Abstract
The outer membrane proteins (OMPs) of bacterial pathogens are essential for their growth and survival and especially for attachment and invasion of host cells. Since the outer membrane is the interface between the bacterium and the host cell, outer membranes and individual OMPs are targeted for development of vaccines against many bacterial diseases. Whole outer membrane fractions often protect against disease, and this protection cannot be fully reproduced by using individual OMPs. Exactly how the interactions among individual OMPs influence immunity is not well understood. We hypothesized that one OMP rich in T-cell epitopes can act as a carrier for an associated OMP which is poor in T-cell epitopes to generate T-dependent antibody responses, similar to the hapten-carrier effect. Major surface protein 1a (MSP1a) and MSP1b1 occur as naturally complexed OMPs in the Anaplasma marginale outer membrane. Previous studies demonstrated that immunization with the native MSP1 heteromer induced strong immunoglobulin G (IgG) responses to both proteins, but only MSP1a stimulated strong CD4+ T-cell responses. Therefore, to test our hypothesis, constructs of CD4+ T-cell epitopes from MSP1a linked to MSP1b1 were compared with individually administered MSP1a and MSP1b1 for induction of MSP1b-specific IgG. By linking the T-cell epitopes from MSP1a to MSP1b1, significantly higher IgG titers against MSP1b1 were induced. Understanding how the naturally occurring intermolecular interactions between OMPs influence the immune response may lead to more effective vaccine design.
Outer membrane proteins (OMPs) mediate interactions between microbial pathogens and their hosts. Within the bacterial outer membrane, proteins exist in homomeric or heteromeric complexes that are dependent on covalent as well as noncovalent interactions. These complexes are essential structural components and also mediate central events in bacterial physiology and pathogenesis. For example, the extensive disulfide cross-linking of chlamydial major outer membrane proteins (MOMPs) (15, 22, 40) provides structural stability in the absence of peptidoglycan (14), and disulfide interactions among MOMPs regulate porin function (3). The Escherichia coli outer membrane contains a complex consisting of a large β-barrel protein, Imp, and a small lipoprotein, RlpB, and both proteins are required for lipopolysaccharide assembly (39). Furthermore, protein complexes within the membrane, including sophisticated macromolecular structures, such as secretion systems and pili, are essential for attachment, invasion, and survival within host cells (33).
How OMP interactions affect induction of immunity is poorly understood. Numerous vaccine studies have been directed at individual OMPs, but these OMPs had limited success compared to immunization using whole bacteria or intact outer membranes. For example, immunization with outer membranes from Chlamydia trachomatis, Francisella tularensis, Haemophilus influenzae, and Anaplasma marginale protects against bacterial challenge, whereas immunization with MOMPs from these bacteria does not protect against bacterial challenge (1, 8, 10-12, 16, 26, 29, 36). The immunologic importance of OMP complexes is demonstrated by the induction of protection against Leptospira by immunization using OmpL1 and LipL41 expressed simultaneously in the context of the E. coli membrane. In contrast, immunization with either protein alone in the membrane context or as part of a mixture of non-membrane-associated proteins is not protective (13). Understanding the basis for differences in the immunogenicity and efficacy of complexed OMPs and individual OMPs would enhance design and development of vaccines for multiple bacterial pathogens.
We have hypothesized that bacterial OMPs act as a carrier-hapten pair, with one OMP containing essential T-cell epitopes that induce CD4+ T-lymphocyte help for antibody production against B-cell epitopes on a different but physically associated OMP. B-lymphocyte somatic hypermutation and class switching, which are necessary for the induction of high-affinity immunoglobulin G (IgG), are driven by cognate and cytokine help provided by CD4+ T cells. This help requires that the peptide recognized by the CD4+ T cell is physically linked to the antigen initially recognized by the B cell, which can then present this CD4+ T-cell epitope to the T cell via the major histocompatibility complex (MHC) class II pathway following internalization of the antigen bound to the B-cell receptor. Thus, the B- and T-cell epitopes can be present in an individual OMP or, alternatively, can be present on different OMPs that are physically associated through covalent or noncovalent interactions within the outer membrane and internalized by the antigen-presenting B cell as a complex.
We determined the potential importance of immunologically linked recognition between OMPs using A. marginale major surface protein 1 (MSP1) as a model. In the St. Maries strain of A. marginale, native MSP1 occurs in the outer membrane as a heteromeric complex of covalently and noncovalently associated MSP1a and MSP1b1 (19, 38). Immunization or infection results in development of antibody to both proteins; however, CD4+ T-cell epitope mapping studies identified epitopes predominantly on MSP1a (6), raising the possibility that the B lymphocytes producing antibody to MSP1b1 receive CD4+ T-cell help via linked recognition. Here we report testing the hypothesis that constructs of CD4+ T-cell epitopes from MSP1a linked to MSP1b1 induce stronger antibody responses against MSP1b1 than an equimolar amount of the two recombinant proteins induces.
MATERIALS AND METHODS
Isolation of A. marginale cells and native MSP1 complex.
Blood collected from an animal infected with the St. Maries strain of A. marginale was washed three times in phosphate-buffered saline (PBS), removing the buffy coat each time, and frozen at −80°C. Infected erythrocytes were washed repeatedly with PBS (pH 7.2), centrifuged at 30,000 × g for 20 min at 4°C to remove hemoglobin, resuspended in PBS containing 1× Complete mini protease inhibitors (Roche, Indianapolis, IN), and stored at −80°C until they were used. A. marginale cells were isolated as previously described (28), with minor modifications. Erythrocyte membranes were disrupted by sonication at an output control setting of 4 and a 100% duty cycle with a Sonifier 350 cell disruptor (VWR Scientific Inc., West Chester, PA), releasing A. marginale cells. After sonication, centrifugation was performed at 30,000 × g for 20 min at 4°C, and the pelleted A. marginale cells were resuspended in PBS with 1× Complete mini protease inhibitors. The protein concentration was determined using a Quick Start Bradford protein assay (Bio-Rad, Hercules, CA). The MSP1 complex was isolated essentially as described previously (27, 28, 38), with some modifications. Sepharose beads were coupled to MSP1a-specific monoclonal antibody (MAb) Ana22B1 (20) through cyanogen bromide chemistry (Sigma, St. Louis, MO). MAb-coupled Sepharose beads were washed three times with TEN (0.02 M Tris HCl, 0.005 M EDTA, 0.1 M NaCl, 0.015 M NaN3; pH 7.6) containing 1% Nonidet P40 (NP-40). A. marginale cells were solubilized in TEN containing 1% NP-40 by sonicating them twice for 30 s at an output control setting of 10 with a 100% duty cycle and were incubated overnight at 4°C with the MAb Ana22B1-coupled Sepharose beads with continuous rotation. The beads were washed at least three times with TEN-1% NP-40 by mixing and centrifugation at 1,000 × g for 5 min at 4°C. Elution was performed in the same way by washing the preparation five times with 1 ml of 3 M potassium thiocyanate (KSCN) in TEN (pH 8.8). The eluted fractions were pooled, dialyzed against PBS, and concentrated using an Amicon Ultra 15 spin column (Millipore, Billerica, MA) with a 10,000-Da cutoff.
To determine the purity of the isolated MSP1 complex, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 4 to 20% gradient mini gel (Bio-Rad) was performed. The gel was either transferred to nitrocellulose for immunoblotting or stained with Coomassie brilliant blue, and excised bands were subjected to liquid chromatography followed by tandem mass spectrometry (LC-MS/MS). Nitrocellulose membranes were blocked overnight in I-Block (Applied Biosystems, Foster City, CA) with 0.5% Tween 20 and incubated for 2 h with MSP1a-specific MAb Ana22B1, MSP1b-specific MAb AmR38A6, MSP2-specific MAb 115/362.17.19, MSP3-specific MAb 115/152.20.19, or MSP5-specific MAb AnaF16C1 (20, 34, 38). Most MAbs were used at a concentration of 2 μg/ml; the only exception was MAb AmR38A6, which was used at a concentration of 10 μg/ml. Chemiluminescent detection was performed with the Western-Star immunodetection system (Applied Biosystems).
Detection of the MSP1 complex by LC-MS/MS.
Coomassie blue-stained gel slices were processed as described previously (35), with minor modifications. Individual gel slices were reduced, alkylated, and digested with trypsin as described previously (18, 19). Peptides were extracted from the gel pieces and submitted to the Proteomics Core Laboratory, Center for Integrated Biotechnology (Washington State University, Pullman, WA), for LC-MS/MS analysis. LC-MS/MS was performed using conditions described previously (19). MASCOT software (www.matrixscience.com) was used for database searches of peptide mass fingerprints against the A. marginale strain St. Maries genome sequence.
Plasmid construction and expression of recombinant proteins.
A strategy of overlapping PCR combined with restriction enzyme cloning was utilized to generate a construct containing two MSP1a repeats, designated J and B (30) (Fig. 1A, region R), and the previously identified CD4+ T-cell epitopes from MSP1a (6) (regions F2 and F3) (Fig. 1A). Through a series of PCRs using overlapping primers and St. Maries strain genomic DNA, the R region was attached to regions F2 and F3; the primers used were JB-F2 (GCTGATAGCTCGTCAGCGGGTG), F2-JB-R (AATCTTCAAACCTTGTAGCCCTCCTAATTGAGACGATGTA), F2-F (GGGCTACAAGGTTTGAAGATTGGCGA), and b-F3R (TGCTTGTCGTCTTCTGTCATCTGTGTAGTAGTGTCCGAAG). The resulting construct was amplified with the same forward primer (JB-F2) and a reverse primer that included a FLAG tag (indicated by bold type) at the 3′ end, primer F2F3FlagR (CTACTATTTGTCGTCGTCGTCTTTATAGTCCTGTGTAGTAGTGTCCGAAG), and it was cloned into the pTrcHis TOPO vector (Invitrogen) for expression and purification (Fig. 1A). This construct encodes what is referred to as the T-cell epitope protein. Recombinant MSP1b1 (Fig. 1B) containing a six-His tag at the N-terminal end and a FLAG tag (DYKDDDDK) (21) at the C-terminal end was expressed as described previously (19). The T-cell epitope protein and MSP1b1 are referred to below as the unlinked proteins.
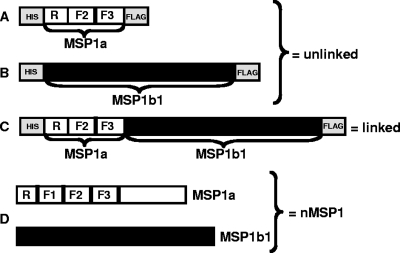
FIG. 1.
Constructs used for immunization. Recombinant proteins contained epitopes used for purification at the N terminus (six-His tag) and the C terminus (FLAG tag) of the recombinant proteins. The R region is the repeat region of MSP1a; the F1, F2, and F3 regions are the areas in MSP1a used to map T-cell epitopes, which localize to F2 and F3 (6). (A) Recombinant T-cell epitope protein containing the F2-F3 region of MSP1a. (B) Recombinant full-length MSP1b1. (C) Recombinant linked protein containing the F2-F3 region of MSP1a linked to full-length MSP1b1. (D) Native MSP1 (nMSP1) consisting of naturally complexed full-length MSP1a and full-length MSP1b1. Open boxes indicate MSP1a sequences, black boxes indicate MSP1b1 sequences, and gray boxes indicate His and FLAG epitopes.
The T-cell epitope construct (Fig. 1A) was amplified with primers that added BamHI sites to both ends of the R-F2-F3 construct (primers BH1-JBF2 [GCGCGGATCCGATGATAGCTCGTTAGCGGGTGGTT] and BH1F3R [GCGCGGATCCCTGTGTAGTAGTGTCCGAAGGCTTCG]), which allowed cloning into the BamHI site upstream from MSP1b1 in the pTrcHis TOPO vector, generating the linked construct (Fig. 1C) which encoded the F2 and F3 T-cell epitopes linked to MSP1b1 and was expressed as a single protein.
All three recombinant proteins were cloned into pTrcHis TOPO, expressed in E. coli, and purified, first by using ProBond nickel resin (Invitrogen) and then by using anti-FLAG immunoaffinity chromatography (Sigma-Aldrich, St. Louis, MO). Protein concentrations were determined using the Quick Start Bradford protein assay (Bio-Rad). The purity of recombinant proteins was determined by SDS-PAGE and Coomassie blue staining, and the product that was the correct size was visualized after immunoblotting using a horseradish peroxidase-labeled FLAG-specific MAb (Sigma-Aldrich) or negative control MAb Tryp-1E1 specific for Trypanosoma brucei. Chemiluminescent detection was performed with the Western-Star immunodetection system (Applied Biosystems).
Full-length MSP1b1 was also cloned into the pBad TOPO vector (Invitrogen) and expressed in E. coli. This protein has a V5 epitope instead of the FLAG epitope and was used in proliferation assays to avoid stimulation due to the FLAG epitope. The MSP1b1-V5 recombinant protein was treated like the MSP1b1-FLAG recombinant protein except that it was purified with a V5-specific MAb instead of a FLAG-specific MAb. A. marginale OMP10 was expressed and purified (23) using the same system and was used as a negative control.
Animals and immunization.
Twelve 6-month-old Holstein calves that were seronegative for A. marginale were used in this study. The bovine lymphocyte antigen-DRB3 alleles of the calves were determined by the PCR-restriction fragment length polymorphism method (37) and sequencing of exon 2 of the DRB3 gene (24, 31). The animals were divided into three groups, and each group consisted of four animals with one of the following DRB3 alleles: DRB3*1501/*1202, DRB3*1501/*14011, DRB3*1501/*0101, or DRB3*1501/*1501. The three groups were immunized four times subcutaneously at 3-week intervals. Each animal received 175 pmol of protein in complete Freund's adjuvant for the first immunization, followed by three immunizations of 175 pmol of protein each in incomplete Freund's adjuvant. Group 1 received 30 μg of native MSP1 complex as a control to detect IgG and CD4 T-cell responses. To ensure that all animals received the same number of epitopes, the amount of protein given to each animal was adjusted to 175 pmol (corresponding to 30 μg of native MSP1 complex). Thus, the amounts of all recombinant proteins were adjusted to 175 pmol, which meant that group 2 received 23 μg of the linked protein (130 kDa) and group 3 received 8 μg of the T-cell epitope protein (45 kDa) plus 17.5 μg of MSP1b1 (100 kDa). For groups 1 and 2, the antigen was divided into two aliquots. One aliquot was administered in the left shoulder, and one aliquot was administered in the right shoulder. Group 3 received one antigen in the left shoulder and the other antigen in the right shoulder. Peripheral blood mononuclear cells (PBMC) and sera were collected at 3-week intervals and stored in liquid nitrogen (PBMC) or at −20°C (sera).
Lymphocyte proliferation assays.
Proliferation assays were carried out in round-bottom 96-well plates (Costar) for 6 days, essentially as described previously (7-9). PBMC (2 × 105 cells) were cultured in triplicate wells with antigen and complete RPMI 1640 medium in a 100-μl (total volume) mixture. The antigens consisted of the A. marginale St. Maries strain bacteria, a panel of overlapping peptides (30 amino acids each, overlapping by 10 amino acids) spanning the F2-F3 region of MSP1a (6), as well as recombinant MSP1b1. Membranes isolated from uninfected red blood cells, an unrelated peptide from Babesia bovis rhoptry-associated protein 1 (RAP-1) (25), and recombinant A. marginale OMP10 (23) were used as negative controls. Antigens were used at concentrations of 0.1 and 1 μg/ml. In a second assay, 0.01, 0.1, and 1 μg/ml of MSP1b1 expressed with a V5 epitope or OMP10 expressed in the same vector were tested with PBMC. PBMC were radiolabeled for the last 18 h of culture with 0.25 μCi [3H]thymidine (Dupont, New England Nuclear, Boston, MA) and harvested onto glass filters. Radionucleotide incorporation was measured with a Beta-plate 1205 liquid scintillation counter (Wallac, Gaithersburg, MD). The results were expressed as the stimulation index,, which was calculated by dividing the mean counts per minute (cpm) of PBMC cultured with antigen by the mean cpm of PBMC cultured with medium alone. A statistical analysis was performed by analysis of variance (ANOVA) using SAS, version 9.1 (SAS Institute Inc., Cary, NC), and a P value of <0.05 was considered statistically significant.
Immunoblotting to determine serum titer.
To determine IgG titers to MSP1a and MSP1b, immunoblotting was performed. Sonicated pellets of A. marginale-infected erythrocytes stored in proteinase inhibitor buffer at −80°C were thawed and mixed with SDS-PAGE sample buffer. A total of 2 × 109 infected erythrocytes were separated using a preparative well 10% Criterion gel (Bio-Rad) and were transferred to nitrocellulose. The nitrocellulose membrane was incubated overnight in I-block (Applied Biosystems), cut into individual strips, and incubated with bovine serum for 2 h at room temperature. Sera from the immunized animals were tested using threefold dilutions from 1:100, to 1:300,000. Horseradish peroxidase-labeled goat anti-bovine IgG (1:4,000; Serotec, Raleigh, NC) was used as the secondary antibody, and blots were incubated for 1 h at room temperature. Blots were washed with I-block and developed using the ECL Western blotting detection system (Amersham Biosciences, Piscataway, NJ). Bands for MSP1a and MSP1b were observed, and the endpoint titer was expressed as the reciprocal of the highest dilution at which a band was visible. This experiment was repeated with frozen and thawed sera diluted 1:5,000, 1:10,000, 1:15,000, 1:20,000, 1:30,000, 1:40,000, 1:50,000, and 1:60,000 (for the native MSP1 and unlinked protein groups) or diluted 1:20,000, 1:40,000, 1:60,000, 1:80,000, 1:100,000, and 1:120,000 (for the linked construct group). Preimmune sera and uninfected red blood cell membranes were used as negative controls. To test for normality of the serum titration data, the Shapiro-Wilk test and the modified Levene equal variance test were performed, which indicated that the data were approximately normal for both experiments. A nonparametric Wilcoxon ranked sum test comparing groups immunized with linked and unlinked recombinant proteins was performed with the antibody titer data. A P value of <0.05 was considered significant.
RESULTS
Isolation of native MSP1 complex, expression of recombinant proteins, and animal immunization.
To determine if linking the MSP1a T-cell epitopes to MSP1b1 enhanced the immune response to MSP1b1, animals were immunized with the linked recombinant protein, unlinked recombinant proteins, or the native MSP1 complex. Schematic diagrams of the proteins used for immunization are shown in Fig. 1. The T-cell epitopes present on MSP1a were identified in a previous study after the hydrophilic domain of MSP1a was expressed as three regions designated F1, F2, and F3 (Fig. 1D) (6). By using a panel of overlapping peptides, the F2-F3 region was identified as a domain with multiple, strong CD4+ T-cell epitopes (6), and therefore the F2-F3 region was used for preparation of our constructs.
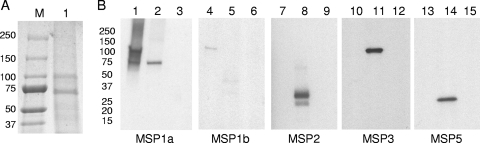
The first group of animals was immunized with the native MSP1 complex. This complex contained full-length MSP1a (Fig. 1D), including the F2-F3 T-cell epitopes, and full-length MSP1b1 (Fig. 1D). The complex was isolated by immunoaffinity chromatography with an MSP1a-specific MAb, MAb Ana22B1, which recognized the repeat region (region R) (2). Coomassie blue staining of the native MSP1 complex separated by SDS-PAGE revealed two distinct bands at 75 and 100 kDa (Fig. 2A). We have previously demonstrated the presence of only MSP1b1 (there is a second transcribed gene, msp1β2) in the MSP1 complex (19). The 75-kDa band was identified by LC-MS/MS as MSP1a with a MASCOT score of 1706, and the 100-kDa band was identified as MSP1b1 with a MASCOT score of 1622. To further rule out the presence of other surface proteins in the purified MSP1 complex, MAbs against three of the major surface proteins of A. marginale were used to determine if any of these proteins were coprecipitated with the native MSP1 complex. Only MSP1a and MSP1b were detected in the native MSP1 complex by immunoblotting with the panel of MAbs (Fig. 2B, lanes 1 and 4). MSP2, MSP3, and MSP5 were detected in the outer membrane fraction (Fig. 2B, lanes 8, 11, and 14) but not in the purified MSP1 complex (Fig. 2B, lanes 7, 10, and 13). Since MSP2 is very abundant and serologically immunodominant and MSP3 and MSP5 are also serologically immunodominant in the outer membrane (18), this result indicates that there was very little contamination with other OMPs.
FIG. 2.
Isolation and detection of native MSP1 complex. (A) Purified native MSP1 complex separated on a 4 to 20% gradient SDS-PAGE gel and stained with Coomassie blue. Lane M, molecular size markers (molecular masses [in kDa] are indicated on the left); lane 1, 23 μg purified native MSP1 complex. (B) Detection of native MSP1 complex with MSP-specific MAbs. Lanes 1, 4, 7, 10, and 13, 0.1 μg of native MSP1 complex; lanes 2, 5, 8, 11, and 14, 10 μg of purified A. marginale strain St. Maries outer membranes; lanes 3, 6, 9, 12, and 15, 0.5 μg of uninfected erythrocyte membranes. The sizes (in kDa) of molecular size markers are indicated on the left. The blot was probed with MAb Ana22B1 specific for MSP1a (lanes 1 to 3), MAb AmR38A6 specific for MSP1b (lanes 4 to 6), MAb 115/362.17.19 specific for MSP2 (lanes 7 to 9), MAb 115/152.20.19 specific for MSP3 (lanes 10 to 12), and MAb AnaF16C1 specific for MSP5 (lanes 13 to 15).
The amount of protein given to each animal was adjusted on a molar basis. To standardize the molar amount for each protein, we assumed that MSP1a and MSP1b1 are present in equimolar amounts, because the actual molar amounts of MSP1a and MSP1b1 in the heteromeric complex have not been determined (19). Taking into account the amounts of MSP1a (75 kDa) and MSP1b1 (100 kDa) in 30 μg of native MSP1 complex, there should have been 200 pmol of MSP1a and 150 pmol of MSP1b1 or an average of 175 pmol per protein. We then adjusted the amounts of all recombinant proteins so that each animal was immunized with 175 pmol of protein.
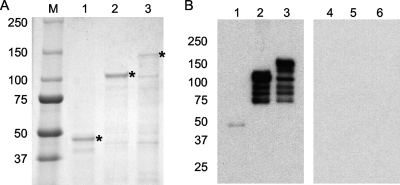
The second group of animals was immunized with 23 μg of linked protein (Fig. 1C), corresponding to 175 pmol of protein. The linked protein contained the CD4+ T-cell epitopes from MSP1a (F2-F3) linked to MSP1b1. After purification and separation by SDS-PAGE, a major 130-kDa band was present (Fig. 3A, lane 3). The linked recombinant protein reacted with a MAb specific for the FLAG epitope (Fig. 3B, lane 3) but not with an unrelated MAb against T. brucei (Fig. 3B, lane 6). The smaller bands observed were due to degradation of the larger protein, as indicated by their reactivity with the anti-FLAG MAb.
FIG. 3.
Purification and detection of recombinant proteins. (A) One microgram of purified recombinant protein per lane separated on a 4 to 20% gradient SDS-PAGE gel and stained with Coomassie blue. Lane M, molecular size markers (molecular masses [in kDa] are indicated on the left); lane 1, recombinant T-cell epitopes; lane 2, recombinant MSP1b1; lane 3, recombinant linked protein. Asterisks indicate the position of the full-length protein. (B) Detection of recombinant proteins. Lanes 1 and 4, recombinant T-cell epitopes; lanes 2 and 5, recombinant MSP1b1; lanes 3 and 6, recombinant linked protein. Lanes 1 to 3 were incubated with a FLAG-specific MAb, and lanes 4 to 6 were incubated with an unrelated MAb specific for T. brucei.
The third group of animals was immunized with the unlinked proteins and received 8 μg of the MSP1a T-cell epitope protein (Fig. 1A), as well as 17.5 μg of MSP1b1 (Fig. 1B). After purification, separation by SDS-PAGE, and Coomassie blue staining, a 45-kDa band was present for the MSP1a T-cell epitope protein (Fig. 3A, lane 1) and a 100-kDa band was present for MSP1b (Fig. 3A, lane 2). Both the MSP1a T-cell epitope protein and MSP1b1 reacted in a Western blot with a MAb specific for the FLAG epitope (Fig. 3B, lanes 1 and 2) but not with the unrelated MAb against T. brucei (Fig. 3B, lanes 4 and 5). The larger proteins were more susceptible to degradation, and smaller bands were also observed for MSP1b1.
MSP1a-specific T-cell responses.
To determine if the constructs effectively primed and induced memory T-cell responses, a panel of overlapping 30-amino-acid peptides spanning the F2-F3 region of MSP1a (6) was used in proliferation assays. The T-cell epitopes present in MSP1a have been mapped previously (6). The F2-F3 region represents a domain that is rich in T-cell epitopes and contains at least five epitopes in the span of 215 amino acids (6). Three weeks after the first immunization most animals responded to A. marginale (data not shown). Negative control B. bovis RAP-1 peptide and MSP1a peptide F2-5 did not elicit a response. T-cell responses to peptide F2-5 have been associated with the bovine lymphocyte antigen DRB3*1101 allele (24), an allele not represented in the vaccinated population. Therefore, both peptides served as negative controls. Animal 4885 (in the linked group) consistently exhibited high background proliferation, and we did not observe significant proliferation against any of the peptides. The remaining vaccinees all responded to peptide F3-3, which is presented by the DRB3*1501-linked DQ gene products (24). There were no statistically significant differences in proliferation against any of the peptides when all three groups were compared using either concentration of antigen, as determined by ANOVA. Table 1 shows data for 1 μg per ml of peptide. Thus, any difference detected in the amount of antibody induced was not due to intrinsic differences in the MSP1a T-cell response between groups.
TABLE 1.
CD4+ T-cell epitopes recognized by immunized animals as determined by proliferation assays
| Protein | Animal | Stimulation indexa
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RAP-1 peptideb | F2-1c | F2-2 | F2-3 | F2-4 | F2-5 | F3-1 | F3-2 | F3-3 | F3-4 | F3-5 | F3-6 | ||
| Native MSP1 | 4841 | 1 | 1 | 13 | 7 | 1 | 1 | 1 | 4 | 9 | 1 | 1 | 2 |
| 4862 | 1 | 1 | 25 | 2 | 11 | 2 | 2 | 1 | 22 | 1 | 2 | 1 | |
| 4882 | 1 | 1 | 73 | 2 | 15 | 1 | 1 | 8 | 11 | 1 | 1 | 1 | |
| 4906 | 1 | 1 | 14 | 3 | 4 | 1 | 2 | 1 | 17 | 1 | 1 | 1 | |
| Linked | 4859 | 1 | 2 | 6 | 1 | 4 | 2 | 3 | 1 | 15 | 8 | 1 | 1 |
| 4904 | 4 | 1 | 27 | 11 | 9 | 2 | 34 | 1 | 116 | 1 | 17 | 2 | |
| 4885 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 4919 | 1 | 1 | 3 | 1 | 4 | 1 | 2 | 1 | 26 | 1 | 1 | 1 | |
| Unlinked | 4911 | 1 | 36 | 33 | 10 | 33 | 2 | 3 | 19 | 65 | 35 | 1 | 4 |
| 4916 | 4 | 17 | 61 | 34 | 43 | 3 | 38 | 6 | 63 | 9 | 37 | 2 | |
| 4914 | 1 | 2 | 8 | 2 | 6 | 2 | 8 | 1 | 5 | 6 | 1 | 1 | |
| 4932 | 1 | 4 | 8 | 1 | 4 | 1 | 1 | 1 | 14 | 1 | 1 | 1 | |
PBMC were obtained 3 weeks after the fourth immunization and used in a 6-day proliferation assay with 1 μg/ml of the indicated peptides. Stimulation indices were calculated by dividing the mean cpm of PBMC cultured with antigen by the mean cpm of PBMC cultured with medium. The data were analyzed by ANOVA using SAS, version 9.1. Significant responses are indicated by bold type.
RAP-1 peptide is a negative control peptide derived from B. bovis RAP-1.
F2-1 to F2-5 and F3-1 to F3-6 are peptides in the F2-F3 region of A. marginale MSP1a (6).
MSP1b-specific B-cell responses.
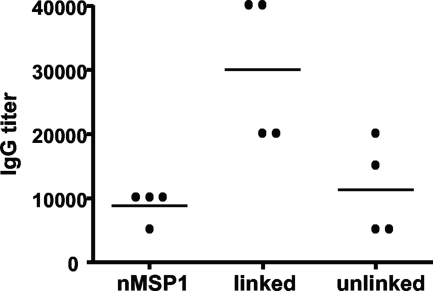
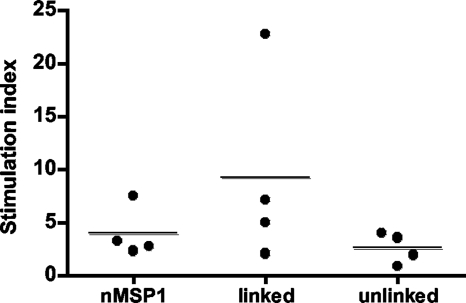
Antibody titers were measured to determine if linking the T-cell epitopes from MSP1a to MSP1b1 would induce a greater MSP1b-specific antibody response using sera diluted 1:100 to 1:300,000. We used a nonparametric ranked sum test to analyze significance because titer data are not continuous. Following the full set of immunizations, the group immunized with the linked construct had significantly greater MSP1b1-specific antibody responses (P = 0.031) than the group that received the unlinked constructs (data not shown). We then repeated this experiment using a narrower range of dilutions (Fig. 4). The same result was obtained; the titers in the group that received the linked construct were significantly higher than those in the group that received the unlinked construct (P = 0.031). Both assays revealed considerable variation between animals, which is often the case with outbred individuals.
FIG. 4.
MSP1b-specific IgG responses in immunized animals. Western blotting was performed to determine the serum titers specific for MSP1b1. Frozen and thawed sera were diluted 1:5,000 to 1:80,000. The IgG titers are expressed as the reciprocal of the lowest dilution at which a band was observed for MSP1b at 100 kDa. Each dot on the graph represents the serum titer for an individual, and the horizontal lines indicate the mean group titers. Significantly higher titers of MSP1b1-specific IgG were obtained for the group immunized with the linked construct than for the group immunized with the unlinked construct, as determined by the Wilcoxon ranked sum test (P = 0.031). nMSP1, native MSP1.
MSP1b1-specific T-cell responses.
To confirm that the higher antibody titer could be attributed to the linkage of the MSP1a T-cell epitopes to MSP1b1 rather than to differences in the T-cell response to MSP1b1 among groups, PBMC were tested by performing proliferation assays with recombinant MSP1b1. MSP1b1 was expressed with a V5 epitope instead of the FLAG epitope to eliminate potential T-cell responses specific for the FLAG epitope. OMP10, which was expressed from the same vector, was used as a negative control. We determined the ratio of stimulation with recombinant MSP1b1 to stimulation with recombinant OMP10 to obtain a stimulation index to control for any background proliferation from contaminating E. coli products. There were no statistically significant differences in proliferation against MSP1b1 when all three groups were compared, at either concentration of antigen. The results for 1 μg/ml of antigen are shown in Fig. 5. Animal 4919 had the highest T-cell response to MSP1b1, although the response did not correlate with the highest antibody titers against MSP1b, which were the titers for animals 4859 and 4904.
FIG. 5.
Proliferative responses of PBMC to recombinant MSP1b1. PBMC obtained 3 weeks after the fourth immunization were cultured for 6 days in triplicate with 0.01, 0.1, and 1 μg/ml recombinant MSP1b1 or recombinant OMP10 expressed from the same vector. To control for any proliferation due to contaminating E. coli products, the stimulation index was determined by dividing the mean cpm induced by MSP1b1 by the mean cpm induced by OMP10. The data for 1 μg/ml antigen are shown, and statistically significant differences were not detected between groups, as determined by ANOVA (P = 0.25).
DISCUSSION
A hapten is a small organic molecule or peptide devoid of T-cell epitopes that is not capable of inducing an antibody response by itself but must be coupled to a larger carrier protein to induce such a response. In contrast, OMPs, which are larger, are more likely to have T-cell epitopes. However, the T-cell epitopes on an individual OMP may not be sufficient to induce a strong T-cell response or cannot be efficiently presented by a given MHC class II haplotype. We hypothesized that one bacterial OMP known to contain multiple T-cell epitopes can act as a carrier for another OMP which is a weak T-cell antigen. The results presented here show that linkage of the MSP1a T-cell epitope protein, which is rich in T-cell epitopes, to MSP1b1 does have a significant immunologic effect and leads to higher levels of production of antibody against MSP1b1. By providing MSP1a-specific T-cell help to B cells, the IgG response to MSP1b1 was significantly higher despite the lack of an intrinsic difference in MSP1a T-cell immunogenicity. In addition, the two unlinked proteins were administered at separate sites to ensure that the MSP1a T-cell epitopes and MSP1b1 would not be taken up by the same antigen-presenting cells and mimic linked recognition. We can thus accept the hypothesis since a higher IgG response to MSP1b1, which cannot be explained by differences in the T-cell response to MSP1b1, was induced after linkage of the MSP1a T-cell epitopes to MSP1b1.
The MSP1b-specific antibody response induced by the linked construct was also greater than the MSP1b-specific response induced by the native MSP1 complex. For the recombinant proteins, the concentration of immunogen was adjusted to obtain similar molar ratios based on the assumption that MSP1a and MSP1b1 are present at equimolar concentrations in the native MSP1. However, this assumption is unlikely to be correct. The immunoblot of native MSP1 incubated with a MAb specific for MSP1a or MSP1b in Fig. 2 and our previous results obtained using LC-MS/MS to analyze the composition of the MSP1 complex (19) suggest that MSP1a is much more abundant than MSP1b1 in the native complex. Furthermore, in the immunoprecipitated native MSP1 complex both covalently linked MSP1a and MSP1b1, as well as unlinked MSP1a, could be present, which would influence the molar ratio of the epitopes. In other studies, the molar ratio of a peptide and its carrier affected the resulting antibody affinity and titer, so that antibody titers increased with increased conjugation (32). Thus, the greater MSP1b-specific antibody titers in the group immunized with the linked construct was likely due a higher MSP1b1-to-MSP1a ratio than that in the native MSP1 complex.
The hapten-carrier effect has been used successfully in vaccine development, but the vaccines often target polysaccharides or small peptides. For example, the flagellin protein of Salmonella can be used as a carrier protein linked to multiple copies of any foreign epitope. However, this approach was used for presentation of small peptides (4, 5). Here we present data that support our hypothesis that one bacterial OMP can act as a carrier for a different OMP by providing multiple T-cell epitopes and expanding the repertoire of T-cell epitope presentation on the MHC class II molecules.
The bacterial outer membrane is essential for host cell invasion and survival and is also the main target of the host immune response. The major surface proteins of A. marginale associate through covalent as well as noncovalent interactions. For example, MSP2 and MSP5 can occur as monomers or disulfide-linked multimers, while MSP1, MSP2, MSP3, and MSP4 are nearest neighbors (38). Recently, our laboratory identified type IV secretion system (TFSS) proteins as immunogenic proteins in animals immunized with A. marginale outer membranes (17). Since CD4+ T cells responding to conjugal transfer protein (CTP) could not be detected even though CTP-specific IgG was present, we hypothesized that, similar to our findings with MSP1a and MSP1b1, CTP is associated with a component of the TFSS or another membrane protein and that CD4+ T-cell help can be provided by this second carrier protein.
To fully understand the protective nature of A. marginale outer membranes, it is important to determine the complexity of interactions among membrane proteins and how the complexes influence immunity. By linking membrane proteins that naturally associate in the bacterial membrane, such as the MSPs from A. marginale or the proteins that form the TFSS complex, or by altering the molar ratio of proteins and thereby increasing the overall immunogenicity of the vaccine antigen, it may be possible to better reproduce the protection induced by whole-outer-membrane immunization.
Acknowledgments
We are grateful to Shelley Whidbee, Stephanie Leach, and Bev Hunter for excellent technical assistance, Gerhard Munske and Bill Siems for performing LC-MS/MS, and Marc Evans for performing the statistical analysis.
This research was supported by National Institutes of Health NIAID grant AI053692.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 17 December 2007.
REFERENCES
- 1.Abbott, J. R., G. H. Palmer, K. A. Kegerreis, P. F. Hetrick, C. J. Howard, J. C. Hope, and W. C. Brown. 2005. Rapid and long-term disappearance of CD4+ T lymphocyte responses specific for Anaplasma marginale major surface protein-2 (MSP2) in MSP2 vaccinates following challenge with live A. marginale. J. Immunol. 1746702-6715. [DOI] [PubMed] [Google Scholar]
- 2.Allred, D. R., T. C. McGuire, G. H. Palmer, S. R. Leib, T. M. Harkins, T. F. McElwain, and A. F. Barbet. 1990. Molecular basis for surface antigen size polymorphisms and conservation of a neutralization-sensitive epitope in Anaplasma marginale. Proc. Natl. Acad. Sci. USA 873220-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bavoil, P., A. Ohlin, and J. Schachter. 1984. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect. Immun. 44479-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Yedidia, T., and R. Arnon. 2006. Flagella as a platform for epitope-based vaccines. Isr. Med. Assoc. J. 8316-318. [PubMed] [Google Scholar]
- 5.Ben-Yedidia, T., and R. Arnon. 2005. Towards an epitope-based human vaccine for influenza. Hum. Vaccines 195-101. [DOI] [PubMed] [Google Scholar]
- 6.Brown, W. C., T. C. McGuire, W. Mwangi, K. A. Kegerreis, H. Macmillan, H. A. Lewin, and G. H. Palmer. 2002. Major histocompatibility complex class II DR-restricted memory CD4+ T lymphocytes recognize conserved immunodominant epitopes of Anaplasma marginale major surface protein 1a. Infect. Immun. 705521-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, W. C., G. H. Palmer, H. A. Lewin, and T. C. McGuire. 2001. CD4+ T lymphocytes from calves immunized with Anaplasma marginale major surface protein 1 (MSP1), a heteromeric complex of MSP1a and MSP1b, preferentially recognize the MSP1a carboxyl terminus that is conserved among strains. Infect. Immun. 696853-6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, W. C., V. Shkap, D. Zhu, T. C. McGuire, W. Tuo, T. F. McElwain, and G. H. Palmer. 1998. CD4+ T-lymphocyte and immunoglobulin G2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect. Immun. 665406-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, W. C., D. Zhu, V. Shkap, T. C. McGuire, E. F. Blouin, K. M. Kocan, and G. H. Palmer. 1998. The repertoire of Anaplasma marginale antigens recognized by CD4+ T-lymphocyte clones from protectively immunized cattle is diverse and includes major surface protein 2 (MSP-2) and MSP-3. Infect. Immun. 665414-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fulop, M., R. Manchee, and R. Titball. 1995. Role of lipopolysaccharide and a major outer membrane protein from Francisella tularensis in the induction of immunity against tularemia. Vaccine 131220-1225. [DOI] [PubMed] [Google Scholar]
- 11.Fulop, M., R. Manchee, and R. Titball. 1996. Role of two outer membrane antigens in the induction of protective immunity against Francisella tularensis strains of different virulence. FEMS. Immunol. Med. Microbiol. 13245-247. [DOI] [PubMed] [Google Scholar]
- 12.Green, B. A., M. E. Vazquez, G. W. Zlotnick, G. Quigley-Reape, J. D. Swarts, I. Green, J. L. Cowell, C. D. Bluestone, and W. J. Doyle. 1993. Evaluation of mixtures of purified Haemophilus influenzae outer membrane proteins in protection against challenge with nontypeable H. influenzae in the chinchilla otitis media model. Infect. Immun. 611950-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haake, D. A., M. K. Mazel, A. M. McCoy, F. Milward, G. Chao, J. Matsunaga, and E. A. Wagar. 1999. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect. Immun. 676572-6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatch, T. P. 1996. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J. Bacteriol. 1781-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatch, T. P., M. Miceli, and J. E. Sublett. 1986. Synthesis of disulfide-bonded outer membrane proteins during the developmental cycle of Chlamydia psittaci and Chlamydia trachomatis. J. Bacteriol. 165379-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khlebnikov, V. S., I. R. Golovliov, D. P. Kulevatsky, N. V. Tokhtamysheva, S. F. Averin, V. E. Zhemchugov, S. Y. Pchelintsev, S. S. Afanasiev, and G. Y. Shcherbakov. 1996. Outer membranes of a lipopolysaccharide-protein complex (LPS-17 kDa protein) as chemical tularemia vaccines. FEMS. Immunol. Med. Microbiol. 13227-233. [DOI] [PubMed] [Google Scholar]
- 17.Lopez, J. E., G. H. Palmer, K. A. Brayton, M. J. Dark, S. E. Leach, and W. C. Brown. 2007. Immunogenicity of Anaplasma marginale type IV secretion system proteins in a protective outer membrane vaccine. Infect. Immun. 752333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez, J. E., W. F. Siems, G. H. Palmer, K. A. Brayton, T. C. McGuire, J. Norimine, and W. C. Brown. 2005. Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect. Immun. 738109-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macmillan, H., K. A. Brayton, G. H. Palmer, T. C. McGuire, G. Munske, W. F. Siems, and W. C. Brown. 2006. Analysis of the Anaplasma marginale major surface protein 1 complex protein composition by tandem mass spectrometry. J. Bacteriol. 1884983-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGuire, T. C., G. H. Palmer, W. L. Goff, M. I. Johnson, and W. C. Davis. 1984. Common and isolate-restricted antigens of Anaplasma marginale detected with monoclonal antibodies. Infect. Immun. 45697-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIlhinney, R. A. 2004. Generation and use of epitope-tagged receptors. Methods Mol. Biol. 25981-98. [DOI] [PubMed] [Google Scholar]
- 22.Newhall, W. J. V. 1987. Biosynthesis and disulfide cross-linking of outer membrane components during the growth cycle of Chlamydia trachomatis. Infect. Immun. 55162-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noh, S. M., K. A. Brayton, D. P. Knowles, J. T. Agnes, M. J. Dark, W. C. Brown, T. V. Baszler, and G. H. Palmer. 2006. Differential expression and sequence conservation of the Anaplasma marginale msp2 gene superfamily outer membrane proteins. Infect. Immun. 743471-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norimine, J., and W. C. Brown. 2005. Intrahaplotype and interhaplotype pairing of bovine leukocyte antigen DQA and DQB molecules generate functional DQ molecules important for priming CD4+ T-lymphocyte responses. Immunogenetics 57750-762. [DOI] [PubMed] [Google Scholar]
- 25.Norimine, J., C. E. Suarez, T. F. McElwain, M. Florin-Christensen, and W. C. Brown. 2002. Immunodominant epitopes in Babesia bovis rhoptry-associated protein 1 that elicit memory CD4+ T-lymphocyte responses in B. bovis-immune individuals are located in the amino-terminal domain. Infect. Immun. 702039-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pal, S., I. Theodor, E. M. Peterson, and L. M. de la Maza. 1997. Immunization with an acellular vaccine consisting of the outer membrane complex of Chlamydia trachomatis induces protection against a genital challenge. Infect. Immun. 653361-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer, G. H., A. F. Barbet, W. C. Davis, and T. C. McGuire. 1986. Immunization with an isolate-common surface protein protects cattle against anaplasmosis. Science 2311299-1302. [DOI] [PubMed] [Google Scholar]
- 28.Palmer, G. H., and T. C. McGuire. 1984. Immune serum against Anaplasma marginale initial bodies neutralizes infectivity for cattle. J. Immunol. 1331010-1015. [PubMed] [Google Scholar]
- 29.Palmer, G. H., D. Munodzana, N. Tebele, T. Ushe, and T. F. McElwain. 1994. Heterologous strain challenge of cattle immunized with Anaplasma marginale outer membranes. Vet. Immunol. Immunopathol. 42265-273. [DOI] [PubMed] [Google Scholar]
- 30.Palmer, G. H., F. R. Rurangirwa, and T. F. McElwain. 2001. Strain composition of the ehrlichia Anaplasma marginale within persistently infected cattle, a mammalian reservoir for tick transmission. J. Clin. Microbiol. 39631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park, Y. H., Y. S. Joo, J. Y. Park, J. S. Moon, S. H. Kim, N. H. Kwon, J. S. Ahn, W. C. Davis, and C. J. Davies. 2004. Characterization of lymphocyte subpopulations and major histocompatibility complex haplotypes of mastitis-resistant and susceptible cows. J. Vet. Sci. 529-39. [PubMed] [Google Scholar]
- 32.Pedersen, M. K., N. S. Sorensen, P. M. Heegaard, N. H. Beyer, and L. Bruun. 2006. Effect of different hapten-carrier conjugation ratios and molecular orientations on antibody affinity against a peptide antigen. J. Immunol. Methods 311198-206. [DOI] [PubMed] [Google Scholar]
- 33.Pizarro-Cerda, J., and P. Cossart. 2006. Bacterial adhesion and entry into host cells. Cell 124715-727. [DOI] [PubMed] [Google Scholar]
- 34.Scoles, G. A., M. W. Ueti, S. M. Noh, D. P. Knowles, and G. H. Palmer. 2007. Conservation of transmission phenotype of Anaplasma marginale (Rickettsiales: Anaplasmataceae) strains among Dermacentor and Rhipicephalus ticks (Acari: Ixodidae). J. Med. Entomol. 44484-491. [DOI] [PubMed] [Google Scholar]
- 35.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 36.Tebele, N., T. C. McGuire, and G. H. Palmer. 1991. Induction of protective immunity by using Anaplasma marginale initial body membranes. Infect. Immun. 593199-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Eijk, M. J., A. Stewart-Haynes, and H. A. Lewin. 1992. Extensive polymorphism of the BoLA-DRB3 gene distinguished by PCR-RFLP. Anim. Genet. 23483-496. [DOI] [PubMed] [Google Scholar]
- 38.Vidotto, M. C., T. C. McGuire, T. F. McElwain, G. H. Palmer, and D. P. Knowles, Jr. 1994. Intermolecular relationships of major surface proteins of Anaplasma marginale. Infect. Immun. 622940-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, T., A. C. McCandlish, L. S. Gronenberg, S. S. Chng, T. J. Silhavy, and D. Kahne. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 10311754-11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yen, T. Y., S. Pal, and L. M. de la Maza. 2005. Characterization of the disulfide bonds and free cysteine residues of the Chlamydia trachomatis mouse pneumonitis major outer membrane protein. Biochemistry 446250-6256. [DOI] [PubMed] [Google Scholar]