Abstract
Borrelia burgdorferi is a flat-wave, motile spirochete that causes Lyme disease. Motility is provided by periplasmic flagella (PFs) located between the cell cylinder and an outer membrane sheath. The structure of these PFs, which are composed of a basal body, a hook, and a filament, is similar to the structure of flagella of other bacteria. To determine if hook formation influences flagellin gene transcription in B. burgdorferi, we inactivated the hook structural gene flgE by targeted mutagenesis. In many bacteria, completion of the hook structure serves as a checkpoint for transcriptional control of flagellum synthesis and other chemotaxis and motility genes. Specifically, the hook allows secretion of the anti-sigma factor FlgM and concomitant late gene transcription promoted by σ28. However, the control of B. burgdorferi PF synthesis differs from the control of flagellum synthesis in other bacteria; the gene encoding σ28 is not present in the genome of B. burgdorferi, nor are any σ28 promoter recognition sequences associated with the motility genes. We found that B. burgdorferi flgE mutants lacked PFs, were rod shaped, and were nonmotile, which substantiates previous evidence that PFs are involved in both cell morphology and motility. Although most motility and chemotaxis gene products accumulated at wild-type levels in the absence of FlgE, mutant cells had markedly decreased levels of the flagellar filament proteins FlaA and FlaB. Further analyses showed that the reduction in the levels of flagellin proteins in the spirochetes lacking FlgE was mediated at the posttranscriptional level. Taken together, our results indicate that in B. burgdorferi, the completion of the hook does not serve as a checkpoint for transcriptional regulation of flagellum synthesis. In addition, we also present evidence that the hook protein in B. burgdorferi forms a high-molecular-weight complex and that formation of this complex occurs in the periplasmic space.
Borrelia burgdorferi, the causative agent of Lyme disease, is a spirochete with a characteristic flat-wave morphology (24, 25; for a review, see reference 12). As a result of their unusual morphology, B. burgdorferi and other spirochete species have a highly specialized ability that allows them to traverse viscous gellike media (8). This unique swimming ability enables these bacteria to penetrate into specific host connective tissues and ecological niches (43). The importance of motility as a virulence factor has been implicated in several spirochete species, including Treponema denticola (42), Brachyspira hyodysenteriae (54), Borrelia garinii (60), and B. burgdorferi (10, 56).
Motility in B. burgdorferi is provided by bundles of between 7 and 11 periplasmic flagella (PFs) that are subterminally attached near the cell ends. These PFs extend inward along the cell cylinder beneath an outer membrane sheath (6, 24, 28, 46). B. burgdorferi PFs have a structure similar to that of flagella of other bacteria; a PF is composed of a basal body, a hook, and a filament containing a single major flagellin (FlaB) and a minor flagellin (FlaA) (6, 21, 28). The location of FlaA in B. burgdorferi is unknown, but preliminary evidence indicates that this protein is on the surface of a PF proximal to the basal body (S. Satoshi, M. Motaleb, S. Aizawa, and N. W. Charon, unpublished data). Not only are the PFs essential for motility, but these structures are also critical in providing the characteristic flat-wave morphology of the intact spirochete (46, 59). Rotation of the PF bundles in opposite directions, as viewed along a PF from its distal end to where it inserts into the cell cylinder, generates backward-moving waves along the cell body that propel the cell forward (12, 24, 25, 38).
The flagellum synthesis described for Escherichia coli and Salmonella enterica serovar Typhimurium is a well-studied paradigm model. It is a finely orchestrated succession of motility gene expression and protein assembly that requires tight regulation. Flagellum synthesis is regulated by a cascade of transcriptional events involving the ordered expression of class 1, class 2, and class 3 motility genes (2, 14). Expression of class 1 genes, which comprise the master operon, directs the transcription of class 2 genes encoding the structural proteins involved in basal body synthesis, as well as specific regulatory proteins. These regulatory proteins control the expression of the class 3 genes, including those encoding flagellin and chemotaxis proteins. Among the class 2 regulatory genes are fliA, encoding the motility-specific transcription factor σ28, and flgM, encoding the anti-σ28 factor FlgM. FlgM and σ28 remain bound as a complex in the cytoplasm during basal body and hook synthesis. Upon completion of the hook, the hook-basal body complex provides an export route for FlgM. As FlgM exits the cell, σ28 is free to initiate the transcription of the class 3 late genes, including the genes encoding flagellin and chemotaxis proteins. Thus, completion of the hook-basal body structure acts as a general assembly checkpoint in the regulation of flagellum synthesis whereby flagellin and other genes are transcriptionally controlled in response to the state of hook completion (30). A similar scheme of cascade control of motility gene expression is found in many species of bacteria, and in some species, σ54 (RpoN) also directly participates in the cascade control of motility gene regulation (1, 4, 16, 44, 50).
The regulation of PF synthesis in B. burgdorferi, however, differs markedly from the regulation of flagellum synthesis in other species of bacteria. Homologs of the class 2 transcriptional regulatory genes fliA and flgM are not apparent, and no σ28 promoter consensus sequences are evident in the B. burgdorferi genome (19, 22, 38). Furthermore, when promoter sequences in B. burgdorferi are analyzed, only σ70 promoters are evident for initiation of motility and chemotaxis gene expression (19, 22, 38).
Little is known about the regulation of flagellum synthesis in B. burgdorferi. Recently, Motaleb et al. described a targeted mutant with a mutation in the major flagellin gene flaB that did not synthesize PFs, was nonmotile, and was rod shaped (46). The only putative motility and chemotaxis protein that did not accumulate at wild-type levels in this mutant was the minor filament protein FlaA; the decrease in the level of FlaA, however, in the flaB mutant was found to occur at the posttranscriptional level (49). As described above for other bacteria, the hook is necessary for flagellum structure and also provides a regulatory checkpoint during flagellum synthesis. We tested these two factors with regard to the importance of the hook in B. burgdorferi by targeting the gene (BB0283) that encodes the hook structural protein FlgE. Our results support the hypothesis that the PFs are important for the motility and morphology of B. burgdorferi. Furthermore, the inhibition of flagellar hook protein synthesis negatively impacted FlaA and FlaB filament protein accumulation, and FlaB synthesis was likely posttranscriptionally controlled. Finally, in the process of carrying out these experiments and producing FlgE antibodies, we found that FlgE forms a high-molecular-weight complex similar to that found in T. denticola and Treponema phagedenis (13, 40). Our results suggest that this complex is present once FlgE is secreted into the periplasmic space.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
High-passage B. burgdorferi sensu stricto strain B31A and the derived flaB mutant MC-1 (referred to as the flaB mutant) have been described previously (9, 46). Strain N40 was kindly provided by E. Fikrig, Yale University. Cells were grown at 34°C in BSK-II liquid medium or on plates in the presence of 3% carbon dioxide. Mutant cells derived from B31A were grown in the presence of kanamycin (300 μg/ml) or streptomycin (80 μg/ml). Swarm plate assays were performed using 0.35% agarose with BSK-II medium diluted 1:10 as previously described (46, 47). E. coli JM109 cells were grown in Luria-Bertani broth with the appropriate antibiotic.
Construction of flgE and fliF mutants and complemented strains.
Standard methods were used to construct flgE mutants (9, 47). Briefly, the flgE gene and the antibiotic resistance genes fused to the B. burgdorferi flgB promoter were amplified by PCR with primers P1 to P6 (Table 1), and the resultant PCR products were cloned into a pGEM-T vector (Promega). The kanamycin (kan) and streptomycin (aad) resistance cassettes were inserted into flgE at its single EcoRI site (9, 18). The resultant flgE-kan and flgE-aad portions of the constructs were amplified by PCR, and approximately 5-μg portions of PCR products were electroporated into competent B. burgdorferi cells. After 28 days of incubation, antibiotic-resistant colonies were picked and grown in liquid BSK-II medium for further analysis. A fliF mutant was isolated using the same technique with primers P7 and P8 (Table 1), with the kan cassette inserted into the NcoI site in the fliF gene. Restriction mapping indicated that the direction of transcription of kan in this insert was opposite that of fliF.
TABLE 1.
Oligonucleotide primers
| Primer | Descriptiona | Sequence |
|---|---|---|
| P1 | flgE (F) | 5′-CCCCACCCAATTATACTAGC-3′ |
| P2 | flgE (R) | 5′-GTCGCGTCACAAACATTAAG-3′ |
| P3 | kan, EcoRI (F) | 5′-GAATTCGGCGAATGAGCAAGCGCCG-3′ |
| P4 | kan (R) | 5′-ACCGGTGGCGAATGAGCTTGCGCCGTCC-3′ |
| P5 | aadA-1 (F) | 5′-GAATTCGCTACCCGAGCTTCAAGGAA-3′ |
| P6 | aadA-2 (R) | 5′-GAATTCGAAGTCGTCCTCCACGAAGT-3′ |
| P7 | fliF (F) | 5′-GTGGGCAAACTAGAGAGAAT-3′ |
| P8 | fliF (R) | 5′-GCCTCACAGAGTTCTTGACC-3′ |
| P9 | flgE, complement, NdeI (F) | 5′-CATATGATGAGGTCTTTATATTC-3′ |
| P10 | flgE, stop, HindIII-SacI (R) | 5′-GAGCTCAAGCTTATTAATTTTTCAATCTTACAAGTTC-3′ |
| P11 | flgB, promoter, HindIII (F) | 5′-AAGCTTTAATACCCGAGCTTCAAG-3′ |
| P12 | flgB, promoter, NdeI (R) | 5′-CATATGGAAACCTCCCTCAT-3′ |
| P13 | flgE, start (F) | 5′-AGGTCTTTATATTCTGG-3′ |
| P14 | flgE, stop (R) | 5′-GAGCTCAAGCTTATTAATTTTTCAATCTTACAAGTTC-3′ |
| P15 | flaA, RT (F) | 5′-TCATCTGCTATGATTATGCCACC-3′ |
| P16 | flaA, RT (R) | 5′-AGAATAAGCATATTCCATGCCAT-3′ |
| P17 | flaB, RT (F) | 5′-CATATTCAGATGCAGACAGAGG-3′ |
| P18 | flaB, RT (R) | 5′-CCCTGAAAGTGATGCTGGTGTG-3′ |
| P19 | eno, RT (F) | 5′-AACAGGAATTAACGAGGCTG-3′ |
| P20 | eno, RT (R) | 5′-AAATTGCATTAGCACCAAGC-3′ |
F, forward; R, reverse; RT, reverse transcription.
For complementation, the flgB promoter was first amplified by PCR with an engineered HindIII site at the 5′ end and an NdeI site at the 3′ end. The intact flgE gene was also amplified by PCR with an engineered NdeI site at 5′ end and a HindIII site at the 3′ end. The resultant PCR fragments were cloned into the pGEM-T Easy vector. The flgE gene was fused to the 3′ end of the flgB promoter at the NdeI site and confirmed by DNA sequence analysis. The HindIII-digested flgB-flgE fragment was further cloned into pBSV2 (62), which resulted in the complementing plasmid flgB-flgE/pBSV2.
PFs and hook-basal body complex purification.
We developed a new method to obtain PFs having attached hook-basal body complexes. Approximately 200 ml of late-logarithmic-phase cells (1 × 108 cells/ml) were centrifuged at 8,000 × g for 20 min, washed in 10 ml of 150 mM phosphate-buffered saline (pH 7.4) (PBS), and centrifuged at 8,000 × g for 10 min at room temperature. The cell pellet was resuspended in 10 ml of PBS with 1% (final concentration) myristate and gently shaken at 37°C for 30 min. Samples were centrifuged at 17,000 × g for 20 min at 4°C. The supernatant fluid (designated S1) containing the PFs was moved to another tube, polyethylene glycol 6000 was added to a final concentration of 2%, and the preparation was kept on ice for several hours. The pellet fraction was resuspended in 5 ml of a sucrose solution (0.5 M sucrose, 0.15 M Tris [pH 8]). Following addition of lysozyme (0.1 mg/ml) and disodium EDTA (2 mM, pH 8.0), the suspension was stirred on ice for 30 min and then at room temperature for 20 min. Myristate was added to a final concentration of 1%, and the preparation was stirred at room temperature for 30 min. Samples were centrifuged at 17,000 × g for 10 min at 4°C. Polyethylene glycol (2%) was added to the supernatant (designated S2), which was then incubated on ice for 30 min. S1 and S2 were centrifuged at 27,000 × g for 20 min at 4°C. The pellets were washed in 10 ml water and ultracentrifuged at 80,000 × g for 30 min at 4°C. Finally, the PFs were resuspended in 10 μl water and stored at 4°C. To obtain purified hook-basal body complexes, intact PFs were diluted in 50 mM glycine (pH 2.5) (total volume, 3 ml) and shaken gently at room temperature for 30 min. The preparation was then centrifuged at 100,000 × g for 30 min at 4°C. The pellet containing the purified hook-basal body structures was resuspended in 10 μl water and stored at 4°C.
Electrophoresis, Western blot, and Southern blot analyses.
Sodium-dodecyl-sulfate polyacrylamide-gel electrophoresis and Western blotting using the enhanced chemiluminescent detection system were performed as previously described (21). All gels were 10% polyacrylamide gels unless otherwise noted. Either labeled secondary antibodies or protein A was used for reaction detection. Whole-cell lysates were prepared by first washing cells in PBS and then boiling them for at least 5 min in Laemmli sample buffer. Polyclonal and monoclonal antibodies were kindly provided by the following investigators. Polyclonal antibodies directed against B. burgdorferi FlaB and MotB and E. coli FliM were provided by M. Caimano (University of Connecticut Health Center, Farmington), J. Carroll (University of Pittsburgh, Pittsburgh, PA), and D. Blair (University of Utah, Salt Lake City), respectively. Monoclonal antibodies to FlaB (H9724), FlaA, and DnaK were provided by A. Barbour (University of California, Irvine), B. Johnson (Centers for Disease Control and Prevention, Atlanta, GA), and J. Benach (State University of New York, Stony Brook), respectively. Polyclonal anti-FliI, anti-CheA2, anti-CheX, and anti-CheY3 were described previously (22, 37, 48). Polyclonal anti-recombinant CheW3 was provided by C. Li (unpublished data). The amounts of immunoreactive protein in the gels were determined using FluorChem spot densitometry as previously described (49). To analyze the stability of the FlgE high-molecular-weight complex, lysates were incubated for 60 min 23°C with either 8 M urea or 8 M guanidine-HCl. The urea-treated lysate was boiled in sample buffer, whereas the guanidine-HCl lysate was not boiled before the gel was loaded. Southern blotting was performed using standard techniques and HindIII-digested genomic DNA, and the blots were probed with a digoxigenin (Roche)-labeled kan cassette.
Generation of polyclonal antiserum directed against FlgE.
The flgE gene from B. burgdorferi was amplified using primers P9 and P10 (Table 1), and the PCR product was cloned into the pCR T7/NT-TOPO expression vector (Invitrogen) that encodes an amino-terminal histidine tag. The recombinant FlgE protein was induced using 0.1 M isopropyl-β-d-thiogalactoside, purified by using a nickel agarose column, and concentrated in 10-kDa-cutoff Amicon Ultra centrifugal concentrators (Millipore). Rabbits were immunized with 200 μg of purified recombinant FlgE protein for 1 month, with the first injection (100 μg) in Freund's complete adjuvant and the other two boosters in incomplete adjuvant. Due to the presence of nonspecific reactive bands in Western blots (see Fig. 6), polyclonal antisera were subsequently preadsorbed with acetone-treated E. coli (26), affinity purified using recombinant FlgE and an AminoLink Plus immobilization kit (Pierce), and eluted as recommended by the manufacturer.
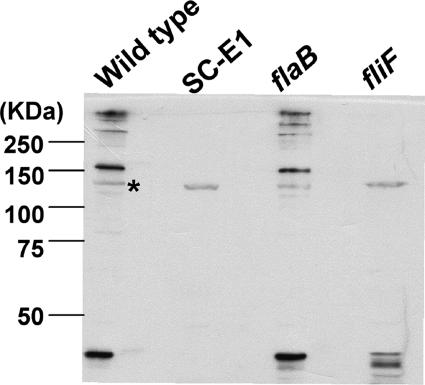
FIG. 6.
Reactivity of SC-E1 with anti-FlgE. Cell lysates of the wild type and the SC-E1, flaB, and fliF mutants were reacted with an antiserum to recombinant FlgE. Nonspecific binding was observed at approximately 140 kDa for each sample, as indicated by the asterisk.
qRT-PCR.
Quantitative reverse transcriptase PCR (qRT-PCR) analysis was carried out as previously described (49). Briefly, total RNA was isolated using an RNeasy kit (Qiagen) and treated with DNase (Turbo DNase I; Ambion) to remove contaminating DNA. Reverse transcription was performed with 100 ng of total RNA using the Reverse Transcription System (Promega) and random hexamers (Roche). Negative controls in which reverse transcriptase was omitted were included for each sample. The cDNA was amplified (ProofStart DNA polymerase; Qiagen) using the flaA P15 and P16 primers or the flaB P17 and P18 primers to generate DNA for use as standards. Sample cDNA, genomic DNA controls, and negative controls (no reverse transcriptase) were run simultaneously with flaA or flaB primers. Quantitative PCR was performed with a LightCycler (Roche) using a QuantiTect SYBR green PCR kit (Qiagen). RNA from the enolase gene (eno, BB0337) was amplified with primers P19 and P20 and was used as an internal control to normalize qRT-PCR data (29). The results were expressed as the threshold cycle at which the measured fluorescence increased above the background level.
Protein turnover assay.
The method used to assay turnover of FlaA and FlaB has been described previously (49). Briefly, cells (2 × 108 cells/ml) were incubated in BSK-II medium supplemented with spectinomycin (100 μg/ml) at 34°C. Five-milliliter samples were withdrawn at the indicated time points and centrifuged (8,000 × g at 4°C for 15 min). The cell pellets were washed by resuspension in 5 ml water and centrifugation at 13,000 × g for 5 min at 23°C. Cell lysates were then analyzed by Western blotting.
Light and electron microscopy.
The cell morphology and motility of wild-type and flgE mutant cells were characterized by light microscopy using previously described methods (46). To assay for the presence of PFs by electron microscopy, wild-type and mutant cells were fixed, embedded, and observed by transmission electron microscopy as previously described (39). Purified PFs and basal bodies were negatively stained on carbon collodion-coated copper grids with 2% phosphotungstate (pH 7.0) and observed with a JEOL JEM 1220 electron microscope using 80-kV acceleration.
RESULTS
Isolation of the SC-E1 mutant.
The flgE gene of B. burgdorferi was first identified by annotation in the published genomic sequence (19) and on the basis of similarity to its counterparts in other bacteria; the B. burgdorferi flgE product shares 41% identity and 63% similarity at the amino acid level to FlgE of E. coli and 44% identity and 59% similarity to FlgE of T. denticola. B. burgdorferi flgE was targeted by insertion mutagenesis using the kan cassette (9). Tiny kanamycin-resistant colonies appeared 28 days after mutagenesis, which is considerably longer than the time required for the appearance of wild-type colonies (diameter, 2 to 3 mm) after plating (10 to 14 days) and the appearance of colonies of the previously isolated chemotaxis and flaB mutants (45; C. Li, M. Motaleb, and N. W. Charon, unpublished). One of the clones, SC-E1, was characterized in detail. Southern blot analysis of SC-E1 indicated that the labeled kan probe hybridized to one unique HindIII fragment that was 3.8 kb long, which is the predicted fragment size for kan inserted into flgE (57). PCR analysis revealed that this cassette was inserted within and transcribed in the same direction as flgE (57). Furthermore, Western blot analysis using antibody to the recombinant FlgE confirmed that SC-E1 was deficient in FlgE synthesis (see Fig. 6). Taken together, these results indicate that kan specifically inserted within flgE at the desired EcoRI location in SC-E1.
Altered motility and morphology of mutant SC-E1.
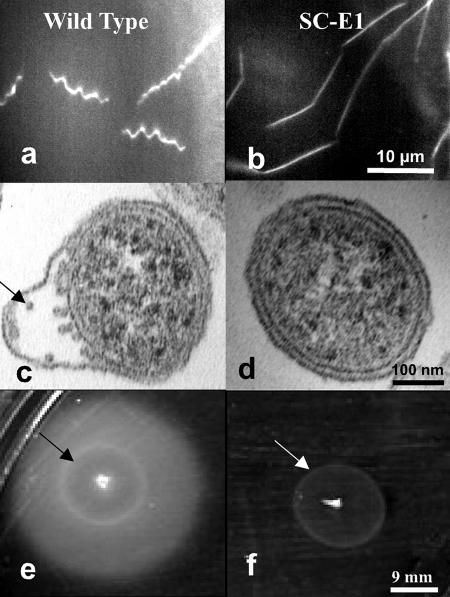
Analysis of targeted mutations in the major flagellin gene, flaB, indicated that loss of PFs in B. burgdorferi influences both cell morphology and motility (46, 59). Because the hook is essential for flagellum assembly in other bacteria, we determined if the cell morphology and motility of SC-E1 were also altered. Dark-field microscopy revealed that whereas wild-type spirochetes had a flat-wave morphology, SC-E1 cells were rod shaped and often grew in chains (Fig. 1a and b). Moreover, electron microscopic examination of thin sections of SC-E1 revealed that the cells completely lacked PFs (Fig. 1c and d). These observations further support the conclusions, drawn from analysis of the flaB mutant, that the PFs influence the shape of cells and have a pronounced skeletal function (46). We also tested whether the motility of SC-E1 was altered. Dark-field microscopy examination of SC-E1 indicated that the cells were completely nonmotile. Furthermore, in contrast to the wild-type cells, SC-E1 cells did not swarm on swarm agar plates (Fig. 1e and f). These results indicate that SC-E1 resembles the previously characterized flaB mutant (46, 59); both strains were deficient in filament synthesis, were rod shaped, and were nonmotile.
FIG. 1.
Cell morphology and swarming of SC-E1. (a and b) Dark-field micrographs showing (a) the flat-wave morphology of the wild type and (b) the rod-shaped morphology of SC-E1. (c and d) Thin-section electron micrographs showing (c) the PFs (arrow) in the wild type and (d) the lack of PFs in SC-E1. (e and f) Plate assays showing (e) the swarming of the wild type and (f) the lack of swarming of SC-E1. The white ring (arrow) indicates the area that was inoculated initially.
Decrease in PF filament proteins in SC-E1.
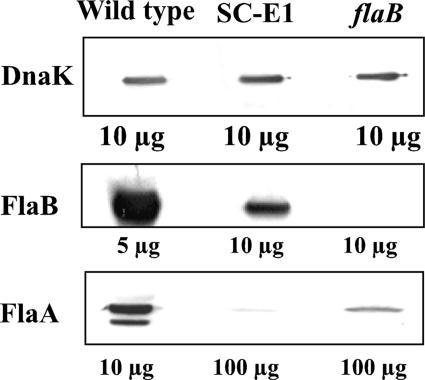
In most bacteria, mutants that do not form the flagellar hook are deficient in flagellar filament proteins (3, 34, 36, 39, 55). Accordingly, we used Western blot analyses to monitor the accumulation of the major FlaB PF filament protein (41 kDa) and the minor FlaA filament proteins (37 and 38 kDa) (21) in SC-E1. To visualize several of the reactions, relatively large amounts of cellular protein from SC-E1 were loaded in the gels. Using densitometry with DnaK as a control (Fig. 2, top panel), we detected a marked reduction (approximately 70 to 82%) in FlaB accumulation in SC-E1 compared to the wild type (Fig. 2, middle panel). FlaA has previously been shown to produce two bands on Western blots: a major band at 38 kDa and a minor band at 37 kDa (49). Although we were unable to detect the 37-kDa FlaA band, we found that there was a >85% reduction in the level of the 38-kDa band of FlaA in SC-E1. The reduction in the level of FlaA in SC-E1 exceeded that previously found in the flaB mutant (Fig. 2, bottom panel). These results indicate that the flgE mutation markedly inhibits FlaA and FlaB accumulation.
FIG. 2.
Analysis of the FlaA and FlaB proteins in SC-E1. Cell lysates from the wild type and the SC-E1 and flaB mutants were analyzed by Western blotting, and DnaK was used as an internal control. The amount of total protein loaded in each well is indicated. The levels of both FlaA and FlaB in the mutants were markedly decreased compared to the levels in the wild type.
Influence of the flgE mutation on the synthesis of select gene products encoded in motility operons.
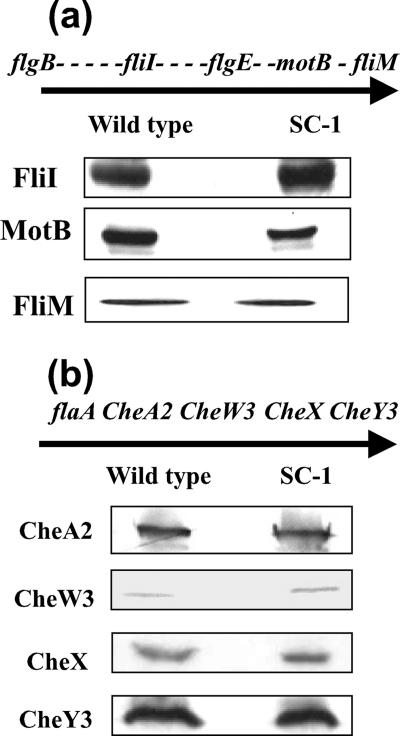
flgE maps near the center of the large flgB motility operon in the B. burgdorferi chromosome (19, 22). This operon is approximately 21 kb long and consists of 26 putative open reading frames. To determine the effect of the flgE mutation on the accumulation of protein products of this operon, we carried out a Western blot analysis of proteins encoded by genes both upstream (fliI) and downstream (motB and fliM) of flgE (Fig. 3a). The flagellum assembly protein FliI (22, 45) and the motor protein FliM (63) were present at similar levels in the wild type and SC-E1, while the level of MotB, which is part of the flagellar stator (33), was slightly reduced in SC-E1 compared to the wild type. Taken together, the Western blot analyses indicated that insertion of kan into flgE did not appreciably alter the amounts of proteins encoded by other select motility genes that were located either upstream or downstream of the mutation.
FIG. 3.
Western blot analysis of gene products of the flgB and flaA operons. Equal amounts of cell lysates from the wild type and SC-E1 were loaded on the gels, and Western blotting was carried out using the indicated antisera; DnaK was used as an internal control (not shown). The relative locations of genes in the (a) flgB operon and (b) flaA operon are shown, and the direction of transcription is indicated by the arrows. Each dash represents an open reading frame between the genes tested by Western blotting. Reverse transcription-PCR analysis indicated that both the flgB and flaA operons are polycistronically transcribed (20, 22).
Because both FlaA accumulation and FlaB accumulation were affected by the flgE mutation (Fig. 2), we tested whether this mutation affects other genes in the flaA operon. flaA is followed by the downstream chemotaxis genes cheA2, cheW3, cheX, and cheY3, which are transcribed as an operon (20). Furthermore, these genes have been shown to be involved in chemotaxis (5, 37, 48; M. Motaleb, C. Li, and N. Charon, unpublished). In contrast to the results for FlaA, blots probed with antisera to CheA2, CheW3, CheX, or CheY3 revealed that these proteins accumulated at wild-type levels in SC-E1 cells (Fig. 3b). These data strongly suggest that within this operon, the mutation in flgE specifically affects the accumulation FlaA. Similar results were obtained for a flaB mutant (49).
Decreases in FlaA and FlaB levels are unrelated to mRNA synthesis.
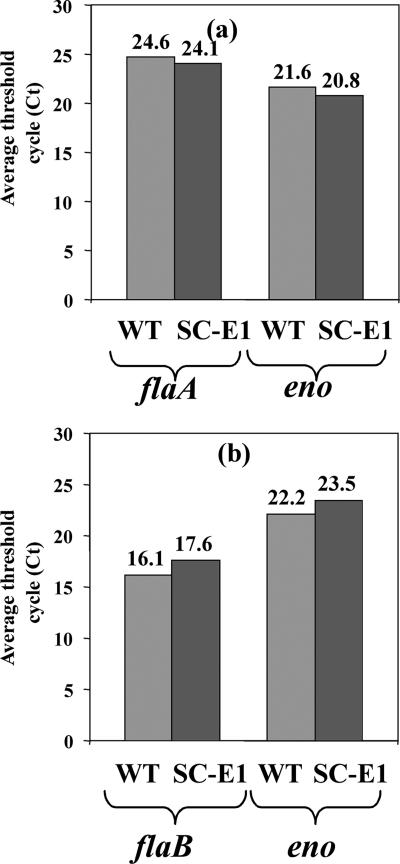
Although flagellin synthesis is regulated at the transcriptional level in other bacteria, the regulation of PF gene expression in B. burgdorferi is poorly understood. We used qRT-PCR to determine if the reduction in FlaB and FlaA levels in SC-E1 was the result of transcriptional regulation. Using the enolase gene as an internal control (29), we found that the levels of flaA (Fig. 4a) and flaB (Fig. 4b) transcripts in SC-E1 were similar to those in the wild type. These results differ from those obtained for other bacteria, in which inhibition of hook protein synthesis results in the cessation of flagellin transcription (3, 34, 36). Thus, in B. burgdorferi, completion of the PF hook is not a checkpoint for transcriptional control of PF synthesis.
FIG. 4.
qRT-PCR. Reverse transcription with random hexamer primers generated cDNA using total RNA from the wild type (WT) and SC-E1. Signals from qRT-PCR using specific primers for flaA or flaB were quantified with SYBR green fluorescent dye. The data from one representative experiment are expressed as the average threshold cycles (Ct) for duplicate samples. The experiment was repeated two more times, and similar results were obtained. The enolase gene, eno, was used as a control for data normalization in each experiment.
PF filament protein turnover in mutant SC-E1.
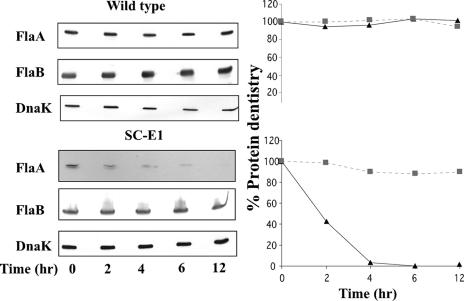
qRT-PCR results indicated that the lack of accumulation of FlaA and FlaB in SC-E1 occurs posttranscriptionally. This lack of accumulation could result from protein turnover or from translational control. To examine these possibilities, spectinomycin was added to growing cells of the wild type and SC-E1 to arrest translation, and the levels of FlaA and FlaB were assayed over time. Gels were loaded with approximately 10 to 20 times more protein from lysates from SC-E1 than protein from lysates from the wild type for detection of FlaA and FlaB. We found that FlaA accumulation in the wild type was stable (Fig. 5, top left panel). However, the level of FlaA in SC-E1 decreased, and the estimated half-life was 2 h; by 12 h, only a trace amount of FlaA was detected (Fig. 5, bottom left panel). These results indicate that the decrease in FlaA accumulation in SC-E1 could be explained, at least in part, by protein degradation. In contrast to FlaA levels, FlaB levels remained stable over 12 h in both wild-type and SC-E1 cells (Fig. 5). Because no turnover of FlaB was detected in SC-E1, translational control is evidently involved in the decrease in FlaB accumulation. These results suggest that the hook, while not a transcriptional checkpoint for filament gene transcription, still plays a key role in the accumulation of both FlaA and FlaB.
FIG. 5.
FlaA and FlaB stability. Spectinomycin (100 μg/ml) was added to wild-type or mutant SC-E1 cells to arrest translation. Samples were withdrawn at the indicated time points and analyzed by Western blotting (left panels). Protein density was determined for FlaA (▴) and FlaB (▪) for the wild type (top right panel) and mutant SC-E1 (bottom right panel). The results are expressed as the percent protein density at zero time compared to subsequent time samples. Gels were loaded with 5 μg of lysate of the wild type, 100 μg of SC-E1 for the FlaA blot, and 50 μg of lysate for the FlaB blot. Five micrograms of lysate was loaded for each strain for the anti-DnaK blot.
Reactivity with FlgE antiserum.
We tested for production of the FlgE protein by performing a Western analysis using a rabbit antiserum raised against recombinant FlgE. Surprisingly, multiple bands from the wild type were found to react to the antiserum (Fig. 6). One band was at approximately 46 kDa, which is the predicted size of monomeric FlgE. A nonspecific band at approximately 140 kDa was present in all samples, and in subsequent experiments affinity-purified antibodies were used. No breakdown of the large complex was detected even after 10 min of boiling in sample buffer, and the complex was stable in the presence of β-mercaptoethanol, dithiothreitol, 8 M guanidine-HCl, and 8 M urea. Formation of a high-molecular-weight FlgE complex was previously reported for T. denticola and T. phagedenis (13, 40), and several lines of evidence indicated that formation of this complex is the result of cross-linking of FlgE. In contrast, Jwang et al. reported that such a complex was absent in B. burgdorferi (31).
Several mutants were tested using this antiserum. As expected, flgE mutant SC-E1 failed to react at 46 kDa (Fig. 6). In addition, the SC-E1 lane did not contain bands corresponding to the high-molecular-mass complex observed in the wild type, although a weak nonspecific band was observed at approximately 140 kDa (see below). These results verify that inactivation of flgE inhibits FlgE synthesis. Cells of the flaB mutant had a pattern of reactivity identical to that of the wild type (Fig. 6). These results suggest that inhibition of filament synthesis did not influence formation of the high-molecular-weight FlgE complex. However, cells of the fliF mutant were quite different. fliF in other bacteria encodes the MS ring, which is essential for flagellar rotation and basal body, hook, and flagellum assembly (7, 35). Consistent with this proposition, the B. burgdorferi fliF mutant was found to be nonmotile, had undetectable amounts of FlaA and FlaB as determined by Western blot analysis, and was rod shaped (M. Motaleb and N. Charon, unpublished). Western blot analysis using the FlgE antiserum indicated that cells of the fliF mutant did not form the high-molecular-weight FlgE complex (Fig. 6). Reactivity was detected at approximately 46 kDa, and new bands appeared in this region compared to the wild type. These results suggest that in a fliF mutant, FlgE is synthesized but formation of the high-molecular-weight complex is inhibited.
Complementation of flgE mutation.
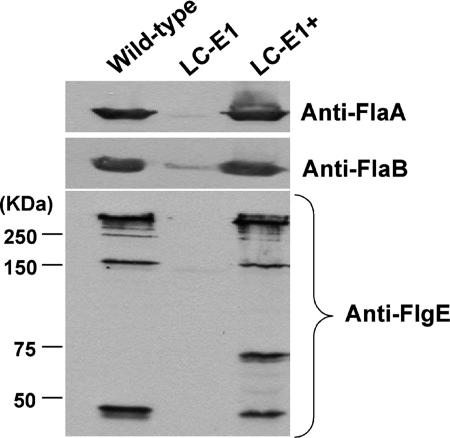
One concern was whether a secondary mutation was responsible for the altered phenotype of SC-E1. To test this possibility, we attempted to genetically complement the SC-E1 mutation. We were unable to successfully complement SC-E1 despite using several different vectors, various methods for preparing competent cells, and different plating techniques (57). Because different antibiotic cassettes influence the plating efficiency of B. burgdorferi on selective media and could have influenced our ability to achieve complementation (18), we constructed a new mutant strain (LC-E1) by replacing the kan cassette at the same EcoRI site in flgE with the aad streptomycin resistance cassette. LC-E1 had the same phenotype as SC-E1; it formed tiny colonies, was rod shaped, and was nonmotile. Furthermore, Western blot analysis of gene products of the flgB operon indicated that there were no polar effects resulting from the aad mutation (not shown) and that the mutation also reduced accumulation of FlaA and FlaB, as found in SC-E1 (Fig. 7, top panel). LC-E1 was successfully complemented with the flgE/pBSV2 vector containing kan and the intact flgE gene preceded by the flgB promoter. PCR analysis indicated that the entire flgE gene was present as a replicating plasmid (not shown). Phenotypic analysis of the complemented strain (designated LC-E1+) revealed that the cells exhibited both the flat-wave morphology and motility based on dark-field microscopy and swarm plate assays (not shown). Furthermore, Western blotting indicated that complementation of flgE resulted in the production and accumulation of FlaA and FlaB at levels similar to wild-type levels (Fig. 7, top panel). We tested lysates of the wild type, LC-E1, and LC-E1+ with the affinity-purified anti-recombinant FlgE antiserum. As observed for SC-E1 (Fig. 6), FlgE accumulation in LC-E1 (Fig. 7, bottom panel) was inhibited. In addition, the nonspecific band at approximately 140 kDa was absent when this purified FlgE antibody was used. The Western blot patterns of the wild type and LC-E1+ were similar (Fig. 7). There was a new band at 70 kDa in LC-E1+ compared to the wild type. The basis for this band is unclear, but it could have been the result of overproduction of a high-molecular-weight FlgE intermediate, followed by degradation. These results verify that inactivation of flgE inhibits FlgE synthesis and results in the altered phenotype.
FIG. 7.
Western analysis of the LC-E1 mutant and complemented strain. Cell lysates were reacted with anti-FlaA, anti-FlaB, and anti-FlgE affinity-purified antibodies. LC-E1 did not react to the FlgE antiserum, and the wild type and LC-E1+ reacted at 46 kDa and at higher molecular masses.
High-molecular-weight complex of FlgE is associated with the basal bodies.
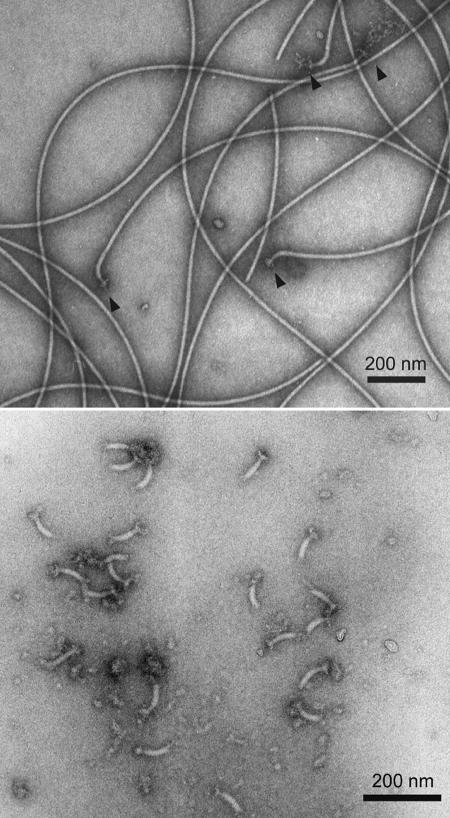
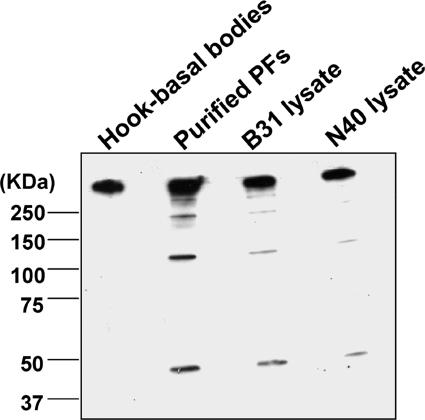
We directly tested if the FlgE antiserum specifically reacts with the flagellum basal body apparatus and another strain previously reported not to form a high-molecular-weight complex of FlgE. We purified PFs containing intact hook-basal body structures using a new gentle lysis procedure (Fig. 8, top panel). We also purified the hook-basal body structures from these PFs (Fig. 8, bottom panel). The hook length was determined to be 61 ± 4.4 nm (mean ± standard deviation), which is slightly longer than the 50 nm reported for other Borrelia species (6). The PFs and purified hook-basal body structures were analyzed by Western blotting using affinity-purified FlgE antibodies. The molecular mass of most of the FlgE protein was found to be greater than 160 kDa in the lanes containing the hook-basal body complexes, PFs, and whole-cell lysates, and there were only small amounts of the monomeric 46-kDa protein in the PFs and whole-cell lysates (Fig. 9). These results indicate that FlgE exists as a stable high-molecular-weight complex within the basal bodies. We also tested the affinity-purified antiserum with B. burgdorferi strain N40, which reportedly did not form the high-molecular-weight complex (31). We found that the analysis of strain N40 yielded results identical to those obtained with strain B31 (Fig. 9). These results suggest that formation of the large FlgE complex is not unique to strain B31.
FIG. 8.
Electron micrographs of (top panel) purified PFs containing hook-basal body complexes (arrowheads) and (bottom panel) purified hook-basal body complexes. Samples were negatively stained with phosphotungstate.
FIG. 9.
Western blot analysis of purified components and strain N40. Hook-basal body complexes, PFs, and cell lysates of different strains were run on an 8% polyacrylamide gel and then reacted with affinity-purified FlgE antibodies after blotting.
DISCUSSION
The flagellar hook acts as a flexible coupling linking the flagellar motor to the filament (58). In addition, it also plays a central role in flagellum assembly, as it acts as a checkpoint for late motility gene transcription (2, 14, 30). In several bacterial species, including the spirochete T. denticola, inactivation of flgE results in a failure to complete flagellum filament synthesis (34, 36, 39, 55). In the studies reported here, we found that inactivation of flgE resulted in cells that were nonmotile and deficient in PFs. In addition, flgE mutant cells were not flat waves but were rod shaped. Because both motility and the flat-wave morphology were restored by complementation, secondary mutations were not involved in these altered phenotypic characteristics. Similar results were obtained for a flaB mutant, which was also nonmotile and rod shaped and lacked the flat-wave morphology of the wild type (46, 59). Our results obtained with the two flgE mutants, along with the results for the fliF mutant also presented here, extend these findings and thus further support the conclusion that the PFs have a skeletal function and that this organelle is essential for motility.
Three lines of evidence strongly indicate that B. burgdorferi has a unique system for controlling its motility genes. First, as mentioned above, there are neither σ28 recognition sequences upstream of the motility genes nor any detectible genes encoding σ28 transcription factors (fliA) and anti-σ28 factors (flgM) (19). In addition, all of the motility promoters identified so far are σ70-like (19, 22, 38). However, discrimination between σS and σ70 promoters in B. burgdorferi based on promoter consensus sequences is problematic (11, 66). Recently, using microarray and transcriptional analysis, Caimano et al. have shown that whereas flagellum biosynthetic genes are controlled by σ70 both in vitro and in vivo, the methyl-accepting chemotaxis genes mcp1, mcp4, and mcp5 and the chemotaxis gene cheW2 are upregulated by σS (RpoS) (11) during growth in vivo. For B. burgdorferi, evidence suggests that σS is directly controlled by σ54 (61, 66). σ54 directly participates in the transcription of motility gene expression in other bacteria, such as Caulobacter crescentus, Vibrio species, Pseudomonas aeruginosa, and Helicobacter species (4, 16, 44, 50). Although it has been speculated that σ54-like promoter sequences are upstream of some of the motility genes in B. burgdorferi (17), there is no definitive proof that transcription is initiated at these sites. Furthermore, B. burgdorferi lacking σS or σ54 remains motile (17). Taken together, these results suggest that B. burgdorferi is not dependent on σ28-, σS-, or σ54-mediated cascade transcriptional control for flagellar gene expression.
Second, as described here, inactivation of flgE dramatically decreases the accumulation of FlaB. The amounts of flaB message in the flgE mutant SC-E1 and the wild type were essentially equivalent. The results of the decreased amount of FlaB in the flgE mutant can best be explained by translational control. Although sigma factor cascade transcriptional control of flagellum synthesis is a major mechanism of control in C. crescentus, Bacillus subtilis, and S. enterica, translation control of flagellin also occurs in these species (1-3, 41, 65). As suggested by Yakhnin et al. for B. subtilis, such control could fine-tune flagellin synthesis in response to the environment (65). In the absence of cascade control of motility gene transcription, B. burgdorferi appears to rely exclusively on translational control of its motility genes. Finally, as found in many other species of bacteria, mutation of flgE results in inhibition of the expression of chemotaxis genes (16, 34, 36). These genes are generally the last genes transcribed in the cascade. We found no inhibition of CheA2, CheX, and CheY3 after inactivation of flgE. These results further reinforce the hypothesis that there is a lack of cascade control of motility gene transcription in B. burgdorferi.
The results obtained for the filament protein FlaA are complex. We found that whereas FlaA accumulation was inhibited, the levels of the downstream genes in the operon encoding CheA2, CheW3, CheX, and CheY3 were the wild-type levels. Similar results were obtained in an analysis of a flaB mutant (49). In this previous study, we speculated that the low rate of turnover noted for FlaA in the flaB mutant is likely the result of excretion of this protein into the periplasmic space by a SecA-dependent mechanism and then degradation due to its inability to complex with FlaB. As reported in this paper, inactivation of flgE resulted in a decrease in the accumulation of FlaA but not in the accumulation of the encoding message. Slow turnover occurred with FlaA in the flgE mutant, as observed for the flaB mutant; both proteins had a half-life of 2 h (49). Because FlaA was turned over in the mutant, it is not possible at this time to sort out whether the decrease can be attributed to translational control, to turnover, or to both of these factors. However, if translational control was responsible, it did not affect the translation of the other genes in the operon. Remarkably, the amount of FlaA that accumulated in the flgE mutant was dramatically less than the amount found in the flaB mutant. These results indicate that the presence of the hook positively influences FlaA accumulation.
Although transcriptional cascade control is not involved in the regulation of B. burgdorferi motility genes, the expression of some of these genes is modulated. Specifically, as shown by microarray analysis, multiple chemotaxis-related genes are upregulated in B. burgdorferi either in vivo or under in vitro conditions that mimic the mammalian or tick hosts (11, 17, 53, 64). qRT-PCR confirmed that there is upregulation of some of these genes (M. Caimano and J. Radolf, personal communication). We previously hypothesized that motility and chemotaxis gene expression is constitutive in B. burgdorferi (12, 22). We based this hypothesis on the lack of σ28-dependent cascade control, the evidence that flaB is transcribed in both the tick and the mammalian host, and the finding that spirochetes in the tissues of these hosts contain PFs (15, 23). We extended this hypothesis by adding the caveat that expression of some of these genes can be modulated depending on the environment.
Western blot analysis indicated that most of the hook protein exists as a high-molecular-weight complex in B. burgdorferi. Furthermore, both the monomeric and high-molecular-weight forms were absent in both flgE mutants but were present in the complemented strain. Using an antiserum specific for recombinant FlgE, we detected bands with molecular weights greater than 160,000 in whole-cell lysates. Most of the reacting protein was present in these high-molecular-weight complexes. Analysis of both purified PFs and purified basal body complexes and the use of affinity-purified antibodies verified that these large protein species were specific to FlgE. A ladder of high-molecular-weight bands corresponding to FlgE has been identified in T. phagedenis and T. denticola, and it was concluded that the hook proteins were covalently cross-linked in these species (13, 40). Our results are similar to those obtained for these Treponema species, indicating that the hook protein in B. burgdorferi is cross-linked, but the results are in contrast to those of Jwang et al., who reported that FlgE in B. burgdorferi was monomeric and did not form high-molecular-weight complexes (31). In an attempt to resolve this difference, we further tested strain N40, which is the strain used by Jwang et al., and we obtained results similar to those obtained for strain B31. One possible explanation is that because the high-molecular-weight complex migrates very slowly, the complex could have remained in the stacking gel. In fact, this has occurred in some of our gels.
The nature of FlgE cross-linking in both Treponema spp. and B. burgdorferi is unknown, but to a large extent this cross-linking resembles the cross-linking of phage head proteins of HK-97 and the recently reported cross-linking of pilus proteins in Streptococcus pyogenes (27, 32, 52). As observed for HK-97 phage head proteins (52), the high-molecular-weight complex was found to be stable when it was boiled and when it was exposed to a number of protein-denaturing agents. Because the PFs necessarily rotate in the confined region between the outer membrane sheath and the cell cylinder, perhaps this cross-linking adds extra strength and flexibility to the hook as a means to promote more efficient swimming. It is interesting that the fliF mutant did not form the high-molecular-weight FlgE complex and only the monomeric FlgE species was detected. Because FliF would be essential for FlgE secretion in B. burgdorferi, as it is in most bacterial species (7, 35), our results suggest that the cross-linking occurs in the periplasmic space. In contrast to other bacteria, in which the flagella are exposed to the ambient medium, in spirochetes the flagellum organelles are located intracellularly. Thus, because of the location of the PFs, perhaps spirochetes are more enzymatically capable of cross-linking their flagellar structures than most bacteria. Because Treponema and Borrelia are phylogenetically divergent genera (51), we speculate that cross-linking of FlgE will be found in other spirochete species as well. Our results, however, do not rule out the possibility that other proteins besides FlgE are part of the high-molecular-weight complex.
The unique cell features and specialized motility of B. burgdorferi make this organism challenging to study. Characterization of B. burgdorferi lacking the hook protein FlgE has reinforced the idea that PFs are essential for the flat-wave morphology, as well as for motility. In addition, the flgE mutant strains have provided important insight into the regulation of PF synthesis in B. burgdorferi. From studies of these mutant strains, we have concluded that the PF hook is not a transcriptional checkpoint for the regulation of PF synthesis. The FlaB flagellin and possibly the flagellin FlaA appear to undergo posttranscriptional regulation beyond simple protein degradation in the absence of the PF hook. The nature of posttranscriptional flagellin regulation in B. burgdorferi and the formation of the complex flagellar hook structure of this organism remain exciting areas to be explored.
Acknowledgments
We thank A. Barbour, J. Benach, D. Blair M. Caimano, J. Carroll, and B. Johnson for antibodies, E. Fikrig for providing a strain, and M. Caimano and D. Yelton for helpful discussions. We also thank Diane Berry for assistance with electron microscopy.
This research was supported by Public Health Service grants AI29743 and P20 RR15574 to the Sensory Neuroscience Research Center at West Virginia University. We are also grateful to CREST programs, Japan Science and Technology Agency, for their financial support to S.-I.A.
Footnotes
Published ahead of print on 11 January 2008.
REFERENCES
- 1.Aizawa, S.-I., I. B. Zhulin, L. M. Marquez-Magana, and G. W. Ordal. 2001. Chemotaxis and motility in Bacillus subtilis, p. 437-452. In A. L. Sonenshein, R. Losick, and J. A. Hoch (ed.), Bacillus subtilis and its relatives: from genes to cells. ASM Press, Washington, DC.
- 2.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5160-165. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, D. K., and A. Newton. 1997. Posttranscriptional regulation of Caulobacter flagellin genes by a late flagellum assembly checkpoint. J. Bacteriol. 1792281-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, D. K., N. Ohta, J. Wu, and A. Newton. 1995. Regulation of the Caulobacter crescentus rpoN gene and function of the purified σ54 in flagellar gene transcription. Mol. Gen. Genet. 246697-706. [DOI] [PubMed] [Google Scholar]
- 5.Bakker, R. G., C. Li, M. R. Miller, C. Cunningham, and N. W. Charon. 2007. Identification of specific chemoattractants and genetic complementation of a Borrelia burgdorferi chemotaxis mutant: a flow cytometry-based capillary tube chemotaxis assay. Appl. Environ. Microbiol. 731180-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour, A. G., and S. F. Hayes. 1986. Biology of Borrelia species. Microbiol. Rev. 50381-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg, H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 7219-54. [DOI] [PubMed] [Google Scholar]
- 8.Berg, H. C., and L. Turner. 1979. Movement of microorganisms in viscous environments. Nature 278349-351. [DOI] [PubMed] [Google Scholar]
- 9.Bono, J. L., A. F. Elias, J. J. Kupko III, B. Stevenson, K. Tilly, and P. Rosa. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 1822445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Botkin, D. J., A. N. Abbott, P. E. Stewart, P. A. Rosa, H. Kawabata, H. Watanabe, and S. J. Norris. 2006. Identification of potential virulence determinants by Himar1 transposition of infectious Borrelia burgdorferi B31. Infect. Immun. 746690-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caimano, M. J., R. Iyer, C. H. Eggers, C. Gonzalez, E. A. Morton, M. A. Gilbert, I. Schwartz, and J. D. Radolf. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 651193-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charon, N. W., and S. F. Goldstein. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu. Rev. Genet. 3647-73. [DOI] [PubMed] [Google Scholar]
- 13.Chi, B., R. J. Limberger, and H. K. Kuramitsu. 2002. Complementation of a Treponema denticola flgE mutant with a novel coumermycin A1-resistant T. denticola shuttle vector system. Infect. Immun. 702233-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das, S., S. W. Barthold, S. S. Giles, R. R. Montgomery, S. R. Telford III, and E. Fikrig. 1997. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and the mammalian host. J. Clin. Investig. 99987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50809-824. [DOI] [PubMed] [Google Scholar]
- 17.Fisher, M. A., D. Grimm, A. K. Henion, A. F. Elias, P. E. Stewart, P. A. Rosa, and F. C. Gherardini. 2005. Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc. Natl. Acad. Sci. USA 1025162-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank, K. L., S. F. Bundle, M. E. Kresge, C. H. Eggers, and D. S. Samuels. 2003. aadA confers streptomycin resistance in Borrelia burgdorferi. J. Bacteriol. 1856723-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, and J. Gocayne. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390580-586. [DOI] [PubMed] [Google Scholar]
- 20.Ge, Y., and N. W. Charon. 1997. Molecular characterization of a flagellar/chemotaxis operon in the spirochete Borrelia burgdorferi. FEMS Microbiol. Lett. 153425-431. [DOI] [PubMed] [Google Scholar]
- 21.Ge, Y., C. Li, L. Corum, C. A. Slaughter, and N. W. Charon. 1998. Structure and expression of the FlaA periplasmic flagellar protein of Borrelia burgdorferi. J. Bacteriol. 1802418-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge, Y., I. G. Old, I. Saint Girons, and N. W. Charon. 1997. Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus σ70 promoter. J. Bacteriol. 1792289-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilmore, R. D., Jr., M. L. Mbow, and B. Stevenson. 2001. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect. 3799-808. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein, S. F., K. F. Buttle, and N. W. Charon. 1996. Structural analysis of Leptospiraceae and Borrelia burgdorferi by high-voltage electron microscopy. J. Bacteriol. 1786539-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstein, S. F., N. W. Charon, and J. A. Kreiling. 1994. Borrelia burgdorferi swims with a planar waveform similar to that of eukaryotic flagella. Proc. Natl. Acad. Sci. USA 913433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harolow, E., and D. Lane. 1988. Adsorption to remove nonspecific binding, p. 632-633. In Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 27.Hendrix, R. W. 2005. Bacteriophage HK97: assembly of the capsid and evolutionary connections. Adv. Virus Res. 641-14. [DOI] [PubMed] [Google Scholar]
- 28.Hovind Hougen, K. 1984. Ultrastructure of spirochetes isolated from Ixodes ricinus and Ixodes dammini. Yale J. Biol. Med. 57543-548. [PMC free article] [PubMed] [Google Scholar]
- 29.Hudson, C. R., J. G. Frye, F. D. Quinn, and F. C. Gherardini. 2001. Increased expression of Borrelia burgdorferi vlsE in response to human endothelial cell membranes. Mol. Microbiol. 41229-239. [DOI] [PubMed] [Google Scholar]
- 30.Hughes, K. T., K. L. Gillen, M. J. Semon, and J. E. Karlinsey. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 2621277-1280. [DOI] [PubMed] [Google Scholar]
- 31.Jwang, B., P. Dwing, E. Fikrig, and R. A. Flavell. 1995. The hook protein of Borrelia burgdorferi, encoded by the flgE gene, is serologically recognized in Lyme disease. Clin. Diag. Lab. Immunol. 2609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang, H. J., F. Coulibaly, F. Clow, T. Proft, and E. N. Baker. 2007. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science 3181625-1628. [DOI] [PubMed] [Google Scholar]
- 33.Kojima, S., and D. F. Blair. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 23393-134. [DOI] [PubMed] [Google Scholar]
- 34.Komeda, Y. 1982. Fusions of flagellar operons to lactose genes on a Mu lac bacteriophage. J. Bacteriol. 15016-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubori, T., S. Yamaguchi, and S. I. Aizawa. 1997. Assembly of the switch complex onto the MS ring complex of Salmonella typhimurium does not require any other flagellar proteins. J. Bacteriol. 179813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kutsukake, K., Y. Ohya, and T. Iino. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, C., R. G. Bakker, M. A. Motaleb, M. L. Sartakova, F. C. Cabello, and N. W. Charon. 2002. Asymmetrical flagellar rotation in Borrelia burgdorferi nonchemotactic mutants. Proc. Natl. Acad. Sci. USA 996169-6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, C., M. A. Motaleb, M. Sal, S. F. Goldstein, and N. W. Charon. 2000. Spirochete periplasmic flagella and motility. J. Mol. Microbiol. Biotechnol. 2345-354. [PubMed] [Google Scholar]
- 39.Li, H., J. Ruby, N. Charon, and H. Kuramitsu. 1996. Gene inactivation in the oral spirochete Treponema denticola: construction of a flgE mutant. J. Bacteriol. 1783664-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Limberger, R. J., L. L. Slivienski, and W. A. Samsonoff. 1994. Genetic and biochemical analysis of the flagellar hook of Treponema phagedenis. J. Bacteriol. 1763631-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Llewellyn, M., R. J. Dutton, J. Easter, D. O'Donnol, and J. W. Gober. 2005. The conserved flaF gene has a critical role in coupling flagellin translation and assembly in Caulobacter crescentus. Mol. Microbiol. 571127-1142. [DOI] [PubMed] [Google Scholar]
- 42.Lux, R., J. N. Miller, N. H. Park, and W. Shi. 2001. Motility and chemotaxis in tissue penetration of oral epithelial cell layers by Treponema denticola. Infect. Immun. 696276-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lux, R., A. Moter, and W. Shi. 2000. Chemotaxis in pathogenic spirochetes: directed movement toward targeting tissues? J. Mol. Microbiol. Biotechnol. 2355-364. [PubMed] [Google Scholar]
- 44.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minamino, T., and K. Namba. 2004. Self-assembly and type III protein export of the bacterial flagellum. J. Mol. Microbiol. Biotechnol. 75-17. [DOI] [PubMed] [Google Scholar]
- 46.Motaleb, M. A., L. Corum, J. L. Bono, A. F. Elias, P. Rosa, D. S. Samuels, and N. W. Charon. 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc. Natl. Acad. Sci. USA 9710899-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motaleb, M. A., M. R. Miller, R. G. Bakker, C. Li, and N. W. Charon. 2007. Isolation and characterization of chemotaxis mutants of the Lyme disease spirochete Borrelia burgdorferi using allelic exchange mutagenesis, flow cytometry, and cell tracking. Methods Enzymol. 422419-437. [DOI] [PubMed] [Google Scholar]
- 48.Motaleb, M. A., M. R. Miller, C. Li, R. G. Bakker, S. F. Goldstein, R. E. Silversmith, R. B. Bourret, and N. W. Charon. 2005. CheX is a phosphorylated CheY phosphatase essential for Borrelia burgdorferi chemotaxis. J. Bacteriol. 1877963-7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Motaleb, M. A., M. S. Sal, and N. W. Charon. 2004. The decrease in FlaA observed in a flaB mutant of Borrelia burgdorferi occurs posttranscriptionally. J. Bacteriol. 1863703-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niehus, E., H. Gressmann, F. Ye, R. Schlapbach, M. Dehio, C. Dehio, A. Stack, T. F. Meyer, S. Suerbaum, and C. Josenhans. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52947-961. [DOI] [PubMed] [Google Scholar]
- 51.Paster, B. J., and F. E. Dewhirst. 2000. Phylogenetic foundation of spirochetes. J. Mol. Microbiol. Biotechnol. 2341-344. [PubMed] [Google Scholar]
- 52.Popa, M. P., T. A. McKelvey, J. Hempel, and R. W. Hendrix. 1991. Bacteriophage HK97 structure: wholesale covalent cross-linking between the major head shell subunits. J. Virol. 653227-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 991562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosey, E. L., M. J. Kennedy, and R. J. Yancey, Jr. 1996. Dual flaA1 flaB1 mutant of Serpulina hyodysenteriae expressing periplasmic flagella is severely attenuated in a murine model of swine dysentery. Infect. Immun. 644154-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruby, J. D., H. Li, H. Kuramitsu, S. J. Norris, S. F. Goldstein, K. F. Buttle, and N. W. Charon. 1997. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J. Bacteriol. 1791628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sadziene, A., D. D. Thomas, V. G. Bundoc, S. C. Holt, and A. G. Barbour. 1991. A flagella-less mutant of Borrelia burgdorferi. Structural, molecular, and in vitro functional characterization. J. Clin. Investig. 8882-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sal, M. 2005. Periplasmic flagella of the spirochetes Borrelia burgdorferi and Brachyspira hyodysenteriae. Ph.D. dissertation. West Virginia University, Morgantown.
- 58.Samatey, F. A., H. Matsunami, K. Imada, S. Nagashima, T. R. Shaikh, D. R. Thomas, J. Z. Chen, D. J. DeRosier, A. Kitao, and K. Namba. 2004. Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature 4311062-1068. [DOI] [PubMed] [Google Scholar]
- 59.Sartakova, M. L., E. Y. Dobrikova, M. A. Motaleb, H. P. Godfrey, N. W. Charon, and F. C. Cabello. 2001. Complementation of a nonmotile flaB mutant of Borrelia burgdorferi by chromosomal integration of a plasmid containing a wild-type flaB allele. J. Bacteriol. 1836558-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sellek, R. E., R. Escudero, H. Gil, I. Rodriguez, E. Chaparro, E. Perez-Pastrana, A. Vivo, and P. Anda. 2002. In vitro culture of Borrelia garinii results in loss of flagella and decreased invasiveness. Infect. Immun. 704851-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, A. H., J. S. Blevins, G. N. Bachlani, X. F. Yang, and M. V. Norgard. 2007. Evidence that RpoS (σS) in Borrelia burgdorferi is controlled directly by RpoN (σ54/σN). J. Bacteriol. 1892139-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stewart, P. E., R. Thalken, J. L. Bono, and P. Rosa. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39714-721. [DOI] [PubMed] [Google Scholar]
- 63.Thomas, D. R., N. R. Francis, C. Xu, and D. J. DeRosier. 2006. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 1887039-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tokarz, R., J. M. Anderton, L. I. Katona, and J. L. Benach. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole-genome DNA array. Infect. Immun. 725419-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yakhnin, H., P. Pandit, T. J. Petty, C. S. Baker, T. Romeo, and P. Babitzke. 2007. CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol. Microbiol. 641605-1620. [DOI] [PubMed] [Google Scholar]
- 66.Yang, X. F., M. C. Lybecker, U. Pal, S. M. Alani, J. Blevins, A. T. Revel, D. S. Samuels, and M. V. Norgard. 2005. Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J. Bacteriol. 1874822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]