Abstract
The rice pathogen recognition receptor, XA21, confers resistance to Xanthomonas oryzae pv. oryzae strains producing the type one system-secreted molecule, AvrXA21. X. oryzae pv. oryzae requires a regulatory two-component system (TCS) called RaxRH to regulate expression of eight rax (required for AvrXA21 activity) genes and to sense population cell density. To identify other key components in this critical regulatory circuit, we assayed proteins expressed in a raxR gene knockout strain. This survey led to the identification of the phoP gene encoding a response regulator that is up-regulated in the raxR knockout strain. Next we generated a phoP knockout strain and found it to be impaired in X. oryzae pv. oryzae virulence and no longer able to activate the response regulator HrpG (hypersensitive reaction and pathogenicity G) in response to low levels of Ca2+. The impaired virulence of the phoP knockout strain can be partially complemented by constitutive expression of hrpG, indicating that PhoP controls a key aspect of X. oryzae pv. oryzae virulence through regulation of hrpG. A gene encoding the cognate putative histidine protein kinase, phoQ, was also isolated. Growth curve analysis revealed that AvrXA21 activity is impaired in a phoQ knockout strain as reflected by enhanced growth of this strain in rice lines carrying XA21. These results suggest that the X. oryzae pv. oryzae PhoPQ TCS functions in virulence and in the production of AvrXA21 in partnership with RaxRH.
Most pathogenic bacteria are nonobligate parasites that must modulate their gene expression patterns in response to environmental cues (75). In some environments, pathogens need to mobilize scarce metabolic resources and vie with numerous competitors in order to survive. In other environments where pathogens are in contact with the host, nutrients are abundant, and competition with other microorganisms is limited, allowing for extensive multiplication of the pathogen, provided it can escape host defenses. Pathogens have therefore developed integrated regulatory systems that control the coordinated expression of one set of genes in one environment and a different set of genes in another environment. In pathogenic bacteria, these regulatory circuits are generally controlled by two-component systems (TCSs), composed of histidine kinases (HKs) and response regulators (RRs). In response to environmental stimuli, the HK phosphorylates the cognate RR, which then regulates gene expression (13).
We have recently shown that the gram-negative bacterium Xanthomonas oryzae pv. oryzae requires a TCS mediated by RaxR and RaxH (called RaxRH) to sense population cell density (44). Remarkably, the activation of rax (required for AvrXA21 activity) gene expression by RaxRH is induced by a small secreted molecule called AvrXa21. The production of AvrXA21, RaxR, and RaxH is regulated in a cell density-dependent manner by RaxRH (44).
Rice lines carrying the host resistance gene product, XA21, which encodes a receptor kinase with a presumed extracellular LRR (leucine-rich repeat) and non-RD (no conserved arginine and aspartate in subdomain VII) cytoplasmic kinase domains (16) are able to mount an effective defense response to Xanthomonas oryzae pv. oryzae strains having AvrXA21 activity but not strains lacking AvrXa21 activity.
The X. oryzae pv. oryzae molecule that activates Xa21-mediated resistance, AvrXa21, has not yet been isolated. Nonetheless, we have shown that it is dependent on eight rax genes that provide clues to its molecular structure, secretion, and regulation. In addition to the raxR and raxH genes (11, 44), three genes, raxA, raxB, and raxC, that encode proteins similar to components of the type I secretion system of gram-negative bacteria are required for AvrXA21 activity (27). Another three genes, raxP, raxQ, and raxST, encode an ATP sulfurylase, an adenosine phosphosulfate kinase, and a putative sulfotransferase, respectively, suggesting that AvrXA21 activity requires sulfation (65).
These data suggest that AvrXa21 is a secreted peptide, which acts as a quorum-sensing (QS) signal molecule whose production is regulated in a cell density-dependent manner in X. oryzae pv. oryzae by the RaxRH TCS (44, 78). On the basis of decreased population sizes of strains that lack AvrXA21 activity in the field (14), AvrXa21 may serve as a signal molecule essential for X. oryzae pv. oryzae cell-cell communication.
Investigation of the roles of the RaxRH system in controlling AvrXa21 activity is still in its infancy. One of the key questions that remains to be answered is what are the signals and responses regulated by the RaxRH two-component system? We hypothesize that X. oryzae pv. oryzae uses the RaxRH TCS to sense AvrXA21, coordinate infection, and produce virulence factors. Support for this hypothesis comes from studies showing that QS has global effects on bacterial growth, survival, and interactions with eukaryotes. One report indicates that QS-responsive genes compose up to 10% of the Pseudomonas aeruginosa transcriptome and that a large portion of QS-activated genes code for membrane proteins and proteins involved in secretion of virulence factors (77). A relationship between QS and virulence has also been described for the human pathogen Vibrio cholerae (51) and the plant pathogen Xanthomonas campestris pv. campestris (18).
The majority of proteins that have been identified as virulence or avirulence factors are known to be delivered via the type three secretion system (TTSS), a broadly conserved structure among gram-negative pathogens of plants and animals. The TTSS is used by many pathogens to deliver effectors into host cells, and some TTSS effectors of plant pathogens share functions with effectors of animal pathogens (55). In pathogenic bacteria, the TTSS is encoded by the hrp genes that are required for the hypersensitive reaction and pathogenicity (for a review, see reference 31).
The hrp gene cluster of X. oryzae pv. oryzae is nearly identical to the well-studied hrp cluster of X. campestris pv. vesicatoria (22, 40, 79-81, 85). hrp genes in Xanthomonas spp. are expressed in planta or in defined minimal media, but not in rich media. Expression is controlled at least in part by a motif upstream of hrp genes known as the plant-inducible promoter (PIP) box (21, 40, 79), which is also present upstream of many effectors and other genes (42, 57). The PIP box is targeted by the transcriptional activator HrpX, an AraC-type transcriptional regulator (40, 58, 79). HrpX regulates the expression of the hrpB operon and several effector proteins (40, 80). Expression of hrpX is up-regulated by HrpG, a member of the OmpR response regulator family of bacterial TCSs. HrpG also activates HrpA expression (80, 81).
HrpG had been shown to assimilate three major signals in the plant pathogen Ralstonia solanacearum: physical contact with the plant host (2), bacterial metabolic status (9), and a QS signal (25). Downstream genes activated by HrpG have recently been characterized using microarray analysis (75). These include previously isolated genes and 10 other genes that encode response regulators. Thus, the HrpG regulator plays a pivotal role in the molecular switch between saprophytic and pathogenic lifestyles, responding to signals perceived during the soil/plant environment transition by shifting the expression of a large set of genes in addition to those driving the biogenesis of the TTSS. Despite its central importance in orchestrating complex functions, a sensor kinase counterpart or other activator of HrpG has not yet been identified.
Here we report that the RaxRH TCS controls expression of another TCS, PhoPQ, that is not only required for AvrXA21 activity but also controls virulence through the regulation of hrpG gene expression. These results reveal the presence of a complex regulatory circuit that integrates responses to multiple environmental signals and suggest that production and perception of AvrXa21 and regulation of genes involved in virulence are intimately linked.
MATERIALS AND METHODS
Biological materials and growth conditions.
Xanthomonas oryzae pv. oryzae Philippine race 6 (PXO99Az kindly provided by Jan Leach and called PXO99 in this study) was used. The bacterial strains and plasmids used in this study are listed in Table 1. Peptone sucrose (PS) medium (73), nutrient broth (NB) (Difco Laboratories), or a modified version of M9 minimal medium containing 20 μg ml−1 of cephalexin (MP Biomedicals) and/or other antibiotics as appropriate were used for growing cultures of X. oryzae pv. oryzae at 28°C. Escherichia coli strains were cultured in Luria-Bertani (LB) medium at 37°C. For E. coli, kanamycin at 50 μg ml−1, spectinomycin at 50 μg ml−1, ampicillin at 100 μg ml−1, and gentamicin at 25 μg ml−1 (15 μg ml−1 for X. oryzae pv. oryzae) were used for selection of transformants. Polymyxin B and purothionin (Sigma), each at 250, 500, 1,000, and 2,000 μg ml−1, were used to test for resistance against the antimicrobial cationic peptides. To establish cell growth curves, strains PXO99 (wild type), PXO99Q, and PXO99P were used to inoculate M9 minimal medium containing different Mg2+ and Ca2+ concentrations (10 mM and 10 μM for each). For assays of gene expression in the wild type and the PhoP mutant strains in response to variation in Mg2+ and Ca2+ concentrations, the cells were cultured in PS medium or NB medium for 3 days, washed three times with sterilized/deionized water, transferred into M9 minimal medium containing different concentrations of the ions, and cultured for an additional 2 days at 28°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Escherichia coli | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 endA1 araΔ139 Δ(ara leu)7697 galU galK λ−rpsL(Smr) nupG λ−tonA | Gibco BRL |
| BL21(DE3)pLysS | F−ompT hsdSB(rB− mB−) gal dcm (DE3) pLysS (Cmr) | Novagen |
| Xanthomonas oryzae pv. oryzae | ||
| PXO99 | Philippine race 6 strain; Cpr | 32 |
| PXO99Q | PXO99 phoQ::aadA; Spr | This study |
| PXO99P | PXO99 phoP::aadA; Spr | This study |
| PXO99P-P* | PXO99P complemented with pML122phoP; Spr Kmr Gmr | This study |
| PXO99R | PXO99 raxR::Km; AvrXA21+/− Kmr | 11 |
| PXO99H | PXO99 raxH::Km; AvrXA21+/− Kmr | 11 |
| PXO99P-G* | PXO99P with pML122hrpG; Spr Kmr Gmr | This study |
| PXO99ST | PXO99 raxST::Km; AvrXA21− Kmr | 27 |
| Plasmids | ||
| pUC18 | pMB1 ori; Apr | Invitrogen Corp., Carlsbad, CA |
| pHP45Ω | pBR322 ori; Apr; containing the Ω cassette (Spr) from R100.1 | 59a |
| pCR2.1 | ColE1 ori; F1 ori; Apr Kmr | Invitrogen Corp., Carlsbad, CA |
| pBluescript II SK | Phagemid; pUC derivative; Apr | Stratagene, La Jolla, CA |
| pET-15b | pBR322 ori; Apr; T7 promoter, lac operator, His6 tag coding sequence | Novagen, Madison, WI |
| pML122 | OriV OriT Gmr pNm (nptII); broad-host-range expression vector | 41a |
| pCRphoP | pCR2.1 containing the PXO99 phoP coding sequence | This study |
| pSKphoP | pBluescript containing an EcoRI and HindIII 683-bp genomic fragment from PXO99 containing the phoP ORF | This study |
| pSKphoQ | pBluescript containing an EcoRI and HindIII 1,415-bp genomic fragment from PXO99 containing the phoQ ORF | This study |
| pSKphoPQ | pBluescript containing an EcoRI and HindIII 2,095-bp genomic fragment from PXO99 containing the phoQ ORF | This study |
| pUCSphoP | pUC18 carrying an NsiI 683-bp fragment from pSKphoP containing part of the phoP ORF, disrupted by a Spr cassette | This study |
| pUCSphoQ | pUC18 carrying a SacII 1,415-bp fragment from pSKphoQ containing part of the phoQ ORF, disrupted by a Spr cassette | This study |
| pUCSphoPQ | pUC18 carrying an XbaI 2,095-bp fragment from pSKphoPQ containing part of the phoPQ ORF, disrupted by a Spr cassette | This study |
| pETphoP | pET-15b containing the phoP coding sequence cloned into the NdeI-BamHI sites, fused to the His6 tag in the N terminus | This study |
| pMLphoP | pML122 carrying an NcoI-BamHI fragment from pETphoP for expression of His6-tagged phoP in X. oryzae pv. oryzae | This study |
Abbreviations: Cpr, cephalexin resistance; Spr, spectinomycin resistance; Kmr, kanamycin resistance; Gmr, gentamicin resistance; AvrXA21+/−, AvrXA21 positive or negative; AvrXA21−, AvrXA21 negative; His6 tag, six-histidine tag; ORF, open reading frame.
For inoculation experiments to test for virulence and avirulence, the Oryza sativa subsp. japonica rice varieties Taipei 309 (TP309, a rice line susceptible to X. oryzae pv. oryzae regardless of the presence of AvrXa21) and 106-17-3-37, a TP309 transgenic line carrying the Xa21 gene (TP309-XA21, resistant to X. oryzae pv. oryzae strain carrying AvrXa21) (69) were used.
Molecular techniques.
Standard methods described by Sambrook et al. (64) were used for DNA manipulations during plasmid preparations and digestions with restriction enzymes. Southern blot analyses were carried out for the purpose of library screening and confirmation of knockout mutants using DNA probes labeled with [32P]dCTP (NEN Life Science Products, Boston, MA) and a random labeling kit (Amersham Life Science, Arlington Heights, IL). Sequencing of the 3.6-kbp fragment containing phoQ and phoP in pUC18 was performed using the dideoxy chain termination method and an automated sequencer (model 400 I; Li-Cor, Lincoln, NE) with M13 forward and reverse primers or specific primers designed based on sequencing data. RNA extractions were carried out with TRIzol reagent (Invitrogen), and most of the enzymes for cDNA synthesis were purchased from Invitrogen. DNA, RNA, and protein concentrations were measured using the Nanodrop ND-1000 from Bio-Rad. The Cell-Porator system (BRL) was used for E. coli (≥600 V) and X. oryzae pv. oryzae (≥700 V) transformations under the following conditions: booster, 4 kΩ; capacitance, 330 μF; charge rate, fast, low Ω. All of the primer sequences used for this study are listed in Table 2. X. oryzae pv. oryzae genomic DNA was prepared using the method developed by Ausubel et al. (3).
TABLE 2.
Primer sets for real-time PCR, RT-PCR, and generation of knockout mutants
| Gene (method or purpose)a | Primer sequence for 5′ sequence | Primer sequence for 3′ sequence |
|---|---|---|
| 16S rRNA | 5′-TAGCTCAGGTGGTTAGAGCGC-3′ | 5′-CAACGCGAACATACGACTCAA-3′ |
| phoP (qPCR, nt 38 to 280) | 5′-GCGAAACCCTTGCGG-3′ | 5′-CCTGCTTGAGGCC-3′ |
| phoP (qPCR, nt 211 to 490) | 5′-CAAGAAATTTCCGG-3′ | 5′-GCATCATCAAGTATTC-3′ |
| phoP (qPCR, nt 421 to 680) | 5′-GTGAGCGTCAACGG-3′ | 5′-CTTCGGTACGCGGAATG-3′ |
| phoP (RT-PCR) | 5′-GCATGCGTATCCTTTTGGTC-3′ | 5′-GGACTCAGCCTTCGGTACG-3′ |
| corA1 (RT-PCR) | 5′-TGTTCGATAGCCCACACAAA-3′ | 5′-AGAAGTTCATGCCCCAGATG-3′ |
| corA2 (RT-PCR) | 5′-AAGCGCGAACTCAACAAGAT-3′ | 5′-GGTGAAATTCATGCCGTACC-3′ |
| mgtE1 (RT-PCR) | 5′-TCTGGCTGAGCATCAATCTG-3′ | 5′-GAGCATGTTCACCGTCAATG-3′ |
| mgtE2 (RT-PCR) | 5′-GTGCAGTGAAATTGCTCGAA-3′ | 5′-TCGAGCAGATAGATCGCGTA-3′ |
| dnaK (RT-PCR) | 5′-GGTGGAAGACCTGGTCAAGA-3′ | 5′-CGATGATCTTGGTGAACACG-3′ |
| groEL (RT-PCR) | 5′-ACACCTCGTCCGATTACGAC-3′ | 5′-TGCCTTCCTTGACCTTGTTC-3′ |
| hrpG (RT-PCR) | 5′-TTCTTGCGCAGCTTGTAG-3′ | 5′-AGCTCCTGCGTTCGTTGC-3′ |
| hrpG (qPCR, nt 18 to 417) | 5′-GATGTGCCGGGCTGAGAC-3′ | 5′-CTATCGCCGGTGGCGCAT-3′ |
| hrpG (qPCR, nt 222 to 765) | 5′-AGACCGCCTTGGCCAGCT-3′ | 5′-GGATCGGCGTTTCTGTTGA-3′ |
| hrpX (RT-PCR/qPCR) | 5′-GCAGGCTTTGAAAGCTAT-3′ | 5′-TAAATCGCTGGGCTCGAA-3′ |
| hrpA (RT-PCR/qPCR) | 5′-CGTGCTCCATTGTTCGCCGT-3′ | 5′-CCGGAGACCACCGCCACAT-3′ |
| xpsD (RT-PCR) | 5′-TCGAATTTACCCAGCAACTG-3′ | 5′-CACATAACCGAGCAGGAACA-3′ |
| phoP (KO) | 5′-GCCGCTCCAAGTGGTATAGA-3′ | 5′-ACTCCCGCTGGTCACAATC-3′ |
| phoQ (KO) | 5′-GCATGCGTATCCTTTTGGTC-3′ | 5′-GGACTCAGCCTTCGGTACG-3′ |
qPCR, quantitative real-time PCR; KO, knockout mutants.
IEF, 2-DE, and protein identification.
Isoelectric focusing (IEF), two-dimensional electrophoresis (2-DE), and the analysis of isolated protein spots were carried out as previously described (59). Total protein was extracted from X. oryzae pv. oryzae PXO99 and PXO99R cultured in PS medium (>108 CFU/ml), resolved using IEF (pH 3 to 10, nonlinear; 13 cm long; Amersham Biosciences), and then separated through a 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel. After SDS-polyacrylamide gel electrophoresis (SDS-PAGE), gels were stained with Coomassie brilliant blue and scanned using a Powerlook III flatbed scanner (UMAX) followed by analysis of protein spots using Image Master 2D Elite software version 4.1 (Amersham Biosciences). Proteins were extracted from gel pieces and treated with trypsin, and then the peptide masses were measured using an Ettan matrix-assisted laser desorption ionization (MALDI)-time of flight pro mass spectrometer (Amersham Biosciences). All MALDI spectra were internally calibrated using a standard peptide mixture of angiotensin III and adrenocorticotrophic hormone fragments 18 to 39 (Amersham Biosciences). The peptide mass fingerprinting data was analyzed using the NCBI database with the ProFound program allowing a 50-ppm mass tolerance. Three biological replicates and two technical replicates of samples were carried out for each strain.
Cloning phoQ and phoP from X. oryzae pv. oryzae.
A cosmid library previously generated from X. oryzae pv. oryzae Philippine race 2 strain PXO86 (32) was screened for the phoP gene. A 453-nucleotide (nt)-long fragment of the PXO99 phoP gene was synthesized using the PCR method and used as the hybridization probe for screening. The specific primers used to generate the 453-nt-long fragment were designed based on the X. oryzae pv. oryzae Korean strain KACC10331 and the X. campestris pv. campestris phoP gene sequences GenBank accession no. (NC_003902); PXO99 genomic DNA was used as the template for PCR. The primer sequences used were 5′-CTTTTGGTCGAAGACGAAGC-3′ for the forward primer and 5′-CGTAGCTGGTCAGATCCACA-3′ for the reverse primer. The PCR product was sequenced to confirm it represented the phoP gene sequence. The library was plated onto LB plates containing 50 μg/ml of spectinomycin; cells were transferred onto nylon membranes by placing the membranes on the plates of cells and incubating overnight at 37°C. The membranes were then serially transferred onto 3 MM paper (Fisher Scientific) saturated with lysis solution (10% SDS) for 5 min, with denaturation solution (0.5 M NaOH) for 5 min, and then with neutralization solution (0.5 M Tris-Cl [pH 7.4], 1.5 M NaCl) for 5 min. After washing with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer, the air-dried membranes were baked in an oven at 120°C for 30 min. Prehybridization and hybridization with [32P]dCTP-labeled probe DNA was performed using the standard protocol of Sambrook et al. (64). A 19.3-kb fragment of X. oryzae pv. oryzae genomic DNA identified during the screening was subcloned into pUC18. Southern blot analysis, carried out using the restriction enzymes HindIII, EcoRI, BamHI, and PstI and the phoP gene probe, was utilized to select the appropriate fragment for subcloning.
Resistance to antimicrobial peptides.
Twenty milliliters of PS medium was inoculated with the wild-type strain (PXO99) or mutant PXO99Q or PXO99P strain and cultured for 3 days at 28°C. Next, 5 × 106 CFU/ml of each cell suspension was transferred to fresh PS medium containing 250, 500, 1,000, or 2,000 μg ml−1 of polymyxin B and purothionin. After 6 h of incubation at the same temperature, 10 μl from each culture was dropped onto PS agar medium plates and incubated at 28°C. The colonies on each plate were counted after 3 days.
Tolerance to acidic conditions.
After culture of PXO99 (wild type) and PXO99Q and PXO99P mutant strains for 3 days at 28°C, 5 × 106 CFU/ml of each strain was used to inoculate 1 ml of PS medium that had been adjusted to pH 7.0, 5.5, 4.5 (see Fig. 3), 3.5, or 3.0 with acetic acid. The population density was measured after 4, 8, and 12 h with a spectrophotometer (optical density at 600 nm), and the number of CFU was determined using the same method described above for testing resistance to antimicrobial peptides.
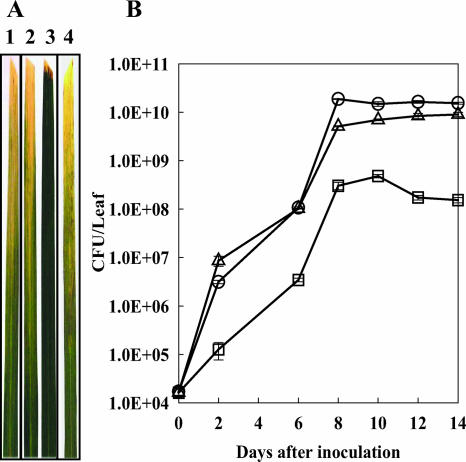
FIG. 3.
(A) Lesions on TP309 leaves inoculated with X. oryzae pv. oryzae strains. The PXO99 (lane 1), PXO99Q (lane 2), PXO99P (lane 3), and Pxo99P-P* (lane 4) strains were inoculated, and then the lesions were measured at 14 days after inoculation. This experiment was repeated three times with more than 10 rice leaves from three individuals each time. (B) Bacterial growth of X. oryzae pv. oryzae strains in inoculated rice leaves (TP309). PXO99 (circles), PXO99Q (triangles), and PXO99P (squares) strains were inoculated onto rice leaves (TP309), and methods were as described in the legend to Fig. 3. Values represent averages ± standard deviations (error bars) of three sampled leaves per treatment.
Activity assay for nonspecific acidic phosphatase.
The para-nitrophenyl phosphate (pNPP) bioassay system kit (BioAssay Systems) was used for activity assays of nonspecific acidic phosphatase. The PXO99, PXO99Q, and PXO99P strains were cultured in PS or NB medium for 3 days at 28°C, washed with water that had been autoclaved and then deionized three times each, and then transferred into M9 minimal medium containing high (10 mM) or low (10 μM) Mg2+ or Ca2+ concentrations. Cells were harvested after centrifugation at 2,500 × g for 20 min, washed, and resuspended in extraction buffer (100 mM Tris-Cl [pH 7.5], 100 mM NaCl, 1 mM EDTA). Total protein was extracted using a Sonic Dismembrator (Fisher Scientific Inc.) on ice. After centrifugation at 2,500 × g for 30 min, the protein concentration in the supernatant was measured using a spectrophotometer. Fifty micrograms of total protein from each sample was added to the reaction buffer (100 mM sodium acetate [pH 5.5], 10 mM MgCl2) containing the substrate, p-nitrophenyl phosphate supplied in the assay kit. The reaction mixtures of each sample were placed at room temperature for 30 min, and then stop solution (supplied in the assay kit) was added to each reaction mixture. The product of the enzyme reaction, p-nitrophenol, was measured using a spectrophotometer at 405 nm.
Inoculation experiments.
Following surface sterilization, TP309 and TP309-XA21 (line 106-17-3-37) seeds were germinated in distilled water at 28°C for 4 days, then planted into soil, and grown for 6 weeks in a greenhouse. Six-week-old plants were transferred to a growth chamber at least 2 days prior to inoculation. The chamber conditions were as follows: 16 h of light at 28°C and 80% relative humidity/8 h of dark at 26°C and 90% relative humidity. X. oryzae pv. oryzae cells were prepared by culturing on PS agar plates containing either cephalexin (for the wild-type strain) or spectinomycin (for the PXO99Q and PXO99P mutant strains) for 3 days at 28°C. Rice leaves were inoculated with the scissor clip method (37), using cells suspended in distilled water at a density approaching 108 CFU/ml. Lesion lengths were measured 14 days after inoculation. X. oryzae pv. oryzae growth in planta was measured using the method reported by Song et al. (69). The results represent an average of three inoculated leaves per strain for each of the eight time points (0, 2, 4, 6, 8, 10, 12, and 14 days after inoculation).
For establishing growth curves, 5-cm sections of the inoculated rice leaves were harvested at each time point, immediately sliced into small pieces, incubated in 1 ml sterile water including 15 μg/ml of cephalexin with shaking for 1 h, and then filtered through two layers of gauze. The filtrates were then plated onto PS agar plates. Colonies on the plates were counted after 3 days of incubation at 28°C.
Construction of null mutants using marker exchange mutagenesis.
Knockout mutants were generated using the marker exchange mutagenesis method (43), a kanamycin or spectinomycin resistance cassette (from pUK-4K and pHM1), and the suicide vector pUC18. DNA fragments for homologous recombination of the phoP and phoQ genes were synthesized using the PCR method with Taq polymerase (Promega Corp., Madison, WI) and with a programmable thermal controller (MJ Research Inc., Watertown, MA). The Spr cassette was inserted into the SacII or NsiI restriction enzyme cleavage site of the X. oryzae pv. oryzae phoQ or phoP gene, respectively. The pUC18 constructs carrying the mutagenized phoP and phoQ genes were then introduced into competent PXO99 wild-type cells. After electroporation, the cells were incubated for 3 h at 28°C and then spread on PS agar plates containing 50 μg/ml spectinomycin. Colonies that grew on those plates were replated onto PS agar plates containing 50 μg/ml spectinomycin and plates containing 50 μg/ml of spectinomycin-ampicillin in order to select for double-crossover events. Colonies which grew on the spectinomycin-only plates were collected and confirmed as insertional mutants using PCR. Primers used to confirm insertion into phoP were PhoP-N-F (5′-GCATGCGTATCCTTTTGGTC-3′) and PhoP-N-R (5′-GGACTCAGCCTTCGGTACG-3′). Primers used to confirm insertion into phoQ were PhoQ-N-F (5′-GCCGCTCCAAGTGGTATAGA-3′) and PhoQ-N-R (5′-ACTCCCGCTGGTCACAATC-3′). Southern blot analyses with probes specifically designed to detect either the phoQ or phoP gene split by the spectinomycin resistance gene were also performed for genomic confirmation that both genes had been “knocked out.”
Generation of the PhoP- and HrpG-expressing mutants.
The X. oryzae pv. oryzae phoP and hrpG genes were synthesized using the PCR method with cloned Pfu polymerase (Stratagene); mutant strains constitutively expressing these genes were generated using the pML122 vector (43). The phoP and hrpG genes were amplified from the genome of the PXO99 wild-type strain and inserted into pCR-Blunt II TOPO (Invitrogen). The inserted sequences were confirmed through sequence analysis and then cut out using the restriction enzymes NdeI and BamHI, ligated into the protein expression vector pET15-b, which had been cut with the same enzymes, and then used to transform E. coli strain BL21. The pET15-b plasmids containing either the phoP or hrpG structural gene were extracted and treated with restriction enzymes HindIII and XbaI, respectively, to obtain versions of the phoP and hrpG genes with coding sequences for six histidines at their 5′ ends. These altered fragments were then inserted into the pML122 vector to promote expression in X. oryzae pv. oryzae and introduced into PXO99P strains using electroporation. After incubation in PS broth (PSB) medium for 3 h, the cells were spread onto PS agar plates containing 15 mg/ml of gentamicin and 50 mg/ml of kanamycin. The mutant strains were confirmed using the PCR method; primer combinations specific for phoP, hrpG, and pML122 vector sequences were used as appropriate. The expression of PhoP and HrpG proteins was verified using Western blot analysis in which the His tag antibody was used for protein detection (data not shown).
Semiquantitative RT-PCR.
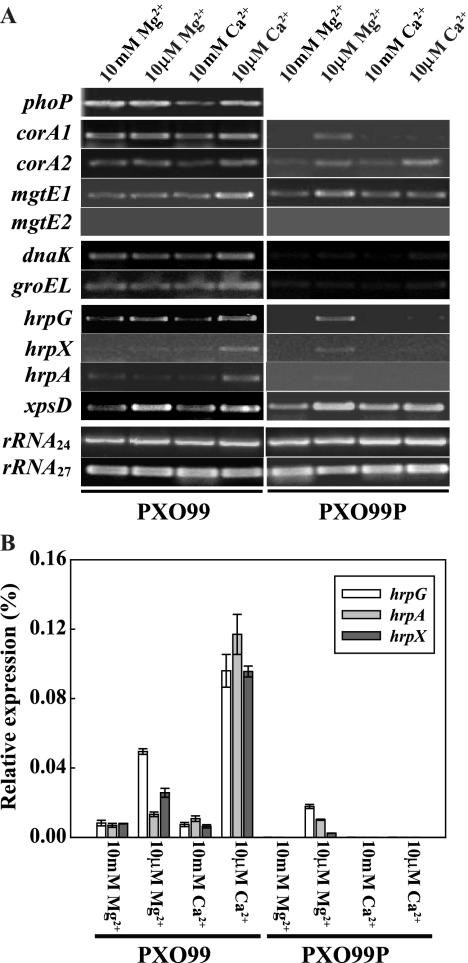
To test expression of the genes which were known to be PhoPQ regulons in bacteria that infect animals, strains PXO99 and PXO99P were cultured in M9 minimal medium containing a high (10 mM) or low (10 μM) concentration of Mg2+ or Ca2+ for 3 days at 28°C. RNA was extracted from the saturated cell cultures (≥108 CFU/ml) using TRIzol reagent following the manufacturer's instructions (Invitrogen) and then quantified using the spectrophotometric method. After the quality of the RNA was confirmed on a 1.5% agarose gel, 5 μg of each RNA sample was used for synthesis of cDNA using a cDNA synthesis kit (Invitrogen Corp., Carlsbad, CA). One microliter of the cDNA prepared from each RNA sample was then treated with RNase (DNase free) (Invitrogen) for 30 min at 37°C and used immediately thereafter as a template for PCR. The annealing temperature for each sample was determined based on the melting temperature of each primer. Other PCR conditions were as follows: denaturation at 92°C for 30 seconds, primer annealing for 30 seconds at the appropriate temperature for each primer pair, and extension at 72°C for 30 seconds. Reactions were run for 27 cycles with the exceptions of those for the hrpG, -A, and -X genes, which ran for 29 cycles. The PCR products were examined after separation through 1% agarose gels and staining with ethidium bromide. The primers were designed for amplification of short and specific parts of the targeted genes (fragments of around 400 bp); primer sequences are listed in Table 2. 16S rRNA was used as a control for this study and was amplified with 24 and 27 cycles to assure equal concentrations (rRNA24 and rRNA27 in Fig. 5). RT-PCR products were quantified using the LabImage 1D software program (Kapelan Bio-Imaging solution) after 1.0% agarose gel electrophoresis. These experiments were repeated at least three times with RNA samples which had been extracted from independently grown cultures.
FIG. 5.
(A) Gene expression analyses with X. oryzae pv. oryzae wild-type and PhoP mutant strains. RT-PCR was carried out with PXO99 and PXO99P strains cultured at high and low Mg2+ and Ca2+ concentrations. The cells were cultured in NB medium for 3 days, washed three times with sterilized and deionized water, transferred into M9 minimal medium containing different concentrations (10 mM or 10 μM) of Mg2+ and Ca2+ ions, and cultured for 2 days at 28°C. cDNA was synthesized from 5 μg of total RNA extracted from the cells, and 1 μl of the synthesized cDNA in each cell strain was used for PCR. This experiment was repeated three times each with three independent replicates. (B) hrpG, hrpA, and hrpX expression analysis using semiquantitative real-time PCR. Wild-type PXO99 and PXO99P mutant strains were cultured as described above for panel A. Five micrograms of total RNA extracted from cells cultured in each condition was subjected to one-step real-time PCR using nonspecific binding chemistry (SYBR green). Standard curves for each gene or rRNA, hrpG, hrpA, and hrpX to quantify transcripts of each gene in each condition were established by the threshold cycle values obtained against the known concentration of 10-fold serially diluted genes (rRNA, hrpG, hrpA, and hrpX) ranging from 105 to 0.001 pg per milliliter. Quantitative determination for each gene was obtained through equations computed from standard curves of each gene, and the data present relative copy numbers of each gene to the copy number of rRNA in each condition. The error bars correspond to the standard deviations from two independent replicates carried out with two sets of primers for the hrpG gene and a set of primers for the hrpA and hrpX genes.
Real-time PCR.
To confirm the differential expression of the X. oryzae pv. oryzae phoP gene in strains PXO99 and PXO99R and to assess the effect of the PhoPQ TCS on expression of hrp genes (hrpG, hrpX, and hrpA), real-time PCR using nonspecific DNA binding chemistry (SYBR green) and the Brilliant SYBR green quantitative RT-PCR master mix kit (Qiagen) was carried out. Three primer sets were designed for amplification of three regions (nt 38 to 280, 211 to 490, and 421 to 680) of the X. oryzae pv. oryzae phoP gene (DNA fragments of approximately 250 nt each). Primer sequences are listed in Table 2. For hrpG, two sets of primers were designed. For hrpA and hrpX, primers previously used for RT-PCR were used in this experiment (Table 2). PXO99 and PXO99R strains were cultured in PS medium for 3 days at 28°C (≥108 CFU/ml). RNA extraction, cDNA synthesis, and PCR methods were the same as the method described above for RT-PCR.
RESULTS
A response regulator encoded by phoP is up-regulated in the raxR gene knockout mutant strain.
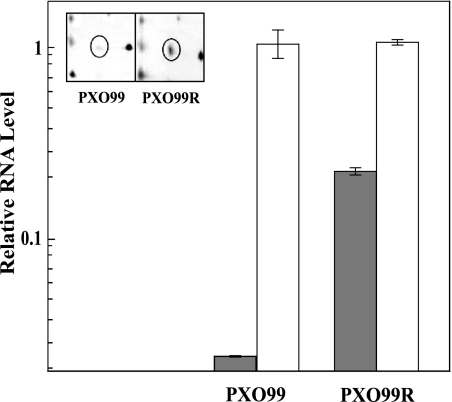
In an effort to identify new genes regulated by the RaxRH TCS, IEF and 2-DE were sequentially carried out with total protein extracts from the X. oryzae pv. oryzae wild-type strain, PXO99, and PXO99R, a strain mutated at the raxR gene locus (11). Forty proteins clearly up- or down-regulated in the mutant RaxR background relative to the wild type were selected, digested with trypsin, and then further analyzed using a MALDI spectrometer (data not shown). Peptide mass fingerprinting data of one of the proteins significantly up-regulated in the PXO99R mutant strain matched a protein annotated as PhoP in the X. oryzae pv. oryzae Korean strain KACC10331 and Xanthomonas campestris pv. campestris (ATCC 33913) translated genomes (Fig. 1, inset). Differential expression of the phoP gene at the transcriptional level was observed using real-time PCR (Fig. 1).
FIG. 1.
Differential expression of PhoP in X. oryzae pv. oryzae strains PXO99 and PXO99R. Total protein (500 μg) extracted from each strain, cultured in PSB for 3 days at 28°C, was processed using isoelectrofocusing (pH 4 to 10), followed by SDS-PAGE (12% acrylamide). IEF and 2-DE were repeated with three biological replicates and two technical replicates. The spot that represents the PhoP protein is circled on the gel photograph (inset). Differential expression of the phoP gene was confirmed using semiquantitative real-time PCR. Gray bars represent phoP RNA levels, and white bars represent rRNA (control) levels. RNA was extracted from strains PXO99 and PXO99R after culture in PS broth. cDNAs were synthesized using 5 μg of the extracted RNAs. One microliter of cDNA was used for quantitative real-time PCR with primers specific for the phoP gene. The relative RNA levels of the phoP gene and rRNA in PXO99 and PXO99R strains were calculated relative to the copy number of rRNA in PXO99. Error bars represent standard deviations from two experiments. For a control, rRNA expression in the strains was measured.
The cognate protein PhoQ (described in detail in the next section) was not identified in this screen. This may be due to a limitation of gel-based proteomics in separation of hydrophobic proteins. The PhoQ protein has two hydrophobic transmembrane domains and is likely localized at the membrane. We did, however, observe a RaxR-dependent expression pattern of phoQ with microarray analysis (PXO99 versus PXO99R [data not shown]).
X. oryzae pv. oryzae phoP is similar to the phoP gene in other species, and X. oryzae pv. oryzae phoQ carries a unique N-terminal domain.
We then cloned phoP from X. oryzae pv. oryzae. Primers were designed using the phoP gene sequences of X. oryzae pv. oryzae KACC10331 and X. campestris pv. campestris ATCC 33913 and used to carry out a PCR in which X. oryzae pv. oryzae genomic DNA was used as the template. The 453-nt DNA fragment synthesized from the PCR was then used as a probe to screen an X. oryzae pv. oryzae cosmid library (32). Three colonies were identified, and the colony expressing the strongest signal was selected for further analysis. Southern blot analysis utilizing the restriction enzymes EcoRI, SalI, PstI, and BamHI was used to determine that the cosmid contained approximately 19.3 kb of X. oryzae pv. oryzae genomic DNA. Sequence analysis of a 3.6-kb subfragment resulting from EcoRI digestion revealed an operon (GenBank accession number DQ531047) containing two open reading frames of 685 (phoP) and 1417 (phoQ) nt. The larger (PhoQ) deduced protein contains a predicted histidine kinase domain A, and the smaller (PhoP) protein contains a predicted signal receiver domain and effector domain, according to the software ScanProsite (http://expasy.org/tools/scanprosite/). These domains are characteristic of HK and RR proteins belonging to the OmpR family of TCSs (29). Alignments between X. oryzae pv. oryzae PhoP and PhoP from Pectobacterium chrysanthemi, Salmonella enterica serovar Typhimurium, and E. coli demonstrated high degrees of relatedness of up to 43% identity and 64% similarity (see Fig. S1A in the supplemental material). Among Xanthomonas strains, the amino acid sequence of the PhoP protein in strain PXO99 exhibits 100% identity to PhoP from X. oryzae pv. oryzae KACC10331 (data not shown) and from X. campestris pv. campestris ATCC 33913 (see Fig. S1A in the supplemental material). These results indicate that PhoP is highly conserved among Xanthomonas spp.
The amino acid sequence of the PhoQ protein in strain PXO99 (see Fig. S1B in the supplemental material) has up to 33% identity and 53% similarity to PhoQ proteins in the P. chrysanthemi and S. enterica serovar Typhimurium, but only in the predicted intracellular domain (from residue 205 to the C terminus) containing a histidine kinase domain A (residues 254 to 311). From the N terminus to residue 205, there is no significant identity to any known PhoQ proteins. Topological prediction software (DAS [http://www.sbc.su.se/∼miklos/DAS/maindas.html]) indicated that this N-terminal domain contains a signal sequence and a periplasmic domain (142 amino acids long) surrounded by two transmembrane domains. These domains are associated with recognition of extracellular stimuli (see Fig. S1B in the supplemental material). These results suggest that the X. oryzae pv. oryzae PhoQ protein is probably membrane localized and that it may interact with different extracellular signals than those perceived by other characterized PhoQ proteins. Alternatively, PhoQ may employ a different mechanism of signal perception.
The PhoPQ two-component system is required for AvrXA21 activity.
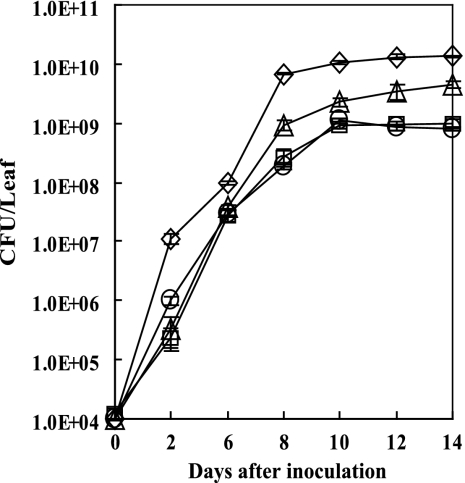
Because PhoP is known to be a component of a TCS in other organisms (52) and in light of the fact that the raxR mutation in strain PXO99R reduced but did not eliminate AvrXA21 activity in that strain (11), we hypothesized that the PhoPQ TCS, in addition to the RaxRH system, might control AvrXA21 activity. To test this hypothesis, null mutant strains for the phoP and phoQ genes (PXO99P and PXO99Q, respectively) were generated and then used to inoculate leaves of TP309-XA21 rice plants (a transgenic rice line of Oryza sativa subsp. japonica cultivar Taipei 309 carrying the Xa21 gene; resistant to PXO99) using the scissor clip method (37). Wild-type PXO99 and PXO99ST, a mutant strain lacking AvrXA21 activity previously generated in our laboratory (27), were used as controls. Slightly longer lesions developed on rice leaves inoculated with PXO99Q than on those inoculated with the PXO99 wild-type strain, but there was no significant difference between the lengths of lesions on PXO99P-inoculated versus PXO99-inoculated leaves (data not shown). Bacterial growth curves established using bacteria isolated from the inoculated rice leaves clearly showed that PXO99Q bacteria grew to a level intermediate between that of the wild-type PXO99 and PXO99ST mutant strains (Fig. 2). These results indicate that the absence of the phoQ gene reduces AvrXA21 activity. Curiously, no difference was observed in the lesion lengths (data not shown) and bacterial growth curves when comparing wild-type bacteria versus the PXO99P mutant strain in Xa21 leaves (Fig. 2), suggesting that the absence of the phoP gene either did not affect AvrXA21 activity or that it reduced overall virulence of the strain.
FIG. 2.
Bacterial growth of PXO99, PXO99Q, PXO99P, and PXO99ST strains in inoculated rice leaves (TP309-Xa21). After culture of PXO99 (circles), PXO99Q (triangles), PXO99P (squares), and PXO99ST (diamonds) strains on NB plates, cells were diluted to 1 × 108 CFU/ml and then were inoculated onto rice leaves using the scissor clip method. Bacteria were extracted from the leaves every 2 days (0, 2, 6, 8, 12, and 14 days after inoculation) and serially diluted for counting. Values represent averages ± standard deviations (error bars) of three sampled leaves per treatment. The experiment was repeated three times with more than 10 rice leaves from three individuals each time.
Virulence is impaired in strain PXO99P.
To distinguish between these two possibilities, the virulence of strain PXO99P was tested in lines not containing Xa21. For this study, we inoculated wild-type TP309 rice plants (susceptible to PXO99) with wild-type PXO99, PXO99P, or PXO99Q strains and measured lesion lengths 2 weeks later (Fig. 3A). Surprisingly, the PXO99P mutant strain caused very short lesions (average, 0.6 ± 0.1 cm) compared to that of the PXO99 wild-type strain (average, 10.1 ± 2.2 cm). In contrast, the PXO99Q mutant strain maintained virulence, although lesion lengths were slightly decreased (average, 7.9 ± 2.0 cm) compared to that of the PXO99 wild-type strain. Bacterial growth curves performed using rice leaves 2, 4, 6, 8, 10, 12, and 14 days after inoculation (Fig. 3B) confirmed the results from the lesion measurement experiment. The PXO99P strain showed a distinct reduction in bacterial growth compared to the wild type, while only a slight decrease in growth of the PXO99Q strain was observed (Fig. 3B). That PhoP contributes to X. oryzae pv. oryzae virulence was confirmed through additional inoculation experiments using PXO99P-P*, a strain of PXO99P that constitutively expresses the PhoP protein (Fig. 3A). The lack of virulence in PXO99P was completely restored in the PXO99P-P* strain. This result demonstrates that the lack of the phoP gene leads to loss of X. oryzae pv. oryzae virulence. Furthermore, this result indicates that any effect of PhoP on AvrXA21 activity would be masked by the loss of virulence in the mutant strain.
Signal transduction mediated by X. oryzae pv. oryzae PhoQ and PhoP affects cell growth under low Mg2+ and Ca2+ conditions.
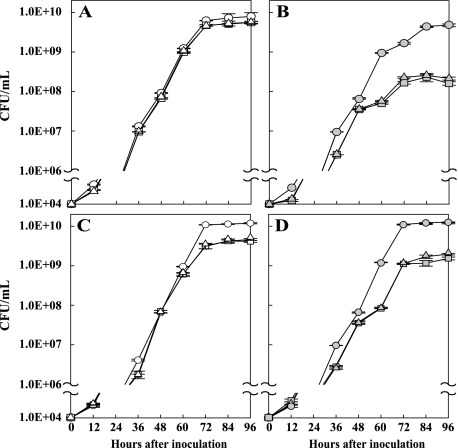
Because the PhoPQ TCS in bacteria that are pathogenic to animals respond to high and low concentrations of Mg2+ and Ca2+ ions (24), we investigated the effects of these divalent cations on X. oryzae pv. oryzae cell growth. The PXO99 (wild-type), PXO99Q, and PXO99P strains were grown in modified M9 minimal medium containing high (10 mM) or low (10 μM) concentrations of Mg2+ and Ca2+, and cell numbers were measured at 0, 12, 24, 36, 48, 60, 72, 84, and 96 h under the experimental conditions (Fig. 4). The growth of the mutant strains was diminished in both high and low Ca2+ concentrations compared to the growth of the wild-type strain (which did not depend on the concentration of this ion [Fig. 4C and D]). While the wild-type strain entered the stationary phase at a cell density of 1 × 1010, the PXO99Q and PXO99P mutant strains entered the stationary phase at cell densities of 5 × 109 (high Ca2+) and 1 × 109 (low Ca2+), respectively (Fig. 4C and D). Similarly, the growth of the mutant strains at the low Mg2+ concentration was remarkably reduced from that of the wild-type strain under the same conditions (Fig. 4B). The growth of the mutant strains at the high Mg2+ concentration, however, was not significantly different than that of the wild-type strain (Fig. 4A). These results indicate that the X. oryzae pv. oryzae PhoPQ regulatory system responds to limiting concentrations of extracellular Mg2+ and Ca2+ ions in a similar manner to the PhoPQ systems in other species.
FIG. 4.
Cell growth with high and low concentrations of Mg2+ (A and B) and Ca2+ (C and D). A single colony of each strain was cultured in NB medium for 3 days. The cells were collected by centrifugation and then washed with sterilized/deionized water three times. The cells (1 × 104) were resuspended in modified M9 minimal medium and incubated at 28°C, and cell growth was monitored at 0, 12, 24, 36, 48, 60, 72, 84, and 96 h after reincubation. The error bars correspond to the standard deviations from three independent cultures. Symbols: circles, PXO99; triangles, PXO99Q; squares, PXO99P; white symbols, high concentration (10 mM) of Mg2+ or Ca2+; gray symbols, low concentration (10 μM) of Mg2+ or Ca2+.
X. oryzae pv. oryzae PhoP regulates gene expression predominantly in response to Ca2+.
Forty different genes in S. enterica serovar Typhimurium have been reported to be regulated by PhoP based on gel-based proteomic analysis (53), and a total of more than 200 genes in S. enterica serovar Typhimurium and nonpathogenic E. coli are predicted to be regulated by PhoP (54). To test whether the X. oryzae pv. oryzae PhoP regulatory activity responds to Mg2+ and Ca2+ starvation, we analyzed the expression of 11 genes shown to be regulated by PhoP by semiquantitative RT-PCR. Expression of the phoP gene itself was tested first because expression of the two genes encoding the HK and RR in many TCSs are often autoregulated by the RR and because expression of an operon containing phoP and phoQ genes is known to be autoregulated in S. enterica serovar Typhimurium (68). Expression of the phoP gene was markedly inhibited by high Ca2+ concentration (10 mM; 29% relative to low Ca2+ concentration [10 μM] according to density quantification by the LabImage 1D software program), but not by high levels of Mg2+ (Fig. 5A). These results are consistent with phoP gene expression in X. oryzae pv. oryzae being autoregulated predominantly in response to high extracellular concentrations of Ca2+ and further suggest that the PhoPQ TCS may control the expression of other genes in response to high Ca2+ concentration.
Next, we analyzed the expression of four genes predicted to encode magnesium transporters in the genome database for X. oryzae pv. oryzae KACC10331 (http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genome&Cmd=ShowDetailView+TermToSearch=631) (Fig. 5A). The four genes were named corA1 (Gi 58583655), corA2 (Gi 58582396), mgtE1 (Gi 58580899), and mgtE2 (Gi 58580093) in this study. Using semiquantitative RT-PCR, we were able to detect expression of the corA1, corA2, and mgtE1 genes under different ion conditions, but we did not detect mgtE2 despite testing various combinations of primers and conditions (Fig. 5A). We suspect that mgtE2 may be nonfunctional or very lowly expressed in X. oryzae pv. oryzae under the conditions tested. The expression of the corA1, corA2, and mgtE1 genes was the same in high and low Mg2+ concentrations. Expression of corA1 and corA2 decreased in high Ca2+ concentrations (35 and 31% relative to low Ca2+ concentration), and conversely, expression of mgtE1 increased at low Ca2+ concentrations (2.9 times than high Ca2+ concentration). In the PXO99P strain, corA1 expression was reduced to nearly undetectable levels under high Mg2+, high Ca2+, and low Ca2+ concentrations, while expression of the gene under low Mg2+ concentrations appeared to be the same in the phoP mutant strain as in the wild type. In contrast, expression of corA2 and mgtE1 under both concentrations was not significantly different from expression of these two genes in the wild-type strain, though corA2 and mgtE1 expression were slightly decreased. The distinct exception was that mgtE1 expression was not enhanced under low Ca2+ concentrations (Fig. 5A) in the PXO99P strain. Taken together, these results indicate that PhoP directly or indirectly controls expression of each of these four genes in response to extracellular ion concentrations.
We also investigated expression of the dnaK (Gi 58581654) and groEL (Gi 58583911) genes in the wild-type PXO99 and PXO99P mutant strains under different divalent ion conditions. The proteins encoded by these genes play important roles in protein folding and promote cell proliferation and survival (28). In addition, the regulation of these genes is known to be controlled by Ca2+ concentration in Streptococcus pneumoniae (41). The expression of both of these genes was slightly higher at lower Ca2+ concentration in strain PXO99 (more than 2.5 times compared to other conditions) but strikingly repressed, down to undetectable levels, in the PXO99P strain under all conditions tested (Fig. 5A). In addition, GroEL protein also accumulated to a much higher level in PXO99 than in PXO99P based on our preliminary studies using SDS-PAGE and mass spectrometric analysis (data not shown). These results clearly indicate that the expression of these genes requires the PhoP protein and is influenced by the Ca2+ concentration.
Ca2+ and Mg2+ perception mediated by X. oryzae pv. oryzae PhoP predominantly modulates hrp gene expression.
Our finding that the virulence of strain PXO99P is impaired in rice plants suggests that one (or more) of the PhoP-regulated gene(s) encodes an X. oryzae pv. oryzae virulence factor. The few virulence factors in Xanthomonas spp. reported so far include xylanase, lipopolysaccharide (LPS), and components of the TTSS (8, 35, 39, 60, 61, 71). To investigate whether genes involved in Xanthomonas virulence are regulated by the X. oryzae pv. oryzae PhoPQ system, another set of semiquantitative RT-PCR experiments was carried out (Fig. 5A). In this experiment, PXO99 or PXO99P RNA was used as the template for cDNA synthesis. Specific primers were designed to amplify the X. oryzae pv. oryzae genes: hrpG (Gi 58581002), which encodes a critical regulator of hrp gene expression (81); xpsD (Gi 58580480), which putatively encodes a component of the type II secretion system involved in secreting xylanase and other enzymes (61); rfb303 (Gi 58581807), which putatively encodes a LPS core biosynthesis protein (76); and the vapI gene (Gi 58584090), which putatively encodes a virulence-associated protein (34). No differences in the expression of the rfb303 and vapI genes between the two strains were observed (data not shown). We found, however, that under low Mg2+ and Ca2+ conditions (10 μM for each), the transcripts of hrpG and xpsD increased in the wild-type strain (Fig. 5A). In the PXO99P strain, hrpG cDNA was undetectable except under low Mg2+ conditions, while xpsD showed wild-type levels of expression in the absence of the phoP gene. These results indicate that the PhoP protein regulates hrpG gene expression in the presence of low concentrations of Ca2+.
The HrpG protein is known to act at the beginning of a signaling cascade, activating gene transcription of hrpF through hrpA and hrpX (81). We therefore used semiquantitative RT-PCR to measure the expression of two additional X. oryzae pv. oryzae hrp genes, hrpA (hrcC in the genome database of strain KACC10331 [Gi 58579717]) and hrpX (Gi 122879096). Expression of these two genes showed significantly higher expression in low Ca2+ concentrations (10 μM), whereas other conditions (high Mg2+, low Mg2+, and high Ca2+) induced minimal expression in the wild-type strain. Expression of hrpG, hrpX, and hrpA was undetectable in the PXO99P strain except under low Mg2+ conditions (10 μM). These results suggest that the X. oryzae pv. oryzae PhoPQ TCS regulates hrp gene expression through HrpG.
To quantify the expression of the hrpG, hrpA, and hrpX genes, we used real-time PCR (Fig. 5B). The semiquantitative RT-PCR data described above (Fig. 5A) indicate that the expression pattern of the hrpA and hrpX genes, known to be targets of HrpG, differed from that of hrpG, which showed higher expression under low Mg2+ and Ca2+ concentrations. In contrast, real-time PCR revealed the same expression patterns for all three genes in the wild-type strain: higher expression under low concentrations of the ions (1.2- to 2-fold-increased expression for hrpA, threefold-increased expression for hrpX, and twofold-increased expression for hrpG) compared with growth in high Mg2+ and Ca2+ concentrations [Fig. 5A and B]). The differences in the results from the semiquantitative RT-PCR and the real-time PCR experiments may be due to the relative insensitivity of semiquantitative RT-PCR compared to real-time PCR. The observed pattern of regulation of these three hrp genes in the wild-type strain was clearly abolished in the PXO99P mutant strain (although some gene expression was detected under the low Mg2+ concentration). The expression of the three genes under low Mg2+ concentration in the PXO99P mutant strain decreased compared to that of the wild-type PXO99 strain (Fig. 5B). These results clearly indicate that X. oryzae pv. oryzae PhoP regulates hrp gene expression in response to Mg2+ and Ca2+ concentrations.
To test whether the deficiency we observed in hrp gene expression in strain PXO99P led to the loss of virulence in that strain, a mutant (PXO99P-G*) of PXO99P that constitutively expresses the HrpG protein was generated. In an inoculation experiment with TP309 (susceptible to PXO99), the development of longer lesions was evident with the HrpG-expressing strain compared to the PXO99P strain (Fig. 6); however, the HrpG-expressing strain did not complement the mutant PXO99P phenotype as fully as constitutive expression of PhoP itself (Fig. 3). Lesions that develop after inoculation of the wild-type PXO99 strain typically develop from the inoculation site, and chlorosis, symptomatic of the disease, is apparent in 2 weeks after inoculation (Fig. 6). The lesions of the strain constitutively expressing HrpG developed more slowly than those of the wild-type strain. Also, chlorosis occurred over a wide area rather than emanating only from the inoculation site (Fig. 6). While the strain constitutively expressing HrpG did not show changes in growth in PS medium or minimal medium containing high (10 mM) or low (10 μM) concentrations of Mg2+ and Ca2+ compared to the PXO99P strain (data not shown), bacterial growth curves established using rice leaves 0, 2, 4, 6, 8, 10, 12, 14, 18, and 22 days after inoculation (see Fig. S3 in the supplemental material) confirmed that the strain constitutively expressing HrpG grew to levels intermediate to those of PXO99 and PXO99P. These results indicate that PhoP is a key regulator of hrp gene expression and virulence and that additional components regulated by the PhoPQ TCS are required for complete virulence. These results are in agreement with reports indicating that the Salmonella, animal-pathogenic bacteria, PhoPQ regulatory system controls the TTSS system (1, 7).
FIG. 6.
Lesions on TP309 leaves inoculated with the PXO99, PXO99P-G*, and PXO99P strains. Each strain was grown on a PSB plate for 3 days at 28°C, diluted with sterilized and deionized water to 108 CFU/ml, and then inoculated using the scissor clip method. Lesion lengths were measured 22 days after inoculation. This picture shows the results of one representative experiment of three experiments carried out with eight individual rice plants for each strain.
Tolerance to antimicrobial peptides is weakened in both strains PXO99P and PXO99Q.
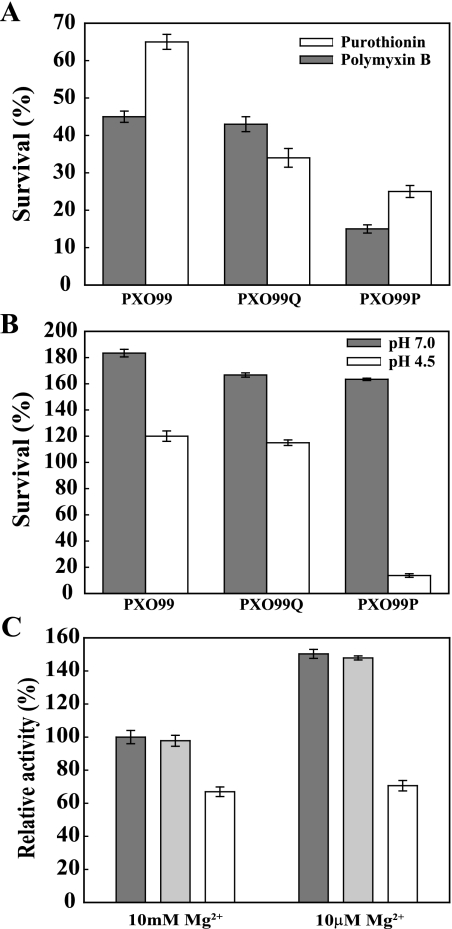
The PhoQ and PhoP proteins are known to enhance tolerance to antimicrobial peptides in pathogenic bacteria (48, 66). To determine whether the X. oryzae pv. oryzae PhoPQ system also regulates tolerance to antimicrobial peptides, we tested the tolerance of strains PXO99, PXO99Q, and PXO99P to polymyxin B, a naturally occurring cyclic AMP found in Bacillus polymyxa (72), and purothionin, an antimicrobial peptide present in plants (23). We added various concentrations (0.25, 0.5, 1, and 2 mg/ml) of the antimicrobial peptides to cultures of PXO99, PXO99Q, and PXO99P and measured cell population size 6, 12, and 18 h later (Fig. 7A; also data not shown). Our results indicate that wild-type X. oryzae pv. oryzae is moderately tolerant to these antimicrobial peptides. The addition of 250 μg/ml of polymyxin B and purothionin into the culture medium reduced the PXO99 cell population to 47% and 28%, respectively, after 6 h (data not shown). When 500 μg/ml of the same antimicrobial peptides was added to the culture medium, the cell populations declined even more to 43% and 65%, respectively (Fig. 7A). Under the same conditions, populations of the phoQ knockout strain were reduced to 42% and 34%, respectively, and populations of the phoP knockout strain showed the most dramatic decreases to 15% and 21%, respectively. These results indicate that X. oryzae pv. oryzae PhoP contributes to tolerance to both antimicrobial peptides tested, while PhoQ provides tolerance mainly to purothionin.
FIG. 7.
Viability of PXO99, PXO99Q, and PXO99P strains after treatment with antimicrobial peptides (A) and acid (B). (A) Polymyxin B (0.5 mg/ml) or purothionin (0.5 mg/ml) was added to saturated cultures of PXO99, PXO99Q, and PXO99P strains and then incubated at 28°C with shaking. To measure survival, after 6 h, 10 μl of each culture was dropped onto PS agar plates and incubated at 28°C, and the colonies were counted after 3 days. (B) PXO99, PXO99Q, and PXO99P strains were inoculated into PS broth adjusted to pH 7.0 or pH 4.5 with acetic acid. After 4 h, the cell population was measured using a UV-visible spectrophotometer at 600 nm. The error bars in panels A and B correspond to the standard deviations from three independent cultures for each study. (C) Activity assay for X. oryzae pv. oryzae nonspecific acid phosphatase. Saturated cultures of PXO99 (dark gray), PXO99Q (light gray), and PXO99P (white) strains were reinoculated into modified M9 minimal medium containing 10 mM or 10 μM MgSO4 after three washes with sterilized, deionized water and incubated at 28°C for 2 days. After a 30-min reaction with the extracted total protein (50 μg) and enzyme reaction mixture, the enzymatic activity of nonspecific acid phosphatase was measured by a UV-visible spectrophotometer at 405 nm. The enzymatic activity of each reaction mixture relative to the activity of the PXO99 wild-type strain at a high concentration of Mg2+ is shown. The error bars correspond to the standard deviations from three independent replicates.
At higher concentrations (1 and 2 mg/ml) and longer incubations (>12 h), all strains succumbed to the antimicrobial peptides, and all cells died (data not shown). These results indicate that X. oryzae pv. oryzae PhoPQ is involved in tolerance to antimicrobial peptides but that an HK other than PhoQ may also be involved.
Strain PXO99P is more susceptible to acidic conditions.
Because the PhoPQ regulatory system in other bacterial species is known to recognize not only the divalent cations Mg2+ and Ca2+ but also to provide tolerance to acidic conditions (6), we tested the tolerance of the PXO99, PXO99P, and PXO99Q strains to acidic conditions. Cells were cultured under neutral pH conditions (pH 7.0) and then transferred to medium with the same or lower pH (pH 7.0, 5.5, 4.5, 3.5, and 3.0). The viability of the strains under the different pH conditions was then measured 4 h after transfer. All of the strains showed a slight decrease in percent survival at pH 5.5 and a dramatic decline at pH 3.5 and 3.0, conditions under which cell populations of all strains were reduced to less than 0.5% (data not shown). However, after 4 h at pH 4.5, a clear difference in tolerance to acidic conditions was evident among the strains (Fig. 7B). The populations of both the wild-type strain and the PXO99Q mutant strain were 120% of what they had been at the time of transfer to pH 4.5. In contrast, the population of the PXO99P mutant strain declined markedly in the acidic medium to less than 20% of its size at the time of transfer (Fig. 7B). These results indicate that the PhoPQ TCS provides tolerance to acidic conditions and suggest that PhoQ function may be partially redundant in X. oryzae pv. oryzae, because mutations in PhoP but not PhoQ eliminated tolerance to acidic conditions.
Activity of a nonspecific acid phosphatase is regulated by the PhoPQ system.
Expression of the phoN gene, which encodes a nonspecific acid phosphatase (EC 3.1.3.2), is associated with Mg2+ sensitivity mediated by the PhoPQ TCS in S. enterica (36, 45). We therefore explored nonspecific acid phosphatase activity in the PXO99, PXO99Q, and PXO99P strains cultured in high and low Mg2+ conditions (Fig. 7C). In the wild-type X. oryzae pv. oryzae strain, enzyme activity was higher (approximately 1.5 times) at low Mg2+ concentrations than at high Mg2+ concentrations, suggesting that a nonspecific acid phosphatase(s) has increased activity at a low concentration of Mg2+ ions. Enzyme activity in PXO99Q was similar to that in PXO99 under both low and high Mg2+ ion concentrations (Fig. 7C). However, enzymatic activity in the PXO99P mutant strain was approximately 60% of the activity level measured in PXO99 and PXO99Q at both high and low Mg2+ concentrations (Fig. 7C). The effects of low Ca2+ concentrations on enzyme activity were similarly tested and resulted in a similar pattern among the three bacterial strains (data not shown). These results suggest that the PhoP protein modulates the expression of a nonspecific acid phosphatase(s) in X. oryzae pv. oryzae.
DISCUSSION
Two-component regulatory systems, present in both gram-negative and gram-positive bacteria, have been shown to regulate a wide variety of biological processes in response to environmental fluctuations. Such processes include the production of bacteriocins, expression of various toxins, and production of other proteins related to virulence and pathogenicity (56). In prokaryotes, TCSs are generally characterized by a simple phosphoryl transfer scheme (70). The histidine kinase carries a sensor domain that is responsible for detection of external stimuli and regulates the kinase activity of the HK. The HK catalyzes ATP-dependent autophosphorylation of a specific histidine residue within its dimerization domain. A cognate response regulator, the other component in this TCS then catalyzes transfer of the phosphoryl group from the phospho-histidine to one of its own aspartate residues in its conserved regulatory (or receiver) domain. This phosphorylation induces a conformational change in the C-terminally located effector domain, which results in its activation and generates the output response of the signaling pathway. Although detailed information on the mechanism of DNA binding is lacking, it has been well established that the majority of RRs function as transcription factors (70).
One of the best characterized examples of a TCS is the one mediated by PhoQ and PhoP (PhoPQ). Since this TCS was first identified in Salmonella enterica serovar Typhimurium (52), it has been extensively studied in S. enterica serovar Typhimurium, Neisseria meningitidis, and Yersinia pseudotuberculosis (50, 52, 63), but it is little known in phytopathogenic bacteria. The name Pho was adopted because the regulatory system controlled expression of a nonspecific acid phosphatase, and it was therefore thought to be involved in phosphate metabolism (38). It is now appreciated, however, that the system regulates a range of genes and, in particular, determines virulence via sensing of divalent cations, such as Mg2+, Ca2+, and Mn2+, while other divalent cationic ions, including Ni2+, Cu2+, and Ba2+, do not affect the function of this TCS (24). The extracellular ions are perceived in the periplasmic domain of the PhoQ protein. In five gram-negative species, S. enterica, E. coli K-12, P. aeruginosa, Erwinia carotovora, and Providencia stuartii, the periplasmic domain has conserved residues necessary for responding to Mg2+ and an acidic cluster (residues 145 to 148) required for response to Mg2+ (12). The domain also is known to have dual binding sites for Mg2+ and Ca2+, suggesting that the mechanism of signal transduction in response to these ions is different (62). Furthermore, a recent paper by Bader et al. (5) indicates that recognition of peptides by PhoQ triggers pathogen virulence.
In this paper we show that the X. oryzae pv. oryzae PhoPQ TCS is negatively regulated by a previously characterized TCS called RaxRH (11). The PhoP protein is highly expressed in the PXO99R mutant strain lacking the RaxRH TCS (Fig. 1). Impairment of the cognate sensor protein, PhoQ, in another mutant strain, PXO99Q, partially reduced AvrXA21 activity in inoculation experiments with a rice line carrying XA21 (Fig. 2). We could not test for AvrXA21 activity specifically in PXO99P because that strain had lost virulence. These results suggest that, in addition to RaxRH, the TCS mediated by PhoPQ is also required for AvrXa21 activity and that these two TCSs might cooperate with regard to AvrXA21 regulation.
Precedents for cooperating TCS phosphotransmission schemes have been reported. For example, two RRs can interact with one HK (e.g., CheA to CheB and CheY [46]), or one RR can interact with two HKs (e.g., ArcB and CpXA to ArcA [33]), or two HKs can control two RRs (46). In addition to these phosphotransmission schemes, it has been reported that a protein induced by a TCS can control the stability of the phosphorylated RR (83). These reports suggest that rather than operating independently to sense and respond to different environmental signals, different TCSs closely coordinate their activities in order to rapidly and effectively respond to environmental changes.
It is not yet clear whether the RaxRH and PhoPQ TCSs are sufficient for regulation of AvrXA21 activity or whether other factors are also needed. In inoculation experiments with double knockout mutant strains for both phoQ and raxH and for phoQ and raxR, partial AvrXA21 activity was retained (data not shown). The inability to completely knock out AvrXa21 activity in these double mutants suggests that additional genes might also mediate AvrXA21 activity. To obtain a clear answer to this question, we would need to test AvrXA21 activity (44) with a double knockout mutant strain for both RRs, RaxR and PhoP. However, despite many attempts, we were not able to isolate such a mutant, suggesting that the lack of both RRs, RaxR and PhoP, is lethal for X. oryzae pv. oryzae.
We have previously shown that RaxRH controls AvrXA21 activity by regulation of rax genes. Our preliminary experiments to test whether raxSTAB expression is also regulated by PhoP under limiting concentrations of Mg2+ and Ca2+ indicate that PhoPQ does not regulate raxSTAB expression (see Fig. S2 in the supplemental material). Although it is not yet clear why the X. oryzae pv. oryzae PhoPQ TCS, negatively regulated by the RaxRH TCS, is required for AvrXA21 activity, a possible explanation is that the X. oryzae pv. oryzae PhoPQ TCS may directly regulate AvrXA21 expression under certain conditions independent of RaxRH regulation of rax gene expression. We are now testing this possibility.
Our results indicate that X. oryzae pv. oryzae PhoP is a critical component of X. oryzae pv. oryzae virulence because strains lacking phoP lose virulence (Fig. 3). As we have also found that tolerance to antimicrobial peptides is weakened in the phoP and phoQ knockout strains, this sensitivity may be a key aspect of the lack of virulence of the phoP mutant. The mechanism for PhoP-controlled virulence in X. oryzae pv. oryzae may be similar to that observed for animal pathogens (52). It is well-known that the PhoPQ TCS in bacteria pathogenic to animals is activated by antimicrobial peptides, α-helical peptides, and aminoglycosides (4, 19, 49). The PhoPQ TCS in the phytopathogen P. chrysanthemi, the only example in phytopathogenic bacteria, is similarly activated by antimicrobial peptides (47, 48). Antimicrobial peptides provide a first line of defense against invading microbes in both plants and animals (84). These peptides are up to 15 to 40 amino acids in length and act mainly at the cell membrane (10). A major component of the outer membrane in gram-negative bacteria, LPSs, is a target of these antimicrobial peptides. The PhoPQ regulatory system in S. enterica serovar Typhimurium, for example, posttranscriptionally activates the products of ugtL and pmrA that, in turn, control lipid A modifications in LPSs (66). The modified LPS in the outer membrane promotes bacterial survival by increasing their resistance to antimicrobial peptides (20). Although no rice antimicrobial peptide has yet been characterized, the rice genome does encode a cysteine-rich peptide called antimicrobial peptide MBP-1 (http://rice.tigr.org/tdb/e2k1/osa1/ca/gene_fams/22_36.shtml). If X. oryzae pv. oryzae can respond to this rice antimicrobial peptide, the absence of PhoP may contribute to the compromised virulence of rice.
Similar to what has been discovered for the role of PhoPQ in animal-pathogenic bacteria, we found that X. oryzae pv. oryzae requires PhoPQ to tolerate acidic conditions (Fig. 7B). This result is interesting in light of the fact that X. oryzae pv. oryzae is confronted with a change in acidic conditions (pH 4.5 to 6.5) after invasion of its host, rice (30). In order for the bacterium to optimize its entire metabolism to survive and proliferate under fluctuating pH, tolerance to acidic conditions is required.
The observation that the X. oryzae pv. oryzae PhoPQ TCS provides resistance to antimicrobial peptides and tolerance to acidic conditions presents a possible explanation for the question: why does the HrpG overexpression mutant not restore complete virulence? Although the overexpressed HrpG protein activates hrp gene expression, the absence of resistance to antimicrobial peptides and to acidic conditions of the mutant strain, PXO99P-G*, restricts full restoration of virulence by complementary HrpG.
Regulation of the components comprising the TTSS is known to involve PhoPQ TCSs in animal-pathogenic bacteria and is a factor critical for pathogenicity. For example, Salmonella PhoP controls expression of the genes encoding the SsrB responsive regulator and SpiR sensor protein of the SsrB/SpiR regulatory system which, in turn, regulates the Salmonella pathogenicity island 2 (SPI-2) (7), a large virulence locus encoding a TTSS. In this case, control of TTSS components by PhoP protein is indirect, and Mg2+ deprivation and phosphate starvation are also required to induce SPI-2 (17). In addition, a PhoP-activated operon in SPI-1 composed of two genes, orgB and orgC in S. enterica, which encode a protein that interacts with InvC ATPase and a TTSS effector protein, respectively, were recently identified (1).
X. oryzae pv. oryzae PhoP also appears to control TTSS regulation, although in X. oryzae pv. oryzae, unlike animal pathogens, this control is exerted through regulation of hrpG gene expression (Fig. 5 and 6). In Xanthomonas spp. and R. solanacearum, HrpG activates hrpX and hrpA expression and then HrpX regulates the expression of the hrpB to hrpF genes, which encode components of the TTSS and several effector proteins. Our results reveal that in X. oryzae pv. oryzae, hrpG, hrpA, and hrpX showed similar expression patterns in response to various levels of ions (Fig. 5A and B). Interestingly, we have found that all 23 hrp and hrp-related genes, including hrpG, hrpA, and hrpX, are down-regulated in RaxR overexpression strains and up-regulated in RaxR knockout strains (data not shown). These results further support the model that X. oryzae pv. oryzae hrp gene expression is under control of PhoP, which is in turn, negatively regulated by the RaxRH TCS (Fig. 8).
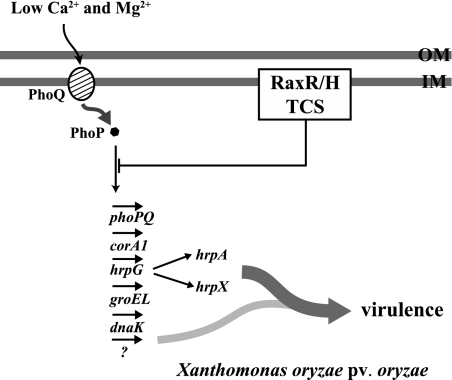
FIG. 8.
Model for integrated circuitry controlling extracellular sensing and response in X. oryzae pv. oryzae. PhoP, which is negatively regulated by RaxR, controls expression of the corA1, groEL, dnaK, and hrp genes. The PhoPQ TCS, also required for AvrXA21 activity in addition to RaxRH, governs X. oryzae pv. oryzae virulence. OM, outer membrane; IM, inner membrane.
Although the regulatory control exerted by the HrpG protein on hrp gene expression has been well studied, the regulation of hrpG itself remains poorly understood. HrpG is considered an orphan regulator because, until now, a cognate sensor protein responding to host conditions had not yet been identified. Our finding that X. oryzae pv. oryzae PhoP controls HrpG and the HrpG-dependent response mainly by the Ca2+ concentration suggests that the PhoPQ TCS regulates downstream hrp genes and TTSS. Interestingly, the Ca2+ concentration is about 100 nM in the plant cytoplasm under normal growth conditions, similar to the conditions used to test the PhoP-mediated HrpG-dependent response to Ca2+ in our studies (82). Although to our knowledge the Ca2+ concentration in extracellular spaces or xylem has not yet been reported, it is possible that low concentrations of Ca2+ in the rice xylem can induce hrp gene expression and the TTSS via PhoPQ TCS. The regulation of hrp gene expression, however, is likely not a simple circuit. A recent paper reports that a putative transcriptional regulator gene, named trh (transcriptional regulator for hrp), is involved in expression of HrpG in X. oryzae pv. oryzae (74). Putting these results together, the orphan regulator HrpG might function as part of multiple regulatory systems rather than interacting with a single cognate HK that senses external stimuli.
Although the X. oryzae pv. oryzae PhoPQ TCS shares many characteristics with animal-pathogenic bacteria, there are several important differences. First, the periplasmic domain of X. oryzae pv. oryzae PhoQ is different in sequence from that of other genera, including the phytopathogen Pectobacterium (47). This difference presents the possibilities that X. oryzae pv. oryzae PhoPQ signal transduction might be stimulated by a different extracellular molecule(s) or that the PhoQ binding to the extracellular signal occurs in a different manner. The X. oryzae pv. oryzae PhoQ protein sequence is highly conserved among Xanthomonas spp., including X. campestris pv. campestris, X. axonopodis pv. citri, and X. axonopodis pv. vesicatoria.
Gene expression analyses carried out with X. oryzae pv. oryzae wild-type and PhoP mutant strains revealed another unusual characteristic of the X. oryzae pv. oryzae PhoPQ system. In animal-pathogenic bacteria, regulation of gene expression by PhoP is known to be sensitive to Ca2+ and Mg2+. Interestingly, regulation by PhoP in X. oryzae pv. oryzae is predominantly sensitive to Ca2+, although both ions have a regulatory effect on the genes (Fig. 5). That is, most of the regulated genes still exhibited expression at low Mg2+ concentrations in strain PXO99P. In light of these results, we suggest that Xanthomonas may have another system for responding to extracellular Mg2+ concentrations and the system might cooperate with PhoPQ TCS.
The discrepancy between the phenotypes of strains PXO99Q and PXO99P is another functional difference between the PhoPQ regulatory systems of X. oryzae pv. oryzae and those characterized from other bacteria. As is well-known, TCSs are generally activated through recognition of extracellular stimuli by a sensor protein, an HK, followed by activation of a responsive regulator, RR. On the basis of this simple signaling scheme, one would expect that knockout strains of the HK and RR would have the same phenotype, the same degree of sensitivity in all characteristics. This is, in fact, the case for the PhoPQ systems of bacteria pathogenic to animals. In X. oryzae pv. oryzae, however, the sensitivity of the phoQ and phoP knockout strains to antimicrobial peptides, acidic conditions, and their requirements for activity of a nonspecific acid phosphatase was curiously different for each strain (Fig. 7). A possible explanation is that the X. oryzae pv. oryzae PhoPQ TCS may have an unusual phosphotransmission scheme in which another HK is able to share the PhoP protein for signal transduction. A precedent for this has been reported; ArcA and CpxA share the ArcB RR protein in E. coli (33). If PhoP can share a sensor, this would explain why defects in the PhoQ protein in strain PXO99Q could be partially complemented.
Figure 8 shows a model for regulation of virulence by the X. oryzae pv. oryzae PhoPQ TCS. The PhoPQ TCS is negatively regulated by the RaxRH TCS and controls gene expression in response to low concentrations of extracellular Ca2+ and Mg2+. These genes include corA1, a putative Mg2+ transporter, as well as groEL and dnaK, genes involved in protein folding, cell proliferation/survival (28), and self regulation. The X. oryzae pv. oryzae PhoPQ TCS also activates hrp gene transcription through the orphan response regulator HrpG, a key regulator of hrp and TTSS effector gene expression. Our results also demonstrate that the X. oryzae pv. oryzae PhoPQ TCS is critical for survival under host conditions. The TCS confers resistance to antimicrobial peptides and tolerance to an acidic environment, conditions that X. oryzae pv. oryzae likely confronts upon entry into a rice plant. These results provide important data for understanding the integrated regulatory system that the bacterium utilizes to respond to environmental fluctuations.
Our previous results suggest that AvrXA21 is likely a small sulfated peptide secreted by the type I system and that AvrXA21 regulates the rax system via its recognition by the RaxRH TCS in a density-dependent manner (44). In support of this hypothesis, we have shown that AvrXA21 is necessary for density-dependent expression of rax genes. These results, combined with the results in this paper, lead to the hypothesis that AvrXa21 triggers a transition from a quiescent or epiphytic state to an invasive or pathogenic state in response to changing extracellular conditions sensed by the two TCSs. This hypothesis would explain why the PhoPQ TCS, which triggers expression of a set of genes, including the hrp genes, through the negative regulation of RaxRH, is also required for AvrXA21 activity. Because X. oryzae pv. oryzae must monitor population size under changing conditions, an integrated and flexible response system would be desirable. In this model, the PhoPQ TCS controls both hrp gene and AvrXA21 gene expression directly. As cell population increases inside the cell, AvrXA21 would accumulate. Sensing of AvrXa21 by the RaxRH TCS would activate another set of genes (including rax genes) required for a second stage of infection. In support of this hypothesis, Daniels et al. report that QS controls both swarming motility in the macerating pathogen Rhizobium etli (15) and tissue maceration of the host plant. These processes are critical to successful infection and also control the ability of R. etli to trigger the hypersensitive response in nonhosts (67). Further studies to mechanistically elucidate the relationship between the integrated circuitry controlled by these two TCSs, RaxRH and PhoPQ, for X. oryzae pv. oryzae virulence and avirulence are in progress.
Supplementary Material
Acknowledgments
We thank Belinda Martineau, Laura Bartley, Rebecca Bart, and Adam Bogdanove for helpful comments and critical reading of the manuscript.
This work was financially supported by USDA grants 2006-01888 and 2007-01531 and NIH grant GM55962. S.-W. L. was partially supported by a grant from the SRC program of MOST/KOSEF (R11-2000-081) through the Plant Metabolism Research Center, Kyung Hee University, Suwon, Republic of Korea. S.-E.L. and K.-S.J. were partially supported by a grant (20050401-034-743-007-04-00) from Bio-Green 21 program, Rural Development Administration, Republic of Korea.
Footnotes
Published ahead of print on 18 January 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aguirre, A., M. L. Cabeza, S. V. Spinelli, M. McClelland, E. Garcia Vescovi, and F. C. Soncini. 2006. PhoP-induced genes within Salmonella pathogenicity island 1. J. Bacteriol. 1886889-6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldon, D., B. Brito, C. Boucher, and S. Genin. 2000. A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 192304-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley, New York, NY.
- 4.Bader, M. W., W. W. Navarre, W. Shiau, H. Nikaido, J. G. Frye, M. McClelland, F. C. Fang, and S. I. Miller. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50219-230. [DOI] [PubMed] [Google Scholar]
- 5.Bader, M. W., S. Sanowar, M. E. Daley, A. R. Schneider, U. Cho, W. Xu, R. E. Klevit, H. Le Moual, and S. I. Miller. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122461-472. [DOI] [PubMed] [Google Scholar]
- 6.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 1802409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bijlsma, J. J., and E. A. Groisman. 2005. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol. 5785-96. [DOI] [PubMed] [Google Scholar]
- 8.Braun, S. G., A. Meyer, O. Holst, A. Puhler, and K. Niehaus. 2005. Characterization of the Xanthomonas campestris pv. campestris lipopolysaccharide substructures essential for elicitation of an oxidative burst in tobacco cells. Mol. Plant-Microbe Interact. 18674-681. [DOI] [PubMed] [Google Scholar]
- 9.Brito, B., M. Marenda, P. Barberis, C. Boucher, and S. Genin. 1999. prhJ and hrpG, two new components of the plant signal-dependent regulatory cascade controlled by PrhA in Ralstonia solanacearum. Mol. Microbiol. 31237-251. [DOI] [PubMed] [Google Scholar]
- 10.Bulet, P., R. Stocklin, and L. Menin. 2004. Antimicrobial peptides: from invertebrates to vertebrates. Immunol. Rev. 198169-184. [DOI] [PubMed] [Google Scholar]
- 11.Burdman, S., Y. Shen, S.-W. Lee, Q. Xue, and P. Ronald. 2004. RaxH/RaxR: a two-component regulatory system in Xanthomonas oryzae pv. oryzae required for AvrXa21 activity. Mol. Plant-Microbe Interact. 17602-612. [DOI] [PubMed] [Google Scholar]
- 12.Chamnongpol, S., M. Cromie, and E. A. Groisman. 2003. Mg2+ sensing by the Mg2+ sensor PhoQ of Salmonella enterica. J. Mol. Biol. 325795-807. [DOI] [PubMed] [Google Scholar]
- 13.Charles, T. C., S. G. Jin, and E. W. Nester. 1992. Two-component sensory transduction systems in phytobacteria. Annu. Rev. Phytopathol. 30463-484. [DOI] [PubMed] [Google Scholar]
- 14.Choi, S. H., S. W. Lee, S. S. Han, D. K. Lee, and T. H. Noh. 2003. Identification of durable resistance genes against Xanthomonas oryzae pv. oryzae in Korea. Kor. J. Breeding 35283-288. [Google Scholar]
- 15.Daniels, R., S. Reynaert, H. Hoekstra, C. Verreth, J. Janssens, K. Braeken, M. Fauvart, S. Beullens, C. Heusdens, I. Lambrichts, D. E. De Vos, J. Vanderleyden, J. Vermant, and J. Michiels. 2006. Quorum signal molecules as biosurfactants affecting swarming in Rhizobium etli. Proc. Natl. Acad. Sci. USA 10314965-14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dardick, C., and P. Ronald. 2006. Plant and animal pathogen recognition receptors signal through non-RD kinase. PLoS Pathog. 2e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 311759-1773. [DOI] [PubMed] [Google Scholar]
- 18.Dow, J. M., L. Crossman, K. Findlay, Y.-Q. He, J. X. Feng, and J.-L. Tang. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 10010995-11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst, R. K., T. Guina, and S. I. Miller. 1999. How intracellular bacteria survive: surface modifications that promote resistance to host innate immune responses. J. Infect. Dis. 179(Suppl. 2)S326-S330. [DOI] [PubMed] [Google Scholar]
- 20.Ernst, R. K., T. Guina, and S. I. Miller. 2001. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 31327-1334. [DOI] [PubMed] [Google Scholar]
- 21.Fenselau, S., and U. Bonas. 1995. Sequence and expression analysis of the hrpB pathogenicity operon of Xanthomonas campestris pv. vesicatoria which encodes eight proteins with similarity to components of the Hrp, Ysc, Spa, and Fli secretion systems. Mol. Plant-Microbe Interact. 8845-854. [DOI] [PubMed] [Google Scholar]
- 22.Furutani, A., S. Tsuge, K. Ohnishi, Y. Hikichi, T. Oku, K. Tsuno, Y. Inoue, H. Ochiai, H. Kaku, and Y. Kubo. 2004. Evidence for HrpXo-dependent expression of type II secretory proteins in Xanthomonas oryzae pv. oryzae. J. Bacteriol. 1861374-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Olmedo, F., A. Molina, J. M. Alamillo, and P. Rodriguez-Palenzuela. 1998. Plant defense peptides. Biopolymers 47479-491. [DOI] [PubMed] [Google Scholar]
- 24.Garcia Vescovi, E., F. C. Soncini, and E. A. Groisman. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84165-174. [DOI] [PubMed] [Google Scholar]
- 25.Genin, S., B. Brito, T. P. Denny, and C. Boucher. 2005. Control of the Ralstonia solanacearum type III secretion system (Hrp) genes by the global virulence regulator PhcA. FEBS Lett. 5792077-2081. [DOI] [PubMed] [Google Scholar]
- 26.Reference deleted.
- 27.Goes da Silva, F., Y. Shen, C. Dardick, S. Burdman, R. Yadav, P. Sharma, and P. Ronald. 2004. Components of a type I secretion system and a sulfotransferase-like protein are required for the Xa21 receptor kinase mediated defense response. Mol. Plant-Microbe Interact. 17593-601. [DOI] [PubMed] [Google Scholar]
- 28.Gottesman, S., S. Wickner, and M. R. Maurizi. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev. 11815-823. [DOI] [PubMed] [Google Scholar]
- 29.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41139-227. [DOI] [PubMed] [Google Scholar]
- 30.Grignon, C., and H. Sentenac. 1991. pH and ionic conditions in the apoplast. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42103-128. [Google Scholar]
- 31.Gurlebeck, D., F. Thieme, and U. Bonas. 2006. Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163233-255. [DOI] [PubMed] [Google Scholar]
- 32.Hopkins, C. M., F. F. White, S. H. Choi, A. Guo, and J. E. Leach. 1992. Identification of a family of avirulence genes from Xanthomonas oryzae pv. oryzae. Mol. Plant-Microbe Interact. 5451-459. [DOI] [PubMed] [Google Scholar]
- 33.Iuchi, S., D. Furlong, and E. C. Lin. 1989. Differentiation of arcA, arcB, and cpxA mutant phenotypes of Escherichia coli by sex pilus formation and enzyme regulation. J. Bacteriol. 1712889-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain, S., B. R. Bloom, and M. K. Hondalus. 2003. Deletion of vapA encoding virulence associated protein A attenuates the intracellular actinomycete Rhodococcus equi. 50115-128. [DOI] [PubMed] [Google Scholar]
- 35.Jha, G., R. Rajeshwari, and R. V. Sonti. 2007. Functional interplay between two Xanthomonas oryzae pv. oryzae secretion systems in modulating virulence on rice. Mol. Plant-Microbe Interact. 2031-40. [DOI] [PubMed] [Google Scholar]
- 36.Kasahara, M., A. Nakata, and H. Shinagawa. 1991. Molecular analysis of the Salmonella typhimurium phoN gene, which encodes nonspecific acid phosphatase. J. Bacteriol. 1736760-6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kauffman, H. E., A. P. K. Reddy, S. P. Y. Hsieh, and S. D. Merca. 1973. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57537-541. [Google Scholar]
- 38.Kier, L. D., R. M. Weppelman, and B. N. Ames. 1979. Regulation of nonspecific acid phosphatase in Salmonella: phoN and phoP genes. J. Bacteriol. 138155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kingsley, M. T., D. W. Gabriel, G. C. Marlow, and P. D. Roberts. 1993. The opsX locus of Xanthomonas campestris affects host range and biosynthesis of lipopolysaccharide and extracellular polysaccharide. J. Bacteriol. 1755839-5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koebnik, R., A. Kruger, F. Thieme, A. Urban, and U. Bonas. 2006. Specific binding of the Xanthomonas campestris pv. vesicatoria AraC-type transcriptional activator HrpX to plant-inducible promoter boxes. J. Bacteriol. 1887652-7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon, H. Y., S. N. Kim, S. N. Pyo, and D. K. Rhee. 2005. Ca2+-dependent expression of the CIRCE regulon in Streptococcus pneumoniae. Mol. Microbiol. 55456-468. [DOI] [PubMed] [Google Scholar]
- 41a.Labes, M., A. Puhler, and S. Reinhard. 1990. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for Gram-negative bacteria. Gene 8937-46. [DOI] [PubMed] [Google Scholar]
- 42.Lee, B. M., Y. J. Park, D. S. Park, H. W. Kang, J. G. Kim, E. S. Song, I. C. Park, U. H. Yoon, J. H. Hahn, B. S. Koo, G. B. Lee, H. Kim, H. S. Park, K. O. Yoon, J. H. Kim, C. H. Jung, N. H. Koh, J. S. Seo, and S. J. Go. 2005. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, S.-W., and P. C. Ronald. 2006. Marker-exchange mutagenesis and complementation strategies for the gram-negative bacteria Xanthomonas oryzae pv. oryzae. Methods Mol. Biol. 35411-18. [DOI] [PubMed] [Google Scholar]
- 44.Lee, S. W., S. W. Han, L. E. Bartley, and P. C. Ronald. 2006. Unique characteristics of Xanthomonas oryzae pv. oryzae AvrXa21 and implications for plant innate immunity. Proc. Natl. Acad. Sci. USA 10318395-18400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lejona, S., A. Aguirre, M. L. Cabeza, E. Garcia Vescovi, and F. C. Soncini. 2003. Molecular characterization of the Mg2+-responsive PhoP-PhoQ regulon in Salmonella enterica. J. Bacteriol. 1856287-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, J., R. V. Swanson, M. I. Simon, and R. M. Weis. 1995. The response regulators CheB and CheY exhibit competitive binding to the kinase CheA. Biochemistry 3414626-14636. [DOI] [PubMed] [Google Scholar]
- 47.Llama-Palacios, A., E. Lopez-Solanilla, C. Poza-Carrion, F. Garcia-Olmedo, and P. Rodriguez-Palenzuela. 2003. The Erwinia chrysanthemi phoP-phoQ operon plays an important role in growth at low pH, virulence and bacterial survival in plant tissue. Mol. Microbiol. 49347-357. [DOI] [PubMed] [Google Scholar]
- 48.Llama-Palacios, A., E. Lopez-Solanilla, and P. Rodriguez-Palenzuela. 2005. Role of the PhoP-PhoQ system in the virulence of Erwinia chrysanthemi strain 3937: involvement in sensitivity to plant antimicrobial peptides, survival at acid pH, and regulation of pectolytic enzymes. J. Bacteriol. 1872157-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macfarlane, E. L., A. Kwasnicka, M. M. Ochs, and R. E. Hancock. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34305-316. [DOI] [PubMed] [Google Scholar]
- 50.Marceau, M., F. Sebbane, F. Ewann, F. Collyn, B. Lindner, M. A. Campos, J. A. Bengoechea, and M. Simonet. 2004. The pmrF polymyxin-resistance operon of Yersinia pseudotuberculosis is upregulated by the PhoP-PhoQ two-component system but not by PmrA-PmrB, and is not required for virulence. Microbiology 1503947-3957. [DOI] [PubMed] [Google Scholar]
- 51.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110303-314. [DOI] [PubMed] [Google Scholar]
- 52.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 865054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller, S. I., and J. J. Mekalanos. 1990. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J. Bacteriol. 1722485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monsieurs, P., S. De Keersmaecker, W. W. Navarre, M. W. Bader, F. De Smet, M. McClelland, F. C. Fang, B. De Moor, J. Vanderleyden, and K. Marchal. 2005. Comparison of the PhoPQ regulon in Escherichia coli and Salmonella typhimurium. J. Mol. Evol. 60462-474. [DOI] [PubMed] [Google Scholar]
- 55.Mudgett, M. B. 2005. New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56509-531. [DOI] [PubMed] [Google Scholar]
- 56.Nes, I. F., and H. H. Eijsink. 1999. Regulation of group II peptide bacteriocin synthesis by quorum-sensing mechanisms. American Society for Microbiology, Washington, DC.
- 57.Noel, L., F. Thieme, D. Nennstiel, and U. Bonas. 2001. cDNA-AFLP analysis unravels a genome-wide hrpG-regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 411271-1281. [DOI] [PubMed] [Google Scholar]
- 58.Oku, T., A. M. Alvarez, and C. I. Kado. 1995. Conservation of the hypersensitivity-pathogenicity regulatory gene hrpX of Xanthomonas campestris and X. oryzae. DNA Seq. 5245-249. [DOI] [PubMed] [Google Scholar]
- 59.Phee, B. K., J. H. Cho, S. Park, J. H. Jung, Y. H. Lee, J. S. Jeon, S. H. Bhoo, and T. R. Hahn. 2004. Proteomic analysis of the response of Arabidopsis chloroplast proteins to high light stress. Proteomics 43560-3568. [DOI] [PubMed] [Google Scholar]
- 59a.Prentki, P., and H. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29303-313. [DOI] [PubMed] [Google Scholar]
- 60.Rajeshwari, R., G. Jha, and R. V. Sonti. 2005. Role of an in planta-expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Mol. Plant-Microbe Interact. 18830-837. [DOI] [PubMed] [Google Scholar]
- 61.Ray, S. K., R. Rajeshwari, and R. V. Sonti. 2000. Mutants of Xanthomonas oryzae pv. oryzae deficient in general secretory pathway are virulence deficient and unable to secrete xylanase. Mol. Plant-Microbe Interact. 13394-401. [DOI] [PubMed] [Google Scholar]
- 62.Regelmann, A. G., J. A. Lesley, C. Mott, L. Stokes, and C. D. Waldburger. 2002. Mutational analysis of the Escherichia coli PhoQ sensor kinase: differences with the Salmonella enterica serovar Typhimurium PhoQ protein and in the mechanism of Mg2+ and Ca2+ sensing. J. Bacteriol. 1845468-5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rustam, T., S. McClean, J. Newcombe, J. McFadden, and L. J. Eales-Reynolds. 2006. Reduced toxicity of lipo-oligosaccharide from a phoP mutant of Neisseria meningitidis: an in vitro demonstration. J. Endotoxin Res. 1239-46. [DOI] [PubMed] [Google Scholar]
- 64.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 65.Shen, Y., P. Sharma, F. G. da Silva, and P. C. Ronald. 2002. The Xanthomonas oryzae pv. oryzae raxP and raxQ genes encode an ATP sulfurylase and adenosine-5′-phosphosulphate kinase that are required for AvrXa21 avirulence activity. Mol. Microbiol. 4437-38. [DOI] [PubMed] [Google Scholar]
- 66.Shi, Y., T. Latifi, M. J. Cromie, and E. A. Groisman. 2004. Transcriptional control of the antimicrobial peptide resistance ugtL gene by the Salmonella PhoP and SlyA regulatory proteins. J. Biol. Chem. 27938618-38625. [DOI] [PubMed] [Google Scholar]
- 67.Smadja, B., X. Latour, D. Faure, S. Chevalier, Y. Dessaux, and N. Orange. 2004. Involvement of N-acylhomoserine lactones throughout plant infection by Erwinia carotovora subsp. atroseptica (Pectobacterium atrosepticum). Mol. Plant-Microbe Interact. 171269-1278. [DOI] [PubMed] [Google Scholar]
- 68.Soncini, F. C., E. G. Vescovi, and E. A. Groisman. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 1774364-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song, W., G. Wang, L. Chen, W. Zhai, H. Kim, T. Holsten, L. Zhu, and P. Ronald. 1995. The rice disease resistance gene, Xa21, encodes a receptor-like protein kinase. Science 2701804-1806. [DOI] [PubMed] [Google Scholar]
- 70.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Phytopathol. 69183-215. [DOI] [PubMed] [Google Scholar]
- 71.Sugio, A., B. Yang, and F. F. White. 2005. Characterization of the hrpF pathogenicity peninsula of Xanthomonas oryzae pv. oryzae. Mol. Plant-Microbe Interact. 18546-554. [DOI] [PubMed] [Google Scholar]
- 72.Tsubery, H., I. Ofek, S. Cohen, and M. Fridkin. 2000. The functional association of polymyxin B with bacterial lipopolysaccharide is stereospecific: studies on polymyxin B nonapeptide. Biochemistry 3911837-11844. [DOI] [PubMed] [Google Scholar]
- 73.Tsuchiya, K., T. W. Mew, and S. Wakimot. 1982. Bacteriological and pathological characteristics of wild types and induced mutants of Xanthomonas campestris pv. oryzae. Phytopathology 7243-46. [Google Scholar]
- 74.Tsuge, S., T. Nakayama, S. Terashima, H. Ochiai, A. Furutani, T. Oku, K. Tsuno, Y. Kubo, and H. Kaku. 2006. Gene involved in transcriptional activation of the hrp regulatory gene hrpG in Xanthomonas oryzae pv. oryzae. J. Bacteriol. 1884158-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Valls, M., S. Genin, and C. Boucher. 2006. Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum. PLoS Pathog. 2e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vorhölter, F.-J., K. Niehaus, and A. Pühler. 2001. Lipopolysaccharide biosynthesis in Xanthomonas campestris pv. campestris: a cluster of 15 genes is involved in the biosynthesis of the LPS O-antigen and the LPS core. Mol. Genet. Genomics 26679-95. [DOI] [PubMed] [Google Scholar]
- 77.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 1852080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang, Y. S., L. Y. Pi, X. Chen, P. K. Chakrabarty, J. Jiang, A. L. De Leon, G. Z. Liu, L. Li, U. Benny, J. Oard, P. C. Ronald, and W. Y. Song. 2006. Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 183635-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wengelnik, K., and U. Bonas. 1996. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 1783462-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wengelnik, K., C. Marie, M. Russel, and U. Bonas. 1996. Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive reaction. J. Bacteriol. 1781061-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wengelnik, K., G. Van den Ackerveken, and U. Bonas. 1996. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol. Plant-Microbe Interact. 9704-712. [DOI] [PubMed] [Google Scholar]
- 82.White, P. J., and M. R. Broadley. 2003. Calcium in plants. Ann. Bot. (London) 92487-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Winfield, M. D., and E. A. Groisman. 2004. Phenotypic differences between Salmonella and Escherichia coli resulting from the disparate regulation of homologous genes. Proc. Natl. Acad. Sci. USA 10117162-17167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zasloff, M. 1992. Antibiotic peptides as mediators of innate immunity. Curr. Opin. Immunol. 43-7. [DOI] [PubMed] [Google Scholar]
- 85.Zhu, W., M. M. Magbanua, and F. F. White. 2000. Identification of two novel hrp-associated genes in the hrp gene cluster of Xanthomonas oryzae pv. oryzae. J. Bacteriol. 1821844-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.