Abstract
Phage display screening with fragmented genomic DNA from the animal pathogen Pasteurella multocida has identified a gene encoding a putative fibronectin binding protein (19). Homologues of this gene (PM1665) are found in all other sequenced members of the Pasteurellaceae. Gene PM1665 has been cloned, and the protein has been expressed. Recombinant PM1665 protein binds to both soluble and immobilized fibronectin and is unique in that it interacts with the integrin-binding fibronectin type III (FnIII) repeats FnIII9-10 and not, as is the case for almost all other fibronectin adhesins, to the N-terminal type I repeats. Surface plasmon resonance analysis revealed a complex binding mechanism with a KD (equilibrium dissociation constant) of 150 nM ± 70 nM. Bioinformatics analysis suggests that the PM1665 protein contains two helix-hairpin-helix (HhH) motifs, and truncation mutation studies have identified the binding site in the protein as a combination of these two HhH motifs in conjunction with a conserved amino acid motif, VNINTA. We have shown that the PM1665 protein is on the cell surface and that binding of P. multocida to fibronectin is almost completely inhibited by anti-PM1665 antiserum. These results support the hypothesis that the PM1665 protein is a member of a new family of fibronectin binding adhesins that are important in the adhesion of P. multocida to fibronectin.
Pasteurella multocida is an important veterinary pathogen that causes a range of animal diseases, including fowl cholera, hemorrhagic septicemia in cattle and buffalo, and atrophic rhinitis in swine (9). The organism can also be a human pathogen; infection with P. multocida, as the result of an animal bite, can result in septic arthritis, meningitis, endocarditis, pneumonia, and sepsis (1, 7). All bacterial infection begins with the process of colonization, and it is important to determine how P. multocida adheres to host cells and tissues.
The ability to cause pathogenesis in such a wide range of host species suggests that P. multocida must have an armamentarium of adhesins to allow tissue colonization. However, to date, only a small number of potential adhesins have been identified. These include hemagglutinins (8), type IV fimbriae (5, 23), and the outer membrane protein OmpA (3). However, the ways in which these molecules interact with the host have not been elucidated.
We have recently identified a novel putative fibronectin binding protein by panning a phage display library of fragmented P. multocida DNA (19). Recombinant phage containing the entire open reading frame of PM1665 (with the exception of the final 3′ 6 base pairs), which codes for a small hypothetical protein, bound to immobilized fibronectin and to collagen (type I) but not to a range of other matrix macromolecules, including fibrinogen, laminin, hyaluronic acid, and plasminogen (19). Bioinformatics analysis of the PM1665 gene predicted a protein of 115 amino acids with a signal peptide of 24 amino acids. The protein was predicted to have two helix-hairpin-helix (HhH) domains in the C terminus, residues 61 to 86 and 92 to 113, respectively. However, the prediction of these HhH motifs by Motifscan had only low certainty, and they were not predicted at all by Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/). The PM1665 protein has sequence homology with the ComEA DNA uptake proteins present in a number of bacteria, and there are homologous proteins in all sequenced members of the Pasteurellaceae (19). The aims of this study were to clone and express this uncharacterized open reading frame from P. multocida, to confirm the fibronectin binding capacity of the recombinant protein, to characterize the interaction of the protein with fibronectin, and to ascertain whether the protein could have a functional role in the interaction of P. multocida with fibronectin.
MATERIALS AND METHODS
Bacterial strains and plasmids.
P. multocida NCTC 10322/ATCC 43137 (serotype A) was obtained from the National Collection of Type Cultures (London, United Kingdom) and cultured on chocolate agar or grown in brain heart infusion (BHI) broth (Oxoid Ltd.) aerobically at 37°C. For expression of recombinant proteins, the glutathione S-transferase (GST) fusion expression vector pGEX6-P-1 (GE Healthcare) was used with the Escherichia coli host Rosetta-gami DE3 (Novagen).
Cloning of the gene for PM1665 protein.
Oligonucleotides containing recognition sequences for the restriction enzymes BamHI and EcoRI (Table 1) were designed to amplify the gene coding for the PM1665 protein without its predicted signal sequence. The PCR fragment obtained was initially cloned into pCR-TOPO (Invitrogen) and transformed into E. coli TOP10. The cloned gene was excised from pCR-TOPO on a BamHI/EcoRI fragment and ligated to BamHI/EcoRI-digested pGEX6-P-1 (GE Healthcare). Three fragments of the PM1665 gene were also cloned and expressed as GST fusion proteins in pGEX-6-P-1. The primer pairs used to amplify DNA fragments of the PM1665 gene from genomic DNA of P. multocida are detailed in Table 1.
TABLE 1.
Primer pairs used in this study
| Recombinant GST fusion protein | Size (bp) | No. of amino acids | Primer paira |
|---|---|---|---|
| rGST-PM1665 | 345 | 94 | 5′-GGATCCAATCAGCAAACATGAC-3′ |
| 5′-GAATTCTTAGAGTTGAAGGCGTG-3′ | |||
| Mature PM1665 | 345 | 115 | 5′-GATCCGAATCAGCAAACATGAC-3′ |
| 5′-GAATTCTTAGAGTTGAAGGCGTG-3′ | |||
| 16S rRNA | 5′-AAGTCGTAACAAGGTA-3′ | ||
| 5′-GGTACTTAGATGTTTCAGTTC-3′ | |||
| rGST-PM1665 54-115 | 161-345 | 54-115 | F 5′-CGCGAATTCGTGAATATTAATACAGCCAGC-3′ |
| R 5′-GAATTCTTAGAGTTGAAGGCGTG-3′ | |||
| rGST-PM1665 61-115 | 181-345 | 61-115 | F 5′-CGCGAATTCGCAACTGACTTGCAACAAAAG-3′ |
| R 5′-GAATTCTTAGAGTTGAAGGCGTG-3′ | |||
| rGST-PM1665 54-86 | 161-256 | 54-86 | F 5′-CGCGAATTCGTGAATATTAATACAGCCAGC-3′ |
| R 5′-CCGCTCGAGATGCTTTTCTCGGTGTTGGAT-3′ |
Restriction sites are underlined.
Expression and purification of PM1665 protein.
For gene expression, positive clones were grown to log phase in nutrient broth no. 2 (Oxoid) containing 200 μg/ml of ampicillin at 30°C. Gene expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h at 30°C. Cells were harvested and then resuspended and lysed for 30 min in 4 ml of B-PER protein extraction reagent (Pierce) containing 750 mM ammonium chloride and 50 μl of protease inhibitor cocktail (Sigma; product number P8465). The lysates were clarified by centrifugation at 15,000 × g for 10 min, diluted 1:1 in phosphate-buffered saline (PBS), and purified on a GSTrap column (GE Healthcare).
Direct binding enzyme-linked immunosorbent assay (ELISA) for measuring binding of PM1665 protein to matrix proteins.
Nunc Maxisorb microtiter plates were coated overnight at 4°C with 100-μl aliquots of 100-μg/ml solutions of the following test ligands dissolved in coating buffer (0.05 M carbonate-bicarbonate buffer, pH 9.6): fibronectin, the 30-kDa N-terminal and 45-kDa α-chymotryptic fragments of fibronectin, collagen (type I, acid soluble, from human placenta), fibrinogen (fraction I from human plasma), hyaluronan, and plasminogen (all of which were purchased from Sigma-Aldrich Co. Ltd.), mouse laminin (Collaborative Biomedical Products, Bedford, MA), and the 120-kDa α-chymotryptic fragment of fibronectin (Chemicon International). The wells were blocked with 200 μl of PBS containing 5% skim milk and 0.1% Tween 20 for 2 h at 37°C. After the plates were washed, serial dilutions (in PBS containing 0.1% Tween 20) of purified recombinant GST-PM1665 (or recombinant GST from Schistosoma japonicam [Sigma] in some experiments) were added, and the mixtures were incubated for 2 h at 37°C. Bound proteins were detected by the addition of anti-GST antibody (1:2,000 dilution; GE Healthcare), followed by mouse anti-goat horseradish peroxidase (HRP)-conjugated antibody (1:40,000 dilution; Sigma-Aldrich). Finally, bound HRP-conjugated antibodies were detected by reading the optical densities at 492 nm after the addition of O-phenylene-diamine-HCl (Sigma-Aldrich). The optical densities at 492 nm for experimental samples were expressed as concentrations of bound recombinant GST-PM1665 (rGST-PM1665) using wells coated with a range of concentrations of rGST-PM1665 to obtain a standard curve. All samples were tested in quadruplicate. The recombinant fibronectin type III (FnIII) repeats FnIII1-2, FnIII4-7, and FnIII9-10 were also tested for binding to the PM1665 protein.
Ligand blotting.
Proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 12% gels and transferred overnight to Immobilon-P nitrocellulose membranes (Sigma-Aldrich) by electroblotting them. The membranes were blocked by incubation in blocking buffer (PBS containing 5% skim milk and 0.1% Tween 20) for 2 h. In some experiments, membranes were incubated with 20 μg/ml fibronectin in blocking buffer for 2 h, then with a 1:1,000 dilution of rabbit anti-fibronectin antibody (Sigma-Aldrich) in blocking buffer for 1 h at room temperature, and finally with mouse anti-rabbit immunoglobulin G (IgG)-peroxidase conjugate (whole molecule; Sigma-Aldrich) at a 1:1,000 dilution. In other experiments, membranes were incubated with a goat anti-GST antibody (GE Healthcare) to detect bound rGST-PM1665, followed by incubation with rabbit anti-goat IgG peroxidase conjugate to detect bound primary antibody, before development. All membranes were developed with Sigma Fast Diaminobenzidine peroxidase substrate (Sigma-Aldrich). For some blotting experiments, biotinylated ligands were used, followed by detection with streptavidin-HRP (Vector Labs) and diaminobenzidine as described above. FnIII1-2 and FnIII9-10 modules were kindly provided by Iain Campbell, University of Oxford. FnIII4-7 was kindly provided by Kim Midwood, Imperial College, United Kingdom. E. Wijelath (University of Washington) and S. Rahman (Kings College, London, United Kingdom) generously provided cDNA coding for FnIII8-10 and FnIII11, which were expressed as GST fusion proteins.
SPR.
The equilibrium dissociation constant (KD) for the interaction of recombinant PM1665 protein with immobilized fibronectin was determined by surface plasmon resonance (SPR) using a BIAcore 3000 system (BIAcore AB International). GST-free PM1665 protein was used for these experiments, as there is evidence that avidity effects due to dimerization of the GST can result in overestimation of KD values (13). The GST tag was removed from rGST-PM1665 by cleavage of the fusion protein with PreScission protease (GE Healthcare). Free GST and the PreScission protease were removed from the PM1665 protein by affinity chromatography on a GSTrap column. Fibronectin was covalently immobilized on a CM5 sensor chip via amine coupling using an amine-coupling kit (BIAcore). A “blank” reference surface was also prepared by activating the dextran surface of a flow cell, followed by blocking of the activated sites with 1 M ethanolamine. Increasing concentrations of the analyte (PM1665 protein) were allowed to flow across both reference and fibronectin-coated flow cells in running buffer (PBS containing 0.05% Tween 20) at 25°C using a flow rate of 5 μl/min for 30 min. Dissociation was then monitored for a further 85 min. Regeneration was performed by treatment with 0.1% SDS, which did not affect subsequent binding levels. Binding was determined by measuring the increase in resonance units after subtraction of the background response obtained from the reference flow cell.
Competition ELISA.
For inhibition ELISAs, recombinant GST-PM1665 was preincubated for 1 h at 37°C with soluble fibronectin, the 120-kDa α-chymotryptic fragment of fibronectin, superfibronectin, or the peptide RGD or RGDSPASSKP (all from Sigma-Aldrich). The reaction mixtures were then added to fibronectin-coated wells, and bound rGST-PM1665 was detected with an anti-GST antibody as described above.
Determination of the binding site for fibronectin in PM1665 protein.
A number of recombinant PM1665 protein fragments were prepared to identify which part(s) of the protein is important in binding to fibronectin. DNA coding for fragments of the PM1665 protein was cloned into pGEX6P1 and expressed as GST fusions. Recombinant proteins were purified as described above. Three smaller PM1665 protein fragments were purchased from Sigma Aldrich as synthetic peptides with an additional biotinylated (Btn) alanine residue, a 27-mer peptide corresponding to the first predicted HhH motif (Btn-AATDLQQKLLGIGAKKAEAIIQHREKH), a 23-mer peptide corresponding to the second predicted HhH motif (Btn-AIEQLRDIQGIGQAILEKNKTRQ), and a 7-mer peptide (Btn-AVNINTAS) corresponding to the motif that is highly conserved among homologous proteins in other members of the Pasteurellaceae. These biotinylated peptides were tested for binding to fibronectin in a direct binding ELISA. The biotinylated peptides were added to wells coated with fibronectin, and bound peptide was detected using a 1:1,000 dilution of streptavidin-HRP (Vector Laboratories).
RT-PCR analysis.
Overnight cultures of P. multocida grown in BHI broth or BHI broth containing 500 μg/ml of fibronectin were diluted 1:1,000 in fresh broth or broth containing 500 μg/ml fibronectin and incubated at 37°C with shaking. Aliquots containing 1 × 109 CFU were removed at various times, and total cellular RNA was extracted using an RNeasy mini kit (Qiagen) according to the manufacturer's instructions. The purified RNA was treated with RNase-free DNase (Promega), and cDNA was prepared from 2 μg of RNA using random hexamers and Superscript II (Invitrogen) according to the manufacturer's instructions. PCR was performed using 2 μl of the cDNA and oligonucleotides designed to amplify the gene coding for mature PM1665 protein (as detailed in Table 1). Control reactions were carried out using the purified RNA as a template for the PCRs to check for DNA contamination. Reverse transcription (RT)-PCR for 16S rRNA was also performed, using the cDNA as a template, as a positive control for the generation of mRNA.
Preparation of antiserum to PM1665 protein.
Antibodies were raised against the PM1665 protein that was produced initially as a recombinant GST fusion protein. Purified rGST-PM1665 was digested with PreScission Protease (GE Healthcare) to cleave the GST tag and then purified on a GSTrap column to remove the protease and the GST. Antibodies against the recombinant PM1665 protein were raised in New Zealand White rabbits by intramuscular injection of 50 μg of protein in Freund's incomplete adjuvant; booster injections (30 μg each) were administered after 6 and 9 weeks. IgG was prepared from antiserum by affinity chromatography on a Protein G HP column (GE Healthcare). The specific activity of the purified IgG from anti-PM1665 protein antiserum compared with IgG purified from preimmune serum was confirmed by ELISA using recombinant PM1665 protein (data not shown).
Transmission electron microscopy and image analysis.
Stationary-phase cultures of P. multocida cells were pelleted and immediately fixed in 0.5% glutaraldehyde in 0.1 M cacodylic acid buffer, pH 7.4; dehydrated to 90% ethanol; and embedded in LR White resin (Agar Scientific Ltd., United Kingdom.). Ultrathin sections of 90 to 100 nm were cut on an ultramicrotome (Reichert, Leica, United Kingdom) with a diamond knife. These sections were mounted onto carbon-Formvar coated grids, stained, and viewed with a JEOL 100CX II transmission electron microscope (JEOL UK, United Kingdom) operating at 80 kV. The images were recorded on Kodak EM 4638 film (TAAB, United Kingdom). Cultures of Porphyromonas gingivalis were prepared in an identical manner. For immunogold labeling, bacteria were rehydrated with distilled water, blocked with blocking buffer (2% skim milk in PBS), and incubated with anti-PM1665 protein IgG or preimmune IgG for 1 h. After being extensively washed, the grids were incubated with a 1-in-100 dilution of goat anti-rabbit antibody conjugated with a 10-nm gold probe (British Biocell, United Kingdom) for 1 h. The grids were then blotted dry and stained with ethanoic uranyl acetate and Reynolds' lead citrate (22).
Bacterial binding assays.
To determine the capacity of P. multocida to bind to immobilized fibronectin or collagen, triplicate wells of a Nunc maxisorb microtiter plate were coated with 10 μg/100 μl of each ligand in PBS overnight at 4°C. After the plates were blocked with 2% bovine serum albumin solution, 3 × 108 CFU of P. multocida was added to each well in a total volume of 100 μl of PBS. The plates were incubated at 37°C for 1.5 h and then washed extensively. Bound bacteria were removed from the wells by the addition of 100 μl of 0.25% trypsin. Serial dilutions were plated in triplicate on chocolate agar and grown at 37°C overnight. For inhibition assays, 100 μl of anti-PM1665 protein antibody or serially diluted rGST-PM1665 was added to wells coated with fibronectin, collagen, or FnIII9-10. The number of P. multocida cells that bound to each well was then determined as described above.
Bioinformatics analysis.
The predicted amino acid sequence of PM1665 protein was analyzed using a number of bioinformatics tools, including SignalP (http://www.cbs.dtu.dk/services/SignalP/), Psipred (http://bioinf.cs.ucl.ac.uk/psipred/), SMART, and ProtParam (http://www.expasy.ch/tools/protparam.html).
Statistical analyses.
Data are expressed as means ± standard errors. Statistical analyses were undertaken using one-way analyses of variance (ANOVAs) with Minitab, or in the case of the competition ELISA data, values were log transformed, logistic regressions were fitted, and the differences between the curves were tested using an F test (GraphPad Prism v4.03). The level for significance was taken as a P value of ≤0.05.
RESULTS
Capacity of recombinant GST-PM1665 to bind to extracellular matrix components determined by ELISA.
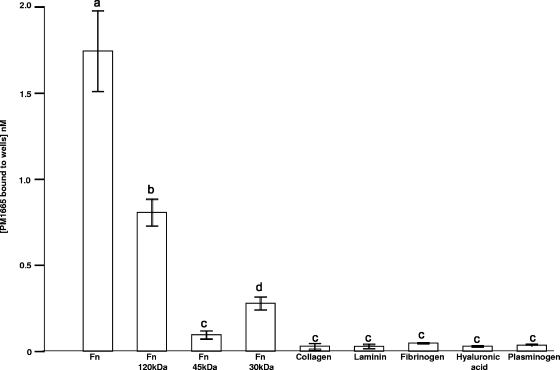
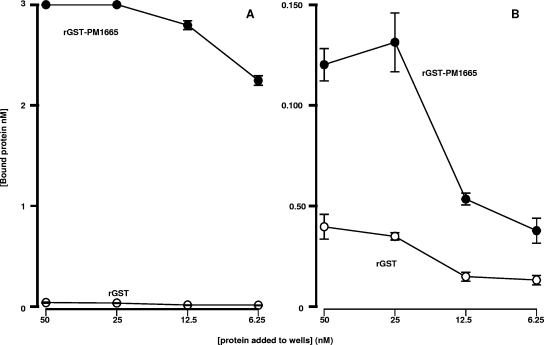
To determine whether the recombinant protein coded for by the PM1665 gene binds to fibronectin and collagen in a manner similar to that of the recombinant phage described previously (19), the PM1665 gene was cloned into the GST fusion expression vector pGEX-6-P-1 (GE Healthcare) and used to produce the mature protein (minus the signal peptide) in fusion with GST (GST-PM1665). The relative capacities of rGST-PM1665 to bind to fibronectin, collagen, and a variety of host extracellular matrix components were determined by direct binding ELISA. The recombinant protein bound only to fibronectin (one-way ANOVA; P ≤ 0.001) and not to type I collagen, fibrinogen, laminin, plasminogen, or hyaluronic acid (one-way ANOVA; P ≥ 0.5) (Fig. 1). A number of commercially available proteolytic fragments of fibronectin were used to determine where the PM1665 protein bound in this multidomain protein. Recombinant GST-PM1665 bound to the central 120-kDa cell-binding region of fibronectin with limited, but significant (one-way ANOVA; P < 0.05), binding to the 30-kDa N-terminal fragment and no significant binding to the 45-kDa gelatin-binding domain of fibronectin (Fig. 1). As the binding characteristics of the recombinant protein differed in some respects from the binding characteristics of the recombinant phage, a second series of direct binding ELISA experiments were performed to determine the effect of the presence of GST on the capacity of the PM1665 protein to bind to fibronectin. In these experiments, the highest concentration of rGST-PM1665 used was 50 nM. The GST showed no binding to fibronectin (Fig. 2A). At high concentrations, 50 nM and 25 nM, rGST-PM1665 did exhibit some binding to collagen (type I), but the extent of this binding was negligible compared to the binding to fibronectin at these concentrations (Fig. 2B).
FIG. 1.
Direct binding ELISA to measure binding of 5 nM of recombinant GST-PM1665 to immobilized mammalian ligands found in the extracellular matrix or in plasma. The amount of rGST-PM1665 bound to each ligand was estimated using a standard curve constructed by coating microtiter plate wells with known amounts of rGST-PM1665. The data are presented as the mean ± standard error of the mean of triplicate wells and are representative of three separate experiments. Bars with different letters are significantly different from each other (one-way ANOVA; see the text for P values).
FIG. 2.
Direct binding ELISA to determine the interaction of 50 nM of rGST-PM1665 with immobilized fibronectin (A) or type I collagen (B). Wells were coated with equimolar concentrations of fibronectin or collagen, and after blocking, doubling dilutions of rGST-PM1665 (closed circles) or rGST (open circles) were added to the wells in quadruplicate. The estimated concentrations of rGST and rGST-PM1665 binding to the fibronectin- or collagen-coated wells are shown as the mean ± standard error of the mean of quadruplicate wells. The data are representative of three such experiments.
Determination of the capacity of recombinant GST-PM1665 to bind to fibronectin by ligand blotting.
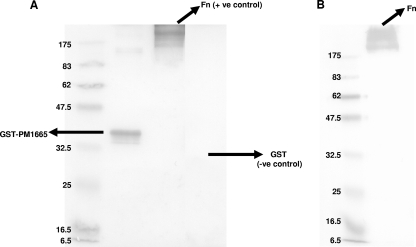
To determine whether rGST-PM1665 could bind to both immobilized and soluble fibronectin, rGST-PM1665 or fibronectin was subjected to SDS-PAGE, blotted from the gel onto nitrocellulose membranes, and detected with an anti-fibronectin antibody (shown in Fig. 3A) or an anti-GST antibody (shown in Fig. 3B), respectively. rGST-PM1665 bound to both soluble and immobilized fibronectin.
FIG. 3.
Ligand blots showing the binding of rGST-PM1665 to immobilized or soluble fibronectin. (A) rGST-PM1665 was subjected to SDS-PAGE, blotted onto a nitrocellulose membrane, and probed with fibronectin and then with an anti-fibronectin antibody. (B) Fibronectin was subjected to SDS-PAGE, blotted onto nitrocellulose, and incubated first with rGST-PM1665 and then with an anti-GST antibody. Molecular mass markers (in kilodaltons) are indicated on the left. +ve control and −ve control, positive and negative controls, respectively.
Analysis of PM1665 protein binding to fibronectin by SPR.
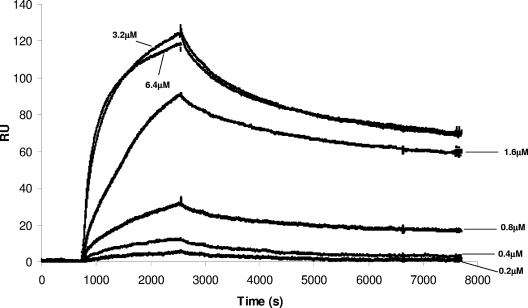
Increasing concentrations of GST-free PM1665 protein (the GST tag having been removed by protease digestion [see Materials and Methods]) were allowed to flow across a flow cell coated with fibronectin, and the binding responses were recorded as changes in response units after subtraction of the binding response for the reference flow cell. A representative sensorgram is shown in Fig. 4. The kinetics of interaction are dominated by a slow dissociation phase. Equilibrium was not reached after 0.5 hour of exposure, and thus, an equilibrium affinity assay was not feasible. In addition, the interaction appeared to be complex, as indicated by unusual spacing of the concentration curves (Fig. 4), and particularly, the two highest concentrations (6.3 and 3.2 μM) had very similar levels of binding. This suggests some kind of concentration-dependent interaction in binding beyond that normally expected for a bimolecular interaction. In addition, the higher-concentration data (3.2, 1.6, and 0.8 μM) showed a significant degree of biphasic behavior consisting of a slow dissociation constant (Kd) (∼0.00005 s−1 ± 0.000001 s−1) that accounted for approximately 75% of the binding and a second, faster rate constant (∼0.001 s−1 ± 0.0002 s−1) that accounted for the remaining 25% of binding. The same behavior was not apparent in the two lower concentrations, but this may be due to a lower signal-to-noise ratio inherent in the lower-concentration curves.
FIG. 4.
SPR analysis of binding of PM1665 protein to fibronectin. Fibronectin was immobilized on a CM5 chip, increasing concentrations of the PM1665 protein (0.2 μM, 0.4 μM, 0.8 μM, 1.6 μM, 3.2 μM, and 6.3 μM) were injected at a flow rate of 5 μl min−1 for 30 min, and the binding responses were recorded as response units (RU). The data are representative of three separate experiments.
To get an approximate idea of the strength of binding, a single, average dissociation rate constant was determined for the three higher concentrations (0.00011 [±0.00002] s−1) and was used to fit their separate association phases, resulting in an average value for association (ka) of 700 M−1 s−1 and a KD of 150 nM (±70 nM).
Determination of the binding site for rGST-PM1665 in fibronectin.
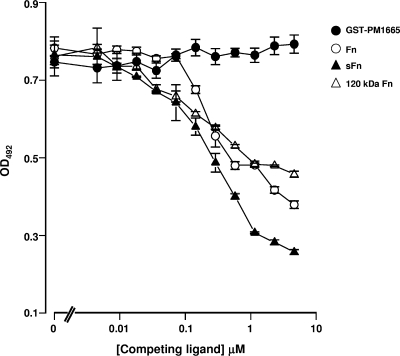
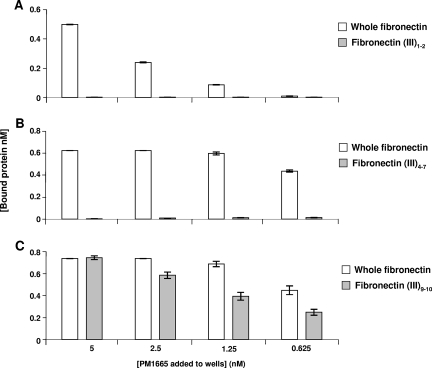
A competition ELISA was used to determine if the binding of rGST-PM1665 to immobilized fibronectin could be inhibited by the addition of different fragments of fibronectin or by the addition of superfibronectin, a form that more closely resembles the structure of the extracellular-matrix form of fibronectin. Soluble fibronectin, the 120-kDa fragment of fibronectin, and superfibronectin were all able to inhibit the binding of rGST-PM1665 to immobilized fibronectin (Fig. 5), with all three of these competitors exhibiting similar levels of inhibition (F test; P > 0.05). The 120-kDa α-chymotryptic fragment of fibronectin is composed of FnIII repeats numbered 2 to 11 (Fig. 6). To determine where the PM1665 protein bound within the 120-kDa fragment, the three biologically active domains within this fragment were tested for binding to the PM1665 protein. The chosen domains were the FnIII1-2 module, which is essential for fibronectin matrix formation; the FnIII4-7 module, which has been shown to bind to DNA; and the FnIII8-10 and FnIII9-10 modules, which both contain the RGD integrin-binding sequence. FnIII11 was also tested. Therefore, the ability of rGST-PM1665 to bind to all of the FnIII modules within the central 120-kDa fragment, with the exception of FnIII3, was tested. The recombinant FnIII fragments were subjected to SDS-PAGE, blotted onto nitrocellulose membranes, and then incubated with biotinylated rGST-PM1665. rGST-PM1665 bound specifically to whole fibronectin and to the fibronectin fragments GST-FnIII8-10 and FnIII9-10 (Fig. 7). These results were confirmed using direct binding ELISAs by immobilizing FnIII1-2, FnIII4-7, or FnIII9-10 fragments onto microtiter plate wells and adding either rGST-PM1665 or biotinylated rGST-PM1665 (Fig. 8). To test the hypothesis that rGST-PM1665 could be interacting with the cell-binding RGD sequence contained within the 120-kDa fragment of fibronectin, a range of concentrations (up to 2 M) of the peptide RGD or the peptide RGDSPASSKP were used in the inhibition ELISA but had no effect on the capacity of rGST-PM1665 to bind to immobilized fibronectin (data not shown).
FIG. 5.
Inhibition ELISA to determine the capacities of fibronectin and fragments of fibronectin to block the binding of rGST-PM1665 to immobilized human serum fibronectin. The competing ligands (soluble serum fibronectin, superfibronectin [sFn], and the 120-kDa α-chymotryptic fragment of fibronectin) were added at concentrations ranging from 1 nM to 4.6 μM. The results are presented as the mean ± standard error of the mean of quadruplicate wells and are representative of at least three experiments. The values for rGST-PM1665 refer to the binding of the protein to fibronectin-coated wells with no competing ligands. OD492, optical density at 492 nm.
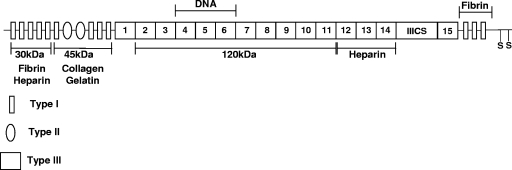
FIG. 6.
Schematic representation of the modular structure of the fibronectin molecule. Fibronectin consists of 12 type I modules (rectangles), 2 type II modules (ovals), and 15 to 17 type III modules (squares). The proteolytic 30-kDa, 45-kDa, and 120-kDa fragments used in this study are indicated.
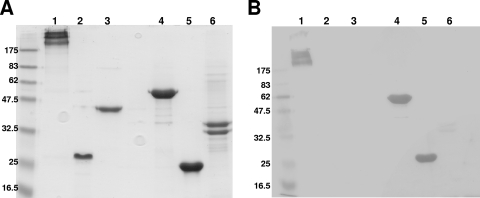
FIG. 7.
Ligand blotting showing binding of biotinylated rGST-PM1665 to fibronectin or fragments of fibronectin. (A) SDS-PAGE gel stained with colloidal blue. Lanes: 1, whole fibronectin; 2, Fn III1-2; 3, Fn III4-7; 4, GST-Fn III8-10;, 5, Fn III9-10; 6, GST-Fn III11. (B) Ligand blot of a gel identical to that shown in panel A. The nitrocellulose membrane was incubated with biotinylated rGST-PM1665, followed by incubation with streptavidin-HRP. Recombinant GST-PM1665 bound preferentially to the GST-FnIII8-10 and FnIII9-10 fragments.
FIG. 8.
Direct binding ELISAs to determine binding of rGST-PM1665 to fibronectin or recombinant fibronectin fragments. Wells were coated with whole fibronectin (control) or FnIII fragments 1 and 2, 4 to 7, or 9 and 10. Biotinylated rGST-PM1665 (A and C) or rGST-PM1665 (B) was added to these coated wells, and the amounts of bound rGST-PM1665 were detected with streptavidin-HRP (A and C) or with anti-GST antibody, followed by an HRP-conjugated anti-goat antibody (B). The error bars indicate standard errors of the mean.
Determination of the binding site for fibronectin in rGST-PM1665.
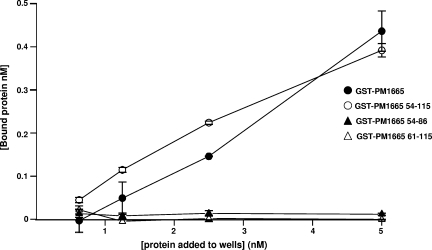
To determine the binding site for fibronectin in the PM1665 protein, a number of fragments of the PM1665 protein were generated either as GST fusions or as synthetic peptides. Sequence comparisons indicated that the homologous proteins in other members of the Pasteurellaceae showed C-terminal primary structures similar to those of the PM1665 protein. There are two weakly predicted HhH domains that are conserved in homologous proteins, both in the Pasteurellaceae and in other bacterial species, as well as a conserved 6-amino-acid sequence (VNINTA). Three synthetic peptides were synthesized commercially, corresponding to each of the two HhH domains and to the conserved VNINTA motif. These peptides were prepared with an N-terminal biotinylated alanine residue to allow easy detection via ELISA. A further three fragments of the PM1665 protein were expressed as GST fusion proteins. The first fragment contained both of the HhH motifs (residues 61 to 115) and the second fragment consisted of the conserved VNINTAS motif and the first HhH motif (residues 54 to 86), while the third fragment consisted of the entire C-terminal half of the protein, i.e., the conserved VNINTAS motif plus both HhH motifs (residues 54 to 115). A direct binding ELISA was used to test the binding capacities of synthetic peptides consisting of the sequence of each of the HhH motifs. Neither of the individual HhH motifs bound to fibronectin (data not shown), nor did a combination of the two motifs expressed as a GST fusion protein (Fig. 9). However, the GST fusion protein consisting of the conserved VNINTA motif plus both HhH motifs bound to fibronectin to a extent similar to that of mature rGST-PM1665 protein (Fig. 9). The VNINTA motif plus the first HhH motif did not bind to fibronectin (Fig. 9), suggesting that all three regions, the VNINTA motif and both HhH motifs, are necessary for binding to fibronectin.
FIG. 9.
Binding of fragments of PM1665 protein (expressed as GST fusion proteins) to fibronectin measured by direct binding ELISA. Increasing concentrations of rGST-PM1665 (open circles), the C-terminal 64 residues of PM1665 protein (closed circles), the two HhH domains of PM1665 protein (closed triangles), or a combination of the conserved VNINTA motif and the first HhH domain (open triangles) were added to wells coated with fibronectin. Optical densities at 492 nm were converted to estimates of the concentration of bound protein by reference to a standard curve for each protein.
Analysis of the expression of the PM1665 protein in P. multocida.
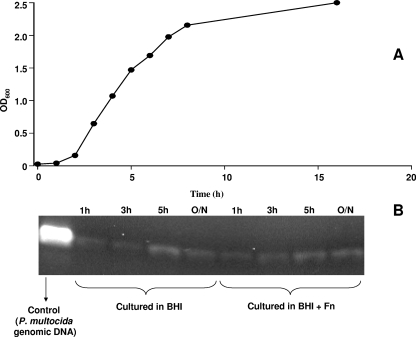
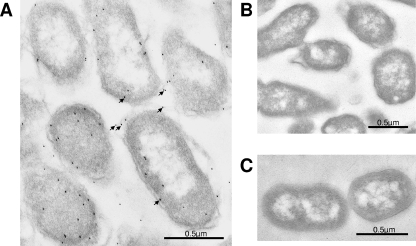
To determine if the gene for the PM1665 protein was transcribed in P. multocida, total cellular RNA was extracted from bacteria at different stages of growth in either BHI broth or BHI broth containing 500 μg/ml of fibronectin and was subjected to RT-PCR analysis. The RNA was reverse transcribed, and the resultant cDNA was used as the template in a PCR. As a control, 16S primers were used with the same cDNA samples and produced a band of about 1 kb in all samples (data not shown). The results shown in Fig. 10B demonstrate that mRNA for the PM1665 protein was transcribed at all growth phases in both the presence and absence of fibronectin. To confirm that the PM1665 protein was produced by P. multocida, a polyclonal antibody was raised against recombinant PM1665 protein and used to detect the PM1665 protein in transmission electron microscopy sections of fixed stationary-phase P. multocida cells. An immunogold-labeled anti-rabbit antibody was used to detect bound anti-PM1665 protein. Immunogold labeling was observed in the P. multocida samples but not in P. gingivitis controls (Fig. 11). While some labeling was located inside the bacteria, there was also a significant amount of labeling on the surfaces of the cells (Fig. 11A), indicating that the PM1665 protein is present on the cell surface. There was no immunogold labeling on the P. multocida cells incubated with the preimmune IgG (Fig. 11B) or on P. gingivitis cells incubated with anti-PM1665 protein (Fig. 11C). Less PM1665 protein was detected on the surfaces of P. multocida cells in the exponential phase of growth (data not shown) than on the cells in the stationary phase shown here.
FIG. 10.
RT-PCR analysis of PM1665 protein mRNA transcription in P. multocida at different time points. (A) Growth curve of P. multocida. OD600, optical density at 600 nm. (B) Cells were harvested at intervals in the growth curve, and the RNA was extracted and reverse transcribed to produce cDNA, which was then amplified by PCR with specific primers to detect transcription of the gene for the PM1665 protein. The cells were cultured in either BHI broth or BHI broth to which 0.5 mg/ml fibronectin had been added.
FIG. 11.
(A) Demonstration of the presence of PM1665 protein in P. multocida stationary-phase cells using anti-PM1665 protein antibody and immunogold labeling. Ten-nanometer colloidal-gold labeling is present on P. multocida cells (the gold probes are indicated by arrows), some of which was within the bacterial cell and some of which was located on or near the cell surface. (B) In contrast, antibody purified from preimmune serum did not react with the bacterial cells. (C) Incubation of P. gingivalis cells with anti-PM1665 protein did not result in any labeling. The procedures for these experiments are described in the text.
Binding of P. multocida to immobilized ligands.
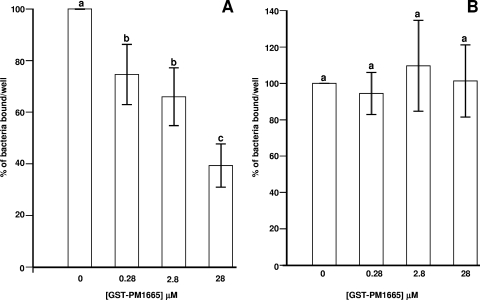
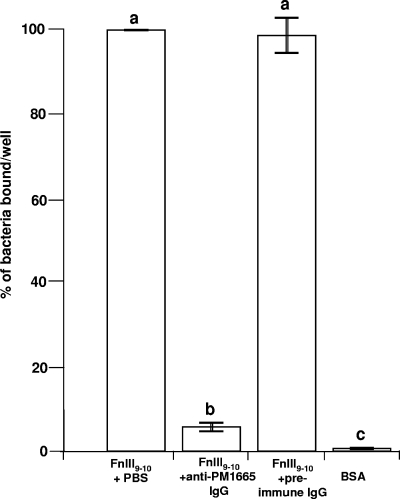
P. multocida exhibited significant binding (compared to bovine serum albumin controls) to fibronectin and collagen (data not shown), but the number of viable cells recovered from wells coated with fibronectin was always greater than the number recovered from wells coated with collagen. To test the hypothesis that the PM1665 protein mediates binding of P. multocida to fibronectin, wells coated with fibronectin, or with collagen as a negative control, were preincubated with serial dilutions of the rGST-PM1665 fusion protein. Figure 12A shows the effect of preincubation of fibronectin-coated wells with rGST-PM1665 on the binding of P. multocida to fibronectin. Recombinant GST-PM1665 inhibited the binding of the bacteria to fibronectin (one-way ANOVA; P ≤ 0.001) in a dose-dependent manner, with a 28 μM solution of rGST-PM1665 inhibiting the binding of P. multocida to fibronectin by about 60% (Fig. 12A). In parallel experiments, collagen-coated wells were preincubated with identical concentrations of rGST-PM1665 prior to the addition of P. multocida cells. rGST-PM1665 did not significantly inhibit the binding of bacteria to collagen-coated wells (one-way ANOVA; P = 1.2) (Fig. 12B). Similar experiments were done to test the capacity of anti-PM1665 antibody to inhibit the binding of P. multocida to FnIII9-10. Prior incubation of P. multocida cells with the anti-PM1665 antibody inhibited binding (one-way ANOVA; P ≤ 0.001) of the bacteria to FnIII9-10 by 94% (Fig. 13). There was no inhibition of binding of P. multocida to FNIII9-10 after incubation with the preimmune IgG (Fig. 13).
FIG. 12.
Binding of P. multocida to fibronectin (A) or collagen (B) preincubated with graded concentrations of rGST-PM1665. The binding value of 100% corresponds to the binding measured in wells preincubated with PBS. The data are the mean ± standard error of the mean of triplicate wells. Bars with different letters are statistically significantly different from each other (one-way ANOVA; P ≤ 0.001). The results are representative of three separate experiments.
FIG. 13.
Binding of P. multocida to FnIII9-10 after preincubation with anti-PM1665 antibody or preimmune antibody. The binding value of 100% corresponds to the binding of bacteria preincubated with PBS, and the other values are expressed as a percentage of that value. The data are the mean ± standard error of the mean of triplicate wells. Bars with different letters are statistically significantly different from each other (one-way ANOVA; P ≤ 0.001). The results are representative of three separate experiments.
DISCUSSION
It is clear that most, if not all, bacteria colonizing mammals possess proteins that bind to fibronectin and that many of these proteins act as adhesins (25). Analysis of the binding site in fibronectin for these proteins has revealed that most bacterial fibronectin binding proteins bind to the N terminus of this host protein. The fibronectin binding proteins of S. aureus and S. pyogenes have received the most attention. These proteins all contain the cell wall-anchoring motif LPX[T,S,A]G and a short positively charged C-terminal intracellular tail. The fibronectin binding site is located in the C-terminal part of the protein, where a varied number of 35 to 40 residue repeats are found. These fibronectin-binding repeats target the N terminus of the fibronectin molecule, binding to FnI modules 1 to 5 (26). Much less information is available for fibronectin binding proteins of gram-negative bacteria. The only gram-negative fibronectin binding protein to be studied in detail is BBK32 from Borrelia burgdorferi, the causative agent of Lyme disease. Surprisingly, this protein shares many structural and functional similarities with the fibronectin binding proteins of S. aureus and S. pyogenes (21), suggesting that this is the general mechanism of bacterial binding to fibronectin.
One family of bacteria for which only limited information on bacterial adhesion is available is the Pasteurellaceae, which includes a wide range of pathogens of humans and domesticated animals. To identify novel adhesins in members of the Pasteurellaceae, we have used a phage display screening strategy with fragmented genomic DNA. By panning libraries against human fibronectin we identified a novel gene (PM1665) encoding a putative fibronectin and type I collagen binding protein in P. multocida (19). Bioinformatics analysis has identified the presence of homologous proteins in all other sequenced members of the Pasteurellaceae. The recently discovered bovine commensal Mannheimia succiniciproducens (16) has two genes encoding protein homologues.
We cloned the PM1665 gene and expressed the gene as a GST fusion protein. Further analysis of recombinant PM1665 protein revealed that it bound only to fibronectin and not to collagen or a range of other connective tissue macromolecules. These differences in observed binding characteristics between the recombinant protein and the phage-displayed protein are likely due to differences in protein folding and conformation. Fibronectin can exist as a soluble protein in body fluids or as part of the extracellular matrix, in which case it is in an insoluble form. We found that the PM1665 protein bound to both soluble and insoluble fibronectin and to an insoluble form of fibronectin, termed superfibronectin, which mimics the form fibronectin takes within the extracellular matrix. To determine where in fibronectin the PM1665 protein was binding, we made use of commercially available proteolytic fragments of fibronectin. We were surprised to find that the PM1665 protein bound to the 120-kDa central cell binding segment of fibronectin and not to the N-terminal 30- or 45-kDa fragments, to which, as discussed above, most other fibronectin binding proteins bind. The 120-kDa region contains 10 FnIII modules (numbered 2 to 11) and is the site of three established biologically active segments. FnIII1-2 is important in fibronectin fibril formation, FnIII4-7 binds to DNA, and FnIII9-10 binds to integrins (10, 20). We have used 9 out of the 10 FnIII modules present in the 120-kDa fragment of fibronectin in binding assays and have concluded that the PM1665 protein binds to FnIII9-10, the cell-binding segment of the protein, but not to any other modules or to type III repeats in another connective tissue protein, tenascin. The FnIII9-10 repeat contains the well-known RGD sequence, which facilitates fibronectin binding to cell surface integrins, such as α5β1 (18). Addition of very high concentrations of RGD peptides failed to inhibit PM1665 protein binding to fibronectin, and the PM1665 protein failed to bind to these peptides, suggesting that this bacterial protein must interact at a site on FnIII9-10 that is removed from the RGD sequence, which forms an extended hairpin-like loop emanating from FnIII10 (15).
While a number of other bacteria have been reported to bind to fibronectin through the 120-kDa region (6, 11), the adhesins involved have not been identified, nor has the binding site in fibronectin been defined. Thus, the PM1665 protein is the first bacterial fibronectin binding protein found to bind to this segment of fibronectin. The nature of the binding site for the PM1665 protein suggests that it must have a novel mechanism of action, either as an adhesin and/or as a cell-modulating protein.
Having established that the PM1665 protein bound to a unique site in fibronectin, the obvious question was whether the protein acted as a bacterial adhesin. Initial studies found that soluble recombinant PM1665 protein could significantly inhibit the binding of P. multocida to insoluble fibronectin. We raised an antiserum to recombinant PM1665 protein in the rabbit and used the IgG fraction to examine the location of this putative adhesin and to see if the antibody could block bacterial binding. Using immunogold labeling of ultrathin sections of P. multocida, it is clear that the PM1665 protein exists on the surface of the bacterium. Furthermore, the antibody was also able to block the binding of P. multocida to fibronectin FnIII9-10, indicating a role for the PM1665 protein in the adhesion of P. multocida to fibronectin. There have been a number of studies on the adhesion of P. multocida to porcine respiratory tract cells (12) and, more recently, to components of the extracellular matrix (4), but the adhesins utilized by the bacterium have not been characterized in any detail. Dabo and colleagues (4) identified five outer membrane proteins as putative fibronectin binding proteins by ligand blotting, but none of them correspond to the PM1665 protein on the basis of apparent molecular mass.
The PM1665 protein is an unlikely fibronectin binding protein, as it has homology to the ComEA DNA binding proteins involved in competence development (14, 17) and has weakly predicted HhH domains, which have been found to be widespread in non-sequence-specific DNA binding proteins (27). DNA is a double helix with a pseudo-twofold structure, so there are problems fitting asymmetric proteins into the symmetric DNA helix. One way in which this can be solved is by domain duplication. In most proteins containing HhH motifs, these motifs are arranged in pairs with each pair generating a five-helical (HhH)2 domain with the two HhH motifs connected by an alpha helix, and this arrangement is believed to display pseudo-twofold symmetry (27). We have used a combination of truncation mutagenesis and synthetic-peptide synthesis to determine which part of the C terminus of this protein binds to fibronectin. As would be expected of a DNA binding protein, neither of the HhH motifs alone bound to fibronectin. However, a combination of both recombinant HhH motifs also failed to bind to fibronectin. It was only when the C-terminal fragment of the PM1665 protein contained the highly conserved sequence VNINTAS that the recombinant truncation mutant bound with high affinity to fibronectin. The HhH-like motif has been suggested to exist in the sterile alpha motif domain (27), which is found in protein kinases and GTPases, and has been proposed to mediate protein-protein interactions and not DNA binding (24). There are also HhH motifs in some DNA glycosylases that have acquired enzymatic activity (2). Therefore, it is possible that the PM1665 protein represents a fourth function for HhH domains, that of binding to specific extracellular matrix macromolecules. Of course, it must be borne in mind that binding requires the presence of the VNINTAS sequence. This sequence contributes significantly to the binding event, but in the absence of a full structure, it is not possible to hypothesize what the mechanism of binding of the protein is.
In conclusion, we have identified a novel fibronectin binding adhesin in P. multocida that binds with relatively high affinity to the FnIII9-10 repeats of fibronectin. Homologous genes exist in all other sequenced members of the Pasteurellaceae, suggesting that the protein is important for the survival of this family of bacteria. The finding that these proteins contain two HhH motifs potentially extends the role that this motif plays in biological systems.
Acknowledgments
B.H., S.P.N., A.N.R., and J.M.W. are grateful to the BBSRC for funding to support L.M.M. (grant BBS/B03238). B.H. and J.M.W. are also grateful to the MRC (grant G0401038) for support for G.R.
Editor: A. Camilli
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.Andersson, S., U. Larinkari, T. Vartia, B. Forsblom, M. Saarela, and M. Rautio. 1994. Fatal congenital pneumonia caused by cat-derived Pasteurella multocida. Pediatr. Infect. Dis. J. 1374-75. [DOI] [PubMed] [Google Scholar]
- 2.Bruner, S. D., D. P. Norman, and G. L. Verdine. 2000. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 403859-866. [DOI] [PubMed] [Google Scholar]
- 3.Dabo, S. M., A. W. Confer, and R. A. Quijano-Blas. 2003. Molecular and immunological characterization of Pasteurella multocida serotype A:3 OmpA: evidence of its role in Pasteurella multocida interaction with extracellular matrix molecules. Microb. Pathog. 35147-157. [DOI] [PubMed] [Google Scholar]
- 4.Dabo, S. M., A. W. Confer, and S. D. Hartson. 2005. Adherence of Pasteurella multocida to fibronectin. Vet. Microbiol. 110265-275. [DOI] [PubMed] [Google Scholar]
- 5.Doughty, S. W., C. G. Ruffolo, and B. Adler. 2000. The type 4 fimbrial subunit gene of Pasteurella multocida. Vet. Microbiol. 7279-90. [DOI] [PubMed] [Google Scholar]
- 6.Eberhard, T., R. Virkola, T. Korhonen, G. Kronvall, and M. Ullberg. 1998. Binding to human extracellular matrix by Neisseria meningitidis. Infect. Immun. 661791-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escande, F., and C. Lion. 1993. Epidemiology of human infections by Pasteurella and related groups in France. Int. J. Med. Microbiol. 279131-139. [DOI] [PubMed] [Google Scholar]
- 8.Fortin, M., and M. Jacques. 2006. Hemagglutination by Pasteurella multocida of porcine origin. J. Clin. Microbiol. 25938-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunt, M. L., B. Adler, and K. M. Townsend. 2000. The molecular biology of Pasteurella multocida. Vet. Microbiol. 723-25. [DOI] [PubMed] [Google Scholar]
- 10.Hynes, R. O. 1987. Integrins: a family of cell surface receptors. Cell 48549-554. [DOI] [PubMed] [Google Scholar]
- 11.Ito, H. O., S. Soutome, K. Nokihara, and M. Inoue. 2004. Identification and characterization of bacterial-binding property in the type III repeat domain of fibronectin. Biochem. Biophys. Res. Commun. 320347-353. [DOI] [PubMed] [Google Scholar]
- 12.Jacques, M., M. Kobisch, M. Belanger, and F. Dugal. 1993. Virulence of capsulated and noncapsulated isolates of Pasteurella multocida and their adherence to porcine respiratory tract cells and mucus. Infect. Immun. 614785-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladbury, J. E., M. A. Lemmon, M. Zhou, J. Green, M. C. Botfield, and J. Schlessinger. 1995. Measurement of the binding of tyrosal phosphopeptides to SH2 domains: A reappraisal. Proc. Natl. Acad. Sci. USA 923199-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lammers, M., H. Nahrstedt, and F. Meinhardt. 2004. The Bacillus megaterium comE locus encodes a functional DNA uptake protein. J. Basic Microbiol. 44451-458. [DOI] [PubMed] [Google Scholar]
- 15.Leahy, D. J., I. Aukhil, and H. P. Erickson. 1996. 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell 84155-164. [DOI] [PubMed] [Google Scholar]
- 16.Lee, P. C., S. Y. Lee, S. H. Hong, and H. N. Chang. 2002. Isolation and characterization of a new succinic acid-producing bacterium, Mannheimia succiniciproducens MBEL55E, from bovine rumen. Appl. Microbiol. Biotechnol. 58663-668. [DOI] [PubMed] [Google Scholar]
- 17.Kramer, N., J. Hahn, and D. Dubnau. 2007. Multiple interactions among the competence proteins of Bacillus subtilis. Mol. Microbiol. 65454-464. [DOI] [PubMed] [Google Scholar]
- 18.Main, A. L., T. S. Harvey, M. Baron, J. Boyd, and I. D. Campbell. 1992. The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions. Cell 71671-678. [DOI] [PubMed] [Google Scholar]
- 19.Mullen, L. M., S. P. Nair, J. M. Ward, A. N. Rycroft, R. J. Williams, and B. Henderson. 2007. Comparative functional genomic analysis of Pasteurellaceae genomes using phage display. Vet. Microbiol. 122123-134. [DOI] [PubMed] [Google Scholar]
- 20.Pierschbacher, M. D., and E. Ruoslahti. 1984. Variants of the cell recognition site of fibronectin that retain attachment-promoting activity. Proc. Natl. Acad. Sci. USA 815985-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raibaud, S., U. Schwarz-Linek, J. H. Kim, H. T. Jenkins, E. R. Baines, S. Gurusiddappa, M. Hook, and J. R. Potts. 2006. Borrelia burgdorferi binds fibronectin through a tandem beta-zipper, a common mechanism of fibronectin binding in staphylococci, streptococci, and spirochetes. J. Biol. Chem. 28018803-18809. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds, E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruffolo, C. G., J. M. Tennent, W. P. Michalski, and B. Adler. 1997. Identification, purification, and characterization of the type 4 fimbriae of Pasteurella multocida. Infect. Immun. 65339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz, J., C. P. Ponting, K. Hoffman, and P. Bork. 1997. SAM as a protein interaction domain involved in developmental regulation. Prot. Sci. 6249-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz-Linek, U., M. Hook, and J. R. Potts. 2004. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 52631-641. [DOI] [PubMed] [Google Scholar]
- 26.Schwarz-Linek, U., M. Hook, and J. R. Potts. 2006. Fibronectin-binding proteins of Gram-positive cocci. Microb. Infect. 82291-2298. [DOI] [PubMed] [Google Scholar]
- 27.Shao, X., and N. V. Grishin. 2000. Common fold in helix-hairpin-helix proteins. Nucleic Acids Res. 282643-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]