Abstract
The intracellular parasite Leishmania causes a wide spectrum of human disease, ranging from self-resolving cutaneous lesions to fatal visceral disease, depending on the species of Leishmania involved. The mechanisms by which different Leishmania species cause different pathologies are largely unknown. We have addressed this question by comparing the gene expression profiles of bone marrow-derived macrophages infected with either Leishmania donovani or L. major promastigotes. We found that the two species had very similar effects on macrophage gene expression. Both species caused a small (<2.5-fold) but statistically significant repression of several hundred genes. In addition, both species strongly induced and repressed about 60 genes. Comparing the effects of the two species showed that only 26 genes were regulated differently by L. major as opposed to L. donovani, including those for metallothioneins 1 and 2, HSP70 and -72, CCL4, Gadd45β, Dsp1, matrix metalloprotease 13, T-cell death-associated gene 51 (Tdag51), RhoB, spermine/spermidine N1-acyl transferase 1 (SSAT), and Cox2. L. donovani-infected macrophages were also found to express higher levels of Cox2 protein and prostaglandin E synthase mRNA than L. major-infected macrophages. While both species have previously been shown to trigger prostaglandin E synthesis by bystander cells, this study suggests that infected macrophages themselves express prostaglandin E-synthesizing genes only in response to L. donovani.
Leishmaniasis is a spectral disease, with pathologies ranging from disfiguring but self-resolving skin lesions (cutaneous leishmaniasis) to a fatal infection of the liver and spleen (visceral leishmaniasis, or kala-azar). In each case, the causative agent is a single-celled intracellular parasite of the genus Leishmania (18, 28). Numerous factors influence disease severity, but the most important determinant of the form of leishmaniasis is the species of Leishmania involved. The lifestyles of all infectious Leishmania species are essentially identical, apart from differences in reservoir host species. They are transmitted by a sandfly vector and are inoculated into the host when an infected sandfly takes a blood meal. Once in the host, they exclusively replicate in macrophage phagolysosomes, from where they modulate macrophage behavior to ensure parasite survival and determine disease progression. During cutaneous leishmaniasis, infected macrophages remain near the original sandfly bite, and pathology results from local inflammation. Other forms of leishmaniasis develop when infected macrophages migrate away from the sandfly bite, with visceral leishmaniasis involving dissemination throughout the body, especially to the liver and spleen (33). While considerable progress is now being made in revealing mechanisms by which Leishmania can manipulate macrophage behavior (reviewed in reference 14), the differences among species, and how they contribute to such markedly different pathologies, are still largely unknown at the molecular level. One study has shown that Leishmania donovani, which causes visceral disease in humans, recruits far fewer leukocytes and proinflammatory cytokines than L. major (which causes only cutaneous lesions) during early infection of the mouse air pouch model (27). A greater inflammatory response may help prevent dissemination of L. major away from the site of infection. Furthermore, amastigotes of cutaneous species, such as L. mexicana and L. panamensis, do not survive well at 37°C in vitro, being better adapted to lower temperatures such as may be found in the skin (16). In contrast, strains of L. donovani amastigotes that can be grown in vitro grow best at 37°C, indicating adaptation to growth at the higher temperatures of the internal organs.
Since Leishmania is an intracellular organism, the clinical form of leishmaniasis depends on the behavior of the infected macrophage. This in turn depends, at least in part, on the genes expressed by the infected macrophage. We therefore addressed the question of how different species cause different pathologies by comparing the gene expression profiles of macrophages identically infected in vitro with either L. major or L. donovani, which cause cutaneous and visceral leishmaniasis, respectively.
MATERIALS AND METHODS
Cell culture and infection.
L. major Friedlin V9 and L. donovani 1S2D promastigotes were cultured at 28°C with 5% CO2 (RPMI 1640 medium with 10% fetal bovine serum, 85 μM minimal essential medium [MEM] nonessential amino acids solution, 0.5× MEM amino acids solution, and 850 μM MEM sodium pyruvate [all from Invitrogen], 10 mM dextrose, 25 mM HEPES, pH 7.3, 55 μg/ml penicillin, and 125 μg/ml streptomycin). L. donovani axenic amastigotes were cultured at 37°C with 5% CO2 in RPMI 1640 medium with 20% fetal bovine serum, 15 mM KCl, 114.6 mM KH2PO4, 10.38 mM K2HPO4, 0.5 mM MgSO4, 24 mM NaHCO3, 1× RPMI 1640 vitamin mixture, RPMI 1640 amino acid mixture, 0.58 mg/ml glutamine, 22 mM d-glucose, 55 μg/ml penicillin, 125 μg/ml streptomycin, 0.25 mM adenosine, 25 mM MES (morpholineethanesulfonic acid), 10 μg/ml folic acid, 5 μg/ml hemin, pH 5.5. Bone marrow-derived macrophages (BMMs) were prepared from the femurs and tibias of female BALB/c mice (Charles River Breeding Laboratories) essentially as described previously (4). The bones were flushed with complete Dulbecco's modified Eagle's medium (DMEM) (Invitrogen catalog no. 11965-092) supplemented with 10% fetal bovine serum, 18 mM HEPES, pH 7.3, 55 μg/ml penicillin, and 125 μg/ml streptomycin, and adherent cell types were removed by growing them at 37°C with 5% CO2 overnight in adherent dishes, in complete DMEM, with the addition of 25% L929-conditioned medium (LCM) as a source of colony stimulating factor-1. Nonadherent, immature macrophages were transferred to fresh, nonadherent dishes (Fisher) and grown for 6 days, with further supplementation with LCM, to induce macrophage differentiation. The cells were grown for a further 24 h in the absence of LCM and scraped, and viable cells were counted following a trypan blue exclusion assay. BMMs (25 × 106) were transferred to each of four 50-ml polypropylene tubes at a density of 1 × 106 cells/ml, and 5 ml DMEM containing nothing, 5 × 108 stationary-phase L. donovani promastigotes, 1 × 109 stationary-phase L. major promastigotes, or 30 μl 10% latex beads (Sigma) was added. Infection was allowed to proceed for 16 h at 37°C with 5% CO2. The cells were then washed four times with 25 ml complete DMEM to remove excess promastigotes and returned to the incubator to recover. Cells were harvested after 24 h of infection. To determine the infection levels, two small aliquots were taken from each culture, spun onto glass microscope slides, and stained with Diff-Quik (Dade Behring) modified Wright-Giemsa stain according the manufacturer's instructions. The percentage of infected cells and the mean number of amastigotes or latex beads within each macrophage was counted by microscopy, in duplicate for each slide.
RNA isolation and microarray analysis.
Macrophages were recovered by centrifugation at 1,800 rpm and lysed directly in 3 ml Trizol reagent (Invitrogen). RNA was purified according to the manufacturer's instructions, resuspended in 50 μl nuclease-free water (Sigma), and quantified by spectrophotometry. Subsequent processing was performed at the McGill University and Genome Quebec Genome Centre, as previously described (31). Briefly, RNA integrity was confirmed using a Bioanalyzer 2100 and Bio Sizing software version A.02.12 (Agilent Technologies). RNA (10 μg) was reverse transcribed to cDNA from a T7-(T)24 primer (Genosys). After second-strand synthesis, the entire cDNA reaction mixture was in vitro transcribed to produce biotin-labeled cRNA, 15 μg of which was hybridized at 45°C overnight to Affymetrix Murine Genome U74Av2 DNA microarrays, which contained 12,488 probe sets, corresponding to 9,322 individual genes. The arrays were then washed using a GeneChip Fluidics Station 400 (Affymetrix). Streptavidin-phycoerythrin was attached to specifically bound probes and detected using a GeneArray Scanner (Agilent). The scanned images were analyzed using Affymetrix Microarray Suite 5.0, and signal intensities were normalized to the mean with a reference of 1,000 units. Each infection and control pair was performed on three different occasions, using different preparations of BMMs, and processed independently to give three replicates. We also performed a single hybridization with RNA derived from L. donovani axenic amastigotes as a control for cross-hybridization.
Statistical analysis.
Microarray data were quantified using Microarray Suite 5.0 software (MAS5) (Affymetrix). Probe sets showing statistically significant variance (P < 0.05) across conditions were identified by one-way analysis of variance (ANOVA), followed by Tukey's honestly significant difference test (HSD) (P < 0.05), to identify the source of variance (e.g., L. major infected versus uninfected or L. major infected versus L. donovani infected). In addition to probes exactly complementary to the target sequences (PM probes), each spot on the Affymetrix arrays used in this study contained oligonucleotides that had a single-base mismatch (MM probes) as a control for nonspecific hybridization. The MAS5 software compares the ratios of signal hybridized to the specific and the mismatched probes to assess whether the signal intensity reliably reflects the abundance of target RNA: the relative PM and MM probe signals are used to exclude data from probes that sense cross-hybridized RNA rather than specifically hybridized RNA and can also be used to identify probe sets showing high levels of cross-hybridization to amastigote RNA. The results are called “P” (present), “M” (marginal), or “A” (absent). Probe sets with statistically significant changes in signal intensity were further filtered to retain only those with at least two “P” calls from the three repeats or one “P” and two “M” calls and a mean signal intensity greater than 250 under the conditions that gave the stronger signal. Thus, for the expression of a gene to count as significantly altered following infection in this study, the corresponding probe set must satisfy the following criteria: (i) significant variance by ANOVA, (ii) significant pairwise difference by HSD, (iii) adequate P calls, (iv) absent cross-hybridization, and (v) sufficient mean signal intensity. For some parts of this study, a minimum difference in mean signal intensity was also required. An alternative analysis using the MAS5 software, which is based on Wilcoxon's signed rank test, gave closely similar results (data not shown). Genes were annotated and classified by function using EASE and DAVID (8, 20) (http://www.david.niaid.nih.gov), with additional input from Ingenuity pathway analysis (Ingenuity Systems).
Quantitative real-time PCR (qRT-PCR).
First-strand synthesis was performed using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. cDNAs were amplified in a Rotor-Gene Real Time PCR machine (Corbett Research), using Taq polymerase (Invitrogen) and the supplied buffer with 3 μM MgCl2, 10 μM of each gene-specific primer, and 1× Sybr green (Sigma). The PCR conditions were 95°C denaturation (20 s), followed by a combined annealing and elongation step (68°C for 20 s) and an additional 15-s data acquisition step at a temperature high enough to denature primer dimers and nonspecific products without affecting the amplified target. The primer sequences and acquiring temperatures are listed in the supplemental material. The data were analyzed using the Rotor-Gene software against a standard curve obtained from serial dilutions of B10R macrophage cDNA. Data from at least three independent infections were normalized to the β2-microglobulin housekeeping gene and expressed as the induction relative to macrophages containing latex beads ± the standard error of the mean.
Western blotting.
Infected macrophages, or macrophages stimulated with 100 ng/ml lipopolysaccharide (LPS) for 6 h, were lysed by incubation on ice in standard buffer (50 mM Tris, pH 8.0, 137 mM NaCl, 1% Triton X-100, and Complete protease inhibitor cocktail [Roche Applied Science]). Soluble fractions were blotted using anti-cyclooxygenase 2 (α-Cox2) polyclonal antibody (Santa Cruz Biotoechnology; sc-1745). The data were quantified using ImageJ (1) (http://rsb.info.nih.gov/ij) and normalized to actin densities.
Microarray data accession number.
The microarray data files have been submitted to Array Express with accession number E-MEXP-1254.
RESULTS
Infection rates and parasite loads were equivalent.
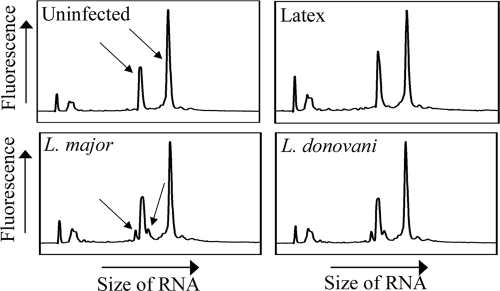
In order to effectively compare the results of infection with two different species of Leishmania, we first established cell culture conditions to ensure that the levels of infection were equivalent. Light microscopy on Diff-Quik-stained aliquots of infected BMMs showed that both the percentages of cells infected and the mean amastigote loads were comparable between L. major- and L. donovani-infected cells in each of the three experimental replicates used in this study (Table 1). Furthermore, comparison of the signal intensities of the Leishmania-specific (16S and 23S) peaks in the RNA extracts themselves revealed similar peak heights (Fig. 1), confirming that the parasite loads were comparable. This was important for such a study, in which reproducibility of experimental conditions was essential. Subsequent microarray analysis was carried out on the three replicate infections described in Table 1.
TABLE 1.
Equivalent parasite loads following infection with L. major and L. donovania
| Repeat no. | % Infection
|
No. of parasites/beads per cell
|
||||
|---|---|---|---|---|---|---|
| Latex | L. major | L. donovani | Latex | L. major | L. donovani | |
| 1 | 71 | 70 | 74 | 6.7 | 4.2 | 4.6 |
| 2 | 87 | 77 | 78 | 11.9 | 4.1 | 4.2 |
| 3 | 92 | 91 | 91 | 17.9 | 8.5 | 6.2 |
BMMs were infected, or allowed to phagocytose latex beads, for 16 h; washed thoroughly; and allowed to recover for a further 8 h. The percentage of cells infected and the mean number of parasites per cell were determined following staining with Diff-Quik.
FIG. 1.
Equivalent infection with L. major and L. donovani. RNA extracts from each condition were analyzed using a Bioanalyzer chip prior to microarray analysis. The arrows indicate the macrophage-derived 18S and 28S rRNAs (shown in uninfected samples) and Leishmania-derived 16S and 23S rRNAs (in L. major-infected samples). The results are representative of three experiments.
Latex beads have very little effect on gene expression.
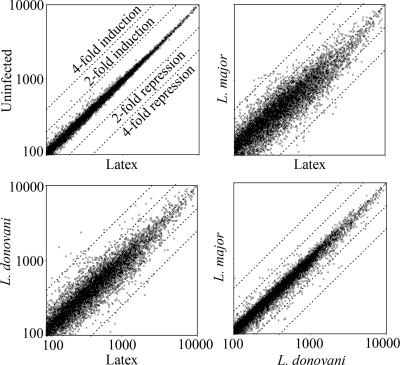
As controls for this study, we used both unstimulated BMMs and macrophages that had been allowed to phagocytose latex beads. While they are phagocytosed effectively by macrophages, latex beads are otherwise inert and so were included to identify changes in gene expression that are a general result of phagocytosis and phagolysosome formation rather than being specific to Leishmania infection. As shown in the scatter plots (Fig. 2), the gene expression profiles of unstimulated macrophages and macrophages that had phagocytosed latex beads were extremely similar. Indeed, we identified only 15 of the 12,488 probe sets that showed a statistically significant difference between no stimulation and phagocytosis of latex beads (see the supplemental material). The difference was greater than twofold for only one of these genes. We conclude that phagocytosis of latex beads does not significantly affect macrophage gene expression at 24 h. This observation strongly supported the validity of the changes in gene expression that we observed following Leishmania infection and allowed us to conclude that the changes we observed following infection are specific to Leishmania and are not simply due to phagocytosis.
FIG. 2.
Variations in global gene expression following infection with L. major or L. donovani or phagocytosis of inert latex beads, as indicated. Each data point represents the fluorescence intensity of an individual probe set (the mean of three experiments, in arbitrary units, normalized to 1,000 without further filtering).
Amastigote RNA does not significantly cross-hybridize to a murine array.
In addition to mammalian RNA, RNA prepared from Leishmania-infected cells contains RNA derived from the intracellular amastigotes (Fig. 1). To assess the effect of cross-hybridized Leishmania RNA, we hybridized RNA from L. donovani axenic amastigotes in culture to a single Affymetrix Mu74Av2 chip. Fewer than 5% of the microarray probe sets returned a “P” or “M” call (indicating significant cross-hybridization). Furthermore, these were generally at weak signal intensities (see the supplemental material). This low level of cross-hybridization indicates that the amastigote RNA did not significantly distort the signal from infected cells. The 569 probe sets that did return “P” or “M” calls were excluded from subsequent analyses.
Infection with either strain results in a general, small repression of gene expression.
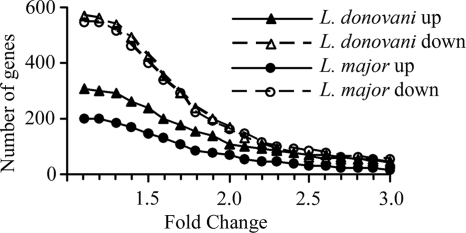
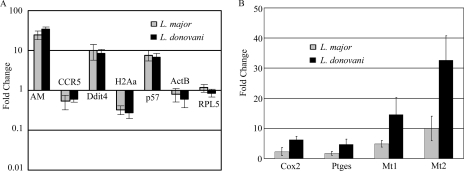
In contrast to the effect of latex beads, macrophages that had been infected with either L. donovani or L. major showed large alterations in overall gene expression compared to unstimulated macrophages (not shown) or macrophages that had phagocytosed latex beads (Fig. 2). More detailed analysis identified 870 probe sets that were significantly affected by L. donovani infection and 748 affected by L. major. This number is consistent with a recent study of L. major-infected THP-1 cells, which showed 741 differentially regulated genes (10). The majority of these genes were inhibited by less than 2.5-fold (Fig. 3). However, when a 2.5-fold change requirement was introduced, approximately equal numbers of genes (∼65) were induced and repressed by L. donovani. In contrast, L. major repressed slightly more (85 probe sets) and induced 30. The change in expression was confirmed by qRT-PCR for a sample of these genes (Fig. 4A). As expected, many of the genes are involved in the immune response, and some have been previously identified by other methods, confirming the validity of this approach. For example, both species repressed the chemokine receptor CCR5 (34) and components of major histocompatibility complex class II (6, 36, 37). However, many genes were identified that had previously not been recognized to play a role during Leishmania infection; they had diverse cellular functions, including intra- and intercellular signaling and regulation of the cell cycle (Table 2; see the supplemental material).
FIG. 3.
Numbers of genes significantly induced or repressed by infection with L. major or L. donovani. Probe sets with significantly changed signal intensities following infection were identified by ANOVA and HSD, and then the mean change was calculated relative to macrophages that had been exposed to latex beads. The graph depicts the numbers of probe sets (y axis) changed by at least the amounts indicated (x axis).
FIG. 4.
Confirmation of altered expression of selected genes by qRT-PCR. (A) mRNAs significantly regulated by both L. major and L. donovani relative to expression in macrophages exposed to latex beads. CCR5, C-C chemokine receptor 5; p57, CDK inhibitor 1C; H2Aa, major histocompatibility complex class II antigen Aα. β-Actin (ActB) and ribosomal large subunit protein 5 (RPL5) were included as negative controls. Note the logarithmic scale: induced mRNAs are shown above the baseline, repressed mRNAs below the baseline. (B) mRNAs regulated differently by L. major and L. donovani. Ptges, PGE synthase; Mt, metallothionein.
TABLE 2.
Selected mRNAs significantly regulated by both L. major and L. donovania
| Gene namea | NCBI GeneID | Baseline expressionb | Fold changec
|
|
|---|---|---|---|---|
| L. major | L. donovani | |||
| Apolipoprotein C2 | 11813 | 4,325 | −.0 | −7.6 |
| Cadherin 1 | 12550 | 862 | 2.9 | 2.9 |
| CCR5 | 12774 | 7,503 | −4.3 | −2.2 |
| Cyclin-dependent kinase inhibitor 1C (p57) | 12577 | 626 | 5.9 | 8.8 |
| DNA-damage-inducible transcript 4 (Ddit4) | 74747 | 8,412 | 14.9 | 26.1 |
| HSP70 a5 (Bip/GRP78) | 14828 | 9,041 | 3.7 | 3.9 |
| MHC class II, antigen Aα (H2-Aa) | 14960 | 8,860 | −5.8 | −5.0 |
| MHC class II, antigen Eα (H2-Ea) | 14968 | 3,025 | −6.8 | −7.3 |
See the supplemental material for the full list.
Baseline expression (the mean normalized intensity from latex bead-treated macrophages, in arbitrary units) is listed as an indicator of how strongly the genes were expressed before infection.
The values represent means relative to macrophages treated with latex beads. Negative numbers indicate repression.
Cox2 is induced by L. donovani but only weakly by L. major.
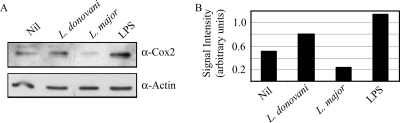
A major goal of this study was to compare the effects of L. donovani and L. major on macrophage gene expression. Statistical comparison of the microarray data from macrophages infected with the two species identified only 26 genes with a statistically significant difference in expression of greater than twofold (Table 3). A sample of these differences were confirmed by qRT-PCR (Fig. 4B). Of particular interest, the microarray data showed that prostaglandin-endoperoxide synthase (Cox2) mRNA was induced strongly by L. donovani but not significantly by L. major, suggesting that the two species may differ in their effects on macrophage prostaglandin metabolism (Table 3). Furthermore, prostaglandin E (PGE) synthase, the enzyme that follows Cox2 in the prostaglandin E2 (PGE2) synthetic pathway, is also induced following infection with L. donovani but only weakly following L. major infection (Fig. 4B). This indicates that L. donovani may induce PGE2 synthesis more strongly than L. major. We therefore investigated Cox2 protein levels in infected cells and found that L. donovani-infected cells expressed significantly more Cox2 protein than L. major-infected macrophages (Fig. 5). LPS, a known inducer of Cox2, was used as a positive control. Interestingly, while L. donovani infection increased Cox2 protein expression, as expected, L. major infection produced a marked reduction in Cox2. This suggests that mechanisms in addition to regulation of mRNA levels contribute to the difference in the Cox2 protein level observed following infection with the two species.
TABLE 3.
Genes with differing expression in L. major- and L. donovani-infected macrophages
| Gene namea | NCBI GeneID | Fold changeb
|
||
|---|---|---|---|---|
| L. donovani relative to latexc | L. major relative to latexc | Expression in L. donovani relative to L. majord | ||
| Tdag51/Phlda1 | 21664 | 12.6 | (0.71) | 17.8 |
| Heat shock protein 1B (HSP70) | 15511 | 8.2 | (1.8) | 4.5 |
| Heat shock protein 1A (HSP72) | 193740 | 15.3 | (4.0) | 3.8 |
| PARP7 | 99929 | 6.2 | (1.7) | 3.7 |
| Gadd45β | 17873 | 8.0 | (2.4) | 3.4 |
| Matrix metalloprotease 13 | 17386 | 2.4 | (0.76) | 3.1 |
| RhoB | 11852 | 2.6 | (0.84) | 3.1 |
| Cox2 | 19225 | 10.3 | (3.5) | 3.0 |
| Ferm domain containing 6 | 319710 | 4.4 | (1.6) | 2.8 |
| Chymase 1 | 17228 | (1.1) | (0.43) | 2.6 |
| Metallothionein 2 | 17750 | 15.5 | (6.25) | 2.5 |
| Plasminogen activator inhibitor 2 | 18788 | 3.0 | (1.3) | 2.4 |
| Connexin 43 | 14609 | 3.3 | (1.4) | 2.4 |
| P34SEI-1 | 55942 | 2.2 | (0.92) | 2.3 |
| Metallothionein 1 | 17748 | 6.3 | (2.8) | 2.3 |
| Dual-specificity phosphatase 1 | 19252 | 2.9 | (1.3) | 2.3 |
| SSAT | 20229 | 1.8 | (0.82) | 2.2 |
| RGC32 | 66214 | 5.4 | (2.5) | 2.2 |
| Mitogen-inducible gene 6 | 74155 | (1.7) | (0.77) | 2.2 |
| Protein phosphatase 2 Cα | 19052 | 2.6 | (1.2) | 2.2 |
| CCL4 (MIP1β) | 20303 | 4.1 | (1.9) | 2.2 |
| 1190002N15Rik | 68861 | (1.3) | (0.63) | 2.0 |
| Kuppel-like factor 4 (Klf4) | 16600 | 2.8 | (1.4) | 2.0 |
| Tubulin α3B | 22147 | 2.8 | (1.4) | 2.0 |
| Estrogen-responsive finger protein | 217069 | (1.7) | (0.82) | 2.0 |
| Linker for activated T cells (LAT2) | 56743 | (0.56) | (1.2) | 0.47 |
Some genes did not show statistically significant differences when macrophages infected with either species were compared with macrophages exposed to latex beads; however, there were significant differences between L. major- and L. donovani-infected macrophages, so they are included in this list.
Parentheses indicate changes that were not statistically significant. Probe sets with statistically significantly different signal intensities between L. major and L. donovani macrophages were identified as described in Materials and Methods.
Changes relative to macrophages exposed to latex beads are included for context only.
Differences between mean signal intensities.
FIG. 5.
(A) Western blot analysis of Cox2 protein abundance following infection of macrophages with L. donovani or L. major or stimulation with LPS, as indicated. Actin protein abundance was used as a loading control. (B) Band intensities were quantified by densitometry, and Cox2 densities are expressed relative to actin intensities.
DISCUSSION
In this study, we investigated the effects of L. major and L. donovani on macrophage gene expression. Our major observation was that, compared to phagocytosis of inert latex beads, the uptake of Leishmania had a significant overall impact on macrophage gene expression, mediating both an increase and a decrease of gene expression. However, the majority of regulated genes were repressed by less than twofold. While some proinflammatory genes were induced, the overall profile did not conform to classical, alternative, or type II macrophage activation, which is consistent with two previous studies showing a “hybrid” gene expression profile in macrophages infected with L. chagasi (38) or L. major (10). Interestingly, there were remarkably few differences between cells infected with L. major and with L. donovani with respect to effects on macrophage gene expression. This is consistent with an earlier study involving a wider variety of pathogens that also observed little difference between the gene expression profiles of macrophages infected for 16 h with L. major or L. donovani promastigotes (5). Although these parasites cause very different diseases, they have very similar effects on macrophages, as they initiate infection within the first 24 h. Future studies should consider later time points, including after several days, when the amastigotes are actively replicating.
Nevertheless, a difference was apparent in the pathway-associated with PGE synthesis, as suggested by the greater increase in Cox2 and PGE synthase gene expression observed following L. donovani infection compared with L. major infection. As discussed below, this may contribute in part to the very different pathologies caused by the two Leishmania species.
General repression of gene expression following infection.
Previous studies have produced contrasting conclusions as to whether Leishmania infection results in a widespread repression of gene expression. An early study from our laboratory, using radioactively labeled probes against a cDNA array of 588 macrophage genes, found that nearly 40% of the detectable expressed genes were repressed at least twofold following infection for 4 days with L. donovani axenic amastigotes (4). However, two subsequent studies, using Leishmania promastigotes and full-genome fluorescence microarrays, have reported similar numbers of genes repressed and induced by at least twofold (38) or fivefold (5). This study agrees with the more recent reports in showing approximately equal numbers of genes induced and repressed by at least 2.5-fold following infection by L. donovani. However, in the absence of a cutoff, we also show that many more genes are repressed than induced. This may provide a resolution to the controversy, since the radiolabeling approach may be able to detect changes that appear small by more current quantitative techniques, and many of the repressed genes we detected in our earlier study were repressed by only around twofold (4). Thus, we propose that a large number of genes are indeed repressed following infection, but by small amounts. Furthermore, it may be that this general repression becomes more pronounced as the infection becomes established, since our earlier study involved a longer infection time than any of the subsequent studies. The general repression is likely distinct from the more dramatic induction or repression of a smaller number of specific genes that is observed in all studies.
We have not addressed here the biological significance of this repression. In particular, our measurements of total mRNA do not allow a distinction between repression/induction at the transcriptional level and alteration in mRNA stability. The repression we observed could simply be a side effect of the inhibitory signaling mechanisms that are known to be activated following infection, such as protein tyrosine dephosphorylation. However, while a small repression of an individual gene may not have a major effect by itself, repression of many genes in the same or related pathways may add up to have a significant effect on macrophage function while maintaining cell viability. In particular, a disproportionate number of the weakly repressed genes are involved in mitochondrial respiration, allowing us to speculate that a constrained ATP supply may help Leishmania gain control of the macrophage, perhaps by preventing ATP-dependent apoptosis.
Potential role of Cox2 in visceralization.
Cox2 is the rate-limiting enzyme induced to allow the synthesis of prostaglandins, including PGE2, by macrophages. Various studies have attempted to address the roles of Cox2 and PGE2 during infection with a single species of Leishmania, but the effects of different species on Cox2 expression have not previously been compared. L. donovani has been shown to trigger macrophage PGE2 production (35) via a protein kinase C-dependent induction of Cox2 (26). However, another study found that in vitro infection of macrophages with L. major did not induce PGE2 (11). Our data support and reconcile these different studies by directly showing the contrasting effects of the two species on Cox2 and PGE synthase induction in the host macrophage.
PGE2 is a multifunctional cytokine and might be expected to play multiple roles during Leishmania infections. Indeed, the ability of PGE2 to repress Th1 cytokines and thus to favor a Th2 response is well established (3), and splenocytes from mice infected with a range of Leishmania species have been shown to secrete PGE2 (7, 11, 24). Furthermore, inhibition of PGE2 production with indomethacin has been shown to increase Th1 cytokine production and decrease susceptibility to cutaneous L. major (7, 11, 24) and L. amazonensis (15) infection. It will be interesting to identify the source of PGE2 during L. major infection, since the present study shows that it is unlikely to be the infected macrophages themselves. Some early studies reported the presence of an adherent, PGE2-secreting suppressor cell in the spleens of L. major-infected mice (11). Interestingly, this suppressor cell, apparently an uninfected macrophage, was also found following infection with L. donovani and L. tropica (29, 39), suggesting that this mechanism of PGE2-mediated immunosuppression is common to all species of Leishmania. Cox2 and PGE2 therefore potentially play at least two roles during Leishmania infection: (i) production by a suppressor cell type, which promotes a Th2 response and parasite survival and which appears to occur regardless of Leishmania species, and (ii) production by the infected macrophages themselves, which occurs following L. donovani but not L. major infection and which therefore may contribute to visceralization. The mechanism by which this might occur is unclear, but it is interesting that PGE2 production has been correlated with the ability of L. donovani to escape the lymph nodes and migrate to the liver and spleen following cutaneous inoculation of malnourished mice (2).
While we have focused on Cox2 for biological reasons, the biggest difference was seen in the expression of Tdag51/Phlda1, which was expressed nearly 18-fold more in L. donovani-infected macrophages than in L. major-infected macrophages. The biological function(s) of the gene and its product have not been intensively studied, but roles have been proposed in promoting (13, 21, 30, 32) or preventing (40) apoptosis and during endoplasmic reticular stress (19, 22, 42), and it may link the two processes. How these functions are linked to visceralization is not immediately clear; however, the proapoptotic function of Tdag51 is neutralized by heat shock proteins (17). Since HSP1A and HSP1B are both induced more strongly by L. donovani than by L. major, this may indicate that an alternative function of Tdag51 is operative in this context. Interestingly, a very recent study of Toll-like receptor (TLR) signaling showed increased induction of Tdag51 by LPS in Trib1 knockout cells (41). Cox2, PAI-2, MMP13, and connexin 43 were also coregulated in this study, while various reports have implicated TLR signaling in the response to Leishmania (9, 12, 23, 25). Taken together, these observations imply that differential TLR usage or costimulation may contribute to the differences in gene expression we observed here.
In this study, we sought to address the question of how L. donovani causes visceral disease while L. major remains cutaneous. Our data suggest that Cox2 induction and PGE2 synthesis may have roles to play in the visceralization of L. donovani. We also observed differences in the expression of 25 other genes between macrophages infected with the two Leishmania species, including metallothioneins 1 and 2, HSP70 and -72, and the less well-characterized Tdag51. It will be of great interest to determine the roles that these genes play in governing the different pathologies caused by the two species.
Supplementary Material
Acknowledgments
We acknowledge the Canadian Institutes of Health Research for supporting this research and Yannick Fortin and Daniel Vincent for technical assistance.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 17 December 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 1136-42. [Google Scholar]
- 2.Anstead, G. M., B. Chandrasekar, W. Zhao, J. Yang, L. E. Perez, and P. C. Melby. 2001. Malnutrition alters the innate immune response and increases early visceralization following Leishmania donovani infection. Infect. Immun. 694709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betz, M., and B. S. Fox. 1991. Prostaglandin E2 inhibits production of Th1 lymphokines but not of Th2 lymphokines. J. Immunol. 146108-113. [PubMed] [Google Scholar]
- 4.Buates, S., and G. Matlashewski. 2001. General suppression of macrophage gene expression during Leishmania donovani infection. J. Immunol. 1663416-3422. [DOI] [PubMed] [Google Scholar]
- 5.Chaussabel, D., R. T. Semnani, M. A. McDowell, D. Sacks, A. Sher, and T. B. Nutman. 2003. Unique gene expression profiles of human macrophages and dendritic cells to phylogenetically distinct parasites. Blood 102672-681. [DOI] [PubMed] [Google Scholar]
- 6.De Almeida, M. C., S. A. Cardoso, and M. Barral-Netto. 2003. Leishmania (Leishmania) chagasi infection alters the expression of cell adhesion and costimulatory molecules on human monocyte and macrophage. Int. J. Parasitol. 33153-162. [DOI] [PubMed] [Google Scholar]
- 7.De Freitas, L. A., L. M. Mbow, M. Estay, J. A. Bleyenberg, and R. G. Titus. 1999. Indomethacin treatment slows disease progression and enhances a Th1 response in susceptible BALB/c mice infected with Leishmania major. Parasite Immunol. 21273-277. [DOI] [PubMed] [Google Scholar]
- 8.Dennis, G., B. Sherman, D. Hosack, J. Yang, W. Gao, H. C. Lane, and R. Lempicki. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 43. [PubMed] [Google Scholar]
- 9.de Veer, M. J., J. M. Curtis, T. M. Baldwin, J. A. DiDonato, A. Sexton, M. J. McConville, E. Handman, and L. Schofield. 2003. MyD88 is essential for clearance of Leishmania major: possible role for lipophosphoglycan and Toll-like receptor 2 signaling. Eur. J. Immunol. 332822-2831. [DOI] [PubMed] [Google Scholar]
- 10.Dogra, N., C. Warburton, and W. R. McMaster. 2007. Leishmania major abrogates gamma interferon-induced gene expression in human macrophages from a global perspective. Infect. Immun. 753506-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell, J. P., and C. E. Kirkpatrick. 1987. Experimental cutaneous leishmaniasis. II. A possible role for prostaglandins in exacerbation of disease in Leishmania major-infected BALB/c mice. J. Immunol. 138902-907. [PubMed] [Google Scholar]
- 12.Flandin, J. F., F. Chano, and A. Descoteaux. 2006. RNA interference reveals a role for TLR2 and TLR3 in the recognition of Leishmania donovani promastigotes by interferon-gamma-primed macrophages. Eur. J. Immunol. 36411-420. [DOI] [PubMed] [Google Scholar]
- 13.Gomes, I., W. Xiong, T. Miki, and M. R. Rosner. 1999. A proline- and glutamine-rich protein promotes apoptosis in neuronal cells. J. Neurochem. 73612-622. [DOI] [PubMed] [Google Scholar]
- 14.Gregory, D. J., and M. Olivier. 2005. Subversion of host cell signalling by the protozoan parasite Leishmania. Parasitology 130(Suppl.)S27-S35. [DOI] [PubMed] [Google Scholar]
- 15.Guimaraes, E. T., L. A. Santos, R. Ribeiro dos Santos, M. M. Teixeira, W. L. C. dos Santos, and M. B. P. Soares. 2006. Role of interleukin-4 and prostaglandin E2 in Leishmania amazonensis infection of BALB/c mice. Microbes Infect. 81219-1226. [DOI] [PubMed] [Google Scholar]
- 16.Gupta, N., N. Goyal, and A. K. Rastogi. 2001. In vitro cultivation and characterization of axenic amastigotes of Leishmania. Trends Parasitol. 17150-153. [DOI] [PubMed] [Google Scholar]
- 17.Hayashida, N., S. Inouye, M. Fujimoto, Y. Tanaka, H. Izu, E. Takaki, H. Ichikawa, J. Rho, and A. Nakai. 2006. A novel HSF1-mediated death pathway that is suppressed by heat shock proteins. EMBO J. 254773-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herwaldt, B. L. 1999. Leishmaniasis. Lancet 3541191-1199. [DOI] [PubMed] [Google Scholar]
- 19.Hinz, T., S. Flindt, A. Marx, O. Janssen, and D. Kabelitz. 2001. Inhibition of protein synthesis by the T cell receptor-inducible human TDAG51 gene product. Cell Signal 13345-352. [DOI] [PubMed] [Google Scholar]
- 20.Hosack, D., G. Dennis, B. Sherman, H. Lane, and R. Lempicki. 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hossain, G. S., J. V. van Thienen, G. H. Werstuck, J. Zhou, S. K. Sood, J. G. Dickhout, A. B. de Koning, D. Tang, D. Wu, E. Falk, R. Poddar, D. W. Jacobsen, K. Zhang, R. J. Kaufman, and R. C. Austin. 2003. TDAG51 is induced by homocysteine, promotes detachment-mediated programmed cell death, and contributes to the development of atherosclerosis in hyperhomocysteinemia. J. Biol. Chem. 27830317-30327. [DOI] [PubMed] [Google Scholar]
- 22.Joo, J. H., G. Liao, J. B. Collins, S. F. Grissom, and A. M. Jetten. 2007. Farnesol-induced apoptosis in human lung carcinoma cells is coupled to the endoplasmic reticulum stress response. Cancer Res. 677929-7936. [DOI] [PubMed] [Google Scholar]
- 23.Kropf, P., M. A. Freudenberg, M. Modolell, H. P. Price, S. Herath, S. Antoniazi, C. Galanos, D. F. Smith, and I. Muller. 2004. Toll-like receptor 4 contributes to efficient control of infection with the protozoan parasite Leishmania major. Infect. Immun. 721920-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, J., U. M. Padigel, P. Scott, and J. P. Farrell. 2002. Combined treatment with interleukin-12 and indomethacin promotes increased resistance in BALB/c mice with established Leishmania major infections. Infect. Immun. 705715-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liese, J., U. Schleicher, and C. Bogdan. 2007. TLR9 signaling is essential for the innate NK cell response in murine cutaneous leishmaniasis. Eur. J. Immunol. 373424-3434. [DOI] [PubMed] [Google Scholar]
- 26.Matte, C., G. Maion, W. Mourad, and M. Olivier. 2001. Leishmania donovani-induced macrophages cyclooxygenase-2 and prostaglandin E2 synthesis. Parasite Immunol. 23177-184. [DOI] [PubMed] [Google Scholar]
- 27.Matte, C., and M. Olivier. 2002. Leishmania-induced cellular recruitment during the early inflammatory response: modulation of proinflammatory mediators. J. Infect. Dis. 185673-681. [DOI] [PubMed] [Google Scholar]
- 28.Murray, H. W., J. D. Berman, C. R. Davies, and N. G. Saravia. 2005. Advances in leishmaniasis. Lancet 3661561-1577. [DOI] [PubMed] [Google Scholar]
- 29.Murray, H. W., S. M. Carriero, and D. M. Donelly. 1986. Presence of a macrophage-mediated suppressor cell mechanism during cell-mediated immune response in experimental visceral leishmaniasis. Infect. Immun. 54487-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neef, R., M. A. Kuske, E. Prols, and J. P. Johnson. 2002. Identification of the human PHLDA1/TDAG51 gene: down-regulation in metastatic melanoma contributes to apoptosis resistance and growth deregulation. Cancer Res. 625920-5929. [PubMed] [Google Scholar]
- 31.Novak, J. P., R. Sladek, and T. J. Hudson. 2002. Characterization of variability in large-scale gene expression data: implications for study design. Genomics 79104-113. [DOI] [PubMed] [Google Scholar]
- 32.Park, C. G., S. Y. Lee, G. Kandala, S. Y. Lee, and Y. Choi. 1996. A novel gene product that couples TCR signaling to Fas(CD95) expression in activation-induced cell death. Immunity 4583-591. [DOI] [PubMed] [Google Scholar]
- 33.Peters, W., and R. Killick-Kendrick. 1987. The leishmaniases in biology and medicine. Academic Press, London, United Kingdom.
- 34.Pinheiro, N. F., Jr., M. D. Hermida, M. P. Macedo, J. Mengel, A. Bafica, and W. L. dos-Santos. 2006. Leishmania infection impairs beta 1-integrin function and chemokine receptor expression in mononuclear phagocytes. Infect. Immun. 743912-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiner, N. E., and C. J. Malemud. 1985. Arachidonic acid metabolism by murine peritoneal macrophages infected with Leishmania donovani: in vitro evidence for parasite-induced alterations in cyclooxygenase and lipoxygenase pathways. J. Immunol. 134556-563. [PubMed] [Google Scholar]
- 36.Reiner, N. E., W. Ng, T. Ma, and W. R. Mcmaster. 1988. Kinetics of gamma interferon binding and induction of major histocompatibility complex class II mRNA in Leishmania-infected macrophages. Proc. Natl. Acad. Sci. USA 854330-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiner, N. E., W. Ng, and W. R. Mcmaster. 1987. Parasite-accessory cell interactions in murine leishmaniasis. 2. Leishmania donovani suppresses macrophage expression of class I and class II major histocompatibility complex gene products. -J. Immunol. 1381926-1932. [PubMed] [Google Scholar]
- 38.Rodriguez, N. E., H. K. Chang, and M. E. Wilson. 2004. Novel program of macrophage gene expression induced by phagocytosis of Leishmania chagasi. Infect. Immun. 722111-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott, P. A., and J. P. Farrell. 1981. Experimental cutaneous leishmaniasis. I. Nonspecific immunodepression in BALB/c mice infected with Leishmania tropica. J. Immunol. 1272395-2400. [PubMed] [Google Scholar]
- 40.Toyoshima, Y., M. Karas, S. Yakar, J. Dupont, H. Lee, and D. LeRoith. 2004. TDAG51 mediates the effects of insulin-like growth factor I (IGF-I) on cell survival. J. Biol. Chem. 27925898-25904. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto, M., S. Uematsu, T. Okamoto, Y. Matsuura, S. Sato, H. Kumar, T. Satoh, T. Saitoh, K. Takeda, K. J. Ishii, O. Takeuchi, T. Kawai, and S. Akira. 2007. Enhanced TLR-mediated NF-IL6 dependent gene expression by Trib1 deficiency. J. Exp. Med. 2042233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, J., S. Lhotak, B. A. Hilditch, and R. C. Austin. 2005. Activation of the unfolded protein response occurs at all stages of atherosclerotic lesion development in apolipoprotein E-deficient mice. Circulation 1111814-1821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.