Abstract
Enteroaggregative Escherichia coli (EAEC) is an emerging enteric pathogen in both developing and industrialized countries. EAEC is defined as a diarrheal pathogen based on its characteristic aggregative adherence to HEp-2 cells in culture and its biofilm formation on the intestinal mucosa. We have reported that the novel protein AatA, which is encoded on the EAEC virulence plasmid pAA2, localizes to the outer membrane and facilitates export of the dispersin Aap across the outer membrane. Because AatA is an E. coli efflux pump TolC homolog, we investigated the role of TolC in the virulence of EAEC. No difference in Aap secretion was observed between the wild type and its tolC mutant (042tolC). However, characteristic aggregation in high-glucose Dulbecco's minimal essential medium for the wild type was diminished for 042tolC. In a microtiter plate assay, there were significantly more planktonic cells for 042tolC than for the wild type, while there were significantly fewer spontaneously precipitated cells on the substratum for 042tolC than for the wild type. In a HEp-2 cell adherence test, 042tolC showed less aggregative adherence than did the wild type. The strong aggregation and aggregative adherence were restored in the complement strain with tolC. In a transwell assay, planktonic cells of 042tolC decreased when cocultured with the wild type or the complement, while precipitated cells of 042tolC increased when cocultured with them. These results suggest that TolC promotes the aggregation and adhesion of EAEC 042 by secreting an assumed humoral factor.
Enteroaggregative Escherichia coli (EAEC) is an emerging enteric pathogen associated with endemic and epidemic diarrheal illness in both developing and industrialized countries (11, 21, 26, 31). Recently, a prospective control study showed that EAEC is an important, unrecognized cause of childhood diarrhea in the United States (3, 23). The defining feature of EAEC remains its distinctive aggregative adherence (AA) pattern observed in the HEp-2 adherence assay, which remains the gold standard for EAEC identification (22). Cells of EAEC strains adhere to the epithelial cell surface, to the glass substratum, and to each other in a distinctive stacked-brick formation. The pathogenesis of EAEC infection is thought to involve the adherence of the bacterium to the intestinal mucosa, followed by the secretion of one or more enterotoxins (7, 8). EAEC's adherence to the substratum and the mucosa is characterized by the presence of a thick, aggregating biofilm (37, 39).
Sheikh et al. have characterized a novel protein, Aap, which is exported from bacteria and forms a protein capsule on the bacterial cell surface (33). Aap promotes the dispersal of EAEC on the intestinal mucosa and therefore has been designated dispersin (33). Nishi et al. reported that the pAA2 region of an EAEC prototype, strain 042, corresponding to the AA probe encodes a putative ABC transporter apparatus which is required for the efficient translocation of the dispersin protein Aap (24). They demonstrated that the locus comprises a cluster of five genes (designated aat-PABCD), including homologs of an inner membrane permease (AatP), an ATP-binding cassette protein (AatC), and the outer membrane protein TolC (AatA) (GenBank accession no. AY351860) (24). E. coli TolC acts as a channel/tunnel in the transport of molecules across the outer membrane and is involved in the export of many molecules both small, such as antibiotics, bile salts, and organic solvents, and large, such as colicin V and α-hemolysin (14). Although the amino acid sequence identity between TolC and AatA is only 25%, TolC was identified as an unambiguous homolog of AatA by alignment analysis (24, 36). It is highly likely that AatA and TolC share a common ancestry and similar three-dimensional structures. We demonstrated that the three nonpolar amino acid residues 381 to 383 in the carboxy terminus of AatA are important for Aap secretion activity (12), as the nonpolar amino acid residue 412 (leucine) at the carboxy terminus of TolC is required to express its activity (43). Thus, AatA was found to be a homolog of TolC in EAEC 042. Like TolC, AatA localizes at the outer membrane independent of its ABC partner and is assumed to export Aap across the outer membrane (24). We also reported that, like that of the dispersin gene, the transcription of the aat cluster is dependent on AggR, a regulator of virulence genes in EAEC (24). Despite a clear contribution of AatA to dispersin translocation through the outer membrane, we were often able to observe some residual dispersin translocation in the absence of a fully functional Aat complex. This could be due to an alternative general translocation mechanism or to the substitution of another TolC homolog, or TolC itself, in dispersin translocation (24). Recently, multidrug resistance efflux pumps including the AcrAB-TolC system have been reported to be associated with the colonization and persistence of bacteria in the host and to have roles in bacterial pathogenicity (28, 29).
In view of the homology of AatA and TolC, we hypothesized that TolC serves as an exporter of Aap like AatA and/or is associated with the virulence of EAEC. To test the hypothesis, we mutated the tolC gene in the prototype strain EAEC 042. We show here that TolC does not export Aap but that it does promote the aggregation and adherence of EAEC 042 and play a role in the pathogenicity of EAEC.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids used in molecular studies are listed in Table 1. Strain 042 was kindly provided by J. P. Nataro at the University of Maryland, Baltimore, MD. This strain was isolated in 1983 from a child with diarrhea in Lima, Peru (18), and has been shown to cause diarrhea in adult volunteers (19). All E. coli strains were grown aerobically at 37°C in Luria-Bertani (LB) medium or Dulbecco's minimal essential medium with 0.45% glucose (high-glucose DMEM) (Invitrogen, Carlsbad, CA). All experiments of aggregation and adhesion were performed in high-glucose DMEM (aggR-inducing condition). All strains were stored at −80°C in Trypticase soy broth with 15% glycerol. Antibiotics were added at the indicated concentrations where appropriate as follows: ampicillin, 100 μg/ml; and kanamycin, 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| 042 | Wild-type EAEC prototype strain | 18 |
| 042aatA | 042 harboring pJP5603 integrated into the aatA gene; Kmr | 24 |
| 042tolC | 042 harboring pJP5603 integrated into the tolC gene; Kmr | This work |
| 042aatAtolC | 042aatA harboring pCVD442 integrated into the tolC gene; Kmr | This work |
| 042tolC(pSEtolC) | 042tolC harboring pSEtolC; Kmr, ABPCr | This work |
| 042tolC(pSEtolCΔ412) | 042tolC harboring pSEtolCΔ412; Kmr, Apr | This work |
| 042tolC(pSE) | 042tolC harboring pSE380; Kmr, Apr | This work |
| 042acrA | 042 harboring pJP5603 integrated into the acrA gene; Kmr | This work |
| DH5αλpir | E. coli K-12 lysogenized for the pir gene, which permits replication of pJP5603 | 6 |
| S17-1λpir | Conjugative K12 lysogenized for pir; Tetr, Kmr | 6 |
| 042aggR | 042 carrying pJP5603 integrated into the aggR gene; Kmr | 35 |
| 042aafA | 042 carrying TnphoA inserted into the gene encoding the AAF/II fimbrial subunit gene; Kmr | 4 |
| 042pet | 042 harboring pJP5603 integrated into the pet gene; Kmr | 10 |
| Plasmids | ||
| pJP5603 | 3.1-kb R6K suicide plasmid; Kmr | 27 |
| pCVD442 | 6.2-kb suicide vector encoding the counterselectable marker sacB; Apr | 17 |
| pSE380 | 4.1-kb expression vector; trc promoter, lacIq; Apr | Invitrogen |
| pINTtolC | 499-bp internal fragment of the tolC gene in pJP5603; Kmr | This work |
| pINT2tolC | 499-bp internal fragment of the tolC gene in pCVD442; Kmr | This work |
| pSEtolC | 1,470-bp fragment encoding tolC cloned into pSE380; Apr | This work |
| pSEtolCΔ412 | 1,467-bp fragment encoding tolCΔ412 cloned into pSE380; Apr | This work |
| pINTacrA | 495-bp internal fragment of the acrA gene in pJP5603; Kmr | This work |
Ap, ampicillin; Km, kanamycin; Tet, tetracycline.
General molecular biology techniques and sequencing procedures.
Plasmid DNA purification, restriction, ligation, transformation, and agarose gel electrophoresis were performed by standard methods (30). Plasmid DNA was introduced into competent-cell E. coli DH5α by heat shock transformation according to the method of Hanahan (7a) or into 042, DH5αλpir, and S17-1λpir by electroporation using the MicroPulser system (Japan Bio-Rad, Tokyo, Japan). The DNA sequence was determined by an ABI Prism 310 sequencer (Applied Biosystems Japan, Tokyo, Japan).
PCR and reverse transcriptase PCR (RT-PCR).
PCR amplifications were performed with 500 ng of purified genomic DNA as the template in a 50-μl reaction mixture containing 2.5 units of Taq DNA polymerase, 0.5 μM of each primer, 0.2 mM of each deoxynucleoside triphosphate, 2 mM MgCl2, and 5 μl of the manufacturer's buffer (Takara, Kyoto, Japan). Amplification reactions were performed in a DNA thermal cycler, PC 707 (Astec, Fukuoka, Japan), for 3 min at 94°C followed by 30 cycles of 94°C for 30 s, 54°C for 40 s, and 72°C for 1 min per kilobase and concluding with extension at 72°C for 10 min unless otherwise stated. The primer sequences used for PCR are listed in Table 1. The products were separated by 1.0% agarose gels, stained with ethidium bromide, and visualized with UV transillumination.
RT-PCR was performed as previously described (35). Total RNA was extracted with an RNeasy mini kit (Qiagen Inc., Valencia, CA) from LB cultures with shaking to the mid-log phase; preparations were treated with RNase-free DNase I (Roche Applied Science, Indianapolis, IN) to eliminate contaminating DNA. The absence of contaminating genomic DNA in RNA preparations was verified by performing PCR for the chromosomal chloramphenicol acetyltransferase (cat) gene. To synthesize cDNA, total RNA (2 μg) was subjected to RT reactions using Thermoscript RT (Invitrogen) and gene-specific reverse primers according to the manufacturer's instructions. Primers used for RT-PCR are listed in Table 2. Amplification reactions were performed in a PC 707 thermal cycler for 3 min at 94°C followed by 28 cycles of 94°C for 30 s, 54°C for 40 s, and 72°C for 1 min per kilobase and concluding with extension at 72°C for 10 min.
TABLE 2.
Primers used in this study
| Primer name | Gene(s) amplified | Map coordinatesa | Sequence (5′ to 3′) | REb |
|---|---|---|---|---|
| tolC-F | tolC | 38-58 | ATGAAGAAATTGCTCCCCATTC | |
| tolC-R | tolC | 1485-1507 | TCAGTTACGGAAAGGGTTATGAC | |
| tolC-int-F | tolC | 375-394 | ACTCTAGACAGGGATTCAGGACGTCACG | XbaI |
| tolC-int-R | tolC | 855-874 | ACGAGCTCCAGAAATCCCGTAGAAGCCG | SacI |
| tolCpSE-F | tolC | 32-59 | ACGGATCCATGCAAATGAAGAAATTGCTCCCCATTC | BamHI |
| tolCpSE-R | tolC | 1485-1507 | ACGCGGCCGCTCAGTTACGGAAAGGGTTATGAC | NotI |
| tolCΔ412-L | tolC | 1294-1357 | GGGTACGTTGAACGAGCAGGATCTGGCACTGAACAATGCGCTGAG | |
| tolCΔ412-R | tolC | 1357-1294 | CTCAGCGCATTGTTCAGTGCCAGATCCTGCTCGTTCAACGTACCC | |
| acrA-F | acrA | 1096-1118 | ATGAACAAAAACAGAGGGTTTAC | |
| acrA-R | acrA | 2268-2289 | TTAAGACTTGGACTGTTCAGGC | |
| acrA-int-F | acrA | 1351-1370 | ACGTCGACGACATCGAAGCAGGTGTCTC | SalI |
| acrA-int-R | acrA | 1825-1845 | ACGGATCCGTCACTGGTGATCAGTGACAC | BamHI |
| aafA-F | aafA | 2620-2639 | ATCAGAATGTTTGCGATTGCTAC | |
| aafA-R | aafA | 3073-3093 | GCATTTAATTTGTCACAAGCTCAGC | |
| cat-F | cap | 14-33 | TCACTGGATATACCACCGTT | |
| cat-R | cap | 626-645 | CCACTCATCGCAGTACTGTT |
Mutagenesis and complementation.
Insertional mutants of the tolC gene were constructed by single-crossover insertion of plasmid pJP5603 as previously described (6, 24). Briefly, an internal portion of the tolC gene was generated by PCR using the primers described in Table 2, and the product was cloned into XbaI and SacI sites of the π-dependent suicide vector pJP5603 (27). The resulting plasmids (pINTtolC) were propagated in E. coli DH5αλpir prior to transformation into the donor E. coli strain in S17-1λpir. The mutant strain was then obtained by conjugal mating between wild-type parent strain 042 and S17-1λpir (pINTtolC). Transconjugants were selected on LB agar supplemented with kanamycin. This process resulted in merodiploid integration of pINTtolC into the homologous site in the tolC gene. Integration of pJP5603 constructs resulted in duplication of predicted codons 375 to 874 of tolC. The acrA gene was mutated in the same way.
A strain with a double knockout of tolC and aatA was constructed from 042aatA. The internal portion of the tolC gene described above was cloned into pCVD442 (17). The resulting plasmids (pINT2tolC) were propagated in E. coli DH5αλpir prior to transformation into the donor E. coli strain in S17-1λpir. The mutant strain was then obtained by conjugal mating between strain 042aatA (kanamycin resistant) and S17-1λpir(pINT2tolC) (ampicillin resistant). Transconjugants were selected on LB agar supplemented with kanamycin and ampicillin.
To complement the tolC mutant, we amplified the tolC construct including both upstream and downstream portions by PCR using the primers listed in Table 2 and cloned them into the expression vector pSE380. Amplifications were performed with 500 ng of purified genomic DNA as templates in a 50-μl reaction mixture containing 2.5 units of Ex-Taq polymerase (Takara, Kyoto, Japan), 0.5 μM of each primer, 0.3 mM of each deoxynucleoside triphosphate, 2 mM MgCl2, and 5 μl of the manufacturer's buffer. Amplification reactions were performed as described above. The PCR products were cloned into the BamHI and NotI sites of the vector pSE380. The resultant plasmid, pSEtolC, was introduced into 042tolC by electroporation using the MicroPulser system (Japan Bio-Rad).
Construction of a deletion mutation at amino acid residue 412 of TolC.
A deletion mutation at amino acid residue 412 of TolC was constructed by PCR-based overlap extension mutagenesis as previously described (41). The complementary primers (tolCΔ412-L and tolCΔ412-R) were designed to delete Leu-412 (Table 2). Two DNA fragments of tolC were prepared by PCR. One was obtained using the tolCpSE-F and tolCΔ412-R primers, and the other was obtained using the tolCΔ412-L and tolCpSE-R primers. They were mixed, heat denatured, and annealed. A small amount of single-strand DNA was formed through interaction at the targeted region. Then, these single-strand DNAs were treated with Ex-Taq polymerase to make the complementary strand. After treatment, the DNA fragments were amplified by PCR with the tolCpSE-F and tolCpSE-R primers. The amplified fragments were digested with BamHI and NotI and cloned into pSE380. The mutation was verified by sequencing. The resultant plasmid, pSEtolCΔ412, was introduced into 042tolC by electroporation.
Aap secretion.
Aap secretion was evaluated as previously described (12, 24, 33). In brief, strains were grown overnight in high-glucose DMEM. Bacteria were harvested from 1.5 ml of culture after centrifugation at 15,000 × g for 5 min and suspended in 100 μl of Laemmli sample buffer. Protein in the supernatant was precipitated with trichloroacetic acid as follows. A 1/4 volume of trichloroacetic acid containing 0.4% (wt/vol) deoxycholate was added to culture supernatants and incubated on ice for 30 min. The pellet was centrifuged at 15,000 × g for 15 min and washed with acetone for 15 min at room temperature. The precipitate was collected by centrifugation at 15,000 × g for 15 min, dried, and suspended in 50 μl of Laemmli sample buffer. One-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on 15% (wt/vol) acrylamide separating gels and 4.0% (wt/vol) acrylamide stacking gels according to standard protocols. The sample of the supernatant (20 μl) or the cells (10 μl) was applied to each lane. Proteins were detected by staining with Coomassie brilliant blue. For immunoblot analyses, protein samples were separated by SDS-PAGE and transferred to Immobilon-P membranes (Millipore, Billerica, MA) by use of standard protocols. For the detection of Aap, anti-Aap antiserum (provided by J. P. Nataro, University of Maryland, Baltimore, MD) (33) was used at dilutions of 1:8,000; antibody binding was detected with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G at a dilution of 1:40,000 and visualized by a chemiluminescence ECL kit (Amersham, Piscataway, NJ). The immunoblotting experiments were performed three times independently.
Flow cytometry.
Flow cytometry analyses were performed with an Epics Altra HyPerSort flow cytometer (Beckman Coulter, Brea, CA) with an air-cooled argon ion laser (488 nm, 15 mW). This standard instrument is equipped with two light scatter detectors that measure forward scatter (FSC) and side scatter. Strains were grown in high-glucose DMEM overnight with shaking at 37°C and mixed by a vortex mixer before examination. At least 20,000 events were recorded for each sample.
Salt aggregation test.
A salt aggregation test was performed as described elsewhere to evaluate hydrophobicity of a bacterial surface (16). In brief, bacterial suspensions and ammonium sulfate [(NH4)2SO4] solutions were prepared in phosphate-buffered saline (PBS), pH 6.8. Molarities of ammonium sulfate used ranged from 1.0 to 0.005 M, differing by serial twofold dilution. Ten-microliter portions of bacterial suspensions were dispensed on glass slide surfaces and mixed with equal volumes of each ammonium sulfate concentration. The glass slide was gently rocked for 2 min at room temperature, and visual reading was performed against a black background. The intensity of agglutination is classified as follows: 2+, aggregation observed immediately (within 10 seconds); 1+, aggregation observed slowly (more than 10 seconds); −, no aggregation.
HEp-2 cell adherence test.
E. coli strains were subjected to a HEp-2 cell adherence test by the method originally described by Nataro et al. with slight modifications (22). Briefly, 20-μl portions of static Luria broth (L-broth) cultures of E. coli (2 × 106 bacteria) after 16 h of incubation at 37°C were incubated with nonconfluent tissue culture HEp-2 cell monolayers (estimated coverage, 25 to 50%) for 3 h on circular glass coverslips in a 24-well microtiter plate in DMEM. The medium was supplemented with 0.5% methyl-alpha-d-mannoside to prevent attachment by type 1 somatic fimbriae.
Microtiter plate assay.
A microtiter plate assay was performed according to the method described previously (35). One milliliter of high-glucose DMEM in 24-well flat-bottom microtiter polystyrene plates (Costar, Corning, NY) with or without glass coverslips was inoculated with 10 μl of overnight L-broth culture grown at 37°C with shaking. The sample was incubated at 37°C for 20 h statically. The culture wells were observed by an SZH-131 stereomicroscope (Olympus, Tokyo, Japan). The medium was collected, and the planktonic cell counts were determined by plating serial dilutions on L-agar plates in triplicate. To evaluate the spontaneous settling, we collected the precipitated cells on the substratum after aspiration of the medium by scraping into 500 μl of PBS. Scraping was performed manually with a blue tip until a film on the substratum visually disappeared. Viable cell number per well was counted as described above. To assess adhesion, we collected the adhesive cells on the substratum after washing three times with PBS and quantified them as described above. Biofilm formation on the substratum was visualized by staining with 0.5% crystal violet for 5 min.
Transwell assay.
A transwell assay was performed using a 24-well polystyrene transwell plate with a 0.4-μm-pore-size polyester membrane (Corning, NY). The diameter of the pores allows cytokines and secreted proteins, but not bacteria, to pass through the membrane. One milliliter of high-glucose DMEM was inoculated with 10 μl of an overnight L-broth culture of 042tolC in the lower chamber and with 10 μl of a culture of 042, 042tolC, or 042tolC(pSEtolC) in the upper chamber. They were cocultured at 37°C for 20 h statically in high-glucose DMEM. The planktonic and spontaneously precipitated cells of 042tolC in the lower chamber were quantified as described above.
SEM.
Scanning electron microscopy (SEM) of 042 and its tolC mutant was performed at the Electron Microscopy Facility (Filgen, Nagoya, Japan) according to the method described previously (33). In brief, specimens were prefixed with 2% glutaraldehyde in CaCo buffer (0.1 M Ca-cacodylate, 3 mM CaC12), postfixed with 2% osmium tetroxide, and then dehydrated for 5 min each in sequential baths of 50%, 70%, 90%, and 100% ethanol. The specimens were then inserted into a critical-point dryer until dry, followed by osmium plasma coating. Specimens were then examined by SEM in a JSM-6320F instrument (Research Resources Center, Chicago, IL) at 5kV. Planktonic cells grown in high-glucose DMEM overnight with shaking at 37°C and adhesive cells grown on glass coverslips in a 24-well flat-bottom microtiter plate in high-glucose DMEM for 20 h at 37°C were analyzed.
RESULTS
Verification of mutagenesis.
To verify successful mutagenesis, we evaluated the transcription of the tolC gene in the mutants by RT-PCR (data not shown). The tolC gene was transcribed in the wild type and 042aatA but not in 042tolC or 042aatAtolC. The transcription of tolC is restored in 042tolC(pSEtolC) to the same level as is found in the wild type. We observed no differences in the transcriptional levels of a housekeeping gene, the chloramphenicol acetyltransferase gene (cat), among strains examined. These results confirmed the success of mutagenesis and the complementation. To check for polar effects of inactivating tolC, we performed RT-PCR on the following four adjacent genes: ygiB and ygiC in the downstream, and yqiB and yqiE in the upstream. These sequences were obtained from the database available at http://www.sanger.ac.uk/Projects/Escherichia_Shigella/. All four genes were found to be successfully transcribed in the tolC mutant (data not shown), suggesting that there were no polar effects of inactivating tolC. To confirm the change of phenotype of the tolC mutant, we evaluated susceptibilities against deoxycholate and Triton X-100. The tolC mutant was found to be more susceptible to both than were the wild type and the complement. MICs to deoxycholate were as follows: for the wild type and the complement, >10 mg/ml; and for the tolC mutant, 0.1 mg/ml. MICs to Triton X-100 were as follows: for the wild type and the complement, >1%; and for the tolC mutant, 0.6%.
TolC does not export Aap to the medium.
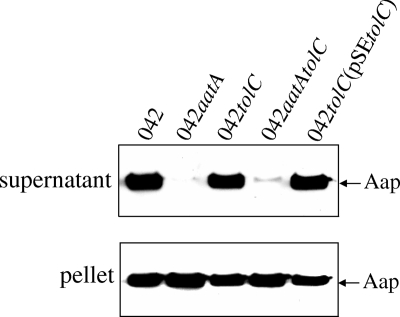
To assess a possible role for TolC in Aap export, we evaluated Aap secretion into supernatants of strains by Western blotting using anti-Aap antiserum. No difference was observed between the wild type and 042tolC in terms of Aap secretion (Fig. 1). A small amount of Aap in the supernatant of 042aatAtolC was not diminished compared to what was seen for 042aatA. We concluded that TolC is not involved in Aap secretion across the outer membrane.
FIG. 1.
Western immunoblot for Aap performed on supernatants or pellets of the wild type (042), 042tolC, 042aatA, 042aatAtolC, and 042tolC(pSEtolC) cultures for 6 h in high-glucose DMEM. Samples were separated using 15% SDS-PAGE and transferred as described in Materials and Methods.
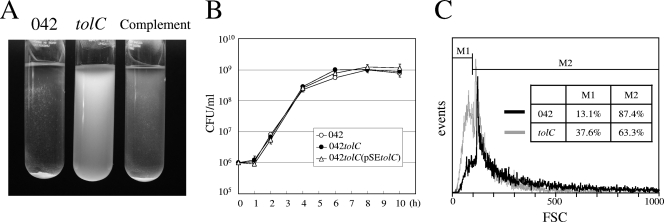
042tolC is less aggregative than the wild type in the liquid phase.
A stationary-phase broth culture of 042tolC showed a different phenotype from the isogenic parent strain 042. Aggregation observed for the wild-type culture in high-glucose DMEM was clearly diminished in the 042tolC culture (Fig. 2A). More floating cells in the medium and fewer precipitated cells were observed for 042tolC than for the wild type. The aggregative phenotype in high-glucose DMEM was restored in the complement strain 042tolC(pSEtolC) (Fig. 2A). The less aggregative phenotype of 042tolC was observed in LB culture, although it was not as prominent as in high-glucose DMEM. We observed no differences among examined strains in the concentration of bacteria after mixing by vortex mixer in both LB medium and high-glucose DMEM. The growth kinetic graph in LB medium is shown in Fig. 2B. Bacteria were analyzed using flow cytometry by measuring FSC to determine their heterogeneity with respect to the size of aggregates (Fig. 2C). A FSC histogram of 042tolC was deviated to the left in comparison with that of the wild type. The percentage of particles of less than 100 FSC was larger in 042tolC (37.6%) than in the wild type (13.1%). Since FSC represents the size of particles, these data showed that the sizes of aggregates or individual cells in 042tolC are smaller than those of the wild type and that 042tolC is less aggregative in the liquid phase than is the wild type.
FIG. 2.
(A) High-glucose DMEM cultures of the wild type (042), 042tolC, and 042tolC(pSEtolC). Strains were grown overnight at 37°C with shaking. tolC, 042tolC; Complement, 042tolC(pSEtolC). (B) Growth curve of the wild type, 042aatA, and 042tolC(pSEtolC) in LB medium. Viable cells in the medium were quantified by plating serial dilutions on L-agar plates in triplicate. Data are presented as means of triplicate experiments, with error bars representing one standard deviation (SD). Where no error bars are visible, the deviation is smaller than the symbol. (C) Flow cytometry analysis; FSC histogram of the wild type (042) (black) and 042tolC (gray) after overnight incubation at 37°C with shaking in high-glucose DMEM. The x axis represents the size of particles, and the y axis represents the cell number.
Hydrophobicity of the surface of 042tolC.
We evaluated the hydrophobicity of bacterial surfaces with the slide glass agglutination test using ammonium sulfate solution (Table 3). The wild type showed strong aggregation even in distilled water, while 042tolC showed mild aggregation in 0.05 M or more but not in 0.025 M or less. 042tolC(pSEtolC) showed mild aggregation in distilled water and strong aggregation in 0.025 M or more. These results suggest that the bacterial surface of 042tolC is less hydrophobic than that of the wild type and that the hydrophobicity was restored in the complement strain.
TABLE 3.
Salt aggregation test
| Strain | Aggregation at indicated concn (M) of ammonium sulfatea
|
||||||
|---|---|---|---|---|---|---|---|
| 1.0 | 0.5 | 0.25 | 0.05 | 0.025 | 0.005 | 0 | |
| 042 | 2+ | 2+ | 2+ | 2+ | 2+ | 2+ | 2+ |
| 042tolC | 1+ | 1+ | 1+ | 1+ | − | − | − |
| 042tolC(pSEtolC) | 2+ | 2+ | 2+ | 2+ | 2+ | 1+ | 1+ |
2+, aggregation observed immediately (within 10 seconds); 1+, aggregation observed slowly (more than 10 seconds); −, no aggregation.
HEp-2 cell adherence test.
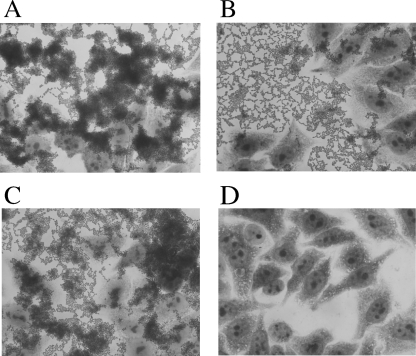
To evaluate the adherence ability to epithelial cells, we performed a standard HEp-2 cell adherence test (Fig. 3). The wild type, 042, strongly adhered to both the epithelial cell surface and the glass substratum in a distinctive stacked-brick formation like a honeycomb, and it formed thick aggregations consisting of multiple layers of cells (Fig. 3A). 042tolC did not form thick aggregations to both the epithelial cell surface and the glass substratum like the wild type did, although it showed a honeycomb formation on the glass (Fig. 3B). In the complement strain 042tolC(pSEtolC), a thick aggregation similar to that of the wild type was partially restored (Fig. 3C).
FIG. 3.
HEp-2 cell adherence test. Bacterial strains were incubated with HEp-2 cell monolayers for 3 h on glass coverslips in a 24-well microtiter plate in DMEM. (A) Wild type (042). (B) 042tolC. (C) 042tolC(pSEtolC). (D) HB101 (as a control).
Microtiter plate assay.
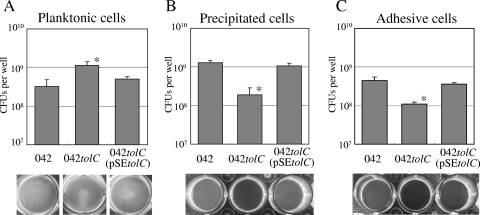
To assess aggregation and adhesion quantitatively, we performed a microtiter plate assay using 24-well polystyrene plates. Strong aggregation in high-glucose DMEM was observed in both the wild type and 042tolC (pSEtolC) after overnight incubation but not in 042tolC (Fig. 4A, bottom). There were more planktonic cells of 042tolC in the medium than wild-type cells or 042tolC(pSEtolC) cells (Fig. 4A). After aspiration of the medium containing planktonic cells, there were significantly fewer spontaneously precipitated cells on the substratum of 042tolC than on that of the wild type or 042tolC(pSEtolC) (Fig. 4B). After washing three times with PBS, there were significantly fewer adhesive cells on the substratum of 042tolC than on that of the wild type or 042tolC(pSEtolC) (Fig. 4C). Biofilm formation was also reduced in 042tolC in compared with what was seen for the wild type or 042tolC(pSEtolC) (data not shown).
FIG. 4.
Microtiter plate assay of the wild type (042), 042tolC, and 042tolC(pSEtolC). Strains were grown in triplicate in high-glucose DMEM in a 24-well microtiter plate for 20 h at 37°C. Wells were observed by a stereomicroscope (bottom). (A) Quantitative analysis of planktonic cells. The planktonic cells in the medium were quantified by plating serial dilutions on L-agar plates in triplicate. (B) Quantitative analysis of spontaneously settling cells. Precipitated cells on the substratum after medium aspiration were collected by scraping into 500 μl of PBS, and viable cells were counted as described above. (C) Quantitative analysis of adhesive cells. Adhesive cells on the substratum, after being washed with PBS three times, were collected and quantified as described above. Data are presented as means of triplicate experiments, with error bars representing one SD. *, P < 0.05 versus strains 042 and 042tolC(pSEtolC) by Student's t test of log-transformed data.
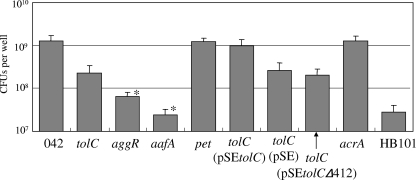
Comparison with other EAEC 042 mutants in microtiter plate assay.
The aggregative phenotype of 042tolC was quantitatively compared with other EAEC 042 mutants by a microtiter plate assay (Fig. 5). The numbers of spontaneously precipitated cells of the transcriptional regulator AggR mutant (042aggR) and the aggregative fimbria mutant (042aafA) were significantly less than that of 042tolC. There were as many cells of the mutant with plasmid-encoded toxin (042pet) as there were cells of the wild type, showing that the mutagenesis by pJP5603 did not affect the aggregative phenotype. There were as many cells of the complement strain with the expression vector pSE380, 042tolC(pSE), as there were cells of 042tolC.
FIG. 5.
Quantitative analysis of precipitated cells in a microtiter plate assay. Strains were grown in triplicate in high-glucose DMEM in a 24-well microtiter plate for 20 h at 37°C. Precipitated cells on the substratum after aspirating the medium were collected by scraping into 500 μl of PBS, and viable cells were counted by plating serial dilutions on L-agar plates in triplicate. tolC, 042tolC; aggR, 042aggR; aafA, 042aafA; pet, 042pet; acrA, 042acrA; tolC(pSEtolC), 042tolC(pSEtolC); tolC(pSE), 042tolC(pSE); tolC(pSEtolCΔ412), 042tolC(pSEtolCΔ412). Data are presented as means of triplicate experiments, with error bars representing one SD. *, P < 0.05 versus strain 042tolC by Student's t test of log-transformed data.
Yamanaka et al. reported that TolC required a nonpolar amino acid residue, Leu, at position 412 to express its activity (43). We constructed a deletion mutant gene, tolCΔ412, and complemented 042tolC with pSE(tolCΔ412). There were almost equal numbers of spontaneously precipitated cells of 042tolC(pSEtolCΔ412) and cells of 042tolC (Fig. 5), showing that complementation with tolCΔ412 did not restore the aggregative phenotype. TolC is known to be linked with an inner membrane protein (e.g., AcrB) and a periplasmic adapter protein (e.g., AcrA). We constructed the acrA mutant to determine whether AcrA was involved in the aggregative phenotype of EAEC 042 as a periplasmic adapter of TolC. There were as many spontaneously precipitated cells of 042acrA as there were cells of the wild type, showing that AcrA does not play a role in the aggregation of EAEC 042 (Fig. 5).
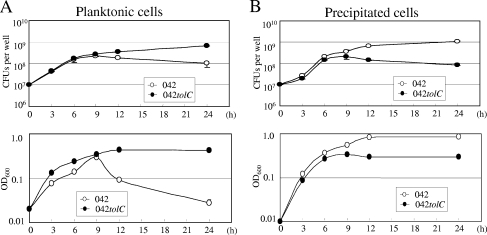
Time courses of planktonic and precipitated cells in microtiter plate assay.
Planktonic cells of the wild type reached their peak after 9 h of incubation and decreased dramatically afterward, whereas those of 042tolC continued to increase and became stationary after 12 h of incubation (Fig. 6A). Spontaneously precipitated cells of the wild type continued to increase and reached the stationary stage after 12 h of incubation, whereas 042tolC became stationary after 6 h of incubation (Fig. 6B). 042tolC appeared to be defective in aggregation and spontaneous settling after the late log phase in high-glucose DMEM culture.
FIG. 6.
Time courses of the planktonic and precipitated cells in a microtiter plate assay. Strains were grown in triplicate in high-glucose DMEM in a 24-well microtiter plate at 37°C for 3 h, 6 h, 9 h, 12 h, and 24 h. (A) The planktonic cells in the medium were measured by plating serial dilutions on L-agar plates in triplicate (top). The concentration of planktonic cells in the medium was measured in terms of optical density at 600 nm (OD600) (bottom). (B) Spontaneously precipitated cells on the substratum after medium aspiration were collected by scraping into 500 μl of PBS, and viable cells were counted as described above (top) and the concentration was measured as described above (bottom). Data are presented as means of triplicate experiments, with error bars representing one SD. Where no error bars are visible, the deviation is smaller than the symbol.
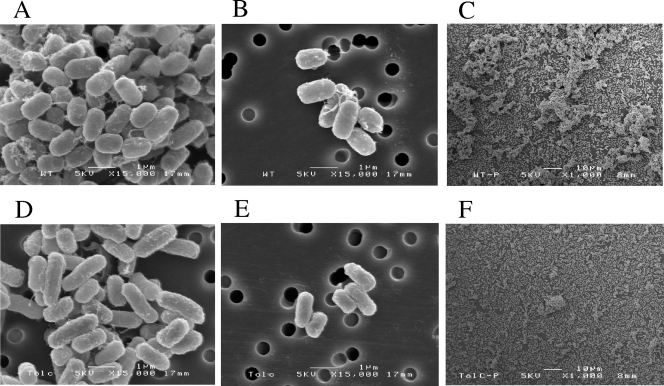
SEM.
SEM revealed that aggregates of the wild type cells in high-glucose DMEM were larger and denser than those of 042tolC (Fig. 7A and D). Fimbriae involved in the cell-cell adhesion were more characteristic in the wild type than in 042tolC (Fig. 7B and E). When cultured on the glass, biofilms of the wild type were thicker and more abundant than those of 042tolC (Fig. 7C and F). Since there were fewer fimbriae in the mutant, we performed RT-PCR of aafA to determine whether the effect is at the level of transcription or downstream thereof. No difference was observed in the transcription levels of aafA between the wild type and the tolC mutant (data not shown).
FIG. 7.
SEM images of planktonic cells of 042 (A and B) and 042tolC (D and E), and adhesive cells of 042 (C) and 042tolC (F). Planktonic cells grown in high-glucose DMEM overnight with shaking and adhesive cells grown on glass coverslips in high-glucose DMEM for 20 h were analyzed. (A, B, D, and E) Magnification, ×15,000; bar = 1 μm. (C and F) Magnification, ×1,000; bar = 10 μm.
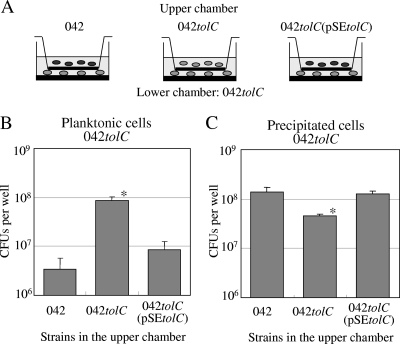
Transwell assay of EAEC 042tolC.
To evaluate whether the AA of the wild type was mediated by a humoral factor secreted by TolC, we performed a transwell assay (Fig. 8A). Planktonic cells of 042tolC when cocultured with the wild type and 042tolC (pSEtolC) were fewer in number than when cocultured with 042tolC (Fig. 8B). Spontaneously precipitated cells of 042tolC when cocultured with the wild type and 042tolC(pSEtolC) were greater in number than when cocultured with 042tolC (Fig. 8C). 042tolC cells were mostly planktonic and less adhesive in late stationary phase when cocultured with 042tolC, while they were rarely planktonic and more adhesive when cocultured with the wild type and the complement. Coculture with the wild type and the complement strain appeared to make 042tolC more aggregative.
FIG. 8.
Transwell assay. (A) Twenty-four-well polystyrene transwell plate with 0.4-μm-pore-size polyester membrane, which allows cytokines and secreted proteins, but not bacteria, to pass through the membrane. Ten microliters of an overnight L-broth culture of 042tolC was added in the lower chamber, and those of 042, 042tolC, and 042tolC(pSEtolC) were in the upper chamber. They were cocultured at 37°C for 20 h statically in high-glucose DMEM. (B) Quantitative analysis of planktonic cells of 042tolC in the lower chamber. The planktonic cells in the medium were measured by plating serial dilutions on L-agar plates in triplicate. (C) Quantitative analysis of precipitated cells. Spontaneously precipitated cells on the substratum after aspiration of the medium were collected by scraping into 500 μl of PBS, and viable cells were measured as described above. Data are presented as means of triplicate experiments, with error bars representing one SD. *, P < 0.05 versus strains 042 and 042tolC(pSEtolC) by Student's t test of log-transformed data.
DISCUSSION
Although TolC did not export Aap, we found that the tolC mutant was less aggregative in the liquid phase. Our results demonstrated quantitatively that tolC mutant cells were mostly planktonic in the medium in comparison with the wild type, 042. The aggregative phenotype was restored in the complement strain with the tolC gene. These data demonstrate that TolC is required for the characteristic aggregation of EAEC 042 in the liquid phase and suggest that TolC is associated with the cell-cell adhesion in aqueous circumstances. Virulence factors associated with aggregation of EAEC in medium include AA fimbriae (AAF), the global regulator AggR, and the dispersin protein Aap. AAF and AggR promote strong aggregation, while the dispersin protein Aap on the cell surface promotes the dispersion of EAEC (8, 33). Besides them, the hydrophobic surface protein layer consisting of a 38-kDa protein was previously reported to play a role in the aggregative phenotype in EAEC strains (38); however, the protein has not been identified. To our knowledge, TolC is considered to be the first chromosomal factor associated with the aggregation of EAEC in aqueous circumstances.
Our results demonstrated that the tolC mutant showed a less aggregative adherence to the substratum and a less abundant biofilm formation. The phenotype was restored in the complement strain with the tolC gene. Thus, TolC is also required for adhesion and biofilm formation of EAEC 042. The diminished adherence to the polystyrene in the tolC mutant might be attributed to the reduced spontaneous settling of cells to the substratum. Virulence factors including Fis, YafK, AAF, AggR, and Aap were reported to be associated with adhesion, colonization, and biofilm formation in EAEC (8, 35). Recently, Sheikh et al. reported that EilA, a HilA-like regulator, and air, encoding the predicted outer membrane protein in the EAEC chromosome, are associated with biofilm formation (34). The relatedness of TolC with these factors remains to be investigated.
One potential mechanism by which the tolC mutant lost the aggregative phenotype is the loss of the hydrophobicity of the bacterial surface. The AAF of EAEC 042 are highly hydrophobic, and this hydrophobicity would favor strong aggregation in an aqueous environment. We found that the tolC mutant was a little more aggregative than the AAF/II null mutant of 042, suggesting that the tolC mutant has the function of AAF to some extent. Indeed, we observed fimbriae in the tolC mutant via SEM, although they were less prominent. Thus, the loss of hydrophobicity of the tolC mutant may be due to hypofunction or reduced expression of the aggregative fimbriae. Since the transcription of the fimbria gene was conserved in the tolC mutant, the hypofunction or reduced expression of fimbriae seems to be at the posttranscriptional level. There is a report of Mg2+ being important for the adherence of some EAEC strains (2). It is possible that TolC might be associated with the regulation of Mg2+ transport.
Although the transwell assay is still preliminary, these results imply that an unknown humoral factor secreted by TolC of the wild type and the complement strain into the medium contributes to the phenotypic change of the tolC mutant. The assumed humoral factor might regulate the function or expression of AAF. Besides AAF, the hydrophobic surface protein layer consisting of a 38-kDa protein reported previously for EAEC (38) may be related to the assumed humoral factor. EAEC 042 may maintain balance between aggregation and dispersion by secreting Aap (dispersin) as an antiaggregative factor through AatA and the assumed aggregative factor through TolC. The identification of this humoral factor is now under investigation. On the other hand, the effect observed in the transwell assay could be via ions or quorum sensing during the adherence process, which could indirectly be affected in the tolC mutant. Further studies are needed to clarify the mechanism.
Anchoring bacteria to the mucosa can be strengthened by adherence not only to epithelial cells but also to each other. Furthermore, interbacterial aggregation is important for virulence even when the aggregates are not fixed to a surface (20). Aggregated bacteria in oral cavity were reported to be more resistant to phagocytes and killing by neutrophils both in vitro and in vivo (25). Factors conferring aggregation of E. coli, such as antigen 43 (Ag43), type I fimbriae, and Curli, have been investigated as important virulence factors thus far (20). Ag43 mediates the agglutination of E. coli in the liquid phase and also promotes E. coli biofilm formation (9). Since Ag43 is translocated across the outer membrane of E. coli by the autotransporter pathway, it may not be a candidate of the assumed aggregative protein secreted through TolC. However, a similar Ag43 homolog has been identified in the genome of EAEC 042 (20). A role of Ag43 in EAEC aggregation remains to be investigated.
Time courses of planktonic and precipitated cells in a microtiter plate assay imply that the wild type, 042, senses some signals to start autoaggregation. The tolC mutant seems not to be able to sense this signal or to start aggregation. Quorum-sensing systems by enteric bacteria have been fully investigated for enterohemorrhagic E. coli and Salmonella (13, 40) but not for EAEC 042. Yang et al. reported that AcrAB or TolC mutants in E. coli grow to higher cell density in stationary phase than their isogenic wild-type strains do (44). They showed that this phenomenon is attributable to the fact that mutants of multidrug transporters emit fewer quorum-sensing signals into the medium (44). Although we did not find different growth rates between the wild type and the tolC mutant of EAEC 042, it may be possible that the tolC mutant emits fewer quorum-sensing signals to start autoaggregation. Further investigations are needed to determine whether TolC is associated with a quorum-sensing system in EAEC 042.
AggR is a transcriptional activator that controls some plasmid-borne factors including AAF adhesins, AggR itself, Aap, and Aat (8). Dudley et al. reported that a pheU pathogenicity island in the chromosome is also regulated by AggR (5). We evaluated the transcriptional dependency of tolC on AggR by use of RT-PCR with the aggR mutant as previously described (24, 35). However, the transcription of tolC was found not to be dependent on AggR (data not shown).
In gram-negative bacteria, transmembrane efflux pumps are composed of three types of proteins: an outer membrane protein (e.g., TolC), an inner membrane protein (e.g., AcrB), and a periplasmic membrane fusion protein (e.g., AcrA) (15, 28, 29). TolC exerts the efflux function with several adapter proteins and inner membrane proteins in E. coli. TolC is also known to function as the protein channel of E. coli for different RND family efflux pumps (for example, MdtABC) and also interact with MFS transporters (for example, EmrAB) and ABC superfamily transporters (for example, MacAB) (29). We showed here that AcrA is not a partner of TolC in playing a role in the aggregation of EAEC. Other candidates, such as MacAB, AcrAD, AcrEF, EmrAB, MdtABC, and EmrAB, remain to be evaluated.
E. coli TolC secretes a heat-stable toxin in enterotoxigenic E. coli (42) and plays a role in the pathogenesis of enterotoxigenic E. coli (32). Recently, the AcrAB-TolC system was reported to be associated with the colonization and persistence of bacteria in the host and to have roles in bacterial pathogenicity (28, 29, 32). For example, Buckley et al. reported that the TolC mutant in Salmonella enterica serovar Typhimurium poorly adhered in vitro and was unable to invade macrophages and that the AcrAB-TolC system is important for the colonization of chickens by S. enterica serovar Typhimurium (1). However, the involvement of TolC in the aggregation, adhesion, and colonization of E. coli has not been reported thus far. We demonstrated here that TolC promotes aggregation and adhesion in EAEC 042. The aggregative phenotype is essential for the virulence of EAEC; therefore, the involvement of TolC in EAEC pathogenicity should be evaluated in the next study. The mechanism by which TolC promotes aggregation of EAEC is a target of ongoing research efforts.
Acknowledgments
This study was financially supported by a grant-in-aid for scientific research (category C), Japan (no. 17591098).
We thank H. Oda and K. Yoshiie of Kagoshima University, Japan, for their helpful discussions and technical support.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.Buckley, A. M., M. A. Webber, S. Cooles, L. P. Randall, R. M. La Ragione, M. J. Woodward, and L. J. Piddock. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell. Microbiol. 8847-856. [DOI] [PubMed] [Google Scholar]
- 2.Chart, H., J. Spencer, H. R. Smith, and B. Rowe. 1997. Magnesium ions are required for HEp-2 cell adhesion by enteroaggregative strains of Escherichia coli O126:H27 and O44:H18. FEMS Microbiol. Lett. 14849-52. [DOI] [PubMed] [Google Scholar]
- 3.Cohen, M. B., J. P. Nataro, D. I. Bernstein, J. Hawkins, N. Roberts, and M. A. Staat. 2005. Prevalence of diarrheagenic Escherichia coli in acute childhood enteritis: a prospective controlled study. J. Pediatr. 14654-61. [DOI] [PubMed] [Google Scholar]
- 4.Czeczulin, J. R., S. Balepur, S. Hicks, A. Phillips, R. Hall, M. H. Kothary, F. Navarro-Garcia, and J. P. Nataro. 1997. Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect. Immun. 654135-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudley, E. G., N. R. Thomson, J. Parkhill, N. P. Morin, and J. P. Nataro. 2006. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 611267-1282. [DOI] [PubMed] [Google Scholar]
- 6.Elias, W. P., Jr., J. R. Czeczulin, I. R. Henderson, L. R. Trabulsi, and J. P. Nataro. 1999. Organization of biogenesis genes for aggregative adherence fimbria II defines a virulence gene cluster in enteroaggregative Escherichia coli. J. Bacteriol. 1811779-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eslava, C., F. Navarro-Garcia, J. R. Czeczulin, I. R. Henderson, A. Cravioto, and J. P. Nataro. 1998. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect. Immun. 663155-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 8.Harrington, S. M., E. G. Dudley, and J. P. Nataro. 2006. Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol. Lett. 254 12-18. [DOI] [PubMed] [Google Scholar]
- 9.Hasman, H., T. Chakraborty, and P. Klemm. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J. Bacteriol. 1814834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson, I. R., S. Hicks, F. Navarro-Garcia, W. P. Elias, A. D. Philips, and J. P. Nataro. 1999. Involvement of the enteroaggregative Escherichia coli plasmid-encoded toxin in causing human intestinal damage. Infect. Immun. 675338-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, D. B., J. P. Nataro, H. L. DuPont, P. P. Kamat, A. D. Mhatre, P. C. Okhuysen, and T. Chiang. 2006. Enteroaggregative Escherichia coli is a cause of acute diarrheal illness: a meta-analysis. Clin. Infect. Dis. 43556-563. [DOI] [PubMed] [Google Scholar]
- 12.Iwashita, M., J. Nishi, N. Wakimoto, R. Fujiyama, K. Yamamoto, K. Tokuda, K. Manago, and Y. Kawano. 2006. Role of the carboxy-terminal region of the outer membrane protein AatA in the export of dispersin from enteroaggregative Escherichia coli. FEMS Microbiol. Lett. 256266-272. [DOI] [PubMed] [Google Scholar]
- 13.Kendall, M. M., and V. Sperandio. 2007. Quorum sensing by enteric pathogens. Curr. Opin. Gastroenterol. 2310-15. [DOI] [PubMed] [Google Scholar]
- 14.Koronakis, V. 2003. TolC—the bacterial exit duct for proteins and drugs. FEBS Lett. 55566-71. [DOI] [PubMed] [Google Scholar]
- 15.Koronakis, V., J. Eswaran, and C. Hughes. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73467-489. [DOI] [PubMed] [Google Scholar]
- 16.Mattos-Guaraldi, A. L., L. C. Formiga, and A. F. Andrade. 1999. Cell surface hydrophobicity of sucrose fermenting and nonfermenting Corynebacterium diphtheriae strains evaluated by different methods. Curr. Microbiol. 38 37-42. [DOI] [PubMed] [Google Scholar]
- 17.Mobley, H. L., K. G. Jarvis, J. P. Elwood, D. I. Whittle, C. V. Lockatell, R. G. Russell, D. E. Johnson, M. S. Donnenberg, and J. W. Warren. 1993. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal(1-4) beta Gal binding in virulence of a wild-type strain. Mol. Microbiol. 10143-155. [DOI] [PubMed] [Google Scholar]
- 18.Nataro, J. P., M. M. Baldini, J. B. Kaper, R. E. Black, N. Bravo, and M. M. Levine. 1985. Detection of an adherence factor of enteropathogenic Escherichia coli with a DNA probe. J. Infect. Dis. 152560-565. [DOI] [PubMed] [Google Scholar]
- 19.Nataro, J. P., Y. Deng, S. Cookson, A. Cravioto, S. J. Savarino, L. D. Guers, M. M. Levine, and C. O. Tacket. 1995. Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J. Infect. Dis. 171465-468. [DOI] [PubMed] [Google Scholar]
- 20.Nataro, J. P., and A. Jansen. 2005. Aggregation and dispersal on mucosal surfaces, p. 253-263. In J. P. Nataro, P. S. Cohen, H. L. T. Mobley, and J. N. Weiser (ed.), Colonization of mucosal surfaces. ASM Press, Washington, DC.
- 21.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nataro, J. P., J. B. Kaper, R. Robins-Browne, V. Prado, P. Vial, and M. M. Levine. 1987. Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr. Infect. Dis. J. 6829-831. [DOI] [PubMed] [Google Scholar]
- 23.Nataro, J. P., V. Mai, J. Johnson, W. C. Blackwelder, R. Heimer, S. Tirrell, S. C. Edberg, C. R. Braden, J. G. Morris, Jr., and J. M. Hirshon. 2006. Diarrheagenic Escherichia coli infection in Baltimore, Maryland, and New Haven, Connecticut. Clin. Infect. Dis. 43402-407. [DOI] [PubMed] [Google Scholar]
- 24.Nishi, J., J. Sheikh, K. Mizuguchi, B. Luisi, V. Burland, A. Boutin, D. J. Rose, F. R. Blattner, and J. P. Nataro. 2003. The export of coat protein from enteroaggregative Escherichia coli by a specific ATP-binding cassette transporter system. J. Biol. Chem. 27845680-45689. [DOI] [PubMed] [Google Scholar]
- 25.Ochiai, K., T. Kurita-Ochiai, Y. Kamino, and T. Ikeda. 1993. Effect of co-aggregation on the pathogenicity of oral bacteria. J. Med. Microbiol. 39183-190. [DOI] [PubMed] [Google Scholar]
- 26.Okeke, I. N., and J. P. Nataro. 2001. Enteroaggregative Escherichia coli. Lancet Infect. Dis. 1304-313. [DOI] [PubMed] [Google Scholar]
- 27.Penfold, R. J., and J. M. Pemberton. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118145-146. [DOI] [PubMed] [Google Scholar]
- 28.Piddock, L. J. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piddock, L. J. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4629-636. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Sarantuya, J., J. Nishi, N. Wakimoto, S. Erdene, J. P. Nataro, J. Sheikh, M. Iwashita, K. Manago, K. Tokuda, M. Yoshinaga, K. Miyata, and Y. Kawano. 2004. Typical enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in Mongolian children. J. Clin. Microbiol. 42133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharff, A., C. Fanutti, J. Shi, C. Calladine, and B. Luisi. 2001. The role of the TolC family in protein transport and multidrug efflux. From stereochemical certainty to mechanistic hypothesis. Eur. J. Biochem. 2685011-5026. [DOI] [PubMed] [Google Scholar]
- 33.Sheikh, J., J. R. Czeczulin, S. Harrington, S. Hicks, I. R. Henderson, C. Le Bouguenec, P. Gounon, A. Phillips, and J. P. Nataro. 2002. A novel dispersin protein in enteroaggregative Escherichia coli. J. Clin. Investig. 1101329-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheikh, J., E. G. Dudley, B. Sui, B. Tamboura, A. Suleman, and J. P. Nataro. 2006. EilA, a HilA-like regulator in enteroaggregative Escherichia coli. Mol. Microbiol. 61338-350. [DOI] [PubMed] [Google Scholar]
- 35.Sheikh, J., S. Hicks, M. Dall'Agnol, A. D. Phillips, and J. P. Nataro. 2001. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol. Microbiol. 41983-997. [DOI] [PubMed] [Google Scholar]
- 36.Shi, J., T. L. Blundell, and K. Mizuguchi. 2001. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310243-257. [DOI] [PubMed] [Google Scholar]
- 37.Tzipori, S., J. Montanaro, R. M. Robins-Browne, P. Vial, R. Gibson, and M. M. Levine. 1992. Studies with enteroaggregative Escherichia coli in the gnotobiotic piglet gastroenteritis model. Infect. Immun. 605302-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wai, S. N., A. Takade, and K. Amako. 1996. The hydrophobic surface protein layer of enteroaggregative Escherichia coli strains. FEMS Microbiol. Lett. 13517-22. [DOI] [PubMed] [Google Scholar]
- 39.Wakimoto, N., J. Nishi, J. Sheikh, J. P. Nataro, J. Sarantuya, M. Iwashita, K. Manago, K. Tokuda, M. Yoshinaga, and Y. Kawano. 2004. Quantitative biofilm assay using a microtiter plate to screen for enteroaggregative Escherichia coli. Am. J. Trop. Med. Hyg. 71687-690. [PubMed] [Google Scholar]
- 40.Walters, M., and V. Sperandio. 2006. Quorum sensing in Escherichia coli and Salmonella. Int. J. Med. Microbiol. 296125-131. [DOI] [PubMed] [Google Scholar]
- 41.Yamanaka, H., N. Morisada, M. Miyano, H. Tsuge, S. Shinoda, E. Takahashi, and K. Okamoto. 2004. Amino-acid residues involved in the expression of the activity of Escherichia coli TolC. Microbiol. Immunol. 48713-722. [DOI] [PubMed] [Google Scholar]
- 42.Yamanaka, H., T. Nomura, Y. Fujii, and K. Okamoto. 1998. Need for TolC, an Escherichia coli outer membrane protein, in the secretion of heat-stable enterotoxin I across the outer membrane. Microb. Pathog. 25111-120. [DOI] [PubMed] [Google Scholar]
- 43.Yamanaka, H., T. Nomura, N. Morisada, S. Shinoda, and K. Okamoto. 2002. Site-directed mutagenesis studies of the amino acid residue at position 412 of Escherichia coli TolC which is required for the activity. Microb. Pathog. 3381-89. [DOI] [PubMed] [Google Scholar]
- 44.Yang, S., C. R. Lopez, and E. L. Zechiedrich. 2006. Quorum sensing and multidrug transporters in Escherichia coli. Proc. Natl. Acad. Sci. USA 1032386-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]