Abstract
Isocitrate dehydrogenase (ICDH) from Hydrogenobacter thermophilus catalyzes the reduction of oxalosuccinate, which corresponds to the second step of the reductive carboxylation of 2-oxoglutarate in the reductive tricarboxylic acid cycle. In this study, the oxidation reaction catalyzed by H. thermophilus ICDH was kinetically analyzed. As a result, a rapid equilibrium random-order mechanism was suggested. The affinities of both substrates (isocitrate and NAD+) toward the enzyme were extremely low compared to other known ICDHs. The binding activities of isocitrate and NAD+ were not independent; rather, the binding of one substrate considerably promoted the binding of the other. A product inhibition assay demonstrated that NADH is a potent inhibitor, although 2-oxoglutarate did not exhibit an inhibitory effect. Further chromatographic analysis demonstrated that oxalosuccinate, rather than 2-oxoglutarate, is the reaction product. Thus, it was shown that H. thermophilus ICDH is a nondecarboxylating ICDH that catalyzes the conversion between isocitrate and oxalosuccinate by oxidation and reduction. This nondecarboxylating ICDH is distinct from well-known decarboxylating ICDHs and should be categorized as a new enzyme. Oxalosuccinate-reducing enzyme may be the ancestral form of ICDH, which evolved to the extant isocitrate oxidative decarboxylating enzyme by acquiring higher substrate affinities.
Isocitrate dehydrogenase (ICDH) (EC 1.1.1.41 and EC 1.1.1.42) is an enzyme that catalyzes the oxidative decarboxylation of isocitrate to 2-oxoglutarate. In prokaryotes, two types of phylogenetically unrelated ICDHs are known: monomeric and oligomeric. Prokaryotic oligomeric ICDH is evolutionarily related to isopropylmalate dehydrogenase (EC 1.1.1.85), tartrate dehydrogenase (EC 1.1.1.93), and homoisocitrate dehydrogenase (EC 1.1.1.87 and EC 1.1.1.286); these enzymes constitute a group called NAD(P)-dependent β-hydroxyacid oxidative decarboxylases or NAD(P)-dependent β-decarboxylating dehydrogenases. The three-dimensional structures of Escherichia coli ICDH (EcICDH), isopropylmalate dehydrogenase from Thermus thermophilus, and homoisocitrate dehydrogenase from T. thermophilus demonstrate that these enzymes share a common fold (11, 12, 17). Prokaryotic ICDH in this group has previously been considered a homodimeric and an NAD(P)-dependent enzyme. However, due to increasing reports of NAD-dependent ICDH in bacteria (Acidithiobacillus, Aquifex, Hydrogenobacter, Methylophilus, and Streptococcus) and archaea (Pyrococcus) and of homotetrameric ICDH in bacteria (Methylococcus and Thermotoga) (3, 6, 7, 9, 13-15, 22-23), prokaryotic oligomeric ICDH is now recognized as an enzyme with various oligomeric states and coenzyme specificities.
Phylogenetic analyses of prokaryotic oligomeric ICDH indicate that this enzyme does not comprise a single lineage but can be divided into many subfamilies (21-23). EcICDH is one of the best analyzed forms and belongs to a distinctive subfamily that also contains ICDH from archaea (Aeropyrum, Archaeoglobus, Caldococcus, and Pyrococcus) and Aquificales (Aquifex and Hydrogenobacter) (3-6, 21-23). These enzymes can be considered a single lineage and can be categorized as EcICDH-type enzymes.
We have previously reported an EcICDH-type enzyme from an organism belonging to the order Aquificales, Hydrogenobacter thermophilus (3). The primary sequence of ICDH from H. thermophilus (HtICDH) is 45.8% identical to that of EcICDH, although its enzymatic characteristics are quite different (3). In particular, the physiological function of HtICDH is distinct from that of EcICDH. While EcICDH is involved in the tricarboxylic acid (TCA) cycle and catalyzes the oxidative decarboxylation of isocitrate, HtICDH is involved in the reductive TCA cycle and catalyzes the reduction of oxalosuccinate (2) (Fig. 1). Thus, differences in the reaction mechanism between these two enzymes were of great interest.
FIG. 1.
Physiological roles of ICDH in E. coli and in H. thermophilus. (A) EcICDH is an enzyme involved in the TCA cycle and catalyzes the oxidative decarboxylation of isocitrate to form 2-oxoglutarate. (B) HtICDH is an enzyme involved in the reductive TCA cycle and catalyzes oxalosuccinate reduction, which corresponds to the second step of the reductive carboxylation of 2-oxoglutarate.
In this study, we analyzed the kinetic mechanism of the oxidation reaction catalyzed by HtICDH. As a result, we clearly demonstrate here that HtICDH is not a conventional decarboxylating ICDH but a novel nondecarboxylating ICDH. Furthermore, we suggest a possible hypothesis concerning the evolutionary history of the prokaryotic oligomeric ICDH where the oxalosuccinate-reducing enzyme is the ancestral form of the decarboxylating ICDHs.
MATERIALS AND METHODS
Enzyme preparation.
Recombinant ICDH from H. thermophilus TK-6 (IAM12695) (HtICDH), E. coli K-12 (EcICDH), and Caldococcus noboribetus (CnICDH) was expressed in E. coli cells and purified as previously described (3, 4). Protein concentrations were determined using bicinchoninic acid protein assay reagent (Pierce) with bovine serum albumin as a standard.
Kinetic analyses.
The steady-state kinetics measurements were performed spectrophotometrically in a solution (total volume of 400 μl) containing 100 mM HEPES-KOH (pH 7.5), 10 mM MgSO4, 100 mM KCl, dl-isocitrate, NAD+, and 7.9 μg of HtICDH at 50°C. The reaction was started by the addition of isocitrate, NAD+, and the enzyme, and the initial velocity was monitored at 340 nm (ɛ = 6.2 mM−1 cm−1) for 30 s.
Kinetic parameters were obtained by varying the dl-isocitrate concentrations (0.4, 0.7, 1.0, and 4.0 mM) at fixed concentrations of NAD+ (2.0, 3.0, and 4.0 mM) and by varying the NAD+ concentrations (0.5, 0.75, 1.0, and 4.0 mM) at fixed concentrations of dl-isocitrate (2.0, 3.0, and 4.0 mM). Product inhibition was analyzed by the addition of different amounts of NADH (0 to 0.02 mM) to the assay mixture while the concentration of NAD+ was fixed at 4 mM, and the dl-isocitrate concentration was varied (0.7, 1.0, 2.0, and 4.0 mM); or the concentration of dl-isocitrate was fixed at 4 mM, and the NAD+ concentration was varied (0.7, 1.0, 2.0, and 4.0 mM).
Nonlinear regression was performed using KaleidaGraph software (Synergy) and the Levenberg-Marquardt algorithm. Kmdl-isocitrate and KmNAD are the Michaelis constants for dl-isocitrate and NAD+, Ksdl-isocitrate and KsNAD are the substrate constants for dl-isocitrate and NAD+, and KiNADH and Ki′NADH are the inhibition constants of NADH toward the enzyme and the binary complex, respectively.
Chromatographic analyses.
The reductive carboxylating activity was confirmed by measuring isocitrate formation using ion chromatography. The reaction was performed in a mixture (total volume of 400 μl) containing 100 mM bicine [N,N-bis(2-hydroxyethyl)glycine]-KOH (pH 6.5), 10 mM MgCl2, 20 mM 2-oxoglutarate, 50 mM NaHCO3, 5 mM NADPH, and ICDH. NADH was used instead of NADPH in the case of HtICDH. The reaction temperatures used were 37°C for EcICDH and 70°C for HtICDH and CnICDH. A total of 7.3 μg of EcICDH, 6.2 μg of CnICDH, and 16 μg of HtICDH was used. After incubation for 5 min, the reaction mixture was cooled on ice-cold water, diluted with MilliQ water (Millipore) if required to yield the optimal chromatogram, and injected onto the column.
Oxalosuccinate released as a product during the oxidation reaction was also detected using ion chromatography. The reaction was performed in a mixture (total volume of 400 μl) containing 100 mM bicine-KOH (pH 9.5), 10 mM MgCl2, 2 mM dl-isocitrate, 2 mM NADP+ (for EcICDH) or 2 mM NAD+ (for HtICDH), and ICDH. Eighteen micrograms of EcICDH and 79 μg of HtICDH were used. After incubation for 20 min at 20°C, the reaction mixture was cooled on ice-cold water, diluted with MilliQ water if required to yield the optimal chromatogram, and injected onto the column.
The chromatographic system and the precise chromatographic conditions used were identical to those described elsewhere (2).
RESULTS
Reductive carboxylation of 2-oxoglutarate.
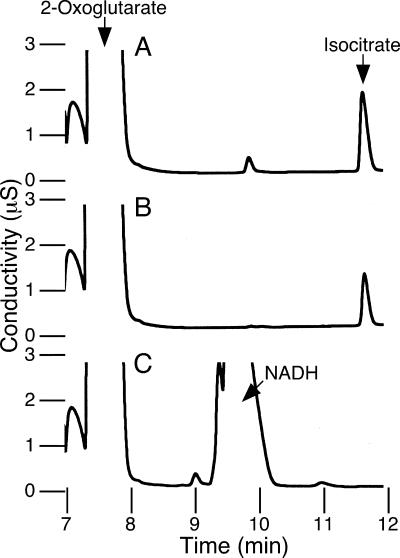
We have previously reported that HtICDH is unable to catalyze the reductive carboxylation of 2-oxoglutarate (3), whereas in the case of EcICDH, by contrast, an ability to catalyze the reductive carboxylation of 2-oxoglutarate has been shown and well analyzed (8). The reversibility of EcICDH was evident from the presence of isocitrate in the chromatogram (Fig. 2A). The ability to catalyze the reductive carboxylation of 2-oxoglutarate seems to be a common feature of the EcICDH-type enzymes since isocitrate formation was also observed when CnICDH, the enzyme from a hyperthermophilic archaeon C. noboribetus (4, 5), was used (Fig. 2B). However, isocitrate formation was not detected when HtICDH was used (Fig. 2C). Therefore, although HtICDH shows high sequence similarity with EcICDH and CnICDH, these results suggest that differences exist between the reaction mechanisms of these enzymes.
FIG. 2.
Reductive carboxylation catalyzed by ICDH. Chromatograms of the mixtures obtained after the reductive carboxylation reaction by EcICDH (incubated for 5 min at 37°C using 7.3 μg of protein) (A), CnICDH (incubated for 5 min at 70°C using 6.2 μg of protein) (B), and HtICDH (incubated for 5 min at 70°C using 16 μg of protein) (C). Reactions were performed in a volume of 400 μl containing 100 mM bicine-KOH (pH 6.5), 10 mM MgCl2, 20 mM 2-oxoglutarate, 50 mM NaHCO3, 5 mM NADPH (for EcICDH and CnICDH) or 5 mM NADH (for HtICDH), and ICDH. After incubation, the reaction mixtures were diluted with water (threefold dilution) before injection onto the column.
Kinetic analysis.
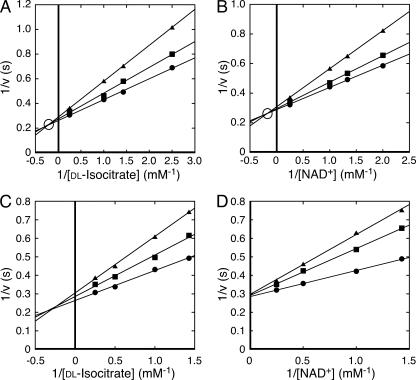
HtICDH utilizes NAD+ instead of NADP+, and the high apparent Km values (for both isocitrate and NAD+) and the low kcat values (3) indicate that it is an ineffective decarboxylating enzyme. In order to elucidate the kinetic mechanism of HtICDH, further precise measurements were carried out. Under the conditions of fixed NAD+ and variable isocitrate concentrations, enzymatic activities were measured by monitoring NAD+ reducing activity. Excess amounts of MgSO4 were added to the assay mixtures so that isocitrate could form a complex with Mg2+ ions. The double-reciprocal plots gave three linear lines with intersections in the second quadrant (Fig. 3A). A similar profile, with intersections in the second quadrant, was also observed when fixed isocitrate and variable NAD+ concentrations were used (Fig. 3B). These observations suggest the possibility of a rapid equilibrium random-order or an ordered mechanism and exclude the possibility of a ping-pong mechanism. Most of the past studies support a random mechanism for this enzyme irrespective of its origin (8). If a random mechanism is also true for this enzyme, it is possible to interpret the profiles obtained as showing nonindependent binding of the two substrates (isocitrate and NAD+). Although the three intersections vary rather widely (indicated by a circle in Fig. 3A), Ksdl-isocitrate was calculated to be 4.0 mM from the average x coordinate value of the intersections. From the secondary plot of 1/kcatapp (kcatapp is an apparent kcat value for each NAD+ concentration) against 1/[NAD+], KmNAD and kcat values were calculated to be 0.47 mM and 4.3 s−1, respectively. From the secondary plot of the slope against 1/[NAD+], Kmdl-isocitrate was calculated to be 0.20 mM. From the average x coordinate value of the intersections of the second profile (indicated by a circle in Fig. 3B), KsNAD was calculated to be 5.4 mM.
FIG. 3.
Kinetics of the oxidation reaction catalyzed by HtICDH. (A) Double-reciprocal plots obtained by varying the isocitrate concentration at several different fixed concentrations of NAD+ (circle, 4 mM; square, 3 mM; triangle, 2 mM). (B) Double-reciprocal plots obtained by varying the NAD+ concentration under several different fixed concentrations of dl-isocitrate (circle, 4 mM; square, 3 mM; triangle, 2 mM). (C) Inhibition by NADH versus isocitrate. The concentration of NAD+ was fixed at 4 mM, and the dl-isocitrate concentration was varied at several different concentrations of NADH (circle, 0 mM; square, 0.01 mM; triangle, 0.02 mM). (D) Inhibition by NADH versus NAD+. The concentration of dl-isocitrate was fixed at 4 mM, and NAD+ concentration was varied at several different concentrations of NADH (circle, 0 mM; square, 0.01 mM; triangle, 0.02 mM). v, velocity.
Product inhibition analysis.
In order to elucidate further the precise kinetic mechanism of HtICDH, product inhibition analysis was carried out. Enzymatic activity in the presence of different concentrations of NADH was measured by monitoring NAD+ reducing activity. When fixed NAD+ and variable isocitrate concentrations were used, the double-reciprocal plots gave three linear lines with intersections in the second quadrant (Fig. 3C). When fixed isocitrate and variable NAD+ concentrations were used, the double-reciprocal plots gave intersections on the y axis (Fig. 3D). The profiles obtained strongly suggest a rapid equilibrium random-order mechanism with abortive complex (isocitrate-enzyme-NADH) formation. NADH is a noncompetitive (or mixed competitive) inhibitor against isocitrate, and from the Dixon plot, KiNADH and Ki′NADH values were calculated to be 0.02 mM and 0.13 mM, respectively. NADH is a competitive inhibitor against NAD+, and from the Dixon plot, a consistent KiNADH value (0.02 mM) was obtained.
Another possible product, 2-oxoglutarate, did not exhibit an inhibitory effect on the enzymatic activity of HtICDH (at least up to 10 mM). This observation indicates that 2-oxoglutarate does not compete with isocitrate and suggests that 2-oxoglutarate does not bind to HtICDH. Thus, it is highly probable that 2-oxoglutarate is not a product of the enzymatic reaction catalyzed by HtICDH.
Oxalosuccinate production.
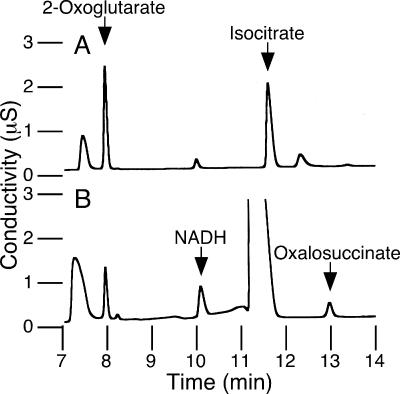
If 2-oxoglutarate is not a product of the enzymatic reaction catalyzed by HtICDH, the genuine product is most likely to be oxalosuccinate. Oxalosuccinate will be produced if HtICDH catalyzes only dehydrogenation and if the following decarboxylation is a nonenzymatic reaction. This is an extraordinary idea since it is well established that, for EcICDH, the dehydrogenation and decarboxylation reactions are closely coupled (10). In the case of NADP-dependent ICDH from pig heart, the hypothetical intermediate, oxalosuccinate, is not released from the enzyme (20). Furthermore, since enzyme-bound oxalosuccinate is not detectable, it is likely that oxalosuccinate is not a true intermediate of the ICDH reaction (19-20). Nevertheless, the hypothesis that HtICDH produces free oxalosuccinate as a reaction product was attractive since it would provide a definite answer to the question of why HtICDH cannot catalyze the reductive carboxylation of 2-oxoglutarate. In order to confirm the hypothesis, we established a detection system for the formation of oxalosuccinate. Since oxalosuccinate is very unstable and readily decarboxylated to yield 2-oxoglutarate, the enzymatic reaction was performed at a low temperature (20°C). In the case of EcICDH, oxalosuccinate was not detected (Fig. 4A), which is consistent with the studies so far reported. By contrast, in the case of HtICDH, the formation of oxalosuccinate was clearly observed (Fig. 4B). Since oxalosuccinate was clearly detectable as a product, HtICDH was shown to be a nondecarboxylating ICDH.
FIG. 4.
Oxalosuccinate formation by the oxidation reaction. Chromatograms of the mixture obtained after the oxidation reaction by EcICDH (using 18 μg of protein) (A) and HtICDH (using 79 μg of protein) (B). The reaction was performed in a volume of 400 μl containing 100 mM bicine-KOH (pH 9.5), 10 mM MgCl2, 2 mM dl-isocitrate, 2 mM NADP+ (for EcICDH) or 2 mM NAD+ (for HtICDH), and ICDH. After incubation at 20°C for 20 min, the reaction mixture was diluted with water (12-fold in A and 2-fold in B) before injection onto the column.
DISCUSSION
Decarboxylating and nondecarboxylating ICDH.
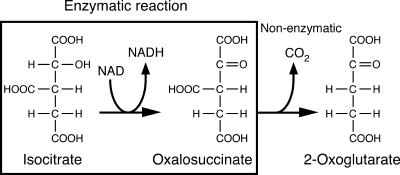
ICDH is known as an enzyme that catalyzes consecutive dehydrogenation and decarboxylation reactions in a closely coupled manner. However, here we clearly demonstrate that HtICDH is not a decarboxylating ICDH but a nondecarboxylating ICDH. HtICDH catalyzes only the oxidation (dehydrogenation) of isocitrate to oxalosuccinate, and the subsequent decarboxylation of oxalosuccinate to 2-oxoglutarate is nonenzymatic (Fig. 5). Physiologically, HtICDH catalyzes the reverse of this reaction, i.e., the reduction of oxalosuccinate to isocitrate, and, thus, it would be more appropriate to call this enzyme oxalosuccinate reductase. As such, HtICDH catalyzes only the conversion between isocitrate and oxalosuccinate by oxidation and reduction (with no decarboxylation or carboxylation), which is completely different from other known conventional ICDHs. Since no nondecarboxylating ICDHs have been reported previously, HtICDH should be assigned to a new category of enzymes with a new EC number.
FIG. 5.
Oxidation reaction catalyzed by HtICDH. HtICDH only catalyzes the oxidation of isocitrate to oxalosuccinate. The following decarboxylation is a nonenzymatic reaction.
We have previously reported that HtICDH is unable to catalyze the reductive carboxylation of 2-oxoglutarate (3). The reason for this phenomenon can now be clearly understood because this reaction is not performed by HtICDH, and 2-oxoglutarate is neither a substrate nor a product of HtICDH. This is supported by the observation that 2-oxoglutarate did not act as a product inhibitor of HtICDH.
In this study, we demonstrated that two types of ICDH exist: decarboxylating and nondecarboxylating. This is analogous to malate dehydrogenase, for which both nondecarboxylating (EC 1.1.1.37 and EC 1.1.1.82) and decarboxylating (EC 1.1.1.38, EC 1.1.1.39, EC 1.1.1.40, and EC 1.1.1.83) enzymes are known. Similarly, both decarboxylating and nondecarboxylating forms of tartrate dehydrogenase exist (EC 1.1.1.93 and EC 1.3.1.7, respectively) (24). Although this is the first report of a nondecarboxylating ICDH, it is possible that this type of ICDH is widely distributed but difficult to distinguish. It is especially difficult to distinguish between the decarboxylating and nondecarboxylating forms when the oxidation reaction is spectrophotometrically analyzed. It may be possible to identify additional nondecarboxylating ICDHs from other organisms by screening for the inability to catalyze the reductive carboxylation of 2-oxoglutarate or to be inhibited by 2-oxoglutarate, as these are the distinguishing features of the nondecarboxylating forms.
Kinetic features of HtICDH.
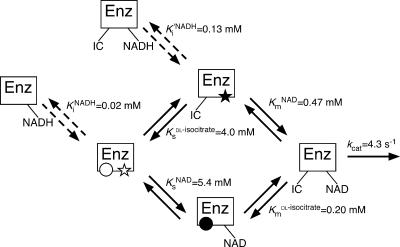
The kinetic analyses performed strongly suggest that the enzymatic reaction catalyzed by HtICDH, in the direction of oxidation, proceeds by a random mechanism where either of the substrates, isocitrate or NAD+, can bind to the enzyme first, as in the case of EcICDH (8). The proposed kinetic mechanism is outlined in Fig. 6. Interestingly, the binding of the two substrates (isocitrate and NAD+) is not independent. In the case of isocitrate, the Michaelis constant (Kmdl-isocitrate, 0.20 mM) is one order of magnitude lower than the substrate constant (Ksdl-isocitrate, 4.0 mM). This observation indicates that the affinity toward isocitrate is increased by the binding of NAD+. The reciprocal effect applies in the case of NAD+. The Michaelis constant (KmNAD, 0.47 mM) is one order of magnitude lower than the substrate constant (KsNAD, 5.4 mM), and thus, the binary complex (isocitrate-enzyme) (Fig. 6, IC-Enz) has a higher affinity for NAD+ than the enzyme with no substrate bound (Fig. 6, Enz). Although the Michaelis constants of HtICDH are lower than the substrate constants, they are both extremely high compared to those of EcICDH (8). Thus, it can be said that HtICDH is quite ineffective as an oxidative enzyme. Taking into account the high substrate constants, the oxidation reaction becomes possible only under conditions where the concentration of at least one of the substrates is extremely high. Because such a high concentration of isocitrate or NAD+ is unlikely to occur in H. thermophilus cells under physiological conditions, it is highly probable that HtICDH cannot function as an oxidative enzyme in this organism. As such, the kinetic parameters obtained in this study strongly suggest that HtICDH is not an oxidative enzyme but a reducing enzyme.
FIG. 6.
Proposed kinetic mechanism of the oxidation reaction catalyzed by HtICDH. Enz, HtICDH; IC, isocitrate; filled circle, isocitrate binding site of the binary complex (high affinity); open circle, isocitrate binding site of HtICDH (low affinity); filled star, NAD binding site of the binary complex (high affinity); open star, NAD binding site of HtICDH (low affinity). Arrows with dotted lines indicate product inhibition by NADH.
Product inhibition analysis performed during this study revealed that NADH is a potent inhibitor of HtICDH, competing for the NAD binding site with NAD+. Inhibitor constants toward the enzyme (KiNADH, 0.02 mM) and toward the binary complex (isocitrate-enzyme; Ki′NADH, 0.13 mM) were similar to the inhibitor constants of NADPH toward EcICDH (8). Although we have not examined the level of cytosolic NADH in H. thermophilus, it may have an inhibitory effect on the oxidation activity of HtICDH.
Since HtICDH is a reducing enzyme in H. thermophilus, the kinetic mechanism analyzed in this study is the reverse of the physiological reaction. If the oxidation reaction pathway analyzed here accurately represents the reverse of the reduction reaction pathway, it may be possible to make deductions about the product-releasing mechanism of the reduction reaction from the substrate-binding mechanism of the oxidation reaction. If this is the case, once one of the products (isocitrate or NAD+) has been released from the enzyme, the other will then be released more readily. This mechanism may also assist in driving the enzymatic reaction in a reductive direction.
Oxalosuccinate reductase as an origin of ICDH.
HtICDH is quite different from other known ICDHs with respect to its reaction mechanism. HtICDH catalyzes a bi-bi reaction (oxidation and reduction between isocitrate and oxalosuccinate) while conventional ICDHs catalyze a bi-ter reaction (in which carbon dioxide is involved). Thus, as we have proposed, it may be more appropriate to redesignate HtICDH an reductase. Nevertheless, a close relationship between HtICDH and other ICDHs is apparent when the structures of these proteins are examined. The primary sequence of HtICDH is 45.8% identical to that of EcICDH, and both sequences show overall similarity (3). This observation indicates that both enzymes share a common fold, and from the phylogenetic viewpoint, they undoubtedly belong to the same subfamily and have evolved from a common ancestral enzyme. Since H. thermophilus has been assigned to the most deeply branching lineage of the domain Bacteria (18), it may be possible that HtICDH is an ancestral form of EcICDH. Therefore, we suggest a possible hypothesis as to how the enzyme evolved from an oxalosuccinate-reducing enzyme to an isocitrate oxidative decarboxylating enzyme.
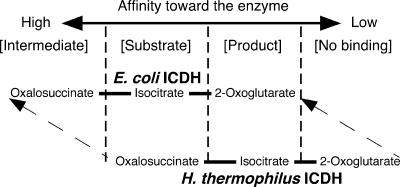
In the case of EcICDH, during the enzymatic reaction, isocitrate is oxidized to oxalosuccinate and then oxalosuccinate is decarboxylated to 2-oxoglutarate. Oxalosuccinate may be an enzyme-bound intermediate, but it is never released from the enzyme. This suggests that oxalosuccinate binds more tightly than isocitrate or 2-oxoglutarate. The affinity of isocitrate is higher than that of 2-oxoglutarate, as shown by their respective Michaelis constants (8). Accordingly, the affinities toward EcICDH can be ranked in the order of the highest to the lowest as oxalosuccinate > isocitrate > 2-oxoglutarate, which corresponds to intermediate > substrate > product (Fig. 7). This would be the preferred order for an isocitrate oxidative decarboxylating enzyme.
FIG. 7.
Hypothetical evolutionary pathway from an oxalosuccinate-reducing enzyme to an isocitrate-oxidative decarboxylating enzyme. The affinities of oxalosuccinate, isocitrate, and 2-oxoglutarate toward the enzyme are shown. H. thermophilus ICDH, an oxalosuccinate-reducing enzyme, is proposed to have evolved to an isocitrate-oxidative decarboxylating enzyme (E. coli ICDH) by acquiring higher substrate affinities.
It is highly probable that, irrespective of the origin of ICDH, the order of the substrate affinities toward the enzyme is always the same (oxalosuccinate > isocitrate > 2-oxoglutarate) and only the absolute affinities of the three vary. Since oxalosuccinate, isocitrate, and 2-oxoglutarate bind to the same site on the enzyme, the affinities of these three ligands may be closely interrelated. This interdependence between affinities is a common feature for multiple ligands that bind to the identical site on enzymes. For example, an interdependence between the affinities of NADP and NADPH has been reported for the engineered isopropylmalate dehydrogenase from E. coli (16).
As shown in Fig. 7, if the affinities of oxalosuccinate, isocitrate, and 2-oxoglutarate are lowered simultaneously, the characteristics of the enzyme change significantly. Oxalosuccinate, although it still binds tightly, can be released from the enzyme. This means that oxalosuccinate is no longer an enzyme-bound intermediate but, rather, a substrate. Isocitrate becomes a product because of its low affinity. 2-Oxoglutarate cannot bind to the enzyme, so it is no longer a substrate or a product. These properties agree exactly with the characteristics of the oxalosuccinate-reducing enzyme, HtICDH. Thus, it can be said that the oxalosuccinate-reducing enzyme corresponds to ICDH with a lowered substrate affinity. If the phylogenetic history of ICDH is traced chronologically, it is likely that the oxalosuccinate-reducing enzyme is the ancestral form, and that this evolved to the isocitrate oxidative decarboxylating enzyme by acquiring higher substrate affinities. Accordingly, it is highly probable that oxalosuccinate reductase is the origin of extant ICDH and that an ancient type of enzyme still remains in H. thermophilus. Since ancestral forms of the TCA cycle enzymes are abundant in H. thermophilus (1), it is a crucial organism for the further investigation of the evolutionary history of the TCA cycle.
As described, the substrate affinities of HtICDH and EcICDH are significantly different. However, residues involved in the substrate (isocitrate-Mg2+) binding in EcICDH are all conserved in the HtICDH sequence (3). As yet, we have not determined which residues in HtICDH are responsible for the low affinity toward the substrates. A preliminary three-dimensional comparison study using a homology modeling tool suggests that the position of D284 in HtICDH has widely diverged from that of the corresponding residue (D283) in EcICDH. Since D283 is one of the substrate binding residues in EcICDH (10), this divergence may be the cause of the low substrate affinity of HtICDH. Further molecular dynamics, crystallographic, and mutagenesis studies are required.
Footnotes
Published ahead of print on 18 January 2008.
REFERENCES
- 1.Aoshima, M. 2007. Novel enzyme reactions related to the tricarboxylic acid cycle: phylogenetic/functional implications and biotechnological applications. Appl. Microbiol. Biotechnol. 75249-255. [DOI] [PubMed] [Google Scholar]
- 2.Aoshima, M., and Y. Igarashi. 2006. A novel oxalosuccinate-forming enzyme involved in the reductive carboxylation of 2-oxoglutarate in Hydrogenobacter thermophilus TK-6. Mol. Microbiol. 62748-759. [DOI] [PubMed] [Google Scholar]
- 3.Aoshima, M., M. Ishii, and Y. Igarashi. 2004. A novel biotin protein required for reductive carboxylation of 2-oxoglutarate by isocitrate dehydrogenase in Hydrogenobacter thermophilus TK-6. Mol. Microbiol. 51791-798. [DOI] [PubMed] [Google Scholar]
- 4.Aoshima, M., and T. Oshima. 1997. Purification and characterization of isocitrate dehydrogenase from a hyperthermophilic archaebacterium, Caldococcus noboribetus. Biochim. Biophys. Acta 1340227-234. [DOI] [PubMed] [Google Scholar]
- 5.Aoshima, M., A. Yamagishi, and T. Oshima. 1996. Eubacteria-type isocitrate dehydrogenase from an archaeon: cloning, sequencing, and expression of a gene encoding isocitrate dehydrogenase from a hyperthermophilic archaebacterium Caldococcus noboribetus. Arch. Biochem. Biophys. 33677-85. [DOI] [PubMed] [Google Scholar]
- 6.Chen, R. D., and S. S. Jeong. 2000. Functional prediction: identification of protein orthologs and paralogs. Protein Sci. 92344-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cvitkovitch, D. G., J. A. Gutierrez, and A. S. Bleiweis. 1997. Role of the citrate pathway in glutamate biosynthesis by Streptococcus mutans. J. Bacteriol. 179650-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean, A. M., and D. E. Koshland, Jr. 1993. Kinetic mechanism of Escherichia coli isocitrate dehydrogenase. Biochemistry 329302-9309. [DOI] [PubMed] [Google Scholar]
- 9.Hampton, M. L., and R. S. Hanson. 1969. Regulation of isocitrate dehydrogenase from Thiobacillus thiooxidans and Pseudomonas fluorescens. Biochem. Biophys. Res. Commun. 36296-305. [DOI] [PubMed] [Google Scholar]
- 10.Hurley, J. H., A. M. Dean, D. E. Koshland, Jr., and R. M. Stroud. 1991. Catalytic mechanism of NADP+-dependent isocitrate dehydrogenase: Implications from the structures of magnesium-isocitrate and NADP+ complexes. Biochemistry 308671-8678. [DOI] [PubMed] [Google Scholar]
- 11.Hurley, J. H., P. E. Thorsness, V. Ramalingam, N. H. Helmers, D. E. Koshland, Jr., and R. M. Stroud. 1989. Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc. Natl. Acad. Sci. USA 868635-8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imada, K., M. Sato, N. Tanaka, Y. Katsube, Y. Matsuura, and T. Oshima. 1991. Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 Å resolution. J. Mol. Biol. 222725-738. [DOI] [PubMed] [Google Scholar]
- 13.Inoue, H., T. Tamura, N. Ehara, A. Nishito, Y. Nakayama, M. Maekawa, K. Imada, H. Tanaka, and K. Inagaki. 2002. Biochemical and molecular characterization of the NAD+-dependent isocitrate dehydrogenase from the chemolithotroph Acidithiobacillus thiooxidans. FEMS Microbiol. Lett. 214127-132. [DOI] [PubMed] [Google Scholar]
- 14.Large, P. J., and G. W. Haywood. 1981. Methylophilus methylotrophus grows on methylated amines. FEMS Microbiol. Lett. 11207-209. [Google Scholar]
- 15.Lloyd, A. J., and P. D. J. Weitzman. 1988. Purification and characterization of NAD-linked isocitrate dehydrogenase from Methylophilus methylotrophus. Biochem. Soc. Trans. 16871-872. [Google Scholar]
- 16.Miller, S. P., M. Lunzer, and A. M. Dean. 2006. Direct demonstration of an adaptive constraint. Science 314458-461. [DOI] [PubMed] [Google Scholar]
- 17.Miyazaki, J., K. Asada, S. Fushinobu, T. Kuzuyama, and M. Nishiyama. 2005. Crystal structure of tetrameric homoisocitrate dehydrogenase from an extreme thermophile, Thermus thermophilus: involvement of hydrophobic dimer-dimer interaction in extremely high thermotolerance. J. Bacteriol. 1876779-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pitulle, C., Y. Yang, M. Marchiani, E. R. B. Moore, J. L. Siefert, M. Aragno, P. Jurtshuk, Jr., and G. E. Fox. 1994. Phylogenetic position of the genus Hydrogenobacter. Int. J. Syst. Bacteriol. 44620-626. [DOI] [PubMed] [Google Scholar]
- 19.Ramakrishna, M., and P. R. Krishnaswamay. 1966. Formation of enzyme bound carbon dioxide in the reductive carboxylation of alpha-ketoglutarate by isocitrate dehydrogenase. Biochem. Biophys. Res. Commun. 25378-382. [DOI] [PubMed] [Google Scholar]
- 20.Siebert, G., M. Carsiotis, and G. W. E. Plaut. 1957. The enzymatic properties of isocitric dehydrogenase. J. Biol. Chem. 226977-991. [PubMed] [Google Scholar]
- 21.Steen, I. H., T. Lien, and N. K. Birkeland. 1997. Biochemical and phylogenetic characterization of isocitrate dehydrogenase from a hyperthermophilic archaeon, Archaeoglobus fulgidus. Arch. Microbiol. 168412-420. [DOI] [PubMed] [Google Scholar]
- 22.Steen, I. H., D. Madern, M. Karlström, T. Lien, R. Ladenstein, and N. K. Birkeland. 2001. Comparison of isocitrate dehydrogenase from three hyperthermophiles reveals differences in thermostability, cofactor specificity, oligomeric state, and phylogenetic affiliation. J. Biol. Chem. 27643924-43931. [DOI] [PubMed] [Google Scholar]
- 23.Stokke, R., D. Madern, A. E. Fedøy, S. Karlsen, N. K. Birkeland, and I. H. Steen. 2007. Biochemical characterization of isocitrate dehydrogenase from Methylococcus capsulatus reveals a unique NAD+-dependent homotetrameric enzyme. Arch. Microbiol. 187361-370. [DOI] [PubMed] [Google Scholar]
- 24.Tipton, P. A., and J. Peisach. 1990. Characterization of the multiple catalytic activities of tartrate dehydrogenase. Biochemistry 291749-1756. [DOI] [PubMed] [Google Scholar]