Abstract
Wzz is a membrane protein that determines the chain length distribution of the O-antigen lipopolysaccharide by an unknown mechanism. Wzz proteins consist of two transmembrane helices separated by a large periplasmic loop. The periplasmic loop of Escherichia coli K-12 Wzz (244 amino acids from K65 to A308) was purified and found to be a monomer with an extended conformation, as determined by gel filtration chromatography and analytical ultracentrifugation. Circular dichroism showed that the loop has a 60% helical content. The Wzz periplasmic loop also contains three regions with predicted coiled coils. To probe the function of the predicted coiled coils, we constructed amino acid replacement mutants of the E. coli K-12 Wzz protein, which were designed so that the coiled coils could be separate without compromising the helicity of the individual molecules. Mutations in one of the regions, spanning amino acids 108 to 130 (region I), were associated with a partial defect in O-antigen chain length distribution, while mutants with mutations in the region spanning amino acids 209 to 223 (region III) did not have an apparent functional defect. In contrast, mutations in the region spanning amino acids 153 to 173 (region II) eliminated the Wzz function. This phenotype was associated with protein instability, most likely due to conformational changes caused by the amino acid replacements, which was confirmed by limited trypsin proteolysis. Additional mutagenesis based on a three-dimensional model of region I demonstrated that the amino acids implicated in function are all located at the same face of a predicted α-helix, suggesting that a coiled coil actually does not exist in this region. Together, our results suggest that the regions predicted to be coiled coils are important for Wzz function because they maintain the native conformation of the protein, although the existence of coiled coils could not be demonstrated experimentally.
Lipopolysaccharide (LPS), a major component of the outer leaflet of the gram-negative bacterial outer membrane (33), consists of lipid A, core oligosaccharide, and O-specific polysaccharide or O antigen (33, 46). LPS biosynthesis involves a large number of enzymes, which are governed by more than 40 genes (for reviews, see references 34, 37, and 42). O antigens are polymers of oligosaccharide repeating units. The chemical compositions, structures, and antigenicities of O antigens vary widely among gram-negative bacteria, giving rise to a large number of O serotypes (19). Lipid A, the membrane-embedded portion of LPS, forms the majority of the outer lipid leaflet of the outer membrane. The core oligosaccharide, made up of hexoses, glycero-manno-heptose, and keto-deoxyoctulosonic acid (17), is assembled on preformed lipid A by sequential transfer of sugar components. In a separate pathway, the O antigen is assembled on undecaprenyl-phosphate (Und-P), forming an Und-PP-linked saccharide. Both pathways converge at the ligation of O antigen to the lipid A-core oligosaccharide, with the concomitant release of Und-PP (17, 18, 34, 42). Und-PP is recycled into Und-P by a poorly characterized pathway, conserved in all cells (5, 15, 41, 42), which involves the hydrolysis of the terminal phosphate.
O-antigen assembly occurs by mechanisms referred to as Wzy (polymerase)-dependent and ATP-binding cassette (ABC)-dependent pathways. In ABC-dependent pathways, the polymeric O antigen is formed on the cytoplasmic side of the plasma membrane (for reviews, see references 34 and 42). The polymer is exported across the membrane by an ABC transporter (4). A similar pathway exists for the synthesis of certain polysaccharide capsules (45). In some systems, Wzm and Wzt are the permease and ATPase components of the ABC transporter, while in other systems a single protein mediates the energy-dependent export (20, 34). In the Wzy-dependent pathway, O repeating subunits are synthesized on the cytosolic side of the plasma membrane. Each subunit is subsequently translocated across the membrane by a process in which there is no apparent direct involvement of ATP hydrolysis and which is mediated by the protein Wzx (21, 34, 35, 42). On the periplasmic side of the plasma membrane, translocated subunits polymerize to form a certain length, unique to each O antigen, by the concerted functions of Wzy (O-antigen polymerase) and Wzz (O-antigen chain length regulator). Finally, the polysaccharide is ligated “en bloc” to the lipid A-core oligosaccharide (23, 28, 32). The Wzy-dependent pathway coordinates the synthesis of many O antigens, especially those with repeating units composed of different sugars (20).
Although not essential for the synthesis and polymerization of O antigen, Wzz regulates the chain length distribution (or modality) of the O-antigen chain (3) by an unknown mechanism. Two models explaining the control of Wzz have been proposed. In one model, Wzz functions as a molecular timer modulating the Wzy polymerase activity by favoring either polymerization or termination through the transfer of the O polysaccharide to the WaaL ligase (2). The other model suggests that Wzz is a molecular chaperone that recruits a protein complex consisting of Wzy, the WaaL O-antigen ligase, and the nascent polysaccharide chain (10, 31). However, WaaL is not directly required for chain length determination, as the modality of the O-polysaccharide chain can be observed in the absence of WaaL (14), and modality appears to be determined before the ligation step (9). Also, there is very strong genetic evidence that the Wzx protein in Escherichia coli forms a putative complex together with Wzz and Wzy (25). More recently, we have shown that the predicted periplasmic loop of the Salmonella enterica WbaP protein, which is involved in the initiation step of O-antigen subunit synthesis, also affects chain length distribution by possibly interacting with Wzz (36). Evidence from in vivo cross-linking and immunoprecipitation demonstrating that the O antigen may physically interact with a putative protein complex (9) and recent observations of purified Wzz interacting with O-antigen polysaccharide in vitro (16, 40) suggest that there are complex interactions between multiple proteins and the nascent O-antigen chain. Still, the molecular mechanism of the Wzz function remains elusive.
Wzz is a membrane protein with two transmembrane helices and a large periplasmic domain that comprises more than 85% of the protein. This protein is a member of a larger family of membrane proteins with similar predicted topology, known as the polysaccharide copolymerases, which are involved in chain length regulation of O antigens and capsular polysaccharides in gram-negative and gram-positive bacteria (30). Low-resolution structural information obtained by cryoelectron microscopy is available for Wzc, a homologue of Wzz involved in chain length determination in group 1 capsular polysaccharides (7, 8). This information suggests that Wzc forms a tetramer. More recently, small-angle X-ray scattering data for purified Wzz from E. coli O86 also suggested that this protein forms a tetramer (40). By using a combination of analytical ultracentrifugation and gel filtration chromatography, we demonstrated in this study that the purified large periplasmic loop of Wzz is a monomer that adopts an extended conformation. Circular dichroism (CD) spectroscopy demonstrated that the periplasmic loop of Wzz is 60% α-helical, which is in good agreement with models constructed for Wzc and Wzz by other workers (7, 8, 40). The hallmarks of the Wzz periplasmic loop are three regions predicted to form coiled coils (30), which are expected to promote protein-protein interactions and might explain the oligomerization of Wzz in vivo. Alternatively, these regions may have functional importance, but their involvement in Wzz function has not been systematically examined. In an attempt to better understand the function of Wzz, we examined whether the predicted coiled coils have a functional role in this protein. A coiled coil is a bundle of α-helices wound into a superhelix (22). Coiled coils consist of repeating patterns of seven residues (heptads), which are labeled a through g (6, 29, 38). There is usually a consensus of hydrophobic residues at the a and d positions, forming an apolar “stripe” on each helix (44). To test our hypothesis, we generated proteins with double amino acid replacements at the a and d positions of each of the three predicted Wzz coiled coils, regions I, II, and III, by introducing pairs of phenylalanine (Phe) residues. Phe is an amino acid that is not preferred at the a and d positions of a coiled coil. Although it would not be expected to affect the helicity of an individual helix, Phe could potentially disrupt helix-helix interactions due to its bulky nature (38). Using this strategy, we demonstrated in this study that perturbation of each of the three predicted coiled-coil regions in the E. coli K-12 Wzz protein has different functional implications. We concluded that perturbation of region I results in a partial functional defect in O-antigen chain length distribution attributable to residues located on the same face of a helix, but the helix is likely not involved in coiled-coil interactions. In contrast, perturbation of region II causes dramatic conformational changes in the native structure, resulting in a nonfunctional protein, while perturbation of region III results in a negligible functional defect.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
Strains and plasmids used in this study are described in Table 1. Bacteria were cultured in Luria broth (LB) supplemented with antibiotics at the following final concentrations: 100 μg/ml ampicillin, 30 μg/ml chloramphenicol, 40 μg/ml kanamycin, and 80 μg/ml spectinomycin. Chemicals and antibiotics were purchased from Sigma Aldrich and Roche Diagnostics. Oligonucleotide primers were purchased from Invitrogen, and they are available from us upon request. Plasmids were introduced into electrocompetent cells by electroporation (12).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| Strains | ||
| BL21(DE3) | F−dcm ompT hsdS(rB− mB−) gal λ(DE3) | Laboratory stock |
| DH5α | F− φ80lacZΔM15 endA recA hsdR(rK− mK−) supE thi gyrA relA? Δ(lacZYA-argF)U169 | |
| EVV16 | W3110 Δwzz::Km | 43 |
| EVV33 | W3110 Δwzz | This study |
| W3110 | rph-1 IN(rrnD-rrnE)1 | Laboratory stock |
| Plasmids | ||
| pBADFlag | pBAD24 for C-terminal Flag fusions, inducible by arabinose, Apr | This study |
| pCM244 | WzzPL6×His (residues K65 to A308) cloned into pET30a | This study |
| pCP20 | FLP+ λ cI857+ Rep(Ts) Apr Cmr λ pR | 11 |
| pEH1 | wzzB cloned into pBADFlag, Apr | This study |
| pEH1-F115A | WzzF115A in pEH1, Apr | This study |
| pEH1-F115P | WzzF115P in pEH1, Apr | This study |
| pEH1-L118F | WzzL118F in pEH1, Apr | This study |
| pEH1-A119F | WzzA119F in pEH1, Apr | This study |
| pEH1-L122A | WzzL122A in pEH1, Apr | This study |
| pEH1-L122F | WzzL122F in pEH1, Apr | This study |
| pEH1-A156L | WzzA156L in pEH1, Apr | This study |
| pEH1-A156F | WzzA156F in pEH1, Apr | This study |
| pEH1-L160F | WzzL160F in pEH1, Apr | This study |
| pEH3 | WzzPL2-6×His (K65 to A308) cloned into pET30a | This study |
| pEH4 | WzzA156F, L160F in pEH1, Apr | This study |
| pEH5 | WzzI212F, L216F in pEH1, Apr | This study |
| pEH6 | WzzL118F, L122F in pEH1, Apr | This study |
| pET30a | Cloning vector, Kmr | Novagen |
| pEV6 | wzzB cloned into pBAD24, Apr | 43 |
| pMF19 | wbbLEcO16 cloned into pEXT21, Spr | 13 |
Ap, ampicillin; Sp, spectinomycin; Km, kanamycin.
Construction of strains and plasmids.
EVV33 was generated by excising the kanamycin gene from EVV16 (W3110 Δwzz::Km) with the FLP recombinase encoded by the thermosensitive plasmid pCP20 (11). pCP20 was introduced by electroporation into EVV16 and cured at 42°C (11). The resulting strain, EVV33, was sensitive to kanamycin due to excision of the antibiotic resistance gene in Δwzz::Km and was also sensitive to ampicillin due to loss of pCP20. Plasmid pEH1, encoding the Flag epitope fused to the C terminus of Wzz (WzzFlag), was constructed by PCR amplification of a 1-kb fragment containing wzz from plasmid pEV6 with primers containing restriction sites (underlined) for NcoI (5′-GACTCCATGGCACGGACTTGGAAATTTCCG-3′) and SalI (5′-GACTGTCGACTTCGCGTTGTAATTGCGT-3′). The PCR product was ligated (Rapid ligation kit; Roche Diagnostics) to pBADFlag also digested with NdeI and SalI, and the ligation mixture was introduced by electroporation into DH5α competent cells. Plasmid pEH3, encoding the Wzz periplasmic loop (WzzPL) fused to one six-His tag at each terminus of the protein (WzzPL2-6×His), was constructed by amplification of a 732-bp fragment encoding 244 amino acids from Lys65 to Ala308 of the parental Wzz protein from E. coli K-12 using pEV6 as the DNA template and primers with SfuI (5′-CAGTTTCGAAAAGGAGAAATGGACGTCAAC-3′) and XhoI (5′-GACTCTCGAGTGCCTTTTTCGGGCTATCACG-3′) restriction sites (underlined). The PCR product and plasmid pET30a were digested with SfuI and XhoI and ligated with T4 DNA ligase, and the ligation mixture was introduced into E. coli DH5α competent cells by transformation. Plasmid pCM244, encoding WzzPL with only an N-terminal six-His tag (WzzPL6×His), was constructed by introducing a termination codon in pEH3 using site-directed mutagenesis (QuickChange site-directed mutagenesis kit; Stratagene) as indicated by the supplier. Further amino acid replacements in WzzFlag (pEH1) were also obtained by site-directed mutagenesis.
Purification and characterization of WzzPL.
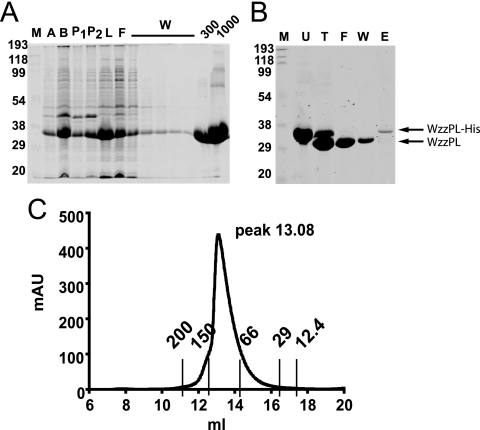
Soluble WzzPL was purified from BL21(DE3) cells carrying pCM244. Cultures were grown in LB at 28°C until the optical density at 600 nm was 0.3, at which point isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.75 mM. Cultures were incubated for additional 3 h, and cells were harvested and suspended in 50 mM Na2PO4 (pH 7) with protease inhibitors (Complete EDTA-free tablets from Roche Diagnostics). The bacterial cell suspension was lysed by two passages with a French press. The lysate was mixed with Sepharose beads (chelating Sepharose Fast Flow; GE Healthcare) previously charged with 0.2 M Ni2SO4 and washed once with binding buffer (50 mM Na2PO4 [pH 7], 300 mM NaCl, 10 mM imidazole). The mixture was incubated with rotation for 2 h at room temperature and loaded on a Poly-Prep chromatography column (Bio-Rad). The loaded column was washed with 10 column volumes of wash buffer (50 mM Na2PO4 [pH 7], 300 mM NaCl, 20 mM imidazole), and the protein was eluted with 1 column volume of elution buffer (50 mM Na2PO4 [pH 7], 300 mM NaCl) containing 300 and 1,000 mM imidazole. Samples were collected at each step and separated by 14% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The purified protein was dialyzed against 50 mM Na2PO4 (pH 7), 100 mM NaCl and concentrated using a Vivaspin 20-ml concentrator (Sartorius AG) with a molecular mass cutoff of 5,000 Da. The N-terminal six-His tag was removed by overnight digestion at room temperature with 3 U of thrombin (Novagen) per mg of protein. Thrombin-treated samples were repurified with Ni2+-charged beads, but this time the flowthrough contained purified WzzPL. To assess the extent of thrombin digestion, the column was washed, and the undigested WzzPL-His6 was eluted as described above. Three milligrams of purified protein was applied to a Superdex 200 10/300 GL (GE Healthcare) column equilibrated with buffer containing 50 mM Na2PO4 (pH 7) and 100 mM NaCl. The column was connected to an AKTA fast protein liquid chromatography apparatus (Amersham Biosciences Corp.) and eluted with the same buffer. The column was calibrated using 12- to 200-kDa gel filtration molecular mass markers (MWGF200; Sigma). Purified WzzPL was used for biophysical analyses and also to raise rabbit polyclonal antiserum that was affinity purified for immunoblot analyses (see below).
Analytical ultracentrifugation.
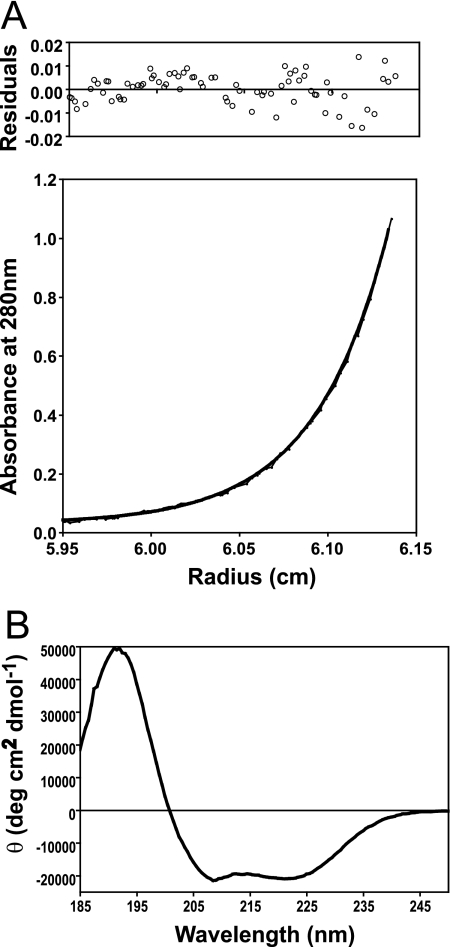
The molecular mass of purified WzzPL was determined by sedimentation equilibrium ultracentrifugation using a Beckman Optima XL-I analytical ultracentrifuge and a 60 Ti analytical rotor with six-channel centerpieces (Biomolecular Interactions and Conformations Facility, University of Western Ontario). The purified protein was dialyzed in 50 mM Na2PO4 (pH 7), 100 mM NaCl. Protein concentrations were determined by measuring the absorbance at 280 nm, and samples containing increasing concentrations (0.127, 0.369, and 0.624 absorbance unit) were used. Centrifugation was performed at 20°C using rotor speeds of 10,000, 15,000, 20,000, 25,000, and 30,000 rpm, and the preparations were equilibrated for a minimum of 18 h before data were collected. For each speed absorbance at 280 or 230 nm was determined in 0.002-cm radial steps, and the values for 10 observations were averaged. Equilibrium was ascertained by comparing scans taken at 1-h intervals. Curve fitting and analysis were performed using software supplied by Beckman (Origin 6.0). Partial specific volumes and densities of solvents were calculated using the program SEDNTERP (http://www.jphilo.mailway.com).
CD spectroscopy.
CD studies were performed with a Jasco J-810 spectropolarimeter equipped with a Peltier temperature control system (Biomolecular Interactions and Conformations Facility, University of Western Ontario). A cell with a path length of 0.1 mm was used. Protein samples were dialyzed into buffer containing 10 mM potassium (pH 7.5) and 50 mM NaCl. CD spectra were collected at 5 and 20°C in 0.5- or 1-nm steps at scan speeds of 5 to 20 nm/min for a range of 185 to 215 nm, using a protein concentration of 1.94 mg/ml. CD values were converted to mean residue ellipticity in units of degree cm2/dmol by using standard procedures based on the intensity of the mean residue ellipticity signals at the maxima and minima at ∼195, 208, and 222 nm.
LPS and protein analysis.
LPS was prepared as previously described from cells grown on LB plates without or with 0.2% (wt/vol) arabinose, and samples were separated by 12.5% (wt/vol) Tris-glycine SDS-PAGE. Gels were stained with silver nitrate as described previously (24, 27). A densitometry analysis of silver-stained gels was performed using the program ImageJ (1). Total membranes were prepared from cells grown using the conditions described above. Bacterial cells were suspended in 20 mM Na2PO4 (pH 7.4) with protease inhibitors (Complete tablets; Roche Diagnostics), and they were lysed by sonic disruption using two 15-s pulses (Branson). Total membrane fractions were obtained by centrifugation of the lysates for 40 min at 40,000 × g, and the pellet was resuspended in the same buffer. The protein concentration was determined by the Bradford assay (Bio-Rad protein assay). Twenty to 40 μg of total membrane proteins was incubated for 30 min at 45°C, separated on a 14% SDS-PAGE gel, and transferred to nitrocellulose membranes that were reacted with either anti-WzzPL affinity-purified polyclonal rabbit antibodies or anti-Flag monoclonal antibodies (Sigma). The reacting bands were detected by fluorescence with an Odyssey infrared imaging system (Li-cor Biosciences) using IRDye800CW affinity-purified anti-rabbit immunoglobulin G (IgG) antibodies (Rockland, Pennsylvania) and Alexa Fluor 680 anti-mouse IgG antibodies (Molecular Probes). Densitometry analysis was performed using the Odyssey software and digital images of the gel membranes, which were incubated simultaneously with both anti-WzzPL and anti-Flag antibodies. This resulted in proteins with two fluorescence colors, and since the anti-Flag antibody cross-reacted with an unknown constitutively expressed membrane protein, we used the density of pixels of this protein band as an internal loading standard.
Limited proteolysis with trypsin.
Forty micrograms of total membrane extract prepared as described above was treated with 30 μg/ml trypsin (Roche Diagnostics) in the presence of 1 mM CaCl2 for 2 h or overnight at 35°C. The digestion was stopped by incubation for 10 min at room temperature with 0.1 mM soybean trypsin inhibitor (Roche Diagnostics). Samples were separated on an 18% SDS-PAGE gel, and proteins were transferred onto a nitrocellulose membrane that was incubated with anti-WzzPL antibodies as described above.
RESULTS AND DISCUSSION
Analysis of purified WzzPL suggests that it is a monomer with an extended conformation.
The periplasmic loop of Wzz accounts for more than 85% of the protein and is absolutely required for its function (10). Therefore, we attempted to purify this loop as a prior step to obtain structural information. Soluble WzzPL was purified from BL21(DE3) cells carrying pCM244 (encoding WzzPL6×His) by Ni2+ affinity chromatography and elution with 1 M imidazole (Fig. 1A). The purified WzzPL6×His was treated with thrombin to remove the N-terminal six-His tag and was repurified as described in Materials and Methods. Figure 1B (lanes U and T) shows that the untreated (WzzPL6×His) and thrombin-treated (WzzPL) polypeptides had apparent masses of 35 and 32 kDa. To assess the efficiency of the thrombin digestion and the degree of purification, the column was washed and the undigested WzzPL6×His was eluted (Fig. 1B, lanes F, W, and E). To further characterize WzzPL, the purified protein was concentrated and analyzed by gel filtration chromatography using a size exclusion column. The results showed that there was a single peak with a broad base eluting at 13.08 ml, which corresponded to a molecular mass of approximately 100 kDa, as calculated by extrapolation from a calibration curve obtained by plotting the elution peaks of molecular mass standards run under the same buffer conditions (Fig. 1C). We interpreted these results as an indication that WzzPL could be aggregated or, alternatively, is a monomer with an extended conformation. The latter possibility was also supported by the presence of a broad elution peak (Fig. 1C). To investigate both possibilities, we analyzed WzzPL by sedimentation equilibrium using an analytical ultracentrifuge (Fig. 2A). Ranges of protein concentrations and rotor speeds were used, and data analysis was carried out by assuming a model with a single ideal species. At initial concentrations between 0.21 and 1.04 mg/ml and rotor speeds between 10,000 and 30,000 rpm, WzzPL gave good fits for a single species with a molecular mass of 38,902 ± 3,590 Da (average ± standard deviation of the mean) (Table 2). A fit of the data collected at 30,000 rpm to the single-species model revealed essentially randomly distributed residuals (Fig. 2A). The results of the analytical centrifugation experiments, combined with the behavior of WzzPL in gel filtration chromatography, strongly suggest that this protein is a monomer that folds into a molecule with an extended configuration. CD spectroscopy was used to analyze the secondary structure of purified WzzPL in solution. The CD spectrum (Fig. 2B) had prominent minima at 222 and 208 nm, with a signal intensity at 222 nm of −20,911° cm2/dmol, indicating the presence of a substantial helical structure. Analysis of the spectra using SELCON 3 (39) indicated that the helical content was 60% and the β-sheet content was 5%. The CD spectrum of WzzPL is practically identical to the spectrum reported for the complete Wzz protein of E. coli O86 (16), suggesting that WzzPL has the same conformation as the native protein.
FIG. 1.
Purification of WzzPL6×His. (A) BL21(DE3)/pCM244 cells were induced with 0.75 mM IPTG, WzzPL6×His was purified using Ni2+-Sepharose beads, and the protein was eluted from the column with imidazole. Aliquots were removed at each step of the purification procedure and separated on a 14% SDS-PAGE gel stained with PageBlue (Fermentas). Lane M, broad-range prestained SDS-PAGE standard (Bio-Rad); lane A, cells boiled after induction; lane B, lysate obtained by French press treatment; lane P1, pellet after the first centrifugation at low speed; lane P2, pellet after the second centrifugation at high speed; lane L, supernatant after centrifugation; lane F, flowthrough; lanes W, washes with 50 mM Na2PO4 (pH 7), 300 mM NaCl, 20 mM imidazole; lane 300, elution with 50 mM Na2PO4 (pH 7), 300 mM NaCl, 300 mM imidazole; lane 1000, elution with 50 mM Na2PO4 (pH 7), 300 mM NaCl, 1,000 mM imidazole. (B) Removal of six-His tag from purified WzzPL6×His. Purified WzzPL6×His was incubated overnight with 3 U thrombin per mg of protein. Aliquots were removed at each step of the purification procedure and separated on a 14% SDS-PAGE gel, and the gels were stained with PageBlue (Fermentas). Lane M, broad-range prestained SDS-PAGE standard (Bio-Rad); lane U, untreated; lane T, treated with thrombin; lane F, flowthrough; lane W, washes with 50 mM Na2PO4 (pH 7), 300 mM NaCl, 20 mM imidazole; lane E, elution with 50 mM Na2PO4 (pH 7), 300 mM NaCl, 1,000 mM imidazole. (C) Size exclusion chromatography profile. Three milligrams of purified WzzPL protein was applied to a Superdex 200 10/300 GL column equilibrated with 50 mM Na2PO4 (pH 7), 100 mM NaCl. The column was calibrated with the following MWGF-200 (Sigma) molecular mass markers: cytochrome c (12.4 kDa; 17.29 ml), carbonic anhydrase (29 kDa; 16.05 ml), albumin (66 kDa; 14.02 ml), alcohol dehydrogenase (150 kDa; 12.47 ml), and β-amylase (200 kDa; 12.5 ml). The height of the peak depended on the relative concentration of the sample loaded into the column. The elution peak (13.08 ml) corresponds to a molecular mass of approximately 100 kDa. The y axis indicates the absorption at 280 mm (in milliabsorbance units [mAU]) as determined with the detector. The x axis indicates the volume (in milliliters) passed over the column.
FIG. 2.
(A) Sedimentation equilibrium analysis of the soluble WzzPL. Sedimentation equilibrium data were collected at 280 nm for a sample with an initial concentration of 0.62 mg/ml at a rotor speed of 30,000 rpm. The data set was fitted to a single ideal species model. Residuals, which are randomly distributed, are shown in the upper graph. (B) CD spectrum of WzzPL at a concentration of 1.94 mg/ml collected at 20°C as described in Materials and Methods. Similar results were obtained at 5°C.
TABLE 2.
Molecular weight of the Wzz loop as determined by sedimentation equilibriuma
| Rotor speed (rpm) | Calculated mol wt |
|---|---|
| 10,000 | 40,057 |
| 15,000 | 42,217 |
| 20,000 | 41,315 |
| 25,000 | 37,670 |
| 30,000 | 33,255 |
Sedimentation equilibrium analytical ultracentrifugation was carried out as described in Materials and Methods. The initial protein concentration of WzzPL was 0.62 mg/ml. The molecular weight that provided the best fit of the data to a single ideal species model is shown.
The demonstration that purified WzzPL is a monomer does not preclude the possibility that Wzz forms multimers in vivo, as has been shown previously by cross-linking experiments (9, 10) involving the complete Wzz protein. Daniels and Morona (10) also demonstrated that the transmembrane helices of Wzz play a role in the ability of this protein to oligomerize.
Functional analysis of three putative coil-coiled regions in the Wzz periplasmic loop.
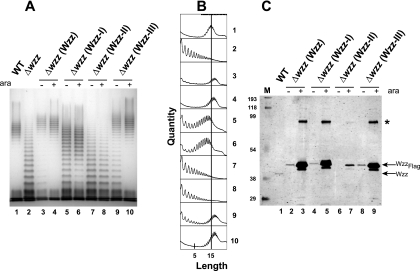
The periplasmic loop of all Wzz proteins has three regions predicted to be coiled coils (30). Coiled coils consist of heptad repeats with residues labeled a through g (6, 29, 38) and consensus specific hydrophobic residues at the a and d positions, which are critical for the coiled-coil interactions (44). In the E. coli K-12 Wzz sequence these predicted regions are at positions 108 to 130 (region I), 153 to 173 (region II), and 209 to 223 (region III) (Fig. 3). To investigate whether the predicted coiled coils are essential for Wzz function, we generated mutants with double Phe replacements in the amino acids at the predicted a and d positions in each of the three putative coiled-coil regions of Wzz (Fig. 3). We anticipated that Phe residues at these positions would retain the hydrophobicity of the coil but would disrupt helix-helix interactions due to their “bulkiness” (38). The following Wzz proteins with double amino acid replacements were constructed as C-terminal Flag epitope fusions and expressed under control of the arabinose-inducible PBAD promoter: Wzz-I (WzzL118F/L122F, encoded by pEH6), Wzz-II (WzzA156F/L160F, encoded by pEH4), and Wzz-III (WzzI212F/L216F, encoded by pEH5). Strain EVV33 (W3110 Δwzz) containing plasmid pMF19 was transformed with each of the plasmids expressing the Wzz derivatives and pEH1 expressing the parental WzzFlag protein. Plasmid pMF19 encodes the rhamnosyltransferase WbbL, which allows reconstitution of O16 antigen synthesis in strains belonging to the W3110 lineage (13, 25, 26). Although the PBAD promoter is sufficiently leaky to express high enough levels of proteins without added arabinose to allow complementation (see below), cultures were grown with and without 0.2% (wt/vol) arabinose to ensure full expression of the recombinant Wzz proteins. Figure 4A shows that deletion of wzz resulted in loss of the characteristic pattern of O-antigen polymerization (Fig. 4, lane 1), resulting in uniform accumulation of O-antigen polymers with lower molecular masses (lane 2). This phenotype could be complemented by plasmid pEH1 expressing wild-type Wzz (lanes 3 and 4). In contrast, expression of Wzz-I produced an O-antigen banding pattern intermediate between that of the parental strain W3110 and that of the wzz mutant (lanes 5 and 6). Expression of Wzz-II did not complement the O-antigen banding pattern of the Δwzz mutant even when cells were grown with 0.2% arabinose (lanes 7 and 8), suggesting that the Phe replacements in this protein compromised the Wzz function. Expression of Wzz-III under arabinose induction conditions restored the wild-type O-antigen banding pattern (lane 10), while no induction still resulted in low-molecular-weight molecules (lane 9), suggesting that the amino acid replacements in region III have a minor effect on Wzz function. The various O-antigen banding patterns detected by silver staining (Fig. 4A) were quantified and confirmed by densitometry (Fig. 4B), taking into account the height of the peaks corresponding to each band on the y axis (quantity) and the number of subunits in the polysaccharide chains on the x axis (chain length distribution). Furthermore, increased numbers of longer O-antigen molecules were observed in cells expressing Wzz and Wzz-III (Fig. 4A, lanes 3, 4, 9, and 10) compared to strain W3110 containing the cloning vector and expressing chromosomal levels of Wzz (Fig. 4A, lane 1). This result indicates that overexpression of a fully functioning Wzz protein leads to longer O-antigen polymers.
FIG. 3.
Amino acid sequences of the predicted coiled-coil regions of E. coli K-12 Wzz. The amino acid positions (positions a through g) of the heptad repeats are indicated below the amino acid sequences. Asterisks indicate the mutated amino acids that correspond to the a and d positions in each predicted seven-amino-acid heptad coil.
FIG. 4.
(A) LPS production in E. coli K-12 strain W3110 (WT) and isogenic mutant EVV33 (Δwzz) complemented with plasmids expressing wild-type Wzz (Wzz) and forms with mutations in each of the coiled-coil regions (Wzz-I, Wzz-II, and Wzz-III). LPS was prepared from cultures not induced and induced with 0.2% (wt/vol) arabinose (ara). Samples were separated on a 12.5% Tris-glycine SDS-PAGE gel, and the gel was stained with silver. Lane 1, W3110(pMF19); lane 2, EVV33(pMF19); lanes 3 and 4, EVV33(pMF19/pEH1); lanes 5 and 6, EVV33(pMF19/pEH6); lanes 7 and 8, EVV33(pMF19/pEH4); lanes 9 and 10, EVV33(pMF19/pEH5). (B) Densitometric analysis of samples shown in panel A. The analysis was performed with the program ImageJ as described in Materials and Methods. The vertical line in all graphs indicates the mean chain length of wild-type strain W3110(pMF19). The numbers on the right correspond to the lane numbers in panel A. (C) Wzz expression in cells cultured under the conditions used for the LPS extracts (see Materials and Methods). Total membranes were separated on a 14% SDS-PAGE gel. Proteins were transferred to a nitrocellulose membrane, and the blot was reacted with affinity-purified rabbit polyclonal anti-WzzPL and anti-Flag antisera. WzzPL-specific bands were detected by fluorescence using IRDye800CW anti-rabbit IgG antibodies, and Flag-specific bands were detected with Alexa Fluor 680 anti-mouse IgG antibodies. Lane M, broad-range prestained SDS-PAGE standard (Bio-Rad); lane 1, W3110(pMF19/pBADFlag); lanes 2 and 3, EVV33(pMF19/pEH1); lanes 4 and 5, EVV33(pMF19/pEH6); lanes 6 and 7, EVV33(pMF19/pEH4); lanes 8 and 9, EVV33 (pMF19/pEH5). The arrows indicate the migration positions of plasmid-encoded WzzFlag and chromosomally encoded Wzz. The asterisk indicates the migration position of a Wzz dimer.
To determine whether the defective O-antigen LPS profiles were due to differences in protein expression, total membranes were examined by Western blotting with WzzPL- and Flag-specific antibodies. It should be noted that the chromosomally encoded Wzz protein has a lower apparent molecular mass than the recombinant Wzz proteins with the mutations in regions I, II, and III, as the latter proteins are fused to the Flag epitope (Fig. 4C). This experiment showed that EVV33 (Δwzz) cells grown with no arabinose expressed WzzFlag, Wzz-I, and Wzz-III at about the same levels as W3110 cells expressing chromosomal levels of Wzz (Fig. 4C, lanes 1, 2, 4, and 8). Densitometry of the protein bands compared to the chromosomal Wzz band (1 relative unit), performed as described in Materials and Methods, resulted in 1.3 relative units for WzzFlag and Wzz-I and 1.1 relative units for Wzz-III. In EVV33 cultures grown with 0.2% (wt/vol) arabinose, the levels of expression of Wzz, Wzz-I, and Wzz-III relative to the level of expression of chromosomally expressed Wzz increased approximately 40-fold (44, 49, and 38 relative densitometric units, respectively). Therefore, we concluded that the O-chain length phenotypes observed with the Wzz-I and Wzz-III proteins were not due to differences in protein expression. In contrast, the level of Wzz-II was drastically reduced; Wzz-II was undetectable without arabinose, and the level increased only threefold relative to the level of chromosomally expressed Wzz with arabinose induction (Fig. 4C, lanes 6 and 7). No additional Wzz-II protein was found in total lysates, ruling out the possibility of a defect in membrane incorporation that could lead to protein accumulation in the cytosol (data not shown). We concluded from these results that the double amino acid replacements in Wzz-II had a drastic effect on protein stability. However, protein instability alone cannot explain the loss of function of this mutant protein, as its induced level of expression was equal to or slightly higher than the level of expression of the chromosomally expressed Wzz (Fig. 4C, compare lanes 1 with 7), yet this protein could not complement the phenotype of the Δwzz strain (Fig. 4A, lanes 7 and 8).
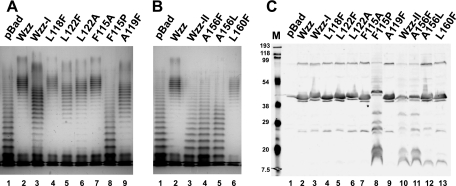
To assess whether the changes in the LPS profile observed with the double Phe substitutions were due to the combined effects of the double mutations, mutants with single replacements at each position were generated, and the LPS patterns determined by plasmids expressing the corresponding mutant Wzz proteins were analyzed. Of the mutations in region I, only L122F (Fig. 5A, lane 5) was responsible for the LPS phenotype observed for the corresponding double mutant (Fig. 5A, lane 3). In contrast, the LPS profile of cells expressing L118F (Fig. 5A, lane 4) resulted in an O-antigen pattern similar to that mediated by the wild-type Wzz (Fig. 5A, lane 2). We also replaced L122 with alanine (Ala) to see if introduction of a smaller hydrophobic amino acid at this position would affect the O-antigen profile. The O-antigen profile mediated by WzzL122A was partially restored to the wild-type phenotype (Fig. 5A, lane 6). These results suggest that L122, but not L118, in Wzz-I is important for the proper function of Wzz. Of the mutations in region II, only A156F altered the Wzz function (Fig. 5B, lanes 1 and 4), while L160F mediated an O-antigen profile identical to that of wild-type Wzz (Fig. 5B, lanes 2 and 6). Replacement by leucine (a larger hydrophobic amino acid) at position 156 (A156L) did not restore the function (Fig. 5B, lane 5). The immunoblot analysis showed that although both substitutions at Ala156 rendered the mutant protein nonfunctional, only the WzzA156F mutant had the same degradation profile as the WzzA156F/L160F double Phe mutant (Fig. 5C, lanes 10 and 11), while the WzzA156L mutant resembled the WzzL160F mutant (Fig. 5C, lanes 12 and 13). This observation further supports the notion that A156 is important for both the stability and function of Wzz.
FIG. 5.
LPS profiles and protein expression for Wzz mutants with single amino acids replacements. LPS was prepared from cultures with 0.2% (wt/vol) arabinose. Samples were analyzed as described in the legend to Fig. 3. All the plasmids were introduced into EVV33(pMF19) by transformation. (A) Lane 1, pBADFlag; lane 2, pEH1 (Wzz); lane 3, pEH6 (Wzz-I); lane 4, pEH1-L118F; lane 5, pEH1-L122F; lane 6, pEH1-L122A; lane 7, pEH1-F115A; lane 8, pEH1-F115F; lane 9, pEH1-A116F. (B) Lane 1, pBADFlag; lane 2, pEH1 (Wzz); lane 3, pEH4 (Wzz-II); lane 4, pEH1-A156F; lane 5, pEH1-A156L; lane 6, pEH1-L160F. (C) Twenty micrograms of total membranes was separated on a 14% SDS-PAGE gel and transferred to a nitrocellulose membrane, and the blot was incubated with anti-Flag monoclonal antibodies. The specific bands were detected by fluorescence using Alexa Fluor 680 anti-mouse IgG antibodies. Lane M, broad-range prestained SDS-PAGE standard (Bio-Rad); lane 1, pBADFlag; lane 2, pEH1 (Wzz); lane 3, pEH6 (Wzz-I); lane 4, pEH1-L118F; lane 5, pEH1-L122F; lane 6, pEH1-L122A; lane 7, pEH1-F115A; lane 8, pEH1-F115F; lane 9, pEH1-A116F; lane 10, pEH4 (Wzz-II); lane 11, pEH1-A156F; lane 12, pEH1-A156L; lane 13, pEH1-L160F.
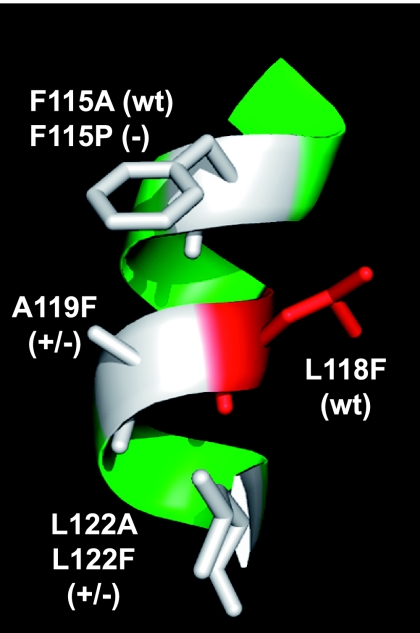
Computer modeling of the Wzz proteins was carried out with a combination of HHPRED and MODELLER using the online Bioinformatics Toolkit of the Max Planck Institute for Developmental Biology (http://toolkit.tuebingen.mpg.de/sections/tertstruct). The model predicted that region I spans an α-helix (Fig. 6). The model also predicted that F115, A119, and L122 are located on the same face of the α-helix, while L118 is on the opposite face (Fig. 6). To analyze whether all the residues predicted to be on the same face of the helix were involved in altering the LPS pattern, we replaced F115 with Ala and proline (Pro) and Ala119 with Phe. The WzzF115A derivative did not mediate any changes in the O-antigen profile compared to the wild type (Fig. 5A, lanes 2 and 7). Moreover, WzzF115P resulted in a loss of function, conferring an O-antigen profile similar to that of mutant strain EVV33 (Fig. 5A, lanes 1 and 8). The F115P substitution also had a dramatic effect on protein expression (Fig. 5C, lane 8), as revealed by the anomalous gel migration of WzzF115P and its higher level of degradation, which was similar to that seen with Wzz substitutions in region II (Fig. 5C, lanes 10 and 11). These results indicate that the F115P substitution causes a significant structural change in Wzz that renders the mutant protein more prone to degradation. Also, the profound effect of the Pro substitution supports the notion that region I is α-helical, since it is well known that proline residues can completely destroy the helicity of a helix. The A119F substitution resulted in only partial restoration of the wild-type phenotype, similar to the LPS pattern observed with L122F, indicating that this residue is also involved in the proper Wzz function (Fig. 5A, lanes 5 and 9). The L118F substitution did not affect either the LPS profile or protein expression, suggesting that this residue lies on the opposite side of the α-helix and might not contribute to Wzz function. Together, our results support the notion that of the three residues on the same face of the predicted α-helix in region I, only A119 and L122 are important for function. Replacement of F115 by a smaller hydrophobic residue (Ala) did not affect the function. Furthermore, L118, which is predicted to occupy the d position of the predicted region I coiled coil, is not involved in Wzz function. This result argues strongly against the existence of a coiled-coil structure in this region. A similar conclusion can be drawn for the predicted coiled coil of region II, since L160 occupying the a position also does not have a functional role.
FIG. 6.
Region I model prediction. The amino acids indicated are all on the same face of the helix and were targeted for mutagenesis. (wt) indicates that the amino acid substitution did not alter the O-antigen chain length distribution; (+/−) indicates that the amino acid substitution resulted in a partially defective O-antigen chain length distribution; (−) indicates that the amino acid substitution eliminated Wzz function.
Limited proteolysis suggests conformational changes in Wzz mutants.
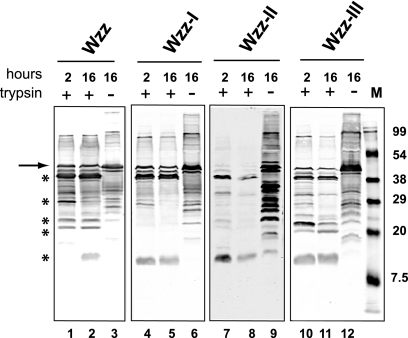
The results obtained with the single and double amino acid replacements suggested that the different LPS profiles observed might have been due to changes in folding of the mutant proteins, especially in the periplasmic loop region. To address this hypothesis, we exposed total membrane proteins extracted from cells expressing Wzz or each of the Wzz-I, Wzz-II, and Wzz-III double-substitution mutants to 30 μg/ml trypsin at 35°C for either 2 or 16 h. As a negative control, untreated membranes were also incubated without trypsin for the same amount of time. Samples were examined by Western blotting using anti-WzzPL antibodies. Some degree of background proteolysis was observed in the untreated control samples (Fig. 7, lanes 3, 6, 9, and 12), which probably resulted from the activity endogenous proteases on the Wzz polypeptide under our experimental conditions. In contrast, a characteristic profile determined by five distinct degradation products was observed in Wzz samples even after 16 h of digestion (Fig. 7, lanes 1 and 2). Analysis of the wild-type Wzz primary sequence revealed 34 predicted trypsin cleavage sites (data not shown). Therefore, we concluded that most of the trypsin cleavage sites in Wzz are not accessible to the protease, as they are likely masked by the secondary and tertiary conformations of the native protein. This also explains the limited but stable proteolytic profile even after long incubations with trypsin. In the preparation containing the Wzz-I double-substitution protein, we detected only three of the five wild-type Wzz degradation products (Fig. 7, lanes 4 and 5), while the remaining two products appeared faintly only when the blot was scanned at a higher density (data not shown). We reasoned that some of the trypsin cleavage sites that were exposed in the wild-type Wzz were not accessible in Wzz-I, possibly reflecting a change in the secondary structure. In contrast, the digestion pattern of the Wzz-III mutant protein was similar to that of the parental Wzz protein, except that the smaller digestion product was present after 2 h of incubation with trypsin (Fig. 7, lanes 10 and 11). This result suggests that the double substitution causes a minor defect in conformation, but as shown previously, this defect cannot alter the function of the protein in a dramatic way. The Wzz-II mutant protein blot was scanned at a higher density to facilitate detection of the trypsin digestion pattern for this protein, since the untreated sample exhibited substantial proteolysis (Fig. 7, lane 9). This result strongly suggests that the amino acid replacements in region II caused dramatic secondary structure changes that compromised protein stability. Together, the results of these experiments support the hypothesis that substitution of critical amino acids in Wzz-I and Wzz-II compromises, to some extent, the proper folding of native Wzz, while only a minor defect occurs in Wzz-III. These observations agree with the functional analysis data based on complementation of O-antigen chain length regulation, as described above. Furthermore, immunoblot analysis showed that none of the mutations in region I compromised the expression or stability of the Wzz protein (Fig. 5C, lanes 2 and 7).
FIG. 7.
Limited trypsin proteolysis of membranes containing Wzz proteins and the double-replacement mutants with mutations in regions I, II, and III. Forty micrograms of total membranes was incubated at 35°C with 30 μg/ml trypsin for 2 or 16 h. Untreated samples were also incubated for 16 h. Lane M contained the broad-range prestained SDS-PAGE standard (Bio-Rad). Asterisks indicate Wzz products, and the arrow indicates undigested Wzz. Samples were separated on an 18% SDS-PAGE gel. Proteins were transferred to a nitrocellulose membrane, and the blot was reacted with affinity-purified rabbit polyclonal anti-WzzPL and developed by fluorescence using IRDye800CW anti-rabbit IgG antibodies.
Conclusions.
Here we describe a functional analysis of predicted coiled-coil regions in the E. coli K-12 Wzz protein. The results obtained with amino acid substitutions, complementation of O-antigen expression profiles, and expression of the mutated proteins themselves strongly argue that the predicted coiled coils in regions I and II probably do not exist as such. However, these regions contain residues that have a functional role and are required to maintain protein conformation. Thus, our results support the conclusion that there are no functional coiled coils in Wzz, as predicted by bioinformatic analysis, just α-helical structures. Attempts were made to purify WzzPL forms in which mutations in regions I and II were recreated, but the proteins could not be purified in their native forms since they were in inclusion bodies (data not shown). These results also support the hypothesis that these regions are critical for proper folding.
Amino acid replacements in region I provided the most interesting results, since the mutated protein was expressed at the same level as the parental Wzz protein, did not have gross defects in conformation as determined by limited trypsin proteolysis, and showed an intermediate defect in O-antigen chain length determination. In contrast, the mutations in region II eliminated the function, and they were clearly associated with gross defects in secondary structure and protein stability. Further confirmation of our results awaits elucidation of the Wzz structure by high-resolution methods.
Acknowledgments
We thank L. K. Briere of the Biomolecular Interactions and Conformations Facility, University of Western Ontario, for help with the analytical centrifugation and CD spectroscopy, as well as R. Morona, J. Orban, K. Patel, and M. S. Saldías for useful discussions.
This study was supported by a grant from the Canadian Institutes of Health Research. M.L. was supported by two University Summer Research Awards from the Natural Sciences and Engineering Research Council of Canada. E.R.H. was supported by an Ontario Graduate Student Award in Science and Technology. M.A.V. holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis.
Footnotes
Published ahead of print on 18 January 2008.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 1136-42. [Google Scholar]
- 2.Bastin, D. A., G. Stevenson, P. K. Brown, A. Haase, and P. R. Reeves. 1993. Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol. Microbiol. 7725-734. [DOI] [PubMed] [Google Scholar]
- 3.Batchelor, R. A., G. E. Haraguchi, R. A. Hull, and S. I. Hull. 1991. Regulation by a novel protein of the bimodal distribution of lipopolysaccharide in the outer membrane of Escherichia coli. J. Bacteriol. 1735699-5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bronner, D., B. R. Clarke, and C. Whitfield. 1994. Identification of an ATP-binding cassette transport system required for translocation of lipopolysaccharide O-antigen side-chains across the cytoplasmic membrane of Klebsiella pneumoniae serotype O1. Mol. Microbiol. 14505-519. [DOI] [PubMed] [Google Scholar]
- 5.Burda, P., and M. Aebi. 1999. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426239-257. [DOI] [PubMed] [Google Scholar]
- 6.Burkhard, P., S. Ivaninskii, and A. Lustig. 2002. Improving coiled-coil stability by optimizing ionic interactions. J. Mol. Biol. 318901-910. [DOI] [PubMed] [Google Scholar]
- 7.Collins, R. F., K. Beis, B. R. Clarke, R. C. Ford, M. Hulley, J. H. Naismith, and C. Whitfield. 2006. Periplasmic protein-protein contacts in the inner membrane protein Wzc form a tetrameric complex required for the assembly of Escherichia coli group 1 capsules. J. Biol. Chem. 2812144-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, R. F., K. Beis, C. Dong, C. H. Botting, C. McDonnell, R. C. Ford, B. R. Clarke, C. Whitfield, and J. H. Naismith. 2007. The 3D structure of a periplasm-spanning platform required for assembly of group 1 capsular polysaccharides in Escherichia coli. Proc. Natl. Acad. Sci. USA 1042390-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels, C., C. Griffiths, B. Cowles, and J. S. Lam. 2002. Pseudomonas aeruginosa O-antigen chain length is determined before ligation to lipid A core. Environ. Microbiol. 4883-897. [DOI] [PubMed] [Google Scholar]
- 10.Daniels, C., and R. Morona. 1999. Analysis of Shigella flexneri wzz (Rol) function by mutagenesis and cross-linking: Wzz is able to oligomerize. Mol. Microbiol. 34181-194. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 166127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldman, M. F., C. L. Marolda, M. A. Monteiro, M. B. Perry, A. J. Parodi, and M. A. Valvano. 1999. The activity of a putative polyisoprenol-linked sugar translocase(Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J. Biol. Chem. 27435129-35138. [DOI] [PubMed] [Google Scholar]
- 14.Feldman, M. F., M. Wacker, M. Hernandez, P. G. Hitchen, C. L. Marolda, M. Kowarik, H. R. Morris, A. Dell, M. A. Valvano, and M. Aebi. 2005. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. USA 1023016-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez, F., J. S. Rush, D. A. Toke, G. Han, J. E. Quinn, G. M. Carman, J.-Y. Choi, D. R. Voelker, M. Aebi, and C. J. Waechter. 2001. The CWH8 gene encodes a dolichyl pyrophosphate phosphatase with a luminally oriented active site in the endoplasmic reticulum of Saccharomyces cerevisiae. J. Biol. Chem. 27641455-41464. [DOI] [PubMed] [Google Scholar]
- 16.Guo, H., K. Lokko, Y. Zhang, W. Yi, Z. Wu, and P. G. Wang. 2006. Overexpression and characterization of Wzz of Escherichia coli O86:H2. Protein Expr. Purif. 4849-55. [DOI] [PubMed] [Google Scholar]
- 17.Heinrichs, D. E., M. A. Valvano, and C. Whitfield. 1999. Biosynthesis and genetics of lipopolysaccharide core, p. 305-330. In H. Brade, D. C. Morrison, S. Vogel, and S. Opal (ed.), Endotoxin in health and disease. Marcel Dekker, Inc., New York, NY.
- 18.Heinrichs, D. E., J. A. Yethon, and C. Whitfield. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 30221-232. [DOI] [PubMed] [Google Scholar]
- 19.Jansson, P.-E. 1999. The chemistry of O-polysaccharide chains in bacterial lipopolysaccharides, p. 155-178. In H. Brade, D. C. Morrison, S. Vogel, and S. Opal (ed.), Endotoxin in health and disease. Marcel Dekker, Inc., New York, NY.
- 20.Keenleyside, W. J., and C. Whitfield. 1999. Genetics and biosynthesis of lipopolysaccharide O-antigens, p. 331-358. In H. Brade, D. C. Morrison, S. Vogel, and S. Opal (ed.), Endotoxin in health and disease. Marcel Dekker, Inc., New York, NY.
- 21.Liu, D., R. A. Cole, and P. R. Reeves. 1996. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J. Bacteriol. 1782102-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21375-382. [PubMed] [Google Scholar]
- 23.Marino, P. A., B. C. McGrath, and M. J. Osborn. 1991. Energy dependence of O-antigen synthesis in Salmonella typhimurium. J. Bacteriol. 1733128-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marolda, C. L., P. Lahiry, E. Vinés, S. Saldías, and M. A. Valvano. 2006. Micromethods for the characterization of lipid A-core and O-antigen lipopolysaccharide. Methods Mol. Biol. 347237-252. [DOI] [PubMed] [Google Scholar]
- 25.Marolda, C. L., L. D. Tatar, C. Alaimo, M. Aebi, and M. A. Valvano. 2006. Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide O antigen. J. Bacteriol. 1885124-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marolda, C. L., J. Vicarioli, and M. A. Valvano. 2004. Wzx proteins involved in O antigen biosynthesis function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology 1504095-4105. [DOI] [PubMed] [Google Scholar]
- 27.Marolda, C. L., J. Welsh, L. Dafoe, and M. A. Valvano. 1990. Genetic analysis of the O7-polysaccharide biosynthesis region from the Escherichia coli O7:K1 strain VW187. J. Bacteriol. 1723590-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGrath, B. C., and M. J. Osborn. 1991. Localization of the terminal steps of O-antigen synthesis in Salmonella typhimurium. J. Bacteriol. 173649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meier, M., A. Lustig, U. Aebi, and P. Burkhard. 2002. Removing an interhelical salt bridge abolishes coiled-coil formation in a de novo designed peptide. J. Struct. Biol. 13765-72. [DOI] [PubMed] [Google Scholar]
- 30.Morona, R., L. Van Den Bosch, and C. Daniels. 2000. Evaluation of Wzz/MPA1/MPA2 proteins based on the presence of coiled-coil regions. Microbiology 1461-4. [DOI] [PubMed] [Google Scholar]
- 31.Morona, R., L. van den Bosch, and P. A. Manning. 1995. Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J. Bacteriol. 1771059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulford, C. A., and M. J. Osborn. 1983. An intermediate step in translocation of lipopolysaccharide to the outer membrane of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 801159-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikaido, H. 1996. Outer membrane, p. 29-47. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology Press, Washington, DC.
- 34.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rick, P. D., K. Barr, K. Sankaran, J. Kajimura, J. S. Rush, and C. J. Waechter. 2003. Evidence that the wzxE gene of Escherichia coli K-12 encodes a protein involved in the transbilayer movement of a trisaccharide-lipid intermediate in the assembly of enterobacterial common antigen. J. Biol. Chem. 27816534-16542. [DOI] [PubMed] [Google Scholar]
- 36.Saldías, M. S., K. Patel, M. Bittner, C. L. Marolda, I. Contreras, and M. A. Valvano. 2008. Distinct functional domains of the Salmonella enterica WbaP transferase that is involved in the initiation reaction for synthesis of the O antigen subunit. Microbiology 154440-453. [DOI] [PubMed] [Google Scholar]
- 37.Samuel, S., and P. Reeves. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 3382503-2519. [DOI] [PubMed] [Google Scholar]
- 38.Smith, T. A., P. M. Steinert, and D. A. Parry. 2004. Modeling effects of mutations in coiled-coil structures: case study using epidermolysis bullosa simplex mutations in segment 1a of K5/K14 intermediate filaments. Proteins 551043-1052. [DOI] [PubMed] [Google Scholar]
- 39.Sreerama, N., and R. W. Woody. 1993. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 20932-44. [DOI] [PubMed] [Google Scholar]
- 40.Tang, K. H., H. Guo, W. Yi, M. D. Tsai, and P. G. Wang. 2007. Investigation of the conformational states of Wzz and the Wzz-O-antigen complex under near-physiological conditions. Biochemistry 4611744-11752. [DOI] [PubMed] [Google Scholar]
- 41.Tatar, L. D., C. L. Marolda, A. N. Polischuk, D. van Leeuwen, and M. A. Valvano. 2007. An Escherichia coli undecaprenyl-pyrophosphate phosphatase implicated in undecaprenyl-phosphate recycling. Microbiology 1532518-2529. [DOI] [PubMed] [Google Scholar]
- 42.Valvano, M. A. 2003. Export of O-specific lipopolysaccharide. Front. Biosci. 8s452-471. [DOI] [PubMed] [Google Scholar]
- 43.Vinés, E., C. L. Marolda, A. Balachandran, and M. A. Valvano. 2005. Defective O-antigen polymerization in tolA and pal mutants of Escherichia coli in response to extracytoplasmic stress. J. Bacteriol. 1873359-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walshaw, J., and D. N. Woolfson. 2001. Socket: a program for identifying and analysing coiled-coil motifs within protein structures. J. Mol. Biol. 3071427-1450. [DOI] [PubMed] [Google Scholar]
- 45.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 311307-1319. [DOI] [PubMed] [Google Scholar]
- 46.Whitfield, C., and M. A. Valvano. 1993. Biosynthesis and expression of cell-surface polysaccharides in gram-negative bacteria. Adv. Microb. Physiol. 35135-246. [DOI] [PubMed] [Google Scholar]