Abstract
One of the key processes in the pathobiology of the malaria parasite is the invasion and subsequent modification of the human erythrocyte. In this complex process, an unknown number of parasite proteins are involved, some of which are leading vaccine candidates. The majority of the proteins that play pivotal roles in invasion are either stored in the apical secretory organelles or located on the surface of the merozoite, the invasive stage of the parasite. Using transcriptional and structural features of these known proteins, we performed a genomewide search that identified 49 hypothetical proteins with a high probability of being located on the surface of the merozoite or in the secretory organelles. Of these candidates, we characterized a novel leucine zipper-like protein in Plasmodium falciparum that is conserved in Plasmodium spp. This protein is expressed in late blood stages and localizes to the rhoptries of the parasite. We demonstrate that this Plasmodium sp.-specific protein has a high degree of conservation within field isolates and that it is refractory to gene knockout attempts and thus might play an important role in invasion.
Malaria is a major public health problem caused by infection with apicomplexan parasites of the genus Plasmodium. These obligatory intracellular parasites infect an estimated 500 million people annually, resulting in over 2 million deaths each year (46, 56). In spite of great advances in molecular medicine, a vaccine is not available, and resistance to antimalarial drugs is widespread.
One of the key processes in the pathobiology of the parasite is the invasion and subsequent modification of the host cell, the human erythrocyte. An unknown number of parasite proteins are involved in this complex process (27, 51). The majority of invasion-related proteins are either located on the surface of the invasive stage of the parasite (merozoite) or stored in secretory organelles that include rhoptries, micronemes, and dense granules (reviewed in reference 11). These organelles have crucial functions in host cell invasion and establishment of the parasitophorous vacuole. During the process of invasion, proteins are released from the secretory organelles into the intercellular space or translocated onto the surface of the invading parasite and are therefore exposed to the human immune system. Consequently, these proteins are of considerable interest as potential vaccine candidates. For instance, the micronemal protein apical membrane antigen 1 (AMA-1), the rhoptry-associated protein 1 (RAP1), and the merozoite surface antigens merozoite surface proteins 1 and 2 (MSP-1/MSP-2) have been explored as blood stage vaccine candidates (3, 18, 20). Most of these invasion-related proteins display some sequence and structural similarities and show some conservation among apicomplexan parasites (11, 60). They are transcribed late in the asexual blood cycle (10, 30), they enter the secretory pathway via a signal peptide, and some display characteristic adhesive domains that are implicated in receptor interaction. For example, members of the erythrocyte-binding-like protein family (1, 2, 41) use so-called Duffy-binding domains to interact with sialoglycoproteins on the surfaces of erythrocytes. Others, like the merozoite TRAP homologue (mTRAP) (7), Plasmodium thrombospondin-related apical merozoite protein (PTRAMP) (59), or the apical Sushi protein (ASP) (36), contain alternative adhesive domains, such as the thrombospondin repeats or a Sushi domain.
Although the number of proteins that are known to be involved in invasion of erythrocytes is continually growing, the understanding of the molecular machinery powering this unique and complex process is only beginning. To deepen our insights into the molecular aspects of erythrocyte invasion by Plasmodium falciparum, we used a bioinformatics screen to identify new proteins potentially involved in the process. From the subset of 89 predicted candidates (49 hypothetical proteins and 40 proteins that have been previously described), we characterize in detail a novel rhoptry protein that is highly conserved within Plasmodium spp. and is unrelated to known proteins involved in invasion.
(This article is based in part on doctoral studies by S.H., N.S., and M.T. in the Faculty of Biology, University of Hamburg.)
MATERIALS AND METHODS
Gene identification and sequence analysis.
Primary sequence data and gene expression profiles were analyzed using the resources implemented at the PlasmoDB website (http://plasmoDB.org). The following query parameters were used to search the database for putative candidates: (i) late transcription (maximum transcription, 42 h ± 6 h; minimum transcription, 16 h ± 8 h; >3-fold induction), (ii) predicted signal peptide, and (iii) a minimum of one transmembrane domain. Predicted protein domains were further analyzed using MyHits (http://myhits.isb-sib.ch/doc/index.html), SOSUIsignal (http://www.expasy.org/tools/#primary), and the GPI Lipid Anchor Project (http://mendel.imp.ac.at/sat/gpi/gpi_server.html).
Nucleic acids.
Genomic DNA was prepared as described previously (67). The gene coding for MAL7P1.119 (http://www.plasmoDB.org) was amplified using genomic DNA with the primers RALP-F and RALP-R (Table 1). The resulting PCR product was cloned into the pCR2.1 TOPO vector (Invitrogen). DNA sequencing was performed using BigDye Terminator cycle sequencing (Perkin Elmer Life Sciences).
TABLE 1.
Oligonucleotide primers used in this study
| Application | Primer | Sequencea |
|---|---|---|
| Sequencing | RALP-F | GCGGGTACCCTTTATAATATAGGAATATAAAAATG |
| RALP-R | GCGCCCTAGGAAAAAGCTCAAATAAGAC | |
| MSP1-F | AAGCTTTAGAAGATGCAGTATTGAC | |
| MSP1-R | AATCATTAA TTTCTTCATATCCATC | |
| Transfection | GFP-F | GCGCGGGTACCAGTAAAGGAGAAGAACTT |
| GFP-R | GCGCCTCGAGGGCCTAGGTTTGTATCGTTCATCCAT | |
| RALP5′-F | GCGCGGTACCGGAA TATAAAAATG | |
| RALP5′-R | GCGCGGTACCTGCCACATTAGG | |
| RALP3′-F | GCGCCCTAGGTCACACGACATATCA | |
| RALP3′-R | GCGCCTCGAGTTAAAAAAGC TC | |
| RALP-TY1-R | GCGCCTCGAGTCAGTCAAGTGGATCCTGGTTAGTATGGACCTCAAAAAGCTCAAATAAGACTTTG | |
| RALP5′KO-F | GCGCGGTACCGGAA TATAAAAATG | |
| RALP5′KO-R | GCGCGGTACCTGCCACATTAGG | |
| RALP3′KO-F | GCGCGAATTCTAAGACCACAAAAATGAGATGACCAC | |
| RALP3′KO-R | GCGCCCATGGTTACATCTATATCAGAATCGTTTATGTC | |
| Expression | RALP-N-F | GCGCGGATCCCCACTTCAACGTGGCGCATC |
| RALP-N-R | GCGCCTCGAGTTAATCAACGTTCACATTCATTTG | |
| RALP-C-F | GCGCGGATCCGGACAAGGAGAATTACTACAATC | |
| RALP-C-R | GCGCCTCGAGTTAGTTATCTTTTTGTTTGTTTCCC | |
| ASP-F | GCGCGGATCCGGACAAGGAGAATTACTACAATC | |
| ASP-R | GCGCCTCGAGTTAGTTATCTTTTTGTTTGTTTCCC |
Restriction endonuclease sites are underlined and in italics, and stop codons are in boldface.
Genotyping of field isolates was performed by PCR amplification of the polymorphic block 2 of the merozoite surface protein 1 gene (msp-1) as previously described (52, 55) with primers MSP1-F and MSP1-R (Table 1). Sequence alignments were carried out using PRALINE (http://ibivu.cs.vu.nl/programs/pralinewww) and ClustalW (http://www.ch.embnet.org/software/ClustalW.html).
Southern blotting was carried out using standard procedures and a Gene Images Random Prime Labeling and Detection kit (Amersham Biosciences).
Transfection plasmids.
To generate rhoptry-associated leucine zipper-like protein 1-green fluorescent protein (RALP1-GFP)-expressing parasites, GFP was amplified using the proofreading Vent polymerase (Stratagene) and the forward/reverse primer pair GFP-F and GFP-R (Table 1), introducing a 5′ KpnI site and a 3′ AvrII/XhoI site. The PCR product was digested with KpnI/XhoI and cloned into pARL1a− (13), resulting in the vector pARL-GFP. The 5′ end of the ralp1 gene (249 bp, including the endogenous start codon) was amplified using the primer pair RALP5′-F and RALP5′-R (Table 1), digested with KpnI, and cloned into the pARL-GFP vector. The 3′ end (798 bp, including the endogenous stop codon) was amplified using the primer pair RALP3′-F and RALP3′-R (Table 1) and inserted into the appropriate restriction sites of pARL-GFP containing the 5′ region of ralp1. The chimeric ralp1-gfp was sequenced to exclude the possibility that unwanted mutations were present.
To overexpress full-length RALP1 in the parasite, the transfection plasmid RALP1-TY1 was generated by amplification of ralp1 with the primers RALP5′-F and RALP-TY1-R (Table 1), introducing a C-terminal TY1 epitope tag (6). The PCR product was digested with KpnI/XhoI, cloned into pARL1a− (13), and sequenced to exclude the possibility that unwanted mutations were present.
Gene disruption was attempted using the pHTK vector system (15). One kilobase of the 5′ end and 0.7 kb of the 3′ end of ralp1 were amplified with additional restriction sites and stop codons and cloned on either side of the human dihydrofolate reductase (hdhfr) cassette of pHTK. The primers RALP5′KO-F and RALP5′KO-R (Table 1) were used to amplify the 5′ ralp1 fragment; the primer pair RALP3′KO-F and RALP3′KO-R (Table 1) was used to amplify the 3′ ralp1 fragment. Both fragments were inserted into the appropriate restriction sites of pHTK.
Parasite culture and transfection.
P. falciparum asexual stages (parasite strain 3D7) were cultured in human erythrocytes (blood group O+) according to standard procedures (62). 3D7 parasites were transfected as described previously (16) with 100 μg purified plasmid DNA (Qiagen). Positive selection for transfectants was achieved using 10 nM WR99210 (16). Stable transfected parasites were either cycled on WR99210 as described by Reed et al. (44) or treated at the completion of each drug cycle with a combination of WR99210 and 20 μM ganciclovir (Roche).
Field isolates.
A total of seven P. falciparum field isolates were analyzed. Two isolates were obtained from the peripheral blood of non-malaria-immune travelers returning from South Africa and Tanzania who were treated in the clinical department of the Bernhard Nocht Institute for Tropical Medicine in Hamburg, Germany. Five isolates were collected from semi-immune children during field trials in the Ashanti Region of Ghana in 2001. Consent for peripheral-blood sampling was obtained from the patients and their parents prior to collection. Ethical clearance was granted by the Committee on Human Research and Ethics, School of Medical Sciences, University of Science and Technology, Kumasi, Ghana.
Recombinant expression and antisera.
In order to generate specific antibodies, two regions of the ralp1 gene were amplified and recombinantly expressed. The 5′ end of the ralp1 gene was amplified using the forward primer RALP-N-F and the reverse primer RALP-N-R (Table 1), encompassing 116 amino acids (aa) (N33 to D148). The C-terminal region encompasses 211 aa (N396 to S606) and was amplified using the forward primer RALP-C-F and the reverse primer RALP-C-R (Table 1). Exon 3 of the gene coding for the ASP (36) was amplified using the forward primer ASP-F and the reverse primer ASP-R (Table 1), encompassing 138 aa (L84 to G221). The PCR fragments were digested with BamHI/XhoI and cloned into the BamHI/XhoI sites of pGEX-4T. Expression of the fusion proteins in Escherichia coli (BL21) was induced with 1 mM isopropyl-β-d-thiogalactopyranosoide (IPTG). The glutathione S-transferase (GST) fusion proteins and GST were purified by affinity chromatography on glutathione-Sepharose 4B (Amersham Biosciences) according to the manufacturer's instructions, dialyzed in phosphate-buffered saline (PBS), and used for immunization of rabbits (against the C-terminal RALP1-GST fusion) and rats (against the N-terminal GST fusion). Antisera were affinity purified using the recombinant GST fusion proteins as ligands. GST or the RALP1 fusion proteins were covalently linked to glutathione-Sepharose 4B (Amersham Bioscience) with a final dimethyl pimelimidate-HCl concentration of 20 mM. Antisera were precleared using the GST column (to remove GST-specific antibodies) and subsequently applied to either a RALP1-N or a RALP1-C column. Specific antibodies to C- and N-terminal RALP1-GST fusion proteins were eluted with 0.1 M glycine-HCl, pH 2.5.
Western analysis.
For immunoblots, parasite proteins from a synchronized culture (28) were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to nitrocellulose membranes (Schleicher & Schuell) in 10 mM CAPS (3-cyclohexylamino-1-propanesulfonic acid), pH 10.8, as described previously (58). Anti-RALP1 rabbit antisera, anti-RALP1 rat antisera, and rabbit anti-ASP antisera were diluted 1:1,000; GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mouse antiserum (14) was diluted 1:3,000; and anti-GFP and anti-TY1 (6) were diluted 1:1,000 in PBS with 5% (wt/vol) skim milk. The secondary antibodies were horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) (1:5,000; Jackson IR), horseradish peroxidase-conjugated goat anti-mouse IgG (1:3,000; Jackson IR), and horseradish peroxidase-conjugated goat anti-rat IgG (1:1,000; Jackson IR). The immunoblots were developed by chemiluminescence using SuperSignal West Pico chemiluminescent substrate (Pierce).
Immunofluorescence.
Immunofluorescence assays were performed on methanol-, acetone-, or formaldehyde/glutaraldehyde-fixed parasites as previously described (19, 58, 61). The following primary antibody dilutions were used: rabbit anti-RALP1-C (1:1,000), rat anti-RALP1-N (1:1,000), mouse anti-EBA-175 (1:2,000) (44), mouse anti-RAP1 (1:1,000) (4), mouse anti-CLAG9 (1:1,000) (17), and mouse anti-GFP (1:1,000; Roche) or anti-TY1 (1:2,000). The following secondary antibodies were used: Alexa-488 goat anti-mouse IgG (1:2,000; Molecular Probes), Alexa-594 goat anti-mouse IgG (1:2,000; Molecular Probes), Alexa-488 donkey anti-rabbit IgG (1:2,000; Molecular Probes), Alexa-594 donkey anti-rabbit IgG (1:2,000; Molecular Probes), and DAPI (4′,6′-diamidino-2-phenylindole) for nuclear staining (1:2,000; Roche). Fluorescence images were captured using a Zeiss Axioskop 2 microscope and Openlab software (Improvision).
Solubility properties of RALP1.
Ten milliliters of late-stage parasite culture expressing cytosolic GFP (57) was harvested by saponin lysis with 1.5 ml 0.03% saponin in PBS for 10 min on ice and centrifuged at 16,100 × g for 5 min. The parasite pellet was washed three times with ice-cold PBS, and the parasites were lysed by hypotonic lysis with 500 μl H2O, frozen at −80°C followed by three “thaw and freeze” cycles, and centrifuged at 16,100 × g. Equal volumes of the soluble protein fraction and the pellet depleted of soluble proteins were subjected to Western analysis with antibodies directed against either RALP1 or GFP.
Immunoprecipitation.
Schizonts from 60 ml of synchronized parasite culture (10 to 12% parasitemia) were harvested by saponin lysis with 0.03% saponin in PBS for 10 min on ice and centrifuged at 16,100 × g for 5 min. Proteins were extracted in 10 ml RIPA buffer/1% Triton X-100 with complete protease inhibitor (Roche). The RALP1-specific antibody (RALP1-C) was added to the parasite protein extract and incubated for 1 h at 4°C. Protein G-Sepharose (GE Healthcare) was added to the antigen-antibody mixture and incubated overnight at 4°C, followed by centrifugation at 600 × g for 2 min 4°C. The supernatant was removed, and the beads were washed three times in RIPA buffer. Bound material was eluted with the addition of an equal bed volume of SDS sample buffer. Proteins were separated by SDS-PAGE and stained with Coomassie stain. In parallel, Western blot analyses with both RALP1-specific antibodies were performed. A band of approximately 90 kDa was cut out of the SDS-PAGE gel and subjected to mass spectrometry (MS).
In-gel digestion and LCMS.
The corresponding SDS band was cut out of the gel, reduced, and alkylated in gel. Proteins were digested overnight with trypsin (Promega). Peptides were eluted from the gel with trifluoroacetic acid. The peptides were sequenced using a nano-high-pressure liquid chromatography (LC) Aligent 1100 nanoflow system connected online to a 7-T linear-quadropole ion-trap Fourier transform mass spectrometer (Thermo Electron) essentially as described previously (37). The corresponding peptides were identified using the Blast search tools of either the NCBI or PlasmoDB database.
RESULTS
Global analysis of candidate genes.
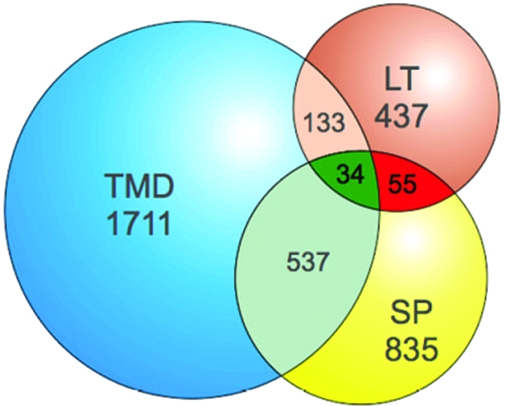
Invasion-related proteins, like those stored in the secretory organelles (micronemes, rhoptries, and dense granules) or located on the surface of the merozoite, are transcribed in late stages during the intraerythrocytic life cycle and have an N-terminal signal peptide, and some are membrane bound via a transmembrane domain or a glycosylphosphatidylinositol (GPI) anchor. Genomewide analysis of the available transcription and primary sequence data using the search tools in PlasmoDB revealed (i) 437 genes with maximum transcription at 42 h ± 6 h and minimum transcription at 16 h ± 8 h, (ii) 835 predicted proteins that possess a putative signal peptide, and (iii) 1,711 predicted proteins that contain at least one transmembrane domain (Fig. 1). Of those that have a signal peptide, 89 are also transcribed in late stages. Thirty-four of these 89 proteins have at least one transmembrane domain. This subset of 89 predictions contained most of the proteins (36 out of 56) known to be located either in the secretory organelles (e.g., members of the erythrocyte-binding-like protein superfamily, RAP, and CLAG proteins) or on the surface of the merozoite (e.g., members of the MSP or the serine repeat antigen [SERA] families). In contrast, only four of the proteins within the group of 89 are known to localize to other parts of the infected red blood cell (see Fig. S1 in the supplemental material). Fifteen known proteins, including the reticulocyte binding proteins Rh1, Rh2a/b, Rh4, and Rh5 (43, 66) and the surface proteins MSP2 and MSP4/5/6 (33, 54, 68, 71), were elusive because of a missing signal peptide annotation in PlasmoDB 5.2 (see Fig. S1 in the supplemental material). Five proteins, two of which contain no signal peptide (PfRom-4 [PFE0340c] and PfRhop148 [PF13_0348]), were missed due to inadequate query parameters, two because of nonmatching transcriptional profiles (Pf41 [PFD0240c] and MSP-8 [PFE0120c]), and one because the sequence was not present in PlasmoDB5.2 (see Fig. S1 in the supplemental material).
FIG. 1.
Global identification for candidate genes involved in invasion. Primary sequence data and gene expression profiles were analyzed using the resources at the PlasmoDB website (http://plasmoDB.org). The following query parameters were used to search the database for putative candidates: (i) transcription in late stages (maximum transcription, 42 h ± 6 h; minimum transcription, 16 h ± 8 h; >3-fold induction), (ii) predicted signal peptide, (iii) minimum of one transmembrane domain. Total numbers and numbers of intersections are given. Blue, proteins with ≥1 transmembrane domain (TMD); yellow, proteins with a predicted signal peptide (SP); brown, proteins transcribed in late stages (LT); red, intersection of proteins with late transcription and predicted signal peptide; green, intersection of proteins fulfilling all three parameters.
This validates the search parameters applied. Most importantly, this analysis identified 49 unknown proteins that might be located in the secretory organelles or on the surface of the merozoite. Seventeen proteins displayed features associated with proteins involved in invasion, such as thrombospondin-like domains (7, 45, 59), epidermal growth factor domains (9), or a predicted GPI attachment site (35, 49). These features strengthen the notion that these proteins might play a role in the process of invasion.
Sequence analysis of a cross-species-conserved leucine zipper-like protein.
From several proteins fulfilling the parameters used in the bioinformatics screen we selected MAL7P1.119 (http://www.plasmoDB.org) for further analysis. MAL7P1.119 was termed RALP1 and was chosen because (i) it is well conserved within Plasmodium spp. but restricted to that genus, (ii) it is unrelated to known proteins involved in invasion, and (iii) it contains a leucine zipper-like domain that might represent a protein-protein interaction domain. The single-copy gene ralp1 codes for a predicted protein of 750 aa. It is defined by a single exon of 2,250 bp. The predicted protein possesses a putative N-terminal signal peptide (SignalP) (8), two negatively charged regions (a glutamic acid-rich region [aa position 92 to 128] and an aspartic acid-rich region [aa position 411 to 505]), and a leucine-rich region (aa position 498 to 604). Within the leucine-rich region, a leucine zipper domain [L511-(X)6-L-(X)6-L-(X)6-L532] (Fig. 2 A; see Fig. S2A in the supplemental material) is predicted by MotifScan (40) and PPsearch (72), but not by the 2ZIP server (http://2zip.molgen.mpg.de). The leucine zipper-like domain lies within a predicted coiled-coil domain (Paircoil [http://paircoil.lcs.mit.edu/cgi-bin/paircoil]) (see Fig. S2A in the supplemental material). These domains are ubiquitous structural features of protein-protein interactions (32).
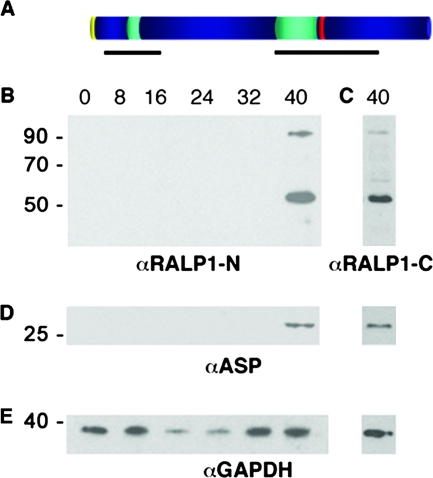
FIG. 2.
Structural features and expression of RALP1 in asexual blood stages. (A) Schematic domain structure of RALP1. Signal peptide, yellow; charged regions, turquoise; leucine zipper-like domain, red. Regions used for raising RALP1-specific antibodies are indicated by black lines. (B and C) Stage-specific Western analysis of wild-type parasites (3D7). Proteins from synchronized parasite cultures were harvested at 8-h intervals and separated by SDS-PAGE on a 10% gel under reducing conditions. Using either the N-terminal (B) or C-terminal (C) anti-RALP1-specific antibodies, two major bands of approximately 55 kDa and 90 kDa were detected in late stages (40 h postinvasion). (D) Stage-specific control. The stage specificity of the parasite material was confirmed using antibodies directed against ASP, which is translated in late-stage parasites. (E) Loading control. Antibodies against the constitutively expressed GAPDH protein were used to demonstrate equal loading.
Putative orthologs of ralp1 can be identified in the available genomes of different Plasmodium species (see Fig. S2D, P. berghei PB000011.03.0, P. chabaudi PC301847.00.0, P. vivax Pv096245, and P. yoelii yoelii PY07382, in the supplemental material). The highest conservation (on average, 85% homology and 63% identity) was within the leucine-rich region (see Fig. S2D in the supplemental material). Blast analysis using the resources of GenBank (http://www.ncbi.nlm.nih.gov) and ApiDB (http://apidb.org) revealed no orthologs in the genomes of any other species.
RALP1 is expressed in late blood stages.
Protein expression was analyzed using stage-specific immunoblots and RALP1-specific antibodies on protein extracts of synchronized parasites. Antisera raised against both the C- and N-terminal regions of RALP1 recognized a protein of the expected mass of 90 kDa in P. falciparum protein extracts in late stages of the parasite (>40 h postinvasion) (Fig. 2B and C), corresponding to the predicted molecular mass of 88 kDa. Additionally, both antisera recognized a 55-kDa protein that might represent a processed form of the RALP1 protein. A stage-specific control was performed using the late-translated ASP (Fig. 2D) (36). To ensure equal loading, the stage-specific immunoblots were probed with GAPDH-specific antibodies (Fig. 2E). GAPDH is detectable as a 36-kDa protein throughout the asexual life cycle.
The endogenous RALP1 protein was immunoprecipitated by using late-stage parasite extract and RALP1-specific antibodies (anti-RALP1-C). Immunoprecipitated proteins were analyzed on SDS-PAGE, and a protein band of about 90 kDa could be visualized by Coomassie staining of the gel and Western blot analysis using either the N- or the C-terminal RALP1-specific antibodies (data not shown). This band was excised from the SDS-PAGE gel and subjected to LCMS. Analysis of the MS data revealed 18 nonoverlapping peptides identical with RALP1 (see Fig. S3 in the supplemental material), covering the protein from the N to the C terminus. The additional 55-kDa protein detected by both RALP1-specific antibodies (Fig. 2B and C) could not be immunoprecipitated in sufficient purity and quantity to allow sequence analysis.
Expression of RALP1-GFP and full-length RALP1-TY1 in transgenic parasites.
To further confirm the specificity of the RALP1-specific antibodies and to localize the protein within the parasite, we generated transgenic cell lines expressing a RALP1-GFP fusion protein and full-length RALP1 with a C-terminal TY1 epitope tag.
The transfection construct RALP1-GFP contained the first N-terminal 83 aa of RALP1 (to ensure proper entry into the secretory pathway via the signal sequence), followed by GFP and finally the C terminus of RALP1 (484 to 749 aa), including the leucine zipper-like domain (Fig. 3 A). The expression of the RALP1-GFP fusion protein was analyzed on Western blots with anti-GFP antibodies using wild-type and transgenic parasite extracts. A protein of approximately 70 kDa (the calculated mass of RALP1-GFP is 66 kDa) could be detected in the transgenic cell line, but not in wild-type cultures (Fig. 3 B). Additionally, a smaller, 24-kDa protein was weakly recognized, presumably representing a GFP breakdown product as previously described for GFP-expressing parasites (70). Antibodies specific to the C terminus of RALP1 (anti-RALP1-C) recognized (in addition to the endogenous proteins) a band of approximately 70 kDa exclusively in the transgenic parasite line matching the molecular mass of the RALP1-GFP chimeric protein (Fig. 3C). Antibodies specific to the N terminus failed to recognize RALP1-GFP (Fig. 3D). This might be due to the fact that the chimeric RALP1-GFP lacks more than half of the region used to raise the antisera and therefore does not display the epitopes recognized by the antibodies.
FIG. 3.
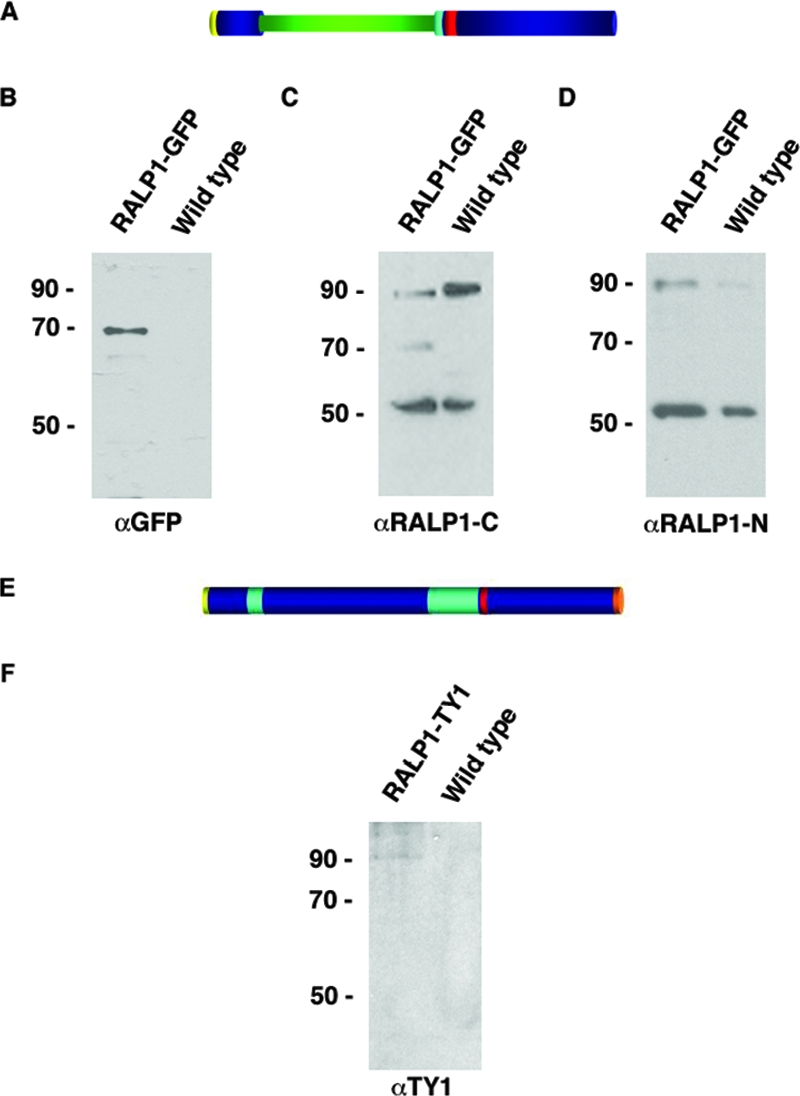
Expression of RALP1-GFP and RALP1-TY1 in transgenic parasites. (A) Schematic domain structure of RALP1-GFP. Signal peptide, yellow; GFP, green; charged region, turquoise; leucine zipper-like domain, red. (B) Immunoblot using GFP-specific antibodies on wild-type and RALP1-GFP-expressing parasites (RALP1-GFP). A band of approximately 70 kDa, representing the GFP fusion protein, is recognized by GFP-specific antibodies in the transgenic, but not in the wild-type, parasite line. (C) Anti-RALP1-specific antibodies (αRALP1-C) recognize the RALP1-GFP fusion protein of approximately 70 kDa, in addition to the endogenous RALP1 protein of 90 kDa and the 55-kDa fragment, in RALP1-GFP-expressing parasites. (D) N-terminal-specific anti-RALP1 antibodies (αRALP1-N) recognize only the endogenous RALP1, but not the fusion protein, in RALP1-GFP-expressing parasites. (E) Schematic domain structure of RALP1-TY1. Signal peptide, yellow; charged region, turquoise; leucine zipper-like domain, red; TY1 epitope tag, orange. (F) Immunoblot using anti-TY1-specific antibodies on wild-type and RALP1-TY1-expressing parasites. A band of approximately 90 kDa, representing RALP1-TY1, is recognized by anti-TY1 specific antibodies in the transgenic, but not in the wild-type, parasite line.
A second transgenic cell line was generated that expressed full-length RALP1 with the 8-aa TY1 epitope at the C terminus (Fig. 3E). A protein of 90 kDa was recognized by the anti-TY1-specific antibody exclusively in the transgenic parasite line corresponding to the calculated molecular mass of 89 kDa for the transgene (Fig. 3F).
Localization of RALP1 in wild-type and transgenic parasites.
We determined the localization of RALP1, the RALP1-GFP chimeric protein, and the full-length RALP1 with a C-terminal TY1 tag by both indirect immunofluorescence in fixed parasites and fluorescence microscopy of live cells.
Using wild-type parasites, the N- and C-terminal RALP1-specific antibodies visualized a protein that was restricted to the apical pole in either schizonts or free merozoites (Fig. 4A). The staining patterns of these antibodies were indistinguishable (Fig. 4A, merge).
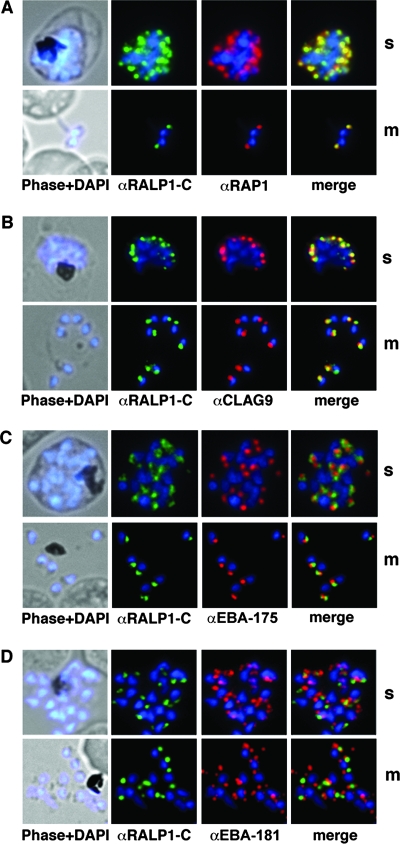
FIG. 4.
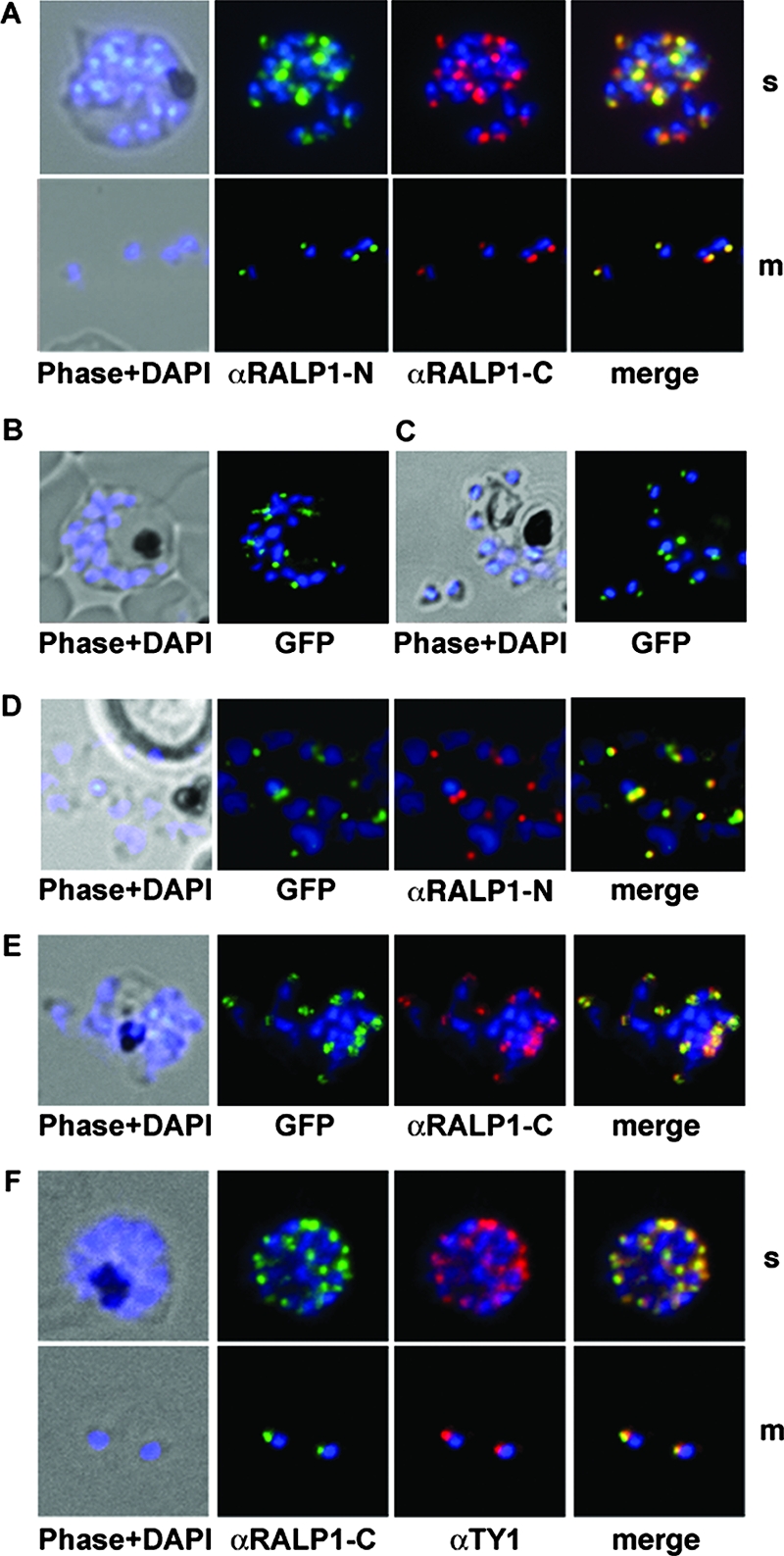
Subcellular distribution of RALP1-GFP fusion protein and immunofluorescence localization of RALP1 and RALP1-TY1. (A) Using methanol-fixed parasites, both RALP1-specific antibodies (αRALP1-N [green] and αRALP1-C [red]) localize the protein in the same subcellular compartment (merged image) in late schizonts (s) and free merozoites (m). The nucleus was stained with DAPI (blue). (B and C) Localization of RALP1-GFP by fluorescence microscopy in unfixed parasites in schizonts (B) and free merozoites (C). Using fluorescence of the GFP reporter protein in live cells, RALP1-GFP distribution (green) was restricted to the apical end of the parasite. The nucleus was stained blue. (D and E) Colocalization of RALP1-GFP with the endogenous RALP1 in free merozoites. N-terminal- and C-terminal-specific anti-RALP1 antibodies (αRALP1-N and αRALP1-C) (red) and the RALP1-GFP fusion protein (green) colocalize, as shown by the merged picture. (F) Colocalization of RALP1-TY1 with the endogenous RALP1 using RALP1-specific antibodies (αRALP1-C) (green) and anti-TY1 antisera (red) resulted in identical staining patterns at the apical end of the parasite.
This localization was further evaluated by the RALP1-GFP fluorescence in unfixed parasites. The fusion protein is concentrated at the apical pole of the parasite (in late schizonts and free merozoites) (Fig. 4B and C). Using the specific anti-RALP1-N antibodies (recognizing only the endogenous RALP1), both RALP1-GFP and the endogenous RALP1 colocalized in the same subcellular compartment at the apical end of the parasite (Fig. 4D). Colocalization with the C-terminal-specific antibody resulted in an identical staining pattern (Fig. 4E). Additionally, the C-terminal TY1-tagged RALP1 was localized to the apical end of the merozoites by indirect immunofluorescence with anti-TY1 antibodies and subsequent colocalization with RALP1-specific antisera (Fig. 4F).
RALP1 colocalizes with the rhoptry marker RAP1.
After establishing the apical localization of RALP1 and the specificity of the RALP1-specific antibodies, we used antisera against several microneme- and rhoptry-specific proteins to test if RALP1 resides in one of these compartments. RAP1, a protein previously localized to the rhoptries (4), was used as a compartment-specific marker. Antibodies against RALP1 and RAP1 (4) showed identical distributions (Fig. 5 A). The gene product of the cytoadherence-linked asexual gene 9 (clag9) (64) was used as a second rhoptry-specific marker. CLAG9 was previously localized exclusively to the bulb of the rhoptry (31). The distribution of CLAG9 predominantly overlaps with that of RALP1 (Fig. 5B). In contrast, the apical distribution of RALP1 was clearly distinct from that of the micronemal marker proteins EBA-175 and EBA-181 (19, 53) (Fig. 5C and D). Together, these findings establish RALP1 as a novel rhoptry-resident protein.
FIG. 5.
Colocalization studies of RALP1 with rhoptry and microneme marker proteins. (A and B) RALP1-C-specific antibodies (green) colocalize with the rhoptry protein RAP1 (red) (A) and predominantly colocalize with CLAG9 (red) (B) in fixed schizonts (s) and free merozoites (m) using RAP1- and CLAG9-specific antibodies, respectively. (C and D) RALP1-C-specific antibodies (green) visualize a different compartment within the parasite than the microneme marker protein EBA-175 (red) (C) and EBA-181 (red) (D), as is evident in the merge of the two fluorescence photomicrographs. Nuclei were stained blue (DAPI).
Solubility properties of RALP1.
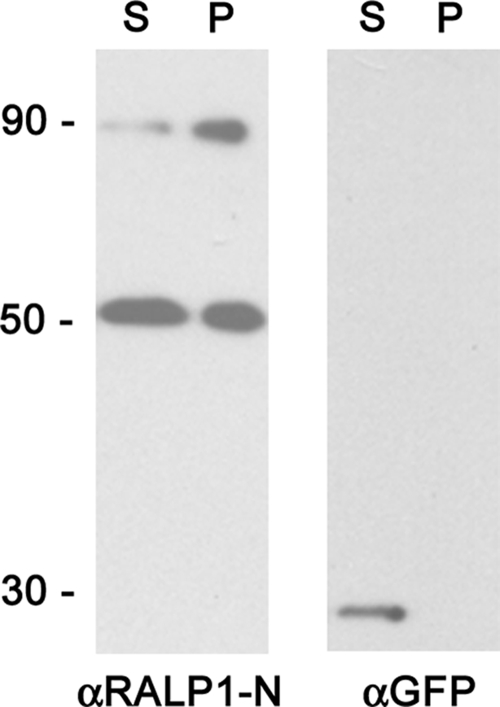
In order to study the solubility properties of RALP1, infected red blood cells were hypotonically lysed, mechanically disrupted, and separated into a soluble and an insoluble fraction. Both fractions were analyzed by Western blotting (Fig. 6). GFP was exclusively detected in the supernatant, whereas full-length RALP1 was found in both fractions, with more than 50% of the protein in the pellet fraction (Fig. 6). Although RALP1 does not contain a predicted transmembrane domain or GPI anchor signal, the protein appears to be only partially soluble.
FIG. 6.
Solubility of RALP1. Parasite material was lysed and fractionated into pellet (P) and supernatant (S). These fractions were separated on 10% SDS-PAGE and analyzed for the presence of RALP1 and GFP using specific antibodies. GFP was exclusively detected in the supernatant, whereas full-length RALP1 was found in both fractions, with more than 50% of the protein in the pellet fraction.
RALP1 is conserved in field isolates and is refractory to gene deletion.
Sequencing of five field isolates from the Kumasi district, Ghana, and single isolates from two other geographically distinct locations in Africa (Tanzania and South Africa) revealed conservation of the RALP1 sequence (see Fig. S2B in the supplemental material). The polymorphic block 2 of the msp-1 gene (52, 55) was used to verify the genetic diversity of these isolates (see Fig. S2C in the supplemental material). Together with the cross-species conservation of RALP1, this might point to an important role of RALP1 in the asexual life cycle of the parasite.
Disruption of the ralp1 gene was attempted using the pHTK vector system (15). This vector allows selection for double-crossover events using the negative selection agent ganciclovir. Further, both flanks were designed with additional stop codons to ensure gene disruption independent of single- or double-crossover events in the endogenous ralp1 locus (see Fig. S4A in the supplemental material). Southern blot analysis revealed that although stable transfectants were obtained, the plasmid remained episomal even after prolonged drug cycling and the addition of the negative selection agent ganciclovir. No integration could be detected (see Fig. S4B in the supplemental material).
DISCUSSION
P. falciparum causes the most lethal form of malaria in humans. The development of novel treatments against the pathogen is a priority (20-22). The understanding of the complex biology of the parasite was recently accelerated with the decoding of the genome, facilitating a detailed analysis of the transcriptome and proteome (reviewed in references 11 and 26).
The invasion of erythrocytes by merozoites is an ordered process requiring sequential steps that involve an unknown number of proteins. An important subset of these proteins are either located on the surface of the merozoite or stored in the secretory apical organelles. Some of these proteins are promising antigens that are currently being tested as vaccine candidates (e.g., MSP1, MSP2, and AMA-1) (reviewed in reference 21). All these proteins appear to be cotranslationally translocated into the endoplasmic reticulum by means of an N-terminal signal peptide and are transported within the secretory pathway to their subcellular locations (13). Additionally, the transcription of the corresponding genes seems to be tightly controlled during intraerythrocytic development (30), featuring minimal transcription in early blood stages (16 h postinfection) and maximal transcription in late stages (42 h postinfection). This profile correlates with the apparent functions of these proteins during the invasion process (10) and also seems to be important for trafficking (25, 63). Using these transcriptional and structural features, we retrieved 89 candidates (including 49 hypothetical proteins) from the annotated P. falciparum genome database that were potentially involved in the invasion process.
Many of the invasion-related proteins described so far share typical sequence features or similarities that facilitated their discovery and characterization. In order to find novel proteins unrelated to previously described proteins involved in invasion and to test our simple query approach, we chose the hypothetical protein MAL7P1.119 from the group of 49 novel uncharacterized proteins. This protein, which we named RALP1, is expressed in the schizont and merozoite stages of the P. falciparum life cycle. Orthologs of RALP1 are restricted to Plasmodium spp., indicating a specific biological function in the parasitic life cycle. Knockout of the ralp1 gene in P. falciparum was attempted, but no gene deletion mutant could be generated (see Fig. S4 in the supplemental material). This is reminiscent of the attempts to target GPI-anchored surface proteins (50), AMA-1 (65), or several SERA (34) proteins. It is recognized that the inability to knock out the ralp1 gene is not conclusive with respect to whether ralp1 is essential to parasite invasion.
An interesting structural feature of RALP1 is the presence of a peptide sequence that contains four heptad repeats of leucines, resembling a leucine zipper. This structural motif is well known as an oligomerization domain for DNA-regulatory proteins (e.g., c-Jun/c-Fos) (69) or signaling proteins (JLP) (29). Leucine zippers in general consist of a stretch of amino acids with a leucine residue in every seventh position in a coiled-coil helix. Coiled-coil regions provide the interface for protein-protein interactions and are ubiquitous assembly motifs found in a wide range of structural and regulatory proteins (reviewed in reference 32). Although it remains unclear whether the leucine heptad repeat itself or the predicted coiled-coil region represents the functional domain involved in protein-protein interactions, it is important to note that this domain lies within a cross-species-conserved region of RALP1 (see Fig. S2D in the supplemental material). Future studies must address the precise function of this domain, identify the putative RALP1 binding partners, and shed light on the putative processing of RALP1 resulting in a 55-kDa fragment. Secondary processing is well known for other secretory proteins, like AMA-1 (24), ASP (36), EBA-175 (44), and MSP-1/7 (9, 23, 38, 39).
We have shown that RALP1 is located at the apical end of the merozoite, where its its distribution is indistinguishable from that of RAP1 (4). This localization was confirmed by transgenic parasites expressing a GFP chimeric protein and by the expression of full-length RALP1 with a C-terminal TY1 epitope tag. The intracellular distributions of both transgenes were indistinguishable from that of the endogenous protein.
The rhoptries discharge their contents upon an appropriate signal during the invasion process. Their biogenesis appears to start around 40 h postinvasion with spheroidal structures growing by progressive fusion of small vesicles derived from the Golgi cisternae (5). Transmembrane proteins, such as members of the Rh family (43, 66), act as binding receptors on the surface of the host cell (12, 42). Others, like the RAP or the CLAG/RhopH proteins, might function as matrix proteins in nascent rhoptries or as escorter proteins ensuring correct trafficking or are involved in remodeling the erythrocyte after invasion (4, 31, 47, 48, 64). RALP1, like the RAP and some of the CLAG/RhopH proteins, displays no obvious transmembrane domain or membrane attachment signal. As RALP1 was only partially soluble after hypotonic lysis, it might be a structural component of the rhoptries and/or part of a densely packed rhoptry matrix.
No RALP1 was detectable in the parasite culture supernatant (data not shown), and the absence of any significant polymorphism (see Fig. S2B in the supplemental material) could argue for a limited immune exposure of RALP1 during invasion. As we were unable to detect RALP1 in young ring stage parasites, its function might be in the development, maintenance, or discharge of the rhoptries or it may play a role in the intimate space between the merozoite and the erythrocyte, aiding the process of invasion.
In summary, we identified a limited pool of novel proteins that might be involved in the process of invasion of erythrocytes and characterized one of them in detail. Investigation of this conserved Plasmodium sp.-specific protein and the elucidation of its precise biological function might help to increase our understanding of the process of erythrocyte invasion and to identify and validate novel therapeutic targets.
Supplementary Material
Acknowledgments
We are grateful to Otto Berninghausen, Julie Healer, Andreas Krüger, and Till Voss for critically reading the manuscript. Special thanks are due to Don Gardiner for providing antibodies directed against CLAG9, to Claudia Daubenberger for providing anti-GAPDH, and to Jacobus Pharmaceuticals for providing WR99120.
This study was supported by the Deutsche Forschungsgemeinschaft (DFG) (GI312) and the BioMalPar network of excellence (European Union Framework Programme). A.C. is the recipient of a DAAD scholarship. A.F.C. is supported by HHMI International Research Scholar Grants. T.S. is the recipient of a Humboldt fellowship. T.-W.G. is the recipient of an Emmy-Noether fellowship (DFG).
Editor: J. F. Urban, Jr.
Footnotes
Published ahead of print on 3 January 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Adams, J. H., P. L. Blair, O. Kaneko, and D. S. Peterson. 2001. An expanding ebl family of Plasmodium falciparum. Trends Parasitol. 17297-299. [DOI] [PubMed] [Google Scholar]
- 2.Adams, J. H., B. K. Sim, S. A. Dolan, X. Fang, D. C. Kaslow, and L. H. Miller. 1992. A family of erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. USA 897085-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16240-247. [DOI] [PubMed] [Google Scholar]
- 4.Baldi, D. L., K. T. Andrews, R. F. Waller, D. S. Roos, R. F. Howard, B. S. Crabb, and A. F. Cowman. 2000. RAP1 controls rhoptry targeting of RAP2 in the malaria parasite Plasmodium falciparum. EMBO J. 192435-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannister, L. H., J. M. Hopkins, R. E. Fowler, S. Krishna, and G. H. Mitchell. 2000. Ultrastructure of rhoptry development in Plasmodium falciparum erythrocytic schizonts. Parasitology 121273-287. [DOI] [PubMed] [Google Scholar]
- 6.Bastin, P., Z. Bagherzadeh, K. R. Matthews, and K. Gull. 1996. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77235-239. [DOI] [PubMed] [Google Scholar]
- 7.Baum, J., D. Richard, J. Healer, M. Rug, Z. Krnajski, T. W. Gilberger, J. L. Green, A. A. Holder, and A. F. Cowman. 2006. A conserved molecular motor drives cell invasion and gliding motility across malaria life cycle stages and other apicomplexan parasites. J. Biol. Chem. 2815197-5208. [DOI] [PubMed] [Google Scholar]
- 8.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 9.Blackman, M. J., I. T. Ling, S. C. Nicholls, and A. A. Holder. 1991. Proteolytic processing of the Plasmodium falciparum merozoite surface protein-1 produces a membrane-bound fragment containing two epidermal growth factor-like domains. Mol. Biochem. Parasitol. 4929-33. [DOI] [PubMed] [Google Scholar]
- 10.Bozdech, Z., M. Llinas, B. L. Pulliam, E. D. Wong, J. Zhu, and J. L. DeRisi. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowman, A. F., and B. S. Crabb. 2006. Invasion of red blood cells by malaria parasites. Cell 124755-766. [DOI] [PubMed] [Google Scholar]
- 12.Cowman, A. F., and M. T. Duraisingh. 2001. An old enemy, a new battle plan. Perspective on combating drug-resistant malaria. EMBO Rep. 277-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crabb, B. S., M. Rug, T. W. Gilberger, J. K. Thompson, T. Triglia, A. G. Maier, and A. F. Cowman. 2004. Transfection of the human malaria parasite Plasmodium falciparum. Methods Mol. Biol. 270263-276. [DOI] [PubMed] [Google Scholar]
- 14.Daubenberger, C. A., E. J. Tisdale, M. Curcic, D. Diaz, O. Silvie, D. Mazier, W. Eling, B. Bohrmann, H. Matile, and G. Pluschke. 2003. The N′-terminal domain of glyceraldehyde-3-phosphate dehydrogenase of the apicomplexan Plasmodium falciparum mediates GTPase Rab2-dependent recruitment to membranes. Biol. Chem. 3841227-1237. [DOI] [PubMed] [Google Scholar]
- 15.Duraisingh, M. T., T. Triglia, and A. F. Cowman. 2002. Negative selection of Plasmodium falciparum reveals targeted gene deletion by double crossover recombination. Int. J. Parasitol. 3281-89. [DOI] [PubMed] [Google Scholar]
- 16.Fidock, D. A., and T. E. Wellems. 1997. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. USA 9410931-10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardiner, D. L., T. Spielmann, M. W. Dixon, P. L. Hawthorne, M. R. Ortega, K. L. Anderson, T. S. Skinner-Adams, D. J. Kemp, and K. R. Trenholme. 2004. CLAG 9 is located in the rhoptries of Plasmodium falciparum. Parasitol. Res. 9364-67. [DOI] [PubMed] [Google Scholar]
- 18.Genton, B., F. Al-Yaman, I. Betuela, R. F. Anders, A. Saul, K. Baea, M. Mellombo, J. Taraika, G. V. Brown, D. Pye, D. O. Irving, I. Felger, H. P. Beck, T. A. Smith, and M. P. Alpers. 2003. Safety and immunogenicity of a three-component blood-stage malaria vaccine (MSP1, MSP2, RESA) against Plasmodium falciparum in Papua New Guinean children. Vaccine 2230-41. [DOI] [PubMed] [Google Scholar]
- 19.Gilberger, T. W., J. K. Thompson, T. Triglia, R. T. Good, M. T. Duraisingh, and A. F. Cowman. 2003. A novel erythrocyte binding antigen-175 paralogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. J. Biol. Chem. 27814480-14486. [DOI] [PubMed] [Google Scholar]
- 20.Girard, M. P., Z. H. Reed, M. Friede, and M. P. Kieny. 2006. A review of human vaccine research and development: Malaria. Vaccine 251567-1580. [DOI] [PubMed] [Google Scholar]
- 21.Good, M. F., D. Stanisic, H. Xu, S. Elliott, and M. Wykes. 2004. The immunological challenge to developing a vaccine to the blood stages of malaria parasites. Immunol. Rev. 201254-267. [DOI] [PubMed] [Google Scholar]
- 22.Greenwood, B. 2005. Malaria vaccines. Evaluation and implementation. Acta Trop. 95298-304. [DOI] [PubMed] [Google Scholar]
- 23.Holder, A. A., M. J. Blackman, P. A. Burghaus, J. A. Chappel, I. T. Ling, N. McCallum-Deighton, and S. Shai. 1992. A malaria merozoite surface protein (MSP1)-structure, processing and function. Mem. Inst. Oswaldo Cruz 87(Suppl. 3)37-42. [DOI] [PubMed] [Google Scholar]
- 24.Howell, S. A., C. Withers-Martinez, C. H. Kocken, A. W. Thomas, and M. J. Blackman. 2001. Proteolytic processing and primary structure of Plasmodium falciparum apical membrane antigen-1. J. Biol. Chem. 27631311-31320. [DOI] [PubMed] [Google Scholar]
- 25.Kocken, C. H., A. M. van der Wel, M. A. Dubbeld, D. L. Narum, F. M. van de Rijke, G. J. van Gemert, X. van der Linde, L. H. Bannister, C. Janse, A. P. Waters, and A. W. Thomas. 1998. Precise timing of expression of a Plasmodium falciparum-derived transgene in Plasmodium berghei is a critical determinant of subsequent subcellular localization. J. Biol. Chem. 27315119-15124. [DOI] [PubMed] [Google Scholar]
- 26.Kooij, T. W., C. J. Janse, and A. P. Waters. 2006. Plasmodium post-genomics: better the bug you know? Nat. Rev. Microbiol. 4344-357. [DOI] [PubMed] [Google Scholar]
- 27.LaCount, D. J., M. Vignali, R. Chettier, A. Phansalkar, R. Bell, J. R. Hesselberth, L. W. Schoenfeld, I. Ota, S. Sahasrabudhe, C. Kurschner, S. Fields, and R. E. Hughes. 2005. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 438103-107. [DOI] [PubMed] [Google Scholar]
- 28.Lambros, C., and J. P. Vanderberg. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65418-420. [PubMed] [Google Scholar]
- 29.Lee, C. M., D. Onesime, C. D. Reddy, N. Dhanasekaran, and E. P. Reddy. 2002. JLP: a scaffolding protein that tethers JNK/p38MAPK signaling modules and transcription factors. Proc. Natl. Acad. Sci. USA 9914189-14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Roch, K. G., Y. Zhou, P. L. Blair, M. Grainger, J. K. Moch, J. D. Haynes, P. De La Vega, A. A. Holder, S. Batalov, D. J. Carucci, and E. A. Winzeler. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 3011503-1508. [DOI] [PubMed] [Google Scholar]
- 31.Ling, I. T., L. Florens, A. R. Dluzewski, O. Kaneko, M. Grainger, B. Y. Yim Lim, T. Tsuboi, J. M. Hopkins, J. R. Johnson, M. Torii, L. H. Bannister, J. R. Yates III, A. A. Holder, and D. Mattei. 2004. The Plasmodium falciparum clag9 gene encodes a rhoptry protein that is transferred to the host erythrocyte upon invasion. Mol. Microbiol. 52107-118. [DOI] [PubMed] [Google Scholar]
- 32.Lupas, A. 1996. Prediction and analysis of coiled-coil structures. Methods Enzymol. 266513-525. [DOI] [PubMed] [Google Scholar]
- 33.Marshall, V. M., A. Silva, M. Foley, S. Cranmer, L. Wang, D. J. McColl, D. J. Kemp, and R. L. Coppel. 1997. A second merozoite surface protein (MSP-4) of Plasmodium falciparum that contains an epidermal growth factor-like domain. Infect. Immun. 654460-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, S. K., R. T. Good, D. R. Drew, M. Delorenzi, P. R. Sanders, A. N. Hodder, T. P. Speed, A. F. Cowman, T. F. de Koning-Ward, and B. S. Crabb. 2002. A subset of Plasmodium falciparum SERA genes are expressed and appear to play an important role in the erythrocytic cycle. J. Biol. Chem. 27747524-47532. [DOI] [PubMed] [Google Scholar]
- 35.Naik, R. S., G. Krishnegowda, and D. C. Gowda. 2003. Glucosamine inhibits inositol acylation of the glycosylphosphatidylinositol anchors in intraerythrocytic Plasmodium falciparum. J. Biol. Chem. 2782036-2042. [DOI] [PubMed] [Google Scholar]
- 36.O'Keeffe, A. H., J. L. Green, M. Grainger, and A. A. Holder. 2005. A novel Sushi domain-containing protein of Plasmodium falciparum. Mol. Biochem. Parasitol. 14061-68. [DOI] [PubMed] [Google Scholar]
- 37.Olsen, J. V., S. E. Ong, and M. Mann. 2004. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell Proteomics 3608-614. [DOI] [PubMed] [Google Scholar]
- 38.Pachebat, J. A., M. Kadekoppala, M. Grainger, A. R. Dluzewski, R. S. Gunaratne, T. J. Scott-Finnigan, S. A. Ogun, I. T. Ling, L. H. Bannister, H. M. Taylor, G. H. Mitchell, and A. A. Holder. 2007. Extensive proteolytic processing of the malaria parasite merozoite surface protein 7 during biosynthesis and parasite release from erythrocytes. Mol, Biochem. Parasitol. 15159-69. [DOI] [PubMed] [Google Scholar]
- 39.Pachebat, J. A., I. T. Ling, M. Grainger, C. Trucco, S. Howell, D. Fernandez-Reyes, R. Gunaratne, and A. A. Holder. 2001. The 22 kDa component of the protein complex on the surface of Plasmodium falciparum merozoites is derived from a larger precursor, merozoite surface protein 7. Molec. Biochem. Parasitol. 11783-89. [DOI] [PubMed] [Google Scholar]
- 40.Pagni, M., V. Ioannidis, L. Cerutti, M. Zahn-Zabal, C. V. Jongeneel, and L. Falquet. 2004. MyHits: a new interactive resource for protein annotation and domain identification. Nucleic Acids Res. 32W332-W335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson, D. S., L. H. Miller, and T. E. Wellems. 1995. Isolation of multiple sequences from the Plasmodium falciparum genome that encode conserved domains homologous to those in erythrocyte-binding proteins. Proc. Natl. Acad. Sci. USA 927100-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rayner, J. C., V. Corredor, D. Feldman, P. Ingravallo, F. Iderabdullah, M. R. Galinski, and J. W. Barnwell. 2002. Extensive polymorphism in the Plasmodium vivax merozoite surface coat protein MSP-3α is limited to specific domains. Parasitology 125393-405. [DOI] [PubMed] [Google Scholar]
- 43.Rayner, J. C., M. R. Galinski, P. Ingravallo, and J. W. Barnwell. 2000. Two Plasmodium falciparum genes express merozoite proteins that are related to Plasmodium vivax and Plasmodium yoelii adhesive proteins involved in host cell selection and invasion. Proc. Natl. Acad. Sci. USA 979648-9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reed, M. B., S. R. Caruana, A. H. Batchelor, J. K. Thompson, B. S. Crabb, and A. F. Cowman. 2000. Targeted disruption of an erythrocyte binding antigen in Plasmodium falciparum is associated with a switch toward a sialic acid-independent pathway of invasion. Proc. Natl. Acad. Sci. USA 977509-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robson, K. J., J. R. Hall, M. W. Jennings, T. J. Harris, K. Marsh, C. I. Newbold, V. E. Tate, and D. J. Weatherall. 1988. A highly conserved amino-acid sequence in thrombospondin, properdin and in proteins from sporozoites and blood stages of a human malaria parasite. Nature 33579-82. [DOI] [PubMed] [Google Scholar]
- 46.Sachs, J., and P. Malaney. 2002. The economic and social burden of malaria. Nature 415680-685. [DOI] [PubMed] [Google Scholar]
- 47.Sam-Yellowe, T. Y., and M. E. Perkins. 1991. Interaction of the 140/130/110 kDa rhoptry protein complex of Plasmodium falciparum with the erythrocyte membrane and liposomes. Exp. Parasitol. 73161-171. [DOI] [PubMed] [Google Scholar]
- 48.Sam-Yellowe, T. Y., H. Shio, and M. E. Perkins. 1988. Secretion of Plasmodium falciparum rhoptry protein into the plasma membrane of host erythrocytes. J. Cell Biol. 1061507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanders, P. R., P. R. Gilson, G. T. Cantin, D. C. Greenbaum, T. Nebl, D. J. Carucci, M. J. McConville, L. Schofield, A. N. Hodder, J. R. Yates III, and B. S. Crabb. 2005. Distinct protein classes including novel merozoite surface antigens in raft-like membranes of Plasmodium falciparum. J. Biol. Chem. 28040169-40176. [DOI] [PubMed] [Google Scholar]
- 50.Sanders, P. R., L. M. Kats, D. R. Drew, R. A. O'Donnell, M. O'Neill, A. G. Maier, R. L. Coppel, and B. S. Crabb. 2006. A set of glycosylphosphatidyl inositol-anchored membrane proteins of Plasmodium falciparum is refractory to genetic deletion. Infect. Immun. 744330-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sargeant, T. J., M. Marti, E. Caler, J. M. Carlton, K. Simpson, T. P. Speed, and A. F. Cowman. 2006. Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 7R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scherf, A., P. Barbot, and G. Langsley. 1989. Sequence and length polymorphism of a major malaria vaccine candidate analysed following DNA amplification. Nucleic Acids Res. 171774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sim, B. K., P. A. Orlandi, J. D. Haynes, F. W. Klotz, J. M. Carter, D. Camus, M. E. Zegans, and J. D. Chulay. 1990. Primary structure of the 175K Plasmodium falciparum erythrocyte binding antigen and identification of a peptide which elicits antibodies that inhibit malaria merozoite invasion. J. Cell Biol. 1111877-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smythe, J. A., M. G. Peterson, R. L. Coppel, A. J. Saul, D. J. Kemp, and R. F. Anders. 1990. Structural diversity in the 45-kilodalton merozoite surface antigen of Plasmodium falciparum. Mol. Biochem. Parasitol. 39227-234. [DOI] [PubMed] [Google Scholar]
- 55.Snewin, V. A., M. Herrera, G. Sanchez, A. Scherf, G. Langsley, and S. Herrera. 1991. Polymorphism of the alleles of the merozoite surface antigens MSA1 and MSA2 in Plasmodium falciparum wild isolates from Colombia. Mol. Biochem. Parasitol. 49265-275. [DOI] [PubMed] [Google Scholar]
- 56.Snow, R. W., C. A. Guerra, A. M. Noor, H. Y. Myint, and S. I. Hay. 2005. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434214-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spielmann, T., M. W. Dixon, M. Hernandez-Valladares, M. Hannemann, K. R. Trenholme, and D. L. Gardiner. 2006. Reliable transfection of Plasmodium falciparum using non-commercial plasmid mini preparations. Int. J. Parasitol. 361245-1248. [DOI] [PubMed] [Google Scholar]
- 58.Spielmann, T., D. J. Fergusen, and H. P. Beck. 2003. etramps, a new Plasmodium falciparum gene family coding for developmentally regulated and highly charged membrane proteins located at the parasite-host cell interface. Mol. Biol. Cell 141529-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson, J., R. E. Cooke, S. Moore, L. F. Anderson, C. J. Janse, and A. P. Waters. 2004. PTRAMP; a conserved Plasmodium thrombospondin-related apical merozoite protein. Mol. Biochem. Parasitol. 134225-232. [DOI] [PubMed] [Google Scholar]
- 60.Tomley, F. M., and D. S. Soldati. 2001. Mix and match modules: structure and function of microneme proteins in apicomplexan parasites. Trends Parasitol. 1781-88. [DOI] [PubMed] [Google Scholar]
- 61.Tonkin, C. J., G. G. van Dooren, T. P. Spurck, N. S. Struck, R. T. Good, E. Handman, A. F. Cowman, and G. I. McFadden. 2004. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol. Biochem. Parasitol. 13713-21. [DOI] [PubMed] [Google Scholar]
- 62.Trager, W., and J. B. Jensen. 1976. Human malaria parasites in continuous culture. Science 193673-675. [DOI] [PubMed] [Google Scholar]
- 63.Treeck, M., N. S. Struck, S. Haase, C. Langer, S. Herrmann, J. Healer, A. F. Cowman, and T. W. Gilberger. 2006. A conserved region in the EBL proteins is implicated in microneme targeting of the malaria parasite Plasmodium falciparum. J. Biol. Chem. 28131995-32003. [DOI] [PubMed] [Google Scholar]
- 64.Trenholme, K. R., D. L. Gardiner, D. C. Holt, E. A. Thomas, A. F. Cowman, and D. J. Kemp. 2000. clag9: a cytoadherence gene in Plasmodium falciparum essential for binding of parasitized erythrocytes to CD36. Proc. Natl. Acad. Sci. USA 974029-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Triglia, T., J. Healer, S. R. Caruana, A. N. Hodder, R. F. Anders, B. S. Crabb, and A. F. Cowman. 2000. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol. Microbiol. 38706-718. [DOI] [PubMed] [Google Scholar]
- 66.Triglia, T., J. Thompson, S. R. Caruana, M. Delorenzi, T. Speed, and A. F. Cowman. 2001. Identification of proteins from Plasmodium falciparum that are homologous to reticulocyte binding proteins in Plasmodium vivax. Infect. Immun. 691084-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Triglia, T., P. Wang, P. F. Sims, J. E. Hyde, and A. F. Cowman. 1998. Allelic exchange at the endogenous genomic locus in Plasmodium falciparum proves the role of dihydropteroate synthase in sulfadoxine-resistant malaria. EMBO J. 173807-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trucco, C., D. Fernandez-Reyes, S. Howell, W. H. Stafford, T. J. Scott-Finnigan, M. Grainger, S. A. Ogun, W. R. Taylor, and A. A. Holder. 2001. The merozoite surface protein 6 gene codes for a 36 kDa protein associated with the Plasmodium falciparum merozoite surface protein-1 complex. Mol. Biochem. Parasitol. 11291-101. [DOI] [PubMed] [Google Scholar]
- 69.Turner, R., and R. Tjian. 1989. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science 2431689-1694. [DOI] [PubMed] [Google Scholar]
- 70.Waller, R. F., M. B. Reed, A. F. Cowman, and G. I. McFadden. 2000. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 191794-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu, T., C. G. Black, L. Wang, A. R. Hibbs, and R. L. Coppel. 1999. Lack of sequence diversity in the gene encoding merozoite surface protein 5 of Plasmodium falciparum. Mol. Biochem. Parasitol. 103243-250. [DOI] [PubMed] [Google Scholar]
- 72.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17847-848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.