Abstract
Expression of two genes of unknown function, Staphylococcus aureus scdA and Neisseria gonorrhoeae dnrN, is induced by exposure to oxidative or nitrosative stress. We show that DnrN and ScdA are di-iron proteins that protect their hosts from damage caused by exposure to nitric oxide and to hydrogen peroxide. Loss of FNR-dependent activation of aniA expression and NsrR-dependent repression of norB and dnrN expression on exposure to NO was restored in the gonococcal parent strain but not in a dnrN mutant, suggesting that DnrN is necessary for the repair of NO damage to the gonococcal transcription factors, FNR and NsrR. Restoration of aconitase activity destroyed by exposure of S. aureus to NO or H2O2 required a functional scdA gene. Electron paramagnetic resonance spectra of recombinant ScdA purified from Escherichia coli confirmed the presence of a di-iron center. The recombinant scdA plasmid, but not recombinant plasmids encoding the complete Escherichia coli sufABCDSE or iscRSUAhscBAfdx operons, complemented repair defects of an E. coli ytfE mutant. Analysis of the protein sequence database revealed the importance of the two proteins based on the widespread distribution of highly conserved homologues in both gram-positive and gram-negative bacteria that are human pathogens. We provide in vivo and in vitro evidence that Fe-S clusters damaged by exposure to NO and H2O2 can be repaired by this new protein family, for which we propose the name repair of iron centers, or RIC, proteins.
Neutrophils and macrophages of the mammalian immune system produce reactive oxygen and reactive nitrogen species that have important roles in killing pathogenic bacteria by damaging such cellular components as DNA, lipids, and proteins. Particularly vulnerable to inactivation are iron-sulfur (Fe-S) proteins, which were among the first catalysts used by nature (19). They participate in numerous cellular processes in virtually all organisms where they fulfill crucial redox, catalytic, and regulatory functions (1, 21, 28). Specialized systems have evolved that facilitate the assembly and insertion of Fe-S clusters into proteins, namely, the products of isc, suf, and csd operons (12, 13, 21). Analysis of bacterial genomes shows that one or more of these systems can be present in any organism for the in vivo maturation of Fe-S proteins. The isc operon encodes several proteins that are necessary for de novo synthesis, and at least one of them, IscS, is proposed to be required for cluster repair (9). The Suf system sustains Fe-S cluster biogenesis during iron starvation and oxidative stress (9, 38, 40), and CSD is proposed to act as a sulfur-generating system (34). Despite their established roles in pathogen survival, little is known about how oxidative and nitrosative damage to Fe-S clusters is repaired since so far only IscS is proposed to have such a function (9, 46, 58).
Transcriptomic studies have shown that nitrosative stress conditions elicit increased expression of not only the isc and suf operons but also various genes of known and unknown function (6, 25, 37, 43, 44, 47). The products of some of these genes are required to detoxify the reactive nitrogen species and are under the control of iron-sulfur regulators. For example, the hmpA gene present in various bacteria encodes an enzyme that catalyzes the oxidation of NO to nitrate in aerobic cultures or the reduction to nitrous oxide during anaerobic growth (14, 15, 29, 42). In Escherichia coli, expression of hmpA is repressed by FNR, the regulator of fumarate and nitrate reduction, which contains an [4Fe-4S]+2/+1 iron-sulfur center that is essential for the binding of FNR to its DNA binding site. FNR, originally identified as an oxygen-sensitive transcription regulator, is also inactivated on exposure to nitric oxide, providing a mechanism by which FNR-repressed genes respond to nitrosative stress (7, 39). Similarly, the repressor activity of NsrR, which from sequence analysis is assumed to contain an [2Fe-2S] iron-sulfur center, is inactivated on exposure to nitric oxide (2, 10, 41). There is overlap between the biological responses to oxidative stress caused by exposure to hydrogen peroxide and to nitrosative stress (4, 18, 48, 59, 60). This overlap includes various iron-sulfur-containing enzymes and the transcription factors that regulate their synthesis, which is a reflection of the fact that iron-sulfur centers are damaged by both reactive oxygen and nitrogen species. In addition, the perturbation of iron homeostasis that occurs under stress conditions causes changes in the transcriptional regulation of a large number of genes involved in iron metabolism, many of which code for iron-containing proteins (28, 36).
Analysis of the data available for the gram-positive pathogen Staphylococcus aureus and for the gram-negative pathogens Neisseria meningitidis and Neisseria gonorrhoeae, organisms that have serious impacts on human health, revealed that exposure to nitric oxide and hydrogen peroxide causes the induction of genes encoding putative iron-containing proteins. Examples include the S. aureus scdA, whose expression was reported to be induced by both NO and hydrogen peroxide (4, 44), and the gonococcal dnrN, which is induced when the NsrR repressor protein is inactivated by NO (41). We therefore investigated whether either of these proteins is implicated in protection against nitric oxide or hydrogen peroxide, reactive nitrogen and oxygen species generated by the human body as part of its defenses against infection by pathogenic bacteria, and in iron metabolism. The results of in vivo and in vitro experiments revealed a role for these proteins in the repair of iron-sulfur centers of both transcription factors and housekeeping enzymes damaged by oxidative and nitrosative stress. Furthermore, the analysis of protein databases emphasizes their importance since related proteins were found in a wide range of prokaryotic and eukaryotic pathogens.
MATERIALS AND METHODS
Strains, plasmids, and primers.
Bacterial strains and plasmids used in this work are listed in Table 1, and oligonucleotides are listed in Table S1 in the supplemental material. To disrupt the scdA gene (SAOUHSC_00229) of S. aureus NCTC 8325, an 820-bp fragment spanning the upstream region and 5′ end of the gene was amplified by PCR using the primers ScdAmutEco and ScdAmutBam, and the fragment was cloned into pSP64D-E (17). The resulting plasmid, pSPScdA, was electroporated into S. aureus RN4220, and transformants were selected on tryptic soy agar (TSA; Difco) plates containing erythromycin (10 μg/ml). The correct integration of pSPScdA into the chromosome of RN4220 in the strain obtained, LMSA0229 (scdA::Ermr), was confirmed by single-colony PCR analysis.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli strains | ||
| BL21 Gold(DE3) | Protein expression cells | Stratagene |
| K-12 ATCC 23716 | Parental strain | Laboratory stock |
| LMS4209 | K-12 (ATCC 23716) ytfE::Cmr | 25 |
| S. aureus strains | ||
| NCTC 8325 | Parental strain | Laboratory stock |
| RN4220 | Restriction-negative derivative of NCTC 8325; transformable by electroporation | Laboratory stock |
| LMSA0229 | RN4220 scdA::Ermr | This study |
| N. gonorrhoeae strains | ||
| F62 | Parental strain | Laboratory stock |
| JCGC704 | dnrN::Kanr | This study |
| JCGC212 | kat::ermC | 51 |
| JCGC705 | kat::ermC dnrN::Kanr | This study |
| Plasmids | ||
| pSP64D-E | Cloning vector carrying an erythromycin resistance cassette | 8 |
| pSPScdA | Upstream region and 5′ end of scdA cloned into pSP64D-E, next to the erythromycin resistance cassette | This study |
| pSUB11 | Epitope tagging plasmid carrying three copies of the FLAG tag and a kanamycin resistance cassette | 55 |
| pGEM-T Easy | Cloning vector | Promega |
| pGEMDnrN | Sequences upstream and downstream of the dnrN gene (NGO0653) cloned into pGEM-T Easy | This study |
| pGEMDnrN-KO | Sequences upstream and downstream of the dnrN gene (NGO0653) flanking a kanamycin resistance cassette cloned into pGEM-T Easy | This study |
| pET28a(+) | T7-based expression vector that inserts a sequence encoding a His6 tag at the N terminus | Novagen |
| pET-ScdA | S. aureus scdA gene cloned into pET28a(+) | This study |
| pGS57 | Fumarase A-expressing plasmid | 57 |
| pUC18 | Cloning vector | Laboratory stock |
| pScdA | pUC18 carrying the scdA gene of S. aureus and its promoter region | This study |
| pYtfE | pUC18 carrying the ytfE gene of E. coli and its promoter region | 24 |
| pRKISC | Plasmid for expression of the E. coli isc operon | 51 |
| pRKSUF | Plasmid for expression of the E. coli suf operon | 52 |
The dnrN gene of N. gonorrhoeae (open reading frame NGO0653) was interrupted with a kanamycin resistance cassette using crossover PCR (31). Primers DnrNA plus DnrNB and DnrNC plus DnrND were used to generate DNA fragments upstream and downstream of the dnrN gene. The flanking fragments were cleaned and combined in a crossover PCR with primers DnrNA and DnrND, yielding a single fragment with an AgeI restriction site between the upstream and downstream sequences. The crossover PCR product was cloned into pGEM T-Easy (Promega, Madison, WI), yielding pGEMDnrN. A kanamycin resistance cassette was amplified from pSUB11 by PCR using primers KanAgeIFwd and KanAgeIRev, which introduced AgeI sites at each end of the resultant fragment, and was ligated into AgeI-digested pGEMDnrN, yielding pGEMDnrN-KO. The dnrN::kan fragment was generated by digestion of pGEMDnrN-KO with EcoRI and was transformed, as previously described (32), into piliated N. gonorrhoeae strain F62, yielding strain JCGC704.
Growth of S. aureus and sensitivity assays.
S. aureus RN4220 and LMSA0229 strains were streaked onto TSA plates and incubated for 16 h at 37°C. Isolated colonies were cultivated aerobically in tryptic soy broth medium (Difco) for 16 h at 37°C and 150 rpm. These were used to inoculate, in duplicate, 20 ml of fresh tryptic soy broth, adjusting the starting optical density at 600 nm (OD600) to 0.1. The cultures, grown aerobically at 37°C, were treated with 10 mM H2O2 or left untreated. After 4 h of growth, 5 μl of serial dilutions of the cultures was spread onto TSA plates and incubated overnight.
Growth of N. gonorrhoeae and sensitivity to hydrogen peroxide.
N. gonorrhoeae was grown on gonococcal agar plates and in gonococcal broth (GCB; BD, Oxford, United Kingdom). Solid and liquid media were supplemented with 1% (vol/vol) Kellogg's supplement (26). For liquid cultures, 2 μl of a stock of N. gonorrhoeae was plated onto a gonococcal agar plate and incubated in a candle jar at 37°C for 24 h. Bacteria from this plate were swabbed onto a second plate and incubated in the same way for a further 16 h. The entire bacterial growth from this second plate was swabbed into 10 ml of GCB and incubated at 37°C in an orbital shaker at 100 rpm for 1 h. This 10-ml preculture was then transferred into 50 ml of GCB in a 100-ml conical flask and incubated in the same way. For growth in the presence of nitrite, 1 mM NaNO2 was added after 1 h, and 4 mM NaNO2 was added 1 h later.
A modified disk diffusion assay was used to compare areas of growth inhibition of various gonococcal strains (54). For growth experiments, different concentrations of H2O2 were added to 60 ml of oxygen-limited cultures of the gonococcal kat mutant and the kat dnrN double mutant in 100-ml conical flasks, and growth was monitored for the following 5 h. Greatest differences between the two strains were observed when the H2O2 concentration added was 0.5 mM.
Complementation assays in E. coli.
A DNA fragment of 955 bp comprising the promoter and coding regions of scdA was amplified by PCR from S. aureus NCTC 8325 genomic DNA, using the primers ScdAHindIII and ScdAEcoRI, and cloned into pUC18 digested with HindIII and EcoRI, generating the plasmid pScdA. The E. coli ytfE mutant strain LMS4209 was transformed with the plasmids pYtfE, pScdA, pRKISC, and pRKSUF that express, respectively, the E. coli ytfE gene, S. aureus scdA gene, E. coli isc operon, and the suf operon from their own promoters. E. coli strains were grown in LB medium under anaerobic conditions (i.e., closed flasks completely filled), from a starting OD600 of 0.1. When cultures reached an OD600 of 0.3, they were treated with 4 mM hydrogen peroxide (Sigma) or left untreated, and the growth was followed for ∼3 h.
Production of the S. aureus recombinant ScdA protein.
The coding region of the scdA gene was amplified by PCR from genomic DNA of S. aureus NCTC 8325 using the primers ScdANheI and ScdAEcoRI and cloned into pET-28a (Novagen) that allows insertion of a nucleotide sequence that encodes a His6 tail at the N terminus. The resulting plasmid, pET-ScdA, was sequenced to ensure the integrity of the cloned sequence. The recombinant protein was overproduced in cells of E. coli BL21 Gold(DE3) (Stratagene) grown aerobically in M9 minimal medium, which was supplemented with 10 mM glucose, 100 μM Fe(NH4)2(SO4)2, and 30 μg/ml kanamycin; cells were cultured at 37°C and 150 rpm. At an OD600 of 0.3, the cultures were induced with 400 μM isopropyl-1-thio-β-D-galactopyranoside (IPTG). After the temperature was lowered to 30°C, cultures were grown for 6 h at 130 rpm and harvested by centrifugation. Cells were resuspended in ice-cold buffer A (20 mM Tris-HCl, pH 7.6), disrupted in a French press, and ultracentrifuged at 100,000 × g for 2 h at 4°C. The soluble extract was loaded onto an immobilized metal affinity chromatography Sepharose Fast Flow column (GE Healthcare), and ScdA was eluted at 300 mM imidazole and immediately dialyzed against buffer A. The protein was found to be pure, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and this sample was used for further characterization.
Protein concentration was determined by a bicinchoninic acid protein assay (Pierce) (49), and the iron content was determined by the TPTZ (2,4,6-tripyridyl-1,2,3-triazine) method (11). Molecular mass determination was performed in a Superdex 200 (10/300) GL column (GE Healthcare) using standard proteins. The electron paramagnetic resonance (EPR) spectrum was obtained in a Bruker EMX spectrometer equipped with an Oxford Instruments continuous-flow helium cryostat and was recorded at a 9.39-MHz microwave frequency with 2.4 mW of microwave power at 10 K.
Repair of the damaged [4Fe-4S] cluster of E. coli fumarase A.
Cells of the E. coli ytfE mutant strain transformed with pGS57 were grown aerobically with 1 mM IPTG to an OD600 of ∼0.5, collected by centrifugation, resuspended (1/100) in fumarase assay buffer (35), and lysed by four freeze-thaw cycles. Two minutes before the stresses were imposed, 100 μg/ml tetracycline was added to the cell extracts to inhibit de novo protein synthesis. After 1 min of incubation with 4 mM H2O2 or 5 min with 150 μM NO, 400 U/ml of catalase or 40 μM hemoglobin (Sigma) was added, respectively, and the fumarase activity was determined at fixed time points. Purified ScdA protein was added at a final concentration of 20 μM immediately after the stresses were removed.
Fumarase activity was determined spectrophotometrically by following the disappearance of fumarate as described by Massey (35). The cell samples were quickly thawed at room temperature, cleared by the addition of 0.5% (wt/vol) sodium deoxycholate, and then diluted in 50 mM sodium phosphate buffer, pH 7.3. The reactions were started by the addition of 10 mM fumarate and followed (at 295 nm, ɛ = 0.07 mM−1 cm−1). Enzyme activities were determined at 25°C and are defined as units (μmol of fumarate consumed per min) per mg of total protein. The enzyme activities were determined in duplicate from two independent cultures and are presented as averaged values, with error bars representing one standard deviation.
Determination of the aconitase activity in S. aureus.
Aconitase activity was determined in cell lysates of S. aureus RN4220 and the scdA mutant that had been grown aerobically in LB medium at 37°C to an OD600 of 0.5. The cells were collected by centrifugation, resuspended (1/200) in assay buffer (50 mM Tris-HCl, pH 7.7, 0.6 mM MnCl2), and lysed by a 10-min incubation at 37°C with 75 μg/ml lysostaphin. Cell lysates were exposed to 100 μM NO or 3 mM H2O2, and at specific times aliquots were frozen in liquid nitrogen and later assayed. To monitor the repair of the damaged enzyme, the lysates were treated with tetracycline (100 μg/ml) prior to exposure to H2O2 for 5 min or NO for 15 min. Upon addition of catalase (400 U/ml) or hemoglobin (40 μM), the aliquots were collected and frozen. Aconitase activity was determined by following the formation of NADPH through the indirect method described by Gardner (16). Samples were quickly thawed at room temperature, cleared by the addition of 0.5% (wt/vol) sodium deoxycholate, and immediately inserted into suba-sealed cuvettes with deaerated assay buffer that contained 0.2 mM NADP+ and 1 U of isocitrate dehydrogenase (Sigma). The reaction was initiated with 50 mM sodium citrate. Aconitase activities determined at 25°C in duplicate from two independent cultures are defined as units (μmol of NADPH formed per min) per mg of total protein and are presented as averaged values, with error bars representing one standard deviation.
Quantitative real-time PCR analysis of gene expression.
Relative gene expression was measured using quantitative reverse transcription-PCR (qRT-PCR) as described previously (41). RNA was stabilized by mixing 500 μl of bacterial culture with 900 μl of RNAlater solution (Ambion). After a 5-min incubation at room temperature, the bacteria were harvested by centrifugation at 3,000 × g for 10 min. RNA was isolated from the pellet using an RNeasy mini kit (Qiagen) using the manufacturer's protocol. Genomic DNA was removed from the purified RNA using Turbo DNase (Ambion). The RNA was reverse transcribed to cDNA using a Superscript first-strand synthesis kit (Invitrogen). For each sample, a control to check for DNA contamination in the RNA preparation was included from which reverse transcriptase was omitted. Transcript levels were measured by quantitative real-time PCR using SensiMix with Sybr green detection (Quantace) and an ABI 7000 sequence analyzer (Applied Biosystems). Primers designed using PrimerExpress (Applied Biosystems) are described in Table S1 in the supplemental material. Transcript levels were quantified using the ΔΔCT (where CT is threshold cycle) method (33) relative to expression of the polA gene. Expression levels were normalized for each strain prior to shock with nitrite. For each experiment, quantitative real-time PCR was used to determine transcript levels on three independent cDNA samples derived from three independent cultures.
Determination of rates of NO reduction by washed bacterial suspensions.
N. gonorrhoeae strain F62 and its dnrN mutant were grown as described above in oxygen-limited cultures supplemented with nitrite and harvested by centrifugation, and the rates of NO reduction were assayed using a Hansatech Instruments oxygen electrode adapted for increased sensitivity to NO (53). All solutions used for these assays were purged of oxygen for at least 10 min using oxygen-free nitrogen gas. The concentration of NO at the start of the assay was 200 μM, and the bacterial density assayed was in the range of 1 to 2 mg of dry mass ml−1.
RESULTS AND DISCUSSION
Effect of a mutation in S. aureus scdA and N. gonorrhoeae dnrN on recovery from oxidative and nitrosative stress.
To assess the function of ScdA in S. aureus, the effects of NO and H2O2 on growth of an scdA mutant and its parent were compared. The scdA mutant strain showed no morphological defects, contrary to what had been previously described (3). Although no differences could be detected between the wild-type and mutant strains in response to exposure to NO (data not shown), the scdA mutant was more sensitive to oxidative conditions than its parent (Fig. 1). Hence, ScdA constitutes an efficient protection system against hydrogen peroxide.
FIG. 1.
The sensitivity of S. aureus to hydrogen peroxide increases in the absence of scdA. (A) Serial dilutions of cultures of S. aureus RN4220 (wt) and the scdA mutant strain after 4 h of growth in the presence (+) or absence (−) of 10 mM H2O2. A representative plate of independent experiments performed in duplicate is shown. (B) Growth of S. aureus RN4220 (squares) and the scdA mutant (circles) monitored by the OD600 nm in cultures untreated (filled symbols) or treated with 10 mM H2O2 (open symbols). Mean values of two independent cultures are given, with error bars showing the standard deviations.
In N. gonorrhoeae, binding sites for the NO-sensitive transcription factor NsrR, a member of the Rrf2 family of transcription factors, were identified at the promoters of aniA, which controls the expression of the gene encoding a copper-containing nitrite reductase similar to NirK in other bacteria; norB, encoding the single subunit nitric oxide reductase; and a gene of unknown function, dnrN. All of these genes are now known to be induced upon exposure to nitric oxide (41, 45). The gonococcal DnrN protein is 16% identical and 31% similar in amino acid sequence to S. aureus ScdA, suggesting that it might be a functional homologue of ScdA. A dnrN deletion mutant was constructed, and the effects of the mutation on recovery from exposure to NO were assessed. As the gonococcal dnrN gene is monocistronic, the possibility of secondary effects of the mutation on downstream genes was discounted. Since gonococci generate NO as the product of nitrite reduction during oxygen-limited growth, it was predicted that the dnrN mutant might be more sensitive to sudden exposure to nitrite, which will be converted rapidly to NO, than its dnrN+ parent. Sudden addition of nitrite to a culture in which AniA has accumulated but NorB synthesis has not been induced will lead to the sudden generation of NO, which would cause damage from which only the parent strain can recover. In contrast, if aniA and norB transcription are gradually induced sequentially because nitrite is available during adaptation to oxygen-limited growth, both the mutant and the parent are able to adapt.
To test these predictions, the mutant and parent strains were first grown in oxygen-limited medium supplemented with 5 mM nitrite. FNR, the regulator of fumarate and nitrate reduction during anaerobic growth, is essential in gonococci for expression of several genes including the nitrite reductase aniA. In the experiment, as the cultures became oxygen-limited, FNR gradually became activated, inducing the transcription of aniA. The consequent production of NO, generated during nitrite reduction, induced synthesis of the gonococcal nitric oxide reductase NorB, which scavenges the NO present in the cell. We propose that, under these conditions, the concentration of NO available is low; therefore, the mutant grew only slightly more slowly than the parent, and growth of both strains stopped at similar cell densities (Fig. 2A).
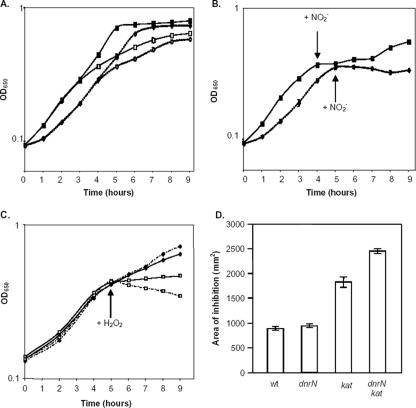
FIG. 2.
Effect of a dnrN mutation on the recovery of N. gonorrhoeae from damage induced by sudden exposure to nitric oxide or hydrogen peroxide. (A and B) Deletion of dnrN results in a growth phenotype in N. gonorrhoeae. The OD650s of oxygen-limited cultures of N. gonorrhoeae strains F62 (dnrN+; solid lines) and JCGC704 (dnrN; dotted lines) were measured at hourly intervals. (A) Growth in the presence (filled symbols) or absence (open symbols) of 5 mM nitrite. (B) Growth in the absence of nitrite until an OD650 of around 0.4 (about 0.16 mg of dry mass ml−1) was reached, followed by shock with 0.5 mM NaNO2 (arrows). Error bars show standard deviations of duplicate cultures. (C and D) Effect of a hydrogen peroxide on the growth of kat and dnrN kat mutants. Oxygen-limited cultures of N. gonorrhoeae JCGC212 (kat; solid lines) and JCGC807 (dnrN kat; dotted lines) were grown in the absence of nitrite (C). Half of the cultures were shocked with 0.5 mM hydrogen peroxide at an OD650 of around 0.4 (shown by arrow; open squares) while the remaining cultures were not treated (filled diamonds). The growth experiment was repeated twice. N. gonorrhoeae strains F62 (wild-type), JCGC704 (dnrN), JCGC212 (kat), and JCGC807 (dnrN kat) were first grown on GC agar plates for 1 day at 37°C (D). A lawn of each strain was spread onto a fresh GC agar plate supplemented with 1 mM sodium nitrite. A 12-mm filter paper disc was seeded in the center of the plate with 10 μl of 30% hydrogen peroxide, and plates were incubated for 4 days at 37°C in an anaerobic jar. The area of growth inhibition was calculated. Error bars are standard deviations of triplicate samples.
Parallel cultures were also grown in the absence of nitrite. Under this condition, expression of the nitrite reductase aniA would still occur (32, 56), but as NO would not be formed, norB would remain repressed by NsrR. When the cultures became oxygen limited, nitrite was added, resulting in a sudden pulse of NO generation. As predicted, after an initial inhibition of growth the parent strain, F62, recovered, but growth inhibition of the dnrN mutant persisted (Fig. 2B). It was concluded that the gonococcal dnrN mutant is more sensitive than its parent to damage induced by a sudden exposure to nitric oxide generated from nitrite.
An NO-sensitive electrode was used to eliminate an alternative possibility, i.e., that the dnrN mutant is defective in its ability to reduce NO compared with the parent strain (50). The rates of NO reduction by bacteria harvested from these cultures were measured. The average values for the two strains were indistinguishable, 162 (±18) nmol of NO reduced min−1· mg of bacterial dry mass−1 for the mutant compared with 164 (±46) nmol of NO reduced min−1· mg of bacterial dry mass−1 for the parental strain. Furthermore, these rates of NO reduction were sufficiently high to exclude the possibility that NO accumulates to a higher concentration in cultures of the mutant, causing more severe or even different types of damage (50).
Pathogenic neisseria synthesize an extremely active catalase that masks any protective functions of other proteins that protect the bacteria from exposure to hydrogen peroxide (22). To reveal whether DnrN plays any role in protection against oxidative stress, the dnrN deletion mutation was transferred into N. gonorrhoeae strain JCGC212, from which the kat gene has been deleted. The effects of exposure to hydrogen peroxide on growth of both the dnrN kat double mutant and its isogenic dnrN+ strain in liquid medium were then compared. Despite the absence of catalase activity, at very low concentrations of H2O2 (<0.5 mM), growth of neither the mutant nor the parental strain was significantly inhibited. Conversely, growth of both strains was completely inhibited at a high concentration of H2O2. However, the dnrN mutant was more sensitive than its parent at an intermediate concentration of H2O2 (Fig. 2C). Disk diffusion assays confirmed that the kat dnrN double mutant was also more sensitive than the kat single mutant to growth in the presence of hydrogen peroxide on solid medium (Fig. 2D). These results implicated DnrN in protection against not only nitrosative stress but also oxidative stress.
Increased sensitivity of the gonococcal dnrN mutant to damage to iron-sulfur centers of the transcription factors FNR and NsrR.
The results presented above established that strains mutated in the gonococcal dnrN and in S. aureus scdA have increased susceptibility to exogenous hydrogen peroxide. This phenotype is frequently correlated with elevated levels of intracellular free iron to which the degradation of iron-sulfur centers contributes (27). In addition, one possible explanation for the sensitivity of the gonococcal dnrN mutant to nitrosative stress is that sudden exposure to NO damaged the iron-sulfur centers of FNR, NsrR, and also many other iron-sulfur proteins. As there is currently no system for overexpressing proteins in the gonococcus, the strategy devised to demonstrate the role for DnrN in repair of nitrosative damage was to monitor by quantitative real-time PCR the accumulation of mRNA synthesized under the control of the two transcription factors, FNR and NsrR, in which iron-sulfur centers are critical for function. First, we exploited the NO-induced damage to the oxygen-sensing [4Fe-4S]+2/+1 iron-sulfur center of FNR that results in loss of DNA binding and transcription activation and consequent loss of aniA expression. The qRT-PCR experiments showed that the loss of aniA expression immediately after exposure to NO was followed by restoration of the accumulation of aniA mRNA in the parental strain but not in the mutant (Fig. 3A). This result confirmed that the damage was repaired more rapidly in a parental strain, F62, than in a dnrN mutant.
FIG. 3.

qRT-PCR analysis of gene expression before and after shock with nitrite. N. gonorrhoeae strains F62 (parent) and JCGC704 (dnrN) were grown under oxygen-limited conditions in the absence of nitrite to an OD650 of ∼0.4 and then shocked with 0.5 mM NaNO2. RNA was isolated preshock and 20, 60, 120, and 180 min after the shock, and quantitative PCR was used to quantify aniA (A), norB (B), and dnrN (C) transcripts. Quantities are normalized against the preshock transcript level for each strain.
The NsrR protein, which is also predicted to contain an Fe-S center (2), represses the expression of the norB gene, but repression is lifted on exposure to low concentrations of NO (20, 41). If the interpretation of the effects of a dnrN mutation on aniA expression is correct, it can be predicted that exposure to NO would result in a rapid increase in norB expression. Repression would be restored rapidly in the parental dnrN+ strain but not in a dnrN mutant. This prediction was confirmed (Fig. 3B). Furthermore, dnrN mRNA also accumulated rapidly in the parental strain following NO exposure, but the level of this transcript also decreased rapidly as NsrR repression was restored (Fig. 3C).
In the absence of S. aureus scdA, the activity of the iron-sulfur enzyme aconitase is decreased.
S. aureus synthesizes a single aconitase, a dehydratase of the tricarboxylic acid cycle, that contains a [4Fe-4S]+2/+1 cluster that is susceptible to damage by NO and oxidants such as hydrogen peroxide. In the absence of scdA, the activity of aconitase was found to be 33% lower than in the S. aureus parent strain. Furthermore, when cell lysates of S. aureus were exposed to NO or to hydrogen peroxide, a faster decrease of the aconitase activity was observed in the scdA mutant than in its parent (Fig. 4A and C). We also tested the influence of ScdA in the recovery of aconitase activity upon damage caused by oxidative or nitrosative stress. The aconitase activities of cell lysates prepared from each culture during subsequent incubation in the absence of NO or H2O2 were then assayed. Tetracycline was added to cultures of each strain to inhibit de novo protein synthesis, and, after a brief exposure to NO or H2O2, hemoglobin or catalase was added to scavenge excess NO or H2O2. Aconitase activity was restored rapidly only in the parental strain (Fig. 4B and D).
FIG. 4.
Nitric oxide and hydrogen peroxide-induced damage to aconitase is more pronounced, and the repair of the damage is severely impaired in the absence of scdA. Cell lysates of the S. aureus RN4220 parent strain (open bars) and the scdA mutant (filled bars) were subjected to 3 mM H2O2 (A and B) or 100 μM NO (C and D). For the time course of damage (A and C) the aconitase activity was monitored for 30 min. To follow the repair of aconitase, after 2 min with H2O2 (B) or 15 min with NO (D), catalase and hemoglobin were added to interrupt the exposures (time zero), and the activity was then monitored. The values are averages of duplicate determinations from two (B and D) or four (A and C) independent experiments, with error bars representing one standard deviation unit. The asterisk represents statistical significance (P < 0.05) using a Student's t test. The values are normalized for the initial activity of each strain (wild type, 17.1 mU/mg protein; scdA, 11.5 mU/mg protein).
Major contribution of S. aureus di-iron ScdA to repair of stress-induced damage to the iron-sulfur center of fumarase.
The phenotype of the S. aureus scdA mutant resembles that recently described for an E. coli ytfE mutant. In both cases, the activities of iron-sulfur-containing enzymes are lower in the mutant (23). To determine whether ScdA and YtfE also showed similar biochemical properties, the recombinant S. aureus ScdA was produced in E. coli and characterized. The purified ScdA protein was isolated as a dimer with a molecular mass of 57 kDa and was found to contain two iron atoms per monomer. The visible spectrum exhibited a broad band at 350 nm, characteristic of iron-containing proteins (data not shown). S. aureus ScdA exhibited an EPR spectrum with g values of 1.96, 1.92, and 1.86 (Fig. 5), which are within the range of values usually observed for proteins containing di-iron centers, including the E. coli YtfE (23, 30).
FIG. 5.

ScdA protein of S. aureus has a di-iron center. EPR spectrum of the as-prepared ScdA protein, recorded at 10 K at a 9.4-MHz microwave frequency with 2.4 mW of microwave power.
The similarity between the E. coli YtfE and S. aureus ScdA proteins led us to investigate whether the recombinant ScdA could support the in vitro repair of a damaged [4Fe-4S] cluster, as shown for E. coli YtfE (24). Indeed, addition of purified ScdA protein to cell lysates of E. coli ΔytfE expressing fumarase A and exposed to hydrogen peroxide (Fig. 6A) or nitric oxide (Fig. 6B) demonstrated that ScdA promotes restoration of the fumarase activity to the levels observed before damage. These results show that S. aureus ScdA is essential for the repair of an [4Fe-4S]+2/+1 protein whose cluster is damaged by oxidative or nitrosative compounds.
FIG. 6.

The ScdA protein of S. aureus repairs the [4Fe-4S] cluster of fumarase A after damage by nitric oxide and hydrogen peroxide. Fumarase activity was monitored in lysates of E. coli K-12 cells (open bars) and E. coli ytfE mutant cells (black bars) expressing fumarase A after treatment with tetracycline and treatment with 4 mM H2O2 for 1 min (A) or 150 μM NO for 10 min (B). Immediately after terminating the stresses by the addition of catalase or hemoglobin, purified ScdA protein was added to ytfE mutant cell lysates (gray bars), and the activity was measured (time zero) and monitored for 30 min. The values are normalized for the initial activity (“before”) of each strain (wild type, 3.7 U/mg protein; ytfE, 2.9 U/mg protein) and are mean values of two experiments analyzed in duplicate. Error bars represent one standard deviation unit.
S. aureus scdA, but not the suf or isc operons, complements the hydrogen peroxide sensitivity of the E. coli ytfE mutant.
Next, we addressed the question of whether S. aureus scdA could replace the function of ytfE. To this end, the E. coli ytfE mutant strain was transformed with plasmids encoding either the E. coli ytfE or the S. aureus scdA genes, and sensitivity of the strains was measured under oxidative stress conditions generated by hydrogen peroxide. The ytfE mutant was more sensitive to hydrogen peroxide than the parent, and hypersensitivity was suppressed by expression in trans of either the E. coli ytfE or the S. aureus scdA gene (Fig. 7).
FIG. 7.

S. aureus scdA, but not the suf or isc operons of E. coli, complement the sensitivity to hydrogen peroxide of the E. coli ytfE mutant. The E. coli K-12 parent strain (wt), ytfE mutant strain (ytfE), ytfE strain expressing E. coli ytfE in trans (ytfE/pytfE), ytfE strain expressing S. aureus scdA in trans (ytfE/pscdA), ytfE strain expressing the E. coli isc operon in trans (ytfE/pisc), and ytfE strain expressing the E. coli suf operon in trans (ytfE/psuf) were grown in LB medium under anaerobic conditions. Cultures were left untreated (filled symbols) or treated with 4 mM H2O2 at an OD600 of ∼0.3 (open symbols). Mean values of two independent experiments with error bars representing the standard deviations are shown.
The E. coli isc and suf operons are proposed to encode proteins that may also be involved in the repair of Fe-S clusters. However, the resistance of the ytfE mutant to hydrogen peroxide was not restored by either the plasmid pRKSUF or pRKISC (Fig. 7), containing the complete sufABCDSE or iscRSUAhscBAfdx operon of E. coli, respectively (51, 52). Hence, the ISC and SUF systems cannot replace YtfE, even though SUF is reported to operate under such stress conditions as oxidative stress and iron starvation (38, 40). Note, however, that the plasmid containing the complete set of suf genes could complement most defects of the ΔiscRSUAhscBAfdx strain (52). We conclude that S. aureus ScdA and E. coli YtfE have similar biochemical roles.
Phylogenetic analysis of ScdA and DnrN homologues.
The amino acid sequences of S. aureus ScdA and gonococcal DnrN share 25 and 31% identity and 46 and 41% similarity to E. coli YtfE, respectively. Moreover, a comprehensive search of the amino acid sequence database revealed that DnrN, ScdA, and E. coli YtfE are members of a large family of proteins that occur widely in the bacterial phyla Proteobacteria, Bacteroidetes, Firmicutes, Actinobacteria, and Acidobacteria. In particular, homologues of these proteins are encoded in the genomes of a significant number of human pathogens, such as Bacillus anthracis, Haemophilus influenzae, and species of the genera Salmonella, Shewanella, Yersinia, and Clostridium. Interestingly, two orthologous sequences were found in the eukaryotic organism Trichomonas vaginalis, which is also a human pathogen.
Since a recent study in the pathogenic yeast Cryptococcus neoformans suggests that this eukaryote contains a homologue of the E. coli YtfE (5), this protein (CNA2870) and other putative fungal homologues were included in the analysis, in spite of the low sequence similarity of CNA2870 to the ScdA, DnrN, and YtfE proteins (7 to 8% identity and 16 to 17% similarity). Using all the above-mentioned amino acid sequences, a dendrogram was constructed (Fig. 8) that shows two main groups, one that includes the ScdA/DnrN/YtfE-like proteins and the other that includes the CNA2870-like proteins, in agreement with the low identity (3 to 11%) and similarity (9 to 22%) values between the sequences from both groups. The group of the ScdA/DnrN/YtfE-like proteins is apparently divided into two other groups, one comprising the majority of the proteobacteria and another containing the sequences of several taxa. The ScdA protein of S. aureus and the DnrN protein of N. gonorrhoeae are clustered separately due to the low amino acid sequence identity between the two proteins (16%).
FIG. 8.
Unrooted dendrogram of ScdA/DnrN family of proteins. The dendrogram was generated with Clustal X and manipulated in TreeView. A total of 102 sequences from S. aureus ScdA and N. gonorrhoeae DnrN homologues were aligned, and the dendrogram was bootstrapped by exclusion gap positions and correcting for multiple substitutions. Shaded boxes distinguish the different taxonomic groups. Abbreviations for the organisms are defined in the legend of Fig. S1 in the supplemental material.
The alignment of the amino acid sequences of the proteins (see Fig. S1 in the supplemental material) that produced the dendrogram in Fig. 8 revealed conservation of some regions (particularly within the ScdA/DnrN/YtfE-like sequences) and a high degree of conservation of the residues His84, His105, His129, Glu133, His160, and His204 (numbering refers to residues in E. coli YtfE). Exceptions are observed for three yeast-like sequences in which Glu133 was replaced by an Asp. Based upon studies with E. coli YtfE, these residues are proposed to constitute the ligand sphere for the di-iron center (our unpublished results). In particular, they are located in conserved α-helix regions of a predicted secondary structure (see Fig. S1 in the supplemental material), corroborating the importance for the function of the four-helix-bundle protein fold that is predicted for ScdA/DnrN/YtfE and characterizes many other di-iron proteins.
RIC, a new family of proteins involved in the repair of iron centers.
The work presented above has revealed the presence in a wide range of human, animal, and plant pathogens of a family of di-iron proteins that have similar functions. Based upon in vivo and in vitro evidence, we have shown that these proteins are present in both gram-positive and gram-negative bacteria and that the two main branches of this protein family can repair Fe-S clusters damaged by exposure to NO and H2O2. Our work corroborates and significantly extends the proposal of Rodionov et al. (45), based on the bioinformatics analysis of complete genome sequences, that DnrN in pathogenic Neisseria is involved in the response to nitrosative stress. Future research must focus on the exact chemical reactions catalyzed by this protein family during the repair process, for example, removal of the nitrosated iron atoms or reinsertion of iron once the primary damage has been removed by other proteins. As it is not known whether the substrates on which these proteins work are limited to those with iron-sulfur centers, we propose the name RIC, for repair of iron centers, for this new and widely distributed protein family.
Supplementary Material
Acknowledgments
Work in the laboratory of J.A.C. was funded by BBSRC project grant P21080 and a Darwin Trust studentship to Y.L. DNA sequencing and quantitative PCR facilities were supported by the Birmingham Functional Genomics Laboratory. Work in the laboratory of L.M.S. was funded by FCT project POCI/SAU-IMI/56088/2004 and an FCT SFRH/BD/13756/2003 studentship to M.C.J.
We are grateful to Yasuhiro Takahashi (Graduate School of Science, Osaka University, Japan) for providing the complementation vectors pRKISC and pRKSUF and to Jeffrey Green (The Krebs Institute for Biomolecular Research, University of Sheffield, United Kingdom) for providing the expression vector pGS57. We thank Miguel Teixeira (Instituto Tecnologia Química e Biológica, UNL, Portugal) for the EPR analysis.
Footnotes
Published ahead of print on 18 January 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Beinert, H., R. H. Holm, and E. Munck. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277653-659. [DOI] [PubMed] [Google Scholar]
- 2.Bodenmiller, D. M., and S. Spiro. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunskill, E. W., B. L. de Jonge, and K. W. Bayles. 1997. The Staphylococcus aureus scdA gene: a novel locus that affects cell division and morphogenesis. Microbiology 1432877-2882. [DOI] [PubMed] [Google Scholar]
- 4.Chang, W., D. A. Small, F. Toghrol, and W. E. Bentley. 2006. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 1881648-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow, E. D., O. W. Liu, S. O'Brien, and H. D. Madhani. 2007. Exploration of whole-genome responses of the human AIDS-associated yeast pathogen Cryptococcus neoformans var. grubii: nitric oxide stress and body temperature. Curr. Genet. 52137-148. [DOI] [PubMed] [Google Scholar]
- 6.Constantinidou, C., J. L. Hobman, L. Griffiths, M. D. Patel, C. W. Penn, J. A. Cole, and T. W. Overton. 2006. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 2814802-4815. [DOI] [PubMed] [Google Scholar]
- 7.Cruz-Ramos, H., J. Crack, G. Wu, M. N. Hughes, C. Scott, A. J. Thomson, J. Green, and R. K. Poole. 2002. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 213235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lencastre, H., I. Couto, I. Santos, J. Melo-Cristino, A. Torres-Pereira, and A. Tomasz. 1994. Methicillin-resistant Staphylococcus aureus disease in a Portuguese hospital: characterization of clonal types by a combination of DNA typing methods. Eur. J. Clin. Microbiol. Infect. Dis. 1364-73. [DOI] [PubMed] [Google Scholar]
- 9.Djaman, O., F. W. Outten, and J. A. Imlay. 2004. Repair of oxidized iron-sulfur clusters in Escherichia coli. J. Biol. Chem. 27944590-44599. [DOI] [PubMed] [Google Scholar]
- 10.Filenko, N., S. Spiro, D. F. Browning, D. Squire, T. W. Overton, J. Cole, and C. Constantinidou. 2007. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J. Bacteriol. 1894410-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, D. S., and D. C. Price. 1964. A simple serum iron method using the new sensitive chromogen tripyridyl-S-triazine. Clin. Chem. 1021-31. [PubMed] [Google Scholar]
- 12.Fontecave, M., S. O. Choudens, B. Py, and F. Barras. 2005. Mechanisms of iron-sulfur cluster assembly: the SUF machinery. J. Biol. Inorg. Chem. 10713-721. [DOI] [PubMed] [Google Scholar]
- 13.Frazzon, J., and D. R. Dean. 2003. Formation of iron-sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Curr. Opin. Chem. Biol. 7166-173. [DOI] [PubMed] [Google Scholar]
- 14.Gardner, A. M., and P. R. Gardner. 2002. Flavohemoglobin detoxifies nitric oxide in aerobic, but not anaerobic, Escherichia coli. Evidence for a novel inducible anaerobic nitric oxide-scavenging activity. J. Biol. Chem. 2778166-8171. [DOI] [PubMed] [Google Scholar]
- 15.Gardner, A. M., R. A. Helmick, and P. R. Gardner. 2002. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J. Biol. Chem. 2778172-8177. [DOI] [PubMed] [Google Scholar]
- 16.Gardner, P. R. 2002. Aconitase: sensitive target and measure of superoxide. Methods Enzymol. 3499-23. [DOI] [PubMed] [Google Scholar]
- 17.Goncalves, V. L., L. S. Nobre, J. B. Vicente, M. Teixeira, and L. M. Saraiva. 2006. Flavohemoglobin requires microaerophilic conditions for nitrosative protection of Staphylococcus aureus. FEBS Lett. 5801817-1821. [DOI] [PubMed] [Google Scholar]
- 18.Hausladen, A., C. T. Privalle, T. Keng, J. DeAngelo, and J. S. Stamler. 1996. Nitrosative stress: activation of the transcription factor OxyR. Cell 86719-729. [DOI] [PubMed] [Google Scholar]
- 19.Huber, C., and G. Wachtershauser. 1998. Peptides by activation of amino acids with CO on (Ni,Fe)S surfaces: implications for the origin of life. Science 281670-672. [DOI] [PubMed] [Google Scholar]
- 20.Isabella, V., L. F. Wright, K. Barth, J. M. Spence, S. Grogan, C. A. Genco, and V. L. Clark. 2008. cis- and trans-acting elements involved in regulation of norB (norZ), the gene encoding nitric oxide reductase in Neisseria gonorrhoeae. Microbiology 154226-239. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, D. C., D. R. Dean, A. D. Smith, and M. K. Johnson. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74247-281. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, S. R., B. M. Steiner, D. D. Cruce, G. H. Perkins, and R. J. Arko. 1993. Characterization of a catalase-deficient strain of Neisseria gonorrhoeae: evidence for the significance of catalase in the biology of N. gonorrhoeae. Infect. Immun. 611232-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Justino, M. C., C. C. Almeida, V. L. Goncalves, M. Teixeira, and L. M. Saraiva. 2006. Escherichia coli YtfE is a di-iron protein with an important function in assembly of iron-sulphur clusters. FEMS Microbiol. Lett. 257278-284. [DOI] [PubMed] [Google Scholar]
- 24.Justino, M. C., C. C. Almeida, M. Teixeira, and L. M. Saraiva. 2007. Escherichia coli di-iron YtfE protein is necessary for the repair of stress-damaged iron-sulfur clusters. J. Biol. Chem. 28210352-10359. [DOI] [PubMed] [Google Scholar]
- 25.Justino, M. C., J. B. Vicente, M. Teixeira, and L. M. Saraiva. 2005. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 2802636-2643. [DOI] [PubMed] [Google Scholar]
- 26.Kellogg, D. S., Jr., W. L. Peacock, Jr., W. E. Deacon, L. Brown, and D. I. Pirkle. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 851274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keyer, K., and J. A. Imlay. 1996. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. USA 9313635-13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiley, P. J., and H. Beinert. 2003. The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 6181-185. [DOI] [PubMed] [Google Scholar]
- 29.Kim, S. O., Y. Orii, D. Lloyd, M. N. Hughes, and R. K. Poole. 1999. Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 445389-394. [DOI] [PubMed] [Google Scholar]
- 30.Kurtz, D. M., Jr. 1997. Structural similarity and functional diversity in diiron-oxo proteins. J. Biol. Inorg. Chem. 2159-167. [Google Scholar]
- 31.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 1796228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lissenden, S., S. Mohan, T. Overton, T. Regan, H. Crooke, J. A. Cardinale, T. C. Householder, P. Adams, C. D. O'Conner, V. L. Clark, H. Smith, and J. A. Cole. 2000. Identification of transcription activators that regulate gonococcal adaptation from aerobic to anaerobic or oxygen-limited growth. Mol. Microbiol. 37839-855. [DOI] [PubMed] [Google Scholar]
- 33.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔCT) method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 34.Loiseau, L., S. Ollagnier-de Choudens, D. Lascoux, E. Forest, M. Fontecave, and F. Barras. 2005. Analysis of the heteromeric CsdA-CsdE cysteine desulfurase, assisting Fe-S cluster biogenesis in Escherichia coli. J. Biol. Chem. 28026760-26769. [DOI] [PubMed] [Google Scholar]
- 35.Massey, V. 1953. Studies on fumarase. III. The effect of temperature. Biochem. J. 5372-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 27829478-29486. [DOI] [PubMed] [Google Scholar]
- 37.Mukhopadhyay, P., M. Zheng, L. A. Bedzyk, R. A. LaRossa, and G. Storz. 2004. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl. Acad. Sci. USA 101745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nachin, L., L. Loiseau, D. Expert, and F. Barras. 2003. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 22427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakano, M. M., H. Geng, S. Nakano, and K. Kobayashi. 2006. The nitric oxide-responsive regulator NsrR controls ResDE-dependent gene expression. J. Bacteriol. 1885878-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Outten, F. W., O. Djaman, and G. Storz. 2004. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52861-872. [DOI] [PubMed] [Google Scholar]
- 41.Overton, T. W., R. Whitehead, Y. Li, L. A. Snyder, N. J. Saunders, H. Smith, and J. A. Cole. 2006. Coordinated regulation of the Neisseria gonorrhoeae-truncated denitrification pathway by the nitric oxide-sensitive repressor, NsrR, and nitrite-insensitive NarQ-NarP. J. Biol. Chem. 28133115-33126. [DOI] [PubMed] [Google Scholar]
- 42.Poole, R. K., and M. N. Hughes. 2000. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36775-783. [DOI] [PubMed] [Google Scholar]
- 43.Pullan, S. T., M. D. Gidley, R. A. Jones, J. Barrett, T. M. Stevanin, R. C. Read, J. Green, and R. K. Poole. 2007. Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: unaltered methionine biosynthesis indicates lack of S nitrosation. J. Bacteriol. 1891845-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson, A. R., P. M. Dunman, and F. C. Fang. 2006. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61927-939. [DOI] [PubMed] [Google Scholar]
- 45.Rodionov, D. A., I. L. Dubchak, A. P. Arkin, E. J. Alm, and M. S. Gelfand. 2005. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol 1e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogers, P. A., L. Eide, A. Klungland, and H. Ding. 2003. Reversible inactivation of E. coli endonuclease III via modification of its [4Fe-4S] cluster by nitric oxide. DNA Repair 2809-817. [DOI] [PubMed] [Google Scholar]
- 47.Sebbane, F., N. Lemaitre, D. E. Sturdevant, R. Rebeil, K. Virtaneva, S. F. Porcella, and B. J. Hinnebusch. 2006. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc. Natl. Acad. Sci. USA 10311766-11771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seib, K. L., H. J. Wu, S. P. Kidd, M. A. Apicella, M. P. Jennings, and A. G. McEwan. 2006. Defenses against oxidative stress in Neisseria gonorrhoeae: a system tailored for a challenging environment. Microbiol. Mol. Biol Rev. 70344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 15076-85. [DOI] [PubMed] [Google Scholar]
- 50.Strube, K., S. de Vries, and R. Cramm. 2007. Formation of a dinitrosyl iron complex by NorA, a nitric oxide-binding di-iron protein from Ralstonia eutropha H16. J. Biol. Chem. 28220292-20300. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi, Y., and M. Nakamura. 1999. Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe-S clusters in Escherichia coli. J. Biochem. (Tokyo) 126917-926. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi, Y., and U. Tokumoto. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 27728380-28383. [DOI] [PubMed] [Google Scholar]
- 53.Thorndycroft, F. H., G. Butland, D. J. Richardson, and N. J. Watmough. 2007. A new assay for nitric oxide reductase reveals two conserved glutamate residues form the entrance to a proton-conducting channel in the bacterial enzyme. Biochem. J. 401111-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner, S., E. Reid, H. Smith, and J. Cole. 2003. A novel cytochrome c peroxidase from Neisseria gonorrhoeae: a lipoprotein from a gram-negative bacterium. Biochem. J. 373865-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uzzau, S., N. Figueroa-Bossi, S. Rubino, and L. Bossi. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA 9815264-15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitehead, R. N., T. W. Overton, L. A. Snyder, S. J. McGowan, H. Smith, J. A. Cole, and N. J. Saunders. 2007. The small FNR regulon of Neisseria gonorrhoeae: comparison with the larger Escherichia coli FNR regulon and interaction with the NarQ-NarP regulon. BMC Genomics 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woods, S. A., S. D. Schwartzbach, and J. R. Guest. 1988. Two biochemically distinct classes of fumarase in Escherichia coli. Biochim. Biophys. Acta 95414-26. [DOI] [PubMed] [Google Scholar]
- 58.Yang, W., P. A. Rogers, and H. Ding. 2002. Repair of nitric oxide-modified ferredoxin [2Fe-2S] cluster by cysteine desulfurase (IscS). J. Biol. Chem. 27712868-12873. [DOI] [PubMed] [Google Scholar]
- 59.Yeo, W. S., J. H. Lee, K. C. Lee, and J. H. Roe. 2006. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol. Microbiol. 61206-218. [DOI] [PubMed] [Google Scholar]
- 60.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 1834562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.