Abstract
Chronic respiratory infections by Burkholderia cenocepacia in cystic fibrosis patients are associated with increased morbidity and mortality, but virulence factors determining the persistence of the infection in the airways are not well characterized. Using a chronic pulmonary infection model, we previously identified an attenuated mutant with an insertion in a gene encoding an RpoN activator protein, suggesting that RpoN and/or components of the RpoN regulon play a role in B. cenocepacia virulence. In this study, we demonstrate that a functional rpoN gene is required for bacterial motility and biofilm formation in B. cenocepacia K56-2. Unlike other bacteria, RpoN does not control flagellar biosynthesis, as evidenced by the presence of flagella in the rpoN mutant. We also demonstrate that, in macrophages, the rpoN mutant is rapidly trafficked to lysosomes while intracellular wild-type B. cenocepacia localizes in bacterium-containing vacuoles that exhibit a pronounced delay in phagolysosomal fusion. Rapid trafficking to the lysosomes is also associated with the release of red fluorescent protein into the vacuolar lumen, indicating loss of bacterial cell envelope integrity. Although a role for RpoN in motility and biofilm formation has been previously established, this study is the first demonstration that the RpoN regulon in B. cenocepacia is involved in delaying phagolysosomal fusion, thereby prolonging bacterial intracellular survival within macrophages.
Burkholderia cenocepacia belongs to a group of closely related gram-negative bacterial species known as the Burkholderia cepacia complex (Bcc) (6). Bcc members are opportunistic pathogens causing severe infections in immunocompromised individuals and in patients with cystic fibrosis (CF) (15, 34). Bcc species can be isolated from many sources, including soil, water, the rhizosphere, and humans, suggesting that they can adapt to and survive in many different niches. Survival under extreme and changing conditions requires adaptive modulation of gene expression in response to environmental cues. These gene expression changes are often controlled by alternative sigma factors (23), which regulate not only effector genes but also other sigma factors and transcriptional regulators (11). Thus, alternative sigma factors play critical roles in the ability of bacteria to adapt to diverse environmental conditions.
Alternative sigma factors also play an important role in the virulence of pathogenic bacteria (23). For example, in Salmonella enterica serovar Typhimurium, RpoE regulates genes that provide resistance to oxidative stress and bacterial survival within macrophages (18). In Vibrio fischeri, RpoN controls motility and biofilm formation and is essential for establishing a symbiotic colonization of the host (52), and in Pseudomonas aeruginosa, RpoN controls the synthesis of alginate, pili, and flagella, which are well-documented virulence factors (14, 47).
RpoN (σN) is unique among the alternative sigma factors because of its absolute requirement for an additional transcriptional activator to initiate RpoN-dependent gene transcription (37, 41). This transcriptional activator is often a response regulator of a two-component system that becomes transiently phosphorylated in response to environmental signals (45). In Escherichia coli, about half of the RpoN-dependent genes are implicated in nitrogen assimilation and metabolism. RpoN also regulates a wide range of processes that are usually not essential for cell survival and growth under favorable conditions (40), including the regulation of genes involved in the utilization of unusual carbon sources (26), flagellar motility (21, 22, 28, 47), O-antigen expression (4), alginate production (14), symbiotic colonization and biofilm formation (52), and the expression of other sigma factors (11).
In a previous study using signature-tagged mutagenesis, we identified several B. cenocepacia genes required for in vivo survival in a rat chronic lung infection model that mimics the CF airways defect (19). One of the attenuated mutants, K56-2 34A1, carried a transposon insertion in an open reading frame encoding an RpoN-specific transcriptional activator, suggesting that RpoN and/or components of the RpoN regulon in this bacterium control the expression of virulence-related factors. In this study, we demonstrate that, indeed, functional RpoN is required for motility and biofilm formation in vitro and, most importantly, for the intracellular trafficking and survival of a clinical isolate of B. cenocepacia within infected macrophages.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacteria and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) medium (Difco) at 37°C. Trimethoprim, chloramphenicol, and tetracycline were used at final concentrations of 50, 30, and 20 μg/ml for E. coli and 100, 100, and 150 μg/ml for B. cenocepacia, respectively. Kanamycin (final concentration, 40 μg/ml) was used to maintain helper plasmid pRK2013. Gentamicin (final concentration, 50 μg/ml) was used during triparental mating to select against the E. coli donor and helper strains. Growth curves were determined with a Bioscreen C automated microbiology growth curve analysis system (MTX Lab Systems, Inc., Vienna, VA). For these experiments, fresh LB medium was inoculated with aliquots of overnight cultures to give a starting optical density at 600 nm (OD600) of 0.05. All chemicals were purchased from Sigma Chemical Co., St. Louis, MO, unless otherwise indicated.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| B. cenocepacia strains | ||
| K56-2 | ET12 clone related to J2315, CF clinical isolate | BCRRCb; 32 |
| MSS13 | K56-2 rpoN::pRW3 Tpr | This study |
| MSS25 | K56-2 fliC::pSM62 Tpr | This study |
| MSS26 | K56-2 ΔrpoN | This study |
| MSS27 | K56-2 ΔrpoN fliC::pSM62 Tpr | This study |
| MSS28 | K56-2 ΔmotA | This study |
| E. coli strains | ||
| DH5α | F− φ80 lacZΔM15 (ΔlacZYA-argF) U169 endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 ΔgyrA96 relA1 | Laboratory stock |
| SY327 | araD Δ(lac pro) argE(Am) recA56 rifrnalA λ pir | 12 |
| Plasmids | ||
| pAP20 | oripBBR1 Cmrmob+Pdhfr | S. Cardona |
| pGPΩTp | oriR6K ΩTpr cassette mob+ | 13 |
| pRedΩCm | oripBBR1 Tpr ΩCmr Pdhfr mRFP1 | R. S. Flannagan |
| pRK2013 | oricolE1 RK2 derivative Knrmob+tra+ | 12 |
| pRF141 | oriR6K ΩTprmob+, including a homing endonuclease site | Flannagan et al. |
| pRF142 | oripBBR1 Tetrmob+Pdhfr expressing a homing restriction endonuclease | Flannagan et al. |
| pRW3 | pGPΩTp, 341-bp internal fragment from rpoN | This study |
| pSM62 | pGPΩTp, 323-bp internal fragment from fliC | This study |
| pSM72 | B. cenocepacia rpoN in pAP20 under control of Pdhfr Cmr | This study |
| pSM63 | pRF141 with fragments flanking rpoN | This study |
| pSM64 | pRF141 with fragments flanking motA | This study |
Tpr, trimethoprim resistance; Cmr, chloramphenicol resistance; Knr, kanamycin resistance; Tetr, tetracycline resistance.
BCRRC, B. cepacia Research and Referral Repository for Canadian CF Clinics.
General molecular techniques.
DNA manipulations were performed as described previously (43). Restriction enzymes, T4 DNA ligase, and alkaline phosphatase were obtained from Roche Diagnostics (Laval, Quebec, Canada). DNA transformations of E. coli DH5α and E. coli SY327 were performed by the calcium chloride method (7). Conjugations were performed by triparental mating (8) with the pRK2013 helper plasmid (12). DNA amplifications by PCR were done with the PTC-0200 or PTC-221 DNA engine (MJ Research, Incline Village, NV) with either Taq DNA polymerase or Pwo polymerase (Roche Diagnostics). DNA sequences were determined by the DNA Sequencing Facility, Robarts Research Institute, London, Ontario, Canada. The computer program BLAST was used to analyze the sequenced genome of B. cenocepacia strain J2315 (http://www.sanger.ac.uk/Projects/B_cenocepacia/).
Quantification of biofilm mass.
The biofilm-forming capacities of wild-type and mutant strains were compared by a crystal violet staining assay. Polystyrene tubes (12 by 75 mm; Falcon) containing 500 μl of LB medium were inoculated in triplicate with the individual strains suspended at an OD600 of 0.005 and incubated at 37°C under static conditions for 24 h. Planktonic bacteria were discarded, and the tubes were gently rinsed with 1 ml of water to remove residual nonadherent bacteria. An 800-μl volume of 1% (wt/vol) crystal violet was added, and the mixture was incubated at room temperature for 1 min. The tubes were rinsed three times with water, and the dye was dissolved with 1 ml of 100% methanol. The absorbance of the solubilized crystal violet was determined at 540 nm with a Beckman DU 530 spectrophotometer. Each experiment was repeated at least three times.
Motility assays.
Bacterial motility was analyzed with soft agar plates (1% [wt/vol] Bacto tryptone, 0.3% [wt/vol] agar) inoculated with 2 μl of culture at an OD600 of 2 and incubated at 37°C. The diameter of the motility zone was measured after 24 h.
Disk diffusion assays.
A disk diffusion assay was used to test the susceptibility of B. cenocepacia strains to oxidative stress. Stationary-phase cells were spread on agar plates with a sterile cotton swab and briefly dried, and 6-mm sterile paper disks were deposited on the agar surface. Filter disks were embedded with 8-μl aliquots of a solution ranging from 0 to 300 mM hydrogen peroxide or 0 to 500 mM methyl viologen. The plates were incubated overnight at 37°C, and the zones of inhibition were measured. Experiments were performed three times with triplicate repeats each time.
Polymyxin B sensitivity assay.
Overnight cultures were diluted to an OD600 of 0.01, and 50-μl aliquots were added to 5 ml of fresh LB medium. A 500-μl volume of this bacterial suspension was aliquoted into Microfuge tubes and mixed with polymyxin B in 0.2% bovine serum albumin-0.01% acetic acid to give final concentrations ranging from 0 to 500 μg/ml. Bacteria were incubated at 37°C for 22 h with constant rotation with a Barnstead Thermolyne LABQUAKE (Barnstead International, Dubuque, IA), and the OD600 was determined.
Transmission electron microscopy.
To visualize flagella, bacterial suspensions were negatively stained with 1% (wt/vol) uranyl acetate and examined in a Philips CM10 transmission electron microscope.
Mutagenesis of B. cenocepacia K56-2.
Insertional inactivation of B. cenocepacia K56-2 genes was performed with pGPΩTp (13). A 341-bp internal fragment of rpoN (BCAL0813) was amplified by PCR with primers 1791 (5′-CGTCTAGAGGATCGCTGATCGCGCAGAC-3′ [XbaI site underlined]) and 1792 (5′-CTGAGAATTCCGTCGTCGTCGAGCGATTCG-3′ [EcoRI site underlined]). The PCR product was digested with XbaI and EcoRI and cloned into suicide vector pGPΩTp, which was similarly digested, resulting in plasmid pRW3 (Table 1). A 323-bp internal fragment of fliC (BCAL0114) was PCR amplified with primers 2812 (5′-CGTCTAGATTGCACAGCAGAACCTCAAC-3′) and 2813 (5′-CTGAGAATTCGATCTGCTGCGAAACTTCCT-3′) and cloned into pGPΩTp as indicated previously, giving rise to plasmid pSM62 (Table 1). Mutagenesis plasmids pRW3 and pSM62 were introduced into B. cenocepacia K56-2 by triparental mating, generating mutants MSS13 (rpoN::pWR3) and MSS25 (fliC::pSM62), respectively. The correct insertion of the integrated plasmid in the K56-2 genome was verified by PCR with chromosome-specific primers for rpoN (5′-CTGAGAATTCGCGCCCCGCTTTGCATCCACG-3′) and fliC (5′-CTGAGAATTCGCTTTCGGCTTATACAGGAG-3′) and a plasmid-specific primer (5′-TAACGGTTGTGGACAACAAGCCAGGG-3′) and also by Southern blot hybridization.
Deletions of rpoN (BCAL0813) and motA (BCAL0126) were performed with a homing endonuclease system that will be described in detail elsewhere (R. S. Flannagan et al., submitted for publication). Briefly, this system allows for the creation of nonpolar and unmarked mutations and comprises two plasmids. One plasmid, pRF141, serves to clone the regions flanking the gene to be deleted and contains a restriction site for a homing endonuclease. Once introduced by conjugation, the mutagenic plasmid is integrated into the B. cenocepacia K56-2 chromosome, giving rise to trimethoprim-resistant mutants. A second plasmid, pRF142 (encoding the homing endonuclease), is introduced by conjugation. Homing endonucleases catalyze site-specific double-strand breaks in the chromosome at the recognition site. As DNA double-strand breaks are lethal, only mutants undergoing second homologous recombination events, including those with a deletion of the gene of interest, can be recovered. PCR amplifications of the 5′ and 3′ regions flanking rpoN and motA were performed with primer pairs 3205 (5′-GCTCTAGACCTCGTGGCTGGCTGCAC-3′ [XbaI site underlined])-3211 (5′-TTTTATCGATCCATCGCGACTTCCTGCTG-3′[ClaI site underlined]) and 3212 (5′-TTTTATCGATCATCCGAGCCCTCATCAAG-3′ [ClaI site underlined])-3206 (5′-GCTTCTCCAAGCGAGTGGCCAC-3′) for rpoN and primer pairs 3210 (5′-GCTCTAGACGAATCGTCTGCGCATTG-3′ [XbaI site underlined])-3207 (5′-TTTTATCGATGTCGGCAGCACGCGCAG-3′ [ClaI site underlined]) and 3209 (5′-TTTTATCGATCACACGATGGCCTCGGC-3′ [ClaI site underlined])-3208 (5′-CGCTCCGCGTCACTTCGCC-3′) for motA. The 5′ amplicons were digested with restriction enzymes XbaI-ClaI, and the 3′ amplicons were digested with ClaI. The DNA fragments were ligated together into pRF141 digested with ClaI and SmaI, giving rise to pSM63 (rpoN mutagenesis plasmid, Table 1) and pSM64 (motA mutagenesis plasmid, Table 1). Mutants MSS26 (ΔrpoN, Table 1) and MSS28 (ΔmotA, Table 1) were confirmed by PCR and Southern blot hybridization. Strain MSS27 (ΔrpoN fliC::pSM62, Table 1) was constructed by insertional inactivation of fliC in strain MSS26 as described before.
Construction of an rpoN-complementing plasmid.
The rpoN gene was PCR amplified from B. cenocepacia K56-2 chromosomal DNA with primers 1793 (5′-CTGAGAATTCGCGCCCCGCTTTGCATCCACG-3′ [EcoRI site underlined]) and 1794 (5′-CGTCTAGAGTGGCCGGGACACGCCTGC-3′ [XbaI site underlined]). The PCR product was digested with EcoRI and XbaI and ligated into pAP20, which was similarly digested, creating plasmid pSM72 (Table 1). The correct insert DNA was verified by DNA sequencing and introduced into the mutant strains by triparental mating.
Macrophage infection assays.
Murine macrophage cell line RAW264.7 (ATCC TIB-71; American Type Culture Collection, Manassas, VA) was maintained in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) at 37°C in a 95% humidified atmosphere and 5% carbon dioxide. Overnight bacterial cultures were washed and resuspended in DMEM-10% FBS. Bacterial suspensions were added to RAW264.7 cells grown on glass coverslips at a multiplicity of infection (MOI) of 50, centrifuged for 1 min at 300 × g, and incubated for 4 h at 37°C under 5% carbon dioxide. Infected macrophages were washed three times with phosphate-buffered saline (PBS) and incubated with 0.5 μM LysoTracker Red DND-99 for 1 min prior to visualization with an Axioscope 2 (Carl Zeiss) microscope with a 100× oil immersion objective. Lysosome labeling was performed by incubating macrophages in the presence of 250 μg/ml of tetramethyl rhodamine (TMR)-dextran for 2 h. External TMR-dextran was removed by serial washes with PBS and chased for 1 h in DMEM with 10% FBS before infection. Infections were carried out as described before. Fluorescence and phase-contrast images were acquired with a Qimaging (Burnaby, British Columbia, Canada) cooled charged-coupled device camera on an Axioscope 2 (Carl Zeiss, Thornwood, NY) microscope with a ×100/1.3 numerical aperture Plan-Neofluor objective and a 50-W mercury arc lamp. Images were digitally processed with the Northern Eclipse version 6.0 imaging analysis software (Empix Imaging, Mississauga, Ontario, Canada). DMEM, PBS, and FBS were purchased from Wisent Inc. (St. Bruno, Quebec, Canada). LysoTracker Red DND-99 and TMR-dextran were from Invitrogen (Eugene, OR). Each experiment was independently repeated at least three times.
RESULTS
Identification of the rpoN gene in the B. cenocepacia J2315 genome.
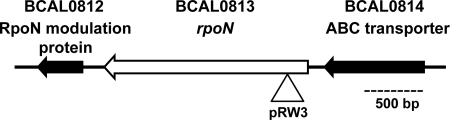
The B. cenocepacia rpoN gene was identified by BLAST search analysis of the B. cenocepacia J2315 translated genome sequence (http://www.sanger.ac.uk/Projects/B_cenocepacia/) with P. aeruginosa PAO1 RpoN (AAG07850.1) as a query. A putative rpoN gene (BCAL0813) was found on chromosome 1 spanning bp 882587 to 884089. Analysis of this genomic region showed a gene encoding the ATPase subunit of an ABC transporter (BCAL0812) terminating 175 bp upstream of rpoN and a gene encoding an RpoN modulation protein (BCAL0814) starting 158 bp downstream of the rpoN termination codon (Fig. 1). This genetic configuration of the rpoN region in B. cenocepacia is widely conserved in many different bacterial genera, including Burkholderia, Ralstonia, Pseudomonas, Salmonella, and Escherichia, among others. Therefore, the bioinformatic analysis strongly supports the assignment of BCAL0813 as rpoN. In other bacteria, rpoN is a monocistronic gene whose transcription initiates within the intervening segment between rpoN and the gene encoding the ABC transporter (Fig. 1). The conservation in the gene organization of the rpoN region strongly suggests that rpoN is also a monocistronic gene in B. cenocepacia (see below).
FIG. 1.
Genetic organization of the rpoN gene and flanking regions of chromosome 1 of B. cenocepacia strains J2315 and K56-2. The rpoN gene is represented by a white arrow, and the insertion site of mutagenesis plasmid pRW3 is indicated by a triangle. Flanking genes BCAL0812 and BCAL0814 are indicated in black, and they encode an RpoN modulation protein and an ABC transporter ATP-binding protein, respectively.
Since B. cenocepacia J2315 is difficult to manipulate genetically, we conducted our research on strain K56-2, which is clonally related to J2315, as demonstrated by macrorestriction and randomly amplified polymorphic DNA methods (32), but much more amenable to genetic analyses. The rpoN gene of B. cenocepacia K56-2 was identical to that of strain J2315, as determined by DNA sequencing (data not shown).
RpoN regulates B. cenocepacia K56-2 motility.
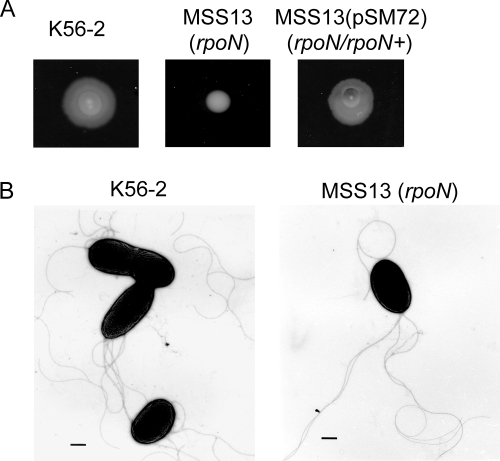
To investigate the role of RpoN in B. cenocepacia K56-2, we constructed isogenic mutant strain MSS13 carrying an insertionally inactivated rpoN gene (see Materials and Methods). The insertion was targeted to the 5′ region of the rpoN coding sequence (Fig. 1). We have previously demonstrated that targeted gene inactivation in B. cenocepacia is possible by the use of suicide vector pGPΩTp (13). This system allows the insertion by homologous recombination of a nonreplicating plasmid carrying strong transcriptional terminators. These terminators prevent readthrough transcription from sequences within the plasmid, as well as from sequences within the targeted gene, thus creating a strong polar effect. Since rpoN is a predicted monocistronic gene, the use of this mutagenesis system was appropriate as we expected the targeted insertion would not compromise transcription of the flaking genes. Considering that RpoN controls the expression of flagellar genes in other bacterial species, including P. aeruginosa, Vibrio cholerae, Legionella pneumophila, and Campylobacter jejuni (21, 22, 28, 47), and that flagellum-mediated motility plays a role in the pathogenesis of B. cenocepacia (46), we examined the motility of mutant strain MSS13 (rpoN::pRW3). Figure 2A shows that MSS13 had reduced motility on soft agar plates compared to that of parental strain K56-2. The motility of MSS13 was restored by complementation with plasmid pSM72, which carries the parental rpoN gene (Fig. 2A). This experiment confirms that the motility defect was due to the inactivation of the rpoN gene. Examination of MSS13 cells by transmission electron microscopy revealed that inactivation of the rpoN gene did not affect the expression of flagella, as MSS13 cells had multiple polar flagella that were indistinguishable from the parental isolate (Fig. 2B). Together, these results suggest that, in contrast to other bacteria, B. cenocepacia RpoN does not control the synthesis of polar flagella but is still required for motility.
FIG. 2.
RpoN controls motility but not flagellar synthesis in B. cenocepacia K56-2. (A) Effect of rpoN insertional inactivation on the motility of B. cenocepacia K56-2. Each panel shows an image of the bacteria grown at 37°C for 24 h on medium solidified with 0.3% agar. (B) Electron micrograph of wild-type and rpoN mutant cells. Bars correspond to 0.5 μm.
Motility is required for B. cenocepacia biofilm formation.
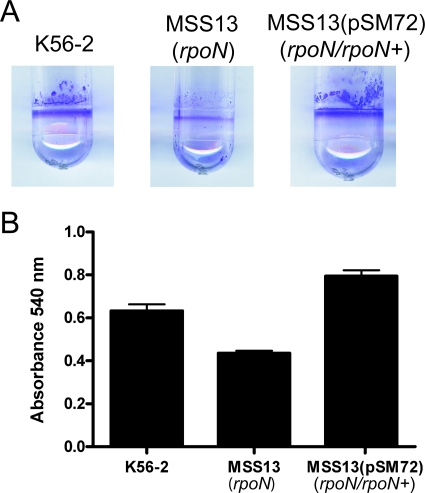
Motility is a well-characterized property associated with biofilm formation (38, 39). Loss of motility perturbs bacterial attachment to abiotic surfaces, affecting the initial stages of biofilm formation (39). Motility is also required for growth and spread of biofilms (39). Thus, we examined the ability of the rpoN mutant and its parental strain to produce biofilm. As shown in Fig. 3, inactivation of rpoN reduced biofilm biomass by approximately 40%, a phenotype that was restored to parental levels after complementation with pSM72 (rpoN+). Reduced biofilm formation by MSS13 was not due to a slower growth rate, since the growth rates of the mutant and parental strains were identical (data not shown). Also, since inactivation of the rpoN gene did not completely abolish biofilm expression, we concluded that additional factors contribute to biofilm production in B. cenocepacia.
FIG. 3.
RpoN controls biofilm production by B. cenocepacia K56-2. (A) Image of crystal violet-stained biofilms formed by K56-2, MSS13, and MSS13(pSM72). Cells were grown in LB medium for 24 h at 37°C under static conditions before crystal violet staining. (B) Quantitative comparison of biofilm formation by B. cenocepacia K56-2, MSS13, and MSS13(pSM72). Each experiment was performed at least three times in triplicate. Error bars represent the standard error of the mean.
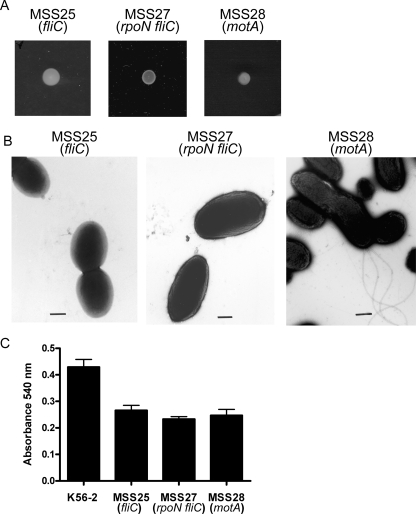
The nonmotile rpoN mutant has intact flagella (Fig. 2B). It has been proposed that flagella are involved in biofilm formation by one of two mechanisms, i.e., (i) by providing motility to enable bacteria to initially reach a surface (39) or (ii) by serving as surface adhesins. To distinguish between these two possibilities, we constructed mutant strains MSS25, carrying a fliC::pSM62 insertion, and MSS27, carrying an rpoN deletion and the fliC::pSM62 insertion (see Materials and Methods). Both mutants were nonmotile and did not produce flagella as detected by transmission electron microscopy (Fig. 4A and B). We compared the abilities of MSS25, MSS27, and the parental strain to produce biofilms. Absence of flagella in MSS25 and MSS27 reduced biofilm biomass by approximately 40% (Fig. 4C), as found with the nonmotile, flagellated rpoN mutant MSS13, demonstrating that the presence of flagella per se has no apparent role in biofilm formation. Moreover, we constructed the mutant MSS28, carrying a motA gene deletion, which is nonmotile but produces intact flagella (Fig. 4A and B), since motA encodes the motor torque generator for flagellar movement (5). The ability of this mutant to produce biofilm was also reduced to the same levels as those found with nonmotile, nonflagellated mutants MSS25 and MSS27 (Fig. 4C). Reduced biofilm formation by mutants MSS25, MSS27, and MSS28 was not due to differences in the growth rate since all of the strains had similar growth rates (data not shown). Together, these experiments demonstrate that flagellum-mediated motility is important for biofilm formation by B. cenocepacia and that the role of RpoN in biofilm formation is restricted to the regulation of motility but not flagellar biogenesis.
FIG. 4.
Flagellum-mediated motility and B. cenocepacia biofilm formation. (A) Each panel shows an image of a bacterial colony grown at 37°C for 24 h on medium solidified with 0.3% agar. (B) Electron micrograph of the fliC, ΔrpoN fliC, and ΔmotA mutants. Representative bacterial cells are shown. Bars correspond to 0.5 μm. (C) Quantitative comparison of biofilms formed by wild-type strain K56-2 and the fliC, ΔrpoN fliC, and ΔmotA mutants under static conditions. Each experiment was performed at least three times in triplicate. Error bars represent the standard error of the mean.
RpoN is required for intracellular trafficking and survival of K56-2 in macrophages.
Previous work in our laboratory demonstrated that B. cenocepacia survives intracellularly within vacuoles in both amoebae and murine macrophages (30, 36, 42). We also established that, in macrophages, B. cenocepacia-containing vacuoles display a maturation delay that temporarily blocks fusion with lysosomes (29). In contrast, B. cenocepacia mutants failing to survive in macrophages behave as heat-killed bacteria, rapidly reaching the lysosome (25, 35). Lack of intracellular bacterial replication and the failure to kill extracellular bacteria prevent us from obtaining viable counts in macrophage infection experiments (29, 42). Therefore, to examine whether disruption of rpoN affects the intracellular behavior of B. cenocepacia, we infected RAW264.7 mouse macrophages with either K56-2 or MSS13 and assessed the colocalization of the bacterium-containing vacuoles with LysoTracker Red by single-cell analysis. LysoTracker Red is an acidotropic dye that preferentially accumulates in the most acidic cell organelles, predominantly lysosomes (1, 51). We previously demonstrated that live wild-type B. cenocepacia cells do not colocalize with LysoTracker Red-containing vacuoles for at least 4 h postinfection, while heat-killed bacteria reach an acidic compartment rapidly after internalization (29).
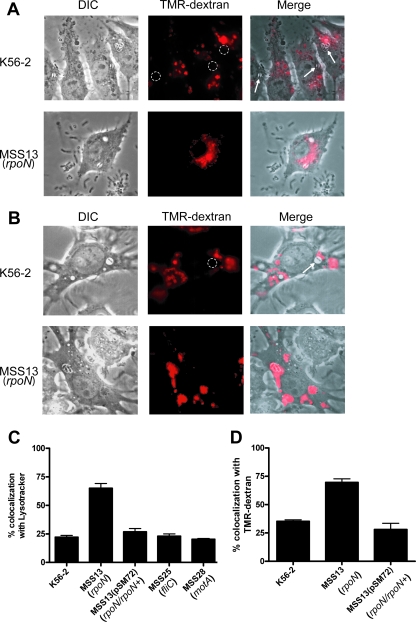
At 4 h postinfection, vacuoles containing MSS13 bacterial cells colocalized with LysoTracker (Fig. 5A) while most of the vacuoles containing the parental K56-2 bacteria did not colocalize with the fluorescent probe (Fig. 5A). Quantitative analysis shows that 69% ± 3.5% of the MSS13-containing vacuoles colocalized with LysoTracker (Fig. 5C). In contrast, only 22.1% ± 4.2% of the vacuoles containing K56-2 colocalized with LysoTracker (P < 0.0001) (Fig. 5C). Introducing plasmid pSM72, which encodes parental RpoN, into MSS13 restored this phenotype. The percentage of LysoTracker colocalization with vacuoles containing MSS13(pSM72) was similar to that of K56-2 (29.3% ± 2.5% and 22.1% ± 4.2%, respectively; Fig. 5C). These results demonstrate that inactivation of rpoN in the K56-2 background causes a significant increase in the number of vacuoles containing bacteria that colocalize with LysoTracker Red.
FIG. 5.
RpoN is required for intracellular survival of B. cenocepacia K56-2. Images of RAW 264.7 macrophage cells infected for 4 h with B. cenocepacia K56-2 or MSS13 at an MOI of 50 by fluorescence and phase-contrast microscopy. (A) Macrophages were incubated with 0.5 μM LysoTracker Red prior to visualization. K56-2 bacteria are within membrane-bound vacuoles that do not colocalize with LysoTracker Red (arrows). (B) Macrophages were incubated with 250 μg/ml of TMR-dextran prior to infection. K56-2 bacteria are within membrane-bound vacuoles that do not colocalize with TMR-dextran (arrows). (C) Percentage of bacterium-containing vacuoles colocalizing with LysoTracker Red. The values correspond to the average and standard error of three experiments in which 21 fields were examined. (D) Percentage of bacterium-containing vacuoles colocalizing with TMR-dextran. The values correspond to the average and standard error of three experiments in which 21 fields were examined. DIC, differential interference contrast.
To confirm that the rpoN mutant was trafficked to the lysosomes, we used TMR-dextran, which is internalized via fluid-phase endocytosis. Prior to bacterial infection, we incubated macrophages with 250 μg/ml of TMR-dextran for 2 h, followed by a 1-h chase in dye-free medium to ensure that the probe is fully delivered from early and recycling endosomes to lysosomes. As reported in the literature, pulse-chase protocols with TMR-dextran are extensively used to label lysosomes (10, 16, 17, 24, 29). We observed that at 4 h postinfection with B. cenocepacia K56-2, the majority of the bacterium-containing vacuoles did not fuse with TMR-dextran-labeled lysosomes (Fig. 5B). In contrast, most of the bacterium-containing vacuoles colocalized with TMR-dextran when the infection was carried out with the MSS13 strain (Fig. 5B). Quantitative analysis shows that only 35.3% ± 2.3% of the K56-2-containing vacuoles colocalized with TMR-dextran, while 69.6% ± 7.1% of the vacuoles containing MSS13 colocalized with the probe (P = 0.0002) (Fig. 5D). This phenotype was restored after complementation with the parental rpoN gene. The percentage of colocalization with TMR-dextran of the vacuoles containing MSS13(pSM72) was similar to that of K56-2 (28% ± 10% and 35.3% ± 2.3%, respectively; Fig. 5D). Together, these results strongly support the notion that the rpoN mutant lost the ability to delay phagolysosomal fusion.
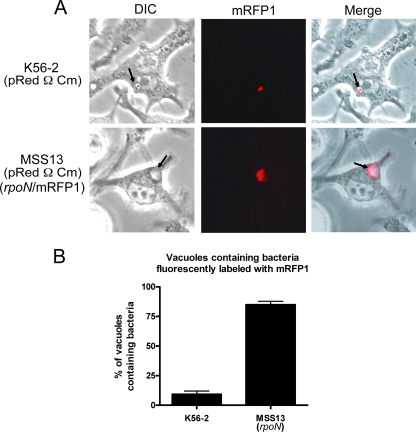
With B. cenocepacia cells expressing monomeric red fluorescent protein 1 (mRFP1), we have previously demonstrated that intracellular B. cenocepacia that traffics into lysosomes rapidly loses cell envelope integrity and is most likely destroyed in this cellular compartment (29). Indeed, while live bacteria retain the fluorescence within the bacterial cytoplasm, heat-killed bacteria, which retained the fluorescence if they were kept in buffer, leaked fluorescence to the vacuolar space once they were phagocytized (29, 35). Thus, dispersal of the fluorescent protein throughout the phagosomal lumen serves as an indication of bacterial cell disruption. We used a similar strategy to assess the viability within RAW264.7 macrophages of the MSS13 mutant containing pRedΩm, which encodes mRFP1 (Table 1). As shown in Fig. 6A, after 4 h postinfection, the majority of the phagosomes containing MSS13(pRedΩCm) bacteria were fluorescently labeled, suggesting that soluble mRFP1 had leaked from the bacterial cytoplasm into the phagosomal lumen. In contrast, K56-2(pRedΩCm) retained the fluorophore within the bacteria cytoplasm (Fig. 6A). Also, in contrast to the apparently normal bacterial morphology of intracellular B. cenocepacia K56-2(pRedΩCm), internalized MSS13(pRedΩCm) exhibited a variety of abnormal morphologies such as rounding and a highly dense cytoplasm (Fig. 6A and data not shown), further suggesting compromise of the cellular envelope. Quantitative analyses demonstrated that after infection with MSS13(pRedΩCm), 84.8% ± 5.1% of the bacterium-containing vacuoles were uniformly fluorescent while only 9.3% ± 4.5% of the vacuoles containing K56-2 where fluorescent (P < 0.0001) (Fig. 6B). Together, the experiments with macrophages infected with the rpoN mutant MSS13, indicating that the majority of the bacterium-containing vacuoles become rapidly acidified, colocalize with a dextran-rich compartment, and accumulate mRFP1 as a result of loss of bacterial envelope integrity, support the notion that RpoN controls the expression of critical factors likely required for the survival of B. cenocepacia K56-2 within macrophages. As an attempt to identify possible factors required for intracellular survival of B. cenocepacia that could be altered in the rpoN mutant, we assessed the ability of MSS13 to resist killing by hydrogen peroxide, superoxide ion, and the antimicrobial peptide polymyxin B (see Materials and Methods). No differences were found compared to the wild-type K56-2 strain (data not shown). To investigate if the defect in intracellular trafficking and survival of the rpoN mutant was due to the loss of flagellar motility, we infected RAW264.7 macrophages with either MSS25 (fliC::pSM62) or MSS28 (ΔmotA) and determined the percentage of colocalization of the vacuole-containing bacteria with LysoTracker Red. As shown in Fig. 5C, quantitative analysis of these experiments demonstrated that a nonflagellated B. cenocepacia strain (MSS25) or one with apparently intact but paralyzed flagella (MSS28) did not have any detectable defect in intracellular trafficking compared with the wild-type strain. Indeed, only 23% ± 4.9% of the vacuoles containing MSS25 and 20.3% ± 1.5% of the vacuoles containing MSS28 colocalized with LysoTracker Red. These values were similar to the 22.1% ± 4.2% colocalization obtained when the infection was carried out with B. cenocepacia K56-2 (Fig. 5C). These results demonstrate that the defect in intracellular trafficking of the rpoN mutant is not due to the RpoN-mediated loss of flagellar motility.
FIG. 6.
RpoN is required for cell envelope integrity of B. cenocepacia within murine macrophages. (A) Images of RAW 264.7 macrophage cells infected for 4 h with B. cenocepacia K56-2(pRedΩCm) or MSS13(pRedΩCm) at an MOI of 50 by fluorescence and phase-contrast microscopy. The rpoN mutant bacteria had compromised cell envelope permeability, as shown by the release of mRFP1 into the vacuolar lumen, while the parental bacteria retained mRFP1 within the cytoplasm. The arrows indicate bacterium-containing vacuoles. (B) Average percentages of B. cenocepacia-containing vacuoles (BcCV) containing released mRFP1 from three independent experiments. Error bars represent the standard error of the mean. DIC, differential interference contrast.
DISCUSSION
Little is known about the mechanisms by which Bcc species cause disease, and the specific bacterial factors involved in the persistence of Bcc strains within the respiratory tract are still unclear. It has been proposed that intracellular survival of Bcc species may explain their persistence in the respiratory systems of patients with CF (49). Burkholderia species have large genomes and a wide-range metabolic potential, which probably explains why these organisms can thrive in many different environmental niches. Therefore, it is conceivable that metabolic adaptation could play a major role in the ability of these bacteria to survive under different environmental conditions and adapt to stress. In this study, we demonstrate for the first time that the alternative sigma factor RpoN plays an important role in controlling the fate of intracellular B. cenocepacia within infected macrophages. Inactivation of the rpoN gene results in a B. cenocepacia mutant unable to delay the maturation of the phagosome, suggesting that RpoN controls the expression of bacterial factors required to delay the acidification of the bacterium-containing vacuoles. Studies in our laboratory have established that a delay in the maturation of bacterium-containing vacuoles is associated with intracellular survival of B. cenocepacia in macrophages (29). Conceivably, such a delay is advantageous for the bacteria, enabling them to activate genes that could confer resistance to the hostile environment of lysosomes, thus providing Bcc strains with an additional survival advantage that may be critical in the context of immunocompromised hosts. This notion is strongly supported by the observation that mutants that cannot cause a maturation delay of the phagosome are not only rapidly cleared from macrophages but also avirulent in the rat agar bead model (19, 35). The role of RpoN in bacterial intracellular survival has not been well studied. A Brucella melitensis mutant with a deletion in the rpoN gene did not show a defect in intracellular survival in vivo (9), while a P. aeruginosa rpoN mutant was associated with poor binding and nonopsonic phagocytosis by macrophages and neutrophils (33). In this work, we show that intracellular bacteria are not only rapidly trafficked to lysosomes but also exhibit loss of cell envelope integrity, as demonstrated by the release of bacterially expressed red fluorescent protein into the vacuolar space. The intracellular phenotype of the rpoN mutant could not be explained by increased susceptibility to hydrogen peroxide, superoxide ion, or the cationic antimicrobial peptide polymyxin B. Therefore, our results reveal a novel role for RpoN in controlling the intracellular trafficking of an opportunistic pathogen within macrophages. RpoN-dependent genes are important in Rhizobium strains to establish symbiotic associations in plants within the root nodules through complex regulatory networks (2). The Burkholderia genus also contains several members that can establish either symbiotic or pathogenic associations with plants and fungi and can also survive in amoebae and macrophages (3, 20, 48). Therefore, it is possible that the requirement of B. cenocepacia RpoN for intracellular trafficking and, potentially, intracellular survival in macrophages may reflect the ability of these microorganisms to interact with eukaryotic cells in several different niches.
We also found that RpoN controls B. cenocepacia motility, a common feature of RpoN that in many other gram-negative bacteria occurs through the regulation of flagellar synthesis (47). However, we show here another unique aspect of Burkholderia RpoN by demonstrating that the synthesis of flagella is not impaired in the B. cenocepacia rpoN mutant, suggesting that RpoN controls motility by a novel mechanism. Differences in established paradigms for flagellar gene regulation have also been reported for other Burkholderia species (27). For example, FlhDC, a master regulator of lateral flagellar synthesis in many bacteria (44), controls polar flagellum gene expression in the rice pathogen B. glumae (27). This suggests that the mechanisms of motility control in Burkholderia species are potentially more complex than those currently described in other bacteria. Furthermore, our experiments show that in B. cenocepacia, as reported for Listeria monocytogenes (31), motility itself, and not the presence of the flagella, plays a role in biofilm formation. Also, loss of motility only results in a 40% reduction in the ability of the mutant to produce biofilm compared to the wild-type strain.
In summary, the results of this study demonstrate that in B. cenocepacia RpoN is required to produce biofilms and to adapt to the intracellular environment within macrophages and possibly other environments in the infected airways. Further work to define the B. cenocepacia genes regulated by RpoN, which are ultimately responsible for the unique phenotypes observed here, is under way in our laboratory.
Acknowledgments
We thank K. Maloney, K. Keith, R. Flannagan, and D. Aubert for critical reading of the manuscript. The electron microscopy was conducted in the Electron Microscopy Research Facility of the Schulich School of Medicine and Dentistry, University of Western Ontario, with the expert assistance of J. Sholdice and S. Koval.
This research was supported in part by grants from the Canadian Cystic Fibrosis Foundation and the Canadian Institutes of Health Research. J.L. was supported by a Studentship from the Canadian Cystic Fibrosis Foundation and a Canada Graduate Doctoral Award from the Canadian Institutes of Health Research. M.A.V. holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis.
Editor: A. Camilli
Footnotes
Published ahead of print on 14 January 2008.
REFERENCES
- 1.Al-Younes, H. M., T. Rudel, and T. F. Meyer. 1999. Characterization and intracellular trafficking pattern of vacuoles containing Chlamydia pneumoniae in human epithelial cells. Cell. Microbiol. 1237-247. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, M. J., C. J. Toman, R. F. Fisher, and S. R. Long. 2004. A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. USA 10116636-16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, G., L. Eberl, and A. Hartmann. 2005. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 71673-1685. [DOI] [PubMed] [Google Scholar]
- 4.Bittner, M., S. Saldías, C. Estevez, M. Zaldivar, C. L. Marolda, M. A. Valvano, and I. Contreras. 2002. O-antigen expression in Salmonella enterica serovar Typhi is regulated by nitrogen availability through RpoN-mediated transcriptional control of the rfaH gene. Microbiology 1483789-3799. [DOI] [PubMed] [Google Scholar]
- 5.Blair, D. F., and H. C. Berg. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60439-449. [DOI] [PubMed] [Google Scholar]
- 6.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5719-729. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, S. N., A. C. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 692110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig, F. F., J. G. Coote, R. Parton, J. H. Freer, and N. J. Gilmour. 1989. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 1352885-2890. [DOI] [PubMed] [Google Scholar]
- 9.Delory, M., R. Hallez, J. J. Letesson, and X. De Bolle. 2006. An RpoH-like heat shock sigma factor is involved in stress response and virulence in Brucella melitensis 16M. J. Bacteriol. 1887707-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eissenberg, L. G., and W. E. Goldman. 1988. Fusion of lysosomes with phagosomes containing Histoplasma capsulatum: use of fluoresceinated dextran. Adv. Exp. Med. Biol. 23953-61. [DOI] [PubMed] [Google Scholar]
- 11.Fang, F. C. 2005. Sigma cascades in prokaryotic regulatory networks. Proc. Natl. Acad. Sci. USA 1024933-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 761648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flannagan, R. S., D. Aubert, C. Kooi, P. A. Sokol, and M. A. Valvano. 2007. Burkholderia cenocepacia requires a periplasmic HtrA protease for growth under thermal and osmotic stress and for survival in vivo. Infect. Immun. 751679-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg, J. B., and T. Dahnke. 1992. Pseudomonas aeruginosa AlgB, which modulates the expression of alginate, is a member of the NtrC subclass of prokaryotic regulators. Mol. Microbiol. 659-66. [DOI] [PubMed] [Google Scholar]
- 15.Govan, J. R., J. E. Hughes, and P. Vandamme. 1996. Burkholderia cepacia: medical, taxonomic and ecological issues. J. Med. Microbiol. 45395-407. [DOI] [PubMed] [Google Scholar]
- 16.Haggie, P. M., and A. S. Verkman. 2007. Cystic fibrosis transmembrane conductance regulator-independent phagosomal acidification in macrophages. J. Biol. Chem. 28231422-31428. [DOI] [PubMed] [Google Scholar]
- 17.Hmama, Z., K. Sendide, A. Talal, R. Garcia, K. Dobos, and N. E. Reiner. 2004. Quantitative analysis of phagolysosome fusion in intact cells: inhibition by mycobacterial lipoarabinomannan and rescue by an 1α,25-dihydroxyvitamin D3-phosphoinositide 3-kinase pathway. J. Cell Sci. 1172131-2140. [DOI] [PubMed] [Google Scholar]
- 18.Humphreys, S., A. Stevenson, A. Bacon, A. B. Weinhardt, and M. Roberts. 1999. The alternative sigma factor, σE, is critically important for the virulence of Salmonella typhimurium. Infect. Immun. 671560-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt, T. A., C. Kooi, P. A. Sokol, and M. A. Valvano. 2004. Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infect. Immun. 724010-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inglis, T. J., and J. L. Sagripanti. 2006. Environmental factors that affect the survival and persistence of Burkholderia pseudomallei. Appl. Environ. Microbiol. 726865-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobi, S., R. Schade, and K. Heuner. 2004. Characterization of the alternative sigma factor σ54 and the transcriptional regulator FleQ of Legionella pneumophila, which are both involved in the regulation cascade of flagellar gene expression. J. Bacteriol. 1862540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jagannathan, A., C. Constantinidou, and C. W. Penn. 2001. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 1832937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazmierczak, M. J., M. Wiedmann, and K. J. Boor. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69527-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keith, K. E., L. Killip, P. He, G. R. Moran, and M. A. Valvano. 2007. Burkholderia cenocepacia C5424 produces a pigment with antioxidant properties using a homogentisate intermediate. J. Bacteriol. 1899057-9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keith, K. E., and M. A. Valvano. 2007. Characterization of SodC, a periplasmic superoxide dismutase from Burkholderia cenocepacia. Infect. Immun. 752451-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kessler, B., S. Marqués, T. Köhler, J. L. Ramos, K. N. Timmis, and V. de Lorenzo. 1994. Cross talk between catabolic pathways in Pseudomonas putida: XylS-dependent and -independent activation of the TOL meta operon requires the same cis-acting sequences within the Pm promoter. J. Bacteriol. 1765578-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, J., Y. Kang, O. Choi, Y. Jeong, J. E. Jeong, J. Y. Lim, M. Kim, J. S. Moon, H. Suga, and I. Hwang. 2007. Regulation of polar flagellum genes is mediated by quorum sensing and FlhDC in Burkholderia glumae. Mol. Microbiol. 64165-179. [DOI] [PubMed] [Google Scholar]
- 28.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28501-520. [DOI] [PubMed] [Google Scholar]
- 29.Lamothe, J., K. K. Huynh, S. Grinstein, and M. A. Valvano. 2007. Intracellular survival of Burkholderia cenocepacia in macrophages is associated with a delay in the maturation of bacterium-containing vacuoles. Cell. Microbiol. 940-53. [DOI] [PubMed] [Google Scholar]
- 30.Lamothe, J., S. Thyssen, and M. A. Valvano. 2004. Burkholderia cepacia complex isolates survive intracellularly without replication within acidic vacuoles of Acanthamoeba polyphaga. Cell. Microbiol. 61127-1138. [DOI] [PubMed] [Google Scholar]
- 31.Lemon, K. P., D. E. Higgins, and R. Kolter. 2007. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 1894418-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahenthiralingam, E., and D. P. Speert. 1995. Nonopsonic phagocytosis of Pseudomonas aeruginosa by macrophages and polymorphonuclear leukocytes requires the presence of the bacterial flagellum. Infect. Immun. 634519-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3144-156. [DOI] [PubMed] [Google Scholar]
- 35.Maloney, K. E., and M. A. Valvano. 2006. The mgtC gene of Burkholderia cenocepacia is required for growth under magnesium limitation conditions and intracellular survival in macrophages. Infect. Immun. 745477-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marolda, C. L., B. Hauröder, M. A. John, R. Michel, and M. A. Valvano. 1999. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology 1451509-1517. [DOI] [PubMed] [Google Scholar]
- 37.Merrick, M. J. 1993. In a class of its own—the RNA polymerase sigma factor sigma 54 (sigma N). Mol. Microbiol. 10903-909. [DOI] [PubMed] [Google Scholar]
- 38.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 5449-79. [DOI] [PubMed] [Google Scholar]
- 39.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30285-293. [DOI] [PubMed] [Google Scholar]
- 40.Reitzer, L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57155-176. [DOI] [PubMed] [Google Scholar]
- 41.Reitzer, L., and B. L. Schneider. 2001. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65422-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saini, L., S. Galsworthy, M. John, and M. A. Valvano. 1999. Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology 1453465-3475. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 44.Soutourina, O. A., and P. N. Bertin. 2003. Regulation cascade of flagellar expression in gram-negative bacteria. FEMS Microbiol. Rev. 27505-523. [DOI] [PubMed] [Google Scholar]
- 45.Studholme, D. J. 2002. Enhancer-dependent transcription in Salmonella enterica Typhimurium: new members of the sigmaN regulon inferred from protein sequence homology and predicted promoter sites. J. Mol. Microbiol. Biotechnol. 4367-374. [PubMed] [Google Scholar]
- 46.Tomich, M., C. A. Herfst, J. W. Golden, and C. D. Mohr. 2002. Role of flagella in host cell invasion by Burkholderia cepacia. Infect. Immun. 701799-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Totten, P. A., J. C. Lara, and S. Lory. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valvano, M. A. 2006. Infections by Burkholderia spp.: the psychodramatic life of an opportunistic pathogen. Future Microbiol. 1145-149. [DOI] [PubMed] [Google Scholar]
- 49.Valvano, M. A., K. E. Keith, and S. T. Cardona. 2005. Survival and persistence of opportunistic Burkholderia species in host cells. Curr. Opin. Microbiol. 899-105. [DOI] [PubMed] [Google Scholar]
- 50.Reference deleted.
- 51.Via, L. E., R. A. Fratti, M. McFalone, E. Pagán-Ramos, D. Deretic, and V. Deretic. 1998. Effects of cytokines on mycobacterial phagosome maturation. J. Cell. Sci. 111897-905. [DOI] [PubMed] [Google Scholar]
- 52.Yip, E. S., B. T. Grublesky, E. A. Hussa, and K. L. Visick. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and sigma-dependent biofilm formation by Vibrio fischeri. Mol. Microbiol. 571485-1498. [DOI] [PubMed] [Google Scholar]