Abstract
The type IV secretion (T4S) system is critical for the virulence of several pathogens. In the rickettsial pathogen Ehrlichia chaffeensis, the virBD genes are split into two operons, the virB3-virB6 (preceded by sodB) and virB8-virD4 operons. Between these two operons, there are duplications of virB4, virB8, and virB9. In this study we found that transcription of all five loci was downregulated prior to the release of E. chaffeensis from host THP-1 cells and was upregulated at the initiation of exponential growth. Electrophoretic mobility shift assays revealed an E. chaffeensis-encoded protein that specifically bound to the promoter regions upstream of the virBD loci. The protein was purified from the bacterial lysate by affinity chromatography using a biotinylated promoter region upstream of sodB. Mass spectrometry identified the protein as an E. chaffeensis 12.3-kDa hypothetical protein, which was designated EcxR. Recombinant EcxR bound to the promoter regions upstream of five individual virBD loci. EcxR also activated transcription of all five virBD loci in lacZ reporter constructs. The expression of ecxR was positively autoregulated by EcxR. These results suggest that the five virBD loci are coordinately regulated by EcxR to allow developmental stage-specific expression of the T4S system in E. chaffeensis.
Ehrlichia chaffeensis is a gram-negative, obligately intracellular bacterium that causes human monocytic ehrlichiosis, an emerging tick-borne zoonosis (10, 23). E. chaffeensis manipulates host monocytes/macrophages throughout its intracellular developmental cycle (25), and this bacterium has a biphasic developmental cycle in mammalian cells that alternates between a small “dense-core cell” (DC) and a large “reticulate cell” (RC), as defined by morphological features, differential surface protein expression, and infectivity (34). The presence of biphasic developmental stages which differ in cell size, infectivity, and the expression of surface protein VirB9 of the type IV secretion (T4S) apparatus has been demonstrated for Anaplasma phagocytophilum, which is closely related to E. chaffeensis (21). The ability to perform the phenotypic switch is likely important for adaptation of the bacterium to harsh extracellular conditions and to initiation of infection, survival, and replication in a new host cell. However, little is known about the mechanisms regulating the developmental cycle of members of the genera Ehrlichia and Anaplasma.
Genes encoding the T4S apparatus, the virBD loci, are found in members of the order Rickettsiales, including E. chaffeensis and A. phagocytophilum (17, 22). The T4S apparatus of gram-negative bacteria is a transmembrane channel composed of multiple conserved proteins that transport macromolecules across the membrane into eukaryotic target cells in an ATP-dependent manner. The T4S system is a critical determinant for virulence in several gram-negative pathogens, such as Agrobacterium tumefaciens, Legionella pneumophila, Helicobacter pylori, and Brucella abortus, because it delivers effector molecules into the host cell cytoplasm or nucleus, which induce tumors, induce inflammatory cytokines, and create an intracellular compartment for bacterial survival and proliferation (4, 12). In recent study we demonstrated that AnkA, a protein rich in ankyrin repeats and important for A. phagocytophilum infection, is translocated to the host cell cytoplasm in a T4S-dependent manner (19).
In several bacteria, it has been shown that the T4S apparatus components and substrates are not constitutively expressed but rather are tightly regulated to ensure proper timing of substrate action. In A. tumefaciens, the expression of the T4S system is regulated by a two-component system, VirA/VirG, which detects chemical signals such as phenolic compounds and particular monosaccharides released by wounded plant cells and subsequently induces transcription of the virBD T4S system (33). In L. pneumophila, the two-component PmrB/PmrA (35) and CpxA/CpxR (13) systems regulate the expression of the icm/dot T4S system. VjbR, a quorum-sensing regulator (9), Rsh, a RelA/SpoT stringent response protein homolog (11), and integration host factor (26) are involved in regulation of the virBD T4S system of Brucella sp. The expression of virBD genes is also regulated during A. phagocytophilum growth in human peripheral blood neutrophils (21). Although genes encoding the integration host factor α and β subunits and three different pairs of two-component systems have been identified in the A. phagocytophilum and E. chaffeensis genomes, genes encoding homologs of the quorum-sensing regulator or the stringent response protein have not been found (6, 17).
Unlike the single locus of clustered virBD genes encoding the T4S apparatus in most bacteria (7), split virBD loci have been found in E. chaffeensis, A. phagocytophilum, and other members of the order Rickettsiales (17, 22). The virBD genes of both E. chaffeensis and A. phagocytophilum are clustered in two primary loci, one consisting of five tandem genes (virB8-1, virB9-1, virB10, virB11, and virD4) and the other consisting of six tandem genes (virB3, virB4-1, and four virB6 paralogs) that are preceded by sodB. Between the primary loci, there are three duplicated genes, virB4-2, virB8-2, and virB9-2 (17, 22). The split virBD loci suggest that a coregulator is required to coactivate the transcription of genes encoding the T4S apparatus to ensure that the complete T4S apparatus is assembled in these bacteria when it is needed.
In the present study, we used quantitative PCR to determine the temporal expression pattern of the two primary loci and the intervening duplicated genes that comprise the other three loci during E. chaffeensis intracellular development in a human acute leukemia cell line, THP-1. We found that all five virBD loci are regulated in a generally synchronous manner. We further identified a previously unknown E. chaffeensis DNA binding protein, EcxR, which binds to promoter regions upstream of virBD loci. Functional studies suggested that EcxR positively autoregulates the expression of ecxR. We further found that EcxR coordinately regulates the expression of the dispersed T4S apparatus loci during E. chaffeensis intracellular development.
MATERIALS AND METHODS
Synchronous culture of E. chaffeensis.
E. chaffeensis Arkansas (1) was propagated in THP-1 cells (ATCC, Manassas, VA) in RPMI 1640 medium supplemented with 10% fetal bovine serum and 2% l-glutamine at 37°C under 5% CO2 as previously described (6). Heavily infected (>90%) THP-1 cells (8 × 107 cells) were harvested by centrifugation at 2,000 × g for 5 min. The pellet was resuspended in 8 ml of culture medium and sonicated on ice twice at setting 2 for 10 s using a W-380 sonicator (Heat Systems-Ultrasonics, Farmingdale, NY), and the unbroken cells and cell debris were removed by centrifugation at 4,000 × g for 5 min. Host-cell-free bacteria in the medium were sonicated on ice twice at setting 4.5 for 30 s to disrupt the fragile bacterial RCs. The sonication-resistant bacterial DCs were harvested by centrifugation at 18,000 × g for 5 min at 4°C. Bacterial viability was determined with a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Eugene, OR). To obtain a synchronous infection, the DC pellet was resuspended in 10 ml of culture medium and incubated with 4 × 107 uninfected THP-1 cells at 37°C for 1 h with shaking every 10 min. The mixture was then washed with cold 2× phosphate-buffered saline (274 mM NaCl, 5.4 mM KCl, 20 mM Na2HPO4, 4 mM KH2PO4; pH 7.4) three times and incubated at 37°C. Samples were collected at this time point (designated 0 h postinfection [p.i.]) and at 24, 48, 72, and 96 h p.i. by centrifugation at 2,000 × g for 5 min.
Quantitative reverse transcription (RT)-PCR.
THP-1 cells synchronously infected with E. chaffeensis were harvested at 0, 24, 48, 72, and 96 h p.i. as described above. One-half of the cells were suspended in RNAlater (Qiagen, Valencia, CA) and stored at −20°C for RNA extraction. The remaining cells were stored at −80°C for DNA extraction. Total DNA and RNA were extracted, and the RNA was reverse transcribed as described previously (6). Samples lacking reverse transcriptase were used to assess DNA contamination for each reaction. Quantitative PCR was performed as described previously (6). Briefly, gene-specific primers were designed to produce 100- to 150-bp amplicons (primer sequences are shown in Table 1). Serially diluted bacterial chromosomal DNA containing known copy numbers of the target genes was used as a standard. Quantitative PCR was performed using an Mx3000P instrument and a Brilliant SYBR green quantitative PCR core reagent kit (both obtained from Stratagene, La Jolla, CA). For each quantitative PCR assay, the dissociation curve was examined to confirm the absence of primer dimers. The log chromosomal DNA amount was plotted versus the cycle threshold value to establish standard curves for each gene. Means and standard deviations of mRNA copy numbers were determined. Samples were normalized by using the E. chaffeensis 16S rRNA gene and the amount of gyrB transcript at 0 h p.i.
TABLE 1.
Oligonucleotide primers used for quantitative RT-PCR
| Gene | Primer
|
Target size (bp) | |
|---|---|---|---|
| Directiona | Sequence (5′-3′) | ||
| gyrB | F | CTGGATTACACCACATGGTA | 141 |
| R | CTACAGGTATACCTCTACCA | ||
| sodB | F | ATTTTGGTAGCGGATGGGTA | 144 |
| R | AAGCATGCTCCCATACATCC | ||
| virB8-1 | F | TTTACAGCATTTGCGTCCAG | 142 |
| R | TTGCCGTTCTTGTTCATTCA | ||
| virB4-2 | F | CAGGTGTGATTTGAGCATGG | 113 |
| R | ACAACGAAAATGCACAAGCA | ||
| virB8-2 | F | AAAACAAATTGCGATAAATGGTG | 115 |
| R | AAACTCTAATGGCATAATTCCTGA | ||
| virB9-2 | F | ACGATGGAGAAGATATCGACA | 146 |
| R | GTAGTATACACGCTTGTTTGTA | ||
F, forward; R, reverse.
EMSA.
DNA fragments that were 351 and 327 bp long and corresponded to sequences upstream of the sodB and virB9-2 start codons, respectively, were amplified by PCR. A 434-bp DNA fragment corresponding to sequence upstream of the virB9-1 start codon was similarly amplified as a negative control (Table 2 shows the primer pairs used). PCR products were biotinylated, and the E. chaffeensis lysate supernatant was prepared as described previously (31). The lysate supernatant (approximately 5 μg protein) was incubated with each DNA probe (0.1 pmol) for 5 min at room temperature in a 20-μl reaction mixture containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 2.5% (vol/vol) glycerol, 0.1% (vol/vol) NP-40, and 50 ng/μl salmon sperm DNA. As a control for binding specificity, a separate reaction mixture containing the components described above plus a 50-fold excess of the corresponding unlabeled DNA probe was prepared. Samples were loaded onto a 6% native polyacrylamide gel in 0.5× Tris-borate-EDTA (0.5× TBE) buffer (0.044 M Tris base, 0.044 M boric acid, 0.001 M EDTA; pH 8.0) that had been prerun for 1 h, electrophoresed at 100 V for 2.5 h at 4°C, and then transferred to a nylon membrane (Amersham Biosciences, Piscataway, NJ) at 380 mA for 1 h. The transferred DNA was cross-linked to the membrane with UV light. The biotinylated DNA was detected using a LightShift chemiluminescent electrophoretic mobility shift assay (EMSA) kit (Pierce, Rockford, IL).
TABLE 2.
Oligonucleotide primers used to amplify promoter regions upstream of virBD loci
| Gene | Primer
|
Target size (bp) | |
|---|---|---|---|
| Directiona | Sequence (5′-3′) | ||
| virB8-1 | F | ATTAAACATACGTATCATCAACC | 258 |
| R | GAATCACGGCATACAAGTTAC | ||
| virB8-2 | F | GTAAAAGTAGTGCTAATCCTCA | 403 |
| R | CATAAAATTCTACATAATACATAATA | ||
| virB4-2 | F | AGACGGTGAATATTTATTGGCTAA | 365 |
| R | CAGGAAACCCTTATTTAACCCTA | ||
| virB9-1 | F | AGTAAACCCTGTATCAGTCA | 434 |
| R | CATCTGCCCTATAGTAAGTTA | ||
| virB9-2 | F | CAGTAGTGAACCTGATGCTA | 327 |
| R | GAACTTGCAACAAATCTATGC | ||
| sodB | F | TTGATAAGATACGATCCTTCC | 351 |
| R | TGTTGATATGGAAGTTCCGGT | ||
| ecxR | F | CAGCTTAGTTGTTGTACAGTAC | 460 |
| R | AGTTATGTACCGTAATCTAGTAA | ||
F, forward; R, reverse.
For EMSAs using purified recombinant EcxR (rEcxR), DNA fragments upstream of the virBD loci and ecxR were amplified by PCR (the primers used and amplicon sizes are shown in Table 2) and were biotinylated as described previously (31). The DNA probes (0.1 pmol each) were incubated in separate reaction mixtures with 25 ng rEcxR purified as described below, and EMSAs were performed as described above.
For supershift experiments, a DNA probe derived from a sequence upstream of sodB (1 pmol) was amplified by PCR, performed as indicated above, and incubated with 0.2 μg rEcxR at room temperature for 5 min. Then 2 μl mouse monoclonal anti-His tag antibody (Sigma, St. Louis, MO) was added, and the reaction mixture was incubated for another 5 min at room temperature. Sample electrophoresis was performed as described above, followed by DNA staining in 0.5× TBE containing 0.5 μg/ml ethidium bromide. Bands were visualized with a lAS-3000 luminescent image analyzer (Fujifilm, Stamford, CT).
Affinity purification of DNA binding proteins and mass spectrometry.
The promoter region upstream of sodB was amplified by PCR using a 5′ biotin-labeled primer as described previously (31) and was purified using a PCR purification kit (Qiagen). Following incubation of the E. chaffeensis lysate supernatant with 15 μg salmon sperm DNA at 4°C for 15 min, 50 pmol of the biotinylated promoter probe upstream of sodB was added to the reaction mixture and incubated for 30 min at 4°C. The DNA-bound proteins were affinity purified using a μMACS streptavidin kit (Miltenyi Biotec, Berglsch Gladbach, Germany), solubilized in 10 μl of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading buffer at 100°C for 5 min, and subjected to 15% SDS-PAGE analysis. After electrophoresis, the gel was fixed as described previously (31), and the resulting bands were excised and digested with trypsin (Promega, Madison, WI). Trypsinized polypeptides were identified by capillary liquid chromatography-nanospray tandem mass spectrometry as described previously (31).
Cloning and expression of rEcxR.
Full-length ecxR was amplified by PCR and directionally cloned into the NdeI and XhoI sites of the pET29a(+) vector (Novagen, San Diego, CA). The PCR primers used are shown in Table 3. The resulting plasmid (pEcxR) was amplified in Escherichia coli Novablue cells (Novagen), and the recombinant protein was expressed in E. coli BL21(DE3) cells (Novagen) as described previously (6). rEcxR was purified by Ni2+ affinity chromatography using a His-Select cartridge (Sigma) and was dialyzed against EMSA binding buffer.
TABLE 3.
Oligonucleotide primers used to construct plasmids
| Gene or region | Primer
|
Target DNA size (bp) | Enzyme | Plasmid | |
|---|---|---|---|---|---|
| Directiona | Sequenceb | ||||
| ecxR | F | TATACATATGACAACAATAAGTAACCAAAATG | 333 | NdeI | pET29a(+) |
| R | GTGCTCGAGATCTTCTTTTTGTATTATTACAAGA | XhoI | |||
| ecxR upstream region | F | GGGAAGCTTCAGCTTAGTTGTTGTACAGTAC | 460 | HindIII | pACYC184 lacZc |
| R | GGGGGATCCAGTTATGTACCGTAATCTAGTAA | BamHI | |||
| virB8-1 upstream region | F | GGGAAGCTTCCAGGATGGTCGTGGTATA | 450 | HindIII | pACYC184 lacZ |
| R | CCCGGATCCCAACCTTATAGTTTTAAGCTATA | BamHI | |||
| virB8-2 upstream region | F | GGGAAGCTTCCGGTATAGGTAAAATGAGTA | 454 | HindIII | pACYC184 lacZ |
| R | CCCGGATCCAAAATTCTACATAATACATAATACA | BamHI | |||
| virB4-2 upstream region | F | GGGAAGCTTAGACGGTGAATATTTATTGGCTAA | 365 | HindIII | pACYC184 lacZ |
| R | CCCGGATCCCAGGAAACCCTTATTTAACCCTA | BamHI | |||
| virB9-2 upstream region | F | GGGTCTAGACAGTAGTGAACCTGATGCTA | 343 | XbaI | pACYC184 lacZ |
| R | CCCGGATCCTCTAACATCAATATTGGAACTTG | BamHI | |||
| sodB upstream region | F | GGGAAGCTTCCTAAGGAACTAACAAGTGCA | 353 | HindIII | pACYC184 lacZ |
| R | CCCGGATCCATGTCCTCTATTAAATAAACACA | BamHI | |||
Construction of lacZ reporter fusions.
lacZ reporter fusions were constructed as described previously (31). Briefly, DNA fragments upstream of the virBD loci (Fig. 2B) and ecxR were amplified by PCR. The PCR primers used are shown in Table 3. A transcriptional fusion was constructed by placing these promoter fragments upstream of a promoterless lacZ gene in pACYC184 (New England Biolabs, Ipswich, MA). BL21(DE3) cells were cotransformed with pEcxR and each of the lacZ reporter constructs individually. The pET29a(+) vector alone was used as a negative control. After overnight culture, transformants were subcultured in LB medium supplemented with 50 μg/ml kanamycin and 25 μg/ml chloramphenicol at 37°C for 2 h, followed by induction with 0.0625 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h. β-Galactosidase activity was measured as described previously (31). rEcxR expression was determined by Western blot analysis using anti-His tag antibody as described previously (18).
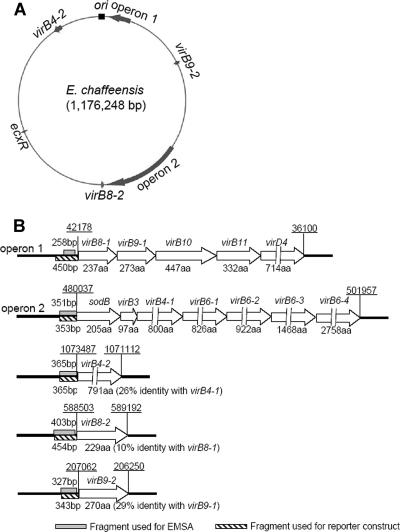
FIG. 2.
virBD gene loci and probe locations. (A) virBD gene loci on the E. chaffeensis genome. The genome map of E. chaffeensis is shown as a circle. The putative origin of replication (ori) is indicated by a black box. The virBD gene loci are indicated by gray arrows. Operon 1 includes virB8-1, virB9-1, virB10, virB11, and virD4. Operon 2 includes sodB, virB3, virB4-1, virB6-1, virB6-2, virB6-3, and virB6-4. The length of each arrow is eight times the length of the locus in the genome. (B) Schematic diagram of virBD gene loci and probe locations. virBD genes are represented by open arrows. The gene designations are above the arrows, and the numbers of amino acids (aa) in the corresponding gene products are indicated below the arrows. The lengths of virD4, virB4-1, virB4-2, virB6-1, virB6-2, virB6-3, and virB6-4 are not proportional, as indicated by discontinuous arrows. The upstream regions amplified by PCR to create DNA probes for EMSAs (shaded boxes) and for lacZ reporter constructs (hatched boxes) are indicated. The levels of amino acid sequence identity of the duplicated genes are indicated in parentheses. Chromosomal coordinates (underlined) are indicated above loci at the start and end of operons or genes.
Statistical analyses.
Statistical analyses were performed using analysis of variance and Tukey's honestly significant difference test or Student's t test, and a P value of <0.001 was considered significant.
RESULTS
Synchronous culture of E. chaffeensis.
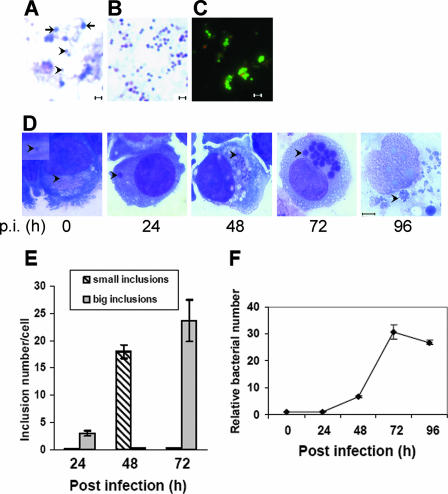
The size of host-cell-free E. chaffeensis cells liberated from heavily infected THP-1 cells by mild sonication (two 10-s pulses on setting 2) varied from 0.2 to 1.0 μm (Fig. 1A). Because the infectious elementary bodies from members of the genus Chlamydia are resistant to sonication (32), we examined whether host-cell-free E. chaffeensis was resistant to more intense sonication (two 30-s pulses on setting 4.5). We determined that approximately 30 to 40% of the bacterial cells remained intact. The sonication-resistant bacteria were smaller (<0.5 μm) and more densely stained with the basic dye in a Diff-Quik staining kit than the sonication-sensitive bacteria (Fig. 1A and 1B). Because the DC and RC forms described previously (34) were morphologically similar to our sonication-resistant and -sensitive forms, these forms were called DCs and RCs, respectively. Approximately 80% of the DC population was viable after more intense sonication (as described above), based on staining with the LIVE/DEAD BacLight bacterial viability kit (Fig. 1C). A synchronous culture of E. chaffeensis was prepared using the E. chaffeensis DC form as the inoculum (Fig. 1C). At 0 h p.i., only one to a few DCs per host cell were found by Diff-Quik staining (Fig. 1D). At 24 h p.i., almost every infected THP-1 cell (>95%) had one to five large (2- to 3-μm) morulae; at 48 h p.i., almost every infected THP-1 cell (>95%) had approximately 20 small (∼1-μm) morulae; at 72 h p.i., almost every infected cell (>99%) had more than 20 large (2- to 3-μm) and dense morulae; and by 96 h p.i., almost every infected THP-1 cell (>99%) began to lyse, and the morulae were loose and swollen (2 to 5 μm) (Fig. 1D and 1E). Quantitative PCR using a primer set specific for the E. chaffeensis 16S rRNA gene (6) revealed an approximately 24-h lag phase for E. chaffeensis growth, followed by an exponential growth phase from 24 to 72 h p.i. and a short stationary phase from 72 to 96 h p.i. (Fig. 1F). These results confirmed that there was synchronous growth of E. chaffeensis for up to 4 days p.i., until the onset of host cell lysis.
FIG. 1.
E. chaffeensis development in synchronously infected THP-1 cells. (A) Mixed developmental forms of E. chaffeensis liberated from infected THP-1 cells. The fragile RCs are indicated by arrows. The sonication-resistant DCs are indicated by arrowheads. Bar = 1 μm. (B) E. chaffeensis DCs are enriched after vigorous sonication. Cells were stained using Diff-Quik stain. Bar = 1 μm. (C) LIVE/DEAD BacLight bacterial viability test of E. chaffeensis DCs. Green indicates live bacteria, and red indicates dead bacteria. Bar = 1 μm. (D) Synchronously cultured E. chaffeensis in THP-1 cells using DCs as the inoculum. The bacteria or morulae are indicated by arrowheads. The inset shows a single bacterium associated with a host cell at 0 h p.i. All images show Diff-Quik staining of cells. Bar = 5 μm. (E) Numbers of small (<2-μm) and large (>2-μm) inclusions at different times after infection. One hundred infected THP-1 cells were scored at each time point. The values are the means ± standard deviations for three specimens. (F) Synchronous growth of E. chaffeensis determined by quantitative PCR. Genomic DNA extracted from infected THP-1 cells at different times after infection was subjected to real-time PCR analysis. The data indicate the numbers of bacteria relative to the number at 0 h p.i. The values are the means ± standard deviations for three specimens.
Temporal expression of five virBD loci.
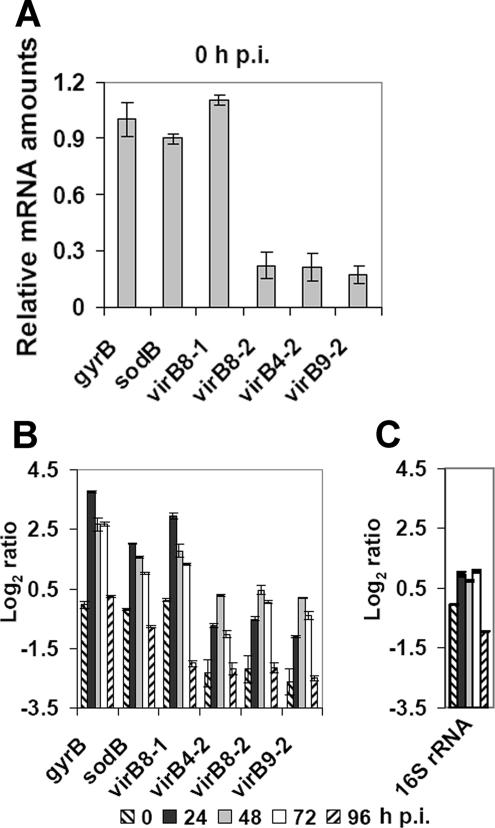
Five E. chaffeensis virBD loci are shown schematically in Fig. 2A and 2B and in Table S1 in the supplemental material. Constitutively expressed E. chaffeensis genes have not been identified, and thus expression of the gyrB gene was examined as a potential normalization control because previous studies have demonstrated that this gene is constitutively expressed in Chlamydia trachomatis (15). Using quantitative PCR, we determined that the expression of the two major virBD operons that start with virB8-1 and sodB was almost equal to that of E. chaffeensis gyrB at 0 h p.i., indicating that the T4S apparatus transcripts were expressed at a low level. Coexpression of each of the two operons as a single long transcript was shown previously (22). In contrast, the transcript levels of the three duplicated genes (virB4-2, virB8-2, and virB9-2) were much lower than that of E. chaffeensis gyrB at 0 h p.i. (Fig. 3A). The expression of the two major operons peaked at 24 h p.i. (the end of the lag phase) (Fig. 3B). The expression of three duplicated genes was significantly increased at 24 h p.i. and peaked at 48 h p.i. (the early exponential phase) (Fig. 3B). All five loci were downregulated at 96 h p.i. (the stationary phase), and the transcript levels were similar to those observed at 0 h p.i. These results suggest that the expression of virBD genes in E. chaffeensis is coordinately regulated during the intracellular development of this bacterium. Unlike transcription of gyrB in C. trachomatis (15), transcription of gyrB in E. chaffeensis was also regulated. The amounts of E. chaffeensis 16S rRNA at different time points were also determined as a measure of the E. chaffeensis total RNA by real-time RT-PCR. The results showed that the 16S rRNA levels changed much less than the virBD locus transcript levels during E. chaffeensis intracellular development (Fig. 3C).
FIG. 3.
Temporal expression of virBD genes in E. chaffeensis. (A) Quantitative RT-PCR to determine the expression of the five virBD loci at 0 h p.i. The transcript levels are the levels relative to the amount of the gyrB transcript at 0 h p.i. The values are the means ± standard deviations for three specimens. (B) Quantitative RT-PCR to determine the temporal expression of the five virBD loci. Transcript levels at different developmental stages were normalized by using the E. chaffeensis 16S rRNA gene. The transcript levels are expressed as the log2 ratio of the amount of a transcript at an indicated time point to the amount of the gyrB transcript at 0 h p.i. The values are the means ± standard deviations for three specimens. (C) Quantitative RT-PCR to determine the temporal amounts of 16S rRNA. The 16S rRNA levels of E. chaffeensis at different developmental stages were normalized by using the E. chaffeensis 16S rRNA gene. The 16S rRNA levels are expressed as the log2 ratio of the amount at an indicated time point to the amount of 16S rRNA at 0 h p.i. The values are the means ± standard deviations for three specimens.
E. chaffeensis protein binds to the promoter regions upstream of sodB and virB9-2.
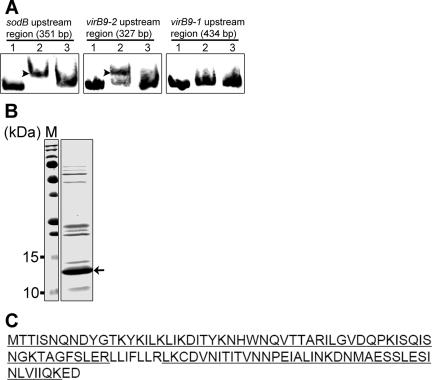
5′ Random amplification of cDNA ends was previously utilized to determine transcriptional start sites and σ70-like promoter elements upstream of both sodB and virB8-1 (22). The transcriptional start sites and σ70-like promoter elements of virB8-2, virB9-2, and virB4-2 upstream of these three genes were predicted by the BPROM program (Softberry, Inc., Mount Kisco, NY) (see Table S2 in the supplemental material). To determine whether an E. chaffeensis native protein binds to the promoter regions upstream of virBD loci, a DNA fragment upstream of the 5′-most proximal gene in the sodB-virB3-virB6 operon (sodB) and a DNA fragment upstream of one of the duplicated genes (virB9-2) were amplified by PCR and biotinylated. A DNA fragment upstream of the second gene in the virB8-1-virD4 operon (virB9-1) was amplified as a negative control. EMSAs revealed that the lysate of E. chaffeensis isolated from heavily infected THP-1 cells contained proteins that bound to the DNA probes derived from the sequences upstream of sodB and virB9-2 but not to the DNA probe derived from the sequence upstream of virB9-1 (Fig. 4A). Binding specificity for each biotinylated probe was demonstrated using a 50-fold excess of the corresponding unlabeled probe.
FIG. 4.
Identification of an E. chaffeensis protein bound to the DNA probes derived from sequences upstream of virBD loci. (A) EMSA of native E. chaffeensis proteins bound to the biotinylated DNA probes derived from sequences upstream of sodB and virB9-2. Native E. chaffeensis proteins bound to the DNA probes (0.1 pmol) upstream of sodB and virB9-2 but not virB9-1. Lane 1, DNA probe; lane 2, DNA probe incubated with E. chaffeensis lysate (5 μg); lane 3, DNA probe incubated with E. chaffeensis lysate in the presence of a 50-fold excess of unlabeled DNA probe. Shifted bands are indicated by arrowheads. Bands were visualized using a LightShift chemiluminescent EMSA kit. (B) Streptavidin affinity chromatography of E. chaffeensis proteins bound to the biotinylated probe derived from a sequence upstream of sodB. Following chromatography, the purified protein sample was subjected to 15% SDS-PAGE analysis, followed by staining. The identity of the protein indicated by the arrow was determined by mass spectrometry. Lane M contained prestained protein size standards. (C) Amino acid sequence identified for the E. chaffeensis native protein that bound to the DNA probe derived from the sequence upstream of sodB. One hypothetical protein (E. chaffeensis Arkansas Ech0795; GenBank accession number YP_507593) was identified by liquid chromatography-nanospray tandem mass spectrometry. Ninety-one percent of the amino acid sequence was determined (sequenced peptides are underlined).
Proteins from the E. chaffeensis lysate that bound to the biotinylated probe derived from the sequence upstream of sodB were purified using streptavidin affinity chromatography. SDS-PAGE analysis of the affinity-purified proteins revealed one predominant approximately 13-kDa polypeptide and several other polypeptides whose yields were significantly lower and which had various molecular masses (Fig. 4B). Tandem mass spectrometry of the predominant band identified the polypeptide as a 108-amino-acid, 12.3-kDa hypothetical protein (E. chaffeensis Arkansas ECH0795; GenBank accession number YP_507593) with 91% coverage of the amino acid sequence (Fig. 4C). We designated this protein EcxR, for E. chaffeensis expression regulator. The identities of the other polypeptides could not be determined by mass spectrometry due to insufficient amounts of the samples.
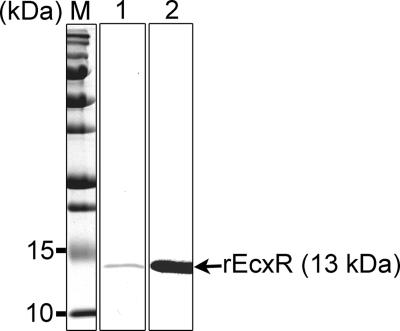
Cloning and expression of rEcxR yielded a 116-amino acid, 13-kDa protein that contained eight C-terminal amino acids derived from the pET29a(+) vector, including a His6 tag. The rEcxR protein was purified to apparent homogeneity as determined by SDS-PAGE analysis followed by Coomassie brilliant blue staining and by Western blot analysis (Fig. 5).
FIG. 5.
Purification of rEcxR. E. chaffeensis ecxR was cloned into the pET29a(+) vector, expressed, and purified using nickel chelate chromatography. The purified protein was subjected to 15% SDS-PAGE analysis, followed by Coomassie brilliant blue staining (lane 1) and Western blot analysis using an anti-His tag antibody (lane 2). Lane M contained prestained protein size standards. Each lane contained 1 μg of recombinant protein.
EMSA analysis with rEcxR.
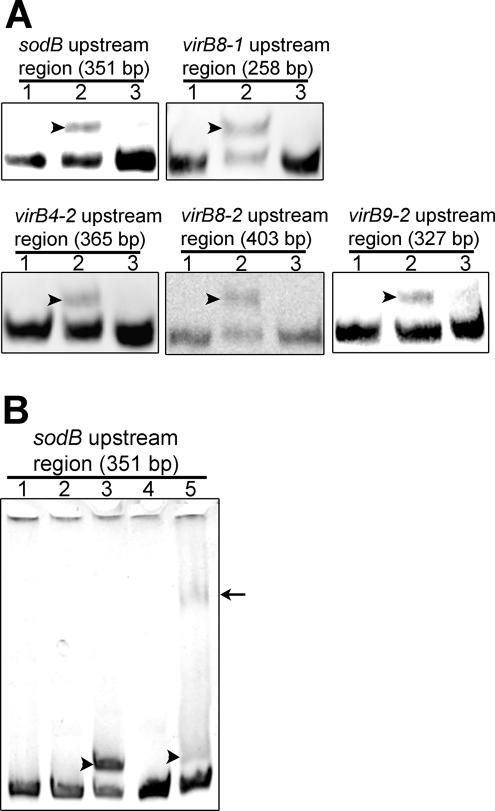
The EMSA analysis showed that the biotinylated DNA probes derived from sequences upstream of all five virBD loci were shifted upon incubation with rEcxR (Fig. 6A). The binding specificity of each DNA probe was demonstrated using a 50-fold excess of the corresponding unlabeled DNA probe. In addition, bovine serum albumin (BSA) did not shift the DNA probe derived from the sequence upstream of sodB (Fig. 6B). When an anti-His tag antibody, which recognized rEcxR, was added to the reaction mixture, the probe was supershifted in the presence of rEcxR but not in the presence of BSA (Fig. 6B).
FIG. 6.
Binding of rEcxR to the DNA probes derived from sequences upstream of virBD loci. (A) EMSA. Lane 1, DNA probe (0.1 pmol); lane 2, DNA probe incubated with rEcxR (25 ng); lane 3, DNA probe incubated with rEcxR in the presence of a 50-fold excess of the corresponding unlabeled DNA probe. Shifted bands are indicated by arrowheads. Bands were visualized using a LightShift chemiluminescent EMSA kit. (B) Antibody supershift of rEcxR bound to the DNA probe derived from the sequence upstream of sodB. Lane 1, DNA probe (1 pmol); lane 2, DNA probe incubated with 0.2 μg BSA; lane 3, DNA probe incubated with 0.2 μg rEcxR; lane 4, DNA probe incubated with 0.2 μg BSA in the presence of 2 μl anti-His tag antibody; lane 5, DNA probe incubated with 0.2 μg rEcxR in the presence of 2 μl anti-His tag antibody. The gel was stained with ethidium bromide. rEcxR-shifted bands are indicated by arrowheads, and the antibody supershifted band is indicated by an arrow.
rEcxR activates lacZ reporter fusions.
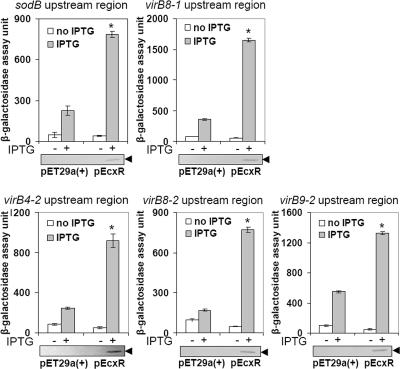
Because EcxR bound to all DNA probes derived from sequences upstream of the virBD loci, we examined whether these regions could be transactivated by EcxR. lacZ reporter fusions were constructed by individually inserting the five virBD promoter fragments (Fig. 2B and Table 3; see Table S1 in the supplemental material) upstream of the translation start site of the promoterless lacZ gene in pACYC184. Each of the lacZ reporter constructs was transformed into E. coli BL21(DE3) containing either pEcxR or empty pET29a(+) vector. For each of the five lacZ reporter constructs, IPTG induction of the pEcxR vector resulted in a significant increase in β-galactosidase activity compared to samples lacking IPTG or compared to IPTG induction of the empty pET29a(+) vector (Fig. 7). The expression of the rEcxR protein upon IPTG induction was confirmed by Western blot analysis (Fig. 7).
FIG. 7.
EcxR activates the transcription of virBD lacZ reporter fusions. β-Galactosidase assays were used to measure the transcriptional activities of lacZ reporter constructs. The values are the means ± standard deviations for three specimens. An asterisk indicates that a value is significantly different (P < 0.001) from the values for samples lacking IPTG or from the values for IPTG induction of the empty pET29a(+) vector as determined by the Tukey honestly significant difference test. Western blot analyses of samples from the β-galactosidase assays were performed using an anti-His tag antibody to verify the expression of rEcxR. The blots below the graphs are representative blots for three independent experiments. The position of rEcxR is indicated by arrowheads.
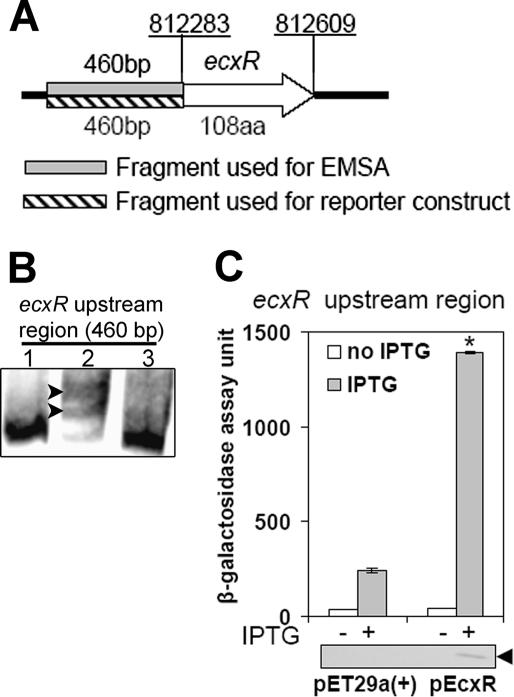
Autoregulation of EcxR.
The fragments used for the EMSA and the reporter construct are shown in Fig. 8A. The EMSA whose results are shown in Fig. 8B indicated that the DNA probe derived from sequence upstream of ecxR was also shifted upon incubation with rEcxR and that the interaction was specific. Therefore, we examined whether EcxR expression is autoregulated. A lacZ reporter construct containing the promoter region upstream of ecxR was prepared as described above, and the resulting plasmid was transformed into E. coli BL21(DE3) containing either pEcxR or the pET29a(+) vector alone. In the presence of this lacZ reporter construct, IPTG induction of the pEcxR vector resulted in a significant increase in β-galactosidase activity compared to samples lacking IPTG or compared to IPTG induction of the empty pET29a(+) vector (Fig. 8C). Expression of rEcxR upon IPTG induction was confirmed by Western blot analysis (Fig. 8C). Taken together, these data suggest that the expression of ecxR is positively autoregulated.
FIG. 8.
Expression of ecxR is autoregulated. (A) Schematic diagram of ecxR gene and probe locations. The ecxR gene is represented by an open arrow. The gene designation is indicated above the arrow, and the number of amino acids (aa) is indicated below the arrow. The upstream regions amplified by PCR to create DNA probes for EMSA (shaded box) and for the lacZ reporter construct (striped box) are indicated. Chromosomal coordinates (underlined) are indicated above the locus at the start and end of the gene. (B) EMSA of rEcxR bound to the DNA probe derived from the sequence upstream of ecxR. Lane 1, DNA probe (0.1 pmol); lane 2, DNA probe incubated with rEcxR (25 ng); lane 3, DNA probe incubated with rEcxR in the presence of a 50-fold excess of unlabeled DNA probe. Shifted bands are indicated by arrowheads. Bands were visualized using a LightShift chemiluminescent EMSA kit. (C) EcxR activates the transcription of an ecxR lacZ reporter construct. β-Galactosidase assays were used to measure the transcriptional activities of the ecxR-lacZ reporter construct. The values are the means ± standard deviations for three specimens. The asterisk indicates that the value is significantly different (P < 0.001) from the value for samples lacking IPTG or from the value for IPTG induction of the empty pET29a(+) vector as determined by the Tukey honestly significant difference test. Western blot analyses of samples from the β-galactosidase assay were performed using an anti-His tag antibody to verify the expression of rEcxR. The blot below the graph is a representative blot for three independent experiments. The position of rEcxR is indicated by an arrowhead.
DISCUSSION
In the present study, we developed a method to synchronize E. chaffeensis cultured in the human myelocytic leukemia cell line THP-1 using the sonication-resistant form of the bacterium. In DH82 cells (a canine histiocytoma cell line) at 1 day p.i., an E. chaffeensis cell is transformed from a small DC (0.4 to 0.6 μm) to a large RC (0.7 to 1.9 μm) and starts to multiply (34). After 3 days of culture, the bacterial population converts back to DCs to initiate new infection of host cells. The infectivity of the DC-rich population has been reported to be 3.5 million times greater than that of the RC-rich population (34). To establish a synchronized culture in THP-1 cells, we used controlled high-intensity sonication to rupture the RC population and enrich the infectious DC population. Morphological observation and real-time PCR analysis demonstrated the reliability of this method for synchronization. It was previously reported that in DH82 cells each E. chaffeensis bacterium progresses through its cycle in a discrete membrane-bound inclusion and is released upon host cell lysis (34). In contrast, in human THP-1 cells, we have found that a large inclusion is formed in the host cell by 24 h p.i., which then appears to disperse into multiple small inclusions at 48 h p.i. These small inclusions continue to expand until 72 h p.i., suggesting that a critical signaling event takes place between 24 and 48 h p.i. to form multiple inclusions in each THP-1 cell. The formation of multiple inclusions at 48 h p.i. coincides with the upregulation of the virBD genes. Future analysis of the molecular basis for this phenomenon may provide important insights into inclusion morphogenesis and development.
The synchronized culture allowed us to characterize the expression pattern of all five virBD loci during intracellular development of E. chaffeensis. At 0 h p.i., transcripts from the two major operons were expressed, suggesting that a low level of the T4S apparatus is present in E. chaffeensis prior to infection. A. phagocytophilum has virBD genes homologous to those of E. chaffeensis (17, 22). In A. phagocytophilum, translocation of AnkA into host cells was shown to commence in a virBD-dependent manner within a few minutes after infection and was shown to play an important role in facilitating intracellular infection by activating the Abl-1 signaling pathway (19). Although the T4S substrates in E. chaffeensis have not been determined, some T4S substrates may also be delivered via the low levels of the preformed T4S apparatus available at the early stage of infection.
The expression of the virB9 and virB6 genes, as well as VirB9 protein, is upregulated in A. phagocytophilum after internalization into neutrophils, whereas the expression of VirB9 is downregulated in human promyelocytic leukemia cell line HL-60 prior to release from the host cells (21). Similarly, here we demonstrated that the expression of the two major E. chaffeensis operons peaked at 24 h p.i. The expression of the three duplicated genes also significantly increased at 24 h p.i. and peaked at 48 h p.i. Legionella has been shown to inhibit phagosome-lysosome fusion in host macrophages (16), and Legionella-containing vacuoles mature into acidic, late endosomes for bacterial replication (27), which require a functional T4S system (8). E. chaffeensis replicates in a slightly acidic early endosome that does not mature into a late endosome (2). Here, we show that expression of the virBD genes was upregulated during the early exponential phase, suggesting that the T4S system contributes to the survival of E. chaffeensis and to the establishment of replicative inclusions. Additionally, our results suggest that two operons and three scattered duplicated virBD genes may be utilized by E. chaffeensis at slightly different stages of its developmental cycle.
In the present study, EcxR, a previously unidentified DNA binding protein, was identified and shown to activate the expression of the virBD genes in E. chaffeensis during intracellular development. The expression of ecxR was autoregulated, like many other transcription factors (20, 29, 30), in response to an unknown signal. The mechanism of the downregulation of the virBD genes has not been determined yet. The rapid degradation of mRNA at the stationary phase might contribute to this downregulation. These results suggest that the expression of the virBD genes is tightly regulated during the intracellular life cycle of E. chaffeensis and that EcxR plays an important role in this regulation. In mouse macrophages, the T4S system of B. abortus is essential for inhibition of lysosomal fusion of a bacterium-containing inclusion and for transformation of the initial inclusion into the replicative niche by acquisition of endoplasmic reticulum membranes (5). Although binding of a transcriptional regulator or a cis-acting element has not been described for regulation of the virB operon in Brucella, several reports have suggested that signaling events regulate virB expression during intracellular development. In Brucella suis, the virB promoter is induced in macrophages within 3 h p.i. (after the bacteria enter cells and phagosome acidification occurs) (3). The stringent response mediator Rsh is required for virB expression, suggesting that a nutrient-poor intracellular environment triggers Brucella melitensis virB expression (11). On the other hand, in B. melitensis, a quorum-sensing pheromone downregulates virB transcription (9, 28). However, an ecxR homolog has not been detected in Brucella, and genes encoding the RelA/SpoT homologs or genes required for biosynthesis of a quorum-sensing pheromone have not been found in the E. chaffeensis genome. Thus, in E. chaffeensis, the temporal regulation of the virBD loci during intracellular development is similar to that in Brucella, but the signaling events that lead to temporal regulation are different.
In Legionella, the icm/dot T4S system is organized into at least 11 transcriptional units that contain monocistronic as well as polycistronic transcripts, several of which contain a conserved sequence (TATAYT) that serves as their putative RpoD (σ70) recognition element and is essential for their expression (14). L. pneumophila contains homologs of at least six sigma factors (RpoD, RpoH, RpoF, RpoE, RpoS, and RpoN) (14). In contrast, E. chaffeensis encodes only two sigma factor homologs: a constitutive σ70 factor and a single alternative σ factor, σ32 (RpoH) (17). The paucity of alternative sigma factors suggests that the intracellular development of E. chaffeensis requires transcription factor regulation of the constitutive σ70-type promoters. The five virBD loci contain putative σ70-type promoters (22; this study), suggesting that EcxR is a common factor regulating T4S system genes. Interestingly the expression of gyrB, which encodes the β-subunit of DNA gyrase, also peaked at 24 h p.i. DNA gyrase is the bacterial type II topoisomerase responsible for introducing negative supercoiling into DNA (24) and is needed to maintain the supercoiling required for bacterial DNA replication, transcription, and recombination.
EcxR homologs have been found in A. phagocytophilum strains, in Anaplasma marginale St. Maries, in Ehrlichia canis Jake, and in Ehrlichia ruminantium Welgevonden and Gardel (31). In the genomes of all of these bacteria virBD loci are split (17). Therefore, the regulation of the virBD genes in these bacteria may be similar to the EcxR regulation of the virBD genes in E. chaffeensis. In A. phagocytophilum, the EcxR homolog ApxR regulates the expression of a putative transcription factor, tr1 (31), and the downstream p44E locus (30). p44E encodes the immunodominant pleomorphic 44-kDa major surface protein (31), suggesting that in E. chaffeensis the expression of tr1 and the downstream omp-1/p28 locus, which encodes the major outer membrane proteins (22), might be also regulated by EcxR.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant R01 AI054476.
We thank Mingqun Lin and Tzung-Huei Lai for their help with drawing the genome map of E. chaffeensis.
Footnotes
Published ahead of print on 11 January 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anderson, B. E., J. E. Dawson, D. C. Jones, and K. H. Wilson. 1991. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J. Clin. Microbiol. 292838-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnewall, R. E., Y. Rikihisa, and E. H. Lee. 1997. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect. Immun. 651455-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. USA 991544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celli, J., C. de Chastellier, D. M. Franchini, J. Pizarro-Cerda, E. Moreno, and J. P. Gorvel. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, Z., Y. Kumagai, M. Lin, C. Zhang, and Y. Rikihisa. 2006. Intra-leukocyte expression of two-component systems in Ehrlichia chaffeensis and Anaplasma phagocytophilum and effects of the histidine kinase inhibitor closantel. Cell. Microbiol. 81241-1252. [DOI] [PubMed] [Google Scholar]
- 7.Christie, P. J. 1997. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 1793085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coers, J., J. C. Kagan, M. Matthews, H. Nagai, D. M. Zuckman, and C. R. Roy. 2000. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38719-736. [DOI] [PubMed] [Google Scholar]
- 9.Delrue, R. M., C. Deschamps, S. Leonard, C. Nijskens, I. Danese, J. M. Schaus, S. Bonnot, J. Ferooz, A. Tibor, X. De Bolle, and J. J. Letesson. 2005. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell. Microbiol. 71151-1161. [DOI] [PubMed] [Google Scholar]
- 10.Demma, L. J., R. C. Holman, J. H. McQuiston, J. W. Krebs, and D. L. Swerdlow. 2005. Epidemiology of human ehrlichiosis and anaplasmosis in the United States, 2001-2002. Am. J. Trop. Med. Hyg. 73400-409. [PubMed] [Google Scholar]
- 11.Dozot, M., R. A. Boigegrain, R. M. Delrue, R. Hallez, S. Ouahrani-Bettache, I. Danese, J. J. Letesson, X. De Bolle, and S. Kohler. 2006. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell. Microbiol. 81791-1802. [DOI] [PubMed] [Google Scholar]
- 12.Economou, A., P. J. Christie, R. C. Fernandez, T. Palmer, G. V. Plano, and A. P. Pugsley. 2006. Secretion by numbers: protein traffic in prokaryotes. Mol. Microbiol. 62308-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gal-Mor, O., and G. Segal. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 1854908-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gal-Mor, O., T. Zusman, and G. Segal. 2002. Analysis of DNA regulatory elements required for expression of the Legionella pneumophila icm and dot virulence genes. J. Bacteriol. 1843823-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hefty, P. S., and R. S. Stephens. 2007. Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J. Bacteriol. 189198-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 1582108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hotopp, J. C., M. Lin, R. Madupu, J. Crabtree, S. V. Angiuoli, J. Eisen, R. Seshadri, Q. Ren, M. Wu, T. R. Utterback, S. Smith, M. Lewis, H. Khouri, C. Zhang, H. Niu, Q. Lin, N. Ohashi, N. Zhi, W. Nelson, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, J. Sundaram, S. C. Daugherty, T. Davidsen, A. S. Durkin, M. Gwinn, D. H. Haft, J. D. Selengut, S. A. Sullivan, N. Zafar, L. Zhou, F. Benahmed, H. Forberger, R. Halpin, S. Mulligan, J. Robinson, O. White, Y. Rikihisa, and H. Tettelin. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumagai, Y., Z. Cheng, M. Lin, and Y. Rikihisa. 2006. Biochemical activities of three pairs of Ehrlichia chaffeensis two-component regulatory system proteins involved in inhibition of lysosomal fusion. Infect. Immun. 745014-5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, M., A. den Dulk-Ras, P. J. Hooykaas, and Y. Rikihisa. 2007. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell. Microbiol. 92644-2657. [DOI] [PubMed] [Google Scholar]
- 20.Maamar, H., and D. Dubnau. 2005. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu, H., Y. Rikihisa, M. Yamaguchi, and N. Ohashi. 2006. Differential expression of VirB9 and VirB6 during the life cycle of Anaplasma phagocytophilum in human leucocytes is associated with differential binding and avoidance of lysosome pathway. Cell. Microbiol. 8523-534. [DOI] [PubMed] [Google Scholar]
- 22.Ohashi, N., N. Zhi, Q. Lin, and Y. Rikihisa. 2002. Characterization and transcriptional analysis of gene clusters for a type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect. Immun. 702128-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paddock, C. D., and J. E. Childs. 2003. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 1637-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reece, R. J., and A. Maxwell. 1991. DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26335-375. [DOI] [PubMed] [Google Scholar]
- 25.Rikihisa, Y. 2006. Ehrlichia subversion of host innate responses. Curr. Opin. Microbiol. 995-101. [DOI] [PubMed] [Google Scholar]
- 26.Sieira, R., D. J. Comerci, L. I. Pietrasanta, and R. A. Ugalde. 2004. Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol. Microbiol. 54808-822. [DOI] [PubMed] [Google Scholar]
- 27.Sturgill-Koszycki, S., and M. S. Swanson. 2000. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 1921261-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taminiau, B., M. Daykin, S. Swift, M. L. Boschiroli, A. Tibor, P. Lestrate, X. De Bolle, D. O'Callaghan, P. Williams, and J. J. Letesson. 2002. Identification of a quorum-sensing signal molecule in the facultative intracellular pathogen Brucella melitensis. Infect. Immun. 703004-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tramonti, A., P. Visca, M. De Canio, M. Falconi, and D. De Biase. 2002. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 1842603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, X., Z. Cheng, C. Zhang, T. Kikuchi, and Y. Rikihisa. 2007. Anaplasma phagocytophilum p44 mRNA expression is differentially regulated in mammalian and tick host cells: involvement of the DNA binding protein ApxR. J. Bacteriol. 1898651-8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, X., T. Kikuchi, and Y. Rikihisa. 2007. Proteomic identification of a novel Anaplasma phagocytophilum DNA binding protein that regulates a putative transcription factor. J. Bacteriol. 1894880-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warford, A. L., T. L. Carter, R. A. Levy, and K. A. Rekrut. 1985. Comparison of sonicated and nonsonicated specimens for the isolation of Chlamydia trachomatis. Am. J. Clin. Pathol. 83625-629. [DOI] [PubMed] [Google Scholar]
- 33.Winans, S. C. 1992. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol. Rev. 5612-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, J. Z., V. L. Popov, S. Gao, D. H. Walker, and X. J. Yu. 2007. The developmental cycle of Ehrlichia chaffeensis in vertebrate cells. Cell. Microbiol. 9610-618. [DOI] [PubMed] [Google Scholar]
- 35.Zusman, T., G. Aloni, E. Halperin, H. Kotzer, E. Degtyar, M. Feldman, and G. Segal. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol. 631508-1523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.