Abstract
Chlamydia trachomatis infection induces a wide array of inflammatory cytokines and chemokines, which may contribute to chlamydia-induced pathologies. However, the precise mechanisms by which Chlamydia induces cytokines remain unclear. Here we demonstrate that the proinflammatory cytokine interleukin-1α (IL-1α) plays an essential role in chlamydial induction of the chemokine IL-8. Cells deficient in IL-1α expression or IL-1α-competent cells treated with IL-1α-specific small interfering RNA failed to produce IL-8 in response to chlamydial infection. However, neutralization of extracellular IL-1α or blockade of or deficiency in type I IL-1 receptor (IL-1RI) signaling did not affect chlamydial induction of IL-8 in cells capable of producing IL-1α. These results suggest that IL-1α can mediate the chlamydial induction of IL-8 via an intracellular mechanism independent of IL-1RI, especially during the early stage of the infection cycle. This conclusion is further supported by the observations that expression of a transgene-encoded full-length IL-1α fusion protein in the nuclei enhanced IL-8 production and that nuclear localization of chlamydia-induced precursor IL-1α correlated with chlamydial induction of IL-8. Thus, we have identified a novel mechanism for chlamydial induction of the chemokine IL-8.
Chlamydia trachomatis is an obligate intracellular bacterial pathogen consisting of more than 15 different serovars, designated A to L, including Ba, L1, L2, and L3 plus various subtypes. Different serovars cause different diseases in humans, with serovars A to C infecting human eyes, potentially leading to preventable blindness (59), and serovars D to K infecting the human urogenital tract, which, if left untreated, can cause severe complications (54). The L serovars, also known as lymphogranuloma venereum serovars, are more invasive than other urogenital serovars and often cause disseminated diseases. The L2 serovar has recently caused several outbreaks in humans (3, 51). Despite their differences in tissue tropism and invasiveness, all C. trachomatis serovars undergo a common intracellular biphasic growth cycle (21). A typical infection starts with the entry of elementary bodies (EBs), the infectious form, into host cells via endocytosis (27). The internalized EBs within the endosomal vacuole can rapidly differentiate into reticulate bodies, the metabolically active but noninfectious form of chlamydial organisms. After numerous rounds of replication, the reticulate bodies can differentiate back into EBs prior to spreading to adjacent cells. All Chlamydia species can accomplish their entire biosynthesis, replication, and differentiation within the cytoplasmic vacuole (also termed inclusion). Depending on the species and serovar/strain, it can take 1 to 3 days to complete an infection cycle. Although the exact molecular mechanisms of chlamydial exit during natural infection are still poorly understood, it is clear that Chlamydia species can use two distinct pathways, including cell lysis and extrusion, to leave infected host cells, at least in in vitro cell culture systems (28).
During intravacuolar growth, chlamydial organisms have to interact with host cells across the inclusion membrane for both import of nutrients and metabolic intermediates from host cells (22, 58) and secretion of chlamydial factors into host cells (66). Furthermore, chlamydiae can actively manipulate host signal pathways (15, 19, 58, 64). As a result of Chlamydia-host interactions, host cell genes coding for inflammatory cytokines and chemokines are often activated. During chlamydial infection in animals and humans as well as in cell culture systems, a wide array of inflammatory cytokines and chemokines, including interleukin-1α (IL-1α), IL-1β, IL-6, IL-18, tumor necrosis factor alpha, beta interferon (IFN-β), IL-8, and CXCL9 and -10 (5, 11, 16, 18, 34, 36, 39, 46), are produced, which is thought to contribute to Chlamydia-induced pathologies (56). However, the precise mechanisms by which Chlamydia species induce each of these cytokines remain unknown. The current study focuses on the mechanism of Chlamydia-induced production of IL-8, a chemokine responsible for the recruitment of leukocytes to the site of infection.
It is known that IL-8 expression can be regulated by IL-1 (1, 9, 29, 30, 50, 63). The IL-1 family consists of 10 different members (41, 60), including the two dominant agonists IL-1α and -β and the IL-1 receptor antagonist (IL-1RA). IL-1α and -β and IL-1RA can compete for binding to the cell surface type I and II IL-1 receptors (IL-1RI and IL-1RII). However, only IL-1RI, not IL-1RII, is functional because IL-1RII lacks a cytoplasmic domain and is thus unable to transmit signals to downstream steps (4). The functional IL-1RI, once triggered by the agonist IL-1α or -β, can induce IL-8 expression via signaling pathways leading to the activation of the IL-8 promoters recognized by various transcriptional factors, such as AP-1, NF-IL-6 (C/EBPβ), and NF-κB (9, 29, 30, 38, 43, 48, 50, 63). The agonistic effects can be blocked by either competitive binding of IL-1RA to the ligand binding site in IL-1RI or neutralization of the agonists with neutralization antibodies or soluble receptors. The agonists IL-1α and -β are synthesized as precursor molecules and then processed into mature/active proteins for secretion into the extracellular space (20, 23, 44). Interestingly, only the IL-1α precursor, not that of IL-1β, is biologically active. A bona fide nuclear localization sequence, KVLKKRR, was identified in the N-terminal prodomain of IL-1α, and full-length precursor IL-1α molecules were frequently detected in the nucleus (62). It has been shown that nucleus-localized IL-1α can indeed exert various biological functions (31, 32, 35, 37, 45, 49, 57, 61). Thus, in addition to activating cell surface IL-1RI-mediated signaling pathways, IL-1α can also function via intracellular pathways, independent of the cell surface receptor.
Although IL-1α is known to participate in the amplification of inflammatory responses and directly contribute to IL-8 production during microbial infection (5, 9, 13, 46), it is not clear whether the intracellular precursor IL-1α signaling mechanism is involved in pathogen infection-induced inflammation. In the current study, we investigated the role of IL-1α intracellular signaling in C. trachomatis induction of IL-8. We found that cells unable to produce IL-1α failed to produce IL-8, while blocking of IL-1R-mediated signaling in cells capable of producing IL-1α did not affect chlamydial induction of IL-8 production. Furthermore, IL-1α was detected in the nuclei of Chlamydia-infected cells, and plasmid-encoded full-length IL-1α expression in the nucleus enhanced IL-8 production. Taken together, these observations suggest that chlamydial induction of IL-8 requires IL-1α to signal via an intracellular pathway independent of IL-1R.
MATERIALS AND METHODS
Chlamydial infection.
C. trachomatis serovar L2 organisms were used throughout the current study. Host cells, including HeLa (human cervical carcinoma epithelial cells; ATCC), SiHa (human cervical carcinoma epithelial cells; HTB35), Hep2 (human cervical epithelial cells with HeLa markers [although initially thought to be derived from the human larynx]; CCL-23), HT-29 (human colorectal adenocarcinoma epithelial cells; HTB-38), HGF (human gingival fibroblast cells; CRL2014), and 293T (human kidney epithelial cells; CRL-11268) cells, were all obtained from ATCC (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (Gibco BRL, Rockville, MD) with 10% fetal calf serum (Gibco BRL) at 37°C in an incubator supplied with 5% CO2. The cells were grown in 6-, 24-, or 48-well plates, with (for immunofluorescence assay) or without (for enzyme-linked immunosorbent assay [ELISA] measurements of cytokines) coverslips, or in tissue culture dishes/flasks (for reverse transcription-PCR [RT-PCR] and ELISA). Wild-type (wt) C57BL/6j mice (stock number 000664), mutant mice deficient in IL-1RI (B6.129S7-IL1r1tm1Imx/J; stock number 003245), wt NOD/LtJ mice (stock number 001976), and mutant mice deficient in IL-1α (NOD.NOR-IL-1α-Pcna/LtJ; stock number 003051) were all obtained from Jackson Laboratory (Bar Harbor, ME). Macrophages (Mφs) were collected from the mouse peritoneal cavity following a previously described protocol (69). Briefly, the mouse peritoneal cavity was injected with 5 to 10 ml cold phosphate-buffered saline, using a 27-gauge needle. After gentle massage, the solution was withdrawn from the mouse peritoneal cavity by use of a 20-gauge needle. After enumeration of the total number of viable cells, the peritoneal cavity-derived cells were resuspended in RPMI 1640 with 10% fetal calf serum, and 2 × 105 cells were added to each well of 48-well plates. The plates were incubated at 37°C for 2 h in a CO2 incubator to allow macrophages to adhere. After nonadherent cells were washed away, fresh medium was added to each well for continued incubation. All cell samples, regardless of the cell type, were cultured overnight prior to chlamydial inoculation. Chlamydial organisms diluted in cell growth medium were inoculated directly onto the cell monolayers. A multiplicity of infection of 5 was used to infect all cell samples, except that a multiplicity of infection of 1 was used to infect cells grown on coverslips for the immunofluorescence assay. The cell samples, with or without infection, were cultured at 37°C in a CO2 incubator. In some cases, the cultures were also transfected with small interfering RNA (siRNA) (see below) or treated with a recombinant human IL-1α antibody (19601V; final concentration, 5 ng/ml) (BD Pharmingen, San Diego, CA), a goat anti-human IL-1α neutralization antibody (AF-200-NA; 2 μg/ml), and/or the IL-1RA (280-RA; 500 ng/ml) (both from R&D Systems, Inc., Minneapolis, MN). Finally, the cell samples were processed at various time points after infection, as indicated in individual experiments, for the various measurements listed below.
RT-PCR.
Host cells with or without chlamydial infection were harvested at various time points postinfection for extraction of RNA, using an RNA extraction and purification kit from Invitrogen (Carlsbad, CA). The RNA samples were reversely transcribed into cDNA by using a random hexamer primer, and the cDNA transcripts were amplified by PCR using the following gene-specific primers: 5′-GTCTCTGAATCAGAAATCCTTCTATC-3′ (forward primer for human IL-1α), 5′-CATGTCAAATTTCACTGCTTCATCC-3′ (reverse primer for human IL-1α to generate a 420-bp product), 5′-AAACAGATGAAGTGCTCCTTCCAGG-3′ (forward primer for human IL-1β), 5′-TGGAGAACACCACTTGTTGCTCCA-3′ (reverse primer for human IL-1β to generate a 390-bp product), 5′-ATGACTTCCAAGCTGGCCGTGGCT-3′ (forward primer for human IL-8), 5′-TCTCAGCCCTCTTCAAAAACTTCTC-3′ (reverse primer for human IL-8 to generate a 289-bp product), 5′-ATGAACTCCTTCTCCACAAGCGC-3′ (forward primer for human IL-6), 5′-GAAGAGCCCTCAGGCTGGACTG-3′ (reverse primer for human IL-6 to generate a 628-bp product), 5′CTACAATGAGCTGCGTGTGG-3′ (forward primer for β-actin), and 5′AAGGAAGGCTGGAAGAGTGC-3′ (reverse primer for β-actin to generate a 528-bp product). All oligonucleotide primers were made by the DNA core facility at the University of Texas Health Science Center at San Antonio (UTHSCSA). PCR was performed using a standard three-step protocol in a thermal cycler from MJ Research, Inc. (Waltham, MA). The PCR products were resolved in 2% agarose gels and visualized after ethidium bromide staining.
ELISA.
At the end of culture, the supernatants were collected for measurements of secreted cytokines, while the remaining cell monolayers were collected, after being washed twice with warm medium, and total cell lysates were made by sonication in an equal amount of medium for measurements of cell-associated or intracellular cytokines by use of commercially available ELISA kits. The kits for human IL-1α, (DY200) and IL-6 (DY206) as well as mouse IL-1α (DY400) and macrophage inflammatory protein 2 (MIP-2) (mouse homolog of IL-8; DY452) were all obtained from R&D Systems, Inc. (Minneapolis, MN). For human IL-8 detection, a mouse monoclonal antibody (MAb) against human IL-8 was used as the capture antibody, and another mouse anti-human IL-8 antibody conjugated with biotin was used as the detection antibody (both from BD Bioscience, San Jose, CA). Recombinant human IL-8 from BD Pharmingen was used as the standard. The ELISA was carried out following the instructions provided by the manufacturer or as described elsewhere (53, 67, 68). Briefly, 96-well ELISA microplates (Nunc, Rochester, NY) were coated with a capture antibody, and after blocking of the plates, the cytokine samples or standards were added to the coated plates, followed by a biotin-conjugated detection antibody. Antibody binding was measured with horseradish peroxidase-conjugated avidin plus a soluble colorimetric substrate [2-2′-azino-di-(3-ethylbenzthiazoline) sulfonic acid (ABTS)]. The absorbance was taken at 405 nm using a microplate reader (Molecular Devices Corporation, Sunnyvale, CA). Sometimes, the culture samples were diluted prior to the ELISA measurements in order to keep the absorbance readings within the linear range. The cytokine concentrations were calculated based on absorbance values, cytokine standards, and sample dilution factors. When the cell numbers were known, the cytokine levels were expressed in ng or pg per 1 million cells, and otherwise they were expressed as per ml or well.
Immunofluorescence staining.
Cell samples grown on coverslips were fixed with 4% paraformaldehyde dissolved in phosphate-buffered saline for 20 min at room temperature, followed by permeabilization with 0.1% Triton X-100 for an additional 4 min. After being washed and blocked, the cell samples were subjected to various combinations of antibody and chemical staining. Hoechst dye (blue; Sigma, St. Louis, MO) was used to visualize nuclear DNA. A rabbit anti-chlamydial CT395 polyclonal antibody (raised with the CT395 fusion protein; CT395 is a chlamydial GrpE-related chaperonin with >70% amino acid sequence identity among all chlamydial species [8]), a mouse anti-human IL-1α MAb (14-7019-83; eBioscience, San Diego, CA), or a mouse anti-human Golgi GM130 MAb (BD Biosciences) plus a donkey anti-rabbit or -mouse immunoglobulin G (IgG) secondary antibody conjugated with Cy2 (green; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) was used to visualize chlamydial inclusions, IL-1α, or the Golgi apparatus. Goat anti-IL-8 (AF-208-NA; R&D) plus donkey anti-goat IgG conjugated with Cy3 (red; Jackson ImmunoResearch) was used to visualize intracellular IL-8. For the transfected cell samples, IL-1α was visualized via the fusion tag red fluorescent protein (RFP). The immunolabeled samples were observed and images were taken with an Olympus AX-70 fluorescence microscope equipped with multiple filter sets (Olympus, Melville, NY) as described previously (15, 19, 65, 66). Briefly, the multicolor-labeled samples were exposed under a given filter set at a time, and single-color images were acquired using a Hamamatsu digital camera. The single-color images were then superimposed with the software SimplePCI to display multicolors. All microscopic images were processed using the Adobe Photoshop program (Adobe Systems, San Jose, CA).
Plasmid/siRNA constructs and transient transfection of mammalian cells.
To construct human IL-1α siRNA, a 21-mer target sequence (5′-AACTTTATGAGGATCATCAAA-3′) from exon 4 of human IL-1α mRNA (NCBI accession no. NM_000575; http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nuccore&id=27894329) was chosen based on a nucleotide sequence selection strategy for targeting the transcript described elsewhere (14). A Silencer siRNA construction kit (Ambion, Austin, TX) was used for making siRNA constructs. Briefly, two 29-mer DNA primers comprising the above 21-mer target sequence (either sense or antisense) and an 8-nucleotide linker sequence complementary to the T7 promoter primer sequence (provided by the siRNA construction kit) were made by the DNA core facility at UTHSCSA. The sequences of the 29-mer primers were 5′-AACTTTATGAGGATCATCAAACCTGTCTC-3′ (antisense) and 5′-AATTTGATGATCCTCATAAAGCCTGTCTC-3′ (sense). To construct the scrambled siRNA, the above 21-mer target sequence nucleotides were jumbled while the base composition was kept constant. The random sequences were searched against the human genome sequence database (http://www.ncbi.nlm.nih.gov/BLAST/). The sequence 5′-AAGTATATCTAGCAGAACTTA-3′, showing no significant homology to any human gene sequences from the database, was selected as the scrambled siRNA. The two 29-mer DNA primers for making the scrambled siRNA were also made by the UTHSCSA DNA core facility and had the sequences 5′-AAGTATATCTAGCAGAACTTACCTGTCTC-3′ (scrambled antisense) and 5′-AATAAGTTCTGCTAGATATACCCTGTCTC-3′ (scrambled sense). Each of the four 29-mer DNA primers was hybridized with the T7 promoter primer, and the overhang regions were filled in using Klenow DNA polymerase enzyme to make double-stranded DNA templates. The double-stranded DNA templates were transcribed into RNA with a T7 RNA polymerase. The corresponding sense and antisense RNA strands were hybridized to make double-stranded RNA (siRNA). The siRNA preps were treated with RNase and DNase to remove DNA and single-stranded RNA and then further purified by passage through a column provided in the siRNA construction kit. The purified siRNA was quantitated, aliquoted, and stored at −20°C until use. Human IL-1α cDNA obtained from Origene (Rockville, MD) was cloned into the pDsRed-C1 monomer vector (Clontech, Mountain View, CA), using KpnI and XmaI restriction sites, and expressed as a fusion protein with an N-terminal RFP tag. The transfection reagent Lipofectamine 2000 (Invitrogen) was used for transfecting both HeLa cells with siRNA constructs (1 μg per well of a 24-well plate) and 293T cells with pDsRed-IL-1α plasmid (1 μg per well of a 24-well plate). For Chlamydia-infected cell samples, transfection was done 2 h after infection. The culture samples were used for either immunofluorescence staining or ELISA measurement of cytokines 24 to 48 h after transfection.
RESULTS
Induction of inflammatory cytokines by C. trachomatis infection.
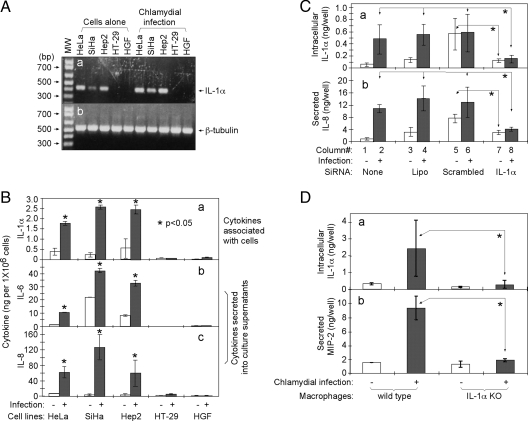
To understand the mechanisms of Chlamydia-induced IL-8 production, we first monitored the expression of IL-1α, IL-6, and IL-8 in HeLa cells infected with C. trachomatis. Chlamydial infection significantly activated all three cytokine genes (Fig. 1A) and induced the corresponding cytokine protein production (Fig. 1B). The cytokine proteins were monitored simultaneously in both the culture supernatants (for secreted cytokines) and the lysates made from the remaining cells (for cytokines associated with cells). Chlamydia-induced IL-1α was associated mainly with cells for up to 48 h after infection, and a sudden increase of IL-1α in the supernatants was noticed at 60 h (Fig. 1B, panels a and b). However, most IL-6 and IL-8 proteins were always secreted into the culture supernatants throughout the infection course (Fig. 1B, panels c to f). These observations suggest a correlation between Chlamydia-induced intracellular accumulation of IL-1α and extracellular secretion of IL-6/8. The rapid increase of IL-1α in the infected culture supernatants 60 h after infection might have been due to lysis of some infected cells, since cell lysis was observed at this time (data not shown). We followed the infection for up to 60 h only when we started to notice cell lysis in the infected cultures. However, under the infection conditions used in the current experiment, no cell lysis occurred at 48 h postinfection, although most organisms were converted into EBs by this time (data not shown). To further determine whether IL-8 was produced by the Chlamydia-infected cells or the adjacent uninfected cells in the infected cultures, we further detected IL-8 at the individual cell level by using an immunofluorescence assay at 48 h postinfection (Fig. 1C). The intracellular IL-8 was detected mainly in chlamydial inclusion-containing cells. Anti-IL-8 staining distributed around the inclusions and overlapped with a Golgi complex marker.
FIG. 1.
Inflammatory cytokine production during chlamydial infection. HeLa cells, with or without C. trachomatis L2 serovar infection for various periods, as indicated in the figure, were processed for the following measurements. (A) RNAs were extracted from whole-cell samples and reversely transcribed into cDNAs by use of random hexamer primers. The cDNAs were amplified by PCRs with primers for human IL-1α (a), IL-6 (b), IL-8 (c), and β-tubulin (d), and the PCR products were loaded into an agarose gel for visualization after ethidium bromide staining. MW, molecular weight in base pairs (bp), as indicated along the left side of the figure. The corresponding DNA bands are indicated along the right side of the figure. (B) Either the culture supernatants (for measuring secreted cytokines) (a, c, and e) or whole-cell lysates (for measuring intracellular cytokines) (b, d, and f) were analyzed for IL-1α (a and b), IL-6 (c and d), and IL-8 (e and f) by using a sandwich ELISA. The cytokine levels were determined based on absorbance readings, sample dilution factors, and corresponding cytokine standards and were expressed in nanograms (ng) per 1 million cells, in the format of means ± standard deviations. Please note the difference in scale between the y axes in the left and right panels. Each data point comes from four to six independent experiments done in duplicate. Asterisks indicate statistically significant differences (P < 0.05) between the infected (hatched bars) and uninfected (open bars) samples from the same time points, using a two-tailed Student t test. (C) HeLa cells grown on coverslips were fixed and permeabilized 48 h after infection. A combination of a goat anti-human IL-8 antibody (red), Hoechst DNA dye (blue; for chlamydial and host nuclear DNA), and a rabbit anti-CT395 antibody (green; for visualizing chlamydial organisms) (a to h) or anti-IL-8, the DNA dye, and a mouse anti-Golgi GM130 antibody (green) (i to l) was used to visualize the corresponding antigens or organelle under a fluorescence microscope. The images were taken one color at a time (a to c, e to g, and i to k), and the single-color images were finally overlaid as tricolor images (d, h, and l). White arrows point to IL-8 staining, and solid white triangles in panels i to l indicate chlamydial inclusions.
IL-1α is required for IL-8 production during chlamydial infection.
We further probed the relationship between IL-1α and IL-8 production during chlamydial infection. Five different human cell lines with various abilities to express IL-1α were evaluated for the ability to secrete IL-8 in response to chlamydial infection. Significant amounts of IL-1α messages were detected in HeLa, SiHa, and Hep2 cells but not in HT-29 and HGF cells, regardless of chlamydial infection (Fig. 2A). Importantly, all three IL-1α-expressing cell lines significantly increased their production of IL-1α, IL-6, and IL-8 proteins after chlamydial infection, while the two IL-1α-negative cell lines failed to produce significant amounts of the cytokines, regardless of the infection (Fig. 2B). The IL-1α-negative cell lines HT-29 and HGF produced significant amounts of IL-8 in response to stimulation with either exogenous IL-1α or tumor necrosis factor alpha (data not shown), indicating that both cells possess the ability to produce IL-8 and suggesting that the failure to produce IL-8 by HT-29 and HGF cells after chlamydial infection might be due to the lack of IL-1α in these cells. While the above observations unraveled a correlation between IL-1α and IL-8 production during chlamydial infection, we further used a siRNA knockdown approach to probe the role of IL-1α in chlamydial induction of IL-8 (Fig. 2C). We found that a siRNA construct specific to IL-1α significantly reduced the production of both IL-1α and IL-8 in HeLa cells after chlamydial infection, while treatment with either Lipofectamine alone or a scrambled siRNA construct failed to do so. It is worth noting that the IL-1α-specific siRNA also blocked the production of both IL-1α and IL-8 induced by the scrambled double-stranded RNA sequence. It is well known that double-stranded RNA preps can induce inflammatory cytokine production (24, 52, 55). Finally, we compared the ability of Mφs from wt versus IL-1α knockout (KO) mice to produce IL-8 in response to chlamydial infection (Fig. 2D). The use of mouse Mφs in this experiment was due mainly to the convenience of harvesting the peritoneal Mφs from mice that were available to us at the time of the experiment. Although both wt and IL-1α KO Mφs were equally susceptible to chlamydial infection, with about 50% infection rates for both groups (data not shown), we found that the wt Mφs produced a significant amount of MIP-2 (mouse homolog of IL-8), while the IL-1α KO Mφs failed to produce IL-8, regardless of chlamydial infection. These observations have demonstrated that IL-1α is essential for IL-8 production induced by chlamydial infection.
FIG. 2.
Role of IL-1α in chlamydial induction of IL-8. Five different cell lines, as indicated in the figure, with or without C. trachomatis L2 serovar infection for 48 h, were processed for either RT-PCR detection of IL-1α and β-tubulin transcripts (A) or ELISA detection of IL-1α, IL-6, and IL-8 proteins (B). The ELISA results were expressed in ng per well, in the format of means ± standard deviations, and each data point comes from three independent experiments done in duplicate. Asterisks indicate statistically significant differences (P < 0.05) between the infected (hatched bars) and uninfected (open bars) samples from the same cell lines, using a two-tailed Student t test. (C) HeLa cells grown in 24-well plates, with or without Lipofectamine-mediated siRNA transfection or chlamydial infection for 48 h, were measured for intracellular IL-1α (a) and secreted IL-8 (b) by ELISA. The cytokine concentrations were expressed in ng per well, in the format of means ± standard deviations. The experiments were repeated three times each, with duplicates. Asterisks indicate statistically significant differences (P < 0.05) between the IL-1α-specific siRNA-treated (column 7) and scrambled siRNA-treated (column 5) samples (uninfected cell samples; open bars) or between the IL-1α-specific siRNA-treated samples (column 8) and each of the three control groups, including no transfection (column 2), Lipofectamine-only treatment (column 4), and scrambled siRNA treatment (column 6) (infected samples; hatched bars). A two-tailed Student t test was used for statistical analysis. (D) Peritoneal macrophages harvested from either wt or IL-1α KO mice were infected with C. trachomatis serovar L2 for 48 h in 48-well plates. Both the intracellular IL-1α (a) and secreted MIP-2 (mouse IL-8 homolog) (b) were measured using a commercially available ELISA kit. The results came from six mice per group and were expressed in ng per well, in the format of means ± standard deviations. Asterisks indicate statistically significant differences (P < 0.05) between the wt and IL-1α KO macrophages (infected samples; hatched bars).
IL-1α-mediated chlamydial induction of IL-8 is independent of extracellular IL-1α and functional IL-1RI.
We next investigated how IL-1α mediated the chlamydial induction of IL-8 by first using an IL-1α-specific neutralization antibody to block extracellular IL-1α (Fig. 3A). The anti-IL-1α neutralization antibody significantly inhibited the production of both IL-6 and -8 induced by exogenously added IL-1α but failed to alter the chlamydial induction of IL-6 and -8 in any significant way 48 h after infection. The chlamydial induction of IL-8 was partially (but significantly) inhibited by the neutralization antibody 72 h after infection, probably due to the fact that the extracellular IL-1α released from the lysed cells at this time after infection also contributed to the IL-8 production. This observation is consistent with a previous report (46). The above results have demonstrated that chlamydial induction of IL-8 does not require extracellular IL-1α during the first 48 h after infection. We next used IL-1RA to further assess the role of cell surface IL-1R in chlamydial induction of IL-8. Treatment with IL-1RA failed to block chlamydial induction of IL-6 and -8 at 48 h and only partially inhibited IL-8 production at 72 h, although the IL-1RA treatment significantly inhibited exogenous IL-1α-induced IL-8 production. This observation is consistent with the result obtained with the neutralization antibody treatment. To further evaluate the contribution of IL-1RI signaling to the chlamydial induction of IL-8, we monitored IL-8 production in Mφs from IL-1RI KO mice (Fig. 3B). We found that the IL-1RI KO Mφs produced both IL-1α and IL-8 regardless of chlamydial infection, demonstrating that IL-1RI-initiated signaling is not required for chlamydial induction of IL-8. The finding that IL-1R1 KO Mφs produce significant amounts of both IL-1α and IL-8 even without chlamydial infection was a surprise and may be due to the compensative production of IL-1α in response to IL-1R1 deficiency. This finding was reproduced in IL-1R1 KO Mφs with a different genetic background (data not shown). Constitutively produced IL-1α may drive IL-8 production in an IL-1R1-independent manner. The question of why IL-1R KO Mφs constitutively produce IL-1α and IL-8 is interesting in itself, and the mechanism is still unclear and under investigation in a separate study. Nevertheless, the above observations together suggest that IL-1α may function intracellularly via a mechanism independent of IL-1RI.
FIG. 3.
IL-1α-mediated chlamydial induction of IL-8 is independent of extracellular IL-1α and IL-1R. (A) HeLa cells, with or without C. trachomatis serovar L2 infection, were treated with 5 ng/ml of IL-1α, 2 μg/ml of an anti-IL-1α neutralizing antibody, or 500 ng/ml of IL-1RA. The treatments commenced at the time of infection and were maintained throughout the infection course by changing the medium with medium containing fresh cytokines every 24 h. Secreted IL-6 and IL-8 were measured 48 (a and c) or 72 (b and d) h after infection by ELISA. The experiments were repeated three or four times each, with duplicates. The results were expressed in ng per well. Asterisks indicate statistically significant differences (P < 0.05) between samples treated with IL-1α (column 2 or 11) and with neutralizing antibody (column 3 or 12) or IL-1RA (column 4 or 13) (uninfected samples; open bars) or between the 72-h infected sample (column 14) and the neutralizing antibody (column 15)- or IL-1RA (column 16)-treated samples (hatched bars). A two-tailed Student t test was used for the statistical analysis. (B) Peritoneal macrophages harvested from either wt or IL-1RI KO mice were infected with C. trachomatis serovar L2 for 48 h. Both the intracellular IL-1α and secreted MIP-2 (mouse homolog of IL-8) were measured using an ELISA. The results came from six mice per group and were expressed in ng per well, in the format of means ± standard deviations. Asterisks indicate statistically significant differences (P < 0.05) between the infected (hatched bars) and uninfected (open bars) samples from the wt macrophages.
Chlamydial induction of IL-8 correlates with intracellular IL-1α expression in host cell nuclei.
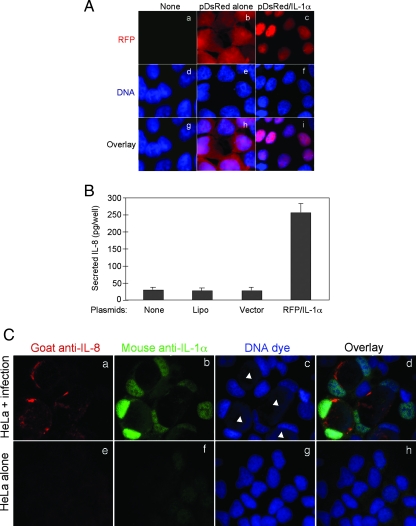
We next transfected 293T cells with a plasmid carrying the human IL-1α gene and monitored the effect of IL-1α gene expression on IL-8 production. We found that plasmid-encoded full-length IL-1α in the form of an RFP-tagged fusion protein migrated into the cell nucleus (Fig. 4A) and induced IL-8 production (Fig. 4B). When endogenous intracellular IL-1α and IL-8 were detected in HeLa cells during chlamydial infection, most of the IL-1α was concentrated in the nuclei, while IL-8 accumulated in the Golgi apparatuses of the same cells infected with chlamydial organisms (Fig. 4C). These observations suggest that Chlamydia-enhanced IL-1α can migrate into the host cell nucleus and induce IL-8 production.
FIG. 4.
Correlation of IL-1α nuclear expression with chlamydial induction of IL-8. 293T cells were transfected with a pDsRed plasmid encoding precursor full-length human IL-1α with RFP as an N-terminal fusion tag (RFP/IL-1α) or with vector alone (vector) or were treated with Lipofectamine alone (Lipo). The culture supernatants were harvested 48 h after infection for measurement of IL-8 by ELISA (B), and the corresponding cells grown on coverslips were processed for visualization of RFP (red) and nuclei (blue) after DNA dye staining (A). Note that the full-length IL-1α-RFP fusion protein was expressed in the nucleus and that a significant level of IL-8 was detected in the sample expressing IL-1α-RFP. (C) HeLa cells, with or without chlamydial infection for 48 h, were processed for immunofluorescence labeling of IL-8 (red), IL-1α (green), and host cell and chlamydial DNA (blue). The filled white triangles indicate chlamydial inclusions. Note that IL-1α was concentrated in the nuclei of the Chlamydia-infected cells, whereas IL-8 accumulated in the Golgi apparatuses of the same cells.
DISCUSSION
C. trachomatis infection induces inflammatory pathologies in the urogenital tracts (10). Many inflammatory cytokines, including IL-1α and IL-8, have been detected frequently in chlamydia-infected tissues and cells. In the current study, we have presented evidence that IL-1α contributes to chlamydial induction of IL-8 via a mechanism independent of the functional IL-1RI. First, IL-1α remained inside cells and was not secreted into the extracellular space for up to 48 h after chlamydial infection when significant amounts of IL-8 were already induced by chlamydial infection (Fig. 1B). Second, Chlamydia-induced IL-8 came mainly from the infected cells, not the adjacent uninfected cells (Fig. 1C), making it possible for intracellular IL-1α to regulate the IL-8 gene in an intracrine fashion. Third, although IL-1α was essential for chlamydial induction of IL-8 (Fig. 2), neither extracellular IL-1α nor IL-1RI was required for IL-8 production during chlamydial infection (Fig. 3). Fourth, we surprisingly found that IL-1R1 KO Mφs produced significant amounts of both IL-1α and IL-8 even without chlamydial infection, which may be due to the compensative production of IL-1α in response to IL-1R1 deficiency. Although the precise mechanism requires further investigation, this observation suggests that constitutively produced IL-1α may drive IL-8 production in an IL-1R1-independent manner. Fifth, expression of the precursor full-length IL-1α in the nucleus indeed enhanced IL-8 production (Fig. 4A and B). Finally, Chlamydia-induced IL-1α was localized in the nuclei of the infected cells and was accompanied by IL-8 production (Fig. 4C).
Previous studies also reported that chlamydial induction of IL-8 was not blocked by treatment with either IL-1α-specific neutralization antibodies or IL-1RA during the early stages of infection and that a partial inhibition of IL-8 production was achieved by these treatments at the late stages of infection (5, 46). These observations led to the conclusions that chlamydial induction of IL-8 during the early stages of infection was independent of IL-1α and that the partial inhibition of IL-8 at the late stages of infection was due to the blockade of extracellular IL-1α released from the lysed cells. Our current study has provided strong evidence demonstrating that it is intracellular IL-1α that plays an essential role in chlamydial induction of IL-8 throughout the infection course, via a mechanism independent of cell surface IL-1R, although the extracellular IL-1α released from lysed cells during the late stages of infection can always contribute to IL-8 production. Similar mechanisms may also exist for other intracellular pathogen infection-induced inflammation, since it was found that during Rickettsia infection, blocking extracellular IL-1α-mediated signaling failed to alter Rickettsia induction of IL-8 during the early stage of infection but partially and significantly suppressed IL-8 production during the late stages of infection (9). Although IL-1β is secreted along the chlamydial infection course and IL-1β can contribute to IL-8 production via the IL-1R1 signaling pathway (34), the finding that blockade of IL-1R1 signaling or IL-1R1 deficiency did not significantly affect Chlamydia-induced IL-8 production has demonstrated that IL-1β is not essential for chlamydial induction of IL-8.
A variety of inflammatory cytokines, including IL-1α (63), IL-33 (7), and high-mobility-group box 1 (HMGB1) (33), have been known to regulate transcription via both cell surface receptor-dependent and -independent pathways. All of these cytokines lack clear secretion signal peptides but possess nuclear localization motif sequences and can accumulate inside cells and migrate into the nucleus, thus allowing them to affect transcription without engaging the cell surface receptors. However, the precise mechanisms by which these cytokines enter the nucleus and participate in gene regulation are still unknown. IL-33 was found to associate with heterochromatin and target to mitotic chromosomes via an evolutionarily conserved homeodomain-like helix-turn-helix motif within its N-terminal portion. Functionally, nuclear IL-33 was found to possess transcriptional repressor properties (7, 17), while extracellular IL-33 is known to promote Th2 cytokine production by activating the ST2 receptor-mediated pathways (17). Nucleus-localized HMGB1 can facilitate gene transcription by stabilizing nucleosomes and allowing bending of DNA, while released HMGB1 has been shown to trigger cell surface receptors to amplify inflammation (33). Nucleus-localized IL-α appears to have multiple functions, including regulation of cell proliferation, migration, and differentiation (6, 35, 37, 57) as well as inflammatory responses (61). For example, the nuclear IL-1α prodomain was found to interact with the RNA processing apparatus and to induce apoptosis in malignant cells (45), while overexpression of the IL-1α prodomain in the nucleus could transform rat glomerular MC cells to a malignant phenotype (57). Like the proinflammatory role of extracellular IL-1α, nuclear IL-1α can also activate inflammatory cytokine genes. Overexpression of precursor IL-1α was sufficient for inducing IL-6 and IL-8 production (61) (Fig. 4B). The fact that full-length precursor IL-1α does not contain any DNA binding domain suggests that IL-1α must interact with other nuclear proteins in order to affect transcription. The observation that intracellular precursor IL-1α can interact with histone acetyltransferase complexes (6) seems to support such a hypothesis. Furthermore, by using a coprecipitation approach, Kawaguchi et al. (31) recently found that intracellular IL-1α can interact with both the intracellular IL-1RII (lacking the cytoplasmic domain) and HAX-1 (an intracellular adaptor molecule known to interact with many other proteins). The interaction of precursor IL-1α with intracellular IL-1RII may help to bring HAX-1 into large protein complexes. However, the relevance of these interaction complexes in the activation of IL-8 by intracellular IL-1α is not clear. It is possible that during microbial infection, intracellular IL-1α may pick up additional binding partners. In the current study, we found that IL-1α accumulated in the nuclei of Chlamydia-infected cells, while IL-8 was detected in the Golgi apparatuses of the same cells (Fig. 4C). Efforts are under way to identify the putative interaction partners of intracellular IL-1α in Chlamydia-infected cells.
Since IL-8 production during C. trachomatis infection is largely dependent on IL-1α, unraveling the mechanisms of chlamydial induction of IL-1α becomes important for further understanding chlamydial pathogenesis. It is known that various chlamydial components can be recognized by host pathogen-associated pattern recognition receptors, which may initiate the inflammatory pathways for inducing IL-1α. Rodriguez et al. (47) reported that infection with C. pneumoniae led to preferential activation of Toll-like receptor 4 (TLR4) but not TLR2 pathways. During C. trachomatis infection, both TLR2 and MyD88 were recruited to the C. trachomatis inclusion membrane, and knocking down either TLR2 or MyD88, but not TLR4, blocked Chlamydia-induced IL-8 (42), suggesting a dominant role of TLR2/MyD88 pathways in C. trachomatis induction of IL-1α, since we now know that chlamydial induction of IL-8 is dependent on IL-1α. However, another study reported that C. trachomatis induction of inflammatory cytokines, including IL-1α, depended only on MyD88, not TLR2 or TLR4 (39). Nevertheless, these studies together have indicated that MyD88-mediated pathways may be essential for chlamydial induction of IL-1α, although the precise receptors responsible for recognizing chlamydial components have still to be defined. Buchholz and Stephens have recently shown that chlamydial infection can activate reporter constructs containing promoter sequences for the transcriptional factors AP-1, NF-κB, and C/EBPβ (5), which may help to draw the link between MyD88 pathway activation and cytokine production during chlamydial infection. However, due to the fact that many inflammatory cytokines and chemokines share similar promoter sequences and are regulated by common inflammatory transcriptional factors as well as being able to activate these transcriptional factors, it is difficult to determine which cytokine gene is activated first and which is the result of secondary activation. A combination of multiple experimental approaches, including monitoring endogenous signaling molecule activation and evaluating the effects of genetic and chemical blockade, may help to pinpoint the molecular pathways required for chlamydial induction of a particular cytokine. The fact that chlamydial infection can activate various kinase pathways related to inflammation (16, 58) suggests that it is possible to further delineate the signaling pathways leading to individual inflammatory cytokine gene activation during chlamydial infection. We are in the process of mapping the pathways required for chlamydial induction of IL-1α.
The duality of IL-1α action has offered IL-1α maximal flexibility for carrying out its multifaceted functions (via both receptor-dependent and -independent pathways). IL-1α is constitutively expressed at a low level and is rapidly up-regulated upon infection in epithelial cells, and epithelial IL-1α can regulate many aspects of host defense (2, 25). Although we have demonstrated in the current study that IL-1α is critical for chlamydial induction of IL-8 in cell culture systems, the role of IL-1α during chlamydial infection in humans or animals is still unclear. A recent study using a human Fallopian tube epithelial tissue culture system demonstrated that C. trachomatis infection caused severe damage to the cultured tissues and that treatment with IL-1RA significantly reduced the tissue damage (26). Although this study failed to evaluate the contribution of intracellular IL-1α-mediated IL-8 production to the tissue damage due to a lack of leukocytes in the experimental system, it did demonstrate that extracellular IL-1α played an important role in Chlamydia-induced epithelial pathologies. The extracellular IL-1α was likely released from the Chlamydia-lysed cells, since the tissue damage was assessed 3 to 5 days after infection, when Chlamydia-induced cell burst was clearly visible under an electron microscope. It is known that C. trachomatis organisms can persist in infected cells for long periods during natural chlamydial infection in humans, which allows IL-1α to enhance inflammation via the intracrine mechanism, as demonstrated in the current study. It is also known that C. trachomatis can be transmitted from one cell to another via cell lysis, which allows IL-1α to activate cell surface IL-1R1 to cause tissue injury. Obviously, both persistently infected cells (intracellular IL-1α) and cell lysis (releasing extracellular IL-1α) can contribute to Chlamydia-induced inflammatory pathologies in humans. It is always preferable to use whole-animal model systems to evaluate the effects of IL-1α's dual actions on chlamydial infection and pathogenesis. Using various TLR and MyD88 KO mouse models, it has been shown that MyD88 is critical for both clearing chlamydial infection and exacerbating pathologies, although direct evidence demonstrating a role of MyD88 in C. trachomatis pathogenesis is still lacking (11, 12, 40, 47). Since MyD88 is a common adaptor molecule required for both the TLR (for inducing cytokines such as IL-1α) and IL-1R (for exerting IL-1α effects) pathways, the above results cannot distinguish whether the lack of immunity and inflammation in MyD88 KO mice is due to a host failure to recognize chlamydial infection for induction of IL-1α or an inability to transmit IL-1α signaling. The results from the current study showing that during chlamydial infection IL-1α can induce inflammatory responses independent of IL-1RI seem to suggest that MyD88, as an adaptor for TLRs to induce inflammatory cytokines such as IL-1α, plays an essential role in chlamydial infection. However, to further determine the role of IL-1α in chlamydial infection, mice deficient in either IL-1α or IL-1RI should be evaluated for the abilities to clear infection and to develop pathologies following a chlamydial challenge infection, and such studies are under way.
Acknowledgments
This work was supported in part by grants (to G. Zhong) from the National Institutes of Health.
We thank Peter Dube for reading the manuscript.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 17 December 2007.
REFERENCES
- 1.Banda, N. K., C. Guthridge, D. Sheppard, K. S. Cairns, M. Muggli, D. Bech-Otschir, W. Dubiel, and W. P. Arend. 2005. Intracellular IL-1 receptor antagonist type 1 inhibits IL-1-induced cytokine production in keratinocytes through binding to the third component of the COP9 signalosome. J. Immunol. 1743608-3616. [DOI] [PubMed] [Google Scholar]
- 2.Bando, M., Y. Hiroshima, M. Kataoka, Y. Shinohara, M. C. Herzberg, K. F. Ross, T. Nagata, and J. I. Kido. 2007. Interleukin-1alpha regulates antimicrobial peptide expression in human keratinocytes. Immunol. Cell Biol. 85532-537. [DOI] [PubMed] [Google Scholar]
- 3.Bauwens, J. E., H. Orlander, M. P. Gomez, M. Lampe, S. Morse, W. E. Stamm, R. Cone, R. Ashley, P. Swenson, and K. K. Holmes. 2002. Epidemic lymphogranuloma venereum during epidemics of crack cocaine use and HIV infection in the Bahamas. Sex. Transm. Dis. 29253-259. [DOI] [PubMed] [Google Scholar]
- 4.Boraschi, D., and A. Tagliabue. 2006. The interleukin-1 receptor family. Vitam. Horm. 74229-254. [DOI] [PubMed] [Google Scholar]
- 5.Buchholz, K. R., and R. S. Stephens. 2006. Activation of the host cell proinflammatory interleukin-8 response by Chlamydia trachomatis. Cell. Microbiol. 81768-1779. [DOI] [PubMed] [Google Scholar]
- 6.Buryskova, M., M. Pospisek, A. Grothey, T. Simmet, and L. Burysek. 2004. Intracellular interleukin-1alpha functionally interacts with histone acetyltransferase complexes. J. Biol. Chem. 2794017-4026. [DOI] [PubMed] [Google Scholar]
- 7.Carriere, V., L. Roussel, N. Ortega, D. A. Lacorre, L. Americh, L. Aguilar, G. Bouche, and J. P. Girard. 2007. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc. Natl. Acad. Sci. USA 104282-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C., D. Chen, J. Sharma, W. Cheng, Y. Zhong, K. Liu, J. Jensen, R. Shain, B. Arulanandam, and G. Zhong. 2006. The hypothetical protein CT813 is localized in the Chlamydia trachomatis inclusion membrane and is immunogenic in women urogenitally infected with C. trachomatis. Infect. Immun. 744826-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clifton, D. R., E. Rydkina, H. Huyck, G. Pryhuber, R. S. Freeman, D. J. Silverman, and S. K. Sahni. 2005. Expression and secretion of chemotactic cytokines IL-8 and MCP-1 by human endothelial cells after Rickettsia rickettsii infection: regulation by nuclear transcription factor NF-kappaB. Int. J. Med. Microbiol. 295267-278. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, C. R., and R. C. Brunham. 1999. Pathogenesis of Chlamydia induced pelvic inflammatory disease. Sex. Transm. Infect. 7521-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darville, T., J. M. O'Neill, C. W. Andrews, Jr., U. M. Nagarajan, L. Stahl, and D. M. Ojcius. 2003. Toll-like receptor-2, but not Toll-like receptor-4, is essential for development of oviduct pathology in chlamydial genital tract infection. J. Immunol. 1716187-6197. [DOI] [PubMed] [Google Scholar]
- 12.Derbigny, W. A., M. S. Kerr, and R. M. Johnson. 2005. Pattern recognition molecules activated by Chlamydia muridarum infection of cloned murine oviduct epithelial cell lines. J. Immunol. 1756065-6075. [DOI] [PubMed] [Google Scholar]
- 13.Dongari-Bagtzoglou, A., and H. Kashleva. 2003. Candida albicans triggers interleukin-8 secretion by oral epithelial cells. Microb. Pathog. 34169-177. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir, S. M., J. Martinez, A. Patkaniowska, W. Lendeckel, and T. Tuschl. 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 206877-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan, T., H. Lu, H. Hu, L. Shi, G. A. McClarty, D. M. Nance, A. H. Greenberg, and G. Zhong. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuda, E. Y., S. P. Lad, D. P. Mikolon, M. Iacobelli-Martinez, and E. Li. 2005. Activation of lipid metabolism contributes to interleukin-8 production during Chlamydia trachomatis infection of cervical epithelial cells. Infect. Immun. 734017-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gadina, M., and C. A. Jefferies. 2007. IL-33: a sheep in wolf's clothing? Sci. STKE 2007pe31. [DOI] [PubMed] [Google Scholar]
- 18.Gervassi, A., M. R. Alderson, R. Suchland, J. F. Maisonneuve, K. H. Grabstein, and P. Probst. 2004. Differential regulation of inflammatory cytokine secretion by human dendritic cells upon Chlamydia trachomatis infection. Infect. Immun. 727231-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greene, W., Y. Xiao, Y. Huang, G. McClarty, and G. Zhong. 2004. Chlamydia-infected cells continue to undergo mitosis and resist induction of apoptosis. Infect. Immun. 72451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gubler, U., A. O. Chua, A. S. Stern, C. P. Hellmann, M. P. Vitek, T. M. DeChiara, W. R. Benjamin, K. J. Collier, M. Dukovich, P. C. Familletti, et al. 1986. Recombinant human interleukin 1 alpha: purification and biological characterization. J. Immunol. 1362492-2497. [PubMed] [Google Scholar]
- 21.Hackstadt, T. 1998. The diverse habitats of obligate intracellular parasites. Curr. Opin. Microbiol. 182-87. [DOI] [PubMed] [Google Scholar]
- 22.Hackstadt, T., M. A. Scidmore, and D. D. Rockey. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. USA 924877-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hazuda, D. J., J. Strickler, P. Simon, and P. R. Young. 1991. Structure-function mapping of interleukin 1 precursors. Cleavage leads to a conformational change in the mature protein. J. Biol. Chem. 2667081-7086. [PubMed] [Google Scholar]
- 24.Hornung, V., M. Guenthner-Biller, C. Bourquin, A. Ablasser, M. Schlee, S. Uematsu, A. Noronha, M. Manoharan, S. Akira, A. de Fougerolles, S. Endres, and G. Hartmann. 2005. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 11263-270. [DOI] [PubMed] [Google Scholar]
- 25.Hurgin, V., D. Novick, A. Werman, C. A. Dinarello, and M. Rubinstein. 2007. Antiviral and immunoregulatory activities of IFN-gamma depend on constitutively expressed IL-1alpha. Proc. Natl. Acad. Sci. USA 1045044-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hvid, M., A. Baczynska, B. Deleuran, J. Fedder, H. J. Knudsen, G. Christiansen, and S. Birkelund. 2007. Interleukin-1 is the initiator of Fallopian tube destruction during Chlamydia trachomatis infection. Cell. Microbiol. 92795-2803. [DOI] [PubMed] [Google Scholar]
- 27.Hybiske, K., and R. S. Stephens. 2007. Mechanisms of Chlamydia trachomatis entry into nonphagocytic cells. Infect. Immun. 753925-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hybiske, K., and R. S. Stephens. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. USA 10411430-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung, Y. D., F. Fan, D. J. McConkey, M. E. Jean, W. Liu, N. Reinmuth, O. Stoeltzing, S. A. Ahmad, A. A. Parikh, N. Mukaida, and L. M. Ellis. 2002. Role of P38 MAPK, AP-1, and NF-kappaB in interleukin-1beta-induced IL-8 expression in human vascular smooth muscle cells. Cytokine 18206-213. [DOI] [PubMed] [Google Scholar]
- 30.Kasahara, T., T. Oda, K. Hatake, M. Akiyama, N. Mukaida, and K. Matsushima. 1998. Interleukin-8 and monocyte chemotactic protein-1 production by a human glioblastoma cell line, T98G in coculture with monocytes: involvement of monocyte-derived interleukin-1alpha. Eur. Cytokine Netw. 947-55. [PubMed] [Google Scholar]
- 31.Kawaguchi, Y., E. Nishimagi, A. Tochimoto, M. Kawamoto, Y. Katsumata, M. Soejima, T. Kanno, N. Kamatani, and M. Hara. 2006. Intracellular IL-1alpha-binding proteins contribute to biological functions of endogenous IL-1alpha in systemic sclerosis fibroblasts. Proc. Natl. Acad. Sci. USA 10314501-14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kupper, T. S., A. O. Chua, P. Flood, J. McGuire, and U. Gubler. 1987. Interleukin 1 gene expression in cultured human keratinocytes is augmented by ultraviolet irradiation. J. Clin. Investig. 80430-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lotze, M. T., and K. J. Tracey. 2005. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat. Rev. Immunol. 5331-342. [DOI] [PubMed] [Google Scholar]
- 34.Lu, H., C. Shen, and R. C. Brunham. 2000. Chlamydia trachomatis infection of epithelial cells induces the activation of caspase-1 and release of mature IL-18. J. Immunol. 1651463-1469. [DOI] [PubMed] [Google Scholar]
- 35.Maier, J. A., M. Statuto, and G. Ragnotti. 1994. Endogenous interleukin 1 alpha must be transported to the nucleus to exert its activity in human endothelial cells. Mol. Cell. Biol. 141845-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maxion, H. K., and K. A. Kelly. 2002. Chemokine expression patterns differ within anatomically distinct regions of the genital tract during Chlamydia trachomatis infection. Infect. Immun. 701538-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMahon, G. A., S. Garfinkel, I. Prudovsky, X. Hu, and T. Maciag. 1997. Intracellular precursor interleukin (IL)-1alpha, but not mature IL-1alpha, is able to regulate human endothelial cell migration in vitro. J. Biol. Chem. 27228202-28205. [DOI] [PubMed] [Google Scholar]
- 38.Mukaida, N., M. Shiroo, and K. Matsushima. 1989. Genomic structure of the human monocyte-derived neutrophil chemotactic factor IL-8. J. Immunol. 1431366-1371. [PubMed] [Google Scholar]
- 39.Nagarajan, U. M., D. M. Ojcius, L. Stahl, R. G. Rank, and T. Darville. 2005. Chlamydia trachomatis induces expression of IFN-gamma-inducible protein 10 and IFN-beta independent of TLR2 and TLR4, but largely dependent on MyD88. J. Immunol. 175450-460. [DOI] [PubMed] [Google Scholar]
- 40.Naiki, Y., K. S. Michelsen, N. W. Schroder, R. Alsabeh, A. Slepenkin, W. Zhang, S. Chen, B. Wei, Y. Bulut, M. H. Wong, E. M. Peterson, and M. Arditi. 2005. MyD88 is pivotal for the early inflammatory response and subsequent bacterial clearance and survival in a mouse model of Chlamydia pneumoniae pneumonia. J. Biol. Chem. 28029242-29249. [DOI] [PubMed] [Google Scholar]
- 41.Nicklin, M. J., J. L. Barton, M. Nguyen, M. G. FitzGerald, G. W. Duff, and K. Kornman. 2002. A sequence-based map of the nine genes of the human interleukin-1 cluster. Genomics 79718-725. [DOI] [PubMed] [Google Scholar]
- 42.O'Connell, C. M., I. A. Ionova, A. J. Quayle, A. Visintin, and R. R. Ingalls. 2006. Localization of TLR2 and MyD88 to Chlamydia trachomatis inclusions. Evidence for signaling by intracellular TLR2 during infection with an obligate intracellular pathogen. J. Biol. Chem. 2811652-1659. [DOI] [PubMed] [Google Scholar]
- 43.Okamoto, S., N. Mukaida, K. Yasumoto, H. Horiguchi, and K. Matsushima. 1993. Molecular mechanism of interleukin-8 gene expression. Adv. Exp. Med. Biol. 35187-97. [DOI] [PubMed] [Google Scholar]
- 44.Perregaux, D. G., and C. A. Gabel. 1998. Post-translational processing of murine IL-1: evidence that ATP-induced release of IL-1 alpha and IL-1 beta occurs via a similar mechanism. J. Immunol. 1602469-2477. [PubMed] [Google Scholar]
- 45.Pollock, A. S., J. Turck, and D. H. Lovett. 2003. The prodomain of interleukin 1alpha interacts with elements of the RNA processing apparatus and induces apoptosis in malignant cells. FASEB J. 17203-213. [DOI] [PubMed] [Google Scholar]
- 46.Rasmussen, S. J., L. Eckmann, A. J. Quayle, L. Shen, Y. X. Zhang, D. J. Anderson, J. Fierer, R. S. Stephens, and M. F. Kagnoff. 1997. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J. Clin. Investig. 9977-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez, N., N. Wantia, F. Fend, S. Durr, H. Wagner, and T. Miethke. 2006. Differential involvement of TLR2 and TLR4 in host survival during pulmonary infection with Chlamydia pneumoniae. Eur. J. Immunol. 361145-1155. [DOI] [PubMed] [Google Scholar]
- 48.Roebuck, K. A. 1999. Regulation of interleukin-8 gene expression. J. Interferon Cytokine Res. 19429-438. [DOI] [PubMed] [Google Scholar]
- 49.Sauder, D. N., B. M. Stanulis-Praeger, and B. A. Gilchrest. 1988. Autocrine growth stimulation of human keratinocytes by epidermal cell-derived thymocyte-activating factor: implications for skin aging. Arch. Dermatol. Res. 28071-76. [DOI] [PubMed] [Google Scholar]
- 50.Sawai, H., H. Funahashi, Y. Okada, Y. Matsuo, M. Sakamoto, M. Yamamoto, H. Takeyama, and T. Manabe. 2005. Interleukin-1alpha enhances IL-8 secretion through p38 mitogen-activated protein kinase and reactive oxygen species signaling in human pancreatic cancer cells. Med. Sci. Monit. 11BR343-BR350. [PubMed] [Google Scholar]
- 51.Schachter, J., and J. Moncada. 2005. Lymphogranuloma venereum: how to turn an endemic disease into an outbreak of a new disease? Start looking. Sex. Transm. Dis. 32331-332. [DOI] [PubMed] [Google Scholar]
- 52.Schlee, M., V. Hornung, and G. Hartmann. 2006. siRNA and isRNA: two edges of one sword. Mol. Ther. 14463-470. [DOI] [PubMed] [Google Scholar]
- 53.Sharma, J., A. M. Bosnic, J. M. Piper, and G. Zhong. 2004. Human antibody responses to a Chlamydia-secreted protease factor. Infect. Immun. 727164-7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherman, K. J., J. R. Daling, A. Stergachis, N. S. Weiss, H. M. Foy, S. P. Wang, and J. T. Grayston. 1990. Sexually transmitted diseases and tubal pregnancy. Sex. Transm. Dis. 17115-121. [DOI] [PubMed] [Google Scholar]
- 55.Sioud, M. 2006. Innate sensing of self and non-self RNAs by Toll-like receptors. Trends Mol. Med. 12167-176. [DOI] [PubMed] [Google Scholar]
- 56.Stephens, R. S. 2003. The cellular paradigm of chlamydial pathogenesis. Trends Microbiol. 1144-51. [DOI] [PubMed] [Google Scholar]
- 57.Stevenson, F. T., J. Turck, R. M. Locksley, and D. H. Lovett. 1997. The N-terminal propiece of interleukin 1 alpha is a transforming nuclear oncoprotein. Proc. Natl. Acad. Sci. USA 94508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su, H., G. McClarty, F. Dong, G. M. Hatch, Z. K. Pan, and G. Zhong. 2004. Activation of Raf/MEK/ERK/cPLA2 signaling pathway is essential for chlamydial acquisition of host glycerophospholipids. J. Biol. Chem. 2799409-9416. [DOI] [PubMed] [Google Scholar]
- 59.Taylor, H. R., S. L. Johnson, J. Schachter, H. D. Caldwell, and R. A. Prendergast. 1987. Pathogenesis of trachoma: the stimulus for inflammation. J. Immunol. 1383023-3027. [PubMed] [Google Scholar]
- 60.Taylor, S. L., B. R. Renshaw, K. E. Garka, D. E. Smith, and J. E. Sims. 2002. Genomic organization of the interleukin-1 locus. Genomics 79726-733. [DOI] [PubMed] [Google Scholar]
- 61.Werman, A., R. Werman-Venkert, R. White, J. K. Lee, B. Werman, Y. Krelin, E. Voronov, C. A. Dinarello, and R. N. Apte. 2004. The precursor form of IL-1alpha is an intracrine proinflammatory activator of transcription. Proc. Natl. Acad. Sci. USA 1012434-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wessendorf, J. H., S. Garfinkel, X. Zhan, S. Brown, and T. Maciag. 1993. Identification of a nuclear localization sequence within the structure of the human interleukin-1 alpha precursor. J. Biol. Chem. 26822100-22104. [PubMed] [Google Scholar]
- 63.Wolf, J. S., Z. Chen, G. Dong, J. B. Sunwoo, C. C. Bancroft, D. E. Capo, N. T. Yeh, N. Mukaida, and C. Van Waes. 2001. IL (interleukin)-1alpha promotes nuclear factor-kappaB and AP-1-induced IL-8 expression, cell survival, and proliferation in head and neck squamous cell carcinomas. Clin. Cancer Res. 71812-1820. [PubMed] [Google Scholar]
- 64.Xiao, Y., Y. Zhong, W. Greene, F. Dong, and G. Zhong. 2004. Chlamydia trachomatis infection inhibits both Bax and Bak activation induced by staurosporine. Infect. Immun. 725470-5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao, Y., Y. Zhong, H. Su, Z. Zhou, P. Chiao, and G. Zhong. 2005. NF-kappa B activation is not required for Chlamydia trachomatis inhibition of host epithelial cell apoptosis. J. Immunol. 1741701-1708. [DOI] [PubMed] [Google Scholar]
- 66.Zhong, G., P. Fan, H. Ji, F. Dong, and Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193935-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhong, G., I. Toth, R. Reid, and R. C. Brunham. 1993. Immunogenicity evaluation of a lipidic amino acid-based synthetic peptide vaccine for Chlamydia trachomatis. J. Immunol. 1513728-3736. [PubMed] [Google Scholar]
- 68.Zhong, G. M., and R. C. Brunham. 1990. Immunoaccessible peptide sequences of the major outer membrane protein from Chlamydia trachomatis serovar C. Infect. Immun. 583438-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhong, G. M., and L. M. de la Maza. 1988. Activation of mouse peritoneal macrophages in vitro or in vivo by recombinant murine gamma interferon inhibits the growth of Chlamydia trachomatis serovar L1. Infect. Immun. 563322-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]