Abstract
Salmonella enterica serovar Typhimurium harbors five pathogenicity islands (SPI) required for infection in vertebrate hosts. Although the role of SPI1 in promoting epithelial invasion and proinflammatory cell death has been amply documented, SPI4 has only more recently been implicated in Salmonella virulence. SPI4 is a 24-kb pathogenicity island containing six open reading frames, siiA to siiF. Secretion of the 595-kDa SiiE protein requires a type I secretory system encoded by siiC, siiD, and siiF. An operon polarity suppressor (ops) sequence within the 5′ untranslated region upstream of siiA is required for optimal SPI4 expression and predicted to bind the antiterminator RfaH. SiiE concentrations are decreased in a SPI1 mutant strain, suggesting that SPI1 and SPI4 may have common regulatory inputs. SPI1 gene expression is positively regulated by the transcriptional activators HilA, HilC, and HilD, encoded within SPI1, and negatively regulated by the regulators HilE and PhoP. Here, we show that mutations in hilA, hilC, or hilD similarly reduce expression of siiE, and mutations in hilE or phoP enhance siiE expression. Individual overexpression of HilA, HilC, or HilD in the absence of SPI1 cannot activate siiE expression, suggesting that these transcriptional regulators act in concert or in combination with additional SPI1-encoded regulatory loci to activate SPI4. HilA is no longer required for siiE expression in an hns mutant strain, suggesting that HilA promotes SPI4 expression by antagonizing the global transcriptional silencer H-NS. Coordinate regulation suggests that SPI1 and SPI4 play complementary roles in the interaction of S. enterica serovar Typhimurium with the host intestinal mucosa.
Salmonella spp. are highly successful pathogens that colonize and cause productive infection in a wide range of animal hosts. Successful infection by Salmonella requires invasion of host intestinal epithelial cells and survival in phagocytic cells (32, 33). Salmonella virulence is highly dependent on horizontally acquired DNA contained within Salmonella pathogenicity islands (SPI); Salmonella enterica serovar Typhimurium carries five pathogenicity islands, designated SPI1 to -5 (58). Coordinated regulation of pathogenicity island genes involves loci situated both within the islands as well as at physically unlinked sites on the chromosome (4, 25, 40).
Both SPI1 and SPI2 encode type III secretion systems (T3SS) that translocate effector molecules into host cells (36). The ability to invade host epithelial cells requires SPI1, and the ability to survive in host phagocytes requires SPI2 (18, 33, 36). SPI3 encodes a specialized magnesium transporter required for virulence and survival in host phagocytes and an adhesin required for intestinal persistence (14, 22, 61), while SPI5 encodes effector proteins that are secreted by the SPI1 and/or SPI2 T3SS (46, 90).
SPI4 was first described as a pathogenicity island by Wong et al. in 1998 and shares characteristics of horizontally acquired gene clusters with other SPI (89). The G+C content of SPI4 is 37 to 44%, compared to 52% for the rest of the S. enterica serovar Typhimurium genome (89). SPI4 was initially thought to carry 18 open reading frames (ORFs), but more-recent annotation shows only 6 ORFs, designated STM4257 to STM4262 (60), or siiA to siiF, for salmonella intestinal infection (Fig. 1A) (62). SPI4 was initially thought to be required for Salmonella survival in murine macrophages (29), but subsequent investigations failed to demonstrate a macrophage survival defect (62, 89). Several recent studies have demonstrated a role for SiiE during the intestinal phase of infection. Morgan et al. demonstrated that mutations in siiD, siiE, or siiF significantly impair the ability of S. enterica serovar Typhimurium to colonize the calf intestine (62, 63). A screen for novel S. enterica serovar Typhimurium virulence factors showed that siiE is required to cause toxic infection in the nematode Caenorhabditis elegans (81). Gerlach et al. showed that SiiE mediates adherence to MDCK cells in vitro that can be blocked by pretreatment with antibody to SiiE (35). These authors also demonstrated that a SPI4 mutant is less able to induce intestinal inflammation in a murine colitis model (35). Finally, siiE is required for long-term survival (47) and orogastric infection of S. enterica serovar Typhimurium during murine infection (45). Collectively, these observations demonstrate a role for SPI4 during the intestinal phase of infection.
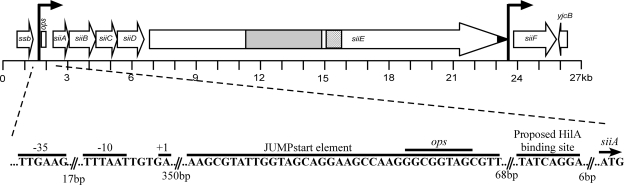
FIG. 1.
Map of the SPI4 region in Salmonella enterica serovar Typhimurium. Annotation is based on sequence data from strain LT2. Arrows denote ORFs, and hatch marks denote 1 kb. The siiA, siiB, siiC, and siiD ORFs are overlapping. Promoters within the operon are depicted as bent arrows. Other features include a region containing 90-amino-acid repeats (gray shading), predicted signal sequence (black shading), and an operon polarity suppressor (ops). The hatched box denotes a region subcloned to create an anti-SiiE polyclonal antiserum. The flanking upstream and downstream genes ssb and yjcB are also shown. The inset details the region upstream of siiA. The transcriptional start site (+1) was experimentally determined. The proposed HilA-binding site was bioinformatically defined, and a fragment containing this region was demonstrated to bind HilA previously (82).
The genetic and functional organization of SPI4 suggests that SiiE is secreted from the cell by a type I secretion system (T1SS) (35, 62). Homology to known T1SS components predicts that SiiC is a TolC homolog that spans the outer membrane, SiiD is a periplasmic adaptor protein, and SiiF forms an inner membrane ATPase (16). SiiA lacks homology to other known proteins, and SiiB is 40% similar to several methyl-accepting chemotaxis and hypothetical proteins. A secretory apparatus consisting of SiiC, SiiD, and SiiF is proposed to secrete SiiE into the culture supernatant. SiiE becomes associated with the outer envelope and can function as an adhesin when in contact with epithelial cells (35, 62).
SiiE is a large protein of approximately 595 kDa which contains more than 10 90-amino-acid repeats (Fig. 1). SiiE exhibits 40 to 60% homology to large proteins of Ralstonia solanacearum, Burkholderia spp., Enterococcus faecalis, Synechocystis spp., Escherichia coli O157:H7, Aeromonas salmonicida, and Vibrio spp., some of which are believed to function as repeats-in-toxin toxins, hemolysins, or adhesins (31, 48, 53, 86). SiiE has retained features common to type I secreted substrates, including repetitive domains and a C-terminal secretion signal (Fig. 1) (88).
Regulation of SPI4 has been recently demonstrated to overlap with that of SPI1 (34). Mutations in hilA and sirA cause a significant reduction in SiiE production and SPI4-mediated adherence to epithelial cells (34). SPI1 expression is known to respond to a complex cascade of transcriptional regulators and a variety of environmental parameters relevant to the intestine, including osmolarity, oxygen tension, pH, presence of bile, Mg2+ concentration, and presence of short-chain fatty acids (4). HilA is a ToxR/OmpR family transcriptional regulator encoded within SPI1 (8) that is required for invasion and destruction of M cells in murine-ligated ileal loops (72). Expression of hilA itself is activated by three AraC family regulators, HilC, HilD, and RtsA (25, 78). HilC and HilD, like HilA, are encoded by SPI1, whereas RtsA is encoded within a separate 15-kb island (24). Additional two-component regulatory systems are involved in the regulation of SPI1, including BarA/SirA, FimY/FimZ, PhoP/PhoQ, and PhoB/PhoR (12, 28, 56). Many of the effects of the two-component regulatory systems are modulated by HilE, a negative regulator found to interact with HilD in a bacterial two-hybrid experiment (11).
Ahmer et al. were the first to demonstrate that SirA regulates the expression of the SPI4 siiE gene in a HilA-dependent manner (1). HilA has been shown to bind within the siiE coding region (20) as well as a site upstream of siiA (83), but the mechanism by which HilA regulates SPI4 gene expression is not known. An additional level of SPI4 regulation may be provided by an operon polarity suppressor (ops) motif upstream of siiA (63, 64). Such motifs can be bound by the RfaH antiterminator protein to facilitate the elongation of long transcripts (7). The goal of the present study is to elucidate the transcriptional organization of regulatory circuits required for SPI4 expression and determine the mechanism by which HilA promotes SPI4 expression.
MATERIALS AND METHODS
Strains and growth conditions.
All strains were constructed or obtained as detailed in Table 1. The phoP::Tn10dCm and sirA::Cm alleles were transduced into SL1344 by using phage P22 HT105/1 int-201 (79), and the phoQ24 allele was transduced into SL1344 as previously described (65). In-frame deletion mutations of hilA, hilC, hilD, hilE, invF, siiA, siiB, siiC, siiD, siiE, siiF, sprB, and rfaH were constructed using a λ-red-based method (19). Primers used are listed in Table 2. Unless otherwise noted, strains were grown in LB (Luria-Bertani) medium.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| SL1344 | Wild type | 80 |
| KM001 | SL1344 ΔsiiA | This study |
| KM002 | SL1344 ΔsiiB | This study |
| KM003 | SL1344 ΔsiiC | This study |
| KM004 | SL1344 ΔsiiD | This study |
| KM005 | SL1344 ΔsiiE | This study |
| KM006 | SL1344 ΔsiiF | This study |
| KM007 | SL1344 siiE-3D (deletion of 1 kb from the 3′ end of siiE) | This study |
| KM008 | SL1344 phoP::Tn10dCm | 13 |
| KM009 | SL1344 phoQ24 | 13 |
| KM010 | SL1344 sirA::Cm (BA746) | 1 |
| KM011 | SL1344 ΔhilA | This study |
| KM012 | SL1344 ΔhilC | This study |
| KM013 | SL1344 ΔhilD | This study |
| KM014 | SL1344 ΔhilE | This study |
| KM015 | SL1344 ΔinvF | This study |
| KM017 | SL1344 rfaH:Km | This study |
| KM018 | SL1344 opsM | This study |
| KM019 | SL1344 ΔSPI1 | This study |
| KM020 | SL1344 ΔinvA | This study |
| KM021 | SL1344 pBAD18-Cm | This study |
| KM022 | KM019 pBAD18-Cm | This study |
| KM023 | KM011 pBAD18-Cm | This study |
| KM024 | KM012 pBAD18-Cm | This study |
| KM025 | KM013 pBAD18-Cm | This study |
| KM026 | SL1344 pKM009 | This study |
| KM027 | SL1344 pKM010 | This study |
| KM028 | SL1344 pKM011 | This study |
| KM029 | SL1344 pKM012 | This study |
| KM030 | KM019 pKM009 | This study |
| KM031 | KM019 pKM010 | This study |
| KM032 | KM019 pKM011 | This study |
| KM033 | KM019 pKM012 | This study |
| KM034 | KM011 pKM009 | This study |
| KM035 | KM012 pKM010 | This study |
| KM036 | KM013 pKM011 | This study |
| s3333 | S. enterica serovar Typhi | 15 |
| SL2154 | S. enterica serovar Enteritidis | Clinical isolate |
| SL2752 | S. enterica serovar Choleraesuis | 41 |
| SL3203 | S. enterica serovar Dublin | 51 |
| SL2323 | S. enterica serovar Arizona | 52 |
| s3041 | S. bongori | 15 |
| SL2371 | E. coli BL21(DE3)/pLysS | Novagen |
| WN153 | 14028s rpoSlowa | 62 |
| WN341 | 14028s rpoSlow Δhns | 62 |
| KM037 | 14028s rpoSlowhilA::Km | This study |
| KM038 | 14028s rpoSlow Δhns hilA::Km | This study |
| KM292 | SL1344 pKM013 | This study |
| Plasmids | ||
| pET16B | Novagen | |
| pKM012 | pET16B-spi4K | This study |
| pTP223 | 18 | |
| pCP20 | 18 | |
| pKD46 | 18 | |
| pKD3 | 18 | |
| pKD4 | 18 | |
| pBAD18-Cm | 36 | |
| pKM009 | pBAD18-Cm-hilA | This study |
| pKM010 | pBAD18-Cm-hilC | This study |
| pKM011 | pBAD18-Cm-hilD | This study |
| pKM013 | pCR2.1-PrsiiF | This study |
14028s rpoSlow, a derivative of S. enterica serovar Typhimurium ATCC 14028s containing an in-frame 15-nucleotide deletion in the rpoS gene.
TABLE 2.
Primers
| Designation | Sequence | Strain made/gene detecteda |
|---|---|---|
| KMp34 | GAAATGAATAGAAGACAAAGCGATCATCTCATGATGATAAGTGTAGGCTGGAGCTGCTTC | KM004 |
| KMp35 | TCAAGGTGTATCTAATCGTTTAGTATTAACTGGTTCTGAACATATGAATATCCTCCTTAG | KM004 |
| KMp40 | GGGTTTACTTATGGATAAAAAACTAGAACCTTATTATTTAGTGTAGGCTGGAGCTGCTTC | KM006 |
| KMp41 | TTACATTAATAATTTATCCGGAGAACAATCACGGATTATTCATATGAATATCCTCCTTAG | KM006 |
| KMp50 | TCGCCGATGGTCAGAAAAC | siiA |
| KMp51 | TCGTAGAGAGTCAGAGAAAGAGATGTC | siiA |
| KMp52 | CCGCATTTGTATCTTCTATCTATGGTG | siiB |
| KMp53 | CTTCGCTGTTTATTATTGGCTCTG | siiB |
| KMp54 | GGCAGCACTTTATCCTACCCTG | siiC |
| KMp55 | TCACTCCAAAATCCGTTATTCG | siiC |
| KMp56 | CTTGATTCTGAGATTAGCGGATTAC | siiD |
| KMp57 | TTCCTTTTTTTACCAGTGGGG | siiD |
| KMp58 | CAAGTATCAAAGGCAATAACCTCG | siiE (5′) |
| KMp59 | CCTGAGCAGAATCAACCAATGAG | siiE (5′) |
| KMp60 | AACGCTGACTCTGGGGATG | siiE (3′) |
| KMp61 | TGCTTTCTGGTTAGTTATCGGC | siiE (3′) |
| KMp62 | TCTTTTTGTCGTTAGGGCTTTC | siiF |
| KMp63 | CCACTCACACACACCATCCAC | siiF |
| KMp64 | CTTGATGAAAGGCGTGGAC | siiDE IGR |
| KMp65 | GGCATTACCCAAAGAAGATAAATC | siiDE IGR |
| KMp66 | CTGTAGATAATCAAGAAGAACACGC | siiEF IGR |
| KMp67 | AGATAATGCCGTTTCCGC | siiEF IGR |
| KMp74 | GCCTTTTCAGAACCAGTTAATACTAAACGATTAGATACACCTTGAGAGTGAATATAATATTGTGTAGGCTGGAGCTGCTTC | KM005 |
| KMp75 | AAGGTTCTAGTTTTTTATCCATAAGTAAACCCCCTCACCCAAAGGTGAGGGGCCTCCGTTACATATGAATATCCTCCTTAG | KM005 |
| KMp77 | AAACAAGGAGGAAAAAAATGAAGATTAAGATGTTTTTTCTGTGTAGGCTGGAGCTGCTTC | KM003 |
| KMp78 | AGATGATCGCTTTGTCTTCTATTCATTTCATTACATTTAACATATGAATATCCTCCTTAG | KM003 |
| KMp294 | ACAAAAACATTTTATTCACAATGTAATATCAGGAGACAACGTGTAGGCTGGAGCTGCTTC | KM001 |
| KMp295 | CAAACAAATAGCGGTAATGATTTATATATTTCACTCTGACCATATGAATATCCTCCTTAG | KM001 |
| KMp112 | CAATCCTGGTATTTACTGTACTGCAAACGCGGGCAACTTCGTGTAGGCTGGAGCTGCTTC | KM017 |
| KMp113 | AATCTTGCGAAAACCGGTGTTTTTTACGCTCTGCTTCACTCATATGAATATCCTCCTTAG | KM017 |
| KMp124 | AACTGAAATCTATAAAGCGTATTGGTAGCAGGAAGCCAAGTTAAGACCCACTTTCACATT | KM018 |
| KMp125 | TAGAAAGGATAAAGGATGCTCAACTTATTCAGAAAGTGAACTAAGCACTTGTCTCCTG | KM018 |
| KMp126 | GATGTAAGAAAACTGAAATCTATAAAGCGTATTGGTAGCAGGAAGCCAAGCCGGGATGCGTTCACTTTC | KM018 |
| KMp127 | TTTTGTGGATTAGAAAGGATAAAGGATGCTCAACTTATTCAGAAAGTGAACGCATCCCGGCTTGGCTTCC | KM018 |
| KMp149 | TATTATAACTTTTCACCCTGTAAGAGAATACACTATTATCGTGTAGGCTGGAGCTGCTTC | KM011 |
| KMp150 | ATGATAAAAAAATAATGCATATCTCCTCTCTCAGATTTTACATATGAATATCCTCCTTAG | KM011 |
| KMp153 | TATAACGATTTTGAGTTCCTTATAGCACACAGGATAAAATGTGTAGGCTGGAGCTGCTTC | KM012 |
| KMp154 | CGCAAACAGATAGTAACGTTTAAAATAATTTCACAAATCACATATGAATATCCTCCTTAG | KM012 |
| KMp157 | CCAGTAAGGAACATTAAAATAACATCAACAAAGGGATAATGTGTAGGCTGGAGCTGCTTC | KM013 |
| KMp158 | ATAAAAATCTTTACTTAAGTGACAGATACAAAAAATGTTACATATGAATATCCTCCTTAG | KM013 |
| KMp161 | ATATAACATAGCAAAGGCTATATTCGATGATTAATTAACCACATTGTTGCGAGGGATACTGTGTAGGCTGGAGCTGCTTC | KM019 |
| KMp162 | AATGCAAAATATGGTCTTAATTATATCATGATGAGTTCAGCCAACGGTGATATGGCCTTACATATGAATATCCTCCTTAG | KM019 |
| KMp165 | GATGGGTTTTCCAGCAGGTATTTC | gyrB |
| KMp166 | AGGTCTGATTGCGGTGGTTTC | gyrB |
| KMp182 | TCCAAATGTTGCATAGATCTTTTCCTTAATTAAGCCCTTAGTGTAGGCTGGAGCTGCTTC | KM020 |
| KMp183 | TGAAAAGCTGTCTTAATTTAATATTAACAGGATACCTATACATATGAATATCCTCCTTAG | KM020 |
| KMp186 | AGCATGGTTTATACAGACGTGTTCCGCGCAAAAGCTGCATGTGTAGGCTGGAGCTGCTTC | KM015 |
| KMp187 | GCACATGCCAGCACTCTGGCCAAAAGAATATGTGTCTTCACATATGAATATCCTCCTTAG | KM015 |
| KMp192 | ATTGTCGGTATTTAATCTGGTATACAGAGACACCAACGAAGTGTAGGCTGGAGCTGCTTC | KM014 |
| KMp193 | GCATCGCCCACTGCGAGTCCGCAAGCTTGTTTTGTCCTCACATATGAATATCCTCCTTAG | KM014 |
| KMp197 | GCTCTAGACTCCTCTCTCAGATTTTACC | pKM009 |
| KMp229 | CGAGCTCGCCCTGTAAGAGAATACACTATTATCATGCC | pKM009 |
| KMp198 | GCTCTAGATCAATGGTTCATTGTACGCATAAAG | pKM010 |
| KMp230 | CGAGCTCGGCACACAGGATAAAATATGG | pKM010 |
| KMp201 | GCTCTAGACTGATAGAGCGTGTTAATGCG | pKM11 |
| KMp228 | CGAGCTCGGGATACCAGTAAGGAACATT | pKM011 |
| KMp215 | GTAAAGCTATTATCACGACTATTAATAAAAAGGTGTCAGAGTGTAGGCTGGAGCTGCTTC | KM002 |
| KMp216 | GCTTTGTGTAATAAAAGCAGTCGTCAGAAAAAACATCTTACATATGAATATCCTCCTTAG | KM002 |
| KMp267 | CCATGTTGTCTCCTGATATTA | PrsiiA |
| KMp107 | CACGGATTACTTTCGTCT | PrsiiA |
| WNp210 | TAGGATCCAGCGAGCTAACGAGAC | PrsiiA |
| WNp213 | GTCTCGTTAGCTCGCTGGATCCTA-inverted T | PrsiiA |
| WNp291 | CATCGCCTCAAGCCTCTGGAATTGACGCAGACACATTGGGGTGTAGGCTGGAGCTGCTTC | KM016 |
| WNp292 | GAATGACCTCTTCCATCTCAGCGATCAGCGCGTCCGCTTTCATATGAATATCCTCCTTAG | KM016 |
| SL483 | CATATGACTAACGACGCCCATCCGCAG | pKM012 |
| SL486 | GGATCCGTCCAGYYGCTGCGTCGTTTTG | pKM012 |
| KMp324 | GCCGATAACTAACCAGAAAG | pKM013 |
| KMp325 | TGTAGAGAAAGCCCTAACGA | pKM013 |
| KMp327 | [FAM]TTAAATTTATATCATCCTCTTTGATGTCAA | PrsiiF |
IGR, intergenic region.
Strains containing an hns mutation were constructed in a derivative of S. enterica serovar Typhimurium ATCC 14028s containing an in-frame 15-nucleotide deletion in the rpoS gene. It has previously been demonstrated that an rpoS mutation does not affect SPI4 expression (34).
Nonquantitative reverse transcription-PCR (RT-PCR) analysis of the SPI4 region.
For RNA preparation, overnight cultures of S. enterica serovar Typhimurium SL1344 were diluted 1:200 and grown with agitation in LB to an optical density at 600 nm (OD600) of 2.0. RNA was extracted from cells treated with RNAprotect bacterial reagent by using an RNeasy mini kit according to the manufacturer's instructions (Qiagen, Valencia, CA). Reverse transcription reactions were performed using Transcriptor reverse transcriptase (Roche, Germany). PCR amplification of the cDNA template was carried out using Taq polymerase (Promega, Madison, WI) according to the manufacturer's protocol. PCR products were visualized by 1% agarose gel electrophoresis and ethidium bromide staining.
Mapping of transcriptional start sites.
RNA was prepared as described above. The 5′ rapid amplification of cDNA ends protocol was adapted from Maruyama et al. (59). Fifteen micrograms of RNA was added to 20 pmol KMp267 for PrsiiA, and the reaction mixture was brought to a total volume of 30 μl. The reaction mixture was heated at 65°C for 10 min and then immediately put on ice. The RT reaction was carried out with Superscript II reverse transcriptase per the manufacturer's instructions (Invitrogen). The cDNA reaction mixture was purified using a PCR cleanup kit (Qiagen, Valencia, CA) and eluted in 50 μl of water. The WNp213 oligonucleotide, which is phosphorylated on the 5′ end and has an inverted T modification on the 3′ end, was ligated to the end of the cDNA as described in Maruyama et al. (59). This reaction mixture was purified and cDNA amplified using internal primers WNp210 and KMp107 for PrsiiA, and the product was visualized by 1.5% agarose gel electrophoresis and ethidium bromide staining. DNA sequencing was subsequently performed (Agencourt, Beverly, MA).
For PrsiiF, a fluorescence-based primer extension protocol was employed as previously described (54). KMp327 is a 5′-6-carboxyfluorescein-labeled primer complementary to the coding region of siiF. RNA was isolated from KM292, which contains a plasmid that expresses the 3′ region of siiE, the siiEF intergenic region, and the 5′ region of siiF. Twenty micrograms of RNA was added to 20 pmol KMp327. The RT reaction was performed as described above. RNA was destroyed with RNase A and cDNA precipitated with ethanol. The cDNA product was dried and resuspended in 20 μl water. Electrophoresis was performed using an ABI3100 BioAnalyzer (Applied Biosystems, Foster City, CA) in the Comparative Genomics Center at the University of Washington. Samples were run with 0.5 μl of a GeneScan 400HD size standard, 15.5 μl formamide, and 1 μl of the sample. DNA fragments were sized using GeneMapper v3.5 analysis software (Applied Biosystems).
Quantitative RT-PCR (qRT-PCR) measurement of mRNA.
Overnight cultures were diluted 1:200 and then grown with agitation in LB to an OD600 of 2.0, except for the experiment whose results are shown in Fig. 8, where cultures were grown to an OD600 of 0.8. As indicated, 0.1% arabinose was added to strains containing the pBAD18-Cm plasmid or derivatives. RNA was isolated as described above, and one-step real-time RT-PCR analysis was used to measure mRNA levels for selected genes by using a Rotor-Gene RG3000 thermal cycler (Corbett Life Science, Sydney, Australia). Each 20-μl reaction mixture contained 40 ng RNA, 10 nM of each primer, 2 μl reverse transcriptase mix, and 10 μl Sybr green RT-PCR master mix from a QuantiTect Sybr green RT-PCR kit (Qiagen, Valencia, CA). All reactions were performed in duplicate. A separate sample was included without the reverse transcriptase mix to assay for the presence of contaminating DNA. The gyrB transcript was used to control for RNA content. Gene-specific standard curves were used to determine amplification efficiency, and the relative abundances of specific mRNAs were determined according to the QuantiTect Sybr green RT-PCR handbook (Qiagen, Valencia, CA).
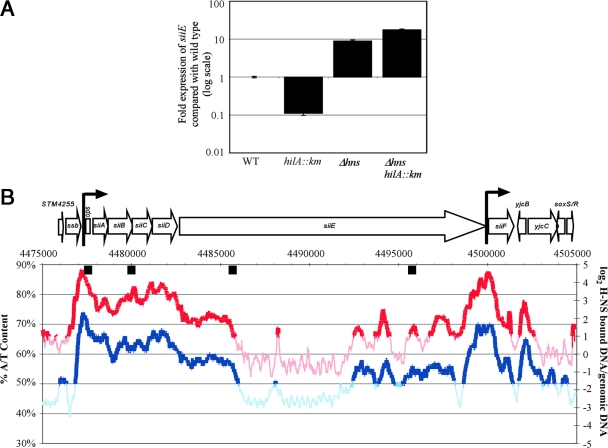
FIG. 8.
H-NS silences SPI4 expression and is antagonized by HilA. (A) qRT-PCR analysis was used to measure siiE transcription levels in wild-type (WT) or isogenic hns, hilA::Km, or hns hilA::Km mutant strains. Expression of siiE is increased in both hns and hns hilA::Km mutant strains. (B) H-NS binding data from reference 67 are plotted against percent AT content. Regions showing H-NS enrichment are shown in red, while regions not enriched in H-NS are pink. Regions with AT content greater than 50% are shown in dark blue, while regions with less than 50% AT are in light blue. Proposed HilA-binding sites are indicated as black boxes. The proposed HilA-binding sites in siiB and the distal region of siiE were identified by bioinformatic analysis.
SiiE antibody production and purification.
Rabbit polyclonal antibodies were raised to a His-tagged internal region of the SiiE protein (Fig. 1). An 820-bp fragment of siiE was amplified by PCR using Pfu polymerase (Stratagene, La Jolla, CA) and primers SL483 and SL486. This fragment was cloned in-frame into pET16B, which encodes an N-terminal 10-histidine tag, and used to transform E. coli BL21(DE3)/pLysS. The His-SiiE protein was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) and purified from crude lysate by using nickel affinity chromatography (Ni-NTA; Qiagen, Valencia, CA) according to the manufacturer's protocol. Fractions containing His-SiiE were pooled and dialyzed in 20 mM Tris-HCl, pH 7.6, 0.1 mM EDTA, and 5 mM dithiothreitol before concentration and storage at −20°C in 50% glycerol. Production of rabbit antiserum directed against purified His-SiiE was performed by Scantibodies (Ramona, CA). The preparation was purified to remove cross-reacting antibodies by serum affinity chromatography as previously described (65).
Western blot analysis of SiiE in whole-cell lysate and supernatants.
Bacteria were grown overnight in LB broth, diluted 1:200 into fresh medium, and grown with agitation to an OD600 of 2.0. One milliliter of cells was collected by centrifugation, resuspended in 1 ml water, and lysed by boiling in 250 μl NuPAGE LDS sample buffer with a sample-reducing agent (Invitrogen). Culture supernatants were filter sterilized with a 0.45-μm polyethersulfone filter (Nalgene, Rochester, NY), added to 0.25 volume NuPAGE LDS sample buffer with the sample-reducing agent (Invitrogen), and boiled. Protein concentration was determined using the RC DC protein assay (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. For the anti-SiiE immunoblot in Fig. 2, protein was added in the following quantities to optimize the detection of protein: Salmonella bongori, 1.5 ng; S. enterica serovar Typhimurium, 0.75 ng; S. enterica serovar Enteritidis, 3 ng; S. enterica serovar Dublin, 15 ng; S. enterica serovar Choleraesuis, 15 ng; S. enterica serovar Arizona, 15 ng; and S. enterica serovar Typhi, 15 ng. For all other anti-SiiE immunoblots, 20 ng of whole-cell lysate and 15 μl supernatant were loaded into each lane of a NuPAGE Novex Tris-acetate 3 to 8% gel (Invitrogen), corresponding to c. 9.25 × 106 CFU. Gels were run according to the manufacturer's instructions by using NuPAGE buffers with antioxidant (Invitrogen). Proteins were transferred to Immobilon-P transfer membranes (Millipore, Bedford, MA) for 1.5 h at 30 V by using the Invitrogen XCell tank transfer system and NuPAGE transfer buffer (Invitrogen). Membranes were blocked in phosphate-buffered saline containing 0.5% Tween and 5% nonfat dried milk and then reacted with purified anti-SiiE antibody. The secondary antibody, an anti-rabbit antibody conjugated to peroxidase, was used at a 1:20,000 dilution (Sigma, St. Louis, MO). Bands were detected using Pierce Super Signal according to the manufacturer's protocol (Pierce, St. Louis, MO).
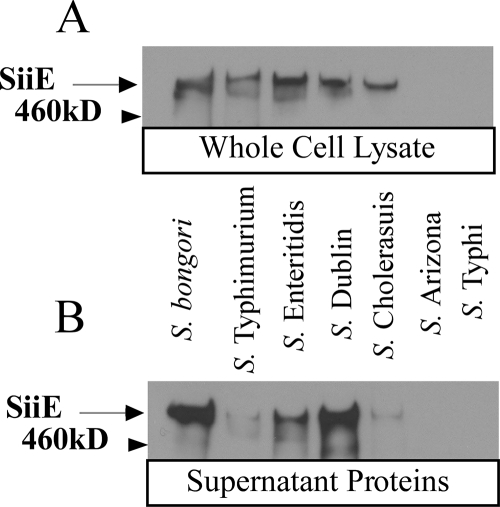
FIG. 2.
Secretion of SiiE by S. bongori and selected S. enterica serovars. (A) Anti-SiiE immunoblot of whole-cell lysates. Total protein was loaded in various amounts as described in Materials and Methods. (B) Anti-SiiE immunoblot of secreted proteins. Supernatant was obtained from the same number of cells as in panel A. SiiE is produced and secreted by S. bongori, S. enterica serovar Cholerasuis, S. enterica serovar Dublin, S. enterica serovar Enteritidis, and S. enterica serovar Typhimurium but not S. enterica serovar Typhi or S. enterica serovar Arizona.
To visualize secreted proteins, 40 ml of culture supernatants was filter sterilized and proteins were precipitated with 10% cold trichloroacetic acid. Pellets were resuspended in 0.3 ml Tris-HCl, and 5 μl of this sample was added to NuPAGE LDS sample buffer before boiling. Proteins were separated on NuPAGE Novex Tris-acetate 3 to 8% gels and then stained with a Bio-Rad silver stain plus kit according to the manufacturer's directions (Bio-Rad, Hercules, CA).
Construction of an opsM mutant.
A mutant containing a scrambled operon polarity suppressor site (designated opsM) was constructed by first replacing the ops region upstream of siiA with a tetRA element by allelic exchange using λ-red-mediated recombination (43, 44). KMp124 and KMp125, the primers used for amplication of the tetRA cassette, have 40 bp of flanking homology to the untranslated region surrounding the ops region. KMp126 and KMp127 are complementary at the 3′ ends, which carry a scrambled ops gene as previously described (6). The opsM fragment was used to replace the tetRA cassette by λ-red-mediated recombination and confirmed by sequencing.
Overexpression of hilA, hilC, and hilD.
To construct hilA, hilC, and hilD overexpression vectors, each gene was amplified with Phusion polymerase (Finnzymes, Finland) and primers described in Table 2. Primers were engineered with a SacI site on the 5′ primer and an XbaI site on the 3′ primer. PCR fragments and pBAD18-Cm were cut with SacI and XbaI, gel purified, and then ligated into pBAD18-Cm with T4 DNA ligase (NEB, Beverly, MA). Constructs were transformed into E. coli DH10B Electomax competent cells (Invitrogen). Plasmids were screened by PCR and DNA sequencing before electroporation into S. enterica serovar Typhimurium SL1344 or KM019 or the appropriate single-mutant strains.
Statistical analysis.
All experiments were performed three times on biological replicates. Each figure is representative of a single biological replicate, with the exception of Fig. 4, which represents the averages of three independent experiments. Error bars represent the standard deviations of triplicate measurements.
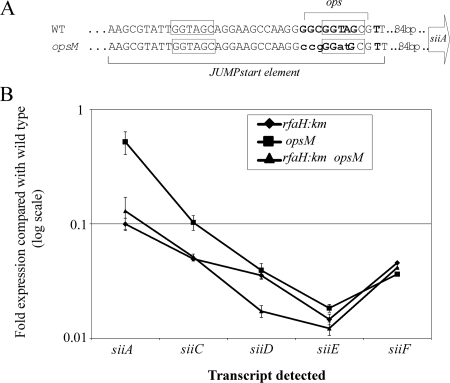
FIG. 4.
Importance of RfaH and the operon polarity suppressor (ops) sequence for SPI4 gene expression. (A) Diagram of the opsM (scrambled ops) construct. Boxes identify the direct repeat sequences characteristic of a JUMPstart element (see text), and boldface identifies residues required for RfaH binding. Lowercase letters denote residues that are mutated in the opsM construct. WT, wild type. (B) qRT-PCR measurement of siiA, siiC, siiD, and siiE transcripts was performed with rfaH::Km, opsM, and rfaH::Km opsM mutant strains. All values are normalized to gyrB transcript levels. Data are expressed as change (n-fold) over wild-type level. Either the loss of RfaH or scrambling of the ops gene reduces expression of SPI4 genes, with progressive effects on distal loci.
RESULTS
Transcriptional organization of SPI4.
The transcriptional organization of SPI4 in S. enterica serovar Typhimurium SL1344 was determined using RT-PCR. Primer sites were chosen to include the recently annotated six ORFs as well as intergenic regions, as described in Table 3. From the genome sequence, siiABCDEF appears to constitute an operon with overlapping coding regions for siiA to siiD. The siiD-siiE and siiE-siiF intergenic regions are 20 bp and 40 bp, respectively (Fig. 1). The results of the nonquantitative RT-PCRs are described in Table 3. All the coding and intergenic regions are transcribed, suggesting that siiABCDEF is a single operon, consistent with observations by Gerlach et al. (34). Rapid amplification of the 5′ cDNA ends was employed to map the transcriptional start site of siiA. A similar method was used to identify transcriptional start sites upstream of siiB, siiC, siiD, and siiE, but no products were obtained, confirming that siiABCDEF is most likely a single transcriptional unit. The transcriptional start site was experimentally determined to be located 470 bp upstream of siiA (Fig. 1). A JUMPstart region containing a direct repeat sequence with an ops site predicted to bind RfaH (62) is located 350 bp downstream of the transcriptional start site. By use of a fluorescence-based primer extension method and computational methods, the transcriptional start site for siiF was determined to be a cytosine residue 128 bp upstream of the siiF start codon (54, 76). A Rho-independent transcriptional terminator is predicted in the intergenic region between siiE and siiF (89); however, a product was generated by RT-PCR using primers flanking the terminator sequence (Table 3). From these analyses, we conclude that SPI4 is composed of two transcriptional units, siiABCDEF and siiF.
TABLE 3.
Presence of product after nonquantitative RT-PCR
| Gene | Primers | Presence of product | Presence of product in no-RT control |
|---|---|---|---|
| siiA | KMp50/KMp51 | + | − |
| siiB | KMp52/KMp53 | + | − |
| siiC | KMp54/KMp55 | + | − |
| siiD | KMp56/KMp57 | + | − |
| siiDE IGRa | KMp64/KMp65 | + | − |
| siiE (5′)b | KMp58/KMp59 | + | − |
| siiE (3′)b | KMp60/KMp61 | + | − |
| siiEF IGRa | KMp66/KMp67 | + | − |
| siiF | KMp62/KMp63 | + | − |
IGR, intergenic region.
siiE (5′) corresponds to the 5′ end of the ORF, and siiE (3′) corresponds to the 3′ end of the ORF.
SiiE is produced and is secreted by Salmonella bongori and several serovars of Salmonella enterica.
The salmonellae have been taxonomically divided into two species, S. bongori and S. enterica (84). The sequential acquisition of pathogenicity islands by Salmonella spp. has been reviewed (10, 73). SPI4 is present in all sequenced subspecies, including the ancestral strain, S. bongori. In S. enterica serovar Typhi CT18, siiE has been annotated as two ORFs, of 9,852 (STY4458) and 6,771 (STY4459) base pairs (70), suggesting that it is a pseudogene, whereas siiE in S. enterica serovar Typhimurium is annotated as a single, 16,680-bp ORF (60). Expression and secretion of SiiE were examined in S. bongori and six serovars of S. enterica: S. enterica serovar Typhi, S. enterica serovar Arizona, S. enterica serovar Choleraesuis, S. enterica serovar Dublin, S. enterica serovar Enteritidis, and S. enterica serovar Typhimurium. Anti-SiiE immunoblots show that SiiE is produced by and secreted from S. bongori, S. enterica serovar Choleraesuis, S. enterica serovar Dublin, S. enterica serovar Enteritidis, and S. enterica serovar Typhimurium (Fig. 2) but not S. enterica serovar Typhi and S. enterica serovar Arizona.
SiiE requires the SPI4-encoded T1SS for secretion.
SiiE is predicted to be secreted by the SPI4-encoded T1SS composed of SiiC, SiiD, and SiiF, which collectively form a membrane pore-secretion complex. SiiC is a TolC homologue that forms a pore in the outer membrane; SiiD is an adaptor protein that spans the periplasm, and SiiF is an inner membrane ATPase. SiiE is predicted to be 595 kDa and runs at a molecular mass greater than 460 kDa, consistent with observations by Morgan et al. and Gerlach et al. (35, 62). To determine the SPI4 genes required for SiiE secretion, defined, nonpolar deletion mutations were constructed in siiA, siiB, siiC, siiD, and siiF. Mutations in siiC, siiD, or siiF did not affect production of SiiE (Fig. 3A) but abolished secretion into the culture supernatant (Fig. 3B). A deletion of the C terminus secretion signal of SiiE resulted in intracellular accumulation of the protein (Fig. 3A) and prevented secretion (Fig. 3B). Both siiA and siiB mutant strains retain the ability to produce and secrete SiiE into the culture supernatant (data not shown and Fig. 3). These data demonstrate that SiiE is a large secreted protein that requires the SPI4-encoded T1SS (SiiC, SiiD, and SiiF) for secretion.
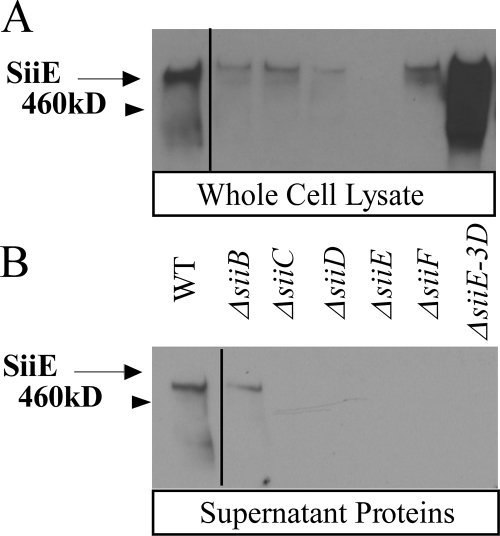
FIG. 3.
Identification of SPI4 genes required for SiiE secretion. (A) Anti-SiiE immunoblot of whole-cell lysates. WT, wild type. (B) Anti-SiiE immunoblot of secreted proteins. The ΔsiiE-3D strain contains a deletion of the 3′ signal sequence of siiE. Protein from the same number of cells was loaded in all lanes. Lower-molecular-weight bands most likely represent degradation of SiiE. Strains carrying nonpolar deletions of the siiC, siiD, and siiF genes fail to secrete SiiE.
RfaH and the operon polarity suppressor (ops) are required for optimal expression of SPI4.
The unusual length (c. 24 kb) of the siiABCDEF transcript suggested that specific mechanisms might be required to prevent premature termination. In some long transcripts, an operon polarity suppressor (ops) sequence within a JUMPstart region (49) is bound by the antiterminator protein RfaH, which reduces RNA polymerase pausing (6, 7). A proposed JUMPstart region with an ops sequence (62) was identified 350 bp downstream of the predicted transcriptional start site and 84 bp upstream of the translational start site of siiA (Fig. 4A).
The transcript levels of siiA, siiC, siiD, and siiE were measured by qRT-PCR in strains carrying an rfaH mutation or a mutated ops site (designated opsM) (Fig. 4B). The transcript levels of siiA were decreased 2-fold and 10-fold in the opsM and rfaH::Km single mutants, respectively, compared to the wild-type level. The effects of RfaH/ops in enhancing transcriptional efficiency became progressively greater for more-distal ORFs, with siiE transcription decreased 55-fold in the opsM mutant and 68-fold in the rfaH mutant strains. Transcription of siiE in an rfaH::Km opsM double-mutant strain was similar to that in strains carrying only an rfaH mutation, suggesting that ops and rfaH are epistatic. Transcript levels of siiF were less influenced by an rfaH mutation, consistent with the expression of siiF from an independent promoter.
Regulatory factors within SPI1 are required for secretion of SiiE.
Morgan et al. (62) previously showed that PrgH, a needle complex protein of the SPI1 T3SS, is not required for SiiE secretion, suggesting that SiiE is not secreted by the SPI1 T3SS. We confirmed this observation with a strain lacking another component of the SPI1 T3SS, InvA (Fig. 5), although an invA mutation reduced the amount of SiiE secreted. A strain lacking the entire SPI1 island secreted no detectable SiiE. We hypothesized that this might have resulted from the loss of a SPI1-encoded regulatory protein required for activation of SPI4 gene expression (20, 34, 62).
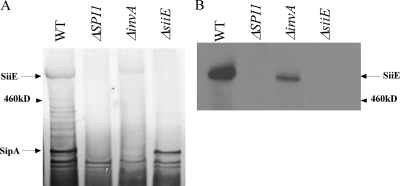
FIG. 5.
SPI1 dependence of SiiE secretion. (A) Silver-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis of secreted proteins. The identities of SiiE and SipA, a SPI1 effector protein, were confirmed by mass spectrometry. WT, wild type. (B) Anti-SiiE immunoblot of the same samples. Strains completely lacking SPI1 exhibit no SiiE secretion, whereas ΔinvA strains deficient in SPI1 function secrete reduced quantities of SiiE.
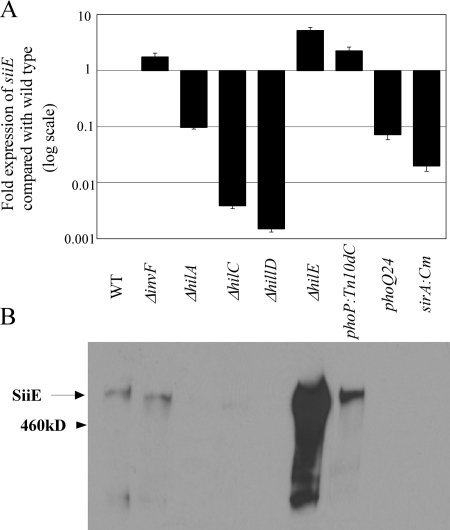
To investigate this possibility, nonpolar mutations were constructed in the genes encoding each of the SPI1-encoded regulators hilA, hilC, hilD, and invF (2, 8, 9, 17, 20, 24, 39, 56, 69, 72, 75, 77, 78). Additional mutations were made in sirA, hilE, and phoPQ (phoP null and phoQ24, which results in constitutive activation of PhoP) since these mutations have previously been shown to influence SPI1 regulation (1, 4, 5, 9, 11, 13, 39, 71, 82). The transcript levels of siiE were measured by qRT-PCR, and SiiE protein in whole cell lysates was measured by immunoblot analysis. For these experiments, bacterial cultures were grown with agitation to an OD600 of 2.0 in LB prior to RNA and protein collection. The results are shown in Fig. 6 and demonstrate that HilA and SirA are each required for expression of siiE, as previously shown for SPI1 and SPI4 (1, 34). We further determined that HilC and HilD are positive regulators of SPI4, while InvF has no effect on siiE expression. Correspondingly, HilE and PhoP were found to negatively regulate the expression of siiE, as has been previously shown for SPI1 (9, 11). SprB is annotated as a putative transcriptional regulator within SPI1 (23). We constructed a nonpolar mutation in sprB and found that it had no effect on invA or siiE expression (data not shown). These observations demonstrate that SPI4 is coregulated with SPI1, and SiiE can be secreted independently of the SPI1-encoded T3SS.
FIG. 6.
Effects of mutations in genes encoding SPI1 regulators on siiE (SPI4) expression. (A) qRT-PCR analysis was used to measure siiE transcript levels in wild-type and isogenic hilA, hilC, hilD, hilE, invF, sirA, phoP, or phoQ24 (phoPc) mutant strains. All values are normalized to gyrB levels. Data are expressed as change (n-fold) over wild-type level. (B) Anti-SiiE immunoblot of whole-cell lysates. An anti-SiiE immunoblot was performed on supernatant proteins, and identical reactivities were observed (data not shown).
Overexpression of hilA, hilC, or hilD fails to activate siiE expression in the absence of SPI1.
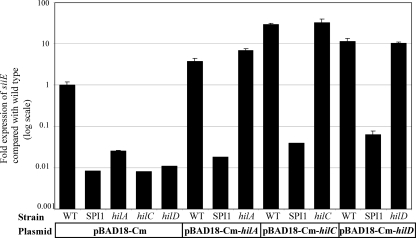
As HilA, HilC, and HilD are DNA-binding regulatory proteins required for SPI1/SPI4 expression, we hypothesized that one of them might directly activate siiABCDEF. To investigate this possibility, hilA, hilC, and hilD were cloned individually into pBAD18-Cm under the control of an arabinose-inducible promoter (37). The plasmids were transformed into wild-type, isogenic SPI1 deletion mutant, or hilA, hilC, or hilD single-mutant strains. The addition of arabinose to induce expression of hilA, hilC, or hilD in the corresponding mutant strains was able to complement expression of siiE (Fig. 7) . Similarly, induction of hilA, hilC, or hilD expression in wild-type S. enterica serovar Typhimurium increased siiE transcription. However, expression of hilA, hilC, or hilD in a SPI1 deletion mutant background had little or no effect on siiE transcript levels. These results suggest that HilA, HilC, and HilD are each required for expression of siiE, but these regulators are likely to act in concert or in combination with additional SPI1-encoded regulatory loci to activate SPI4 gene expression.
FIG. 7.
Overexpression of hilA, hilC, or hilD requires SPI1 for activation of siiE expression. qRT-PCR analysis was used to measure siiE transcript levels in wild-type (WT) or isogenic SPI1, hilA, hilC, or hilD mutant strains with overexpression of hilA, hilC, or hilD as indicated. Plasmids expressing hilA, hilC, or hilD could complement their cognate mutations with regard to siiE activation, but only in strains carrying an intact SPI1 island.
H-NS silences SPI4 expression and is antagonized by HilA.
The nucleoid-associated protein H-NS silences AT-rich sequences that are characteristic of horizontally acquired DNA (57, 67). We have observed that H-NS binds to all SPI, including SPI4. The expression of H-NS-repressed genes can be achieved through countersilencing by sequence-specific transcription factors (66). Examples of these interactions include ToxT at the ctx and tcpA promoters in Vibrio cholerae (68, 91), RovA at the inv promoter of Yersinia enterocolitica (27), and SsrB activation of SPI2 (87). HilA is a transcriptional regulator that has been shown to bind its own promoter (20), other promoters within SPI1 (55), and two regions within SPI4 (20, 83). Previous investigations of SPI4 found that HilA binds a region within siiE and a region upstream of siiA located downstream of the promoter (20, 83). While these binding sites are not compatible with a conventional transcriptional activation mechanism, DNA binding downstream of promoters is able to antagonize transcriptional silencing by H-NS (66).
To determine whether HilA promotes SPI4 expression by relieving H-NS-mediated silencing, siiE expression levels were measured in wild-type and isogenic hns, hilA::Km, and hns hilA::Km mutant strains. Deletion of hns resulted in a 10-fold derepression of siiE (Fig. 8A). A hilA::Km mutation caused a ninefold reduction in siiE expression. However, HilA was not required for siiE expression in an hns mutant strain. Moreover, putative HilA-binding sites with the sequence (T)cATCAGgA (20, 55), where capitalized bases are more highly conserved, were found to correspond to regions of SPI4 bound by H-NS (Fig. 8B). While the sites upstream of siiA and in the proximal region of siiE have been shown to bind HilA (20, 83), the sites within siiB and the distal region of siiE are bioinformatically predicted. Collectively, these observations suggest that HilA enhances the expression of SPI4 by antagonizing the silencing of SPI4 transcription by H-NS.
DISCUSSION
Following ingestion in contaminated food or water, Salmonella enterica invades the intestinal epithelium to produce enteritis and gain access to deeper tissues (30). Epithelial cell invasion and the elicitation of intestinal inflammatory responses require the virulence genes of SPI1 (42, 85). Recently, another genetic locus, designated SPI4, has also been implicated in the ability of Salmonella to cause enterocolitis (35, 47, 63, 81). SPI4 encodes a T1SS and a large secreted protein called SiiE. SiiE has been proposed to mediate adherence to epithelial cells in a variety of hosts, including mice and calves (35, 62). This study confirms that S. enterica serovar Typhimurium secretes SiiE in a T1SS-dependent process and provides novel insights into the regulation of SPI4 gene expression.
The secretion of SiiE specifically requires the TolC homolog SiiC, the periplasmic adaptor SiiD, and the putative inner membrane ATPase SiiF. Deletion of the SiiE C terminus abolishes secretion, confirming that SiiE contains the C-terminal signal sequence conserved among type I secreted proteins. These observations corroborate and extend earlier studies (35, 62). We have further determined that the ability to secrete SiiE is conserved among enteritis-causing serovars of Salmonella but not the human-adapted serovar S. enterica serovar Typhi, which causes enteric fever rather than enterocolitis (74).
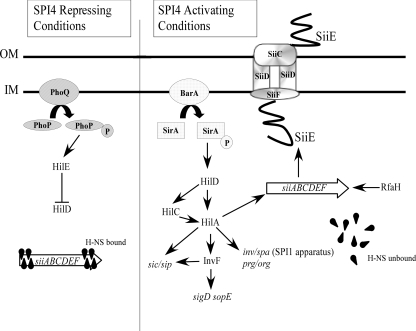
Previous observations have indicated that SPI1 and SPI4 have certain regulatory features in common (1, 20, 34, 62, 83), but the degree of overlap and mechanism of coregulation have not been established. Our observations demonstrate an absolute requirement of SPI1 for the activation of SPI4 genes. As shown in Fig. 9, which incorporates findings from the present study as well as previous work on SPI1 and SPI4 regulation (4, 9, 26, 35), SirA activates hilD within SPI1, which in turn leads to activation of hilC and hilA, also contained within SPI1. HilA subsequently activates the expression of selected SPI1 genes and antagonizes H-NS at SPI4. The complex network of activators controlling SPI1 expression has been proposed to constitute a “feed forward loop” (3, 24) that optimizes the temporal dynamics of the SPI1 response to environmental conditions encountered within the host intestine.
FIG. 9.
Working model of SPI1/SPI4 regulation. See text for details. Under noninducing conditions, PhoP/PhoQ activation leads to production of HilE, which antagonizes the HilD activator required for SPI1/SPI4 expression. Additional negative regulation of SPI4 is provided by H-NS. Under inducing conditions, activation of BarA/SirA initiates a regulatory cascade involving HilD, HilC, and HilA, which leads both directly and indirectly to the expression of SPI1 and SPI4 genes. HilA promotes SPI4 expression by countering H-NS-mediated silencing. RfaH further enhances SPI4 expression by preventing premature termination of transcription. HilA alone is insufficient to activate SPI4 transcription, implicating an additional SPI1-encoded factor in SPI4 activation. OM, outer membrane; IM, inner membrane.
H-NS is a nucleoid-associated protein that recognizes and transcriptionally represses horizontally acquired sequences in enteric bacteria in a process known as xenogeneic silencing (66, 67). H-NS binds and represses genes carried by the Salmonella virulence plasmid and pathogenicity islands, including SPI4 (Fig. 8B). Gene silencing by H-NS can be countered by competition with high-affinity, sequence-specific DNA-binding proteins (27, 68, 87). The SPI1-encoded DNA-binding protein HilA is required for SPI4 expression and has been shown to bind a site upstream of siiA (83). Here, we show that HilA is required for SPI4 expression only in the presence of H-NS, suggesting that HilA promotes SPI4 expression by antagonizing silencing by H-NS. This provides an explanation for the ability of HilA to activate SPI4 by binding DNA downstream of the siiABCDEF promoter, as H-NS silencing appears to involve local polymerization along DNA with bridging of adjacent helices (21).
In addition to the aforementioned HilA-binding site upstream of siiA, the siiB and siiE coding regions each contain predicted HilA boxes based on the proposed consensus sequences tN3TgCAtCAGga (55) and (T)cATCAGgA (20, 83), and HilA binding to the siiE region has been verified experimentally. These putative HilA boxes are each situated within areas of H-NS binding (Fig. 8B), but their roles in SPI4 expression remain to be demonstrated.
Unexpectedly, the overexpression of HilA alone is unable to stimulate SPI4 transcription in the absence of SPI1 (Fig. 7), indicating that HilA countersilencing must be accompanied by additional SPI1-encoded factors to achieve SPI4 expression. InvF, the only other characterized transcriptional activator encoded by SPI1 (17), does not appear to play a role in SPI4 activation. Further studies are under way to identify the additional SPI1 loci required for SPI4 expression. The antiterminator protein RfaH can bind to operon polarity suppressor (ops) sequences of mRNA to reduce RNA polymerase pausing and facilitate the expression of long transcripts (6, 7). Our study demonstrates that RfaH is required for optimal transcription of siiABCDE, in particular the distal gene siiE. In the absence of RfaH binding to the ops, transcription of siiE is severely abrogated. RfaH is similarly required for the transcription of LPS biosynthetic genes in S. enterica serovar Typhimurium (64) and for capsule and hemolysin biosynthesis in E. coli (50, 80). The existence of an independent promoter for siiF might reflect inabilities of RfaH and the ops sequence to preserve sufficient expression of the most distal gene carried by the c. 22-kb transcript.
During infection, SPI1 promotes cytoskeletal rearrangements within intestinal epithelial cells and plays an essential role in Salmonella enteritis (38, 85). SPI4 also appears to be required for intestinal infection in calves (62), mice (35, 45), and the nematode C. elegans (81), although the mechanism by which SPI4 contributes to Salmonella pathogenesis is incompletely understood. One possible role is the promotion of epithelial cell adherence by SiiE (35). Since SPI4 expression is reduced in the absence of SPI1, it is also conceivable that SPI4 may contribute to SPI1-associated phenotypes observed in earlier studies. The coregulation of SPI1 and SPI4 suggests that these virulence loci play complementary roles in the interaction of Salmonella enterica with host intestinal epithelial cells.
Acknowledgments
We are grateful to William Navarre and Anthony Richardson for thoughtful discussions and critical reading of the manuscript. We also thank Chris Allen for initial characterization of SPI4 and Brian Ahmer for strains used during the initial characterization of SPI4.
This work was supported in part by grants from the National Institutes of Health (AI048622 to S.L. and AI44486 to F.F.) and the National Science Foundation (graduate research fellowship to K.M.H.).
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.Ahmer, B. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31971-982. [DOI] [PubMed] [Google Scholar]
- 2.Akbar, S., L. M. Schechter, C. P. Lostroh, and C. A. Lee. 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47715-728. [DOI] [PubMed] [Google Scholar]
- 3.Alon, U. 2007. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8450-461. [DOI] [PubMed] [Google Scholar]
- 4.Altier, C. 2005. Genetic and environmental control of Salmonella invasion. J. Microbiol. 4385-92. [PubMed] [Google Scholar]
- 5.Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35635-646. [DOI] [PubMed] [Google Scholar]
- 6.Artsimovitch, I., and R. Landick. 2002. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109193-203. [DOI] [PubMed] [Google Scholar]
- 7.Bailey, M. J., C. Hughes, and V. Koronakis. 1997. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26845-851. [DOI] [PubMed] [Google Scholar]
- 8.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18715-727. [DOI] [PubMed] [Google Scholar]
- 9.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22703-714. [DOI] [PubMed] [Google Scholar]
- 10.Baumler, A. J. 1997. The record of horizontal gene transfer in Salmonella. Trends Microbiol. 5318-322. [DOI] [PubMed] [Google Scholar]
- 11.Baxter, M. A., T. F. Fahlen, R. L. Wilson, and B. D. Jones. 2003. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 711295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baxter, M. A., and B. D. Jones. 2005. The fimYZ genes regulate Salmonella enterica serovar Typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility. Infect. Immun. 731377-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behlau, I., and S. I. Miller. 1993. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J. Bacteriol. 1754475-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanc-Potard, A. B., F. Solomon, J. Kayser, and E. A. Groisman. 1999. The SPI-3 pathogenicity island of Salmonella enterica. J. Bacteriol. 181998-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyd, E. F., F. S. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 1391125-1132. [DOI] [PubMed] [Google Scholar]
- 16.China, B., and F. Goffaux. 1999. Secretion of virulence factors by Escherichia coli. Vet. Res. 30181-202. [PubMed] [Google Scholar]
- 17.Darwin, K. H., and V. L. Miller. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 1814949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darwin, K. H., and V. L. Miller. 1999. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12405-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Keersmaecker, S. C., K. Marchal, T. L. Verhoeven, K. Engelen, J. Vanderleyden, and C. S. Detweiler. 2005. Microarray analysis and motif detection reveal new targets of the Salmonella enterica serovar Typhimurium HilA regulatory protein, including hilA itself. J. Bacteriol. 1874381-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorman, C. J. 2007. Probing bacterial nucleoid structure with optical tweezers. Bioessays 29212-216. [DOI] [PubMed] [Google Scholar]
- 22.Dorsey, C. W., M. C. Laarakker, A. D. Humphries, E. H. Weening, and A. J. Baumler. 2005. Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol. Microbiol. 57196-211. [DOI] [PubMed] [Google Scholar]
- 23.Eichelberg, K., W. D. Hardt, and J. E. Galan. 1999. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol. Microbiol. 33139-152. [DOI] [PubMed] [Google Scholar]
- 24.Ellermeier, C. D., J. R. Ellermeier, and J. M. Slauch. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57691-705. [DOI] [PubMed] [Google Scholar]
- 25.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1855096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellermeier, J. R., and J. M. Slauch. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 1024-29. [DOI] [PubMed] [Google Scholar]
- 27.Ellison, D. W., and V. L. Miller. 2006. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J. Bacteriol. 1885101-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fahlen, T. F., N. Mathur, and B. D. Jones. 2000. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 2825-35. [DOI] [PubMed] [Google Scholar]
- 29.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 835189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finlay, B. B., and J. H. Brumell. 2000. Salmonella interactions with host cells: in vitro to in vivo. Philos. Trans. R. Soc. Lond. B 355623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fullner, K. J., and J. J. Mekalanos. 2000. In vivo covalent cross-linking of cellular actin by the Vibrio cholerae RTX toxin. EMBO J. 195315-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galan, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 1753-86. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-del Portillo, F. 2001. Salmonella intracellular proliferation: where, when and how? Microbes Infect. 31305-1311. [DOI] [PubMed] [Google Scholar]
- 34.Gerlach, R. G., D. Jackel, N. Geymeier, and M. Hensel. 2007. Salmonella pathogenicity island 4-mediated adhesion is coregulated with invasion genes in Salmonella enterica. Infect. Immun. 754697-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerlach, R. G., D. Jackel, B. Stecher, C. Wagner, A. Lupas, W. D. Hardt, and M. Hensel. 2007. Salmonella pathogenicity island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell. Microbiol. 91834-1850. [DOI] [PubMed] [Google Scholar]
- 36.Groisman, E. A., and H. Ochman. 1997. How Salmonella became a pathogen. Trends Microbiol. 5343-349. [DOI] [PubMed] [Google Scholar]
- 37.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardt, W. D., L. M. Chen, K. E. Schuebel, X. R. Bustelo, and J. E. Galan. 1998. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93815-826. [DOI] [PubMed] [Google Scholar]
- 39.Johnston, C., D. A. Pegues, C. J. Hueck, A. Lee, and S. I. Miller. 1996. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22715-727. [DOI] [PubMed] [Google Scholar]
- 40.Jones, B. D. 2005. Salmonella invasion gene regulation: a story of environmental awareness. J. Microbiol. 43110-117. [PubMed] [Google Scholar]
- 41.Kaneko, A., M. Mita, K. Sekiya, H. Matsui, K. Kawahara, and H. Danbara. 2002. Association of a regulatory gene, slyA with a mouse virulence of Salmonella serovar Choleraesuis. Microbiol. Immunol. 46109-113. [DOI] [PubMed] [Google Scholar]
- 42.Kaniga, K., J. C. Bossio, and J. E. Galan. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13555-568. [DOI] [PubMed] [Google Scholar]
- 43.Karlinsey, J. E. 2007. lambda-Red genetic engineering in Salmonella enterica serovar Typhimurium. Methods Enzymol. 421199-209. [DOI] [PubMed] [Google Scholar]
- 44.Karlinsey, J. E., and K. T. Hughes. 2006. Genetic transplantation: Salmonella enterica serovar Typhimurium as a host to study sigma factor and anti-sigma factor interactions in genetically intractable systems. J. Bacteriol. 188103-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiss, T., E. Morgan, and G. Nagy. 2007. Contribution of SPI-4 genes to the virulence of Salmonella enterica. FEMS Microbiol. Lett. 275153-159. [DOI] [PubMed] [Google Scholar]
- 46.Knodler, L. A., J. Celli, W. D. Hardt, B. A. Vallance, C. Yip, and B. B. Finlay. 2002. Salmonella effectors within a single pathogenicity island are differentially expressed and translocated by separate type III secretion systems. Mol. Microbiol. 431089-1103. [DOI] [PubMed] [Google Scholar]
- 47.Lawley, T. D., K. Chan, L. J. Thompson, C. C. Kim, G. R. Govoni, and D. M. Monack. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee, J. H., M. W. Kim, B. S. Kim, S. M. Kim, B. C. Lee, T. S. Kim, and S. H. Choi. 2007. Identification and characterization of the Vibrio vulnificus rtxA essential for cytotoxicity in vitro and virulence in mice. J. Microbiol. 45146-152. [PubMed] [Google Scholar]
- 49.Leeds, J. A., and R. A. Welch. 1997. Enhancing transcription through the Escherichia coli hemolysin operon, hlyCABD: RfaH and upstream JUMPStart DNA sequences function together via a postinitiation mechanism. J. Bacteriol. 1793519-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leeds, J. A., and R. A. Welch. 1996. RfaH enhances elongation of Escherichia coli hlyCABD mRNA. J. Bacteriol. 1781850-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Libby, S. J., L. G. Adams, T. A. Ficht, C. Allen, H. A. Whitford, N. A. Buchmeier, S. Bossie, and D. G. Guiney. 1997. The spv genes on the Salmonella dublin virulence plasmid are required for severe enteritis and systemic infection in the natural host. Infect. Immun. 651786-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Libby, S. J., M. Lesnick, P. Hasegawa, M. Kurth, C. Belcher, J. Fierer, and D. G. Guiney. 2002. Characterization of the spv locus in Salmonella enterica serovar Arizona. Infect. Immun. 703290-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin, W., K. J. Fullner, R. Clayton, J. A. Sexton, M. B. Rogers, K. E. Calia, S. B. Calderwood, C. Fraser, and J. J. Mekalanos. 1999. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc. Natl. Acad. Sci. USA 961071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lloyd, A. L., B. J. Marshall, and B. J. Mee. 2005. Identifying cloned Helicobacter pylori promoters by primer extension using a FAM-labelled primer and GeneScan analysis. J. Microbiol. Methods 60291-298. [DOI] [PubMed] [Google Scholar]
- 55.Lostroh, C. P., and C. A. Lee. 2001. The HilA box and sequences outside it determine the magnitude of HilA-dependent activation of PprgH from Salmonella pathogenicity island 1. J. Bacteriol. 1834876-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1832733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lucchini, S., G. Rowley, M. D. Goldberg, D. Hurd, M. Harrison, and J. C. Hinton. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marcus, S. L., J. H. Brumell, C. G. Pfeifer, and B. B. Finlay. 2000. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2145-156. [DOI] [PubMed] [Google Scholar]
- 59.Maruyama, I. N., T. L. Rakow, and H. I. Maruyama. 1995. cRACE: a simple method for identification of the 5′ end of mRNAs. Nucleic Acids Res. 233796-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413852-856. [DOI] [PubMed] [Google Scholar]
- 61.Moncrief, M. B., and M. E. Maguire. 1998. Magnesium and the role of MgtC in growth of Salmonella typhimurium. Infect. Immun. 663802-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morgan, E., A. J. Bowen, S. C. Carnell, T. S. Wallis, and M. P. Stevens. 2007. SiiE is secreted by the Salmonella enterica serovar Typhimurium pathogenicity island 4-encoded secretion system and contributes to intestinal colonization in cattle. Infect. Immun. 751524-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54994-1010. [DOI] [PubMed] [Google Scholar]
- 64.Nagy, G., V. Danino, U. Dobrindt, M. Pallen, R. Chaudhuri, L. Emody, J. C. Hinton, and J. Hacker. 2006. Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect. Immun. 745914-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Navarre, W. W., T. A. Halsey, D. Walthers, J. Frye, M. McClelland, J. L. Potter, L. J. Kenney, J. S. Gunn, F. C. Fang, and S. J. Libby. 2005. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol. Microbiol. 56492-508. [DOI] [PubMed] [Google Scholar]
- 66.Navarre, W. W., M. McClelland, S. J. Libby, and F. C. Fang. 2007. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 211456-1471. [DOI] [PubMed] [Google Scholar]
- 67.Navarre, W. W., S. Porwollik, Y. Wang, M. McClelland, H. Rosen, S. J. Libby, and F. C. Fang. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313236-238. [DOI] [PubMed] [Google Scholar]
- 68.Nye, M. B., J. D. Pfau, K. Skorupski, and R. K. Taylor. 2000. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J. Bacteriol. 1824295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olekhnovich, I. N., and R. J. Kadner. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 1844148-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413848-852. [DOI] [PubMed] [Google Scholar]
- 71.Pegues, D. A., M. J. Hantman, I. Behlau, and S. I. Miller. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17169-181. [DOI] [PubMed] [Google Scholar]
- 72.Penheiter, K. L., N. Mathur, D. Giles, T. Fahlen, and B. D. Jones. 1997. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol. Microbiol. 24697-709. [DOI] [PubMed] [Google Scholar]
- 73.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 998956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raffatellu, M., D. Chessa, R. P. Wilson, C. Tukel, M. Akcelik, and A. J. Baumler. 2006. Capsule-mediated immune evasion: a new hypothesis explaining aspects of typhoid fever pathogenesis. Infect. Immun. 7419-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rakeman, J. L., H. R. Bonifield, and S. I. Miller. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 1813096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reese, M. G. 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2651-56. [DOI] [PubMed] [Google Scholar]
- 77.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32629-642. [DOI] [PubMed] [Google Scholar]
- 78.Schechter, L. M., and C. A. Lee. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 401289-1299. [DOI] [PubMed] [Google Scholar]
- 79.Schmieger, H. 1971. The fate of the bacterial chromosome in P22-infected cells of Salmonella typhimurium. Mol. Gen. Genet. 110238-244. [DOI] [PubMed] [Google Scholar]
- 80.Stevens, M. P., B. R. Clarke, and I. S. Roberts. 1997. Regulation of the Escherichia coli K5 capsule gene cluster by transcription antitermination. Mol. Microbiol. 241001-1012. [DOI] [PubMed] [Google Scholar]
- 81.Tenor, J. L., B. A. McCormick, F. M. Ausubel, and A. Aballay. 2004. Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr. Biol. 141018-1024. [DOI] [PubMed] [Google Scholar]
- 82.Teplitski, M., R. I. Goodier, and B. M. Ahmer. 2003. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J. Bacteriol. 1857257-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thijs, I. M., S. C. De Keersmaecker, A. Fadda, K. Engelen, H. Zhao, M. McClelland, K. Marchal, and J. Vanderleyden. 2007. Delineation of the Salmonella enterica serovar Typhimurium HilA regulon through genome-wide location and transcript analysis. J. Bacteriol. 1894587-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tindall, B. J., P. A. Grimont, G. M. Garrity, and J. P. Euzeby. 2005. Nomenclature and taxonomy of the genus Salmonella. Int J. Syst. Evol. Microbiol. 55521-524. [DOI] [PubMed] [Google Scholar]
- 85.Tsolis, R. M., L. G. Adams, T. A. Ficht, and A. J. Baumler. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 674879-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Urban, T. A., J. B. Goldberg, J. F. Forstner, and U. S. Sajjan. 2005. Cable pili and the 22-kilodalton adhesin are required for Burkholderia cenocepacia binding to and transmigration across the squamous epithelium. Infect. Immun. 735426-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walthers, D., R. K. Carroll, W. W. Navarre, S. J. Libby, F. C. Fang, and L. J. Kenney. 2007. The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol. Microbiol. 65477-493. [DOI] [PubMed] [Google Scholar]
- 88.Welch, R. A. 2001. RTX toxin structure and function: a story of numerous anomalies and few analogies in toxin biology. Curr. Top. Microbiol. Immunol. 25785-111. [DOI] [PubMed] [Google Scholar]
- 89.Wong, K. K., M. McClelland, L. C. Stillwell, E. C. Sisk, S. J. Thurston, and J. D. Saffer. 1998. Identification and sequence analysis of a 27-kilobase chromosomal fragment containing a Salmonella pathogenicity island located at 92 minutes on the chromosome map of Salmonella enterica serovar Typhimurium LT2. Infect. Immun. 663365-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wood, M. W., M. A. Jones, P. R. Watson, S. Hedges, T. S. Wallis, and E. E. Galyov. 1998. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol. Microbiol. 29883-891. [DOI] [PubMed] [Google Scholar]
- 91.Yu, R. R., and V. J. DiRita. 2002. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol. Microbiol. 43119-134. [DOI] [PubMed] [Google Scholar]