Abstract
Polyamines (putrescine, spermidine, and spermine) are major organic polycations essential for a wide spectrum of cellular processes. The cells require mechanisms to maintain homeostasis of intracellular polyamines to prevent otherwise severe adverse effects. We performed a detailed transcriptome profile analysis of Pseudomonas aeruginosa in response to agmatine and putrescine with an emphasis in polyamine catabolism. Agmatine serves as the precursor compound for putrescine (and hence spermidine and spermine), which was proposed to convert into 4-aminobutyrate (GABA) and succinate before entering the tricarboxylic acid cycle in support of cell growth, as the sole source of carbon and nitrogen. Two acetylpolyamine amidohydrolases, AphA and AphB, were found to be involved in the conversion of agmatine into putrescine. Enzymatic products of AphA were confirmed by mass spectrometry analysis. Interestingly, the alanine-pyruvate cycle was shown to be indispensable for polyamine utilization. The newly identified dadRAX locus encoding the regulator alanine transaminase and racemase coupled with SpuC, the major putrescine-pyruvate transaminase, were key components to maintaining alanine homeostasis. Corresponding mutant strains were severely hampered in polyamine utilization. On the other hand, an alternative γ-glutamylation pathway for the conversion of putrescine into GABA is present in some organisms. Subsequently, GabD, GabT, and PA5313 were identified for GABA utilization. The growth defect of the PA5313 gabT double mutant in GABA suggested the importance of these two transaminases. The succinic-semialdehyde dehydrogenase activity of GabD and its induction by GABA were also demonstrated in vitro. Polyamine utilization in general was proven to be independent of the PhoPQ two-component system, even though a modest induction of this operon was induced by polyamines. Multiple potent catabolic pathways, as depicted in this study, could serve pivotal roles in the control of intracellular polyamine levels.
Agmatine, a cationic compound derived from arginine decarboxylation, serves as the precursor of three major polyamines, putrescine, spermidine, and spermine. These polyamines are the major organic polycations found in all living cells. Polyamines have pleiotropic effects on several cellular processes. In more complex organisms, these compounds are required for cell proliferation and differentiation (2). In Escherichia coli, these polycations play significant roles in the structural and functional organization of the chromosome (35). They are implicated in RNA synthesis through the stimulation of the activity of RNA polymerase and in protein synthesis through the stabilization of ribosomal structure and modulation of translational fidelity (9). In addition, polyamines are involved in the induction of recA in E. coli in response to UV or γ irradiation (14). Polyamines are thought to protect DNA from oxidative damage by serving as free radical scavengers (7, 13). Some microorganisms also use polyamines for the synthesis of secondary metabolites (35). Recently, we also reported that exogenous polyamines exert a significant effect on antibiotic susceptibility in bacteria (16-18). While intracellular polyamines play pivotal roles in ensuring optimal growth, their accumulation inhibits protein synthesis and decreases cell viability in the stationary phase and at low temperatures (5, 19). Thus, a balance between polyamine synthesis and catabolism appears to be required to adjust the optimal concentration of these compounds in accordance with the growth environment.
Synthesis of spermidine and spermine requires the addition of an aminopropyl group from decarboxylated S-adenosylmethionine (dSAM) to putrescine and spermidine, respectively. These reactions are catalyzed by SAM decarboxylase (speD) for the synthesis of the aminopropyl donor and by spermidine synthase (speE) for transfer of the aminopropyl group from dSAM to putrescine. Although it is not available in bacteria, a similar reaction catalyzed by spermine synthase is responsible for spermine synthesis in eukaryotes.
Putrescine biosynthesis can occur by three routes (33): route 1, from ornithine by ornithine decarboxylase (speC); route 2, from arginine and agmatine, through the actions of arginine decarboxylase (speA) and agmatine ureohydrolase (speB); and route 3, from arginine, agmatine, and N-carbamoylputrescine (N-CP) via arginine decarboxylase, agmatine deiminase (aguA), and N-carbamoylputrescine amidohydrolase (aguB). While it is assumed that E. coli and many other microorganisms utilize both routes 1 and 2 (35), both routes 1 and 3 are used by Pseudomonas aeruginosa (24).
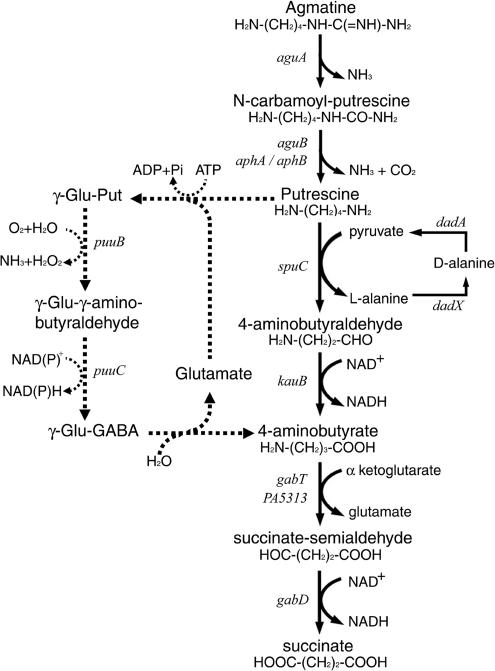
Agmatine and putrescine also serve as good sources of carbon and nitrogen for P. aeruginosa. As shown in Fig. 1, exogenous agmatine was first converted into putrescine by AguA and AguB, and the carbon skeleton of putrescine was preserved as succinate via two sets of transamination and dehydrogenation reactions before being channeled into the tricarboxylic acid (TCA) cycle. The aguBA operon is induced by agmatine and controlled by the AguR repressor (25). While the aguA mutant was unable to grow on agmatine as the sole source of carbon and nitrogen, a leaky growth phenotype of the aguB mutant on agmatine suggested an alternative route yet to be identified.
FIG. 1.
Catabolic pathways of agmatine and putrescine in P. aeruginosa PAO1. Dashed arrows indicate the proposed γ-glutamylation pathway for putrescine catabolism in E. coli.
SpuC, a putrescine-pyruvate aminotransferase that converts putrescine to 4-aminobutyraldehyde, is essential for putrescine utilization in P. aeruginosa (20). The subsequent dehydrogenation of 4-aminobutyraldehyde to 4-aminobutyrate (GABA) is catalyzed by a bifunctional enzyme, KauB, possessing both 4-guanidinobutyraldehyde dehydrogenase and 4-aminobutyraldehyde dehydrogenase activities (Fig. 1). The KauB activity is induced by putrescine or by agmatine (11). Conversion of GABA into succinate was proposed to require the function of two successive enzymes, succinic semialdehyde transaminase (GabT) and succinic semialdehyde dehydrogenase (GabD), respectively; however, no molecular information regarding the encoding genes gabDT has been reported for P. aeruginosa.
Putrescine catabolism involving γ-glutamylated intermediates (Fig. 1) in the conversion of putrescine into GABA was recently reported for E. coli (15). The putrescine utilization (puu) genes of this novel pathway were clustered in a single locus of several transcriptional units in E. coli (15). P. aeruginosa also possesses several possible homologues of the Puu proteins scattered on the chromosome (www.pseudomonas.com), but it was not clear whether these proteins participate in polyamine utilization. To exploit the catabolic capacity of P. aeruginosa, we conducted transcriptome analysis to identify and characterize genes that are differentially induced by exogenous agmatine and putrescine.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown with Luria-Bertani (LB) medium supplemented with ampicillin, 50 μg/ml; tetracycline, 12.5 μg/ml; gentamicin, 10 μg/ml; chloramphenicol, 35 μg/ml; kanamycin, 50 μg/ml; and 5-bromo-3-indolyl-β-d-galactoside (X-Gal) at 0.03% (wt/vol); and the P. aeruginosa strain was grown with LB medium supplemented with carbenicillin, 200 μg/ml; streptomycin, 300 μg/ml; tetracycline, 100 μg/ml; and gentamicin, 50 μg/ml. The minimal medium P (8) containing the indicated carbon and nitrogen sources at 20 mM was used for the growth of P. aeruginosa.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or descriptiona | Source |
|---|---|---|
| E.coli strains | ||
| DH5α | F− φ80dlacΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK−) supE44 λ−thi-1 gyrA96 relA | Bethesda Research Laboratories |
| Rosetta (DE3)pLysS | F−ompT hsdSB (rB− mB−) gal dcm (DE3)pLysSRARE (Camr) | EMD Bioscience |
| SM10 | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu (Kmr) | 6 |
| P. aeruginosa strains | ||
| PAO1 | Wild type | 8 |
| PAO1-Sm | Spontaneous Smr mutant strain of PAO1 | 26 |
| PAO5001 | aguA::Tcr | 25 |
| PAO5002 | aguB::Tcr | 25 |
| PAO5003 | aguR::Tcr | 25 |
| PAO5701 | gabD::Tcr | This study |
| PAO5702 | gabD::Tcr/PA5313::Gmr | This study |
| PAO5703 | gabT::Tcr | This study |
| PAO5704 | gabT::Tcr/PA5313::Gmr | This study |
| PAO5008 | spuC::Tcr | 20 |
| PAO5706 | spuC::Tcr/PA5313::Gmr | This study |
| PAO5707 | kauB::Gmr | This study |
| PAO5708 | PA5313::Gmr | This study |
| PAO5709 | PA5314::Gmr | This study |
| PAO5710 | dadA::Tcr | This study |
| PAO5711 | dadX::Gmr | This study |
| PAO5712 | PA5303::Gmr | This study |
| PAO5713 | dadR::Gmr | This study |
| PAO5714 | phoP::Gmr | This study |
| PAO5715 | phoQ::Gmr | This study |
| Plasmids | ||
| pRTP1 | Ampr Sms conjugation vector | 34 |
| pUCP18 | Escherichia-Pseudomonas shuttle vector (Ampr) | 32 |
| pGMΩ1 | Gentamicin cassette with omega loop on both ends (Ampr) | 31 |
| pHC5302 | pUCP18 derivative carrying the dadX gene | This study |
| pHC5303 | pUCP18 derivative carrying the PA5303 gene | This study |
| pRSET | pUC-derived expression vectors; T7 promoter N-terminal six-His tag (Ampr) | Invitrogen Life Technologies |
| pHE1409 | pRSET derivative for six-His-tagged AphA protein expression (Ampr) | This study |
| pQF50 | bla lacZ transcriptional fusion vector (Ampr) | 3 |
| pQF52 | bla lacZ translational fusion vector derived from pQF50 (Ampr) | 26 |
| pHT0322 | aphB::lacZ transcriptional fusion of pQF50 | This study |
| pHT1409 | aphA::lacZ transcriptional fusion of pQF50 | This study |
| pHT1178 | oprH::lacZ translational fusion of pQF52 | This study |
| pHT5312 | kauB::lacZ transcriptional fusion of pQF50 | This study |
| pHT5313 | PA5313::lacZ transcriptional fusion of pQF50 | This study |
| pHT0265 | gabD::lacZ translational fusion of pQF52 | This study |
| pGU102 | spuA::lacZ translational fusion of pQF52 | 20 |
| pZY6 | dadA::lacZ translational fusion of pQF52 | 36 |
Camr, chloramphenicol resistance; Kmr, kanamycin resistance; Smr, streptomycin resistance; Sms, streptomycin sensitive; Tcr, tetracycline resistance; Gmr, gentamicin resistance; Ampr, ampicillin resistance.
RNA preparation and DNA microarray analyses.
Two independent sets of P. aeruginosa PAO1 cultures were grown aerobically in minimal medium P, with 350 rpm shaking at 37°C, in the presence of l-glutamate alone or with the addition of putrescine, agmatine, or GABA at 20 mM. Cells were harvested when the optical density at 600 nm reached 0.5 to 0.6 by centrifugation for 5 min at 4°C. Total RNA samples were isolated by using an RNeasy purification kit following the instructions of the manufacturer (Qiagen). Reverse transcription for cDNA synthesis, fragmentation by DNase I treatment, and cDNA probe labeling and hybridization were performed according to the instructions of GeneChip (Affymetrix). Data were processed by Microarray Suite 5.0 software, normalizing the absolute expression signal values of all chips to a target intensity of 500. GeneSpring software (Silicon Genetics) was used for the expression pattern analysis and comparison. Only genes showing consistent expression profiles in duplicates were selected for further analysis.
Construction of lacZ fusions.
For the construction of lacZ fusions, the regulatory regions of interest were amplified by PCR with specific primers (see Table S1 in the supplemental material) from the genomic DNA of P. aeruginosa PAO1. After restriction digestion of the purified PCR products, these DNA fragments were cloned into the corresponding restriction sites of pQF50 or pQF52 (Table 1) before they were transformed into E. coli DH5α. The positive clones were selected on LB plates containing ampicillin and X-Gal. The nucleotide sequence of the inserts was confirmed by DNA sequencing.
Construction of mutant strains.
For knockout mutants created by insertion, DNA fragments covering the genes of interest were PCR amplified (see Table S2 in the supplemental material) from PAO1 genomic DNA and cloned into the conjugation vector pRTP1 (34). The tetracycline resistance cassette was introduced by using an EZ-Tn5 (TET-1) insertion system (Epicenter). The mutation sites were mapped by restriction endonuclease digestion and subsequently by nucleotide sequencing with a transposon-specific flanking primer. For deletion mutants, two flanking regions of a targeted gene were amplified by PCR with specific primers (see Table S2 in the supplemental material). The PCR products were purified after agarose gel electrophoresis and applied to spin columns (Qiagen). Following restriction digestions, the DNA fragments were cloned into pRTP1. A cassette carrying the gentamicin resistance gene from pGMΩ1 was purified after restriction digestion and inserted into the conjunction of the two DNA fragments, which was introduced by primer sequence designs. For gene replacement, E. coli SM10 served as the donor in biparental mating with PAO1-Sm (6). The desired knockout mutants were selected on LB plates containing streptomycin and either gentamicin or tetracycline, and the mutation was confirmed by PCR.
Overexpression and purification of the recombinant AphA.
Recombinant His-AphA was expressed in E. coli Rosetta (DE3)(pLysS), following instructions of a pRSET expression system (Invitrogen Life Technologies). Logarithmically growing cells were obtained from a single-colony inoculum consisting of LB medium containing ampicillin and chloramphenicol at 20°C with shaking. The induction for protein expression was triggered by adding 1 mM of isopropylthiogalactopyranoside into the medium when the optical density at 600 nm reached 0.5. Recombinant cells were harvested after 16 h of induction by centrifugation at 4°C. The cell pellet was suspended in a 5× volume of phosphate buffer (20 mM sodium phosphate, 500 mM NaCl, 10 mM imidazole [pH 7.4]) supplemented with EDTA-free protease inhibitor cocktail (Roche). Cell extract was obtained by passage through a French pressure cell at 8,500 lb/in2, and cell debris was removed by centrifugation at 25,000 × g for 30 min at 4°C. The soluble His-tagged AphA protein was purified over nickel-nitrilotriacetic acid agarose resin in batch mode for native proteins, following the instructions of the manufacturer (Qiagen). The protein fraction eluted by 150 mM imidazole in phosphate buffer was subjected to anion exchange chromatography using a Mono Q FF 5/5 column (Pharmacia). Before the protein sample application, the column was equilibrated after a blank run with 20 mM Tris-HCl (pH 7.4). A linear gradient of 0 to 1 M KCl over 20 column volumes was used for protein elution. Active fractions detected by UV and sodium dodecyl sulfate-polyacrylamide gel electrophoresis were pooled together and concentrated by using an Amicon Ultra-15 centrifugal filter unit (Millipore).
Measurements of enzyme activities.
The cells were grown in minimal medium P containing the carbon and nitrogen sources as indicated above. Cells in the mid-log phase were harvested by centrifugation and then passed through a French press cell at 8,500 lb/in2 The cell debris were removed by centrifugation at 20,000 × g for 10 min at 4°C, and protein concentrations in the crude extracts were determined by the Bradford method (1) using bovine serum albumin as the standard.
The activity of β-galactosidase was measured at 37°C, using o-nitrophenyl-β-galactopyranoside as the reaction substrate, and the formation of o-nitrophenol was determined by spectrophotometry at 420 nm.
The succinic-semialdehyde dehydrogenase activity was assayed as described previously (29). The cell pellet was washed in 100 mM sodium phosphate buffer (pH 7.0) containing 9% glycerol, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride. The reaction was carried out at 30°C in a mixture containing 100 mM HEPES (pH 8.0), 1.5 mM succinic semialdehyde, 0.35 mM NADP, and 5 mM 2-mercaptoethanol. The reaction was initiated by the addition of crude extract, and the activity of succinic-semialdehyde dehydrogenase was determined by monitoring the generation of NADPH at 340 nm.
To demonstrate the acetylpolyamine amidohydrolase activity of AphA, acetylputrescine was used as the substrate in the reaction mixture. Purified recombinant AphA protein was further activated by incubation with 5 mM MnCl2 or CoCl2 on ice for 2 h preceding the enzyme assay. The composition of the reaction mixture was as follows (0.5 ml total volume): 20 μg of purified enzyme, 20 mM acetylputrescine, l00 mM Tris-HCl (pH 7.8), 8 mM MgSO4, and 60 mM KCl. After 16 h at 37°C, the reaction was stopped by boiling the samples for 10 min. Denatured proteins were precipitated and removed by centrifugation at 25,000 × g for 30 min at 4°C. The supernatant containing small molecules was submitted for mass spectrometry analysis to the core facilities of Georgia State University. Samples were diluted 10 times (vol/vol) in 50% methanol in water with the addition of 0.1% formic acid for positive ion analysis by electrospray ionization quadrupole-time-of-flight mass spectrometry.
Microarray data accession numbers.
The raw data from GeneChip experiments have been deposited in the Gene Expression Omnibus (GEO) database at NCBI with the accession number GSE9926.
RESULTS
DNA microarrays were employed to analyze the transcriptional profiles of P. aeruginosa PAO1 in response to agmatine and putrescine, with an emphasis on possible catabolic routes. As shown in Table 2, cells were grown in glutamate minimal medium in the absence and presence of agmatine or putrescine, and genes that were induced by either of these two compounds were identified based on the criteria described in Materials and Methods. We have reported characterization of some of these genes in the following aspects: aguBA for agmatine catabolism (25), spuA-I for polyamine utilization and uptake (20), and oprH-phoPQ as well as PA3552 to PA3559 for polyamine-dependent antibiotic susceptibility (18). The following sections would therefore focus mainly on other genes that participate in the proposed catabolic pathway(s) of agmatine and putrescine (Fig. 1).
TABLE 2.
Genes induced by agmatine and putrescine in P. aeruginosa PAO1
| Straina | Gene name | Cell growth in medium supplemented with (absolute signal value)b:
|
Fold change in expression with the addition ofc:
|
Description | |||
|---|---|---|---|---|---|---|---|
| Glu | Glu plus Agm | Glu plus Put | Agm | Put | |||
| PA0265d | gabD | 448 | 6,310 | 6,923 | 14.4 | 15.8 | Succinate-semialdehyde dehydrogenase |
| PA0266d | gabT | 721 | 9,263 | 9,244 | 12.9 | 12.8 | 4-Aminobutyrate aminotransferase |
| PA0292 | aguA | 705 | 10,862 | 681 | 15.4 | 1.0 | Agmatine deiminase |
| PA0293 | aguB | 74 | 8,564 | 94 | 115.5 | 1.3 | N-CP amidohydrolase |
| PA0295 | 203 | 743 | 522 | 3.6 | 2.5 | Periplasmic polyamine binding protein | |
| PA0296 | spuI | 1,113 | 8,354 | 7,613 | 7.4 | 6.8 | PA2040 homolog, glutamine synthetase |
| PA0297 | spuA | 275 | 2,451 | 1,663 | 10.3 | 6.8 | Glutamine amidotransferase |
| PA0298 | spuB | 582 | 3,931 | 2,625 | 6.8 | 4.5 | Glutamine synthetase |
| PA0299 | spuC | 1,203 | 7,953 | 6,179 | 6.7 | 5.1 | Putrescine-pyruvate aminotransferase, patase |
| PA0321 | aphB | 37 | 241 | 16 | 7.1 | 0.5 | Acetylpolyamine aminohydrolase |
| PA0322 | 55 | 253 | 40 | 4.9 | 0.9 | Small molecule transporter | |
| PA0534 | 26 | 525 | 683 | 28.4 | 28.7 | Putative d-amino acid oxidases (deaminating) | |
| PA0535 | 77 | 338 | 447 | 4.8 | 5.1 | Probable transcriptional regulator | |
| PA1178d | oprH | 1,863 | 14,493 | 10,917 | 9.4 | 6.7 | Outer membrane porin oprh |
| PA1179d | phoP | 646 | 2,471 | 1,680 | 3.7 | 2.5 | Two-component response regulator phop |
| PA1180d | phoQ | 455 | 1,262 | 966 | 2.7 | 2.1 | Two-component sensor kinase phoq |
| PA1409 | aphA | 69 | 1,513 | 32 | 22.1 | 0.6 | Acetylpolyamine aminohydrolase |
| PA1410 | 99 | 1,563 | 89 | 16.8 | 0.9 | Periplasmic spermidine/putrescine-binding protein | |
| PA1540 | 38 | 271 | 213 | 7.3 | 5.6 | Conserved hypothetical protein | |
| PA1541 | 58 | 667 | 406 | 54.7 | 32.7 | Drug efflux transporter | |
| PA1742 | 366 | 2,038 | 2,335 | 5.8 | 6.5 | Amidotransferase | |
| PA2041 | 107 | 1,629 | 1,245 | 15.5 | 11.9 | Amino acid permease | |
| PA2268 | 60 | 292 | 647 | 4.9 | 10.8 | Hypothetical protein | |
| PA2776 | 194 | 3,655 | 3,674 | 18.5 | 18.6 | Conserved hypothetical protein | |
| PA3355 | 96 | 289 | 271 | 3.1 | 2.9 | Hypothetical protein | |
| PA3356 | 591 | 3,301 | 3,232 | 5.6 | 5.5 | Glutamine synthetase | |
| PA3552d | pmrH | 299 | 1,870 | 794 | 6.2 | 2.5 | Putative enzyme for lipopoly saccharide modification |
| PA3553d | pmrF | 138 | 924 | 362 | 6.3 | 2.7 | Putative glycosyl transferase |
| PA3554d | pmrI | 288 | 1,154 | 626 | 4.0 | 2.3 | Putative formyl transferase |
| PA3555d | pmrJ | 77 | 194 | 118 | 2.8 | 1.5 | Putative deacetylase |
| PA3556d | pmrK, arnT | 189 | 672 | 324 | 7.1 | 2.9 | Inner membrane l-Ara4N transferase arnt |
| PA3557d | pmrL | 60 | 265 | 144 | 4.5 | 2.4 | Putative membrane protein |
| PA3558d | pmrM | 140 | 597 | 303 | 4.5 | 2.3 | Putative permease |
| PA3559d | ugd | 117 | 540 | 264 | 4.6 | 2.2 | UDP-glucose dehydrogenase |
| PA3766 | 91 | 978 | 985 | 12.5 | 11.7 | Aromatic amino acid transporter | |
| PA4358d | feoB | 91 | 462 | 315 | 5.3 | 3.5 | Ferrous iron transport protein B |
| PA4359d | feoA | 96 | 381 | 218 | 3.9 | 2.3 | Ferrous iron transport protein A |
| PA5302 | dadX | 40 | 2,177 | 2,770 | 65.2 | 91.7 | Alanine racemase |
| PA5303 | 105 | 3,657 | 3,934 | 36.7 | 42.6 | Putative endoribonuclease | |
| PA5304 | dadA | 165 | 5,126 | 5,682 | 34.6 | 43.5 | d-Alanine dehydrogenase |
| PA5308 | dadR | 552 | 635 | 501 | 1.2 | 0.9 | Transcriptional regulator of the AsnC family |
| PA5309 | 398 | 1,369 | 1,363 | 3.4 | 3.4 | Oxidoreductase | |
| PA5312 | kauB | 922 | 7,906 | 7,744 | 8.5 | 8.3 | NAD-dependent aldehyde dehydrogenase |
| PA5313 | 144 | 2,847 | 4,226 | 19.9 | 29.4 | Transaminase | |
| PA5314 | 91 | 1,834 | 2,298 | 19.4 | 24.1 | Hypothetical protein of unknown function | |
| PA5521 | 292 | 873 | 910 | 3.1 | 3.2 | Short-chain dehydrogenase | |
| PA5522 | 116 | 585 | 591 | 4.7 | 5.2 | Glutamine synthetase | |
| PA5523 | 122 | 832 | 894 | 6.7 | 7.2 | Aminotransferase | |
PA gene numbers are annotated according to the Pseudomonas Genome Project (www.pseudomonas.com).
GeneChip raw data are mean values from two independent sets of cultures. Cells were grown in minimal medium P supplemented with 20 mM of the following supplements as indicated: Glu, glutamate; Agm, agmatine; Put, putrescine.
Expression change (n-fold) in cultures with the addition of agmatine or putrescine.
Genes proposed to be regulated by magnesium limitation and by the oprH-phoPQ operon, according to McPhee at al. (22).
The agmatine-specific module.
With few exceptions, the list of genes induced by the presence of agmatine overlaps completely with the list of putrescine-responsive genes. Only six genes in three possible transcriptional units were considered agmatine specific: the aguBA operon and the two putative PA1409-PA1410 and PA0322-PA0321 operons (Table 2 and Fig. 1). We have reported characterization of the aguBA operon and its agmatine-specific regulation by AguR (25). The other two operons were quite similar in their genetic contents; PA1409 and PA0321 encode two putative acetylpolyamine amidohydrolases, while PA1410 and PA0322 are open reading frames for a probable periplasmic binding protein and a transporter, respectively. PA1409 and PA0321 were designated acetylpolyamine amidohydrolase A(aphA) and aphB based on the demonstrated enzymatic activity (see description below) and the 69% sequence identity between the encoded polypeptides.
Induction of aphA and aphB by exogenous agmatine and acetylputrescine.
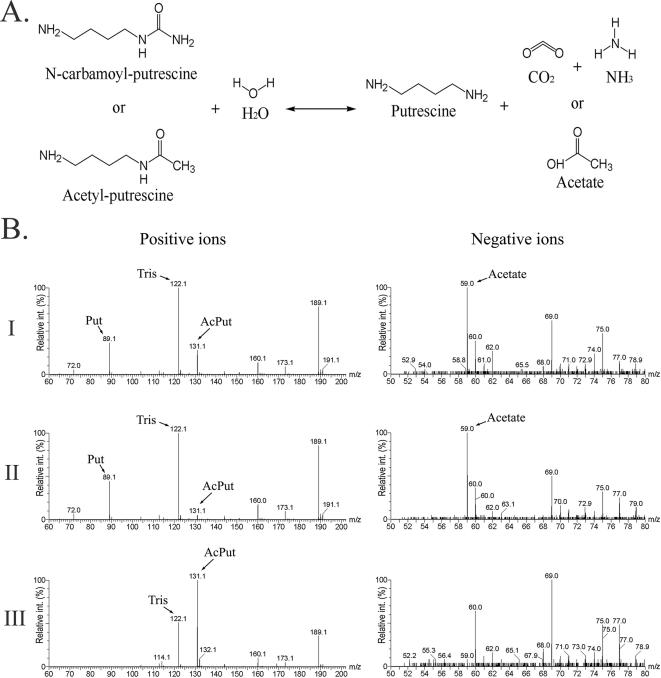
The aguBA operon encodes two enzymes with activity in the conversion of agmatine into putrescine (Fig. 1). We have reported that the aguB mutant exhibited a leaky growth phenotype, while the aguA mutant showed no growth on agmatine as the sole source of carbon and nitrogen (25). In the presence of agmatine, the aguB mutant was expected to accumulate N-CP, which has a chemical structure (Fig. 2a) similar to that of acetylputrescine, a substrate of AphA and an inducer signal of aphA and aphB (see below). We fortuitously proposed that aphA and aphB might be induced by N-CP (and hence agmatine) and that the leaky growth of the aguB mutant on agmatine was due to the conversion of N-CP to putrescine by the induced AphA and AphB.
FIG. 2.
Formation of putrescine by acetylpolyamine amidohydrolases. (A) Reactions catalyzed by AphA and/or AphB. Either N-CP or acetylputrescine can be converted into putrescine based on the similarities in chemistry. (B) Mass spectrometry analysis of AphA enzymatic activity. Purified AphA was applied to reaction mixtures containing acetylputrescine as substrate. Reaction products were subjected to electrospray ionization quadrupole-time-of-flight mass spectrometry analysis for positive and negative ions. Panel I, reactions with purified enzyme; panel II, reactions with MnCl2 or CoCl2 preactivated enzymes; panel III, negative control with heat-inactivated enzyme.
To test this hypothesis, an aphA::lacZ promoter fusion was constructed and introduced into the aguA and aguB mutants and the parent strain PAO1. As shown in Table 3, the aphA promoter in PAO1 was only slightly induced (sixfold) by exogenous agmatine but not by putrescine. In contrast, acetylputrescine exerted a strong induction effect (74-fold). In comparison, agmatine showed no induction in the aguA mutant (no N-CP synthesis), but it exerted an enhanced level of induction in the aguB mutant (N-CP accumulation). On the other hand, agmatine induction was sustained in the aguR mutant strain. Similar results were also obtained when an aphB::lacZ fusion was applied in the same set of experiments. These results were consistent with the hypothesis that acetylputrescine is the authentic inducer molecule of aphA and aphB and that the agmatine induction of aphA/aphB is mediated by N-CP and is independent of the AguR regulatory system.
TABLE 3.
Expression profile of aphA and aphB promotersa
| Strain | Genotype | Supplement | β-galactosidase activity (nmol/min/mg) induced by:
|
|
|---|---|---|---|---|
| PaphA | PaphB | |||
| PAO1 | WT | Glu | 10 | 145 |
| Glu plus Agm | 57 | 323 | ||
| Glu plus AcPut | 736 | 2,262 | ||
| Glu plus Put | 14 | 67 | ||
| Glu plus GABA | 16 | 52 | ||
| PAO5001 | aguA | Glu | 4 | 148 |
| Glu plus Agm | 2 | 84 | ||
| PAO5002 | aguB | Glu | 9 | 169 |
| Glu plus Agm | 634 | 1,991 | ||
| PAO5003 | aguR | Glu | 10 | 141 |
| Glu plus Agm | 109 | 461 | ||
Cells were grown in minimal medium P supplemented with 20 mM of the following supplements: Glu, glutamate; Agm, agmatine; AcPut, acetyl-putrescine; Put, putrescine; WT, wild type. Specific activities (nmol/min/mg) represent the averages from two measurements, with standard errors below 5%.
The acetylputrescine amidohydrolase activity of AphA.
The AphA protein of P. aeruginosa PAO1 exhibits 70% sequence similarity to acetylpolyamine amidohydrolase of Mycoplana ramosa, which requires metals (zinc or cobalt) for its catalytic reaction on a variety of acetylpolyamines, including acetylputrescine (28). To demonstrate the proposed enzymatic activity of AphA, a recombinant form of AphA was overexpressed in E. coli and purified to homogeneity, as described in Materials and Methods. The recombinant AphA was applied to a reaction mixture containing acetylputrescine as substrate, and the presence of the anticipated reaction products was analyzed by mass spectrometry. As shown in Fig. 2B, a comparison of the mass spectra of the samples versus that of the control revealed the appearance of sample signals with m/z values characteristic of putrescine and acetate, the products expected for acetylputrescine hydrolysis by AphA. Although the values were not quantitative, the putrescine/acetylputrescine ratio signals were significantly higher in samples of metal-containing reactions (Fig. 2B, compare panels I and II). These results support AphA as a metalloenzyme of acetylpolyamine amidohydrolase.
The dadRAX locus of alanine catabolism.
A three-gene locus containing the putative genes dadA and dadX in alanine catabolism and the PA5303 gene of an unknown function was induced by agmatine and putrescine (Table 2). The DadA and DadX polypeptides were annotated by the genome project as d-alanine dehydrogenase and alanine racemase, respectively, based on the highly conserved amino acid sequence similarities for bacteria. To substantiate the results of transcriptome analyses and the proposed physiological functions of dadAX in alanine utilization, we constructed a dadA::lacZ promoter fusion construct to study its expression profile and conducted a growth phenotype analysis of dadA and dadX mutants.
As shown in Table 4, the dadA promoter was induced with an order of strength of alanine that was greater than putrescine utilization in the wild-type strain PAO1 grown in glutamate minimal medium. A mutation in either dadA or dadX severely hampered alanine utilization as the sole source of carbon and nitrogen but showed only a moderate effect on agmatine/putrescine (Table 6). When the dadX mutant was complemented with a functional dadX gene expressed from multicopy plasmids, the growth of this recombinant strain on l-alanine was even enhanced in comparison to that of the wild-type PAO1. These results are consistent with the function proposed for dadAX in alanine utilization and support the hypothesis that the induction of dadAX by agmatine/putrescine is mediated by alanine as a product of a transamination reaction in the proposed catabolic pathway (Fig. 1).
TABLE 4.
Expression profile of the dadA promotera
| Strain | Genotype | β-galactosidase activity (nmol/min/mg) in:
|
||
|---|---|---|---|---|
| Glu | Glu plus Put | Glu plus Ala | ||
| PAO1 | WT | 184 | 1,628 | 2,783 |
| PAO5713 | dadR | 15 | 16 | NG |
| PAO5008 | spuC | 99 | 137 | 3,352 |
| PAO5706 | spuC/PA5313 | 118 | 114 | 2,879 |
| PAO5708 | PA5313 | 249 | 716 | 1,742 |
Cells were grown in minimal medium P supplemented with 20 mM of the following supplements: Glu, glutamate; Put, putrescine; Ala, alanine. Specific activities (nmol/min/mg) represent the averages from two measurements, with standard errors below 5%. NG, no growth. WT, wild type.
TABLE 6.
Growth phenotypea
| Strain | Genotype | Growth response in medium plus:
|
|||||
|---|---|---|---|---|---|---|---|
| Glu | Agm | Put | GABA | Ala | Arg | ||
| PAO1 | WT | ++ | ++ | ++ | ++ | ++ | ++ |
| PAO5710 | dadA | ++ | + | + | + | − | NT |
| PAO5711 | dadX | ++ | + | + | + | − | NT |
| PAO5713 | dadR | ++ | ++ | ++ | ++ | − | NT |
| PAO5707 | kauB | ++ | − | − | ++ | NT | NT |
| PAO5708 | PA5313 | ++ | ++ | ++ | ++ | ++ | NT |
| PAO5703 | gabT | ++ | ++ | ++ | + | + | ++ |
| PAO5704 | gabT/PA5313 | ++ | ++ | ++ | − | + | NT |
| PAO5701 | gabD | ++ | + | + | − | + | ++ |
| PAO5702 | gabD/PA5313 | ++ | + | + | − | + | NT |
| PAO5008 | spuC | ++ | − | − | ++ | ++ | NT |
| PAO5706 | spuC/PA5313 | ++ | − | − | ++ | ++ | NT |
| PAO5714 | phoP | ++ | ++ | ++ | ++ | ++ | ++ |
| PAO5715 | phoQ | ++ | ++ | ++ | ++ | ++ | ++ |
Cell growth was tested after 24 h of incubation at 37°C in minimal medium P with 20 mM concentrations of the following supplements: Glu, glutamate; Agm, agmatine; Put, putrescine; Ala, l-alanine; Arg, l-arginine. ++, prominent growth; +, retarded growth; −, no growth; NT, not tested; WT, wild type. Consistent growth phenotypes were observed from three independent culture sets.
Between dadA and dadX, the PA5303 gene encodes a putative endoribonuclease. Insertion of a gentamicin-omega loop cassette on PA5303 also caused a severe negative effect on alanine utilization, and this growth defect can be complemented by a recombinant plasmid carrying dadX but not by the one carrying PA5303. Since the omega loop structure on the flanking regions of the inserted cassette could exert a polar effect on the downstream gene, these results indicated that PA5303 may not be essential for alanine utilization and that the growth defect on alanine of the PA5303::GmΩ mutant was most likely due to a polar effect on the downstream dadX.
The PA5308 gene upstream of dadA encodes a putative transcriptional regulator of the AsnC family. Although the expression of PA5308 was not affected by agmatine or putrescine, its close proximity to dadAX prompted us to investigate the potential role of PA5308 in the control of dadAX expression. The PA5308 mutant cannot grow on l-alanine as the sole source of carbon and nitrogen (Table 6), and this mutation completely alleviated the induction effect of agmatine and putrescine on the dadA promoter (Table 4). On the basis of these results, PA5308 was designated dadR (dadAX regulator), which is essential for the transcriptional activation of dadAX.
It was interesting to note that mutations of the dadRAX genes abolished l-alanine utilization and also that the growth of the resulting mutants on other carbon/nitrogen sources became extremely sensitive to the presence of exogenous l-alanine. For example, the growth of these mutants, but not the parent strain PAO1, in the minimal medium supplemented with either glutamate or glucose and ammonium, was severely retarded by 5 mM of l-alanine. A similar case has been reported with Klebsiella aerogenes, and it was proposed that this was the result of the allosteric inhibition by l-alanine on glutamine synthetase (10). We tested this hypothesis by the addition of 5 mM of glutamine in the growth medium and still found no sign of alleviation of growth inhibition by exogenous l-alanine in these mutants. These results suggest that the dadRAX genes are essential to maintaining alanine homeostasis and that exogenous l-alanine could exert a growth inhibition effect on targets yet to be identified in the dad mutants.
Linkage of alanine catabolism to putrescine utilization via a pyruvate transaminase.
As described above, the dadA promoter was found to be inducible by putrescine (and hence agmatine). We proposed that it was due to alanine synthesis from putrescine transamination catalyzed by SpuC, which employs pyruvate as the amino acceptor (20). This hypothesis was supported by a drastic reduction of putrescine-dependent induction of the dadA promoter in the spuC mutant (Table 4). As we reported previously that the spuC mutants exhibited a severe defect in putrescine utilization (20), this result further supports the conclusion that SpuC is the major putrescine-pyruvate transaminase.
The kauB-PA5313 locus.
The induction profile of the kauB (initially identified as an essential gene for ketoarginine utilization) and PA5313 genes was confirmed by transcriptional lacZ fusions. As shown in Table 5, the kauB-PA5313 divergent promoters were both induced by exogenous agmatine and putrescine. The kauB gene and the KauB protein were reported to possess an aldehyde dehydrogenase activity that can utilize 4-aminobutyraldehyde (derived from putrescine; Fig. 1) and 4-guanidinobutyraldehyde (derived from ketoarginine) as substrates (11). The results of growth phenotype analyses (Table 6) indicated that kauB is essential for the utilization of agmatine and putrescine but not of GABA.
TABLE 5.
Expression profiles of kauB, PA5313, and gabD promotersa
| Strain | Genotype | Promoter | β-galactosidase activity (nmol/min/mg) in:
|
|||
|---|---|---|---|---|---|---|
| Glu | Glu plus Agm | Glu plus Put | Glu plus GABA | |||
| PAO1 | WT | PkauB | 263 | 907 | 1,013 | NT |
| PPA5313 | 136 | 904 | 961 | NT | ||
| PGabD | 3,868 | 42,944 | 53,911 | 45,436 | ||
Cells were grown in minimal medium P supplemented with 20 mM of the following supplements: Glu, glutamate; Agm, agmatine; Put, putrescine. NT, not tested. WT, wild type. Specific activities (nmol/min/mg) represent the averages from two measurements, with standard errors below 5%.
Both PA5313 and its downstream gene PA5314 were induced by agmatine and putrescine (Table 2), and these two genes encode a putative pyruvate transaminase and a hypothetical protein of unknown function. The PA5313 and PA5314 mutants grew normally on either agmatine, putrescine, or GABA, but growth on spermidine was significantly retarded.
The gabDT locus.
Conversion of GABA to succinate requires a pair of deamination and oxidation reactions (Fig. 1). In the genome annotations, PA0265 (gabD) and PA0266 (gabT) were proposed to code for succinic semialdehyde dehydrogenase (SSAD) and GABA transaminase, respectively, which exhibit high sequence similarities to GabD and GabT of E. coli (30). In P. aeruginosa PAO1, the induction of gabDT by exogenous agmatine and putrescine was first revealed by GeneChip analyses (Table 2). This induction effect of agmatine and putrescine was confirmed by the measurements of β-galactosidase activities from PAO1 harboring pHT0265, a PgabD::lacZ fusion, as shown in Table 5. In addition, exogenous GABA also exerted a strong induction effect on the gabD promoter activity.
The gabD and gabT knockout mutants were constructed as described in Materials and Methods, and the growth phenotype of these mutants was tested on arginine, agmatine, putrescine, GABA, and glutamate as the sole sources of carbon and nitrogen (Table 6). These mutants did not differ from the parent strain in terms of glutamate and arginine utilization. However, the gabD mutant showed retarded growth on agmatine and putrescine and was unable to grow on GABA. The growth of the gabT mutant on GABA was also affected; the generation time of the gabT mutant was significantly longer than that of the wild-type PAO1 (87 min versus 42 min). These results suggested that GabD is essential for GABA utilization and the presence of at least one more GABA transaminase other than GabT.
As described above, PA5313 was proposed to encode a putative transaminase. To test whether PA5313 may serve a role in GABA utilization, a gabT PA5313 double mutant was constructed, and its growth on GABA as the sole source of carbon and nitrogen was completely abolished (Table 6). These results support GabT and PA5313 as two redundant transaminases in GABA catabolism.
The proposed SSAD activity of GabD was measured for a gabD mutant and its parent strain PAO1, grown in glutamate minimal medium in the presence or absence of GABA. In the presence of GABA, the SSAD activity was induced ninefold in PAO1 (140 versus 1,281 nmol min−1 mg−1). In comparison, only a negligible level of SSAD activity could be detected in the gabD mutant in both growth conditions. These results support the idea that gabD encodes a GABA-inducible SSAD.
Effects of PhoP and PhoQ.
As shown in Table 2, the oprH-phoP-phoQ operon was induced by agmatine and putrescine, which encodes an outer membrane porin OprH and the response regulator and sensor kinase of the PhoPQ two-component system (21). To investigate whether the PhoPQ system plays a role in agmatine and putrescine catabolism, we constructed the phoP and phoQ mutants and tested the growth phenotype of these two mutants on glutamate, agmatine, putrescine, and GABA as the sole sources of carbon and nitrogen. As shown in Table 6, the phoP and phoQ mutants grew well on all of these tested compounds.
Employing an oprH::lacZ fusion, exogenous putrescine exerted a 2.4-fold induction on the oprH promoter activities in the wild-type strain PAO1 as shown in Table 7. In the phoP mutant, the oprH promoter remained constitutively low in the presence or absence of putrescine. In comparison, the phoQ mutant retained a constitutively high level of oprH promoter activity but also showed no response to exogenous putrescine. These results supported the data from the GeneChip analysis and were consistent with the functions reported for PhoP and PhoQ on the autoregulation of the oprH-phoP-phoQ operon (21).
TABLE 7.
Effects of PhoPQ on expression profiles of oprH, gabD and spuA promotersa
| Strain | Promoter | Genotype | β-galactosidase activity (nmol/min/mg) in:
|
|
|---|---|---|---|---|
| Glu | Glu plus Put | |||
| PAO1 | PoprH | WT | 801 | 1,953 |
| PAO5714 | phoP | 409 | 160 | |
| PAO5715 | phoQ | 4,878 | 3,497 | |
| PAO1 | PspuA | WT | 154 | 833 |
| PAO5714 | phoP | 139 | 947 | |
| PAO1 | PgabD | WT | 3,868 | 53,911 |
| PAO5714 | phoP | 3,582 | 39,088 | |
Cells were grown in minimal medium P supplemented with 20 mM of the following supplements as indicated: Glu, glutamate; Put, putrescine. Specific activities (nmol/min/mg) represent the averages from two measurements, with standard errors below 5%.
The potential effects of PhoP on putrescine-dependent induction of the spuABCD and gabDT operons was also investigated. As shown in Table 7, the expression of β-galactosidase from either the spuA::lacZ or gabD::lacZ fusion was still subjected to induction by exogenous putrescine in the phoP mutant. These data strongly suggested that the PhoPQ system may not play a significant role in the control of putrescine or GABA catabolism.
DISCUSSION
Participation of acetylpolyamine amidohydrolases in agmatine catabolism.
In this report, we identified aphA and aphB as agmatine-inducible genes, by DNA microarrays analyses. With purified recombinant AphA proteins, a metal-dependent acetylpolyamine amidohydrolase activity of AphA was demonstrated (Fig. 2a). This enzymatic property of AphA is consistent with that reported for AphA from Mycoplana ramosa (28). The promoters of aphA and aphB were both induced by exogenous acetylputrescine to a much higher level than that of agmatine in the wild-type strain PAO1. However, in the aguB mutant, exogenous agmatine did exert a significant induction effect on the aphA and aphB promoters. N-CP, an intermediate in agmatine/putrescine conversion (Fig. 1), is anticipated to accumulate in the aguB mutant when it is fed with agmatine. Considering the fact that N-CP and acetylputrescine have very similar chemical structures (Fig. 2a), it is very likely that N-CP can serve as the signal compound of aphA and aphB induction as well as the substrate of the encoded enzymes. These fortuitous events could explain the leaky growth phenotypes of aguB mutants on agmatine in the previous reports (20, 25).
From human cells to many types of bacteria including E. coli, the acetylation of polyamines was reported as a reaction important to the conversion of excess polyamines into a physiologically inert form and hence in keeping with the homeostasis of intracellular polyamines. In E. coli, polyamine acetylation is catalyzed by SpeG (4, 5, 19), and acetylated polyamine is readily excreted (27). E. coli might not recycle acetylated polyamine, and no apparent AphA homologue can be found in this organism as revealed by a BLAST search against all available genome sequences of E. coli strains in the NCBI database. In contrast, P. aeruginosa possesses AphA and AphB, but no promising SpeG homologue can be found by sequence comparison. By taking advantage of its enormous capability of polyamine catabolism, P. aeruginosa may be able to keep polyamine homeostasis through catabolic pathways instead of acetylation.
Putrescine catabolic pathways.
Putrescine catabolism into GABA can be mediated by two routes in bacteria: the conventional transamination/dehydrogenation route and a recently reported γ-glutamylation route (15). In P. aeruginosa, the abolishment of putrescine utilization in the spuC and kauB mutants (Table 6) supports the importance of the conventional route for putrescine catabolism. When this major pathway is blocked, putrescine might be catabolized through the γ-glutamylation pathway. Alternatively, since putrescine is the precursor compound of spermidine, it may be channeled into spermidine catabolic pathways yet to be exploited.
Enzymes and the corresponding genes of the γ-glutamylation pathway were first proposed in E. coli as essential elements in putrescine utilization. The PuuA protein of E. coli catalyzes γ-glutamylation as the first step in this route. In E. coli, PuuA is the only member of its kind that exhibited significant similarities to GlnA, the glutamine synthetase. In comparison, there are seven PuuA homologues in P. aeruginosa that show up to 44% identity (SpuI, SpuB, PA1566, PA2040, PA3356, PA5508, and PA5522), and four of them (except PA2040, PA1566, and PA5508) are listed in Table 2 as putrescine-inducible genes in this study. For an unknown reason, the PA2040 gene was not included in the GeneChip analysis. However, considering the fact that SpuI and PA2040 share 96% sequence identity (nucleotide as well as protein) with an identical length of 458 amino acids and that the downstream PA2041 gene was inducible by putrescine, we proposed that PA2040 should also be included in the list. The spuB and spuI genes have been reported in our previous study (20); the spuB mutant exhibited a specific growth deficiency on spermidine but not on putrescine or agmatine, while the spuI mutant showed no apparent effect on polyamine utilization. Although γ-glutamylation seems important for spermidine catabolism, whether it plays a role in putrescine utilization was not clear due to the redundancy of these enzymes.
Another major difference between these two routes is the enzymatic reactions for putrescine deamination. The spuC gene encodes a putrescine-pyruvate transaminase (20), which plays a pivotal role in the putrescine-dependent induction of dadAX for alanine catabolism (Table 4). In the case of γ-glutamylation, the terminal amino group of glutamylputrescine was proposed to be released as ammonia by oxidoreductase (Fig. 1, PuuB), which also generates hydrogen peroxide in this reaction. Four puuB homologues (PA0534, PA1566, PA5309, and PA2776) were identified as putrescine-inducible genes in this study.
The KauB and SpuA proteins (Table 2) are the most prominent homologues of PuuC and PuuD, respectively, of the γ-glutamylation pathway. KauB was proposed to play an essential role in the transaminase pathway, as the kauB mutant lost the capability to grow on agmatine and putrescine but not on GABA (Table 6). SpuA is the only candidate of the PuuD homologue in PAO1 based on sequence comparison; however, we have reported that the spuA mutant grew normally on agmatine and putrescine but did exhibit a server growth defect on spermidine (20). It provides another piece of evidence to support the transaminase pathway as the major route of putrescine utilization and strongly suggests spermidine catabolism via γ-glutamylation in P. aeruginosa.
Apparently, the γ-glutamylation route is not energy efficient, as it takes ATP to drive the first reaction (Fig. 1). In E. coli, putrescine can be utilized only as a sole source of nitrogen and not of carbon, and the expression of the puu gene possesses a very distinct pattern of temporal induction in the early stationary phase, needs aeration (presumably for the deamination reaction by oxidoreductase), and is subject to repression by its end products succinate and ammonia. In addition, GABA catabolism, as described below, may also put another layer of restrictions to the γ-glutamylation pathway in E. coli. As reflected by its regulatory systems, perhaps the physiological functions of this pathway in E. coli are to serve mainly as a defense mechanism against any toxic effect by polyamine accumulation when the cells reach the stationary phase and secondarily as a source of nitrogen or carbon under nutrient limitation conditions. Regardless, it was very intriguing to have multiple sets of PuuA/PuuB homologues in P. aeruginosa. More studies are needed to elucidate the potential role of γ-glutamylation in putrescine and spermidine catabolism in P. aeruginosa.
Alanine catabolism.
In this study, we provided genetic evidence to support the physiological function of dadRAX in alanine and putrescine catabolism (Table 4 and 6) and established DadR as a transcriptional activator of dadAX. For putrescine catabolism, l-alanine is generated from the transamination reaction catalyzed by SpuC. A dadAX operon induced in response to exogenous putrescine would ensure recycling of pyruvate to the transamination reaction and hence can optimize the metabolic flux of putrescine into the TCA cycle as well as gluconeogenesis.
GABA utilization.
Conversion of GABA into succinate also requires a pair of deamination and oxidation reactions. When GABA was supplied as the sole source of carbon and nitrogen, the gabD mutant and the gabT PA5313 double mutant lost the capability to grow on this compound. The proposed succinyl semialdehyde dehydrogenase activity of GabD was induced by exogenous GABA in the wild-type PAO1 and was absent in the gabD mutant. The gabT gene encoding a GABA transaminase most likely forms an operon with the upstream gabD gene. As revealed by DNA microarrays analyses and gabD::lacZ fusion studies, expression of the gabDT operon is induced by GABA as well as by agmatine and putrescine and, presumably, by any compounds that can increase intracellular concentration of GABA.
In comparison, the gabDT genes of E. coli are part of the csiD-ygaF-gabDTP operon (23), which is transcribed from two σS promoters in response to carbon starvation and environmental stresses and one σ70 promoter regulated by Nac in response to nitrogen limitation (30). GabD/T-mediated GABA catabolism is considered a general stress adaptation (12, 23), and it is not inducible by exogenous GABA.
PhoPQ two-component system.
It has been reported that the PhoPQ system is required for autoregulation of the oprH-phoP-phoQ operon under divalent cation-limiting growth conditions and is also involved in resistance to cationic antimicrobial peptides (21). Furthermore, an extensive study of microarray transcriptional profiling reported a PhoP-dependent induction of gabDT under Mg2+-limited conditions and suggested the presence of a putative PhoP-binding site in the promoter region of the spuABCD operon (22). As shown in Table 2, exogenous agmatine and putrescine induce the expression level of the oprH-phoP-phoQ operon, and we have demonstrated this induction effect on the oprH promoter activity (18). In addition, we have reported the function of the PhoPQ two-component system in antibiotic resistance triggered by exogenous agmatine, putrescine, and spermidine (18). Due to the effect of agmatine and putrescine on PhoPQ, it was not surprising to find that some genes listed in Table 2 have been reported as members of the PhoP regulon (22). However, the PhoPQ system did not seem to play a role in the catabolism of these compounds, as phoP and phoQ mutants grew normally on these nutrients as the sole sources of carbon and nitrogen (Table 6). Studies of the other putative regulatory genes in the control of putrescine and GABA utilization are in progress.
Supplementary Material
Acknowledgments
We thank Siming Wang and the Georgia State University MS facility for ESI-MS analysis.
This work was supported by the National Science Foundation (MCB-0415608).
Footnotes
Published ahead of print on 11 January 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 2.Casero, R. A., Jr., and A. E. Pegg. 1993. Spermidine/spermine N1-acetyltransferase—the turning point in polyamine metabolism. FASEB J. 7653-661. [PubMed] [Google Scholar]
- 3.Farinha, M. A., and A. M. Kropinski. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 1723496-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuchi, J., K. Kashiwagi, K. Takio, and K. Igarashi. 1994. Properties and structure of spermidine acetyltransferase in Escherichia coli. J. Biol. Chem. 26922581-22585. [PubMed] [Google Scholar]
- 5.Fukuchi, J., K. Kashiwagi, M. Yamagishi, A. Ishihama, and K. Igarashi. 1995. Decrease in cell viability due to the accumulation of spermidine in spermidine acetyltransferase-deficient mutant of Escherichia coli. J. Biol. Chem. 27018831-18835. [DOI] [PubMed] [Google Scholar]
- 6.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 1733000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ha, H. C., N. S. Sirisoma, P. Kuppusamy, J. L. Zweier, P. M. Woster, and R. A. Casero, Jr. 1998. The natural polyamine spermine functions directly as a free radical scavenger. Proc. Natl. Acad. Sci. USA 9511140-11145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas, D., B. W. Holloway, A. Schambock, and T. Leisinger. 1977. The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa. Mol. Gen. Genet. 1547-22. [DOI] [PubMed] [Google Scholar]
- 9.Huang, S. C., C. A. Panagiotidis, and E. S. Canellakis. 1990. Transcriptional effects of polyamines on ribosomal proteins and on polyamine-synthesizing enzymes in Escherichia coli. Proc. Natl. Acad. Sci. USA 873464-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janes, B. K., and R. A. Bender. 1998. Alanine catabolism in Klebsiella aerogenes: molecular characterization of the dadAB operon and its regulation by the nitrogen assimilation control protein. J. Bacteriol. 180563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jann, A., H. Matsumoto, and D. Haas. 1988. The fourth arginine catabolic pathway of Pseudomonas aeruginosa. J. Gen. Microbiol. 1341043-1053. [DOI] [PubMed] [Google Scholar]
- 12.Joloba, M. L., K. M. Clemmer, D. D. Sledjeski, and P. N. Rather. 2004. Activation of the gab operon in an RpoS-dependent manner by mutations that truncate the inner core of lipopolysaccharide in Escherichia coli. J. Bacteriol. 1868542-8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan, A. U., P. Di Mascio, M. H. Medeiros, and T. Wilson. 1992. Spermine and spermidine protection of plasmid DNA against single-strand breaks induced by singlet oxygen. Proc. Natl. Acad. Sci. USA 8911428-11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, I. G., and T. J. Oh. 2000. SOS induction of the recA gene by UV-, gamma-irradiation and mitomycin C is mediated by polyamines in Escherichia coli K-12. Toxicol. Lett. 116143-149. [DOI] [PubMed] [Google Scholar]
- 15.Kurihara, S., S. Oda, K. Kato, H. G. Kim, T. Koyanagi, H. Kumagai, and H. Suzuki. 2005. A novel putrescine utilization pathway involves gamma-glutamylated intermediates of Escherichia coli K-12. J. Biol. Chem. 2804602-4608. [DOI] [PubMed] [Google Scholar]
- 16.Kwon, D. H., and C. D. Lu. 2007. Polyamine effects on antibiotic susceptibility in bacteria. Antimicrob. Agents Chemother. 512070-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon, D. H., and C. D. Lu. 2006. Polyamines increase antibiotic susceptibility in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 501623-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon, D. H., and C. D. Lu. 2006. Polyamines induce resistance to cationic peptide, aminoglycoside, and quinolone antibiotics in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 501615-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Limsuwun, K., and P. G. Jones. 2000. Spermidine acetyltransferase is required to prevent spermidine toxicity at low temperatures in Escherichia coli. J. Bacteriol. 1825373-5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu, C. D., Y. Itoh, Y. Nakada, and Y. Jiang. 2002. Functional analysis and regulation of the divergent spuABCDEFGH-spuI operons for polyamine uptake and utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 1843765-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macfarlane, E. L., A. Kwasnicka, M. M. Ochs, and R. E. Hancock. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34305-316. [DOI] [PubMed] [Google Scholar]
- 22.McPhee, J. B., M. Bains, G. Winsor, S. Lewenza, A. Kwasnicka, M. D. Brazas, F. S. Brinkman, and R. E. Hancock. 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 1883995-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metzner, M., J. Germer, and R. Hengge. 2004. Multiple stress signal integration in the regulation of the complex sigma S-dependent csiD-ygaF-gabDTP operon in Escherichia coli. Mol. Microbiol. 51799-811. [DOI] [PubMed] [Google Scholar]
- 24.Nakada, Y., and Y. Itoh. 2003. Identification of the putrescine biosynthetic genes in Pseudomonas aeruginosa and characterization of agmatine deiminase and N-carbamoylputrescine amidohydrolase of the arginine decarboxylase pathway. Microbiology 149707-714. [DOI] [PubMed] [Google Scholar]
- 25.Nakada, Y., Y. Jiang, T. Nishijyo, Y. Itoh, and C. D. Lu. 2001. Molecular characterization and regulation of the aguBA operon, responsible for agmatine utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 1836517-6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, S. M., C. D. Lu, and A. T. Abdelal. 1997. Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol. 1795300-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parry, L., J. Lopez-Ballester, L. Wiest, and A. E. Pegg. 1995. Effect of expression of human spermidine/spermine N1-acetyltransferase in Escherichia coli. Biochemistry 342701-2709. [DOI] [PubMed] [Google Scholar]
- 28.Sakurada, K., T. Ohta, K. Fujishiro, M. Hasegawa, and K. Aisaka. 1996. Acetylpolyamine amidohydrolase from Mycoplana ramosa: gene cloning and characterization of the metal-substituted enzyme. J. Bacteriol. 1785781-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez, M., M. A. Alvarez, R. Balana, and A. Garrido-Pertierra. 1988. Properties and functions of two succinic-semialdehyde dehydrogenases from Pseudomonas putida. Biochim. Biophys. Acta 953249-257. [DOI] [PubMed] [Google Scholar]
- 30.Schneider, B. L., S. Ruback, A. K. Kiupakis, H. Kasbarian, C. Pybus, and L. Reitzer. 2002. The Escherichia coli gabDTPC operon: specific gamma-aminobutyrate catabolism and nonspecific induction. J. Bacteriol. 1846976-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schweizer, H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15831-834. [PubMed] [Google Scholar]
- 32.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97109-121. [DOI] [PubMed] [Google Scholar]
- 33.Sekowska, A., A. Danchin, and J. L. Risler. 2000. Phylogeny of related functions: the case of polyamine biosynthetic enzymes. Microbiology 1461815-1828. [DOI] [PubMed] [Google Scholar]
- 34.Stibitz, S., W. Black, and S. Falkow. 1986. The construction of a cloning vector designed for gene replacement in Bordetella pertussis. Gene 50133-140. [DOI] [PubMed] [Google Scholar]
- 35.Tabor, C. W., and H. Tabor. 1985. Polyamines in microorganisms. Microbiol. Rev. 4981-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, Z., and C. D. Lu. 2007. Functional genomics enables identification of genes of the arginine transaminase pathway in Pseudomonas aeruginosa. J. Bacteriol. 1893945-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.