Abstract
Previous studies have established that members of the Mycobacterium tuberculosis complex exhibit variable production of the antigenic proteins MPT70 and MPT83 due to mutations in their positive regulator, SigK (sigma factor K), and their negative regulator, RskA (regulator of sigma K). To further understand this highly specific SigK-controlled regulon, we have undertaken evolutionary studies to determine the presence of homologues of SigK-regulated genes in other organisms and to predict its transcriptional network. Evolutionary analysis indicates that the positive and negative regulators are conserved across many organisms, but that the genes under their control are variable. Moreover, the addition, loss, and movement of various genes in the mpt70/83 locus suggest that these genes are unlikely to be cotranscribed. To test predictions from sequence analysis, we have used promoter luciferase fusions and Northern blots to show that the majority of genes in this locus have their own promoters, of which a subset are SigK regulated (mpt83, dipZ, mpt70, and Rv0449c). Next, we have shown that the intracellular inducibility of mpt70 and mpt83 is a conserved property, shared between M. tuberculosis and Mycobacterium marinum. In addition, we have shown that SigK and RskA from an environmental mycobacterium isolate (M. gilvum PYR-GCK) complemented the regulatory activity of M. tuberculosis ΔsigK rskA. Together, our data indicate that the regulatory system SigK/RskA is conserved across the Mycobacterium genus, whereas the regulon under its control varies considerably across species.
Mycobacterium tuberculosis is an extremely successful pathogen of humans, infecting an estimated one-third of the world's population. This capacity to persist in the host, despite the dynamic alterations effected by the host immune response, testifies to an evolved capacity of the organism to regulate its genetic expression to the conditions encountered during infection. Indeed, a number of studies have demonstrated that disruption of M. tuberculosis regulatory elements, such as sigma factors, results in reduced virulence in experimental models (21, 28, 30).
The determination of genomic sequences for a number of nonpathogenic mycobacteria has revealed that homologues of many genes, including regulatory elements, are often present in organisms that are not virulent to humans. For instance, comparative analysis has demonstrated conservation of biochemical pathways and a secretory system important for full virulence of M. tuberculosis (7, 10). This indicates that genetic elements and the proteins they encode that exist in other mycobacteria either serve an important function in M. tuberculosis pathogenicity or have evolved a specific role in M. tuberculosis complex organisms. As a number of predicted regulatory elements are also conserved between environmental mycobacteria and M. tuberculosis, it follows that an evolutionary analysis may guide predictions about the origins of such elements, the transcriptional networks they control, and the stimuli that drive gene expression.
Among regulatory genes, extracytoplasmic sigma factors serve in the transduction of signals extracellular to the bacteria to effect targeted alterations in bacterial transcription. In the specific case of sigma factor K, we have previously shown microevolution of its regulon within the M. tuberculosis complex, with sigK mutated in certain Mycobacterium bovis BCG strains and its negative regulator rskA mutated in virulent M. bovis (9, 26). As a result, M. tuberculosis produces small quantities of the antigens MPT70 and MPT83 in vitro and only induces expression during infection, while in M. bovis expression of both is constitutively high. Given the profound differences in expression between M. tuberculosis complex organisms, we were interested in looking toward other mycobacteria for clues as to the evolution of this regulon. We therefore performed a comparative study across mycobacterial species, looking for homologues of the positive and negative regulators, as well as genes under their control. Our goal was to determine the transcriptional control of genes within this regulon and to test for conservation of function between M. tuberculosis genes and their homologues in other organisms.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. tuberculosis H37Rv, M. tuberculosis H37Ra, M. bovis 68799 (gift from Louise Thibert), BCG Russia, BCG Pasteur, BCG Pasteur::sigK, Mycobacterium marinum ATCC BAA-535, and Mycobacterium kansasii ATCC 12478 were grown under rotation conditions in Middlebrook 7H9 medium (Difco Laboratories, Detroit, MI) containing 0.05% Tween 80 (Sigma-Aldrich, St. Louis, MO) and 10% albumin-dextrose-catalase (Becton Dickinson and Co., Sparks, MD). Middlebrook 7H10 medium was used as the solid medium supplemented with 10% oleic acid-albumin-dextrose-catalase (Becton Dickinson and Co., Sparks, MD). Exceptionally, M. marinum was cultured at 30°C, whereas the other organisms were grown at 37°C. For cloning experiments, Escherichia coli Top10 was grown at 37°C in Luria-Bertani medium (Difco). As required, antibiotics were added as follows: kanamycin, 50 μg ml−1 for E. coli and 25 μg ml−1 for mycobacteria; hygromycin, 100 μg ml−1 for E. coli and 50 μg ml−1 for mycobacteria.
Genomic DNA.
A protocol based upon lysozyme and proteinase K (31) was used to extract genomic DNA from mycobacteria. DNA from Mycobacterium gilvum PYR-GCK (ATCC 700033) was a gift from Charles Miller at Utah State University. DNA from Mycobacterium canetti was a gift from Debby Cousins and was amplified using the GenomiPhi DNA amplification kit (Amersham Biosciences).
Construction of BCG Russia, BCG Pasteur, and M. tuberculosis H37Ra strains expressing luciferase.
The pMind-Lx vector (4), coding for luxAB from Vibrio harveyi under control of tetRO, is a gift from Brian Robertson. All putative promoters have been amplified, using Pfu cloned polymerase (Stratagen Inc.), from M. tuberculosis H37Rv genomic DNA using primers listed in Table 1. As an example, the putative promoter of Rv2871 has been amplified using pRv2871F and pRv2871R primers. All amplicons have been cloned in pMind-Lx using XbaI and BamHI restriction enzymes. Of note, this digestion completely removes the tetRO module. After transformation in E. coli, verification by PCR, and restriction digestion, plasmids were electroporated into M. tuberculosis H37Ra, BCG Pasteur, and BCG Russia as previously described (3). Hygromicin- and kanamycin-resistant clones were selected for further experiments.
TABLE 1.
Oligonucleotide primers used in this study
| Primer name | Sequence (5′→3′)a |
|---|---|
| pRv2871F | CGTTCTAGAGGATGTCGCCGACC |
| pRv2871R | TCGGGATCCGCACGCATGGCATCT |
| pMPB83F | GGGTCTAGAAGATAGCGTGCTCG |
| pMPB83R | CGTGGATCCTCGCTTCGATTCCT |
| pATF | AACTCTAGACTGACGCCAAGCTG |
| pATR | CCGGGATCCGCTTGGGATATCAC |
| pDipZF | TGCTCTAGAACACCGCCGGGCTG |
| pDipZR | GAGGGATCCGACTAGTGCGAGGG |
| pMPB70F | TACTCTAGAGACGTACTTCGGAGT |
| pMPB70R | CCGGGATCCTGTTCTTTACCTTCAT |
| pRv2876F | CAATCTAGAGTTGACAACGCTGAC |
| pRv2876R | GATGGATCCGGTTGGACTGACGTC |
| pMPB53F | CGGTCTAGACGGTGGGTCTGGC |
| pMPB53R | CGGGGATCCCAGGCGAAGACTC |
| pRv0448F | CCATCTAGACCAGGCGCTGCTG |
| pRv0448R | TCGGGATCCACGTACCAGCTGTA |
| pRv0449F | CCATCTAGATTGGCTCGTGCTTGC |
| pRv0449R | CGGGGATCCCACTTCCGACAAC |
| pSigAF | ACATCTAGAGGTCGGCGGCAAG |
| pSigAR | TTTGGATCCTGCCACATACACCCC |
| pHSP60F | CGTTCTAGATTAGCACGTCGATGC |
| pHSP60R | CGGGATCCGCAATTGTCTTGGCCAT |
| Rv0444OF | GTGCTGCAGGGTGCGGCCAACG |
| Rv0444OR | GGAAAGCTTGATAACGGCGACATC |
| SigKflavF | GCAGGTACCCTCCCCCGTGGC |
| SigKflavR | GCAAAGCTTACGGCGTCGCCAG |
| ATSigKflavR | GAAAAGCTTAGTAATGCCCTGCCG |
| EcoRI1 | GCATTTGAGCTCCACGTCAGCGACTG |
| EcoRI2 | AATTCAGUCGCUGACGUGGAG |
| EcoRIF | GAGCTCCACGTCAGCGACTGAA |
| Uni70F | CCCGCGTTCGACAAGCTGCCG |
| Uni70R | CCACCGGAACCTGCGACATTCC |
| dxrUniF | GCTCTACGCCCTTGTTGACCAG |
| Rv2876R | GTGCTCCACCTCGGTACTTG |
Underlining indicates differences in primers with respect to the template sequence. Boldface indicates restriction sites.
Complementation of M. tuberculosis H37Rv ΔsigK.
To complement the strain M. tuberculosis H37Rv ΔsigK (26), we used pMV306-hyg plasmids (26). Inserts were amplified using Pfu-cloned polymerase (Stratagene, Inc.). To construct pMV::dyad bovis, a part of the sigK-rskA region has been amplified, using Rv0444OF and Rv0444OR primers, from genomic DNA of M. bovis 68799. This amplicon was ligated to pMV306-Hyg::dyad (26) digested with XhoI and HindIII. The sigK and sigK-rskA genes from M. gilvum PYR-GCK were amplified from genomic DNA using, respectively, SigKflavF-SigKflavR and SigKflavF-ATSigKflavR. These amplicons were digested with HindIII and KpnI and ligated separately to pMV306-Hyg::SigK (26), digested with the same enzymes to generate pMV::SigKflav and pMV::SigKRskAflav. These plasmids, pMV::SigKflav, pMV::SigKRskAflav, pMV::dyad bovis, and pMV306-Hyg::SigK, were electroporated in M. tuberculosis H37Rv ΔsigK. Hygromycin-resistant clones were verified by PCR and sequencing.
To determine expression of mpt70 and mpt83, normalized to the amount of sigA RNA (20), in M. tuberculosis H37Rv ΔsigK and complemented strains, we performed quantitative reverse transcription-PCR (qRT-PCR). RNA was extracted from cultures with an optical density at 600 nm (OD600) of 0.4 to 0.6 by a modified phenol-chloroform extraction method (9). The qRT-PCR methods as well as the primers used for mpt70, mpt83, and sigA have already been described (9).
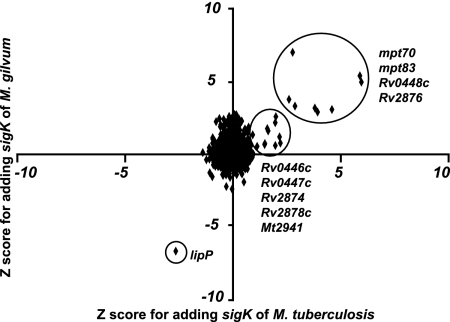
To determine the specificity of the transcriptional effect of heterologous complementation, the set of genes induced by the introduction of the sigK homologue from M. gilvum into M. tuberculosis H37Rv ΔsigK was determined by transcriptome analysis. Microarray studies were performed and analyzed as previously described (9, 26). The arrays were done in duplicate, and mean Z-scores for each spot (across two arrays) were calculated to quantify how many standard deviations that spot lay from the mean spot result for the experiment. As each gene is represented by a probe that has been spotted twice on the same array, transcriptional alteration was considered significant when both spots had a Z-score of ≥2. To compare genes induced by sigK from M. tuberculosis to those induced by sigK of M. gilvum, Z-scores were represented as a scatter plot, with the homologous complement (average of two arrays) on the x axis and the heterologous complement (average of two arrays) on the y axis.
Northern blot.
To determine the size and the relative amount of RNA, a modified version of a previously published mycobacterial RNA Northern blot protocol was used (22). Briefly, 15 μg of RNA from BCG Russia, BCG Pasteur, and BCG Pasteur::sigK and 1.5 μg of RNA marker (0.2 to 10 kb of RNA; Sigma) were fractionated on a 1% agarose-formaldehyde gel. Each sample was loaded with 2× RNA loading dye solution (Fermentas) containing ethidium bromide. RNA was transferred from the gel to a Hybond-N filter membrane (Amersham) using a model 785 vacuum blotter (Bio-Rad) in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer, at 12.5 cm of Hg for 1.5 h. To ensure that the same amount of RNA had been transferred, the resulting membranes were photographed under UV light. After UV treatment of 1,200 μJ with the Stratalinker 100 (Stratagen), the membrane and tubes were washed with 4× SSC and prehybridized (1 h at 42°C) and hybridized (16 h at 42°C) as described in the manufacturer's instructions for the ECL enhanced chemiluminescence direct nucleic acid labeling and detection systems (Amersham Biosciences). The probes for mpt83, dipZ, and mpt70 were PCR products obtained with the following primer pairs: Rv2873iF-Rv2873i2R, Rv2873i3F-Rv2874R, and MPB70L-MPB70R. After hybridization, the membrane was washed three times (10 min, 5 min, and 5 min) in primary wash buffer (0.4% of 20× SSC, 0.4% sodium dodecyl sulfate) at 55°C and two times (10 min) in 2× SSC at 42°C. Detection was by the ECL system, with the membrane exposed to a Hyperfilm (Amersham Biosciences).
Luciferase assays.
For the in vitro luciferase assay, bacteria were grown in 7H9 medium with hygromycin until the cultures reached an OD600 of 0.5. After centrifugation, cultures were resuspended in phosphate-buffered saline (PBS) and 10 μl of 1% n-decyl aldehyde (Sigma) in ethanol was added to 90 μl (9 × 106 bacteria) in 96-well plates (Becton Dickinson Labware). Light was measured in the Victor 3 Wallac 1420 multilabel counter (Perkin-Elmer) over a 10-s period. For each experiment, three to five transformants were measured two times and luminescence output was expressed in relative light units (RLU).
For the ex vivo luciferase assay, peritoneal macrophages were collected by lavage from C57BL/6 mice 72 h after injection with 3% of BBL thioglycolate medium brewer modified (BD). Subsequently, the cells were counted, centrifuged, and diluted at a concentration of 0.625 × 106 cells/ml in prewarmed RPMI 1640 medium supplemented with 10% fetal bovine serum and 2.5% 1 M HEPES solution (Multicell). In six-well plates (Falcon), 3 ml of RPMI containing macrophages was added to half of the wells. As our expression results were normalized with expression in RPMI alone, we also add 3 ml of RPMI medium to the other half of each well. The plates were incubated for 18 h at 37°C in 5% CO2. Following this incubation, 1 × 106 bacteria from fresh cultures (OD600 of 0.5) of M. marinum Lx or H37Ra-Lx were added to the wells, aiming for a multiplicity of infection of 1 bacterium for 2 macrophages. At 24 h of incubation, the supernatant was recovered and 1 ml of 1% Triton in PBS was added to all wells. After scraping the wells, the lysate was added to the corresponding supernatant. After a centrifugation at 4,000 rpm in an Allegra 25R centrifuge (Beckman Coulter), the pellet was resuspended in 180 μl of 1% Triton in PBS. Subsequently, in 96-well plates (Becton Dickinson Labware), 90 μl of this suspension was mixed with 10 μl of 1% n-decyl aldehyde (Sigma). The light was measured as described above. The results represent the mean luciferase activity of triplicates measured two times.
Determination of homologues and sequence analysis.
To look for homologues of the proteins of interest, we have searched the NCBI protein database via the BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/) looking for hits with more than 50% similarity. To supplement these results with organisms not being sequenced, we have performed targeted sequencing. Because of the ancestral situation of M. canetti in the M. tuberculosis complex phylogeny (6), we have amplified overlapping fragments of the mpt70 and sigK loci in M. canetti using primers already described (9). These fragments were sequenced at the McGill University and Genome Quebec Innovation Center (http://genomequebec.mcgill.ca/.)
In addition, we have determined the sequence of homologues in M. kansasii, based on deposited sequence data indicating the presence of at least one MPT70 homologue in this organism (33). To amplify the flanking region of putative mpk70 and/or mpk83, we did mixed-linker PCR by modifying a previously described method (15, 24). To obtain larger fragments, we used EcoRI-digested M. kansasii genomic DNA. The linkers and primers are listed in Table 1. Briefly, EcoRI1 and EcoRI2 linkers were mixed in equimolar amounts (6.7 pmol μl−1) in water and heated at 95°C for 15 min, followed by three cycles of 58°C for 10 min and 70°C for 5 min, before cooling to 4°C. The ligation step was performed in 20 μl, by adding 1 ng of digested genomic DNA, annealing primers (final concentration, 0.67 pmol/μl), 5× T4 DNA ligase buffer, and 1 U of T4 DNA ligase (Invitrogen). After incubation for 16 h at 15°C, 0.5 U of uracil DNA glycosylase and the corresponding 5× buffer (Gibco Life Technologies) were added and incubated for 20 min at 37°C. This solution served as template for PCR, using the AccuPrime Taq DNA high-fidelity polymerase (Invitrogen), following the manufacturer's instructions, using Uni70F-EcoRIR or Uni70R-EcoRIR primers. The PCR products were purified by gel extraction kit (Qiagen) and sequenced.
As we detected two different mpk70 homologues, we verified if they are located in the same region by amplifying the region between mpk70 and mpk83 with the primers Uni70F and Uni70R, using AccuPrime Taq DNA high-fidelity polymerase (Invitrogen). The resulting ∼2,600-bp fragment was sequenced, revealing the presence of an intervening dipZ homologue. However, the sequences so obtained failed to show evidence that these genes were adjacent to dxr or Rv2876 homologues. To verify this, we amplified the region between dxr and Rv2876 homologues, using dxrUniF and Rv2876R primers, exploiting conserved sequences between Mycobacterium avium, Mycobacterium leprae, and M. tuberculosis H37Rv, and using AccuPrime Taq DNA high-fidelity polymerase (Invitrogen). The resulting ∼1,900-bp fragment was then sequenced.
After identification of homologues, their genomic localizations were determined from NCBI (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). To determine open reading frames (ORFs) from our sequences and nonanalyzed sequences downloaded from the Sanger Institute (http://www.sanger.ac.uk/) and JGI (http://genome.jgi-psf.org/mic_home.html), the BLASTX program at NCBI was used. To determine whether MPT70/83 homologues contain a predicted lipid branching signal, we used the program LipoP (http://www.cbs.dtu.dk/services/LipoP/) (18). In order to determine the putative sequence of the SigK binding site, promoter sequences have been aligned using the AlignX program from VectorNTI (Invitrogen). Subsequently, this short alignment of similar sequences was used as a matrix to detect other putative binding sites in sequenced mycobacterial genomes using the programs PredictRegulon (34) and PREDetector (16).
Nucleotide sequence accession number.
The sequences of the overlapping mpt70 and sigK fragments have been deposited in GenBank under accession no. EU122437 and EU122438, respectively. The sequence of the ∼2,600-bp fragment obtained by amplifying the region between mpk70 and mpk83 has been submitted to GenBank under accession no. EU122436. The sequence of the ∼1,900-bp fragment resulting from amplification of the region between dxr and Rv2876 has been submitted to GenBank under accession no. EU122435.
RESULTS
Evolutionary analysis.
To derive a model for evolution of the SigK regulon, we generated an alignment plot of homologues of these genes across selected bacterial species (Fig. 1). This also served to predict sites of SigK promoter activity, in order to determine whether the set of genes under control of SigK functions as an operon, as previously published (17). For uniformity in gene designations, where possible, genes were labeled with the corresponding name in M. tuberculosis H37Rv.
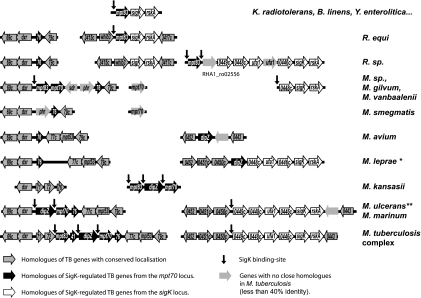
FIG. 1.
Alignment plot of genome organization of SigK regulon homologous genes across Actinomycetales. The strains used for this alignment were Kineococcus radiotolerans SRS30216, Brevibacterium linens BL2, Yersinia enterocolitica strain 8081, Rhodococcus equi strain 103, Rhodococcus (R.) sp. strain RHA1, Mycobacterium (M.) sp. strain JLS, Mycobacterium vanbaalenii PYR-1, Mycobacterium gilvum PYR-GCK, Mycobacterium smegmatis strain MC2 155, Mycobacterium avium 104, Mycobacterium leprae TN (*, majority pseudogenes), M. kansasii ATCC 12478, Mycobacterium ulcerans Agy99 (**, mpt70 interrupted by an IS element), Mycobacterium marinum ATCC BAA-535, Mycobacterium tuberculosis H37Rv, and M. canetti. Of note, for visual reasons, genes from the mpt70/83 locus have been labeled with the two last numbers (e.g., Rv2869c has been named 69c), while other genes have been labeled with the four last numbers (e.g., Rv0452 has been labeled as 0452).
The first observation is that in several bacteria, whether more related to mycobacteria (e.g., Rhodococcus equi, Kineococcus radiotolerans, or Brevibacterium linens) or less (e.g., Yersinia enterocolitica), the same minimum locus was observed as mpt83-sigK-rskA. From previous work, SigK is known to serve as the positive regulator and RskA as the negative regulator of mpt83 (9, 26). Therefore, the triad of mpt83-sigK-rskA could represent the minimal set of genes (effector-regulator-sensor) that serves these bacteria in response to a specific condition.
In Rhodoccocus sp. strain RHA1, the cassette mpt83-sigK-rskA is interrupted by six genes inserted between mpt83 and sigK. Four of these six genes (hypothetically implicated in cyclopropane fatty acid synthesis) are conserved in the same order in M. tuberculosis, M. marinum, and M. ulcerans. The addition of these genes in the SigK regulon appears to have occurred before the divergence of Rhodococcus and Mycobacteria.
Next, during the evolution of mycobacteria, the locus appears to have separated to two sites, the sigK-rskA locus and the mpt70/83 locus. Of note, dxr and Rv2876 are adjacent in most Actinomycetales, with the exception of mycobacteria, where they are separated by the mpt70/83 locus. Furthermore, some mycobacteria have two homologues of mpt70/83 inserted between dxr and Rv2876. In M. flavescens, for example, the homologous proteins of MPT70 share 76% identity and 85% similarity (excluding the signal peptide), strongly suggestive of a gene duplication. Using the LipoP program (18), it is observed that homologues of MPT83 have a signal of lipid branching predicted but for MPT70 this signal is absent. This argues that, during the duplication, MPT70 has lost the lipid branching signal, resulting in a different protein localization (secreted versus membrane protein).
In slow-growing pathogenic mycobacteria (M. tuberculosis complex, M. marinum, Mycobacterium ulcerans, M. canetti, and M. kansasii), there is a further gene, dipZ, inserted between the two mpt83 paralogs. Surprisingly, using the BLASTP program, we observe that this full-length protein is not present in most bacteria, where instead one finds two separated proteins, CcdA and TlpA-like, homologous to respective halves of DipZ. This finding suggests that DipZ is a fused protein. Further support for this hypothesis comes from Ralstonia eutropha H16, where there is both a dipZ homologue (H16_A1760) and a separate locus where ccdA (H16_B1487) and tlpA-like (H16_B1488) are in a putative operon but annotated as distinct genes.
In more recent evolutionary terms, one encounters only in M. canetti and other M. tuberculosis complex organisms the addition of a small gene, annotated in CDC 1551 as MT2941 (not annotated in H37Rv). This gene codes for a predicted antitoxin from the VapC family and is highly similar to Rv0626.
Beyond genes added to this locus during evolution, it is notable that certain pathogenic mycobacteria have lost either discrete members of the regulon or the entire set of genes. In M. leprae, all remnants of this regulon are found as pseudogenes. In M. avium, only dipZ remains. Both M. marinum and M. ulcerans lack mpt83, and mpt70 is replaced by an insertion sequence in M. ulcerans.
Together, evolutionary considerations indicate that genes of this regulon have been duplicated, separated, and relocated around the genome. From this, we hypothesize that many of the genes of this regulon will have their own distinct promoters. Furthermore, we hypothesize that the observed intracellular inducibility of mpt70/mpt83 in M. tuberculosis should be a conserved feature of this regulon that can be experimentally verified in nontuberculous mycobacteria. Finally, we hypothesize that sigK and rskA homologues in nontuberculous mycobacteria will control the expression of the genes regulated by M. tuberculosis sigK and rskA.
Transcriptional studies: mpt70, mpt83, and dipZ have their own SigK-regulated promoter.
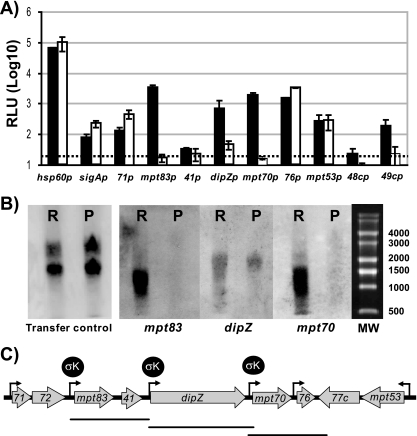
In order to determine the mechanism by which SigK affects the expression of genes in the mpt70/83 locus, different putative promoters have been cloned into a luciferase reporter system. In M. smegmatis, which does not have a sigK homologue, light was generated for Rv2871p, MT2941p, dipZp, Rv2876p, and mpt53p, but not for mpt83p, mpt70p, Rv0448cp, and Rv0449cp (data not shown). Next, we introduced these plasmids into BCG Russia (where sigK is wild type and intact) and BCG Pasteur (where there is a mutation in the start codon of sigK). The differential production of light in these different BCG strains should therefore represent the SigK-attributed effect in this system. As a control, to show that any observed differences in luciferase expression are not an artifact of these strains, we also introduced plasmids coding for the luciferase under control of hsp60p and sigAp. As seen in Fig. 2A, for the two controls, luciferase expression was on the same order in BCG Russia and BCG Pasteur, with ratios of Russia to Pasteur of 0.6 and 0.4 for hsp60p and sigAp, respectively. These minor differences in luciferase activity are probably attributable to differences in growth rate of these two BCG, but serve as a control for SigK-associated effects, where we expect to see increased light in BCG Russia. As already observed for M. smegmatis, we detected promoter activity with Rv2871p, Rv2876p, and mpt53p at levels for BCG Russia and Pasteur comparable with that of the control plasmids (respectively, 0.3, 0.5, and 1). In striking contrast, when the luciferase was fused with mpt70p or mpt83p, the changes (fold) were 2 orders of magnitude (111 and 144, respectively). This indicates that there are distinct promoters before mpt70 and mpt83 that are under very strong control of SigK. Interestingly, some light could be detected for dipZp in BCG Pasteur, but the same promoter in BCG Russia was significantly more active, with a change (fold) of 15. This suggests that dipZ is regulated by two sigma factors, and one of these is SigK. For the promoter before MT2941p, for which activity was detected in M. smegmatis, we were not able to detect any activity in either BCG strain.
FIG. 2.
Transcriptional study of the mpt70/83 locus. (A) Mean luciferase activity, at early exponential phase, of three to five transformants with plasmid coding for luxAB under control of corresponding promoters. The black bars represent the luciferase activities of transformed BCG Russia, whereas the white bars represent those of BCG Pasteur. The dashed line corresponds to minimal level of confidence. (B) Northern blot results of RNA from BCG Russia (R) and BCG Pasteur (P) probed alternatively with mpt70-, dipZ-, or mpt83-specific DNA and detected by ECL methods. The transfer control corresponds to the membrane photographed under UV light. (C) Schematic representation of the results obtained for the mpt70 locus regulation with functional promoters (black arrow), SigK-regulated promoters, and Northern blot-deducted RNA species. Abbreviations: 71, Rv2871; 41, MT2941; 76, Rv2876; 48c, Rv0448c; and 49c, Rv0449c.
Additionally, we also explored promoter activity in the sigK locus using Rv0448cp and Rv0449cp. In neither M. smegmatis nor BCG strains were we able to detect activity for Rv0448cp, indicating that there is probably no promoter at this site. In the case of Rv0449cp, we did not detect activity in BCG Pasteur, unlike in BCG Russia where there was eightfold more activity. From this, we conclude that the SigK-regulated promoter in the sigK locus seems to at least include Rv0449cp.
Together, the luciferase assays suggested that Rv2871p, mpt83p, dipZp, mpt70p, Rv2876p, mpt53p, and Rv0449cp are functional promoters, but under our conditions we could detect a significant effect of SigK only for Rv0449cp, mpt83p, dipZp, and mpt70p. To confirm that mpt83, dipZ, and mpt70 are not part of an operon, but rather that each gene has its own SigK-regulated promoter, we performed a Northern blot, probing RNA from BCG Russia and BCG Pasteur sequentially for mpt83, dipZ, and mpt70. As shown in Fig. 2B, RNA for mpt83 and mpt70 is clearly reduced in BCG Pasteur compared to BCG Russia. Furthermore, the sizes of the transcripts are approximately 1,000 bp to 1,500 bp, arguing against this region being a 5-kb operon (Fig. 2C). When RNA was probed with dipZ DNA, we detected an RNA species of approximately 2,250 bp, which is on the same order as the predicted ORF (2,000 bp). For dipZ, the SigK effect was less marked than for mpt70 and mpt83 transcripts, consistent with findings by the luciferase assay. In addition, we also analyzed BCG Pasteur and BCG Pasteur::sigK by Northern blot analysis (data not shown). Again, RNA for mpt83, mpt70, and dipZ presented as different sized transcripts, with a strong SigK effect for mpt83 and mpt70 and a less-marked SigK effect for dipZ transcript. Therefore, these Northern blots confirm the luciferase results and indicate that mpt83, mpt70, and dipZ have their own SigK-regulated promoters.
Inducibility in macrophages is conserved between M. marinum and M. tuberculosis.
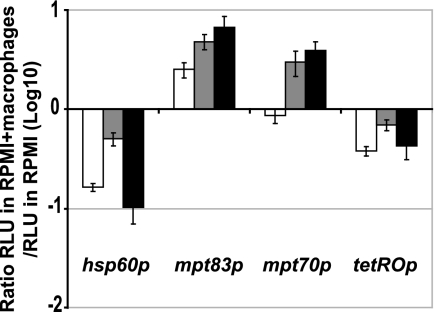
In M. tuberculosis, while basal expression of mpt70 and mpt83 is low, there is strong induction of these genes during infection of macrophages (26, 27). We therefore asked whether this inducibility is M. tuberculosis specific or rather a conserved feature of this regulon. To test this, the plasmids pLx-mpt70 and pLx-mpt83 containing the promoter, and thus the SigK binding site, were introduced in M. marinum. Additionally, because the activity of the hsp60p and tetROp is tightly correlated to the number of live bacteria (4, 29), the plasmids pLx-hsp60 and pMind were also electroporated into M. marinum, as a plasmid control. The bacteria so obtained were then used for infection of peritoneal macrophages, with results expressed as the ratio of promoter activity in RPMI medium containing macrophages divided by the activity observed in RPMI medium alone. As a control of induction, the same experiment has been done with M. tuberculosis H37Ra electroporated with the same plasmids.
From the results, presented in Fig. 3, for the M. marinum controls (tetROp and hsp60p), one notes that the ratio of macrophages to RPMI alone was less than 1, probably indicative of some loss of bacteria in RPMI medium containing macrophages. In contrast, for M. marinum with mpt70p and mpt83p, addition of macrophages to RPMI medium resulted in strong induction, at both 32°C and 37°C. These results prove that these M. tuberculosis promoters are recognized by SigK from M. marinum and that the macrophage-dependent signaling mediated via RskA and SigK to induce expression of mpt70/83 is conserved in M. marinum.
FIG. 3.
Activation of mpt70p and mpt83p during infection of macrophages with M. tuberculosis and M. marinum. Shown are the ratios of the RLU of peritoneal macrophages infected during 24 h with transformed strains of M. marinum at 37°C (white) or 32°C (gray) and M. tuberculosis H37Ra at 37°C (black) divided by the RLU of the same strains in RPMI alone at the same temperature. The data represent the means of triplicate experiments.
Function of RskA and SigK is conserved between M. gilvum and M. tuberculosis.
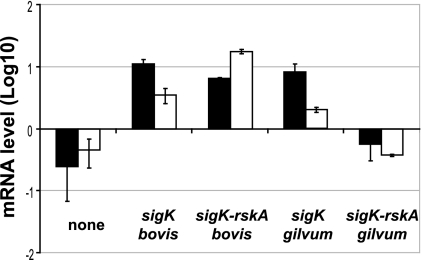
In previous work, it has been established that expression of MPT83 and MPT70 varies between M. tuberculosis and M. bovis, due to point mutations in rskA (26). Because other mycobacteria share the inducibility phenotype of M. tuberculosis (as demonstrated above), but differ in their RskA sequence, this suggested to us that RskA variations in nontuberculous mycobacteria would permit functional protein activity in our M. tuberculosis deletion mutant. In order to test this, we introduced sigK or sigK-rskA homologues into M. tuberculosis ΔsigK (with polar effect on rskA) from the most distantly related mycobacterium for which a genome sequence has been determined and a sigK homologue is present. For this, we used M. gilvum (95% identity in 16S rRNA), best known for its capacity to degrade polycyclic aromatic hydrocarbons (5).
As seen in Fig. 4, complementation with sigK from M. tuberculosis H37Rv (which is identical to sigK of M. bovis) increased the expression of mpt70, as measured by the ratio of mpt70 to sigA. The same effect was observed when measuring mpt83 levels. Interestingly, the same induction of expression was seen with sigK of M. gilvum. When sigK/rskA from M. bovis was introduced, mpt70/mpt83 expression was also high, due to the presence of a functioning SigK paired with a nonfunctional anti-Sigma factor. In contrast, the sigK/rskA pair from M. gilvum suppressed expression of both mpt70 and mpt83. This indicates that the SigK from M. gilvum is able to recognize the same binding site as its M. tuberculosis orthologue, despite only 65% identity (81% similarity) between their protein sequences. Likewise, the RskA of M. gilvum is able to serve as the negative regulator, despite only 59% identity (72% similarity) to RskA of M. tuberculosis.
FIG. 4.
Heterologous complementation of M. tuberculosis H37Rv ΔsigK (with polar effect on rskA). Quantification of mpt70 (black) and mpt83 (white) expression by semiquantitative RT-PCR, normalized to sigA, of M. tuberculosis ΔsigK rskA and strains complemented with pMV::sigKbovis (same as H37Rv), pMV::sigK rskAbovis, pMV::sigKgilvum, and pMV::sigK-rskAgilvum (from Mycobacterium gilvum). The data represent the mean quantification of RNA of two transformants in early exponential phase.
The SigK binding site is conserved and specific.
As we observed that the M. gilvum orthologue of SigK is able to regulate expression of mpt70 and mpt83, we performed a microarray experiment to determine the number of genes induced by the introduction of this gene. To compare the activity of M. gilvum sigK to that of M. tuberculosis, we plotted differential expression (shown as Z-scores) for the M. tuberculosis knockout complemented with sigKRv (x axis) against the same strain complemented with sigKM. gilvum (y axis). As shown in Fig. 5, in both cases, the genes which showed the greatest up-regulation with the complementation of SigK were the same: Rv2873 (mpt83), Rv2875 (mpt70), Rv2876, and Rv0448c. Also induced in both, with slightly lower Z-scores, were Rv0446c, Rv0447c (ufa1), Rv2874 (dipZ), Rv2878c, and MT2941. These results extend the findings above to show that SigKM. gilvum induces expression of other genes of the M. tuberculosis SigK regulon. Furthermore, these data confirm than that the regulon under control of SigK is restricted to just these two areas of the genome.
FIG. 5.
Comparison of SigKRv and SigKM. gilvum activity assessed by microarray. Z-scores for M. tuberculosis ΔsigK complemented with pMV::sigKRv (x axis, representing average of two arrays) have been plotted against Z-scores obtained when M. tuberculosis ΔsigK has been complemented with pMV::sigKgilvum (y axis, representing the average of two arrays). Genes for which expression was increased by the presence of sigK are reported with a positive Z-score. Inversely, genes for which expression was decrease by the presence of sigK are reported with a negative Z-score. Genes whose expression is up-regulated in both cases are exclusively part of the sigK or mpt70 locus. Of note, some genes from the Rv2463c (lipP) region are down-regulated, in both cases, due to the presence at this locus of the pMV integration site.
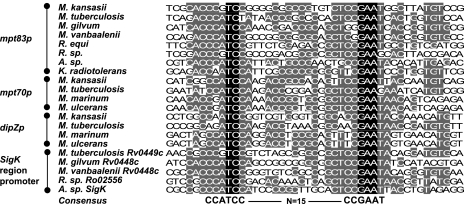
By chromatin immunoprecipitation assay, Rodrigue and coworkers have shown a direct interaction of SigK with the promoters of mpt70 and mpt83 (25). Furthermore, we have shown that SigKRv acts on the promoters of mpt70, dipZ, mpt83, and Rv0449c. To determine the binding site, we performed an alignment, using VectorNTI-AlignX, of the upstream promoter regions of mpt70, dipZ, mpt83, and Rv0449c from M. tuberculosis. In addition, since we have shown that SigKM. gilvum and SigKM. marinum are able to regulate the mpt70 and mpt83 promoters, this suggested that the binding site between species should be conserved. Consequently, we added the promoters of homologous genes from several Actinomycetales which have a SigK in this alignment. By these methods, we detected a strong consensus sequence, which is CCATCC-N15-CCGAAT (Fig. 6). This indicates that not only is the activity of SigK and RskA conserved across species, but the SigK binding site is strongly conserved. To verify that this consensus sequence is not random, we also aligned the upstream sequence of the mpt70 homologue from Yersinia enterocolitica, where we find a strongly similar sequence with a spacer that is 1 base longer (data not shown).
FIG. 6.
Consensus SigK-binding sequence. Alignment of promoter sequences of mpt83p, mpt70p, dipZp, and Rv0449cp with promoters of corresponding homologues in Actinomycetales. The DNA sequences come from promoters indicated in the Fig. 1; additionally, promoters for homologues from Arthrobacter sp. strain FB24 have been added. The black-highlighted letters correspond to nucleotides conserved in all sequences, whereas the gray-highlighted letters correspond to nucleotides conserved in at least in half of the 21 sequences.
As we have detected an effect of SigK for only two loci in the genome, the mpt70/83 and sigK loci (9), we wanted to verify if other SigK-regulated promoters could be present but not up-regulated in our in vitro experimentation (for example, under the control of repressor). To test this possibility, we performed a bioinformatic search looking for similar sequences in different genomes that contain SigK. Despite searching with different programs, using different strategies and parameters, we have been unable to detect other strongly similar sequences in intergenic regions of different mycobacterial genomes. This finding, supported by the microarray data, suggests that SigK strictly controls only the mpt70/83 locus and the sigK locus.
DISCUSSION
Organisms of the M. tuberculosis complex comprise a closely related set of subspecies or ecotypes that are associated with different hosts. Postgenomic analyses have determined the amount of genomic variation between these organisms (2, 14) and have begun to uncover the genetic basis of their phenotypic differences (19). The expression of MPT70 and MPT83 represents a further example of phenotypic variability between M. tuberculosis complex subspecies; in M. tuberculosis, expression is induced during infection of macrophages (26, 27), whereas M. bovis and the Oryx bacillus have constitutively high expression of these antigenic proteins due to mutations in the RskA, the anti-sigma K (26). As different RskA mutations were independently selected in M. bovis and the Oryx bacillus, there would appear to be an adaptive advantage of overexpression, at least in certain hosts. In support of this, a mutation in SigK during the prolonged laboratory passage of M. bovis BCG resulted in reduced expression of these proteins (9), likely reflecting the absence of a selective pressure to produce these proteins in vitro. This established history of variable expression among closely related organisms prompted us to examine the evolution of this regulon across the genus, to determine whether SigK homologues in mycobacteria serve similar roles, both in terms of the genes under their control and the dynamics of induction.
Using comparative gene alignments, we have shown that sigK and rskA are conserved in environmental mycobacteria and that macrophage-dependent inducibility of the SigK regulon is not specific to M. tuberculosis. Moreover, the capacity of SigKM. gilvum and RskAM. gilvum to perform the same function as their orthologues in M. tuberculosis indicates a conservation of the regulatory control. Given the presence of homologues of these genes in organisms that are not classified as pathogens, further study will be needed to determine whether the same or a different signal is sensed via RskA for gene induction. One possibility is that mycobacteria in close contact with free-living phagocytic organisms, such as Acanthamoeba (1), employ this signaling pathway during environmental infections of eukaryotic hosts, but this remains to be experimentally verified.
A further utility of evolutionary analysis was to derive a prediction for control of expression by SigK. Gene alignments indicated insertion, loss, or movements of certain SigK-regulated genes across different Mycobacteria (e.g., mpt83, dipZ, and mpt70 in M. kansasii and dipZ in M. avium are found in other loci). This suggested transcriptional independence and, by extension, the presence of distinct promoters. By luciferase assay and Northern blots, we present evidence for these distinct promoters. While the majority of genes in the mpt70/83 locus (Rv2871, mpt83, dipZ, mpt70, Rv2876, and mpt53) have functional promoters, under our conditions mpt83p, mpt70p, and Rv0449cp were under the control of SigK. Additionally, dipZp showed evidence of SigK regulation, with indirect evidence of another sigma factor effect. Thus, the mpt70/83 locus is not an operon from Rv2871 to Rv2875, as has been proposed (17). We suspect that the difference between our findings and previous studies is explained by the fact that Mycobacterium smegmatis, now known to not have a SigK homologue, was used to study regulation of these genes (17). In addition to mpt83, dipZ, and mpt70, we observed that MT2941 and Rv2876 are under transcriptional control of SigK, even through we have not shown the presence of a SigK-regulated promoter. In the case of Rv2876, a study employing chromatin immunoprecipitation reported a direct interaction of SigK with Rv2876p, although this was considerably less pronounced than the interaction observed with mpt70p or mpt83p (25). While our results do not demonstrate a significant effect of SigK on this promoter fused to luxAB, the observation that Rv2876 is strongly induced in our array experiments, as well as others comparing the transcriptome of virulent M. bovis to M. tuberculosis (23), indicates that SigK has some effect on Rv2876 expression, whether through a promoter before Rv2876 or by cotranscription with mpt70.
The evolution of other processes across mycobacterial species has proven informative for understanding the specificity, or lack thereof, of certain conserved systems. While such conservation could occur if the encoded proteins serve a useful role in M. tuberculosis pathogenesis, an alternative possibility is that a conserved system has evolved a specific function for M. tuberculosis complex organisms. For instance, in silico comparisons and heterologous complementation studies have established that a major virulence secretion system of M. tuberculosis, encoded by the RD1 region of the genome, can be found in environmental bacteria (11, 12). However, protein-protein interaction studies have shown M. tuberculosis-specific alterations in the C terminal portion of CFP-10, where the secretory signal is found (8). For SigK and RskA, our analysis also points to a conserved system with species-specific idiosyncrasies. While our results indicate that the core signaling system has been maintained across bacteria, alignment plots reveal that the number and identity of SigK-regulated genes are variable. From a minimal module of mpt83-rskA-sigK, a gene duplication first resulted in two MPT70/83 paralogues. Of these, MPT83 shares a membrane lipid anchor with a number of protein from other Actinomycetales, unlike MPT70, which lacks this feature and is secreted (32). As another example of how this regulon has evolved, the addition of dipZ is interesting on a number of counts. The protein DipZ appears to represent a fusion of at least two proteins, and it is principally found in host-associated gram-negative bacteria (Burkholderiaceae or Rhizobiaceae family). In the phylum Actinobacteria, full-length DipZ is present exclusively in bacteria reported to cause disease (Corynebacterium diphtheriae, Corynebacterium jeikeium, M. avium, M. kansasii, M. marinum, M. ulcerans, and M. tuberculosis complex). This distribution of DipZ contrasts with that of other members of the SigK regulon, which are present across a greater range of organisms. However, despite the crystallization of the C-terminal part of this protein (13), the function of this gene is not yet known, so it remains difficult to predict how and why dipZ came to reside between mpt70 and mpt83, where its expression in part depends on SigK.
Together these findings indicate that the conserved SigK-RskA motif enables the induction of expression of a small number of genes, variable between mycobacterial species, many of which have their own SigK promoter. Important next steps will to determine the trigger or triggers that induce expression of this regulon and the role of members of this regulon in M. tuberculosis pathogenicity.
Acknowledgments
We thank members of the Behr and Schurr laboratories for their input.
F.V. was supported through a CIHR training grant to Erwin Schurr. M.B. is Chercheur Boursier Senior of the FRSQ. This work was funded by an operating grant from the Canadian Institutes for Health Research, MOP-79309.
Footnotes
Published ahead of print on 18 January 2008.
REFERENCES
- 1.Adékambi, T., S. Ben Salah, M. Khlif, D. Raoult, and M. Drancourt. 2006. Survival of environmental mycobacteria in Acanthamoeba polyphaga. Appl. Environ. Microbiol. 725974-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 2841520-1523. [DOI] [PubMed] [Google Scholar]
- 3.Belley, A., D. Alexander, T. Di Pietrantonio, M. Girard, J. Jones, E. Schurr, J. Liu, D. R. Sherman, and M. A. Behr. 2004. Impact of methoxymycolic acid production by Mycobacterium bovis BCG vaccines. Infect. Immun. 722803-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blokpoel, M. C., H. N. Murphy, R. O'Toole, S. Wiles, E. S. Runn, G. R. Stewart, D. B. Young, and B. D. Robertson. 2005. Tetracycline-inducible gene regulation in mycobacteria. Nucleic Acids Res. 33e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brezna, B., A. A. Khan, and C. E. Cerniglia. 2003. Molecular characterization of dioxygenases from polycyclic aromatic hydrocarbon-degrading Mycobacterium spp. FEMS Microbiol. Lett. 223177-183. [DOI] [PubMed] [Google Scholar]
- 6.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 993684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burguiere, A., P. G. Hitchen, L. G. Dover, L. Kremer, M. Ridell, D. C. Alexander, J. Liu, H. R. Morris, D. E. Minnikin, A. Dell, and G. S. Besra. 2005. LosA, a key glycosyltransferase involved in the biosynthesis of a novel family of glycosylated acyltrehalose lipooligosaccharides from Mycobacterium marinum. J. Biol. Chem. 28042124-42133. [DOI] [PubMed] [Google Scholar]
- 8.Champion, P. A., S. A. Stanley, M. M. Champion, E. J. Brown, and J. S. Cox. 2006. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science 3131632-1636. [DOI] [PubMed] [Google Scholar]
- 9.Charlet, D., S. Mostowy, D. Alexander, L. Sit, H. G. Wiker, and M. A. Behr. 2005. Reduced expression of antigenic proteins MPB70 and MPB83 in Mycobacterium bovis BCG strains due to a start codon mutation in sigK. Mol. Microbiol. 561302-1313. [DOI] [PubMed] [Google Scholar]
- 10.Gao, L. Y., S. Guo, B. McLaughlin, H. Morisaki, J. N. Engel, and E. J. Brown. 2004. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol. Microbiol. 531677-1693. [DOI] [PubMed] [Google Scholar]
- 11.Gey van Pittius, N. C., J. Gamieldien, W. Hide, G. D. Brown, R. J. Siezen, and A. D. Beyers. 2001. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gey van Pittius, N. C., S. L. Sampson, H. Lee, Y. Kim, P. D. van Helden, and R. M. Warren. 2006. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstone, D., E. N. Baker, and P. Metcalf. 2005. Crystallization and preliminary diffraction studies of the C-terminal domain of the DipZ homologue from Mycobacterium tuberculosis. Acta Crystallograph. Sect. F Struct. Biol Cryst. Commun. 61243-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32643-655. [DOI] [PubMed] [Google Scholar]
- 15.Haas, W. H., W. R. Butler, C. L. Woodley, and J. T. Crawford. 1993. Mixed-linker polymerase chain reaction: a new method for rapid fingerprinting of isolates of the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 311293-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiard, S., R. Maree, S. Colson, P. A. Hoskisson, F. Titgemeyer, G. P. van Wezel, B. Joris, L. Wehenkel, and R. Sebastien. 2007. PREDetector: a new tool to identify regulatory elements in bacterial genomes. Biochem. Biophys. Res. Commun. 357861-864. [DOI] [PubMed] [Google Scholar]
- 17.Juarez, M. D., A. Torres, and C. Espitia. 2001. Characterization of the Mycobacterium tuberculosis region containing the mpt83 and mpt70 genes. FEMS Microbiol. Lett. 20395-102. [DOI] [PubMed] [Google Scholar]
- 18.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 121652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keating, L. A., P. R. Wheeler, H. Mansoor, J. K. Inwald, J. Dale, R. G. Hewinson, and S. V. Gordon. 2005. The pyruvate requirement of some members of the Mycobacterium tuberculosis complex is due to an inactive pyruvate kinase: implications for in vivo growth. Mol. Microbiol. 56163-174. [DOI] [PubMed] [Google Scholar]
- 20.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31715-724. [DOI] [PubMed] [Google Scholar]
- 21.Manganelli, R., L. Fattorini, D. Tan, E. Iona, G. Orefici, G. Altavilla, P. Cusatelli, and I. Smith. 2004. The extra cytoplasmic function sigma factor σE is essential for Mycobacterium tuberculosis virulence in mice. Infect. Immun. 723038-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parish, T., and N. G. Stoker (ed.). 1998. Methods in molecular biology, vol. 101. Mycobacteria protocols. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 23.Rehren, G., S. Walters, P. Fontan, I. Smith, and A. M. Zarraga. 2007. Differential gene expression between Mycobacterium bovis and Mycobacterium tuberculosis. Tuberculosis 87347-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reisig, F., K. Kremer, B. Amthor, D. van Soolingen, and W. H. Haas. 2005. Fast ligation-mediated PCR, a fast and reliable method for IS6110-based typing of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 435622-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrigue, S., J. Brodeur, P.-É. Jacques, A. L. Gervais, R. Brzezinski, and L. Gaudreau. 2007. Identification of mycobacterial σ factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 1891505-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Said-Salim, B., S. Mostowy, A. S. Kristof, and M. A. Behr. 2006. Mutations in Mycobacterium tuberculosis Rv0444c, the gene encoding anti-SigK, explain high level expression of MPB70 and MPB83 in Mycobacterium bovis. Mol. Microbiol. 621251-1263. [DOI] [PubMed] [Google Scholar]
- 27.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, I. 2003. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 16463-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snewin, V. A., M.-P. Gares, P. Ó. Gaora, Z. Hasan, I. N. Brown, and D. B. Young. 1999. Assessment of immunity to mycobacterial infection with luciferase reporter constructs. Infect. Immun. 674586-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun, R., P. J. Converse, C. Ko, S. Tyagi, N. E. Morrison, and W. R. Bishai. 2004. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 5225-38. [DOI] [PubMed] [Google Scholar]
- 31.van Soolingen, D., P. W. M. Hermans, P. E. W. de Haas, D. R. Soll, and J. D. A. van Embden. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 292578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vosloo, W., P. Tippoo, J. E. Hughes, N. Harriman, M. Emms, D. W. Beatty, H. Zappe, and L. M. Steyn. 1997. Characterisation of a lipoprotein in Mycobacterium bovis (BCG) with sequence similarity to the secreted protein MPB70. Gene 188123-128. [DOI] [PubMed] [Google Scholar]
- 33.Woolford, A. J., R. G. Hewinson, M. Woodward, and J. W. Dale. 1997. Sequence heterogeneity of an mpb70 gene analogue in Mycobacterium kansasii. FEMS Microbiol. Lett. 14843-48. [DOI] [PubMed] [Google Scholar]
- 34.Yellaboina, S., J. Seshadri, M. S. Kumar, and A. Ranjan. 2004. PredictRegulon: a web server for the prediction of the regulatory protein binding sites and operons in prokaryote genomes. Nucleic Acids Res. 32W318-W320. [DOI] [PMC free article] [PubMed] [Google Scholar]