Abstract
The regulatory locus sae is a two-component system in Staphylococcus aureus that regulates many important virulence factors, including alpha-toxin (encoded by hla) at the transcriptional level. The SarA homologs Rot and SarT were previously shown to be repressors of hla in selected S. aureus backgrounds. To delineate the interaction of rot and sae and the contribution of sarT to hla expression, an assortment of rot and sae isogenic single mutants, a rot sae double mutant, and a rot sae sarT markerless triple mutant were constructed from wild-type strain COL. Using Northern blot analysis and transcriptional reporter gene green fluorescent protein, fusion, and phenotypic assays, we found that the repression of hla by rot is dependent on sae. A rot sae sarT triple mutant was not able to rescue the hla defect of the rot sae double mutant. Among the three sae promoters, the distal sae P3 promoter is the strongest in vitro. Interestingly, the sae P3 promoter activities correlate with hla expression in rot, rot sae, and rot sae sarT mutants of COL. Transcriptional study has also shown that rot repressed sae, especially at the sae P3 promoter. Collectively, our data implicated the importance of sae in the rot-mediated repression of hla in S. aureus.
Staphylococcus aureus is an important community- and nosocomially acquired pathogen that can cause both local and systemic infections in humans. The pathogenicity of this microorganism depends largely on its successful adaptation to the human host and hence requires the environmentally coordinated expression of virulence factors. The expression of virulence factors in S. aureus is regulated by a network of interacting regulators, including two-component regulatory systems and the SarA protein family (2, 19).
The sae locus is a two-component regulatory system first described for a Tn551 insertional mutant with an exoprotein-defective phenotype (5). Subsequent works showed that the sae locus consists of four open reading frames (ORFs), with two encoding the response regulator (SaeR) and the sensor kinase (SaeS) and the other two encoding the two hypothetical proteins designated ORF3 and ORF4 (6, 20, 28). Transcriptional analysis revealed four overlapping transcripts driven by three promoters (P3, P1, and P2) (20, 28). The sae locus was found to be a key element in the regulatory cascade governing the staphylococcal virulon. In vitro, it up-regulates many virulence factors, including alpha-hemolysin (encoded by hla), beta-hemolysin, DNase, coagulase, the protease SspA, thermonuclease, protein A, extracellular adherence protein (Eap), extracellular matrix binding protein (Emp), and FnbpA, and down-regulates capsular polysaccharide at the transcriptional level (4, 5, 8, 10, 24). More importantly, sae is an important element for the expression of virulence genes in vivo (7, 8, 23).
The rot locus was first identified via transposon mutagenesis as a repressor of alpha-hemolysin synthesis in S. aureus strain PM614 by partially restoring hla expression in an agr null mutant, presumably via an agr-independent mechanism (17). The rot gene product (Rot) is a member of the SarA protein family that shares homology with the smaller SarA homologs (e.g., SarA, SarR, SarT, SarV, and SarX). Although originally perceived as a 166-residue protein (17), Rot was recently found to be only 133 residues long (16) and is likely a winged-helix protein, as with other members of the SarA protein family (2).
Several studies have shown that hla is one of the major virulence factors produced by most S. aureus strains (9, 13, 22). Alpha-toxin is a pore-forming toxin that has cytolytic, hemolytic, and dermonecrotic activities. Although alpha-toxin has been shown to be up-regulated by sae and down-regulated by rot and sarT, both of which are repressors of hla expression, the exact pathways of hla regulation by these three regulators have not been defined. To address this issue, we constructed sae and rot single- and double-deletion mutants as well as a sae rot sarT triple-deletion mutants in the sigma B-positive strain COL to examine hla expression at the phenotypic and transcriptional levels. Our results clearly showed that down-modulation of the sae P3 promoter, the strongest sae promoter among the three promoters in vitro, correlated with decreased hla expression in sae, sae rot, and sae rot sarT mutants compared to that of the parent. We also demonstrated that rot represses sae to control hla expression. Our studies here thus revealed an intricate network between sae and rot in the regulation of hla.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. aureus cells were grown at 37°C with aeration in Mueller-Hinton broth (MHB; Difco) supplemented with antibiotics as indicated below. Luria-Bertani (LB) broth was used for cultivating Escherichia coli. Antibiotics used for S. aureus were erythromycin (5 μg/ml), tetracycline (3 μg/ml), and chloramphenicol (10 μg/ml). For E. coli, ampicillin was used at 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Comment | Reference |
|---|---|---|
| Strains | ||
| E. coli XL-1 Blue | General-purpose host strain for cloning | 14 |
| S. aureus | ||
| RN4220 | Mutant strain of 8325-4 that accepts foreign DNA | 18 |
| COL | Methicillin-resistant laboratory strain | 21 |
| ALC4481 | COL sae single-deletion mutant | This study |
| ALC4909 | COL rot single-deletion mutant | This study |
| ALC4988 | COL sae rot double-deletion mutant | This study |
| ALC5245 | COL sae rot sarT triple-deletion mutant | This study |
| ALC3227 | COL with pALC1740 | This study |
| ALC5097 | ALC4481 with pALC1740 | This study |
| ALC5098 | ALC4909 with pALC1740 | This study |
| ALC5099 | ALC4988 with pALC1740 | This study |
| ALC5256 | ALC5245 with pALC1740 | This study |
| ALC5018 | COL with pALC4989 | This study |
| ALC5019 | COL with pALC4990 | This study |
| ALC5020 | COL with pALC4991 | This study |
| ALC5370 | ALC4481with pALC4991 | This study |
| ALC5371 | ALC4909 with pALC4991 | This study |
| ALC5373 | ALC4988 with pALC4991 | This study |
| ALC5376 | ALC5245 with pALC4991 | This study |
| Plasmids | ||
| pALC1484 | Modified pALC236 shuttle vector with a promoterless gfpuvr reporter gene preceded by an S. aureus ribosome binding site | 11 |
| pALC1740 | pALC1484 with the hla promoter | 15 |
| pALC4989 | pALC1484 with the sae P1 promoter fused with the gfpuvr reporter gene at the EcoRI and XbaI sites | This study |
| pALC4990 | pALC1484 with the sae P2 promoter fused with the gfpuvr reporter gene at the EcoRI and XbaI sites | This study |
| pALC4991 | pALC1484 with the sae P3 promoter fused with the gfpuvr reporter gene at the EcoRI and XbaI sites | This study |
| pMAD | Vector for allelic replacement in gram-positive bacteria | 1 |
Construction of single-, double-, and triple-deletion sae, rot, and sarT mutants.
To introduce a single deletion of the rot, saeR, or saeR gene, DNA fragments corresponding to the upstream and downstream regions of the gene were amplified by PCR, using chromosomal DNA from strain COL as a template. The PCR products were purified, digested with BamHI and NcoI or EcoRI, and ligated into the temperature-sensitive shuttle plasmid pMAD, containing a temperature-sensitive S. aureus origin of replication, an erythromycin resistance cassette, and the β-galactosidase gene (1). The resulting plasmids (Table 1) containing the upstream and downstream fragments in tandem were then amplified in E. coli XL1-Blue. The recombinant pMADs were then extracted from E. coli and transformed into S. aureus RN4220 by electroporation. Plasmids obtained from RN4220 were then transformed into S. aureus strain COL. Transformants of S. aureus were selected at 30°C on Trypticase soy agar containing 2.5 μg/ml erythromycin and 150 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The allelic exchange in the absence of a selection marker was performed as previously described (1). Briefly, recombinant pMAD was integrated into the chromosome through homologous recombination at the nonpermissive temperature (42°C). From the 42°C plate containing erythromycin and X-Gal, one light-blue colony was picked into 10 ml of Trypticase soy broth and incubated overnight at 30°C without antibiotic. Tenfold serial dilutions of this culture were plated on Trypticase soy agar plates containing X-Gal. White colonies, which were sensitive to erythromycin and hence no longer contained the pMAD plasmid, were selected, and the gene deletion was confirmed by PCR and DNA sequencing. We used a similar strategy to sequentially construct sae rot double mutants and sae rot sarT triple mutants, using the pMAD plasmid containing the fragments flanking the deleted genes, and then confirmed the sequences with PCR and DNA sequencing.
Isolation of RNA and Northern blot hybridization.
Overnight cultures of S. aureus were diluted 1:100 in MHB and grown in 18-mm borosilicate tubes to early log phase (optical density at 650 nm [OD650] = 0.7), late exponential phase (OD650 = 1.2), and post-exponential phase (OD650 = 1.7). The cells were harvested and processed with Trizol (Invitrogen, Gaithersburg, MD) in combination with 0.1-mm sirconia-silica beads in a Biospec reciprocating shaker to yield RNA as described previously (3). RNA concentrations in the extracts were measured by determining absorbance at 260 nm using an Eppendorf BioPhotometer (Brinkmann, Westbury, NY). Twenty to 40 μg of each sample was electrophoresed in a 1.5% agarose-0.66 M formaldehyde gel in 4-morpholinepropanesulfonic acid (MOPS) running buffer (20 mM MOPS, 10 mM sodium acetate, 2 mM EDTA, pH 7.0) and blotted onto Hybond N+ membranes (Amersham, Arlington Heights, IL), as previously described (3). Prior to being blotted, the gel was viewed under UV light to ensure that equivalent amounts of ethidium bromide-stained rRNA bands were present for each sample. After being blotted, the gel was again viewed under UV light to confirm complete RNA transfer. For the detection of specific transcripts (hla, saeRS, and rot), digoxigenin (DIG)-labeled probes (the oligonucleotides listed in Table 2) generated by PCR were prepared by using the DIG labeling PCR kit according to the manufacturer's instructions (Roche Biochemicals, Mannheim, Germany). The blotted membrane was prehybridized in 25 ml Dig-Easy-Hyb buffer (Dig High Prime DNA labeling and detection starter kit II; Roche) for 2 h at 50°C with rotation and hybridized in the same Dig-Easy-Hyb buffer containing 25 ng/ml DIG-labeled probe overnight at 50°C. The hybridized membrane was washed first with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 sodium citrate)-0.1% sodium dodecyl sulfate (SDS) for 30 min at 37°C, second with 0.5× SSC-0.1% SDS for 30 min at 37°C, and finally twice with 0.5× SSC-0.1% SDS for 15 min at 37°C, all with rotation. After being washed with 1× washing buffer (Roche) for 5 min, the membrane was incubated with blocking solution for 60 min and antibody solution (anti-DIG-alkaline phosphatase, 75 mU/ml) for 60 min at 37°C with rotation. The membrane was then washed, covered with 1 ml chemiluminescent substrate for alkaline phosphatase, and incubated for 15 min at 37°C according to the manufacturer's protocol. The membrane was immediately exposed to film (Kodak Biomax MR film) for 3 to 30 min.
TABLE 2.
Primers used in this study
| Use | Primera |
|---|---|
| sae deletion | 5′GTCAGGATTCGTACGGATACCACTATAGAT3′ |
| 5′TCCGATTTATTTATAAAATAAAATGCAAAGACTAAAAAGAAGCTC3′ | |
| 5′CATTTTATTTTATAAATAAATCGGACTATTTTTTCACCTCTGTTCTTACGA3′ | |
| 5′GATCCCATGGTCCAGATTTATACGTCTACCTAACA3′ | |
| rot deletion | 5′GATCGGATCCCACGAGGTTCACAATGAGC3′ |
| 5′CCCAACAATCCCAAAACTTGTATGTGCT3′ | |
| 5′ATACAAGTTTTGGGATTGTTGGGGTTTAATAGCATAAAAAGAGGT3′ | |
| 5′GATCCCATGGTGACTCAAGAAGAGTACACAAAC3′ | |
| sarT deletion | 5′GTCAGGATTCATCCTTTCATCTGCAAGGGATCGT3′ |
| 5′CACCAAGATATTAAAATCTCGCAAATCATTCATCAAGTCTTC3′ | |
| 5′GAAGACTTGATGAATGATTTGCGAGATTTTAATATCTTGGTC3′ | |
| 5′GTCAGAATTCATGGTTATTTGCCACTCTAAC3′ | |
| sae P1 promoter-gfp fusion | 5′GTCAGAATTCTCGCAATGGTTGACTACGAT3′ |
| 5′GTCATCTAGAATTTATTGTGTGTAATTTATATAAACA3′ | |
| sae P2 promoter-gfp fusion | 5′GTCAGAATTCTTAGTACCAGTCATCGCTAAC3′ |
| 5′GTCATCTAGACTTACGACCTCTAAAGTAATTAATGAT3′ | |
| sae P3 promoter-gfp fusion | 5′GTCAGAATTCTTATTGTGGCAAAAGGTTTATAAA3′ |
| 5′GTCATCTAGACAATTTGATAAGTTAAGTTTAAAAT3′ |
Restriction sites are in bold and underlined.
Transcriptional fusion studies of different promoters linked to the gfpuvr reporter gene.
Promoter fragments of hla, sae, and rot were cloned into the shuttle vector pALC1484 upstream of the gfpuvr reporter gene to generate transcriptional fusions. Restriction analysis and DNA sequencing confirmed the orientation and authenticity of the promoter fragments. The recombinant plasmids containing these promoters were first amplified in E. coli and transformed into S. aureus strain RN4220 by electroporation (26). Plasmids purified from RN4220 transformants were then electroporated into COL and their isogenic mutants.
For the assay, overnight cultures of S. aureus strains harboring the recombinant plasmids were diluted 1:100 and grown at 37°C with shaking in MHB with chloramphenicol (10 μg/ml). The control strains contained only the vector pALC1484. Aliquots (200 μl) were transferred hourly or every 2 h to microtiter wells to assay for cell density (OD650) and fluorescence for 8 h and then overnight in a model FL600 fluorescence spectrophotometer (BioTek Instruments, Winooski, VT). Promoter activities were plotted as mean fluorescence divided by the OD650 from triplicate samples to minimize variations due to cell density.
The production of alpha-hemolysin.
After overnight growth in MHB, 2 μl of supernatant from each culture was placed on 2.5% defibrinated rabbit blood agar for 24 h at 37°C. The clear zone around bacterial growth represented alpha-hemolysis. The titer of each culture supernatant was also determined for hemolytic activity against rabbit erythrocytes. The method was modified from the work of Kehoe et al. (12). Briefly, the rabbit erythrocytes were washed and resuspended in phosphate-buffered saline to a final concentration of 1% (vol/vol). Erythrocytes were mixed with culture supernatants at equal proportions and incubated at 37°C for 1 h in microtiter wells. The highest dilution giving rise to visibly detectable lysis was defined as the hemolytic titer (12).
RESULTS
Effects of sae, rot, sae rot, and sae rot sarT mutations on hla expression in COL.
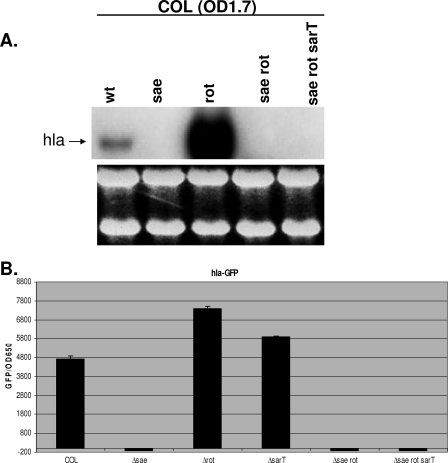
Previous studies have shown rot and sae to be negative and positive regulators, respectively, of hla expression. To determine the relationship between these two regulators, we constructed sae and rot single-deletion mutants and also sae rot double-deletion mutants in the COL background. COL was chosen because methicillin resistance in this strain is clinically relevant. In addition, this strain has an intact sigB operon with a functional rsbU gene, which has been shown to be important in down-modulating hla expression. To minimize the issues of the ectopic promoter and disruption of gene transcription within the same operon, we created in-frame deletions of saeRS and rot with pMAD, while leaving promoters and the transcription termination sites intact. In concordance with previous observations (8, 20, 23), hla transcription was completely absent in the sae mutant of strain COL compared with that in the isogenic parent (Fig. 1A). To our surprise, hla transcription was significantly up-regulated in the rot deletion mutant compared with that in the parent. This contrasts with data from previous studies showing that a rot mutation by itself, in the absence of an agr mutation, does not have any effect on hla expression (17, 25). To determine if rot expression is dependent on sae, we constructed a rot sae double mutant in the COL background. Northern blot analysis revealed that the sae rot double mutant displayed significant down-regulation of hla transcription in comparison to the rot single mutant (Fig. 1A). This indicated that the effect of rot on hla transcription is likely dependent on sae and not vice versa since the up-regulation of hla was not observed in the sae rot double mutant of COL. Previous studies have shown that sarT is a repressor of hla expression (27). To determine if sarT impacts hla expression via the sae pathway, we constructed the sae rot sarT triple-deletion mutant of strain COL. In comparison to what occurred with the sae rot double mutants, introduction of a sarT mutation into the sae rot double mutant in the COL background did not increase hla expression in the double mutants, thus indicating that sarT may act upstream of sae or rot and not downstream of sae in hla repression.
FIG. 1.
Transcription of hla in COL and its isogenic mutants. (A) Northern blot analysis of hla. RNA was harvested from cells grown to an OD650 of 1.7 (OD1.7) at a time when hla expression was expected to be the strongest. One typical blot with RNA with an OD650 of 1.7 is shown. An RNA gel below shows equal sample loadings. wt, wild type. (B) The expression of GFP driven by the hla promoter was measured. The fluorescence and cell density (OD650) were measured hourly for 10 h and then overnight by transferring aliquots (200 μl) to microtiter plates in triplicate. The results from one time point (8 h; OD650 of 1.7) of promoter activation are shown and are plotted as mean fluorescence divided by the OD650 using average values from triplicate readings.
To confirm the effect of sae on hla transcription on Northern blots, we performed hla promoter fusion assays. For this assay, we transformed strain COL and its isogenic sae mutant, rot mutant, sae rot double mutant, and sae rot sarT triple mutant with the shuttle plasmid carrying the gfpuvr reporter gene as driven by the hla promoter (15). Green fluorescent protein (GFP) levels, expressed as fluorescence units per OD650 unit to minimize the effect of bacterial cell densities, also confirmed the effect of rot and rot sae mutations on hla expression in the COL background, thus confirming our data from Northern blots (Fig. 1B). As a positive control, we also included a sarT mutant of COL, which displayed elevated hla promoter expression, as expected.
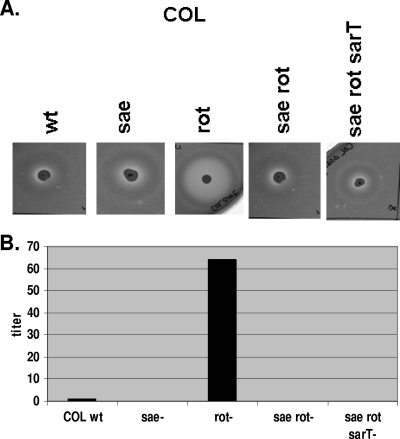
The phenotypic expression of alpha-hemolysin in parental strain COL and its isogenic mutants was measured on rabbit blood agar containing 2.5% rabbit erythrocytes as shown in Fig. 2A. The expression of alpha-hemolysin in the parent COL and the sae mutant on the rabbit erythrocyte agar plate was low. In agreement with the data on hla transcription as determined by Northern blot analysis and GFP reporter fusion assays, the hemolytic zone was noticeably bigger in the rot mutant but was reduced in the sae rot double mutant to the level in the sae single mutant. Likewise, a sarT mutation did not alter the hemolytic profile of the sae rot double mutant. Quantitation of the titers, defined as the highest dilution from overnight cultures giving rise to erythrocyte lysis, correlated well with those observed on the rabbit erythrocyte agar plates (Fig. 2B).
FIG. 2.
Production of alpha-hemolysin on 2.5% rabbit blood agar in COL and its isogenic mutants. (A) Two microliters each of supernatant from an overnight culture was placed on rabbit blood agar plates for 24 h at 37°C. wt, wild type. (B) One percent rabbit erythrocytes were mixed with culture supernatants in equal proportions, and the mixture was incubated at 37°C for 1 h in microtiter wells. The highest dilution giving rise to visibly detectable lysis was defined as the hemolytic titer.
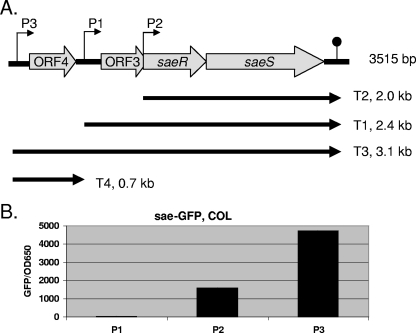
Differential effects of the three different promoters of the sae locus.
Previous studies have shown that the sae locus is composed of three promoters, resulting in four major transcripts (Fig. 3A) (20, 28). Besides containing the two-component system involving saeR and saeS, the 3.1-kb transcript bears two ORFs, ORF3 and ORF4, the functions of which are not yet defined. As we have shown above that rot may have an impact upon sae in the regulation of hla, we wanted to determine if differential levels of expression exist for each of the sae promoters. For this assay, we cloned each of the sae promoters, based on the transcription start sites derived by Steinhuber et al. (28), upstream of the GFPuvr reporter gene into shuttle plasmid pALC1484. Recombinant plasmids containing the sae P1, P2, and P3 promoters (designated pALC4989, pALC4990, and pALC4991, respectively) were then introduced into parental strain COL. Fluorescence assays of bacterial cells every 2 h and overnight disclosed that the sae P3 promoter driving both the 0.4- and the 3.1-kb transcript was the strongest promoter in strain COL, and the results of one typical GFP experiment from post-exponential-phase cultures (OD650 = 1.7) are shown in Fig. 3B. The sae P3 promoter was at least 2 to 5 times stronger than the P2 promoter and more than 10 times stronger than the sae P1 promoter in strain COL. Interestingly, the sae P1 promoter was silent in both wild-type strains. Thus, it is conceivable that the sae P1 promoter may not be a true promoter and hence may represent an mRNA degradation product in the primer extension experiment (communication from Christaine Wolz, Tuebingen, Germany). To confirm that our results were not unique to strain COL, we also introduced pALC4989, pALC4990, and pALC4991 into strains Newman, SH1000, and clinical isolate MW2, with similar results with the sae P3 and P1 promoters (data not shown).
FIG. 3.
Transcription of sae. (A) Schematic drawing of the sae locus modified from the work of Steinhuber et al. (28). Three promoters yielding four overlapping transcripts are indicated. (B) Expression of GFP driven by three different sae promoters (P1, P2, and P3) in wild-type strain COL. Fluorescence as the indicator of promoter activity was measured hourly for 10 h and then overnight. The results of one typical assay (OD650 = 1.7; 8 h) were plotted as mean fluorescence divided by the OD650, using average values of triplicate readings from experiments with three clones from each transformant. These experiments were repeated at least three times with similar results.
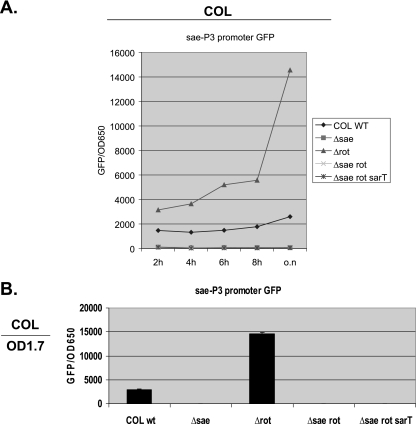
Impact of rot and sae on the sae P3 promoter.
Since we have shown that the effect of rot on hla expression is dependent on sae and that the sae P3 promoter is the strongest promoter within the sae locus, we wanted to examine whether rot represses sae P3 promoter activation, thereby down-modulating hla expression in rot and rot sae mutants (Fig. 1). For this purpose, we introduced pALC4991 containing the sae P3 promoter fused to the gfpuvr reporter gene into wild-type strain COL and its isogenic sae, rot, and sae rot deletion mutants as well as the sae rot sarT triple-deletion mutant. As shown in Fig. 4, a deletion of rot in strain COL resulted in elevated expression of the sae P3 promoter compared with that in the respective parents. In rot sae double mutants, the sae P3 promoter became silent, similarly to what occurred with the vector control in the parental background (data not shown). Deletion of the sarT gene in the triple mutants did not alter sae P3 promoter activity as assessed by GFP-mediated fluorescence, thus indicating that sarT does not play a major regulatory role with the sae P3 promoter in the absence of rot and sae. These data, in conjunction with those on hla expression (Fig. 1 and 2), indicated that rot may repress the sae P3 promoter to down-regulate hla expression.
FIG. 4.
Expression of GFP driven by the sae P3 promoter in COL and its isogenic mutants. (A) Promoter activation was measured every 2 h for 8 h and then overnight (o.n) and plotted as mean fluorescence divided by the OD650, using average values of triplicate readings from experiments with three clones of each transformant. wt, wild type. (B) Representative readings at an OD650 of 1.7 (OD1.7) for one of the experiments are shown.
Interestingly, a deletion of sae alone also resulted in significant down-modulation of sae P3 promoter activity compared with the activity in the parent COL. This finding implied the existence of an autoregulatory circuit in the sae locus, with the major effect being primarily on the strongest sae P3 promoter.
rot is a repressor of sae but not vice versa.
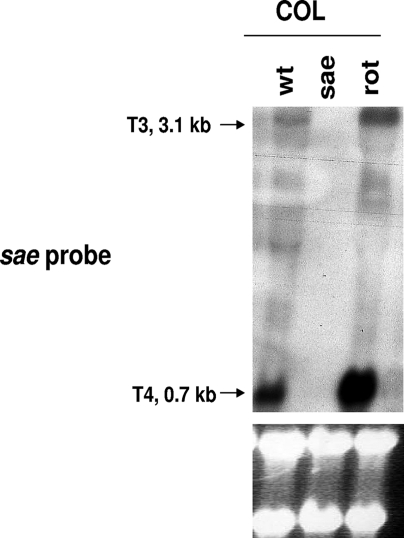
To further confirm the repressive effect of rot on sae promoters, we analyzed sae transcription in both wild-type COL and its isogenic sae and rot mutants by Northern blotting. As shown in Fig. 5, expression of the monocistronic T4 transcript (0.7 kb) and, to a much lesser extent, the 3.1-kb sae T3 transcripts, both driven by the strongest sae P3 promoter, was increased in the rot mutant of COL. Therefore, these data indicated that rot likely represses the sae P3 promoter to down-regulate hla expression.
FIG. 5.
Northern blot analysis of sae transcripts in COL (wild type [wt]) and its isogenic sae and rot mutants. The probe for sae was a 402-bp fragment containing ORF4 of sae, which yielded two transcripts (T3 and T4) driven only by the sae P3 promoter. RNA was harvested from cells grown to an OD650 of 1.7. The RNA gels below show equivalent sample loadings.
To determine if sae regulates rot in hla expression, we detected rot transcripts in sae mutants by Northern blot analysis. The rot-hybridizing bands were preserved in both the parents and the isogenic sae mutant (data not shown). We also transduced pALC5552, a shuttle plasmid containing a 338-bp rot promoter fragment fused with the gfpuvr reporter, into strain COL and its isogenic sae and rot mutants. The GFP values of rot promoter activity were similar between the parents and the isogenic sae mutant, thus confirming that sae does not regulate rot (data not shown). We also confirmed that rot promoter activity was not diminished in the rot mutant, thus indicating that the rot locus was not autoregulatory.
DISCUSSION
The rot locus was first identified by the restoration of hla expression in a transposon mutant with an agr null mutation (17). Remarkably, a transposon-mediated rot mutation by itself was reported to have no effect on alpha-hemolysin expression in strain RN6390, which is partially SigB deficient by virtue of an rsbU nonsense mutation (17). A subsequent microarray study of rot transcription confirmed that rot is a negative regulator of toxins, but only in an agr-negative background (25). In this report, we showed that the rot mutant was able to express hla at a higher level than the parental strain, COL, and that an agr mutation was not necessary to elicit the increased hla response, contrary to the data from McNamara et al. (17). In support of our Northern blot data, we discerned corroborative data on hla expression with GFP transcriptional fusion and also hemolytic titer assays, indicative of increased alpha-toxin activity in rot mutants (Fig. 1 and 2). This discrepancy in hla expression in rot mutants between our data and those of previous studies may be due to differences in the deletion or in the genetic background since our mutants have an in-frame rot deletion in a strain with an intact sigB operon, whereas previous studies with rot were performed with a sigB-deficient strain with a Tn917 insertion in which activation of a cryptic transposon promoter or polar effect on downstream genes may occur.
As the effect of rot on hla expression is likely indirect (25), we hence ascertained if sae, one of the major regulators of hla expression in vitro and in vivo (8, 20, 24), contributes to this regulatory effect. Our results showed that the repression of hla by rot was sae dependent since the rot sae double mutant expressed alpha-hemolysin at a very low level, similar to that of the sae single mutant. This conjecture was supported by the finding that rot repressed sae transcription (Fig. 5).
The sae transcription pattern is complex, with four overlapping transcripts (3.1, 2.4, 2.0, and 0.7 kb) arising from three promoters (20, 28). The sae P3 promoter drives two transcripts: the 3.1-kb transcript containing saeRS and two additional ORFs and the monocistronic 0.7-kb transcript containing ORF4 only (Fig. 3A). Among the three promoters within the sae locus, transcriptional fusion data disclosed that the sae P3 promoter was the strongest in the parental strain, COL, followed by the sae P2 promoter (Fig. 3B). Similar trends were also discerned in other laboratory strains, including SH1000, Newman, and methicillin-resistant S. aureus clinical strain MW2 (data not shown). Importantly, the two sae transcripts of 3.1 and 0.7 kb, originating from the P3 promoter and as detected by a probe comprising ORF4, were augmented in the rot mutant, compared with their levels in the parent, but not in the sae mutant (Fig. 5 and 6). Notably, levels of sae P2 expression as detected by a saeRS probe on Northern blots did not differ significantly between the rot mutant and the parent (data not shown). These results demonstrated that the rot gene product specifically represses the sae P3 promoter, the strongest promoter in the sae locus.
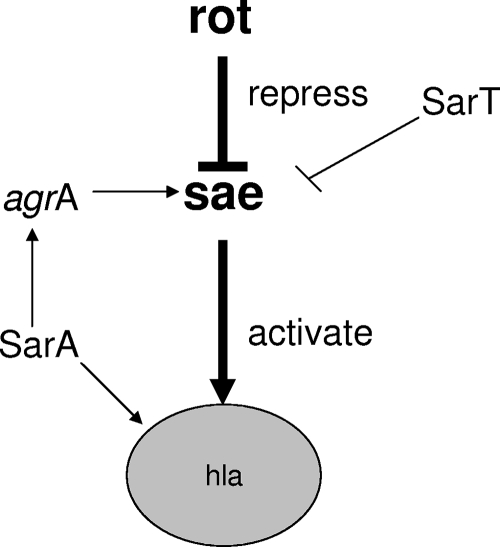
FIG. 6.
Proposed regulation scheme of hla by sae, rot, and sarT.
The rot gene was recently found to encode a 113-residue protein (16) rather than a 166-residue protein as initially thought (17). Previous Northern blots with rot have shown one and possibly two transcripts (25) that were not well delineated. However, recent data by Manna and Ray indicated that the rot gene is driven by three and possibly four different promoters, as deduced by primer extension analysis (16). Our Northern blot analysis with a 338-bp probe containing most of the rot ORF clearly revealed three distinct transcripts, with one at 1.8 kb and two at 0.7 and 0.8 kb (data not shown). An examination of the S. aureus COL genome reveals that there are two ORFs upstream of the rot gene that are transcribed in the same orientation, with one coding for a pseudogene that bears resemblance to a truncated transposase in the 8325-4 genome and the other coding for a 828-bp putative phospholipase gene in the 8325-4 and COL genomes. It is conceivable that the 1.8-kb transcript may comprise both ORFs upstream and rot downstream. Thus, these results revealed that rot may be transcribed in a more complex pattern than has been revealed until now. The finding that rot is flanked by a truncated transposase suggests that rot may be horizontally transferred to S. aureus from a related species.
Another gene, sarT, which is homologous to sarA, was initially identified by genomic scanning and was subsequently shown to be a repressor of hla (27). To define the contribution of sarT to hla expression with regard to sae and rot, we constructed sarT sae rot triple-deletion mutants of strain COL. This triple mutant did not rescue the low level of hla expression seen in the sae rot double mutants (Fig. 1). This finding and the fact that we did not find strong experimental evidence that sarT regulates rot or vice versa (data not shown) suggest that sarT may impact hla expression in a sae-dependent but rot-independent manner (Fig. 1).
It has been suggested that the sae locus is autoregulatory (20). In our promoter activation assay, expression from the P3 promoter, the strongest sae promoter, was almost completely silent in all saeRS deletion mutants of COL (Fig. 4) and SH1000 (data not shown), similar to what occurred in the negative-control vector, thus implying autoregulation of saeRS on the sae P3 promoter. We also checked the native sae P3-P1-P2 promoters in isogenic saeRS strains in COL and RN6390 and also found them to be silent in the sae mutants (data not shown). Accordingly, we confirmed that saeRS likely has a positive feedback on its own promoter.
In this study, we showed that the sae locus is autoregulatory by having an impact upon its own promoter. We found that a mutation in rot on its own can lead to the up-regulation of hla in the absence of any agr mutation, which is contrary to previous data. A similar rot mutation in strain Newman also led to elevated hla expression compared with that in the parental strain. Accordingly, it will not be feasible to explain our finding based only on differences in RNAIII expression levels between strains (unpublished data). Based on our study, it is highly likely that sae is an important checkpoint for hla expression. It has been established previously that both rot and sarT are repressors of hla (17, 27). We have now provided evidence that the repression of hla by rot and, possibly, by sarT is sae dependent. More specifically, Rot represses the sae P3 promoter, the strongest promoter within the sae locus, to control hla expression. As the sae P3 transcript also encodes ORF4 (Fig. 3), it will be of interest to evaluate the role of ORF4 in regulating SaeRS expression. Additionally, investigations on whether the effect of Rot on sae is direct or indirect will be of interest.
Editor: A. Camilli
Footnotes
Published ahead of print on 3 January 2008.
REFERENCES
- 1.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 706887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y.-Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Microbiol. Lett. 16491-9. [DOI] [PubMed] [Google Scholar]
- 3.Cheung, A. L., K. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222511-514. [DOI] [PubMed] [Google Scholar]
- 4.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 16853-58. [DOI] [PubMed] [Google Scholar]
- 5.Giraudo, A. T., C. G. Raspanti, A. Calzolari, and R. Nagel. 1994. Characterization of a Tn551 mutant of Staphylococcus aureus defective in the production of several exoproteins. Can. J. Microbiol. 40677-681. [DOI] [PubMed] [Google Scholar]
- 6.Giraudo, A. T., A. Calzolari, A. A. Cataldi, C. Bogni, and R. Nagel. 1999. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 17715-22. [DOI] [PubMed] [Google Scholar]
- 7.Goerke, C., U. Fluckiger, A. Steinhuber, V. Bisanzio, M. Ulrich, M. Bischoff, J. M. Patti, and C. Wolz. 2005. Role of Staphylococcus aureus global regulators sae and σB in virulence gene expression during device-related infection. Infect. Immun. 733415-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 401439-1447. [DOI] [PubMed] [Google Scholar]
- 9.Grimminger, F., F. Rose, U. Sibelius, M. Meinhardt, B. Potzsch, R. Spriestersbach, S. Bhakdi, N. Suttorp, and W. Seeger. 1997. Human endothelial cell activation and mediator release in response to the bacterial exotoxins Escherichia coli hemolysin and staphylococcal alpha-toxin. J. Immunol. 1591909-1916. [PubMed] [Google Scholar]
- 10.Harraghy, N., J. Kormanec, C. Wolz, D. Homerova, C. Goerke, K. Ohlsen, S. Qazi, P. Hill, and M. Herrmann. 2005. sae is essential for expression of the staphylococcal adhesins Eap and Emp. Microbiology 1511789-1800. [DOI] [PubMed] [Google Scholar]
- 11.Kahl, B., M. Goulian, W. Van Wamel, M. Herrmann, S. Simon, G. Kaplan, G. Peters, and A. L. Cheung. 2000. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line derived from a cystic fibrosis patient. Infect. Immun. 685385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kehoe, M., J. Duncan, T. Foster, N. Fairweather, and G. Dougan. 1983. Cloning, expression, and mapping of the Staphylococcus aureus α-hemolysin determinant in Escherichia coli K-12. Infect. Immun. 411105-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kernodle, D. S., R. K. Voladri, B. E. Menzies, C. C. Hager, and K. M. Edwards. 1997. Expression of an antisense hla fragment in Staphylococcus aureus reduces alpha-toxin production in vitro and attenuates lethal activity in a murine model. Infect. Immun. 65179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 15.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manna, A. C., and B. Ray. 2007. Regulation and characterization of rot transcription in Staphylococcus aureus. Microbiology 1531538-1545. [DOI] [PubMed] [Google Scholar]
- 17.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 1823197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novick, R. P. 1990. The staphylococcus as a molecular genetic system, p. 1-40. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH, New York, NY.
- 19.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 481429-1449. [DOI] [PubMed] [Google Scholar]
- 20.Novick, R. P., and D. Jiang. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 1492709-2717. [DOI] [PubMed] [Google Scholar]
- 21.Oshida, T., M. Sugai, H. Komatsuzawa, Y. M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel, A. H., P. Nowlon, E. D. Weavers, and T. Foster. 1987. Virulence of protein A-deficient and alpha-toxin-deficient mutants of Staphylococcus aureus isolated by allele replacement. Infect. Immun. 553103-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rampone, H., G. L. Martinez, A. T. Giraudo, A. Calzolari, and R. Nagel. 1996. In vivo expression of exoprotein synthesis with a Sae mutant of Staphylococcus aureus. Can. J. Vet. Res. 60237-240. [PMC free article] [PubMed] [Google Scholar]
- 24.Rogasch, K., V. Ruhmling, J. Pane-Farre, D. Hoper, C. Weinberg, S. Fuchs, M. Schmudde, B. M. Broker, C. Wolz, M. Hecker, and S. Engelmann. 2006. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 1887742-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Said-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 94133-138. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT: a repressor of alpha-hemolysin synthesis in Staphylococcus aureus. Infect. Immun. 694748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinhuber, A., C. Goerke, M. G. Bayer, G. Doring, and C. Wolz. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 1856278-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]