Abstract
Clavibacter michiganensis subsp. michiganensis is a plant-pathogenic actinomycete that causes bacterial wilt and canker of tomato. The nucleotide sequence of the genome of strain NCPPB382 was determined. The chromosome is circular, consists of 3.298 Mb, and has a high G+C content (72.6%). Annotation revealed 3,080 putative protein-encoding sequences; only 26 pseudogenes were detected. Two rrn operons, 45 tRNAs, and three small stable RNA genes were found. The two circular plasmids, pCM1 (27.4 kbp) and pCM2 (70.0 kbp), which carry pathogenicity genes and thus are essential for virulence, have lower G+C contents (66.5 and 67.6%, respectively). In contrast to the genome of the closely related organism Clavibacter michiganensis subsp. sepedonicus, the genome of C. michiganensis subsp. michiganensis lacks complete insertion elements and transposons. The 129-kb chp/tomA region with a low G+C content near the chromosomal origin of replication was shown to be necessary for pathogenicity. This region contains numerous genes encoding proteins involved in uptake and metabolism of sugars and several serine proteases. There is evidence that single genes located in this region, especially genes encoding serine proteases, are required for efficient colonization of the host. Although C. michiganensis subsp. michiganensis grows mainly in the xylem of tomato plants, no evidence for pronounced genome reduction was found. C. michiganensis subsp. michiganensis seems to have as many transporters and regulators as typical soil-inhabiting bacteria. However, the apparent lack of a sulfate reduction pathway, which makes C. michiganensis subsp. michiganensis dependent on reduced sulfur compounds for growth, is probably the reason for the poor survival of C. michiganensis subsp. michiganensis in soil.
The genus Clavibacter belongs to the family Microbacteriaceae in the high-G+C-content branch of the gram-positive bacteria (11, 47). This genus currently includes only one species divided into five subspecies, all of which are plant pathogens of specific hosts. All subspecies cause systemic infections of the xylem which may be latent without visible symptoms, seem to be able to invade seeds, and show only poor survival capabilities in soil (14, 60). They may have an epiphytic lifestyle (25). The subspecies Clavibacter michiganensis subsp. michiganensis causes bacterial wilt and canker of tomato (Solanum lycopersicum, formerly Lycopersicon esculentum), an economically important disease causing yield losses worldwide (2). The other subspecies cause diseases of potato (Clavibacter michiganensis subsp. sepedonicus), alfalfa (Clavibacter michiganensis subsp. insidiosus), maize (Clavibacter michiganensis subsp. nebraskensis), and wheat (Clavibacter michiganensis subsp. subsp. tessellarius).
Infection of tomato by C. michiganensis subsp. michiganensis occurs through wounds and stomata or by infection of seeds, leading to a systemic infection that culminates in wilt and canker (55). The main habitat of C. michiganensis subsp. michiganensis inside the plant is the slightly acidic, nutrient-poor xylem fluid. Latent infections with no or only mild symptoms also occur. Plants with latent infections can be the source of contaminated seed, which is the major cause of outbreaks of C. michiganensis subsp. michiganensis infections in agriculture (59). C. michiganensis subsp. michiganensis can be considered a mainly biotrophic and moderately necrotrophic plant pathogen. However, C. michiganensis subsp. michiganensis is not a true soil bacterium, since survival of C. michiganensis subsp. michiganensis in soil for long periods of time is possible only when the bacteria are associated with plant debris (18).
For C. michiganensis subsp. michiganensis NCPPB382, it was shown that the essential pathogenicity determinants are plasmid borne (40). One of these determinants is the celA gene encoding an β-1,4-endocellulase carried by the 27-kb plasmid pCM1 (26). The second known pathogenicity factor, Pat-1, is a putative serine protease encoded on the 70-kb plasmid pCM2 (13). Further genes homologous to pat-1 have been identified both in plasmid pCM2 and in the chromosome (9). All the gene functions required for infection, successful colonization, and evasion or suppression of plant defenses are carried on the chromosome. This is shown by the fact that CMM100, a plasmid-free cured derivative of C. michiganensis subsp. michiganensis NCPPB382, is able to colonize tomato plants without causing any wilting symptoms (40). With the development of a random transposon mutagenesis system (34) and directed gene replacement experiments (32), a functional investigation of chromosomal genes recently became possible.
Currently, our knowledge of pathogenic bacterium-plant interactions is based mainly on studies of proteobacteria, while there is little information about the molecular mechanisms of actinomycete pathogenesis. The first genome sequence of an actinomycete plant pathogen, the sugarcane pathogen Leifsonia xyli subsp. xyli CTCB07, was recently published (43). L. xyli subsp. xyli is closely related to the genus Clavibacter, in which it was formerly included (15). Here we report the complete genome sequence of C. michiganensis subsp. michiganensis NCPPB382, while the genome sequence of the closely related potato pathogen C. michiganensis subsp. sepedonicus is described in the accompanying paper. Thus, a comparison of these three pathogens that have similar lifestyles on the genomic level is now possible.
MATERIALS AND METHODS
Whole-genome sequencing.
DNA shotgun clone libraries with average insert sizes of 1, 2 to 3, and 8 kb were constructed using the pSMART vector (Lucigen Corp., Middleton, WI) by GATC Biotech AG (Konstanz, Germany). Plasmid clones were end sequenced with ABI 3700 sequencing machines (ABI, Weiterstadt, Germany) by GATC Biotech AG. Base calling was carried out using PHRED (16, 17). High-quality reads were defined by a minimal length of 250 bp with an averaging quality value of ≥phred20. Finally, 48,072 high-quality reads and 38,289 (5.87 genome equivalents), 6,850 (1.03 genome equivalents), and 2,933 (0.40 genome equivalent) end sequences from the libraries with 1-, 2- to 3-, and 8-kb inserts, respectively, were established.
Sequence assembly and assembly validation.
Base calling, quality control, and elimination of vector DNA sequences of the reads were performed using the software package BioMake as previously described (30). Sequence assembly was performed with the PHRAP assembly tool (www.phrap.org), resulting in 157 contigs containing ≥10 reads. For finishing of the genome sequence, the CONSED/AUTOFINISH software package (21, 22) supplemented with the tool BACCardI (4) was used. For gap closure and assembly validation, a bacterial artificial chromosome (BAC) library with approximately 100-kb BamHI inserts was constructed by Genoscope (Evry, France) as described by Tauch et al. (56). Sequencing of BAC ends was carried out with ABI 3100 and ABI 377 sequencing machines by Integrated Genomics GmbH (Jena, Germany) and IIT GmbH (Bielefeld, Germany), respectively. Gaps between contigs of the whole-genome shotgun assembly were closed by sequencing of shotgun and BAC clones carried out by IIT GmbH and GBF GmbH (Braunschweig, Germany) with LI-COR 4200L (LI-COR Inc., Lincoln, NE) and ABI 377 sequencing machines. To obtain a high-quality genome sequence, all bases of the consensus sequence were polished to at least phred40 quality by primer walking. Collectively, 967 sequencing reads were added to the shotgun assembly for finishing and polishing of the genomic sequence. The number of rrn operons was determined by hybridization with a probe derived from the 16S rRNA gene. The two rRNA operons were sequenced completely by primer walking of BAC clones. For assembly validation, BAC end sequences were mapped onto the genome sequence using BACCardI.
Genome analysis and annotation.
In the first annotation step, automatic annotation was performed using the GenDB 2.0 genome annotation system (41) as previously described by Thieme et al. (57). In the second annotation step, all predicted open reading frames (ORFs) were manually reinspected to correct start codon and function assignments. Intergenic regions were checked for ORFs missed by the automatic annotation using the BLAST programs (1). For analysis of putative proteases and transporters the MEROPS (http://merops.sanger.ac.uk/) and TCDB (http://www.tcdb.org/) databases were used, respectively. Only transporters in categories 1.A (channels), 2.A (secondary transporters), 3.A. (primary transporters), and 9.A (uncharacterized transporters) were analyzed, while phosphotransferase systems (PTS), ATPases involved in generation of membrane gradients, transporters involved in protein secretion and DNA uptake, and proteins with predicted transmembrane helices without predicted functions (conserved hypothetical and hypothetical proteins) were excluded. Multiple alignments were constructed using ClustalX 1.83 (58). Searches for putative DNA binding sites were conducted using PATSCAN of the Staden package (53).
Genomic comparison.
For comparative analyses, the annotated genome sequences of the following bacteria were imported into GenDB: C. michiganensis subsp. sepedonicus (accession no. AM849034) and L. xyli subsp. xyli CTCB07 (accession no. AE016822). Sequence comparisons were carried out using the Artemis comparison tool (10), and the gene content comparisons were carried out by performing a reciprocal BLAST analysis. Two genes were considered orthologs if the reciprocal hit on the protein level had an e-value of at least e−30 and a level of sequence identity of at least 50%. Pseudogenes were not considered. Obvious mistakes (e.g., ribosomal proteins, which tend to be short and are missed using this e-value cutoff) were corrected manually.
PFGE.
DNA for pulsed-field gel electrophoresis (PFGE) was isolated from C. michiganensis subsp. michiganensis strains NCPPB382 and CMM30-18 as described by Redenbach et al. (48). Undigested DNA was analyzed to exclude the presence of linear plasmids. After digestion with VspI and DraI, the sizes of the DNA fragments were determined for C. michiganensis subsp. michiganensis NCPPB382 and used to validate the final assembly. To determine the number of rrn operons, PFGE gels were blotted (52) and hybridized with a 16S rRNA gene probe as specified by the manufacturer (Roche Diagnostics, Mannheim, Germany).
DNA techniques.
Plasmid DNA of Escherichia coli was prepared by the alkaline lysis method (50). Plasmid DNA was isolated and purified by using Qiagen columns as specified by the manufacturer (Qiagen, Hilden, Germany). Preparation of total DNA for hybridization experiments and electroporation of C. michiganensis subsp. michiganensis were performed as described by Kaup et al. (32). DNA restriction, ligation, and transformation were carried out by using standard procedures (50). DNA fragments used as hybridization probes were isolated from agarose gels and labeled by random priming as specified by the manufacturer (Roche Diagnostics, Mannheim, Germany).
Nucleotide sequence accession numbers.
The sequences reported here have been deposited in the GenBank database under accession numbers AM711867, AM711865, and AM711866 for the chromosome, pCM1, and pCM2, respectively.
RESULTS AND DISCUSSION
General features of the C. michiganensis subsp. michiganensis NCPPB382 chromosome.
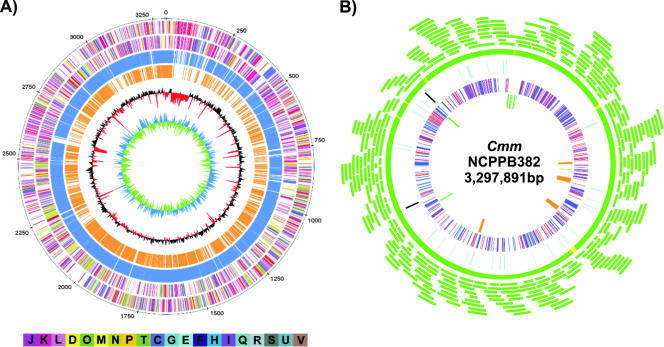
The genome sequence of C. michiganensis subsp. michiganensis was established by employing a whole-genome shotgun approach. The assembly of the high-quality sequence was validated by a complete BAC map (Fig. 1B) and PFGE analysis of VspI- and DraI-hydrolyzed C. michiganensis subsp. michiganensis DNA (data not shown).
FIG. 1.
Circular diagrams of the C. michiganensis subsp. michiganensis strain NCPPB382 chromosome displaying relevant genome features and validation of the sequence assembly by a BAC map. (A) Circles providing the following information (from the outermost circle to the innermost circle): circle 1, position (in kilobases); circles 2 and 3, predicted CDS on the forward (circle 2) and reverse (circle 3) strands, with the colors indicating the assigned COG classes; circles 4 and 5, CDS with homologues in the chromosomes of C. michiganensis subsp. sepedonicus (blue) and L. xyli subsp. xyli (orange), respectively; circle 6, G+C content, showing deviations from the average value (72.66%); circle 7, GC skew. The bar at the bottom explains the colors used to indicate the functional COG groups (C, energy production and conversion; D, cell cycle control, mitosis, and meiosis; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation; K, transcription; L, replication, recombination, and repair; M, cell wall and membrane biogenesis; N, cell motility; O, posttranslational modification, protein turnover, and chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolite biosynthesis, transport, and catabolism; R, general function prediction; S, unknown function; T, signal transduction mechanisms; U, intracellular trafficking and secretion; V, defense mechanisms). (B) BAC map of the C. michiganensis subsp. michiganensis (Cmm) chromosome. Each green arc represents a single BAC clone mapped on the assembled sequence. From the outer circle to the inner circle: circle 1, coverage of the C. michiganensis subsp. michiganensis chromosome with BAC clones (green, covered by more than one BAC clone; yellow, covered by one BAC clone); circle 2, genes coding for tRNAs (cyan) and rRNAs (black); circle 3, predicted transporter proteins (blue) and transcriptional regulators (red); circle 4, genes putatively involved in pathogenicity (green) and EPS clusters (orange).
C. michiganensis subsp. michiganensis has a single, circular chromosome consisting of 3,297,837 bp (Fig. 1A and Table 1). The G+C content of the chromosome is 72.66%. No clear GC skew was detectable, which was also observed for other actinomycetes, including Streptomyces coelicolor (5) and Bifidobacterium longum (51). Consequently, neither the origin of replication nor the termination region was detectable by a strong bias of G toward the leading strand. Hence, the start codon of the dnaA gene was defined as the zero point of the chromosome, as usual for most bacterial genomes. The genetic organization of the origin region is conserved among actinomycetes and contains several DnaA boxes located up- and downstream of the dnaA gene (27). The 544-bp intergenic region between dnaA and dnaN containing five DnaA boxes (see Fig. S1 in the supplemental material) probably represents the origin of replication, as is the case in Mycobacterium and Streptomyces (27).
TABLE 1.
Characteristics of the C. michiganensis subsp. michiganensis NCPPB382 genome
| Characteristic | Chromosome | pCM1 | pCM2 |
|---|---|---|---|
| Size (bp) | 3,297,891 | 27,357 | 69,989 |
| G+C content (%) | 72.66 | 67.56 | 66.50 |
| CDS | |||
| Predicted no. | 2,984 | 28 | 68 |
| No. with function assigned | 2,029 | 7 | 14 |
| No. of conserved hypothetical proteins | 530 | 3 | 15 |
| No. of hypothetical proteins | 425 | 18 | 39 |
| Coding % | 90.5 | 80.78 | 73.62 |
| Average length (bp) | 986 | 796 | 759 |
| Maximal length (bp) | 6,888 | 2,241 | 4,479 |
| % ATG initiation codons | 65.64 | 67.86 | 67.57 |
| % GTG initiation codons | 32.27 | 28.57 | 32.43 |
| No. of rRNA operons | 2 | ||
| No. of tRNAs | 45 | ||
| No. of small RNAs | 3 | ||
| No. of riboswitches | 6 | ||
| No. of pseudogenes | 24 | 2 | |
| No. of insertion sequence elements | 3 |
The chromosome of C. michiganensis subsp. michiganensis contains a total of 2,984 predicted coding sequences (CDS) (Table 1). Based on the manual annotation, biological functions were assigned to 2,029 CDS (68.0%). The remaining 955 CDS comprise 530 conserved hypothetical CDS and 425 hypothetical CDS (see Table S1 in the supplemental material). The chromosome shows an average coding capacity of 90.5%. Two rRNA operons were identified, which are organized in the order 16S-23S-5S and are located in a region between bp 2,240,000 and 2,870,000 (Fig. 1B). Altogether, 45 genes for tRNAs representing all 20 amino acids were identified (Table 1; see Table S2 in the supplemental material). The C. michiganensis subsp. michiganensis chromosome harbors only 24 pseudogenes, about half of which are within or associated with low-G+C-content regions (see Table S3 in the supplemental material). Insertion elements (transposases) occur only rarely and seem to be nonfunctional. Two ORFs encoding transposases are pseudogenes (CMM_PS_03 and CMM_PS_06). Only ORFs CMM_1940 and CMM_1939 may encode a functional transposase belonging to the IS481 family, which also includes IS1122, IS1121, and ISLxx4 from C. michiganensis subsp. insidiosus, C. michiganensis subsp. sepedonicus, and L. xyli subsp. xyli, respectively. However, only one copy of an insertion element occurs in C. michiganensis subsp. michiganensis, and ribosomal frameshifting is required to produce a functional transposase. Otherwise, the ORFs may also represent a pseudogene. In contrast, homologous intact high-copy-number insertion elements and several other insertion elements occur in both C. michiganensis subsp. sepedonicus and L. xyli subsp. xyli. Apart from genes potentially encoding an integrase (CMM_2286) and an invertase/recombinase (CMM_2297) associated with a putative prophage, only one other putative invertase/recombinase gene (CMM_2068) was found.
The evaluation of the best BLAST hits for each C. michiganensis subsp. michiganensis protein (excluding C. michiganensis subsp. sepedonicus) showed the taxonomic position of C. michiganensis subsp. michiganensis among the actinomycetes (see Table S4 in the supplemental material). As expected, the closest relatives are other members of the Microbacteriaceae, the sugarcane pathogen L. xyli subsp. xyli and the marine actinobacterium PHSC20C1, a putative Agreia sp. A low number of best protein matches (5.4%) are matches with different Proteobacteria. These proteins include several transporters, the phn gene products, which may be involved in the utilization of phosphonates (CMM_0380 to CMM_0371), several proteins involved in extracellular polysaccharide (EPS) biosynthesis, some surface proteins, and a few proteins which may play a role in the virulent interaction, like a putative phospholipase C (CMM_0504), a perforin (CMM_2382), and a number of serine proteases (belonging to the Chp and Ppa families [see below]). The best matches for other proteins that are potentially important for virulence are matches with other actinomycetes; however, these proteins show very high levels of similarity to plant or fungal enzymes (e.g., the serine proteases belonging to the subtilase subfamily and the tomatinase).
Of the three pathogenic members of the Microbacteriaceae, C. michiganensis subsp. michiganensis and C. michiganensis subsp. sepedonicus have similar genome sizes (3.4 Mb) and similar numbers of CDS (about 3,100), while the genome of L. xyli subsp. xyli is one-fourth smaller (2.6 Mb, 2,326 CDS). Chromosomal synteny between the three species is obscured by the fact that a lot of rearrangements have occurred in the genomes, probably caused by recombination between copies of different, although related, insertion elements in both C. michiganensis subsp. sepedonicus and L. xyli subsp. xyli (data not shown). At the level of individual genes or operons there is a high degree of similarity. The number of pseudogenes increases from C. michiganensis subsp. michiganensis (26 pseudogenes) to C. michiganensis subsp. sepedonicus (104 pseudogenes) to L. xyli subsp. xyli (296 pseudogenes). A more detailed comparison at the individual gene level is described in the accompanying paper.
Plasmids pCM1 and pCM2 of C. michiganensis subsp. michiganensis NCPPB382.
The genome of C. michiganensis subsp. michiganensis strain NCPPB382 contains two circular plasmids, which are 27,357 bp (pCM1) and 69,989 bp (pCM2) long (Table 1). Compared to the G+C content of the chromosome, the G+C contents of the two plasmids are lower, 67.56% for pCM1 and 66.50% for pCM2. The known virulence factors, celA on plasmid pCM1 and pat-1 on pCM2 (13, 26), were confirmed to map on the plasmids. No other genes relevant for pathogenicity (based on homology to such genes in other bacterial pathogens) were found on the plasmids. The observation that curing of plasmids pCM1 and pCM2 has no detectable effect on growth in planta (40) also indicates that the plasmids seem to carry no other genes conferring a selective advantage. On plasmid pCM2 the location of the pat-1 homologous genes phpA and phpB in the vicinity of pat-1 (9) was also confirmed. Both plasmids contain a gene cluster (about 10 kb in pCM1 and 18.5 kb in pCM2) which may encode proteins engaged in conjugative transfer of the plasmids. Also, the replicon regions of both plasmids which were used for construction of cloning vectors for the pDM and pHN families (36, 39) were identified. These replicon regions contain genes for both replication control and partitioning. In addition, two pseudogenes are located on pCM2, but there are no pseudogenes on pCM1 (see Table S3 in the supplemental material).
Genomic islands.
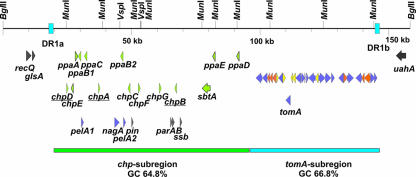
Excluding the rrn operons, about 20 regions with a significantly lower G+C content are distributed all over the chromosome (Table 2). All these low-G+C-content regions are C. michiganensis subsp. michiganensis specific and do not occur in C. michiganensis subsp. sepedonicus. These regions account for a total of 203 kb (∼6% of the chromosome). The largest of these regions, the chp/tomA region, is located close to the origin (∼129 kb; positions 38,000 to 167,000). Near its borders are two 1.9-kb large direct repeats with 99% nucleotide sequence identity. Based on structural features, the region can be divided into two subregions (Fig. 2). The first subregion, designated the chp subregion, has an average G+C content of only 64.8%, with high local variations. The second subregion, designated the tomA subregion, has a more uniform composition, with an average G+C content of 66.8%. For the chp subregion (79 kb), only 44 genes could be predicted (coding capacity, 45.3%). The codon usage of most of these genes differs from the normal codon usage of C. michiganensis subsp. michiganensis. Additionally, 11 pseudogenes were identified. However, a few genes (e.g., CMM_0069 and CMM_0070 possibly encoding an NADH oxidase and a subtilase, respectively) have ordinary G+C contents and codon usage. In contrast, the tomA subregion has a high coding density (96.3%) and an ordinary codon usage pattern compared to the bulk genome and contains no pseudogenes. Most of the genes in this subregion encode proteins predicted to be involved in uptake or metabolism of carbohydrates. This subregion contains genes coding for 12 different glycosidases and the only cytochrome P450 (CMM_0094 to CMM_0096) and the tomA gene (CMM_0090). The latter gene encodes a tomatinase involved in the detoxification of α-tomatine, a growth-inhibiting alkaloid produced by tomato (32). Furthermore, some transporter and sugar-responsive regulatory genes are present. The chp/tomA region seems to be involved in the C. michiganensis subsp. michiganensis-tomato interaction (see below). Interestingly, homologues of tomA and a few other genes located in the tomA subregion were recently shown to be present on a mobile pathogenicity island in the potato pathogen Streptomyces turgidiscabies (33). On the other hand, a corresponding island is missing in the C. michiganensis subsp. sepedonicus genome, and many of the genes harbored on the island have no closely related counterparts in C. michiganensis subsp. sepedonicus.
TABLE 2.
Regions with low G+C contents and rrn operons in the C. michiganensis subsp. michiganensis chromosome
| Region | Position | ORF(s) | Size (kb) | Avg G+C content (%) | Commentsa |
|---|---|---|---|---|---|
| chp | 38100-117150 | CMM_0034-CMM_0077 | 79.05 | 64.6 | Putative pathogenicity island, 11 pseudogenes; VspI (2×), DraI |
| tomA | 117150-166800 | CMM_0078-CMM_0112 | 49.65 | 66.8 | Putative pathogenicity island |
| low GC1 | 193650-196300 | CMM_0135, CMM_0136 | 2.65 | 55.0 | |
| low GC2 | 306300-310200 | CMM_0246 | 4.1 | 54.1 | Bordered by direct repeat (69 bp) |
| low GC3 | 504400-506400 | 2.1 | 57.7 | VspI | |
| low GC4 | 552200-556000 | 3.8 | 54.9 | VspI | |
| low GC5 | 656600-658600 | CMM_0574, CMM_0575 | 2 | 61.0 | Putative restriction-modification system; bordered by 29-bp direct repeat |
| low GC6 | 708300-716650 | CMM_0627 | 8.35 | 61.1 | Putative rhs element, one pseudogene; DraI |
| low GC7 | 745100-748700 | CMM_661 | 3.6 | 53.1 | One pseudogene; DraI |
| low GC8 | 830520-833300 | CMM_0738 | 2.8 | 53.3 | |
| low GC9 | 942950-946600 | CMM_0828 | 3.65 | 58.4 | Internal in EPS cluster II; DraI |
| low GC10 | 1270450-1274400 | CMM_1110 | 3.95 | 58.1 | Downstream tRNAGln |
| low GC11 | 1285200-1286600 | 1.4 | 54.7 | ||
| low GC12 | 2226650-2228350 | CMM_1971 | 1.7 | 54.8 | Upstream tRNAPro |
| rrnB | 2253517-2247622 | 54.7 | rRNA operon; VspI, DraI | ||
| low GC13 | 2514300-2516050 | CMM_2221, CMM_2222 | 1.75 | 53.8 | Hypothetical pilin gene |
| low GC14 | 2587100-2608380 | CMM_2286-CMM_2304 | 21.3 | 58.9 | Putative prophage remnant; bordered by duplicated gene |
| low GC15 | 2614420-2615500 | 1.1 | 59.9 | One pseudogene; bordered by partially duplicated gene | |
| low GC16 | 2684300-2686750 | CMM_2382 | 2.45 | 64.8 | Putative perforin |
| rrnA | 2873849-2867954 | 54.7 | rRNA operon; VspI, DraI | ||
| low GC17 | 3018000-3019600 | CMM_2688 | 1.6 | 58.2 | Putative acetyl xylan esterase; bordered by 29-bp direct repeat |
| low GC18 | 3172450-3174400 | CMM_2846, CMM_2847 | 1.95 | 56.3 | Toxin/antitoxin system |
| low GC19 | 3224800-3228500 | CMM_2901, CMM_2902 | 3.7 | 56.2 | Downstream tRNAGly; VspI |
The numbers of sites for the rare cutters (VspI and DraI) used in PFGE are indicated.
FIG. 2.
Physical map of the chp/tomA region of C. michiganensis subsp. michiganensis NCPPB382. The 1.9-kb direct repeats flanking the region are indicated by blue boxes. Selected genes are indicated by colors as follows: green, genes encoding serine proteases; yellow, genes encoding regulators; orange, genes encoding transporters; violet, genes encoding glycosidases. Pseudogenes are underlined.
Most of the other low-G+C-content regions (Table 2) are small (between 1 and 5 kb). They may have been acquired by horizontal gene transfer. For example, CMM_2688 in low-G+C-content region 17 encodes a putative acetyl xylan esterase that shows the highest similarity to fungal enzymes. Some of these low-G+C-content regions are bordered by repeats or partially duplicated genes. However, the potential mobility of these low-G+C-content regions remains unclear as neither integrases nor insertion elements were found to be associated with them. The only exception is low-G+C-content region 14, which is ∼21 kb long and contains a gene encoding an integrase. This region seems to represent the remains of a prophage containing only a few predicted genes. Low-G+C-content region 15, which is bordered by a partially duplicated gene, may constitute a part of this prophage. Region 15 is separated from low-G+C-content region 14 by only six genes with an ordinary G+C content. Four of these genes may encode a modification system for an extracellular polysaccharide.
Metabolic functions of C. michiganensis subsp. michiganensis.
After infection, C. michiganensis subsp. michiganensis spreads systemically in the xylem vessels of tomato, where it lives as a biotrophic phytopathogen at least in the first stages of infection and grows to high titers in the xylem sap of the host plant before disease symptoms develop. Thus, C. michiganensis subsp. michiganensis has to obtain sufficient nutrients from the acidic, nutrient-poor xylem fluid. Sugars normally occur only at low concentrations in the xylem sap of tomato, while carboxylic acids, like malate, citrate, fumarate, and succinate, are present at higher concentrations (7). A number of genes encoding transporters putatively specific for carboxylates may enable C. michiganensis subsp. michiganensis to utilize these compounds (e.g., CMM_1885 to CMM_1887 encoding a putative DAACS transporter and two-component system, CMM_2878 to CMM_2876 encoding a CitMHS transporter and two-component system, and CMM_2051 encoding a DASS family member). The pathways for glycolysis, the pentose phosphate pathway, and gluconeogenesis (phosphoenolpyruvate carboxykinase encoded by CMM_1473 and fructose-1,6-bisphosphatase encoded by CMM_2735) are apparently complete. The anaplerotic reaction enzymes predicted include a putative phosphoenolpyruvate carboxylase (encoded by CMM_0383) and pyruvate carboxylase (encoded by CMM_1880). The citrate cycle of C. michiganensis subsp. michiganensis seems to be complete and contains the flavoprotein malate:quinone oxidoreductase (encoded by CMM_0904) instead of malate dehydrogenase. This may be an adaptation to growth on carboxylates as the energetically unfavorable malate dehydrogenase reaction is driven by the subsequent citrate synthase step (42). The glyoxylate pathway is missing, but genes putatively encoding enzymes for the 2-methylcitrate cycle allowing growth on propionate were found.
At a later stage, when C. michiganensis subsp. michiganensis has reached a high titer in the xylem, the cell walls of the xylem vessels and the surrounding parenchyma cells may be attacked by cellulases (encoded by pCM1_0020 and CMM_2443) and by other plant cell wall-hydrolyzing extracellular enzymes (a predicted polygalacturonase encoded by CMM_2871, pectate lyase encoded by CMM_0043 and CMM_0051, xylanases encoded by CMM_1673 and CMM_1674, and endoglucanase encoded by CMM_2691 and CMM_2692), leading to tissue maceration and canker. For C. michiganensis subsp. sepedonicus a similar set of extracellular enzymes is predicted, but the homologues of CMM_2691 and CMM_2692 are pseudogenes and the putative xylanase of C. michiganensis subsp. sepedonicus is unrelated to the C. michiganensis subsp. michiganensis enzymes. In this late phase of infection, a higher proportion of sugars derived from plant cell wall degradation may become available as nutrients for C. michiganensis subsp. michiganensis. Genes encoding about 20 ABC transporters that are putatively sugar specific (e.g., a transporter encoded by CMM_0086 to CMM_0084 putatively specific for cellobiose and a transporter encoded by CMM_2438 to CMM_2436 putatively specific for arabinose) are present in the chromosome. Many of these genes are associated with genes encoding transcriptional regulators and/or glycosidases. C. michiganensis subsp. michiganensis seems to be able to use a variety of sugar polymers and sugars as carbon and energy sources. The useable compounds include the polysaccharides cellulose, xylan, arabinan, galactan, mannan, and starch. Thirty-eight glycosidases (α and β) were predicted, indicating that oligosaccharides derived from plant material by the action of extracellular enzymes can be transformed to mono- and disaccharides. Sugars may also be the most important nutrients for C. michiganensis subsp. michiganensis associated with plant debris in soil.
Genes for at least three PTS seem to be present. One operon (CMM_1503 to CMM_1505) coding for a putative phosphofructokinase and PTS components is probably specific for fructose (CMM_1754 encoding the IIA component for fructose). A second operon (CMM_2594 to CMM_2589 and the divergently transcribed CMM_2595 gene encoding a putative regulatory component) contains two sugar-specific components for mannitol and cellobiose. A third operon (CMM_1827) may be used for uptake of glycerol since it seems to form an operon with a gene encoding dihydroxyacetone kinase, followed downstream by two divergently transcribed genes coding for a glycerol uptake protein and a glycerol kinase. A number of other genes encode further PTS components (CMM_0968, CMM_0984, and CMM_0985). No homologue of an adenylate cyclase was found, indicating that the regulation of carbon metabolism (catabolite repression) differs from that in proteobacteria.
C. michiganensis subsp. michiganensis displays aerobic metabolism with no indication of enzymes involved in fermentation and anaerobic respiration using nitrate, sulfate, or fumarate as an electron acceptor. The respiratory chain is formed by genes encoding a single-subunit NADH dehydrogenase of the ndh-2-type (CMM_2259), a putative Na+-translocating NADH dehydrogenase consisting of six subunits (CMM_1087 to CMM_1092), a menaquinone:cytochrome c reductase (CMM_1835 to CMM_1837), a cytochrome bd complex (CMM_1541 and CMM_1542), and the terminal cytochrome c oxidase (CMM_1842, CMM_1841, and CMM_1834). An F0F1-type ATP synthase (encoded by CMM_1163 to CMM_1170) is used for ATP generation.
While complete pathways were predicted for the biosynthesis of menaquinones, coenzyme A, riboflavin, folate, thiamine, and vitamin B6, the pathways for nicotinate, biotin, and cobalamin biosynthesis are not present or incomplete. Nicotinamide may be taken up, since a putative transporter for nicotinamide mononucleotide is encoded by CMM_1524. The antioxidant glutathione is not produced in actinomycetes; instead, other thiol compounds, like mycothiol (Micrococcus, Mycobacterium, and Streptomyces) or coenzyme A (Arthrobacter and Agromyces), serve the same function (46). The thiol composition of Clavibacter is unknown, but all of the genes necessary for the biosynthesis of a mycothiol were annotated.
C. michiganensis subsp. michiganensis contains the genes required for production of a type B lantibiotic (CMM_1967 and CMM_1968). This mersacidin-type lantibiotic putatively acts on cell wall biosynthesis. A lantibiotic with an identical amino acid sequence was recently purified from a different C. michiganensis subsp. michiganensis strain and characterized (23). No counterpart has been found in C. michiganensis subsp. sepedonicus. A gene encoding a nonribosomal peptide synthethase with an unknown function (CMM_0624) is present; this gene is a fragmented pseudogene in C. michiganensis subsp. sepedonicus. However, a type III polyketide synthase was predicted in both subspecies (encoded by CMM_1534 and CMS1773). A complete functional operon for the synthesis of a carotenoid is present (CMM_2884 to CMM_2889). Cloning of these genes into E. coli leads to a yellow-pigmented E. coli strain (R. Eichenlaub, unpublished data). The production of carotenoids may be advantageous when organisms are exposed to light during epiphytic growth (25).
Iron metabolism.
Pathogens compete with their hosts for trace metals, especially iron. Accordingly, numerous genes encoding proteins putatively involved in the uptake of metals are present in C. michiganensis subsp. michiganensis. Five ABC transporters for the uptake of different iron compounds and, in addition, a putative Fe2+ permease (encoded by CMM_2175) were predicted. Genes encoding three putative siderophore-interacting proteins required for the release of Fe3+ from siderophores are present (only two such genes are present in C. michiganensis subsp. sepedonicus). Furthermore, genes which may encode two ABC transporters for manganese or zinc, two members of the NRAMP transporter family, and one member of the ZIP family are present.
In most bacteria, iron uptake is regulated tightly by regulator proteins belonging to either the Fur or DtxR family, which may also be involved in the regulation of genes for manganese and zinc metabolism or oxidative stress (44). In C. michiganensis subsp. michiganensis, two members of both metal-dependent regulator families were found (furA and furB; dtxR and sirR). A search for putative binding sites for DtxR (37) and Fur (3) identified a number of genes or operons potentially regulated by these proteins (see Table S7 in the supplemental material). C. michiganensis subsp. michiganensis may produce two different types of siderophores. The proteins encoded by the CMM_2095 to CMM_2093 genes may be involved in the biosynthesis of a hydroxamate siderophore similar to alcaligin (31). A Fur box was predicted upstream of this operon (see Table S7 in the supplemental material). In C. michiganensis subsp. sepedonicus a similar operon is present (CMS1133 to CMS1135). The second putative siderophore of the catecholate type has no counterpart in C. michiganensis subsp. sepedonicus. Genes organized in two operons may participate in the biosynthesis of this siderophore. One operon contains genes encoding two nonribosomal peptide synthetases (CMM_0330 and CMM_0331) and several downstream genes (CMM_0329 to CMM_0324) which seem to be required for modifications of the basic structure and export of the siderophore. The second operon (CMM_0332 to CMM_0334) is transcribed divergently and may encode proteins for the biosynthesis of an organic acid like salicylate, which is incorporated into the siderophore. Between the two operons a putative Dtx box was identified.
Missing pathways which may impede growth in soil.
Since C. michiganensis subsp. michiganensis seems to lack the ability to reduce nitrate (genes encoding a nitrate or nitrite reductase were not identified), it has to obtain reduced nitrogen compounds from the plant or from the soil. Ammonia may be taken up by the product of CMM_1669 (AmtB transporter), which may be most important in soil. In the plant xylem nitrogen is available mainly as nitrate, but ammonia also occurs (24). Furthermore, higher concentrations of the amino acids glutamate, glutamine, aspartate, and asparagine have been detected in the xylem fluid of tomato (7). For uptake of amino acids, several transporters are predicted; these include seven members of the APC family and four ABC transporters, probably specific for methionine (encoded by CMM_2283 to CMM_2281), branched-chain amino acids (encoded by CMM_2562 to CMM_2566), polar amino acids (encoded by CMM_2628 to CMM_2626), and glutamate (encoded by CMM_2004 to CMM_2007). Amino acids can serve as nitrogen sources for C. michiganensis subsp. michiganensis either by transamination reactions or by release of ammonia by enzymes like glutaminase, asparaginase, and aspartate ammonia-lyase, which may be encoded by CMM_0029, CMM_1146, and CMM_2077. Finally, urea may provide ammonia due to the action of a urea amidolyase (encoded by CMM_0120). Genes coding for glutamine synthetases of both the prokaryotic and eukaryotic types are present. As in other actinomycetes (19), nitrogen regulation is clearly different from nitrogen regulation in proteobacteria as no homologues of either a σ54 sigma factor or a PII protein were identified. Instead, the glnR homologue (CMM_2501) may be involved in nitrogen regulation.
The biosynthetic pathways for all amino acids seem to be complete, with the exception of the tryptophan biosynthesis pathway, which lacks trpF, a gene also missing in other actinomycetes. Genes encoding tRNA synthetases for glutamine and asparagine have not been found, so the gatABC system seems to be used to produce glutamine and asparagine from tRNAs charged with glutamate and aspartate. However, C. michiganensis subsp. michiganensis was shown to require methionine when it was grown on minimal medium. This auxotrophy can be explained by the lack of genes responsible for the reduction of sulfate, resulting in a dependence on reduced sulfur compounds like H2S or methionine. The lack of sulfate reduction is probably also the reason for the poor survival of C. michiganensis subsp. michiganensis in soil when sulfate is the most common source of sulfur.
Complex repertoire of transporter and regulatory genes.
In general, the number of transporters and regulatory genes is correlated to genome size and probably to the variability of the environment in which the bacteria live (35). In many cases, obligate pathogenic bacteria show a reduction in genome size, which may be explained by the more stable environment that the bacteria face in association with their hosts. Predicted transporters of C. michiganensis subsp. michiganensis were classified according to the TCDB database using the nomenclature of Saier (49). The number of predicted transporters was quite high, with 190 transport systems specified by 357 genes. This accounts for 6.10% of the total CDS and 61 transport systems/Mb (see Table S5 in the supplemental material). The most common families are the ABC transporters (70 apparently functional transporters encoded by 231 genes), MFS permeases (44 members), the APC transporters (nine members, putatively responsible for amino acid uptake), and the DMT transporters (eight members and one pseudogene). In conclusion, the number of transport systems is relatively high in C. michiganensis subsp. michiganensis, especially compared to Xylella fastidiosa CVC, a gram-negative plant pathogen that is of similar size (2.7 Mb), also lives mainly in xylem sap, and lacks a type III secretion system (TTSS). Xylella has only 49 secondary transporters, 9 of which are members of the MFS superfamily, and 32 primary transporters, including 23 ABC transporters (38). This indicates that C. michiganensis subsp. michiganensis still has the ability to adapt to a variety of environmental conditions, a trait which is lost over time in many pathogens after adaptation to a stable host environment by the process of genome reduction.
For export of proteins, genes for the general secretory pathway, the signal recognition particle pathway (ffh and ftsY [CMM_1362 and CMM_1363]), and the twin-arginine pathway (tatABC [CMM_1182, CMM_1686, and CMM_1687]) are present. As in Mycobacterium and Corynebacterium, a TTSS used by many proteobacterial plant and animal pathogens to translocate proteins directly into host cells is missing. Furthermore, no type IV secretion system involved in conjugation and virulence in some bacteria was predicted. However, the presence of transport systems with functions similar to those of type III and IV systems cannot be ruled out. The sequence similarity to such systems of proteobacteria is certainly low due to the completely different architecture of the cell envelope in actinomycetes.
As is the case for transporters, the number of genes encoding transcriptional regulators is high; 215 such genes were predicted, which is about 6.9% of all identified CDS (see Table S6 in the supplemental material). Stover et al. (54) described a correlation between the percentage of genes encoding regulators and genome size. About 7% of the proteins encoded by C. michiganensis subsp. michiganensis are involved in regulation, a higher percentage than the percentages in the larger genomes of Bacillus subtilis and E. coli, indicating that there is overrepresentation of regulatory proteins.
In total, genes encoding nine putative sigma factors, three anti-sigma factors, and two anti-anti-sigma factors were identified (see Table S6 in the supplemental material). ECF sigma factors account for six of the nine genes. ECF sigma factors often play a role at the beginning of regulatory networks, reacting to signals like stress and activating corresponding gene sets necessary to react to environmental stimuli. Both C. michiganensis subsp. michiganensis and C. michiganensis subsp. sepedonicus have one specific ECF sigma factor that does not occur in the other subspecies. Other high-level regulators involved in adaptation to changing environmental conditions include the two-component systems, and a relatively high number of such systems (∼30) are also present in C. michiganensis subsp. michiganensis.
Genes involved in the interaction with the host plant.
EPS are involved in a number of different functions, like protection against desiccation, scavenging of cations, adhesion to surfaces, and host interactions (12). In the C. michiganensis subsp. michiganensis-tomato interaction adhesion to the xylem vessels by formation of a biofilm may occur. However, the EPS of C. michiganensis subsp. michiganensis is not directly involved in virulence since neither colonization of tomato nor the development of disease symptoms was affected in an EPS− mutant produced by chemical mutagenesis (6). C. michiganensis subsp. michiganensis has at least four gene clusters putatively involved in the production of different EPS (see Table S8 in the supplemental material). The wcm cluster seems to encode all the proteins required to produce the major EPS of C. michiganensis subsp. michiganensis NCPPB382 when it is growing on rich medium. This EPS has been characterized, and its composition is known (6). Directed mutagenesis experiments with three genes in the wcm cluster confirmed the loss of EPS production (data not shown). The expression pattern and function of the three other clusters are unknown, but these clusters may be required in special environments or growth conditions (for example, during infection). The wco cluster is special as it encodes a protein putatively involved in fatty acid synthesis and several putative surface proteins, suggesting that there is a modified EPS anchored to the membrane. All four clusters identified are also present in C. michiganensis subsp. sepedonicus, but there are some differences, especially in the genes for glycosyl transferases, as the main EPS of the two subspecies have different compositions (see Table S8 in the supplemental material). However, at least two of the clusters seem to be nonfunctional in C. michiganensis subsp. sepedonicus (see the accompanying paper), while no obvious defects were found in C. michiganensis subsp. michiganensis.
Two genes necessary for the development of symptoms in tomato have been described previously (40). Both of these genes are plasmid encoded. pCM1 carries celA encoding a β-1,4-endoglucanase (26). CelA consists of three domains, an N-terminal catalytic glycosyl hydrolase domain, a cellulose-binding domain, and a C-terminal domain which is similar to α-expansins from plants. The third domain was shown to be essential both for development of wilting symptoms and for degradation of crystalline cellulose (26). The cellulase may be involved in the provision of nutrients derived from plant cell walls and in the tissue maceration that occurs in later stages of infection. A homologue of CelA lacking the third domain was predicted to be on the chromosome (celB [CMM_2443]). Additionally, a gene putatively encoding an expansin was identified (CMM_1480). In C. michiganensis subsp. sepedonicus both CelA and Cel5B are present and very similar. However, celB is a pseudogene, and C. michiganensis subsp. sepedonicus contains no expansin homologue. In L. xyli subsp. xyli only a homologue of Cel5B is present, while CelA and expansin homologues are not present.
A prominent feature of C. michiganensis subsp. michiganensis is the occurrence of numerous genes encoding extracellular proteases, although when C. michiganensis subsp. michiganensis is grown on agar plates containing casein or skim milk, it shows no proteolytic activity (9). At least 28 serine proteases belonging to three different families were identified. The first group, containing the second known virulence factor, Pat-1, was designated the Chp family and can be placed into serine protease family S1A. The catalytic triad consists of the amino acids HDS. Ten members were found in C. michiganensis subsp. michiganensis (see Figure S2 and Table S9 in the supplemental material). All of them have an atypical low G+C-content (between 52 and 65%), which may indicate a “foreign” origin. Three of them are located on plasmid pCM2, while the other seven are clustered in the chromosomal chp region near the origin. The plasmid-encoded Pat-1 was shown to be necessary for symptom development (wilting), but not for colonization of the tomato plant (13). Analysis of a mutant in which the serine in the catalytic triad was replaced by a threonine via site-specific mutagenesis suggested that pat-1 probably encodes a functional serine protease (9). The other genes in pCM2, phpA and phpB, are not essential for pathogenicity because the plasmid-free strain CMM100 colonizes normally and cloning of these genes into CMM100 did not affect the nonvirulent phenotype of the strain (9, 40). The seven chromosomal chp members include three pseudogenes (chpA, chpB, and chpD) containing frameshifts and/or in-frame stop codons. The other four genes appear to be intact. Inactivation of chpC and chpG by gene replacement was recently conducted. The chpG mutant was found to be unable to cause a hypersensitive response on the nonhost plant Mirabilis jalapa (Eichenlaub, unpublished data). The colonization of tomato plants by the chpC mutant was drastically reduced (Eichenlaub, unpublished data). This indicates that serine proteases have an important role in the C. michiganensis subsp. michiganensis-plant interaction. Genes homologous to chp genes have also been found in C. michiganensis subsp. sepedonicus and L. xyli subsp. xyli (see Fig. S2 in the supplemental material). While L. xyli subsp. xyli has only one homologous protein, which is most closely related to ChpC, the C. michiganensis subsp. sepedonicus genome contains 11 members of the chp family.
A second family of chymotrypsin-related serine proteases was designated the Ppa family (see Fig. S3 and Table S10 in the supplemental material). There are 11 members of this family in C. michiganensis subsp. michiganensis. One member is encoded on plasmid pCM1 (ppaJ), the genes encoding six members are located in the chp region (ppaA to ppaE), and the genes encoding the remaining four members (ppaF and ppaGHI) are located at two different loci on the chromosome. The encoded proteins consist of approximately 330 amino acids. A signal peptide has been predicted for all of them. The three residues of the catalytic triad of serine proteases (HDS) are conserved. Comparisons to the MEROPS database placed the members of the Ppa family in serine protease subfamily S1X. Homologous peptidases were identified in C. michiganensis subsp. sepedonicus (six members), L. xyli subsp. xyli, Xanthomonas spp., and X. fastidiosa. The G+C content of the ppaA to ppaE genes, located in the chp region, and the plasmid-borne gene ppaJ, which is around 65%, is remarkably lower than the G+C content of the ppaF to ppaI genes (∼75%).
Finally, three members of the subtilase family having the catalytic triad DHS were predicted; one gene is located in the chp region (CMM_0070), and two proteins are encoded elsewhere on the chromosome (CMM_2536 and CMM_2535). In C. michiganensis subsp. sepedonicus only two homologs are present (CMS0598 and CMS0597). The encoded proteins are interesting as they have high similarity to the SBT1, SBT2, and P69 subtilases of tomato (28, 29). The exact functions of these tomato proteases are unknown, but some of them have been implicated in wound and pathogen responses (29).
At the moment, the substrates of all C. michiganensis subsp. michiganensis proteases are unknown. It has been shown that in proteobacteria containing a TTSS proteases translocated into the plant cell interfere with plant signaling pathways (45). Although C. michiganensis subsp. michiganensis has no TTSS, the proteases may serve a similar function.
Although certain genes affecting virulence have been identified, the annotation of the C. michiganensis subsp. michiganensis genome and the comparison to the genome of C. michiganensis subsp. sepedonicus did not provide any clues concerning genes which may determine host specificity. The virulence and colonization of the tomato host plant of tomA and EPS mutants of C. michiganensis subsp. michiganensis are not altered (6, 32), and thus tomA and EPS apparently are not involved in determination of host specificity. The role of other potential factors, like surface proteins, should be investigated.
chp/tom region of the C. michiganensis subsp. michiganensis chromosome is involved in pathogenicity.
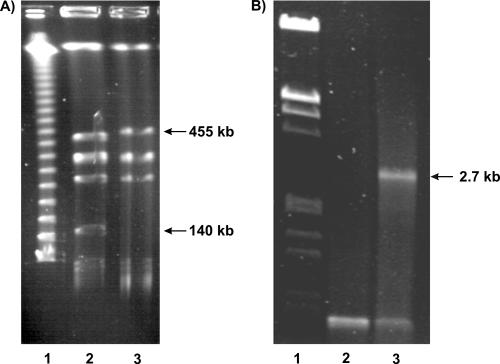
The importance of the chp/tomA region for the C. michiganensis subsp. michiganensis-tomato interaction is demonstrated by the fact that a derivative of C. michiganensis subsp. michiganensis NCPPB382, strain CMM30-18, which lacks the complete chp/tomA region, is nonvirulent and unable to colonize tomato effectively. Mutant CMM30-18 was obtained after transposon mutagenesis of C. michiganensis subsp. michiganensis NCPPB382 using Tn1409Cβ (20). The insertion site of Tn1409Cb in CMM30-18 was cloned and found to be in the CMM_0135 gene encoding a conserved hypothetical protein. However, complementation of the mutant with the intact gene cloned in the shuttle vector pDM302 (40) did not lead to restoration of the virulent phenotype, indicating that there is a second mutation. Analysis of strain CMM30-18 by PFGE of VspI-digested DNA showed a large chromosomal deletion in this mutant (Fig. 3A). The deletion was mapped by Southern hybridization of MunI-digested DNA with specific probes for all MunI fragments of the region (Fig. 2). Only the bordering fragments containing the 1.9-kb direct repeats gave positive hybridization signals (data not shown), indicating that almost the complete chp/tomA region was deleted. Finally, a PCR experiment with two oligonucleotide primers, one binding outside the left repeat and the other binding outside the right repeat, was conducted, which resulted in a 2.7-kb amplificate with CMM30-18 (Fig. 3b). This result confirmed that the intervening DNA region was lost by homologous recombination within the direct repeats. After cloning and sequencing of the amplificate, the recombination site could be mapped to an approximately 300-bp region in the middle of the direct repeats due to base exchanges, which allowed the left and right repeats to be distinguished (data not shown). Further characterization of the deleted region by directed gene replacement experiments indicated that some of the serine proteases encoded in the region are involved in the interaction with the host plant. CMM30-18 does not induce wilting symptoms in tomato, and the bacterial titer in planta reaches only 2.8 × 104 bacteria/g of plant homogenate, compared to 8 × 109 bacteria/g in infections with wild-type strain NCPPB382.
FIG. 3.
(A) PFGE analysis of VspI-hydrolyzed DNA. The following conditions were used: 14°C, 24 h, 6 V/cm, pulse changing from 10 to 90 s, and 1% agarose. For CMM30-18, the positions of the missing 140-kb fragment and the enlarged 455-kb fragment are indicated by arrows. Additionally, an 8-kb fragment is missing. Lane 1, λ concatemers; lane 2, C. michiganensis subsp. michiganensis NCPPB382; lane 3, CMM30-18. (B) PCR analysis showing the deletion of the chp/tomA region in CMM30-18. PCR was conducted using primers DR1a_up (5′-GTT GCT ACA GCC ATC C-3′) and DR1b_down (5′-GTG CAG CTG CAT GGG TC-3′) and 30 cycles of 1.5 min at 94°C, 1.5 min at 60°C, and 2 min at 72°C. Lane 1, λEcoRI/HindIII; lane 2, C. michiganensis subsp. michiganensis NCPPB382; lane 3, CMM30-18.
Conclusions.
In many pathogenic species genome reduction occurs as an adaptation to a stable environment provided by the host. In C. michiganensis subsp. michiganensis such genome reduction is not very pronounced or has not occurred yet. C. michiganensis subsp. michiganensis apparently possesses most biosynthetic pathways except the pathways for nitrate and sulfate reduction and for some vitamins leading to several auxotrophies (especially methionine and nicotinic acid). The high number of genes encoding transporters and transcriptional regulators indicates that the versatility of C. michiganensis subsp. michiganensis is similar to that of soil bacteria, although the survival of this organism in soil seems to be severely affected due to the defects mentioned above. The number of pseudogenes is low, and their location is mainly restricted to low-G+C-content regions. Thus, it is tempting to speculate that C. michiganensis subsp. michiganensis is a “recent” pathogen which evolved from plant-associated Microbacteriaceae and is still in the process of proper adaptation to the plant host. The genome size (3.4 Mb) is similar to that of the potato pathogen C. michiganensis subsp. sepedonicus, although roughly 0.2 kb of the C. michiganensis subsp. sepedonicus genome is comprised of insertion elements. The number of pseudogenes in C. michiganensis subsp. sepedonicus is higher, and the differences may be even more pronounced on the functional level. The genome of the sugarcane pathogen L. xyli subsp. xyli is about one-third smaller and also contains lots of insertion elements and has a high proportion of pseudogenes (13%). If grown on agar plates, C. michiganensis subsp. michiganensis grows slightly faster than C. michiganensis subsp. sepedonicus, while L. xyli subsp. xyli has a much longer generation time. This slower growth in vitro may be the result of adaptation to the plant host accompanied by decay and subsequent loss of unnecessary gene functions, rendering the bacteria more unable to grow outside the plant. Thus, it would be interesting to compare these genomes to that of the alfalfa pathogen C. michiganensis subsp. insidiosus (which infects a host plant that has not been changed as much by human breeding) or to those of epiphytic species of the Microbacteriaceae.
The genomes of the two Clavibacter subspecies contain a number of low-G+C-content regions (C. michiganensis subsp. michiganensis, ∼200 kb; C. michiganensis subsp. sepedonicus, ∼150 kb) which are different in the two subspecies. There are some indications that the low-G+C-content islands are foreign, and at least the chp/tomA region resembles a pathogenicity island. However, in C. michiganensis subsp. michiganensis these regions harbor no genes like genes encoding transposases which may confer mobility. Macrorestriction of DNA of several C. michiganensis subsp. michiganensis strains with VspI or DraI showed little conservation of the band pattern (data not shown). The genome data show that the sites for these rare cutters are located in the low-G+C-content regions (six out of nine VspI sites and four of seven DraI sites; two more sites for each restriction enzyme occur in the rrn operons). Thus, this apparently high genome variability may indicate that there has been exchange, mobility, or frequent loss and reacquisition of low-G+C-content regions in the strains, probably with no effect on the chromosomal backbone.
The virulence factors of C. michiganensis subsp. michiganensis seem to be extracellular enzymes, especially proteases. No TTSS or associated Avr proteins, which are prominent virulence factors in most proteobacteria, are present. Many of the potential virulence genes are clustered in the chp/tomA region, which is essential for effective colonization. It was shown previously that three of the proteases (Pat-1, ChpC, and ChpG) participate in the plant interaction, but the exact target of the proteases remains unknown (13; Eichenlaub, unpublished data). Whether they modulate plant defense reactions by interference with plant signaling pathways remains to be investigated. A similar function has been described for some cysteine proteases that serve in proteobacteria as suppressors of plant defense (redirecting the salicylate-induced defense reaction to the jasmonate pathway normally activated as a wound response) (45). A further candidate for interference with plant signaling may be the tomatinase since breakdown products of α-tomatine were described as suppressors of plant defense reactions in a fungal system (8).
Certainly the sequence information for the genomes of C. michiganensis subsp. michiganensis and the closely related organism C. michiganensis subsp. sepedonicus should provide a better understanding of the interaction with the host plants and eventually allow new approaches for control of these important plant-pathogenic microorganisms.
Supplementary Material
Acknowledgments
We thank all the people involved in this project. We thank C. A. Ishimaru (University of Minnesota) and S. Bentley (Sanger Institute, United Kingdom) for sharing data for C. michiganensis subsp. sepedonicus before publication. We are grateful to M. Redenbach, University of Kaiserslautern, Kaiserslautern, Germany, for help with the PFGE experiments.
This work was supported by grants in the GenoMik framework of the German Federal Ministry of Education and Research (grants FKZ 031U213D and 0313105) and by the Deutsche Forschungsgemeinschaft (grant GZ: EI 535/12-1).
Footnotes
Published ahead of print on 11 January 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 2005. PM 7/42 Clavibacter michiganensis subsp. michiganensis. Bull. OEPP/EPPO Bull. 35275-283. [Google Scholar]
- 3.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 1845826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartels, D., S. Kespohl, S. Albaum, T. Drüke, A. Goesmann, J. Herold, O. Kaiser, A. Pühler, F. Pfeiffer, G. Raddatz, J. Stoye, F. Meyer, and S. C. Schuster. 2005. BACCardI—a tool for the validation of genomic assemblies, assisting genome finishing and intergenome comparison. Bioinformatics 21853-859. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, S. D., K. F. Chater, A.-M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrel, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417141-147. [DOI] [PubMed] [Google Scholar]
- 6.Bermpohl, A., J. Dreier, R. Bahro, and R. Eichenlaub. 1996. Exopolysaccharides in the pathogenic interaction of Clavibacter michiganensis subsp. michiganensis with tomato plants. Microbiol. Res. 151391-399. [Google Scholar]
- 7.Bialczyk, J., Z. Lechowski, and D. Dziga. 2004. Composition of the xylem sap of tomato seedlings cultivated on media with HCO3− and nitrogen sources as NO3− or NH4+. Plant Soil 263265-272. [Google Scholar]
- 8.Bouarab, K., R. Melton, J. Peart, D. Boulcombe, and A. Osbourn. 2002. A saponin-detoxifying enzyme mediates suppression of plant defences. Nature 418889-892. [DOI] [PubMed] [Google Scholar]
- 9.Burger, A., I. Gräfen, J. Engemann, E. Niermann, M. Pieper, O. Kirchner, K.-H. Gartemann, and R. Eichenlaub. 2005. Identification of homologues to the pathogenicity factor Pat-1, a putative serine protease of Clavibacter michiganensis subsp. michiganensis. Microbiol. Res. 160417-427. [DOI] [PubMed] [Google Scholar]
- 10.Carver, T. J., K. M. Rutherford, M. Berriman, M.-A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis comparison tool. Bioinformatics 213422-3423. [DOI] [PubMed] [Google Scholar]
- 11.Davis, M. J., A. G. Gillaspie, A. K. Vidaver, and R. W. Harris. 1984. Clavibacter: a new genus containing some phytopathogenic coryneform bacteria, including Clavibacter xyli subsp. xyli sp. nov., subsp. nov. and Clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratoon stunting disease of sugarcane and bermudagrass stunting disease. Int. J. Syst. Bacteriol. 34107-117. [Google Scholar]
- 12.Denny, T. P. 1995. Involvement of bacterial polysaccharides in plant pathogenesis. Annu. Rev. Phytopathol. 33173-197. [DOI] [PubMed] [Google Scholar]
- 13.Dreier, J., D. Meletzus, and R. Eichenlaub. 1997. Characterization of the plasmid-encoded virulence region pat-1 of phytopathogenic Clavibacter michiganensis subsp. michiganensis. Mol. Plant-Microbe Interact. 10195-206. [DOI] [PubMed] [Google Scholar]
- 14.Eichenlaub, R., K.-H. Gartemann, and A. Burger. 2006. Clavibacter michiganensis, a group of Gram-positive phytopathogenic bacteria, p. 385-422. In S. S. Gnanamanickam (ed.), Plant-associated bacteria. Springer, Dordrecht, The Netherlands.
- 15.Evtushenko, L. I., L. V. Dorofeeva, S. A. Subbotin, J. R. Cole, and J. M. Tiedje. 2000. Leifsonia poae gen. nov., sp. nov., isolated from nematode galls on Poa annua, and reclassification of ‘Corynebacterium aquaticum ’ Leifson 1962 as Leifsonia aquatica (ex Leifson 1962) gen. nov., nom. rev., comb. nov. and Clavibacter xyli Davis et al. 1984 with two subspecies as Leifsonia xyli (Davis et al. 1984) gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 50371-380. [DOI] [PubMed] [Google Scholar]
- 16.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8186-194. [PubMed] [Google Scholar]
- 17.Ewing, B., L. Hillier, M. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8175-185. [DOI] [PubMed] [Google Scholar]
- 18.Fatmi, M., and N. W. Schaad. 2002. Survival of Clavibacter michiganensis subsp. michiganensis in infected tomato stems under natural field conditions in California, Ohio and Morocco. Plant Pathol. 51149-154. [Google Scholar]
- 19.Fink, D., N. Weiβschuh, J. Reuther, W. Wohlleben, and A. Engels. 2002. Two transcriptional regulators GlnR and GlnRII are involved in nitrogen metabolism of Streptomyces coelicolor A3(2). Mol. Microbiol. 46331-347. [DOI] [PubMed] [Google Scholar]
- 20.Gartemann, K.-H., and R. Eichenlaub. 2001. Isolation and characterization of IS1409, an insertion element of 4-chlorobenzoate-degrading Arthrobacter sp. strain TM1 and development of a system for transposon mutagenesis. J. Bacteriol. 1833729-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8195-202. [DOI] [PubMed] [Google Scholar]
- 22.Gordon, D., C. Desmarais, and P. Green. 2001. Automated finishing with autofinish. Genome Res. 11614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holtsmark, I., D. Mantzillas, V. G. H. Eijsink, and M. B. Brurberg. 2006. Purification, characterization, and gene sequence of michiganin A, an actagardine-like lantibiotic produced by the tomato pathogen Clavibacter michiganensis subsp. michiganensis. Appl. Environ. Microbiol. 725814-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Husted, S., C. A. Hebbern, M. Mattson, and J. K. Schjoerring. 2000. A critical experimental evaluation of methods for determination of NH4+ in plant tissue, xylem sap and apoplastic fluid. Physiol. Plant. 109167-179. [Google Scholar]
- 25.Jacobs, J. L., and G. W. Sundin. 2001. Effect of solar UV-B radiation on a phyllosphere bacterial community. Appl. Environ. Microbiol. 675488-5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jahr, H., J. Dreier, D. Meletzus, R. Bahro, and R. Eichenlaub. 2000. The endo-β-1,4-glucanase of Clavibacter michiganensis subsp. michiganensis is a pathogenicity determinant required for the induction of bacterial wilt of tomato. Mol. Plant-Microbe Interact. 13703-714. [DOI] [PubMed] [Google Scholar]
- 27.Jakimowicz, D., J. Majka, W. Messer, C. Speck, M. Fernandez, M. C. Martin, J. Sanchez, F. Schauwecker, U. Keller, H. Schrempf, and J. Zakreska-Czerwinska. 1998. Structural elements of the Streptomyces oriC region and their interactions with the DnaA protein. Microbiology 1441281-1290. [DOI] [PubMed] [Google Scholar]
- 28.Janzik, I., P. Macheroux, N. Amrhein, and A. Schaller. 2000. LeSBT1, a subtilase from tomato plants. J. Biol. Chem. 2755193-5199. [DOI] [PubMed] [Google Scholar]
- 29.Jorda, L., A. Coego, V. Conejero, and P. Vera. 1999. A genomic cluster containing four differentially regulated subtilisin-like processing protease genes is in tomato plants. J. Biol. Chem. 2742360-2365. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser, O., D. Bartels, T. Bekel, A. Goesmann, S. Kespohl, A. Pühler, and F. Meyer. 2003. Whole genome shotgun sequencing guided by bioinformatics pipelines—an optimized approach for an established technique. J. Biotechnol. 106121-133. [DOI] [PubMed] [Google Scholar]
- 31.Kang, H. Y., T. J. Brickman, F. C. Beaumont, and S. K. Armstrong. 1996. Identification and characterization of iron-regulated Bordetella pertussis alcaligin siderophore biosynthesis genes. J. Bacteriol. 1784877-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaup, O., I. Gräfen, E.-M. Zellermann, R. Eichenlaub, and K.-H. Gartemann. 2005. Identification of a tomatinase in the tomato-pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382. Mol. Plant-Microbe Interact. 181090-1098. [DOI] [PubMed] [Google Scholar]
- 33.Kers, J. A., K. D. Cameron, M. V. Joshi, R. A. Bukhalid, J. E. Morello, M. J. Wach, D. M. Gibson, and R. Loria. 2005. A large, mobile pathogenicity island confers plant pathogenicity on Streptomyces species. Mol. Microbiol. 551025-1033. [DOI] [PubMed] [Google Scholar]
- 34.Kirchner, O., K.-H. Gartemann, E.-M. Zellermann, R. Eichenlaub, and A. Burger. 2001. A highly efficient transposon mutagenesis system for the tomato pathogen Clavibacter michiganensis subsp. michiganensis. Mol. Plant-Microbe Interact. 141312-1318. [DOI] [PubMed] [Google Scholar]
- 35.Konstantinidis, K. T., and J. Tiedje. 2004. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA 1013160-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laine, M. J., H. Nakhei, J. Dreier, K. Lehtilä, D. Meletzus, R. Eichenlaub, and M. C. Metzler. 1996. Stable transformation of the gram-positive phytopathogenic bacterium Clavibacter michiganensis subsp. sepedonicus with several cloning vectors. Appl. Environ. Microbiol. 621500-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, J. H., and R. K. Holmes. 2000. Characterization of specific nucleotide substitutions in DtxR-specific operators of Corynebacterium diphtheriae that dramatically affect DtxR binding, operator function, and promoter strength. J. Bacteriol. 182432-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meidanis, J., M. D. V. Braga, and S. Verjoski-Almeida. 2002. Whole-genome analysis of transporters in the plant pathogen Xylella fastidiosa. Microbiol. Mol. Biol. Rev. 66272-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meletzus, D., and R. Eichenlaub. 1991. Transformation of the phytopathogenic bacterium Clavibacter michiganense subsp. michiganense by electroporation and development of a cloning vector. J. Bacteriol. 173184-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meletzus, D., A. Bermpohl, J. Dreier, and R. Eichenlaub. 1993. Evidence for plasmid-encoded virulence factors in the phytopathogenic bacterium Clavibacter michiganensis subsp. michiganensis NCPPB382. J. Bacteriol. 1752131-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer, F., A. Goesmann, A. C. McHardy, D. Bartels, T. Bekel, J. Clausen, J. Kalinowski, B. Linke, O. Rupp, R. Giegerich, and A. Pühler. 2003. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 312187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molenaar, D., M. E. van der Rest, A. Drysch, and R. Yucel. 2000. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Corynebacterium glutamicum. J. Bacteriol. 1826884-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monteiro-Vitorello, C. B., L. E. A. Camargo, M. A. Van Sluys, J. P. Kitajima, D. Truffi, A. M. do Amaral, R. Harakava, J. C. F. de Oliveira, D. Wood, M. C. de Oliveira, C. Miyaki, M. A. Takita, A. C. R. da Silva, L. R. Furlan, D. M. Carraro, G. Camarotte, N. F. Almeida, Jr., H. Carrer, L. L. Coutinho, H. A. El-Dorry, M. I. T. Ferro, P. R. Gagliardi, E. Giglioti, M. H. S. Goldman, G. H. Goldman, E. T. Kimura, E. S. Ferro, E. E. Kuramae, E. G. M. Lemos, M. V. F. Lemos, S. M. Z. Mauro, M. A. Machado, C. L. Marino, C. F. Menck, L. R. Nunes, R. C. Oliveira, G. G. Pereira, W. Siqueira, A. A. de Souza, S. M. Tsai, A. S. Zanca, A. J. G. Simpson, S. M. Brumbley, and J. C. Setúbal. 2004. The genome sequence of the Gram-positive sugarcane pathogen Leifsonia xyli subsp. xyli. Mol. Plant-Microbe Interact. 17827-836. [DOI] [PubMed] [Google Scholar]
- 44.Moore, C. M., and J. D. Helmann. 2005. Metal ion homeostasis in Bacillus subtilis. Curr. Opin. Microbiol. 8188-195. [DOI] [PubMed] [Google Scholar]
- 45.Mudgett, M. B. 2005. New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56509-531. [DOI] [PubMed] [Google Scholar]
- 46.Newton, G. L., K. Arnold, M. S. Price, C. Sherrill, S. B. Delcardayre, Y. Aharonowitz, G. Cohen, J. Davies, R. C. Fahey, and C. Davis. 1996. Distribution of thiols in microorganisms: mycothiol is a mayor thiol in most actinomycetes. J. Bacteriol. 1781990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park, Y.-H., K.-I. Suzuki, D.-G. Yim, K.-C. Lee, E. Kim, J.-S. Yoon, S.-J. Kim, Y.-H. Kho, M. Goodfellow, and K. Komogata. 1993. Suprageneric classification of peptidoglycan group B actinomycetes by nucleotide sequencing of 5S ribosomal RNA. Antonie van Leeuwenhoek 64307-313. [DOI] [PubMed] [Google Scholar]
- 48.Redenbach, M., J. Scheel, and U. Schmidt. 2000. Chromosome topology and genome size of selected actinomycetes species. Antonie van Leeuwenhoek 78227-235. [DOI] [PubMed] [Google Scholar]
- 49.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 51.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M.-C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaption to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 9914422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, G. E., and M. D. Summers. 1980. The bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl-paper. Anal. Biochem. 109123-129. [DOI] [PubMed] [Google Scholar]
- 53.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5233-241. [DOI] [PubMed] [Google Scholar]
- 54.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406959-964. [DOI] [PubMed] [Google Scholar]
- 55.Strider, D. L. 1969. Bacterial canker of tomato caused by Corynebacterium michiganense. A literature review and bibliography. N. C. Agric. Exp. Stn. Tech. Bull 193.
- 56.Tauch, A., I. Homann, S. Mormann, S. Rüberg, A. Billault, B. Bathe, S. Brand, O. Brockmann-Gretza, C. Rückert, N. Schischka, C. Wrenger, J. Hoheisel, B. Möckel, K. Huthmacher, W. Pfefferle, A. Pühler, and J. Kalinowski. 2002. Strategy to sequence the genome of Corynebacterium glutamicum ATCC13032: use of a cosmid and a bacterial artificial chromosome library. J. Biotechnol. 9525-38. [DOI] [PubMed] [Google Scholar]
- 57.Thieme, F., R. Koebnik, T. Bekel, C. Berger, J. Boch, D. Büttner, C. Caldana, L. Gaigalat, A. Goesmann, S. Kay, O. Kirchner, C. Lanz, B. Linke, A. McHardy, F. Meyer, G. Mittenhuber, D. H. Nies, U. Niesbach-Klösgen, T. Patschkowski, C. Rückert, O. Rupp, S. Schneiker, S. C. Schuster, F.-J. Vorhölter, E. Weber, A. Pühler, U. Bonas, D. Bartels, and O. Kaiser. 2005. Insights into genome plasticity and pathogenicity of the plant-pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 1877254-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 244876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsiantos, J. 1987. Transmission of Corynebacterium michiganense pv. michiganense by seeds. J. Phytopathol. 119142-146. [Google Scholar]
- 60.Vidaver, A. K. 1982. The plant pathogenic corynebacteria. Annu. Rev. Microbiol. 36495-517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.