Abstract
The extreme metabolic dormancy and resistance properties of spores formed by members of the Bacillus and Clostridium genera are lost upon exposure to a variety of small-molecule germinants. Germinants are known to interact in an as yet undefined manner with cognate receptor complexes that reside in the inner membrane that surrounds the spore protoplast. The receptor itself is a complex of at least three proteins, and in this study we identify amino acid residues, predicted to lie in loop regions of GerVB on the exterior aspect of the membrane, that influence the Bacillus megaterium spore germination response. Three consecutive residues adjacent to putative transmembrane domain 10 (TM10) were demonstrated to mediate to various degrees the proline germinative response while also influencing germination in response to leucine, glucose, and inorganic salts, suggesting that this region may be part of a ligand binding pocket. Alternatively, substitutions in this region may affect the conformation of associated functionally important TM regions. Leucine- and KBr-mediated germination was also influenced by substitutions in other outer loop regions. These observations, when considered with accompanying kinetic analyses that demonstrate cooperativity between germinants, suggest that binding sites for the respective germinants are in close spatial proximity in the receptor but do not overlap. Additionally, proline recognition was conferred to a chimeric receptor when TM regions associated with the putative binding loop were present, indicating that residues in TM9 and/or TM10 of GerVB are also of functional importance in the proline-induced germinative response.
Endospores formed in response to nutrient starvation by members of the genera Bacillus and Clostridium display remarkable properties of resistance and dormancy that permit their survival in the environment for extended periods. Despite this extreme dormancy, spores retain the ability to initiate vegetative metabolism rapidly upon exposure to appropriate nutrients via the process of germination (20, 30). Significant advances have been made in recent years through the application of mainly genetic techniques to characterize the molecular apparatus involved in spore germination, identifying structural genes that encode germinant receptors (4, 8, 14, 23, 37), ion channels (31, 35), and hydrolytic enzymes involved in the degradation of the cortical peptidoglycan that surrounds the spore protoplast (6, 11, 16, 21). Cytological observations on spore coat degradation during germination (29) have also recently been extended by the application of atomic force microscopy to provide insights to the structural basis for the disassembly of the outer layers that provide the primary barrier to environmental insult (26).
Despite these advances, however, little is known of the molecular mechanism of the primary event of spore germination—the interaction of the germinant, typically an amino acid, sugar, or riboside, with its cognate receptor—and how this interaction triggers the subsequent cascade of germination events. Biochemical and structural analyses of the germinant receptor have been hindered by an inability to express key receptor proteins at preparative levels, which, consistent with their status as receptors for environmental stimuli, have been identified as integral or membrane-associated proteins (12, 22). Thus, virtually all information regarding functionality of the germinant receptor has been accumulated from genetics-based analyses. Evidence that the receptor comprises a complex of at least three different proteins, for example, is provided by the conserved tricistronic receptor operon structure observed in all sporeformer genomes, suggesting that genes within the operon have coevolved, and, where tested, by the absolute requirement for all three receptor components for functionality (20, 23). Indirect genetic evidence for physical interaction between the respective A, B, and C proteins that comprise the receptor (13) has been complemented recently by yeast two-hybrid experiments (34), which, in addition to providing evidence for the interaction between receptor subunits, also suggest that some receptor proteins interact with SpoVA proteins thought to be involved in the release of calcium dipicolinate during germination. Similarly, while some germinant receptors can function independently to trigger the spore germination response, for example, the GerA-mediated l-alanine response in Bacillus subtilis, most germinant receptors appear to work in concert to initiate germination, either in response to single germinants (e.g., the GerQ and GerI response to inosine in Bacillus cereus [4]) or to mixtures of germinants (e.g., the GerB- and GerK-mediated response to AGFK in B. subtilis [9, 15]). Again, genetic evidence employing mutant B. subtilis constructs has been presented suggesting that different receptors can physically interact (3, 5).
While it is generally accepted that germination is triggered by the binding or interaction of a germinant with its cognate receptor, relatively little experimental data have been presented regarding the identification of the receptor protein(s) and/or key amino acids that form the germinant binding site(s). B. subtilis strains carrying point mutations in the structural gene for GerAB, which necessitate a higher concentration of l-alanine to initiate germination, were described a number of years ago (28), providing the first indication that the B protein of the receptor forms the ligand binding site. Further evidence was presented recently when our laboratory demonstrated that two close protein B homologues could interact with A and C proteins of the Bacillus megaterium GerU receptor to form receptors with different specificities (7). Thus, spores expressing GerUB are responsive to glucose and leucine, whereas those expressing GerVB are responsive to proline and KBr in addition to glucose and leucine.
The high degree of shared identity between these B proteins and the hitherto largely unrecognized interchangeability between receptor components suggested that this system might be amenable to a site-directed mutagenesis (SDM) approach to identify residues that are crucial in determining the range of germinants recognized. We describe such an approach in this communication and, via construction of mutant constructs that display conditional germination responses, identify a number of functionally important residues that may participate in binding of the main B. megaterium QM B1551 germinants.
MATERIALS AND METHODS
Bacterial strains and media.
B. megaterium strains employed in this study (Table 1) are essentially all derivatives of strain PV361, a plasmidless variant of the wild-type QM B1551 strain that lacks the GerU operon and GerVB structural gene required for efficient spore germination. B. megaterium strains were routinely cultured at 30°C on LB agar or broth containing antibiotics, where appropriate (1 μg/ml erythromycin and 25 μg/ml lincomycin for macrolide-lincosamide-streptogramin B resistance [MLSr]). Preparation of B. megaterium protoplasts and polyethylene glycol-mediated transformation of plasmid DNA was performed as described previously (19), with direct selection for transformants on RHAF agar plates containing 1 μg/ml erythromycin and 25 μg/ml lincomycin. RHAF medium consists of the following (per liter): NH4Cl, 1.0 g; Tris base, 12.0 g; KCl, 35 mg; NaCl, 58 mg; Na2SO4·10H2O, 300 mg; KH2PO4, 140 mg; MgCl2·5H2O, 4.26 g; yeast extract, 5.0 g; tryptone (Oxoid), 5.0 g; sucrose, 68.46 g; and glucose, 2.0 g. The pH was adjusted to 7.5 with HCl before addition of MgCl2·5H2O. Solid medium contained 1% agar. Escherichia coli strains used for SDM (XL1-Blue and XL10-Gold [Stratagene]) or preparation of plasmid for transformation of B. megaterium (E. coli NovaBlue [Novagen]) were cultured in LB medium at 37°C supplemented with 75 μg/ml carbenicillin.
TABLE 1.
Strains and plasmids used in this study
| B. megaterium strain or plasmid | Receptor characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| PV361 | Nongerminative derivative of QM B1551 | 33 |
| PV361 transformants | ||
| GC-GerU | GerU (containing gerUA, gerUC, and gerUB) | 7 |
| GC-GerU* | GerU* (containing gerUA, gerUC, and gerVB) | 7 |
| GC-OL | GerVB outer loops changed to corresponding GerUB outer loops | This study |
| GC-IL | GerVB inner loops changed to corresponding GerUB inner loops | This study |
| GC-L1 | GerVB with substituted OL1 | This study |
| GC-L2 | GerVB with substituted OL2 | This study |
| GC-L3 | GerVB with substituted OL3 | This study |
| GC-L4 | GerVB with substituted OL4 | This study |
| GC-L5 | GerVB with substituted OL5 | This study |
| GC-L5K334Y | GerVB K334Y | This study |
| GC-L5V335A | GerVB V335A | This study |
| GC-L5E336A | GerVB E336A | This study |
| GC-L5M337K | GerVB M337K | This study |
| GC-GerUloop5 | GerUB Y334 → K337 changed to KVEM | This study |
| GC-GerUVfusion | GerUB TM1 to TM8 fused with GerVB TM9 and TM10 | This study |
| Plasmids | ||
| pGEM-3Z | E. coli cloning vector; Ampra | Promega |
| pHT315 | B. megaterium host plasmid; MLSrb | 2 |
Ampr, ampicillin (β-lactam) resistance.
MLSr, macrolide-lincosamide-streptogramin B resistance.
Spore preparation.
B. megaterium spores were prepared in 200-ml aliquots of supplemented nutrient broth (10) in 2-liter flasks (at 30°C and 200 rpm) supplemented with 1 μg/ml erythromycin to provide selective pressure for maintenance of the plasmid-borne receptor operon. Spores were harvested by centrifugation after 72 h of culture and then subjected to repeated rounds of washing in sterile cold water before storage on ice. All spore suspensions used in this work were observed to be free of vegetative cells and debris.
SDM.
SDM procedures were conducted with Stratagene QuikChange II and QuikChange Multi SDM kits, as directed by the manufacturer. Primers for SDM, sequences of which are available upon request, were designed both manually and by using the QuikChange primer design program (Stratagene). The main parental plasmids subjected to SDM comprised gerU* (gerUA, gerUC, and gerVB) and gerU (gerUA, gerUC, and gerUB) receptor operons cloned into the BamH1 site of pGEM-3Z. Construction of the gerU* fusion operon was described previously (7). Transformant E. coli harboring mutagenized plasmids was identified by DNA sequence analysis of the appropriate B cistron amplified by colony PCR using appropriate primers prior to purification of plasmid. Plasmids identified as carrying the correct mutation(s) then served as templates for PCRs employing primers containing BamH1 restriction sites spanning the entire receptor operon. The receptor amplicons were then digested and ligated with plasmid pHT315 (2) restricted with the same enzyme, and the ligation mixture was used to transform E. coli, from which recombinant plasmid was prepared. Plasmid pHT315 is less likely to be cured from the B. megaterium population during growth/sporulation than the pUCTV2 vector employed in previous complementation analyses, permitting 100% germination responses to be recorded. The fidelity of the entire receptor operon cloned into pHT315 was assessed by sequence analysis prior to transformation of B. megaterium PV361 to MLSr. Final B. megaterium constructs are detailed in Table 1.
GerUB/VB fusion.
An overlapping PCR technique was used to construct a gerU fusion operon encompassing gerUA, gerUC, and a chimeric gerUB/VB cistron, which was predicted to encode a protein comprising transmembrane domain 1 (TM1) to TM8 of GerUB and TM9 and TM10 (and outer loop 5 [OL5]) of GerVB. PCR was used to prepare two fragments of DNA, the first a 4,197-bp amplicon encompassing the coding and putative regulatory sequences for GerUA, GerUC, and the first 296 codons of GerUB. The second fragment comprised a 435-bp amplicon encompassing codons 289 to 366 of GerVB and a potential rho-independent terminator sequence. These products, which included 24 bp of overlapping sequence at the 3′ end of fragment 1 and 5′ end of fragment 2 were purified and mixed to provide a template for a subsequent round of PCR using primers with BamH1 sites, resulting in the creation of a fragment of DNA encompassing gerUA, gerUC, and the chimeric gerUB/VB gene arranged as a GerA-type receptor operon with appropriate regulatory sequences. This product was subsequently digested with BamH1, ligated with pHT315 restricted with the same enzyme, and used to transform E. coli. The resultant recombinant plasmid was used to transform PV361 to MLSr, yielding the GC-GerUVfusion strain.
Germination assays.
Spores at 5 to 10 mg/ml (dry weight) in water were heat shocked at 60°C for 10 min and then cooled on ice. Spores were resuspended at an optical density at 600 nm (OD600) of 0.95 to 1.0 in germination buffer (5 mM Tris-HCl, pH 7.8, plus 10 mM germinant [50 mM for KBr]) at 30°C. Control experiments with heat-shocked spores in buffer alone were included for each experiment. Spore germination was monitored by measurement of the A600 of the suspension over a 40-min period using a 1-ml cuvette in a Hewlett Packard 8452A diode array spectrophotometer. All values reported are the averages of at least duplicate experiments utilizing independently prepared batches of spores. Where presented, maximum rates of spore germination are given relative to the A600 loss observed for B. megaterium QM B1551 spores incubated with 10 mM glucose, where a 65% loss of the OD600 correlates to approximately 100% spore germination, as determined by loss of heat resistance.
Experiments designed for germination kinetic analyses were conducted in a 96-well plate format in a total volume of 200 μl/well using a Tecan Infinite-200 series shaking-incubating plate reader. Experiments were conducted in triplicate with two different spore preparations. Heat-activated spores (OD600 of 0.4) were exposed to various concentrations of leucine (0.1, 0.175, 0.25, 0.5, 1, 2.5, and 5 mM) at specific constant proline concentrations (75, 100, 150, and 200 μM), and germination was monitored by measuring the decrease in A600 at 30°C every minute for 60 min. Germination rates (v [OD units/min]) were calculated as the slope of the linear segment of OD changes over time that follows the initial lag phase. Data were subsequently plotted as double-reciprocal plots of 1/v versus 1/[leucine concentration]. The Michaelis-Menten function of SigmaPlot, version10 (Systat Software Inc.), was employed to determine apparent Km and Vmax values. The effect of varying individual germinant concentrations was also assessed in a 96-well plate format (total volume, 300 μl/well), where heat-activated spores (OD600 of 0.4) were incubated at 30°C in 5 mM Tris-HCl (pH 7.8) with germinant concentrations ranging from 0.1 to 50 mM, and germination was monitored by measuring the decrease in the A600 value every minute for 60 min. Germination rates and apparent Vmax values were calculated as described above.
Molecular biology methods and bioinformatics analyses.
All PCR procedures were performed by standard methodologies using Kod Hot Start Polymerase (Novagen). Plasmid DNA was prepared using a QIAprep Spin miniprep kit (Qiagen). DNA sequencing was performed by the Department of Biochemistry sequencing facility (University of Cambridge), while DNA sequence analysis was performed using CLC free-workbench software (CLC bio). Protein topology and transmembrane helix predictions were made using the HMMTOP program (32), which is available on the ExPASy server (Swiss Institute of Bioinformatics) (http://expasy.org/).
RESULTS
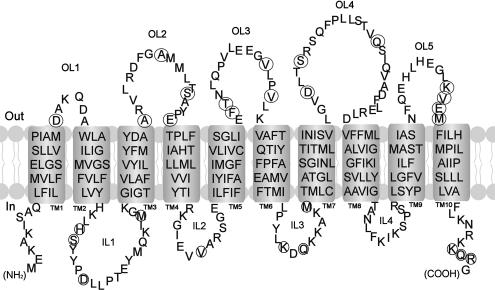
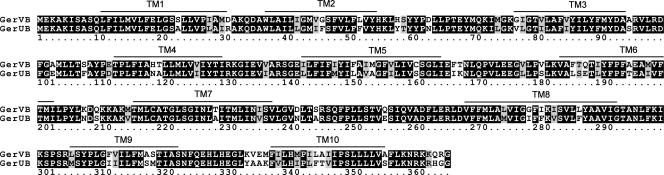
We have previously demonstrated that phenotypic differences in the germination properties of spores carrying the respective GerU and GerU* germinant receptors are due to two closely related B protein homologues that may present different ligand binding sites (7). These proteins, GerUB and GerVB, share 80% amino acid identity; therefore, phenotypic differences are presumably conferred by the remaining 20% of relatively randomly distributed amino acids that differ between the two proteins. As a first step toward identifying residues potentially involved in the recognition of defined germinants, we decided to examine the role of amino acids that are predicted to lie in the hydrophilic loop regions of GerVB (Fig. 1) by replacing putative outer-facing (integument) and inner-facing (protoplast) residues with those in the corresponding positions of GerUB, as adjudged by ClustalW alignment (Fig. 2). The assignment of loops as protoplast- or integument-facing was via HMMTOP analysis. We decided initially to examine these residues collectively, preparing plasmid-encoded receptors with 16 amino acid substitutions to the putative outer loop regions and 11 changes to the inner loops via multiple rounds of SDM (Table 2). Strains carrying the respective mutagenized receptor plasmids (GC-OL and GC-IL) were observed to sporulate normally; these were then prepared and assayed for germination properties as described in the Materials and Methods section. Spores carrying the parental GerU and GerU* receptors were also prepared for comparative purposes.
FIG. 1.
A model of GerVB with 10 putative TM domains predicted by the HMMTOP program. Circled residues in loop regions were subjected to replacement with the corresponding residues in GerUB, as adjudged by ClustalW alignment (Fig. 2).
FIG. 2.
ClustalW alignment of GerVB and GerUB. Predicted TM regions, based on hydropathy profiling of GerVB by HMMTOP analysis, are highlighted.
TABLE 2.
Amino acid substitutions
| Region | Amino acid substitution(s) (n)a
|
|
|---|---|---|
| Inner loop | Outer loop | |
| N terminus | None (0) | NA |
| Loop 1 | H58Y, S59T, D63N, M74L (4) | D31R (1) |
| Loop 2 | V137I (1) | A95S, A103E, S108F, E112D (4) |
| Loop 3 | K208N, Q210S, M215V (3) | F166I, T167K, V178L, V181S (4) |
| Loop 4 | None (0) | D242N, S245A, Q256E (3) |
| Loop 5 | NA | K334Y, V335A, E336A, M337K (4) |
| C terminus | K363R, Q364H, R365G (3) | NA |
n, number of substitutions; NA, not applicable.
Phenotypic differences in the germination properties of spores carrying the GerU and GerU* receptors were as expected, with only GerU* spores showing a strong germination response to proline and KBr (Table 3). Despite the apparent reduced efficiency of the GerU receptor to single germinant compounds, GerU demonstrated a stronger germination response to leucine than GerU*, which is in agreement with previous data. Spores of strain GC-IL, in which the predicted inner loops of the GerVB protein have been replaced with their GerUB equivalents, show strong germination responses to all single-trigger germinant compounds, suggesting that residues involved in recognition of proline and KBr have not been significantly modified. Conversely, spores carrying the GerVB protein with substituted outer loops (GC-OL) germinate efficiently only in response to glucose. The loss of response to proline and KBr indicates that B protein outer loops may be involved in recognition of germinants; however, the loss of germinative response to leucine, where a gain-of-function phenotype might have been expected—since GerU mediates the stronger response to leucine—was not anticipated. The mutations may introduce subtle differences to the composition and spatial arrangement of the helical bundles, resulting in conformational changes to the connecting loops and putative ligand binding sites that permit the GC-OL mutant to germinate in response to glucose but not to leucine. Regardless, this observation provides the first evidence of the close association between the composition of the TM and outer loop regions and receptor functionality.
TABLE 3.
Rates of germination of spores complemented with pHT315 plasmid-borne receptor variantsa
| Strain | Rate of spore germination with indicated germinantb
|
||||
|---|---|---|---|---|---|
| Buffer | Glucose | Proline | Leucine | KBr | |
| GC-GerU | 3 | 85 | 3 | 76 | 10 |
| GC-GerU* | 1 | 99 | 100 | 57 | 100 |
| GC-IL | 1 | 97 | 100 | 41 | 88 (± 9) |
| GC-OL | <1 | 100 | <1 | <1 | 7 |
Spores were germinated in 5 mM Tris-HCl, pH 7.8, for 40 min with 10 mM germinant (50 mM KBr). Spore germination was measured as described in Materials and Methods.
Rates of spore germination are given relative to the A600 loss (65%) for B. megaterium QM B1551 spores in 10 mM glucose, which was set at 100. This value is equivalent to 100% germination after a 40-min incubation. Values are the means of at least duplicate experiments; standard deviation was ≤5% of the mean, except where indicated in parentheses.
In order to identify specific outer loops involved in the spore response to individual germinants, we decided to construct a series of mutants carrying the GerVB protein with single outer loops changed to match those of the corresponding region of GerUB. Substitutions comprised a single amino acid change to OL1, three changes to OL4, and four changes to OL2, OL3, and OL5 (Table 2).
The germinative response of this series of mutants to proline is the most notable (Table 4), where a germination rate of 100% is observed for strains complemented with receptors with mutations in OL1, OL2, OL3, and OL4. Spores carrying substitutions in OL5, however, which comprises four consecutive amino acid changes (K334Y, V335A, E336A, and M337K), show no germinative response to this trigger compound, indicating that some of these residues might participate in ligand binding.
TABLE 4.
Rates of germination of spores complemented with GerU* receptors with substitutions to outer loop regions of GerVBa
| Strain | Rates of spore germination with indicated germinantb
|
||||
|---|---|---|---|---|---|
| Buffer | Glucose | Proline | Leucine | KBr | |
| GC-GerU* | 1 | 99 | 100 | 57 | 100 |
| GC-L1 | 4 | 100 | 100 | 71 (± 24) | 100 |
| GC-L2 | 3 | 100 | 100 | 100 | 10 |
| GC-L3 | <1 | 100 | 100 | 85 | 100 |
| GC-L4 | <1 | 100 | 100 | 18 (± 10) | 67 |
| GC-L5 | 2 | 97 | 2 | 10 (± 6) | 13 |
Spores were germinated in 5 mM Tris-HCl, pH 7.8, for 40 min with 10 mM germinant (50 mM KBr). Spore germination was measured as described in Materials and Methods.
Rates of spore germination are given relative to the A600 loss (65%) for B. megaterium QM B1551 spores in 10 mM glucose, which was set at 100. This value is equivalent to 100% germination after a 40-min incubation. Values are the means of at least duplicate experiments; standard deviation was ≤5% of the mean, except where indicated in parentheses.
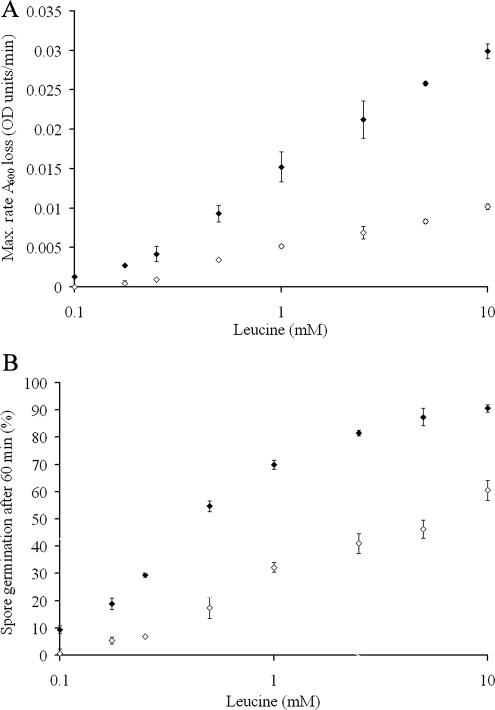
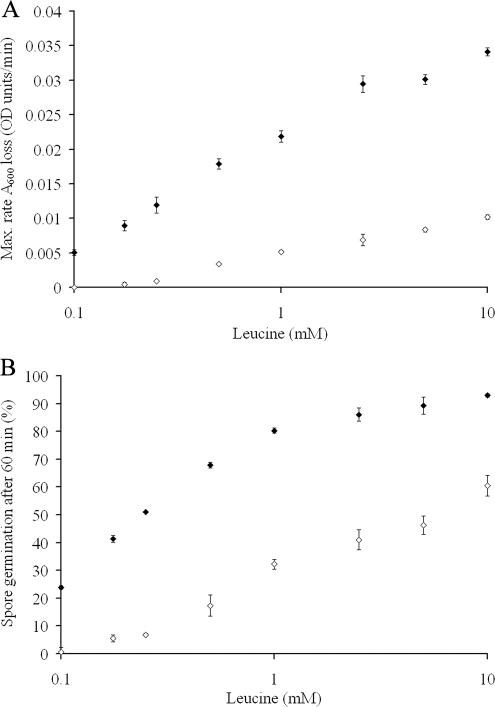
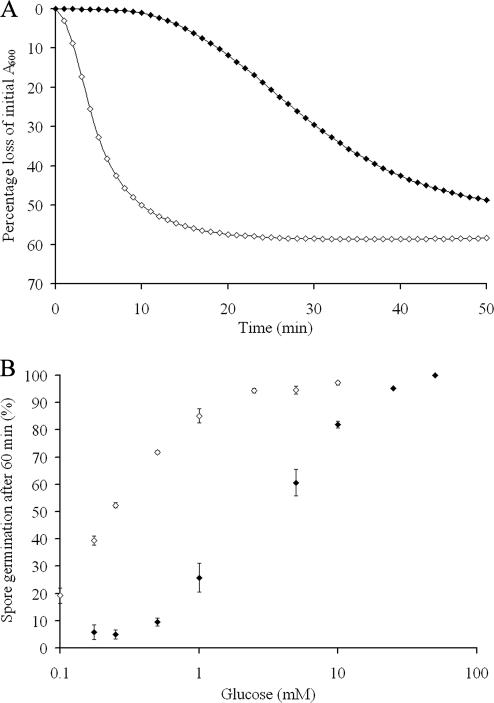
While mutations to a single outer loop appear to be primarily responsible for the differences in receptor recognition to proline, the response to other germinants is more complicated (Table 4). The germination response to leucine is actually enhanced with respect to GerU* and even the GerU receptor when OL1 and, in particular, OL2 and OL3 are modified. Further analysis of the concentration dependence on leucine-induced germination (Fig. 3) reveals that spores carrying receptors with OL2 substitutions (strain GC-L2) display both an increased germinative rate (Vmax of 0.034 OD/min) and sensitivity to leucine (50% spore germination at approximately 0.44 mM) compared to spores carrying the GerU* receptor (Vmax of 0.011 OD/min and 50% germination at approximately 6.30 mM leucine). Spores of strain GC-L2 were also observed to show increased germinative rates and sensitivity to proline (Vmax 0.078 versus 0.043 OD/min; 50% spore germination at approximately 0.30 mM versus 0.53 mM proline) and glucose (Vmax of 0.117 versus 0.098 OD/min; 50% spore germination at approximately 0.13 mM versus 0.23 mM glucose) compared to strain GC-GerU* (data not shown). Contrary to these gain-of-efficiency observations, mutations in both OL4 and OL5 severely impact the germination response to leucine (Table 4). In either case, differences in the efficiency of germination may be explained by direct changes to residues involved in ligand binding—which may involve interactions between multiple loops—or by introduced conformational changes that promote or inhibit the ability to accommodate the germinant and/or transduce the signal upon ligand binding.
FIG. 3.
Effects of l-leucine concentration on spore germination rates of strains GC-GerU* and GC-L2. Spores carrying receptors with mutations in the putative OL2 of GerVB (strain GC-L2 [⧫]) display an increased germinative rate (A) and sensitivity, in terms of minimal germinant requirement, to leucine (B) compared to spores carrying the parental GerU* receptor (⋄). Spores were germinated in 96-well plates in a total volume of 300 μl of 5 mM Tris-HCl, pH 7.8, at 30°C with variable leucine concentrations. Maximum rates of germination were calculated from the slope of the linear segment of A600 loss over time that follows the initial lag phase. Under these experimental conditions, a 60% loss in A600 correlates with 100% spore germination, as determined by loss of heat resistance of strain GC-GerU* after a 60-min incubation in 10 mM glucose. This value was used subsequently to estimate the proportion of germinated spores after incubation for 60 min under the range of germinant concentrations tested. Plotted values are averages of experiments conducted in triplicate (error bars represent standard deviations from the mean).
The GerVB-mediated germination response to inorganic salts, exemplified by KBr in these experiments, is also influenced by substitutions on more than one outer loop (Table 4). Spores carrying receptors with substitutions in OL2, OL4, and OL5 all show decreased germination efficiency upon exposure to KBr, while mutations to OL1 and OL3 have no impact. KBr is the only germinant that is adversely affected by OL2 substitutions, which demonstrably enhance the spore response to nutrient-type germinants, as described above. As with leucine, we cannot yet distinguish between conformational changes to the protein and/or direct changes to residues involved in ligand binding. However, since both the cation and anion have been demonstrated to influence the strength of the germinative response (27), then perhaps different loops are involved in binding of the respective ions.
Since initiation of germination upon exposure to proline appeared to be primarily affected by substitutions to OL5, we decided to characterize further the residues involved by constructing mutants with single amino acid changes. Collectively, these data demonstrate that even single mutations can exert considerable influence on the functionality of the receptor (Table 5). The E336A mutation, for example, completely prohibits the response to proline while conferring an enhanced response to leucine with respect to the native GerVB protein. Additionally, strain GC-L5 with the mutation E336A (GC-L5E336A) displays increased sensitivity to leucine in terms of the minimal germinant requirement for stimulation of germination, with 50% spore germination occurring at approximately 0.24 mM leucine in comparison to 6.30 mM for spores carrying the GerU* receptor (Fig. 4). In contrast, the V335A mutation essentially prohibits the germinative response to leucine, while also adversely impacting the proline and KBr responses. Analysis of A600 loss upon exposure to glucose reveals that spores carrying the V335A mutation are also subject to an extended lag phase and reduced germinative rate, suggesting that this residue is also involved in the germinative response to glucose (Fig. 5). Spores of strain GC-L5V335A also require increased concentrations of glucose to stimulate half-maximal levels of germination (50% germination at 3.70 mM versus 0.23 mM for GerU* spores). Of the other residues in this region, the M337K mutation is also observed to impact adversely on proline-, leucine-, and KBr-induced germination, while the K334Y substitution is observed to reduce the germinative response to inorganic salts (Table 5).
TABLE 5.
Rates of germination of spores complemented with GerU* receptors with single substitutions to putative OL5 residuesa
| Strain | Rates of spore germination with indicated germinantb
|
||||
|---|---|---|---|---|---|
| Buffer | Glucose | Proline | Leucine | KBr | |
| GC-GerU* | 1 | 99 | 100 | 57 | 100 |
| GC-L5K334Y | 2 | 100 | 100 | 63 | 66 |
| GC-L5V335A | 1 | 97 | 45 (± 6) | 2 | 41 (± 15) |
| GC-L5E336A | 2 | 100 | 1 | 87 (± 8) | 87 (± 12) |
| GC-L5M337K | <1 | 99 | 7 | 20 | 15 |
Spores were germinated in 5 mM Tris-HCl, pH 7.8, for 40 min with 10 mM germinant (50 mM KBr). Spore germination was measured as described in Materials and Methods.
Rates of spore germination are given relative to the A600 loss (65%) for B. megaterium QM B1551 spores in 10 mM glucose, which was set at 100. This value is equivalent to 100% germination after a 40-min incubation. Values are the means of at least duplicate experiments; standard deviation was ≤5% of the mean, except where indicated in parentheses.
FIG. 4.
Effects of l-leucine concentration on spore germination rates of strains GC-GerU* and GC-L5E336A. Spores of strain GC-L5E336A (⧫), in which the glutamate residue at position 336 of GerVB is replaced by alanine, germinate at an increased rate (A) and display a lower minimal germinant requirement for leucine (B) than spores carrying the parental GerU* receptor (⋄). Spores were germinated in 96-well plates in a total volume of 300 μl of 5 mM Tris-HCl, pH 7.8, at 30°C with variable leucine concentrations. Maximum rates of germination were calculated from the slope of the linear segment of A600 loss over time that follows the initial lag phase. Under these experimental conditions, a 60% loss in A600 correlates with 100% spore germination, as determined by loss of heat resistance of strain GC-GerU* after a 60-min incubation in 10 mM glucose. This value was used subsequently to estimate the proportion of germinated spores after incubation for 60 min under the range of leucine concentrations tested. Plotted values are averages of experiments conducted in triplicate (error bars represent standard deviations from the mean).
FIG. 5.
Effect of mutation V335A on the glucose-mediated spore germination response. Spores of strain GC-L5V335A (⧫) display an extended lag phase and slower loss of A600 when incubated in 10 mM glucose (A) than the parental strain GC-GerU* (⋄). The mutant spores also require an increased glucose concentration to stimulate a significant degree of spore germination (50% germination at approximately 3.7 mM) (B). Spores were germinated in 96-well plates in a total volume of 300 μl of 5 mM Tris-HCl, pH 7.8, at 30°C with variable glucose concentrations. Under these experimental conditions, a 60% loss in A600 correlates with 100% spore germination, as determined by loss of heat resistance of strain GC-GerU* after a 60-min incubation in 10 mM glucose. This value was used subsequently to estimate the proportion of germinated spores after incubation for 60 min under the range of glucose concentrations tested. Experiments were conducted in triplicate. In panel B, error bars represent standard deviations from the mean.
In view of the importance of these four consecutive OL5 residues to the functionality of the receptor, particularly in respect to their role in initiating strong proline and KBr germinative responses, we decided to substitute the corresponding residues in the GerUB protein to ascertain whether gain-of-function germinative responses could be observed. Additionally, spores carrying a plasmid-encoded receptor where the B protein comprises a fusion of TM1 to TM8 of GerUB and TM9 and TM10 (and OL5) of GerVB were also prepared. Germinative analysis of these mutant strains (GC-GerUloop5 and GC-GerUVfusion) reveals that while substitution of residues required for the recognition of proline in OL5 results in a slight increase in the response to leucine and KBr (Table 6), germination in response to proline is still weak (10% versus 8% for incubation in buffer alone). However, spores carrying the TM fusion protein display a relatively efficient germinative response to proline (∼39% spore germination) as well as improved germinative rates to the other trigger compounds. The observation that residues spanning TM9 and TM10 of GerVB can confer novel gain of function to a noncognate parental receptor suggests that binding sites for proline and probably the other germinants can be attributed to this region of the protein, while also demonstrating the importance of residues in the TM regions in addition to the previously identified OL5 residues.
TABLE 6.
Rates of germination of spores complemented with GerU receptor variantsa
| Strain | Rates of spore germination with indicated germinantb
|
||||
|---|---|---|---|---|---|
| Buffer | Glucose | Proline | Leucine | KBr | |
| GC-GerU | 3 | 85 | 3 | 76 | 10 |
| GC-GerUloop5 | 8 | 88 | 10 | 95 | 28 |
| GC-GerUVfusion | 14 | 100 | 39 | 92 | 36 |
Spores were germinated in 5 mM Tris-HCl, pH 7.8, for 40 min with 10 mM germinant (50 mM KBr). Spore germination was measured as described in Materials and Methods.
Rates of spore germination are given relative to the A600 loss (65%) for B. megaterium QM B1551 spores in 10 mM glucose, which was set at 100. This value is equivalent to 100% germination after a 40-min incubation. Values are the means of at least duplicate experiments; standard deviation was ≤5% of the mean, except where indicated in parentheses.
Kinetic analysis of spore germination.
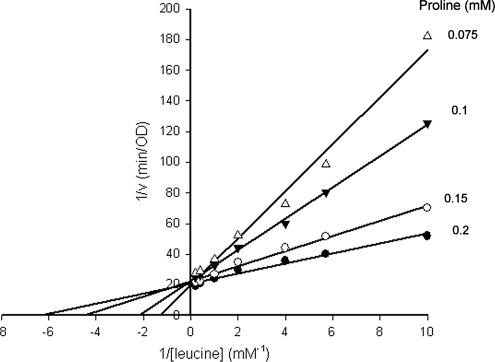
The observation that substitutions to a localized region of the putative OL5 influence, to various degrees, the rate of spore germination with all of the major germinants suggests that the respective ligand binding sites may be in close spatial proximity in the tertiary receptor structure or may perhaps even overlap. We decided to investigate the latter by performing a kinetic study of germination whereby spores were exposed to varying concentrations of leucine in the presence of different fixed concentrations of proline. Analysis of double-reciprocal plots of 1/v versus 1/[leucine] at increasing proline concentrations yields plots that intersect the x axis at different points (Fig. 6), indicating that the affinity of spores for leucine increases upon proline binding. Indeed, from calculated Km values (Fig. 6), the affinity of the spore for leucine is observed to increase by a factor of approximately 2.5 over a similar proline concentration range tested, reflecting cooperativity between the germinants. Apparent germination Vmax values are also observed to increase, albeit modestly, with increasing proline concentrations. Considered together, these data strongly suggest that proline and leucine bind to different sites on the receptor, although the potential for germination to proceed via activation of alternative pathways upon exposure to mixtures of germinants should also be acknowledged, particularly since at least one other functional germinant receptor has been recognized in B. megaterium QM B1551 (7). Kinetic analyses conducted with combinations of other germinants indicate similar cooperativity in stimulating germination (data not shown).
FIG. 6.
Kinetics of GC-GerU* spore germination in the presence of leucine and proline. Germination rates were calculated from the linear segment of OD changes over time to produce Lineweaver-Burk plots of spore germination at variable leucine concentrations (0.1, 0.175, 0.25, 0.5, 1, 2.5, and 5 mM) and different fixed proline concentrations (0.075, 0.1, 0.15 and 0.2 mM). Apparent Km values ranged from 210 μM to 527 μM in the presence of concentrations of proline that varied from high to low, respectively. Vmax values ranged from 0.052 to 0.041 OD/min for concentrations of proline that varied from high to low, respectively.
DISCUSSION
Data presented in this communication represent the first extensive application of SDM to study the function of the Bacillus spore germinant receptor. The construction of a series of strains carrying mutagenized receptor B protein variants that display conditional germinative responses (where initiation of germination is observed to at least one known germinant but not to others) substantiates the hypothesis that this component of the receptor determines the range of compounds to which the spore germinates.
While virtually all membrane protein helix prediction programs tested predicted GerVB to comprise 10 TM domains with N and C termini localized to the protoplast side of the membrane, as is characteristic for germinant receptor B proteins (17), agreement on residues that mark the boundaries between helices and connecting loops was not observed. However, the HMMTOP program appeared to provide a reasonable consensus on helix-loop boundaries and was therefore employed in this study to define loop regions for SDM experiments. The subsequent observation that, of the residues tested, only modified outer loops appear to influence the range of compounds that initiate germination suggests that the general model of topology predicted by HMMTOP is probably correct, since one would expect residues involved in receptor sensing of germinants to be on the exterior aspect of the spore inner membrane.
The rationale behind the SDM approach undertaken in this study—substitution of residues for those in the corresponding position of a close homologue—is perhaps vindicated by the observation that in all cases a significant degree of functionality is maintained by the modified receptors. However, while a number of residues have been identified as influencing to various degrees the germinative response to specific trigger compounds, the key issue concerns discriminating between, on the one hand, residues that participate directly in ligand binding and are therefore components of the germinant binding site and, on the other hand, those that are indirectly involved by influencing the ability to accommodate or mediate access of the germinant to the binding site.
In the absence of tertiary structural information or complementary biochemical data, it is difficult to assign residues to either of these categories. However, we can state with relative confidence that functionally important residues reside in both the outer loop and TM domains. This is best exemplified by the observation that TM domains associated with OL5 (TM9 and TM10 of GerVB), which includes key residues for proline recognition, are required in addition to the outer loop residues to confer novel gain of function to a noncognate parental receptor. Similarly, spores of strain GC-OL, in which all five outer loops have been modified to match those of GerUB, a cognate receptor for glucose and leucine, germinate only in response to glucose. Thus, modification of putative binding sites of the outer loop region alone appears to be insufficient for efficient initiation of germination since additional TM-associated residues are required to mediate presumed conformational changes that trigger the cascade of germination reactions.
Potential candidates for sites of ligand binding include the cluster of four nonconserved residues (K334 through M337) predicted to reside in OL5 of GerVB, immediately adjacent to TM10, since substitutions to this region, both collective and individual, were observed to influence to various degrees the germinative response to all four germinants. The construction of mutant receptors with differing affinities for germinants perhaps indicates that the various substitutions affect the size, shape, and charge status of ligand binding sites. The E336A mutation, for example, results in the complete loss of proline recognition with a concomitant enhancement of the leucine response. Perhaps, then, the glutamate side chain carboxyl group participates in the binding of proline, while the alanine residue is more favorable for hydrophobic interactions with the leucine side chain. Thus, amino acids in this region may participate in a binding pocket that encompasses binding sites for all germinants, rendering them close in spatial proximity in the tertiary structure of the protein. This is certainly consistent with the cooperativity observed upon kinetic analysis of the spore response to combinations of germinants, which indicates a sequential binding mechanism such that binding of one germinant induces a conformational change that increases the affinity of the receptor for a second germinant. Synergy upon germinant binding has been observed in spores of other Bacillus species (1, 3, 4) although this typically involves interaction between two or more cognate germinant receptors.
Information regarding nearby binding sites for substrates and ions in prokaryotic secondary amino acid transporters, thought to ensure close cooperativity during transport, is extremely well established (18), and it is tempting to draw parallels between the two systems. Although speculative, such inferences may be appropriate since receptor B proteins show distant homology to these transporters, leading to their classification as a subgroup of the amino acid-polyamine-organocation superfamily (17). If the molecular basis of transport in prokaryotic amino acid transporters does represent a functional precedent for germinant-receptor B proteins, then it seems likely that residues that participate directly in germinant binding are also to be found in TM domains, since this is where ligand binding sites in a number of amino acid transporters are often located (24, 25, 36). The observation that residues spanning TM9 and TM10 in GerVB confer proline recognition to the GerUVfusion protein while OL5 residues alone do not would appear to substantiate this hypothesis. Previously described B. subtilis mutants that carry point mutations in putative TM regions of the GerAB structural gene, necessitating higher concentrations of alanine to initiate germination, further support this idea (28).
Further SDM studies should reveal the identity of residues predicted to lie in TM domains that may participate in germinant binding, perhaps as part of a ligand binding pocket comprising both loop and TM residues. Mutagenesis-led studies may also permit the as yet undefined role of the receptor A and C proteins in germinant recognition to be assessed. Integration of these analyses with biochemical and structural studies—assuming protein expression and purification problems can be surmounted—should ultimately increase our understanding of the molecular basis for spore germinant receptor function.
Acknowledgments
We thank Didier Lereclus (INRA/La Miniere, France) for the gift of plasmid pHT315.
Work in our laboratory is funded by grants awarded to C.R. Lowe by the BBSRC and the Home Office (CBRN).
Footnotes
Published ahead of print on 18 January 2008.
REFERENCES
- 1.Akoachere, M., R. C. Squires, A. M. Nour, L. Angelov, J. Brojatsch, and E. Abel-Santos. 2007. Identification of an in vivo inhibitor of Bacillus anthracis spore germination. J. Biol. Chem. 28212112-12118. [DOI] [PubMed] [Google Scholar]
- 2.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108115-119. [DOI] [PubMed] [Google Scholar]
- 3.Atluri, S., K. Ragkousi, D. E. Cortezzo, and P. Setlow. 2006. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J. Bacteriol. 18828-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlass, P. J., C. W. Houston, M. O. Clements, and A. Moir. 2002. Germination of Bacillus cereus spores in response to l-alanine and to inosine: the roles of gerL and gerQ operons. Microbiology 1482089-2095. [DOI] [PubMed] [Google Scholar]
- 5.Cabrera-Martinez, R. M., F. Tovar-Rojo, V. R. Vepachedu, and P. Setlow. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J. Bacteriol. 1852457-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chirakkal, H., M. O'Rourke, A. Atrih, S. J. Foster, and A. Moir. 2002. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology 1482383-2392. [DOI] [PubMed] [Google Scholar]
- 7.Christie, G., and C. R. Lowe. 2007. Role of chromosomal and plasmid-borne receptor homologues in the response of Bacillus megaterium QM B1551 spores to germinants. J. Bacteriol. 1894375-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clements, M. O., and A. Moir. 1998. Role of the gerI operon of Bacillus cereus 569 in the response of spores to germinants. J. Bacteriol. 1806729-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corfe, B. M., R. L. Sammons, D. A. Smith, and C. Mauel. 1994. The gerB region of the Bacillus subtilis 168 chromosome encodes a homologue of the gerA spore germination operon. Microbiology 140471-478. [DOI] [PubMed] [Google Scholar]
- 10.English, J. D., and P. S. Vary. 1986. Isolation of recombination-defective and UV-sensitive mutants of Bacillus megaterium. J. Bacteriol. 165155-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster, S. J., and K. Johnstone. 1987. Purification and properties of a germination-specific cortex-lytic enzyme from spores of Bacillus megaterium KM. Biochem. J. 242573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hudson, K. D., B. M. Corfe, E. H. Kemp, I. M. Feavers, P. J. Coote, and A. Moir. 2001. Localization of GerAA and GerAC germination proteins in the Bacillus subtilis spore. J. Bacteriol. 1834317-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Igarashi, T., and P. Setlow. 2005. Interaction between individual protein components of the GerA and GerB nutrient receptors that trigger germination of Bacillus subtilis spores. J. Bacteriol. 1872513-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ireland, J. A., and P. C. Hanna. 2002. Amino acid- and purine ribonucleoside-induced germination of Bacillus anthracis DeltaSterne endospores: gerS mediates responses to aromatic ring structures. J. Bacteriol. 1841296-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irie, R., Y. Fujita, and M. Kobayashi. 1996. Nucleotide sequence and gene organisation of the gerK spore germination locus of Bacillus subtilis 168. J. Gen. Appl. Microbiol. 42141-153. [Google Scholar]
- 16.Ishikawa, S., K. Yamane, and J. Sekiguchi. 1998. Regulation and characterization of a newly deduced cell wall hydrolase gene (cwlJ) which affects germination of Bacillus subtilis spores. J. Bacteriol. 1801375-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jack, D. L., I. T. Paulsen, and M. H. Saier. 2000. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 1461797-1814. [DOI] [PubMed] [Google Scholar]
- 18.Jung, H., T. Pirch, and D. Hilger. 2006. Secondary transport of amino acids in prokaryotes. J. Membr. Biol. 213119-133. [DOI] [PubMed] [Google Scholar]
- 19.McCool, G. J., and M. C. Cannon. 2001. PhaC and PhaR are required for polyhydroxyalkanoic acid synthase activity in Bacillus megaterium. J. Bacteriol. 1834235-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moir, A., B. M. Corfe, and J. Behravan. 2002. Spore germination. Cell Mol. Life Sci. 59403-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriyama, R., A. Hattori, S. Miyata, S. Kudoh, and S. Makino. 1996. A gene (sleB) encoding a spore cortex-lytic enzyme from Bacillus subtilis and response of the enzyme to l-alanine-mediated germination. J. Bacteriol. 1786059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paidhungat, M., and P. Setlow. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J. Bacteriol. 1833982-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paidhungat, M., and P. Setlow. 2000. Role of Ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 1822513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pirch, T., S. Landmeier, and H. Jung. 2003. Transmembrane domain II of the Na+/proline transporter PutP of Escherichia coli forms part of a conformationally flexible, cytoplasmic exposed aqueous cavity within the membrane. J. Biol. Chem. 27842942-42949. [DOI] [PubMed] [Google Scholar]
- 25.Pirch, T., M. Quick, M. Nietschke, M. Langkamp, and H. Jung. 2002. Sites important for Na+ and substrate binding in the Na+/proline transporter of Escherichia coli, a member of the Na+/solute symporter family. J. Biol. Chem. 2778790-8796. [DOI] [PubMed] [Google Scholar]
- 26.Plomp, M., T. J. Leighton, K. E. Wheeler, H. D. Hill, and A. J. Malkin. 2007. In vitro high-resolution structural dynamics of single germinating bacterial spores. Proc. Natl. Acad. Sci. USA 1049644-9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rode, L. J., and J. W. Foster. 1962. Ionic germination of spores of Bacillus megaterium QM B1551. Arch. Mikrobiol. 43183-200. [DOI] [PubMed] [Google Scholar]
- 28.Sammons, R. L., A. Moir, and D. A. Smith. 1981. Isolation and properties of spore germination mutants of Bacillus subtilis 168 deficient in the initiation of germination. J. Gen. Microbiol. 124229-241. [Google Scholar]
- 29.Santo, L. Y., and R. H. Doi. 1974. Ultrastructural analysis during germination and outgrowth of Bacillus subtilis spores. J. Bacteriol. 120475-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6550-556. [DOI] [PubMed] [Google Scholar]
- 31.Thackray, P. D., J. Behravan, T. W. Southworth, and A. Moir. 2001. GerN, an antiporter homologue important in germination of Bacillus cereus endospores. J. Bacteriol. 183476-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17849-850. [DOI] [PubMed] [Google Scholar]
- 33.Vary, P. S., and Y. P. Tao. 1988. Development of genetic methods in Bacillus megaterium, p. 403-407. In A. T. Ganesan and J. A. Hoch (ed.), Genetics and biotechnology of bacilli, vol. 2. Academic Press, New York, NY. [Google Scholar]
- 34.Vepachedu, V. R., and P. Setlow. 2007. Analysis of interactions between nutrient germinant receptors and SpoVA proteins of Bacillus subtilis spores. FEMS Microbiol. Lett. 27442-47. [DOI] [PubMed] [Google Scholar]
- 35.Vepachedu, V. R., and P. Setlow. 2007. Role of SpoVA proteins in release of dipicolinic acid during germination of Bacillus subtilis spores triggered by dodecylamine or lysozyme. J. Bacteriol. 1891565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamashita, A., S. K. Singh, T. Kawate, Y. Jin, and E. Gouaux. 2005. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437215-223. [DOI] [PubMed] [Google Scholar]
- 37.Zuberi, A. R., A. Moir, and I. M. Feavers. 1987. The nucleotide sequence and gene organization of the gerA spore germination operon of Bacillus subtilis 168. Gene 511-11. [DOI] [PubMed] [Google Scholar]