. [DOI] [PubMed] [Google Scholar]
- 10.Goodhew, C. F., A. B. elKurdi, and G. W. Pettigrew. 1988. The microaerophilic respiration of Campylobacter mucosalis. Biochim. Biophys. Acta 933114-123. [DOI] [PubMed] [Google Scholar]
- 11.Grant, K. A., and S. F. Park. 1995. Molecular characterization of katA from Campylobacter jejuni and generation of a catalase-deficient mutant of Campylobacter coli by interspecific allelic exchange. Microbiology 1411369-1376. [DOI] [PubMed] [Google Scholar]
- 12.Guerry, P., R. Yao, R. A. Alm, D. H. Burr, and T. J. Trust. 1994. Systems of experimental genetics for Campylobacter species. Methods Enzymol. 235474-481. [DOI] [PubMed] [Google Scholar]
- 13.Hadden, R. D., H. Karch, H. P. Hartung, J. Zielasek, B. Weissbrich, J. Schubert, A. Weishaupt, D. R. Cornblath, A. V. Swan, R. A. Hughes, and K. V. Toyka. 2001. Preceding infections, immune factors, and outcome in Guillain-Barre syndrome. Neurology 56758-765. [DOI] [PubMed] [Google Scholar]
- 14.Henderson, M. J., and F. H. Milazzo. 1979. Arylsulfatase in Salmonella typhimurium: detection and influence of carbon source and tyramine on its synthesis. J. Bacteriol. 13980-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrixson, D. R. 2006. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 611646-1659. [DOI] [PubMed] [Google Scholar]
- 16.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40214-224. [DOI] [PubMed] [Google Scholar]
- 17.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52471-484. [DOI] [PubMed] [Google Scholar]
- 18.Hendrixson, D. R., and V. J. DiRita. 2003. Transcription of σ54-dependent but not σ28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50687-702. [DOI] [PubMed] [Google Scholar]
- 19.Herren, C. D., E. R. Rocha, and C. J. Smith. 2003. Genetic analysis of an important oxidative stress locus in the anaerobe Bacteroides fragilis. Gene 316167-175. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman, P. S., and T. G. Goodman. 1982. Respiratory physiology and energy conservation efficiency of Campylobacter jejuni. J. Bacteriol. 150319-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korlath, J. A., M. T. Osterholm, L. A. Judy, J. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152592-596. [DOI] [PubMed] [Google Scholar]
- 22.Makarova, O., E. Kamberov, and B. Margolis. 2000. Generation of deletion and point mutations with one primer in a single cloning step. BioTechniques 29970-972. [DOI] [PubMed] [Google Scholar]
- 23.Myers, J. D., and D. J. Kelly. 2005. A sulphite respiration system in the chemoheterotrophic human pathogen Campylobacter jejuni. Microbiology 151233-242. [DOI] [PubMed] [Google Scholar]
- 24.Nachamkin, I., B. M. Allos, and T. W. Ho. 2000. Campylobacter jejuni infection and the association with Guillain-Barre syndrome, p. 155-176. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. ASM Press, Washington, DC.
- 25.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403665-668. [DOI] [PubMed] [Google Scholar]
- 26.Pittman, M. S., and D. J. Kelly. 2005. Electron transport through nitrate and nitrite reductases in Campylobacter jejuni. Biochem. Soc Trans. 33190-192. [DOI] [PubMed] [Google Scholar]
- 27.Purdy, D., S. Cawthraw, J. H. Dickinson, D. G. Newell, and S. F. Park. 1999. Generation of a superoxide dismutase (SOD)-deficient mutant of Campylobacter coli: evidence for the significance of SOD in Campylobacter survival and colonization. Appl. Environ. Microbiol. 652540-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson, D. J., and S. J. Ferguson. 1995. Competition between hydrogen peroxide and nitrate for electrons from the respiratory chains of Thiosphaera pantotropha and Rhodobacter capsulatus. FEMS Microbiol. Lett. 132125-129. [Google Scholar]
- 29.Sellars, M. J., S. J. Hall, and D. J. Kelly. 2002. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 1844187-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skirrow, M. B., and M. J. Blaser. 2000. Clinical Aspects of Campylobacter infection, p. 69-88. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. ASM Press, Washington, DC.
- 31.Sommerlad, S. M., and D. R. Hendrixson. 2007. Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J. Bacteriol. 189179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner, S., E. Reid, H. Smith, and J. Cole. 2003. A novel cytochrome c peroxidase from Neisseria gonorrhoeae: a lipoprotein from a gram-negative bacterium. Biochem. J. 373865-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130127-130. [DOI] [PubMed] [Google Scholar]
- 34.Yao, R., and P. Guerry. 1996. Molecular cloning and site-specific mutagenesis of a gene involved in arylsulfatase production in Campylobacter jejuni. J. Bacteriol. 1783335-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]