Abstract
Certain bacteria use a type III secretion system (TTSS) to deliver effector proteins that interfere with cell function into host cells. While transcription of genes encoding TTSS components has been demonstrated, studies to date have failed to identify TTSS effector proteins in Bordetella pertussis. Here we present the first evidence of a functionally active TTSS in B. pertussis. Three known TTSS effectors, Bsp22, BopN, and BopD, were identified as TTSS substrates in B. pertussis 12743. We found expression of Bsp22 in a significant proportion of clinical isolates but not in common laboratory-adapted strains of B. pertussis. We generated a TTSS mutant of B. pertussis 12743 and showed that it induced significantly lower respiratory tract colonization in mice than the wild-type bacteria. Respiratory infection of mice with the mutant bacteria induced significantly greater innate proinflammatory cytokine production in the lungs soon after challenge, and this correlated with significantly higher antigen-specific interleukin-17, gamma interferon, and immunoglobulin G responses later in infection. Our findings suggest that the TTSS subverts innate and adaptive immune responses during infection of the lungs and may be a functionally important virulence factor for B. pertussis infection of humans.
Bordetella pertussis is the causative agent of whooping cough or pertussis, a respiratory disease that is most severe in infants and young children. Although vaccination has significantly reduced morbidity and mortality, pertussis remains an endemic disease and is one of the major causes of vaccine-preventable deaths today, with WHO estimates of 45 million cases and 409,000 deaths each year. In recent years a resurgence of pertussis was observed in a number of vaccinated populations (6, 29). Furthermore, it has become increasingly clear that pertussis is not only a childhood disease but also is highly prevalent among adults (21). This has called into question the level of protection provided by current pertussis vaccines and highlighted the need for a better understanding of the molecular mechanisms underlying the pathogenesis of B. pertussis infection.
Bacteria produce a complex array of virulence factors, including toxins and adhesins, which facilitate colonization and/or suppress immune responses and allow the bacteria to establish infection in the host. One of these virulence factors, the type III secretion system (TTSS), is a specialized secretory apparatus that allows gram-negative bacteria to inject proteins, known as effectors, directly into the eukaryotic cell cytosol. In laboratory conditions bacteria can be induced to secrete TTSS substrates, which include effectors and proteins involved in the delivery process, into the extracellular milieu in the absence of eukaryotic cells. TTSSs have been shown to be important mediators of virulence of a range of animal pathogens, including Yersinia spp., Salmonella spp., Shigella spp., Escherichia coli, and Bordetella bronchiseptica (15, 39). Yuk and colleagues have reported that the TTSS of B. bronchiseptica modulates dendritic cell (DC) maturation (31, 33), enhancing production of the anti-inflammatory cytokine interleukin-10 (IL-10) and promoting bacterial persistence (32).
Despite reports describing transcription of genes encoding components of the Bordetella bsc TTSS machinery in B. pertussis Tohama I (14, 22), the B. pertussis isolate chosen for genome sequencing, studies to date have failed to demonstrate TTSS effector secretion by B. pertussis in vitro or in vivo (9, 22). The sequencing of the B. pertussis Tohama I genome has revealed an extraordinary high level of genetic flexibility (28), and this raises concerns about the adequacy of laboratory-adapted strains for the study of natural clinical pathogenesis. Differences in gene expression have been shown to affect virulence characteristics of laboratory-adapted versus corresponding low-passage clinical isolates of E. coli, Staphylococcus aureus, and Pseudomonas aeruginosa (11, 34, 37).
Here we demonstrate secretion of the Bordetella TTSS effector, Bsp22, by a significant portion of low-passage clinical isolates of B. pertussis, but not by common laboratory-adapted strains, such as Tohama I and Wellcome 28. Mutation of bscN, which encodes an essential component for TTSS secretion across bacterial membranes, abolished in vitro secretion of TTSS substrates by a clinical isolate of B. pertussis, resulting in a reduced ability of the bacteria to colonize the respiratory tracts of mice, and this was associated with enhanced local inflammatory and antigen-specific cellular and humoral immune responses. Our data suggest that expression of a functional TTSS is a feature of natural infection of humans with B. pertussis and that this may confer virulence to the bacteria by subverting the protective innate and adaptive immunity of the host.
MATERIALS AND METHODS
Bacterial strains and growth media.
Low-passage isolates B. pertussis ATCC 12743 (5375 [3865]), ATCC 12742 (5374 [3747]), and ATCC 9340 (5 [17921]), hereafter referred to as B. pertussis 12743, 12742, and 9340, respectively, were obtained from the ATCC. B. pertussis 12743 and 12742 were from cultures made by E. K. Anderson and deposited in the ATCC by G. Eldering (8), and B. pertussis 9340 was from a culture made by P. Kenrick and deposited in the ATCC by M. Pittman in the 1950s. Sixteen clinical isolates were cultivated from the sputum, noses, nasopharynges, or throats of infants or adults with whooping cough in The Netherlands between 1949 and 2005. Wild-type (WT) B. pertussis and B. bronchiseptica were grown at 37°C on Bordet-Gengou (BG) agar and in Stainer-Scholte (SS) broth. Gentamicin-resistant ΔbscN derivatives of B. pertussis 12743 and B. bronchiseptica RB50 were grown on BG agar or SS broth supplemented with 10 μg/ml gentamicin (Gibco, United Kingdom). For allelic exchange WT B. pertussis 12743 was first rendered streptomycin resistant by subculture in increasing sublethal concentrations of streptomycin (final concentration, 100 μg/ml). For routine cloning and conjugation, E. coli XL1-Blue and SM10λpir were grown at 37°C on Luria-Bertani (LB) agar or LB broth (BD Difco) supplemented with the appropriate antibiotics (ampicillin, 150 μg/ml; gentamicin, 10 μg/ml; kanamycin, 25 μg/ml).
Generation of ΔbscN bacteria.
Gentamicin-resistant ΔbscN derivatives of B. pertussis 12743 and B. bronchiseptica RB50, in which a 0.5-kb internal portion of the bscN gene was removed and replaced with a 0.7-kb fragment containing a gentamicin resistance cassette, were constructed as follows. Primers PAB20 (5′-GCCCTGCGGATCCCGCG-3′) and NF5 (5′-TACTGACGCATGCCCCTATCC-3′), annealing to bp 65 to 81 of bscL (5′ flanking gene to bscN) and bp 1 to 12 of bscN, respectively, were used to amplify a 0.5-kb fragment from Tohama I. The resulting PCR product was digested with BamHI and SphI (recognition sequences underlined in primer sequences) and inserted into the corresponding sites of the cloning vector pQE-80 (Qiagen, United Kingdom) to produce pNF2. Primers NF4 (5′-GCTGGGCATGCTGGTCAAGGGC-3′) and PAB21 (5′-GCCGGCTCGCGATGCATCG-3′), annealing to bp 560 to 582 of bscN and bp 470 to 489 of bscO (3′ flanking gene to bscN), respectively, were used to amplify a 1.3-kb fragment from Tohama I. The resulting PCR product was digested with SphI and AgeI (recognition sequence underlined in primer sequence) to yield a 0.5-kb fragment and inserted into the corresponding SphI and XmaI sites on pNF2 to create pNF7, containing a mutant bscN allele. The gentamicin resistance cassette was amplified from pSS1129 using primers Gmr_for_2 (5′-ATAGCATGCTGACGCACACCG-3′) and Gmr_rev (5′-GCATGCTTAGGTGGCGGTAC-3′) with SphI sites engineered at the 5′ and 3′ ends, respectfully, and inserted into the unique SphI site at the center of the mutant bscN allele on pNF7 to create pNF12. BamHI (bp 1 to 6 of the bscN::Gmr mutant allele) and NheI (138 bases downstream) sites flanking the bscN::Gmr mutant allele on pNF12 were then used to subclone the bscN::Gmr mutant allele into the allelic exchange vector pEGBR (1) to create pNF13. pNF13 was then introduced into streptomycin-resistant B. pertussis 12743 and B. bronchiseptica RB50 by conjugation, and gentamicin-resistant double recombinants of B. pertussis 12743 and B. bronchiseptica RB50 generated by allelic exchange were screened by PCR for the presence of the bscN::Gmr mutant allele.
Recombinant His-Bsp22 and Ab production.
Primers PAB37 (5′-CGGAAGCTTTTAGCGCATGTTGCTGGTG-3′), which binds bp 596 to 615 of the Bsp22 gene, and PAB38 (5′-AGCGGATCCAGCATTGATCTCGGAGTTCAC-3′), which binds bp 4 to 25 of the Bsp22 gene, were used to amplify the Bsp22 gene from B. pertussis 18323. The resulting PCR fragment was cloned in frame with the six-His tag located on pQE-80 (Qiagen, United Kingdom), creating pAPB15, from which recombinant His-tagged Bsp22 was expressed in E. coli XL1-Blue. His-Bsp22 was purified by affinity chromatography on Ni2+ columns. For antibody (Ab) production, a rabbit was immunized subcutaneously with recombinant His-Bsp22 (230 μg) emulsified in complete Freund's adjuvant (Difco) and boosted on days 14 and 28 with His-Bsp22 (115 μg) in incomplete Freund's adjuvant. The rabbit was exsanguinated 12 days after the last immunization, and serum was prepared and shown by Western blotting to contain an Ab that specifically recognized Bsp22.
Protein identification by MALDI-TOF mass spectrometry and immunoblotting.
For matrix-assisted laser desorption-ionization-time of flight (MALDI-TOF) mass spectrometry analysis of protein samples, culture supernatants were harvested from Bordetella spp. following 24 h of growth in liquid culture. Filtered supernatants were precipitated with 30% ammonium chloride. Protein pellets were analyzed by MALDI-TOF mass spectrometry, and identified peptide fragments were searched against the predicted proteins from the complete B. bronchiseptica RB50 and B. pertussis Tohama I genomes (www.sanger.ac.uk/Projects/B_pertussis/). For Western blots, cultures were harvested for each bacterial strain at the same stage of bacterial growth, as determined by measurements of optical density at 600 nm (OD600), and total protein from a 1-ml supernatant fraction was precipitated with 10% (wt/vol) trichloroacetic acid and resuspended in sample buffer. Proteins were separated on 10% (wt/vol) sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes (Sigma) prior to Western blotting with a polyclonal Ab raised against B. pertussis Bsp22, pertussis toxin (PT), or filamentous hemagglutinin (FHA), followed by the appropriate secondary Ab.
In vitro growth curves.
B. pertussis 12743 and ΔbscN B. pertussis 12743 were grown in SS medium in 100-ml cultures inoculated at a starting OD600 of 0.2. Samples (1 ml) were removed at 4, 8, 12, 23, 27, 37, and 49 h, OD600 was determined, and CFU counts were performed by plating neat and diluted samples on BG agar plates.
In vitro cytotoxicity and adherence assays.
DC were expanded in vitro from bone marrow of BALB/c mice by culture for 10 days with granulocyte-macrophage colony-stimulating factor as described previously (13). Murine DC, J774 macrophages, and human HEK-293T epithelial cells (1 × 106/ml) were incubated in Dulbecco's modified Eagle's medium supplemented with 8% fetal calf serum in 24-well plates. Stationary-phase cultures of Bordetella spp. were harvested, resuspended in 1% casein, and added to triplicate wells at a multiplicity of infection (MOI) of 20, 100, or 500. Cytotoxicity was determined after 4 h using the Cytotox 96 nonradioactive cytotoxicity assay (Promega), which measures lactate dehydrogenase release as a measure of cell lysis. For detection of bacterial adherence to macrophages and of internalization, J774 cells were incubated with Bordetella spp. at an MOI of 100. Following incubation at 37°C for 2 h, the contents of each well were harvested, washed to remove unbound bacteria, and resuspended in either 1 ml of medium supplemented with kanamycin (100 μg/ml) to kill extracellular bacteria or 1 ml medium alone. The cell suspensions were incubated at 37°C for a further 1 h and then centrifuged at 1,200 rpm for 5 min. The cells were washed and resuspended in 1 ml phosphate-buffered saline with 1% (wt/vol) Triton X-100 and left at room temperature for 5 min. One hundred-microliter samples of the appropriate dilutions of the lysed cells were then plated in triplicate onto BG agar plates and incubated at 37°C for 2 days for B. bronchiseptica and 3 to 5 days for B. pertussis.
B. pertussis infection of mice.
Mice were infected with WT or ΔbscN B. pertussis 12743 by exposure to an aerosol of bacteria for 15 min as described previously (20). The standard inoculum was 2 × 1010 bacteria/ml (based on OD600), and this was reduced or increased by two- to threefold to achieve a lower or higher initial colonization. Four mice from each experimental group were sacrificed 3 h and 3, 7, 14, 21, and 28 days after challenge to assess the number of viable bacteria in the lungs. The numbers of CFU in lung homogenates were estimated as described previously (20). Results are given as the mean numbers of B. pertussis CFU for individual lungs from four mice per experimental group. The sensitivity of the assay was 0.56 log10 CFU per lung.
Quantification of cytokine concentrations and neutrophil infiltration into the lungs.
Lung homogenates were centrifuged at 13,000 rpm for 5 min, and the concentrations of IL-10, IL-12p70, IL-1β, tumor necrosis factor alpha (TNF-α), macrophage inflammatory protein 1 (MIP-1) and MIP-2 (R&D Systems), IL-12p40, IL-6, and transforming growth factor β (BD Pharmingen) were determined by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions. Neutrophil infiltration into the lungs was quantified by performing differential counts on hematoxylin-and-eosin-stained cytospin preparations of cells recovered by bronchoalveolar lavage as described previously (23).
Detection of antigen-specific immune responses.
Spleens and serum samples were recovered from mice 14, 21, and 28 days after bacterial challenge. Spleen cells (2 × 106 cells/ml) were stimulated with sonicated B. pertussis, PT, FHA, or medium alone and incubated at 37°C for 72 h. Supernatants were removed, and gamma interferon (IFN-γ) and IL-17 concentrations were determined by ELISA. Lung T cells were purified on T-cell isolation columns as described previously (23). Lung T cells were stimulated with heat-inactivated B. pertussis antigen and splenic antigen-presenting cells (2 × 106 irradiated spleen cells). After 5 days the surviving T cells were stimulated with phorbol myristate acetate and ionomycin for 1 h, and then brefeldin A was added for 4 h at 37°C. Cells were stained with fluorescently labeled Abs specific for CD4 (eBioscinces). Cells were then fixed, permeabilized, and incubated with fluorescently labeled anti-IFN-γ or anti-IL-17 Abs (BD Pharmingen) according to the manufacturer's instructions (Fix & Perm cell permeabilization kit; Invitrogen). Immunofluorescence was analyzed using Summit software on a Cyan ADP flow cytometer (Dako). Anti-B. pertussis immunoglobulin G (IgG), IgG2a, and IgG1 Ab titers were determined by ELISA as described previously (13). Results are expressed as log10 end point Ab titers, determined by extrapolation of the straight part of the dilution curve, versus the OD492 value of the control serum for naive mice.
Statistical analysis.
Statistical analysis was performed using Graphpad Prism. Student's t test was used to compare the mean values between two groups. Statistical differences in mean values between more than two experimental groups were determined by analysis of variance. Linear regression was used to examine the correlation between bacterial growth estimated by CFU counts and OD600. A nonlinear regression analysis was performed on the growth curves for B. pertussis 12743 and ΔbscN B. pertussis 12743. An Akaike's information criterion test with a confidence interval of 95% was applied to the data, with a supplemental t test, to determine if the growth curves of bacterial strains are different.
RESULTS
Secretion of TTSS effector proteins by clinical but not laboratory-adapted isolates of B. pertussis.
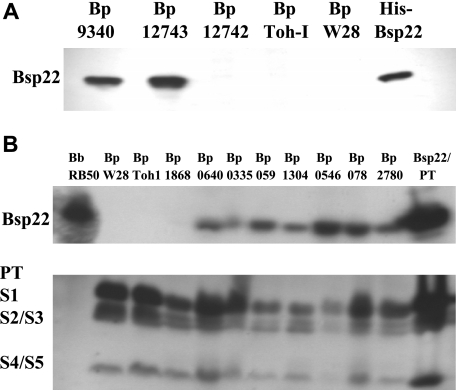
B. pertussis Tohama I is well characterized and was chosen for genome sequencing (28) but has been through extensive laboratory passage since its isolation in Japan in 1954 (16). Since the TTSS expression and secretory function of Yersinia is regulated and activated in vivo by eukaryotic cell contact and can be altered by prolonged laboratory passage (4, 36), we examined low-passage B. pertussis isolates from the ATCC (B. pertussis 12743, 12742, and 9340) and compared these with the laboratory-adapted strains (Tohama I and Wellcome 28) for expression of Bsp22. Bsp22, whose gene is located in the TTSS operon in B. pertussis and B. bronchiseptica (www.sanger.ac.uk/Projects/B_pertussis/), is secreted by B. bronchiseptica (39) and is a useful marker of TTSS secretory function. In the present study, His-tagged Bsp22 was cloned and expressed in E. coli and the purified protein was used to generate polyclonal anti-Bsp22 Abs. Western blotting with anti-Bsp22 revealed significant quantities of secreted Bsp22 protein in culture supernatants of B. pertussis 12743 and 9340, but not in 12742, Tohama I, or Wellcome 28 (Fig. 1A). These data demonstrate that two of the three early isolates from the ATCC that were examined expressed TTSS proteins in vitro, but this may have been lost in the common laboratory-adapted strains Tohama I and Wellcome 28.
FIG. 1.
Clinical isolates but not common laboratory-adapted stains of B. pertussis express Bsp22. Protein samples from supernatants of stationary-phase cultures of B. pertussis (Bp) 9340, 12743, 12742, Tohama I (Toh-I), and Wellcome 28 (W28) (A) or B. bronchiseptica (Bb) RB50, B. pertussis W28 and Toh-I, and eight clinical isolates of B. pertussis recovered from respiratory tracts of patients with whooping cough (B), prepared by precipitation with 10% trichloroacetic acid or purified His-Bsp22 or PT (2 μg), were resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and probed with Abs specific for Bsp22 or PT (S1 to S5 subunits) by Western blotting.
We next examined supernatants from stationary-phase cultures of 16 clinical isolates, cultivated from infants or adults with whooping cough in The Netherlands between 1949 and 2005. Western blotting with anti-Bsp22 Abs revealed secretion of Bsp22 in 15 of 16 isolates (Fig. 1; results are shown for 8 clinical isolates; an additional 8 [not shown] all expressed Bsp22). Bsp22 was also detected in B. bronchiseptica RB50 but not B. pertussis Tohama I or Wellcome 28. PT was detected in all strains except B. bronchiseptica RB50, confirming that the clinical isolates were B. pertussis and not B. bronchiseptica. These results demonstrate that a large proportion of the low-passage clinical isolates of B. pertussis examined have a functional TTSS.
In order to confirm these findings and to facilitate identification of additional Bordetella TTSS substrates, we carried out mass spectroscopy analysis on total secreted proteins from culture supernatants. MALDI-TOF analysis revealed a number of known Bordetella virulence factors, including FHA, adenylate cyclase toxin, and pertactin in B. pertussis Tohama I, B. pertussis 12743, and B. bronchiseptica RB50 (Table 1), as well as other proteins not yet implicated in virulence (data not shown). Tracheal colonization factor, a virulence factor of B. pertussis but not B. bronchiseptica (10), was detected in B. pertussis Tohama I and B. pertussis 12743 but not B. bronchiseptica RB50. Serum resistance protein (BrkA) was detected in B. pertussis Tohama I and B. pertussis 12743 but not B. bronchiseptica RB50; BrkA expression has been demonstrated for some strains of B. bronchiseptica, but not for strain RB50 (30). PT, a B. pertussis-specific virulence factor, was also detected in B. pertussis 12743; this required a separate analysis from that shown in Table 1, using more concentrated culture supernatant (not shown). As well as Bsp22, the TTSS translocator protein, BopD, and the sensor/plug protein, BopN, were detected in supernatants from B. pertussis 12743 but not B. pertussis Tohama I. The Bordetella TTSS effector protein, BteA/BopC, was detected in B. bronchiseptica RB50 but not in B. pertussis 12743. These results provide further evidence that B. pertussis 12743 has a functional TTSS in vitro and demonstrate for the first time that BopD and BopN, in addition to Bsp22, are substrates of the B. pertussis TTSS.
TABLE 1.
Identification of secreted TTSS substrates and other virulence factors from Bordetella spp. by MALDI-TOF mass spectrometrya
| Mascot accession no. | Identified protein | Result for:
|
||||
|---|---|---|---|---|---|---|
| B. pertussis Toh I | B. pertussis 12743 | ΔbscN B. pertussis 12743 | B. bronchiseptica RB50 | ΔbscN B. bronchiseptica RB50 | ||
| BP1879/BB2993 | FHA | + (141) | + (8,869) | + (3,776) | + (567) | + (434) |
| BP0760/BB0324 | Bifunctional hemolysin-adenylate cyclase toxin (CyaA) | + (116) | + (2,423) | + (1,074) | + (2,551) | + (2,571) |
| BP1054/BB1366 | Pertactin (Prn) | + (111) | + (2,066) | + (759) | + (450) | + (350) |
| BP3494/BB0961 | Serum resistance protein (BrkA) | + (234) | + (2,261) | + (1,893) | − | − |
| BP1201BB3291 | Tracheal colonization factor (TcfA) | + (412) | + (1,530) | + (3,117) | − | − |
| BP2256/BB1617 | Putative secreted protein (Bsp22) | − | + (470) | − | + (500) | − |
| BP2253/BB1620 | Putative outer protein D (BopD) | − | + (781) | − | + (193) | − |
| BP2257/BB1616 | Putative outer protein N (BopN) | − | + (204) | − | − | − |
| BP0500/BB4228 | Hypothetical protein (BteA/BopC) | − | − | − | + (67) | − |
Quantitative MALDI-TOF tandem mass spectrometry analysis was carried out on extracted proteins from culture supernatant of B. pertussis Tohama I (Toh I), B. pertussis 12743, ΔbscN B. pertussis 12743, B. bronchiseptica RB50, and ΔbscN B. bronchiseptica RB50. Identified peptides were searched against the complete B. bronchiseptica and B. pertussis genome sequences (www.sanger.ac.uk/Projects/B_pertussis/) using the Mascot search engine. Numbers in parentheses represent the individual Mascot ion scores. Individual ion scores >33 indicate identity or extensive homology (P < 0.05).
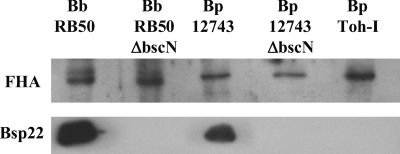
Mutation of bscN abolishes expression of TTSS effector proteins by B. pertussis 12743 but does not affect bacterial growth in vitro.
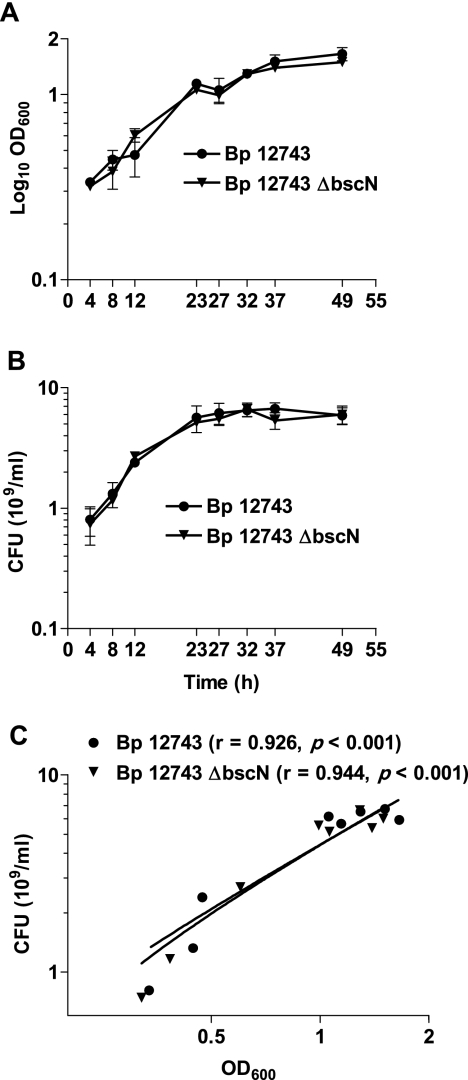
In order to assess the contribution of the TTSS to the virulence of BP 12743, we constructed a strain carrying an insertional mutation of the bscN gene. Mutation of bscN in B. bronchiseptica or its homologs, yscN in Yersinia and invC in Salmonella, has been shown to abolish in vitro TTSS secretion and in vivo TTSS-mediated effects on host cells (7, 38-40). For comparative purposes, we also mutated bscN in B. bronchiseptica RB50. Bsp22 protein was detected by Western blotting in the culture supernatants of WT B. pertussis 12743 and B. bronchiseptica RB50 but not in ΔbscN derivatives (Fig. 2). Furthermore, Bsp22, BopN, and BopD were detected by mass spectrometry analyses of secreted proteins of WT but not ΔbscN derivatives (Table 1). Mutation of bscN did not affect secretion of other virulence factors, including FHA, adenylate cyclase toxin, and pertactin (Table 1 and Fig. 2). These results demonstrate that mutation of the bscN gene abolishes secretion of TTSS substrates by B. pertussis 12743 and B. bronchiseptica RB50 and demonstrate that BscN is an essential component of the B. pertussis TTSS. Furthermore, mutation of bscN did not significantly affect bacterial growth rate. The in vitro growth curves based on OD600 (Fig. 3A) and CFU (Fig. 3B) were almost superimposable for B. pertussis 12743 and ΔbscN B. pertussis 12743. Nonlinear regression analysis with Akaike's information criterion test determined that there was a 93% probability that the curves for the two bacterial strains were identical. Furthermore linear regression analysis demonstrated a highly significant (P < 0.001) correlation between OD600 and CFU for B. pertussis 12743 and ΔbscN B. pertussis 12743. These findings demonstrate that OD600 can be used as an accurate estimate of the number of viable bacteria and suggest that equivalent numbers of viable WT and TTSS mutant bacteria were employed for experiments to compare the virulence levels of and immune responses to B. pertussis 12743 and ΔbscN B. pertussis 12743 in vitro and in vivo.
FIG. 2.
Mutation of bscN in Bordetella spp. abolishes Bsp22 production but does not affect secretion of the virulence factor FHA. Proteins from culture supernatants of WT and ΔbscN B. pertussis (Bp) 12743 and B. bronchiseptica (Bb) RB50 or B. pertussis Tohama I (Toh-I) were examined for the presence of Bsp22 and FHA by Western blotting.
FIG. 3.
Correlation between in vitro growth curves for B. pertussis (Bp) 12743 and ΔbscN B. pertussis 12743. B. pertussis 12743 and ΔbscN B. pertussis 12743 were cultured for 2 days in SS medium, and samples were removed at the indicated time points. (A) OD600 was determined. (B) The number of viable bacteria was determined by performing CFU counts after plating on BG agar. (C) CFU counts were plotted against OD600 for each strain separately, and linear regression analysis was performed. The correlation (r value) and level of significance (P value) are shown.
The TTSS of B. pertussis 12743 promotes adherence to macrophages but does not mediate cytotoxicity in vitro.
Consistent with previous reports (39, 40), B. bronchiseptica RB50 was significantly cytotoxic to cultured epithelial cells, macrophages, and DC (Fig. 4A) and this activity was lost in the ΔbscN mutant. In contrast, no significant cell lysis was detected in any cell type incubated with WT or ΔbscN B. pertussis 12743. The TTSS of Yersinia spp. plays a crucial role in preventing bacterial uptake during infection (4), while the TTSS of Salmonella spp. mediates bacterial uptake to facilitate tissue invasion and intracellular replication (12). Here we investigated the role of the Bordetella TTSS in bacterial uptake. Significantly higher CFU were detected in kanamycin-treated macrophages cultured with ΔbscN B. bronchiseptica than in those cultured with WT B. bronchiseptica, indicative of a higher number of internalized viable mutant bacteria (Fig. 4B). After correcting for cell death induced by B. bronschiseptica, there was still significantly greater uptake of the mutant than the WT B. bronschiseptica (data not shown). In contrast, the numbers of CFU recovered following treatment with medium only were similar following culture with WT or ΔbscN B. bronchiseptica, suggesting that the TTSS of B. bronchiseptica did not facilitate binding to the cell surface. The numbers of viable WT and ΔbscN B. pertussis 12743 CFU recovered from kanamycin-treated macrophages were very similar, indicative of similar numbers of internalized B. pertussis CFU. In contrast, the number of viable B. pertussis CFU recovered following treatment with medium alone from cells cultured with ΔbscN B. pertussis 12743 was significantly lower than the number of WT bacteria, indicating a lower number of cell-associated bacteria following incubation with the mutant bacteria (Fig. 4B). These results demonstrate that the TTSS of B. bronchiseptica RB50 inhibits bacterial uptake by murine macrophages, whereas the TTSS of B. pertussis 12743 promotes adherence of the bacteria to macrophages in vitro.
FIG. 4.
The TTSS of B. pertussis 12743 promotes bacterial adherence to macrophages but does not mediate cytotoxicity. (A) Epithelial cells, J774 macrophages, or bone marrow-derived DC were cultured with WT or ΔbscN B. pertussis (Bp) 12743 or B. bronchiseptica (Bb) RB50 at MOIs of 20, 100, and 500 for 4 h. Cytotoxicity was measured by a lactate dehydrogenase release assay. (B and C) J774 macrophages were cultured with WT or ΔbscN B. bronchiseptica RB50 (B) or B. pertussis 12743 (C) at an MOI of 100:1. Adherence was assessed by assessing CFU counts following 2 h of incubation followed by 1 h of treatment with kanamycin or medium only. *, P < 0.05; **, P < 0.01 (ΔbscN versus WT). Results are representative of three experiments.
Enhanced innate and adaptive immune responses in mice infected with ΔbscN B. pertussis 12743.
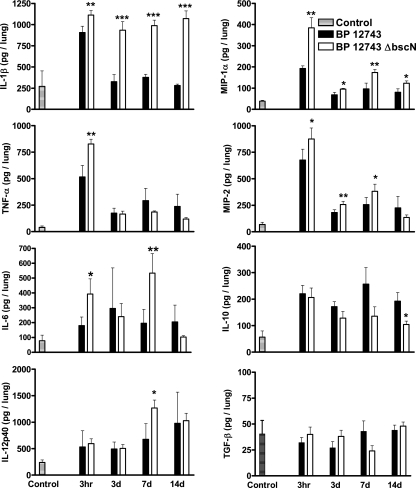
Protective immunity to B. pertussis involves a combination of local innate inflammatory responses in the lungs (13, 24) and adaptive immune responses, mediated by Th1 and Th17 cells, which help to clear bacteria from the respiratory tract (13, 20). Here, we examined local innate inflammatory cytokine production in the lungs of mice infected with WT and ΔbscN B. pertussis 12743. We detected significantly higher concentrations of the proinflammatory cytokines, TNF-α (P < 0.01), IL-1β (P < 0.01) and IL-6 (P < 0.05), and proinflammatory chemokines MIP-1α (P < 0.01) and MIP-2 (P < 0.05) in the lungs 3 h after challenge with the ΔbscN bacteria compared with WT B. pertussis 12743 (Fig. 5). Furthermore, the concentrations of IL-1β, MIP-1α, and MIP-2 remained significantly higher in the lungs of mice infected with the ΔbscN B. pertussis 12743 up to 7 to 14 days postchallenge. We also detected significantly higher concentrations of IL-12p40 and IL-6 in the lungs of mice 7 days after challenge with ΔbscN B. pertussis 12743. In contrast, the concentrations of the immunosuppressive cytokine IL-10 were consistently lower in mice infected with ΔbscN B. pertussis than in those infected with WT bacteria. The concentration of transforming growth factor β in the lungs was not significantly changed over the course of infection with B. pertussis 12743 or ΔbscN B. pertussis 12743 (Fig. 5). These results suggest a role for the TTSS of B. pertussis 12743 in suppressing innate proinflammatory cytokine and chemokine responses in the lungs, especially early in infection.
FIG. 5.
Enhanced inflammatory cytokine and chemokine induction in the lungs of mice infected with ΔbscN B. pertussis (BP) 12743. BALB/c mice were challenged with an aerosol of WT or ΔbscN B. pertussis 12743. Cytokine and chemokine concentrations were determined by ELISA on lung homogenates from mice 3 h and 3, 7, and 14 days (d) after aerosol challenge and in uninfected control mice. Results are means ± standard deviations for four mice per group at each time point. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (ΔbscN versus WT).
Analysis of the cellular content of bronchoalveolar lavage samples 7 days postchallenge revealed a nonsignificant increase in the number of neutrophils in the lungs of mice infected with ΔbscN B. pertussis 12743 (4.6 × 104 ± 1.5 × 104/lung) compared with that in lungs of mice infected with B. pertussis 12743 (2.6 × 104 ± 1.5 × 104/lung). However, histological analysis of lungs did not reveal evidence of increased pathology in the lungs of mice infected with the mutant versus WT bacteria (data not shown).
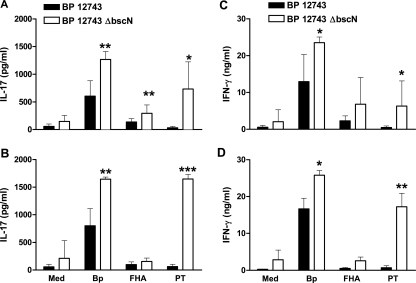
We examined antigen-specific cytokine production in mice infected with WT or ΔbscN B. pertussis 12743 by stimulation of spleen cells with B. pertussis sonicate, purified PT, or FHA. We detected B. pertussis-specific IL-17 and IFN-γ production by spleen cells recovered 14 or 21 days after infection of mice with WT B. pertussis 12743 (Fig. 6). However, antigen-specific IL-17 and IFN-γ production was significantly stronger in mice infected with ΔbscN B. pertussis 12743. The enhanced IL-17 production in mice infected with the mutant bacteria in response to PT and also to B. pertussis sonicate was particularly striking, and this was a consistent finding at days 14, 21 (Fig. 6), and 28 (data not shown) postchallenge. Preliminary experiments to determine the frequency of antigen-specific T cells in the lungs of infected mice (based on a single time point, day 14 postchallenge) revealed a higher frequency of B. pertussis-specific Th1 and Th17 cells in mice infected with ΔbscN B. pertussis 12743 (IFN-γ secreting, 2.86%; IL-17 secreting, 1.98%) than in mice infected with the WT bacteria (IFN-γ secreting, 1.92%; IL-17 secreting, 0.88%). Collectively, these results suggest that the TTSS may suppress antigen-specific IFN-γ and IL-17 production during infection of mice with B. pertussis 12743. Significantly higher B. pertussis-specific serum IgG levels in mice infected with ΔbscN B. pertussis 12743 than in mice infected with WT bacteria were also observed (Fig. 7). Consistent with the stronger Th1 responses, we detected significantly higher titers of serum IgG2a in mice infected with ΔbscN B. pertussis 12743 (Fig. 7). In contrast, there were no differences in serum IgG1 between the mutant and WT bacteria. These results suggest that the Bordetella TTSS may suppress Ab responses, especially IgG2a, as a consequence of suppressing the induction of Th1 cells.
FIG. 6.
Enhanced antigen-specific IL-17 and IFN-γ production in mice infected with ΔbscN B. pertussis (BP) 12743. BALB/c mice were challenged with an aerosol of WT or ΔbscN B. pertussis 12743. Spleen cells recovered 14 (A and C) and 21 (B and D) days postchallenge were stimulated with sonicated B. pertussis 12743 (Bp), FHA, PT, or medium (Med) only as a control. Supernatants were removed after 3 days, and IL-17 (A and B) and IFN-γ (C and D) concentrations were determined by ELISA. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (ΔbscN versus WT).
FIG. 7.
Enhanced B. pertussis-specific IgG and IgG2a responses in mice infected with ΔbscN B. pertussis (BP) 12743. BALB/c mice were challenged with an aerosol of WT or ΔbscN B. pertussis 12743. Serum was recovered from infected mice 28 days postchallenge or from uninfected control mice and assayed for anti-B. pertussis IgG, IgG1, and IgG2a by ELISA. Results are means ± standard deviations for four mice per group. *, P < 0.05; **, P < 0.01 (ΔbscN versus WT).
The TTSS of B. pertussis 12743 enhances bacterial persistence in the lungs of mice.
In order to examine the role of a functional TTSS in virulence of Bordetella in vivo, BALB/c mice were infected by exposure to an aerosol containing equivalent concentrations (2 × 1010/ml) of either WT or ΔbscN B. pertussis 12743. After an initial rise in bacterial numbers, peaking at day 7, WT B. pertussis 12743 began to be steadily cleared from the lungs between days 7 and 21, with complete clearance of the bacteria 28 days postinfection (Fig. 8A). In contrast, ΔbscN B. pertussis 12743 colonized the lungs to a significantly lower degree at each time point examined and began to be cleared from the lung earlier in infection and was completely cleared by day 21. The lower number of mutant bacteria recovered 3 h after challenge was a consistent finding in three experiments using our standard challenge inoculum. Evidence from over 100 challenge experiments with B. pertussis has shown that this method of infection results in highly reproducible lung colonization of mice within an experiment. Furthermore, we have also shown a highly significant correlation between protection in this model and vaccine efficacy in children (26). We used OD600 to estimate the number of bacteria in the challenge inoculum, and, since there was a highly significant correlation between OD600 and CFU counts (Fig. 3), it is unlikely that the difference in colonization at 3 h reflects exposure to a lower number of viable bacteria. Furthermore, the in vitro growth curves, as determined by measuring either OD600 or CFU, for the TTSS mutant and WT bacteria were almost identical (Fig. 3). Nevertheless, in order to provide further evidence of the impact of the TTSS on virulence in vivo, we examined the course of infection when differing concentrations of the challenge inocula resulted in higher or lower initial lung colonization. In a challenge experiment where the initial colonization was 3.8 log10 CFU per lung for the WT B. pertussis, ΔbscN B. pertussis 12743 was cleared within 7 days and there were significantly greater numbers of CFU recovered at 3, 7, and 14 days postchallenge from the lungs of mice infected with the WT bacteria than with mutant bacteria (Fig. 8B). In contrast, when the initial colonization was 4.8 log10 CFU per lung for the WT B. pertussis, the effect of the mutation was less obvious. Although CFU recovered from the mice infected with the TTSS mutant were 4- to 10-fold (0.6 to 1 log10) lower than CFU of WT B. pertussis on days 3 and 7, these differences were not statistically significant (Fig. 8C). The absence of the TTSS and the corresponding loss of its cell binding and immunosuppressive effects may be less obvious when the lungs are overwhelmed with a high number of bacteria in the aerosol. Nevertheless the dramatically reduced CFU recovered from mice infected with the TTSS mutant in mice with lower initial colonization (which may be more relevant to the mode of exposure in humans) suggests that the TTSS does play an important role both in colonization and persistence of B. pertussis 12743 in the lungs following aerosol challenge of mice. Despite the lower bacterial load, we detected significantly higher concentrations of innate proinflammatory cytokine (Fig. 5) and antigen-specific Th1 and Th17 cells (Fig. 6) in mice infected with the mutant than in mice infected with WT B. pertussis (the experiments shown in Fig. 5 to 7 correspond to a level of bacterial colonization shown in Fig. 8A). This provides indirect evidence that the lower lung colonization observed with the mutant bacteria is unlikely to be due to exposure to a lower dose of viable bacteria but may reflect reduced adherence and a stronger host immune response due to the absence of the immune-subversive properties of the TTSS.
FIG. 8.
ΔbscN B. pertussis 12743 has reduced ability to colonize lungs of mice. BALB/c mice were challenged with an aerosol of WT or ΔbscN B. pertussis (Bp) 12743, where the challenge inoculum resulted in intermediate (A), low (B), or high (C) initial colonization with the WT B. pertussis. Groups of four mice were sacrificed 3 h and 3, 7, 14, 21, and 28 days later, and CFU counts were performed on lung homogenates. The dashed line represents the limit of detection. **, P < 0.01; ***, P < 0.001 (ΔbscN versus WT). Results are representative of three experiments for panel A and one experiment each for panels B and C.
DISCUSSION
This study has demonstrated for the first time that B. pertussis expresses a functionally active TTSS, which may be lost following prolonged in vitro culture of the bacteria. We have identified several substrates of the B. pertussis TTSS and shown that the TTSS of B. pertussis is an important virulence factor and immunomodulatory apparatus for subverting protective immune responses and prolonging survival in the host. It appears that the TTSS may facilitate colonization and survival of B. pertussis by promoting bacterial adherence and by inhibiting local innate inflammatory responses and the consequent induction of antigen-specific Th1, Th17, and Ab responses that function to clear the bacteria from the respiratory tract.
The closely related pathogens B. pertussis and B. bronchiseptica produce a range of common virulence factors, including adhesins and toxins, which function to establish and maintain the infection, but have also evolved distinct strategies for survival in the host. The relatively short-lived and acute severity of B. pertussis infection of humans, which favors a high rate of host-to-host transmission, contrasts with the relatively asymptomatic and often lifelong persistence of B. bronchiseptica infection in animals. This was thought to be consistent with the functional activity of the TTSS in B. bronchiseptica, which suppresses DC maturation and migration (32, 33), and the lack of TTSS function in B. pertussis Tohama I. However, the present study has revealed that clinical isolates of B. pertussis do express a functionally active TTSS, which plays a role in promoting more efficient bacterial colonization and persistence through immunosuppression following respiratory infection of mice. We demonstrated significant secretion of Bsp22, a reliable marker of a functional TTSS in Bordetella spp. (39), by 15 of 16 clinical isolates of B. pertussis, in addition to 2 isolates obtained from the ATCC, B. pertussis 12743 and 9340, which had not undergone extensive laboratory passage. We also demonstrated secretion of two additional Bordetella TTSS substrates, BopN and BopD, by B. pertussis 12743. In contrast, secretion of TTSS substrates could not be detected in two well-studied laboratory-adapted strains of B. pertussis, Tohama I and Wellcome 28. While the TTSS loci are conserved and have been shown to be transcribed in B. pertussis Tohama I during in vitro growth, it has been suggested that protein translation is prevented by a posttranscriptional block (22). The sequencing of the B. pertussis Tohama I genome revealed extensive expansion of the insertion sequence element IS481, indicative of large-scale genome rearrangements and thus a high level of genome plasticity. Long-term laboratory passage can lead to significant changes in gene expression and virulence factor production in E. coli and Pseudomonas and Staphylococcus spp. (11). Compared with that in laboratory-cultured strains, transcription of genes encoding the TTSS and its effector proteins was upregulated in a highly adherent P. aeruginosa strain isolated from the lung of a cystic fibrosis patient, and this correlated with increased cytotoxicity in vitro and enhanced virulence in the respiratory tracts of mice (37). Furthermore, it has been reported that the Yersinia YopT TTSS effector protein is not expressed by serotype O3 strains of Y. pseudotuberculosis that have been extensively passaged in vitro (36). Significant changes in gene transcription by clinical isolates of B. pertussis were reported after as few as 12 laboratory passages (2), indicating that B. pertussis, like other bacterial species, can alter gene expression when introduced into a new environment. It is therefore possible that the absence of a functional TTSS in B. pertussis Tohama I and Wellcome 28 is a consequence of long-term laboratory culture in the absence of eukaryotic cell contact.
The present study demonstrated that the TTSS of B. bronchiseptica may facilitate bacterial persistence through subversion of bacterial uptake by macrophages, a strategy in the pathogenesis of Yersinia spp. (5). We found that B. pertussis 12743 did not induce cytotoxicity in macrophages, DC, and epithelial cells. However, consistent with previous reports (39, 40), B. bronchiseptica was cytotoxic to a range of cultured cells. Although other TTSS effector proteins may also contribute to cytotoxicity, it has been demonstrated that BteA, also called BopC, is required for the induction of necrotic cell death (19, 27). Interestingly, BteA/BopC was not detected in secreted proteins from B. pertussis 12743. In contrast, we found evidence that the TTSS of B. pertussis facilitates bacterial binding to macrophages in vitro. In addition, compared with the WT bacteria, the TTSS mutant of B. pertussis 12743 had significantly reduced colonization in the lungs 3 h after respiratory challenge of mice. Taken together, these data suggest that the TTSS of B. pertussis may function as a host adherence factor, as has been demonstrated for enteropathogenic E. coli (17).
Significantly, we found evidence that the TTSS facilitates persistence of the bacteria in the respiratory tract by subverting innate and adaptive immune responses. Protective immunity to B. pertussis is mediated through recruitment of neutrophils and macrophages to the lungs, local secretion of inflammatory cytokines, and the induction of B. pertussis-specific Th1 cells, Th17 cells, and Ab responses (18, 20, 24). IL-12-induced IFN-γ production by NK cells and Th1 cells prevents bacterial dissemination from the respiratory tract and activates production of opsonizing and complement-fixing IgG2a Abs in the mouse (3, 20). IL-23, IL-1, TNF-α, and IL-6 promote the differentiation and expansion of Th17 cells, and these cells have been implicated in inflammatory responses that mediate autoimmunity and also function in the recruitment of neutrophils to the site of infection, where they may help contain the pathogen until a subsequent clearing IFN-γ-producing Th1 response can be generated (25, 35). We have recently reported that Th17 cells have a protective role in vaccine-induced immunity to B. pertussis by activating bacterial killing by macrophages (13). In the present study we found evidence that deletion of a functional TTSS from B. pertussis 12743 resulted in enhancement of local inflammatory responses in the lungs of infected mice. Despite the lower bacterial burden in the lungs, mice infected with the TTSS mutant had significantly greater local IL-1β, IL-12p40, MIP-1α, and MIP-2 production. We also observed a modest reduction in IL-10 in mice infected with the TTSS mutant, and this was significant at day 14. This is consistent with the report on B. bronchiseptica where antigen-specific IL-10 production in mice infected with the TTSS mutant is lower than that in mice infected with WT bacteria (32). In contrast, we demonstrated significantly greater B. pertussis-specific IFN-γ, IL-17, and IgG2a responses of mice infected with the TTSS mutant than with WT B. pertussis 14 to 28 days after B. pertussis challenge. The augmentation of the IL-17 response to PT in mice infected with the mutant bacteria was particularly striking. Although we do not know the precise mechanisms, this may reflect enhancement of innate IL-1, which together with IL-23 is known to promote the differentiation of Th17 cells (13). The enhanced cellular immune response in mice infected with the TTSS mutant correlated with earlier respiratory clearance of TTSS-defective bacteria than of WT bacteria. Thus, it appears that the TTSS contributes to persistence of B. pertussis by suppressing innate inflammatory responses, which not only allows greater bacterial colonization, but also delays clearance due to significant suppression of Th1, Th17, and Ab responses.
Our study provides the first evidence that B. pertussis uses the TTSS as a means of colonization and survival in the host and may in particular target the innate immune system. Furthermore, we have demonstrated that one of the substrates, Bsp22, is secreted in significant quantities by clinical isolates of B. pertussis and is immunogenic in animals.
Acknowledgments
This work was supported by The Irish Research Council for Science, Engineering & Technology and Science Foundation Ireland.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 14 January 2008.
REFERENCES
- 1.Akerley, B. J., P. A. Cotter, and J. F. Miller. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80611-620. [DOI] [PubMed] [Google Scholar]
- 2.Brinig, M. M., C. A. Cummings, G. N. Sanden, P. Stefanelli, A. Lawrence, and D. A. Relman. 2006. Significant gene order and expression differences in Bordetella pertussis despite limited gene content variation. J. Bacteriol. 1882375-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrne, P., P. McGuirk, S. Todryk, and K. H. Mills. 2004. Depletion of NK cells results in disseminating lethal infection with Bordetella pertussis associated with a reduction of antigen-specific Th1 and enhancement of Th2, but not Tr1 cells. Eur. J. Immunol. 342579-2588. [DOI] [PubMed] [Google Scholar]
- 4.Cornelis, G. R. 2002. The Yersinia Ysc-Yop ‘type III’ weaponry. Nat. Rev. Mol. Cell Biol. 3742-752. [DOI] [PubMed] [Google Scholar]
- 5.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M. P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 621315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowcroft, N. S., C. Stein, P. Duclos, and M. Birmingham. 2003. How best to estimate the global burden of pertussis? Lancet Infect. Dis. 3413-418. [DOI] [PubMed] [Google Scholar]
- 7.Eichelberg, K., C. C. Ginocchio, and J. E. Galan. 1994. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J. Bacteriol. 1764501-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eldering, G., C. Hornbeck, and J. Baker. 1957. Serological study of Bordetella pertussis and related species. J. Bacteriol. 74133-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fauconnier, A., A. Veithen, P. Gueirard, R. Antoine, L. Wacheul, C. Locht, A. Bollen, and E. Godfroid. 2001. Characterization of the type III secretion locus of Bordetella pertussis. Int. J. Med. Microbiol. 290693-705. [DOI] [PubMed] [Google Scholar]
- 10.Finn, T. M., and L. A. Stevens. 1995. Tracheal colonization factor: a Bordetella pertussis secreted virulence determinant. Mol. Microbiol. 16625-634. [DOI] [PubMed] [Google Scholar]
- 11.Fux, C. A., M. Shirtliff, P. Stoodley, and J. W. Costerton. 2005. Can. laboratory reference strains mirror “real-world” pathogenesis? Trends Microbiol. 1358-63. [DOI] [PubMed] [Google Scholar]
- 12.Galan, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 866383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins, S. C., A. G. Jarnicki, E. C. Lavelle, and K. H. Mills. 2006. TLR4 mediates vaccine-induced protective cellular immunity to Bordetella pertussis: role of IL-17-producing T cells. J. Immunol. 1777980-7989. [DOI] [PubMed] [Google Scholar]
- 14.Hot, D., R. Antoine, G. Renauld-Mongenie, V. Caro, B. Hennuy, E. Levillain, L. Huot, G. Wittmann, D. Poncet, F. Jacob-Dubuisson, C. Guyard, F. Rimlinger, L. Aujame, E. Godfroid, N. Guiso, M. J. Quentin-Millet, Y. Lemoine, and C. Locht. 2003. Differential modulation of Bordetella pertussis virulence genes as evidenced by DNA microarray analysis. Mol. Genet. Genomics 269475-486. [DOI] [PubMed] [Google Scholar]
- 15.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasuga, T., Y. Nakase, K. Ukishima, and K. Takatsu. 1954. Studies on Haemophilus pertussis. V. Relation between the phase of bacilli and the progress of the whooping-cough. Kitasato Arch. Exp. Med. 2757-62. [PubMed] [Google Scholar]
- 17.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91511-520. [DOI] [PubMed] [Google Scholar]
- 18.Kirimanjeswara, G. S., P. B. Mann, M. Pilione, M. J. Kennett, and E. T. Harvill. 2005. The complex mechanism of antibody-mediated clearance of Bordetella from the lungs requires TLR4. J. Immunol. 1757504-7511. [DOI] [PubMed] [Google Scholar]
- 19.Kuwae, A., T. Matsuzawa, N. Ishikawa, H. Abe, T. Nonaka, H. Fukuda, S. Imajoh-Ohmi, and A. Abe. 2006. BopC is a novel type III effector secreted by Bordetella bronchiseptica and has a critical role in type III-dependent necrotic cell death. J. Biol. Chem. 2816589-6600. [DOI] [PubMed] [Google Scholar]
- 20.Mahon, B. P., B. J. Sheahan, F. Griffin, G. Murphy, and K. H. Mills. 1997. Atypical disease after Bordetella pertussis respiratory infection of mice with targeted disruptions of interferon-gamma receptor or immunoglobulin mu chain genes. J. Exp. Med. 1861843-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattoo, S., and J. D. Cherry. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin. Microbiol. Rev. 18326-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattoo, S., M. H. Yuk, L. L. Huang, and J. F. Miller. 2004. Regulation of type III secretion in Bordetella. Mol. Microbiol. 521201-1214. [DOI] [PubMed] [Google Scholar]
- 23.McGuirk, P., B. P. Mahon, F. Griffin, and K. H. Mills. 1998. Compartmentalization of T cell responses following respiratory infection with Bordetella pertussis: hyporesponsiveness of lung T cells is associated with modulated expression of the co-stimulatory molecule CD28. Eur. J. Immunol. 28153-163. [DOI] [PubMed] [Google Scholar]
- 24.McGuirk, P., and K. H. Mills. 2000. A regulatory role for interleukin 4 in differential inflammatory responses in the lung following infection of mice primed with Th1- or Th2-inducing pertussis vaccines. Infect. Immun. 681383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenzie, B. S., R. A. Kastelein, and D. J. Cua. 2006. Understanding the IL-23-IL-17 immune pathway. Trends Immunol. 2717-23. [DOI] [PubMed] [Google Scholar]
- 26.Mills, K. H., M. Ryan, E. Ryan, and B. P. Mahon. 1998. A murine model in which protection correlates with pertussis vaccine efficacy in children reveals complementary roles for humoral and cell-mediated immunity in protection against Bordetella pertussis. Infect. Immun. 66594-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panina, E. M., S. Mattoo, N. Griffith, N. A. Kozak, M. H. Yuk, and J. F. Miller. 2005. A genome-wide screen identifies a Bordetella type III secretion effector and candidate effectors in other species. Mol. Microbiol. 58267-279. [DOI] [PubMed] [Google Scholar]
- 28.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 3532-40. [DOI] [PubMed] [Google Scholar]
- 29.Poynten, M., P. B. McIntyre, F. R. Mooi, K. J. Heuvelman, and G. L. Gilbert. 2004. Temporal trends in circulating Bordetella pertussis strains in Australia. Epidemiol. Infect. 132185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rambow, A. A., R. C. Fernandez, and A. A. Weiss. 1998. Characterization of BrkA expression in Bordetella bronchiseptica. Infect. Immun. 663978-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reissinger, A., J. A. Skinner, and M. H. Yuk. 2005. Downregulation of mitogen-activated protein kinases by the Bordetella bronchiseptica type III secretion system leads to attenuated nonclassical macrophage activation. Infect. Immun. 73308-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skinner, J. A., M. R. Pilione, H. Shen, E. T. Harvill, and M. H. Yuk. 2005. Bordetella type III secretion modulates dendritic cell migration resulting in immunosuppression and bacterial persistence. J. Immunol. 1754647-4652. [DOI] [PubMed] [Google Scholar]
- 33.Skinner, J. A., A. Reissinger, H. Shen, and M. H. Yuk. 2004. Bordetella type III secretion and adenylate cyclase toxin synergize to drive dendritic cells into a semimature state. J. Immunol. 1731934-1940. [DOI] [PubMed] [Google Scholar]
- 34.Somerville, G. A., S. B. Beres, J. R. Fitzgerald, F. R. DeLeo, R. L. Cole, J. S. Hoff, and J. M. Musser. 2002. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J. Bacteriol. 1841430-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutton, C., C. Brereton, B. Keogh, K. H. Mills, and E. C. Lavelle. 2006. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J. Exp. Med. 2031685-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viboud, G. I., and J. B. Bliska. 2005. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 5969-89. [DOI] [PubMed] [Google Scholar]
- 37.von Gotz, F., S. Haussler, D. Jordan, S. S. Saravanamuthu, D. Wehmhoner, A. Strussmann, J. Lauber, I. Attree, J. Buer, B. Tummler, and I. Steinmetz. 2004. Expression analysis of a highly adherent and cytotoxic small colony variant of Pseudomonas aeruginosa isolated from a lung of a patient with cystic fibrosis. J. Bacteriol. 1863837-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woestyn, S., A. Allaoui, P. Wattiau, and G. R. Cornelis. 1994. YscN, the putative energizer of the Yersinia Yop secretion machinery. J. Bacteriol. 1761561-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuk, M. H., E. T. Harvill, P. A. Cotter, and J. F. Miller. 2000. Modulation of host immune responses, induction of apoptosis and inhibition of NF-κB activation by the Bordetella type III secretion system. Mol. Microbiol. 35991-1004. [DOI] [PubMed] [Google Scholar]
- 40.Yuk, M. H., E. T. Harvill, and J. F. Miller. 1998. The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol. Microbiol. 28945-959. [DOI] [PubMed] [Google Scholar]