The IX International Conference on Bacterial Locomotion and Signal Transduction (BLAST IX) was held from 14 to 19 January 2007 in Laughlin, NV, a town in the Mojave Desert on the Nevada-Arizona border near old Route 66 and along the banks of the Colorado River. This area is a home to rattlesnakes, sagebrush, abandoned gold mines, and compulsive gamblers. What better venue could scientists possibly dream of for a professional meeting? So there they were, about 190 scientists gathered in the Aquarius Casino Resort, the largest hotel and casino in Laughlin, discussing the latest advances in the field. Nothing, not the bustling sounds from countless slot machines and poker tables or the unusually cold and windy weather, dampened the atmosphere of the conference or distracted those zealots from their science. Aside from a brief excursion to an abandoned gold mine and a dinner cruise on the Colorado River, the scientists focused on nothing but their data and hypotheses, in spirited arguments and rebuttals, and outlined their visions and future plans in a friendly and open environment. The BLAST IX program was dense, with nearly 50 talks and over 90 posters. For that reason, this meeting report will not attempt to be comprehensive; instead it will first provide general background information on the central topics of the meeting and then highlight only a few talks that were of special interest to us and hopefully to the wider scientific community. We will also attempt to articulate some of the future directions or perspectives to the best of our abilities.
The best known and understood bacterial motility mechanism is swimming powered by flagella. The rotation of bacterial flagella drives this form of bacterial movement in an aqueous environment. A bacterial flagellum consists of a helical filament attached to the cell body through a complex structure known as the hook-basal body, which drives flagellar rotation. The essential components of the basal body are the MotA-MotB motor-stator proteins bound to the cytoplasmic membrane (7). These stator proteins interact with proteins that comprise the supramembrane and cytoplasmic rings, which are components of the motor imbedded in the cytoplasmic membrane. The interaction causes the supramembrane and cytoplasmic rings to rotate along with the flagellar filaments. The energy for flagellar rotation comes from proton motive force or other ions, especially sodium in marine bacteria, which generate an electrochemical gradient across the cell membrane. Three proteins, FliM, FliN, and FliG, located at the base of the motor act as switches that control the direction of flagellar rotation (4, 20, 33). As exemplified by the enteric bacteria Escherichia coli and Salmonella enterica serovar Typhimurium, changes in the direction of flagellar rotation affect the swimming behavior of the bacterial cell. Counterclockwise (CCW) rotation of the flagella causes the flagellar filaments to form a bundle that pushes the cell forward in a “run.” In contrast, clockwise (CW) rotation causes the flagellar bundle to fly apart, and the cell tumbles to reorient to a new direction for the ensuing run upon the return of CCW rotation. The interchanging pattern of CCW and CW rotations produces a random walk, composed of relatively long runs with occasional direction changes or turns (5, 38, 63). By modulating the lengths of the runs or the frequency of tumbling, bacteria can regulate their motile behavior to move in a desirable direction.
Many bacteria can also move on surfaces. Except for flagellum-driven swarming motility (23), all the other forms of known bacterial surface movement involve no flagella. The flagellum-independent surface motility, known as gliding, is observed in cyanobacteria (6), Mycoplasma species (3), Cytophaga-Flexibacterium species (43), and Myxococcus species (75). Without a doubt, the most thoroughly studied model gliding bacterium is Myxococcus xanthus, which also serves as a prokaryotic model for developmental biology due to its ability to develop multicellular fruiting bodies.
M. xanthus cells use gliding motility both to hunt for food during vegetative growth and to aggregate during fruiting body formation (75). When nutrients are present, groups of cells or swarms propagate and move outward like hunting wolf packs in search of additional macromolecules or prey. Upon starvation, cells aggregate at discrete foci to form mounds and then macroscopic fruiting bodies, each with hundreds of thousands of cells. The rod-shaped cells in the fruiting bodies eventually morph into spherical spores that are metabolically inactive and partially resistant to desiccation and temperature. When nutrients become available, spores can germinate and reenter the vegetative cell cycle. Two talks highlighted below in this meeting review will tackle the mysteries of the gliding motility of M. xanthus in greater detail.
In addition to M. xanthus, Caulobacter crescentus has extensively been investigated as a bacterial model of cell differentiation and development (27, 67). C. crescentus is a gram-negative bacterium with a dimorphic life cycle that includes swarmer and stalked cells. A motile swarmer cell possesses a single flagellum and several pili at one pole. The swarmer cell swims for about one-third of the cell cycle, and then, during the differentiation process, the polar flagellum is shed, the pili are lost, and a stalk tipped by an adhesive holdfast is synthesized at the pole where the flagellum and pili were originally located. The stalked cell will eventually divide asymmetrically to give rise to another pair of swarmer and stalked daughter cells.
Caulobacter differentiation is controlled in part by two-component regulatory systems which function as the primary signaling pathways in bacteria. A prototypical two-component system is comprised of a sensor histidine protein kinase and its cognate response regulator (53). A relevant signal may induce the autophosphorylation of a conserved histidine on the sensor kinase, which subsequently transfers the phosphoryl group to a conserved aspartate residue on the response regulator protein. Upon modification of their phosphorylation state, response regulators go on to modulate cellular physiology or development by acting either directly as transcription factors or indirectly through interaction with downstream signaling partners (59). One such two-component system of C. crescentus is the subject of further discussion below in this meeting review.
As alluded to earlier, swimming bacteria bias their random walks to move in more-favorable directions, thereby enhancing their survival and growth. Biased movement in a chemical gradient is known as chemotaxis, which enables a cell to move away from repellents or toward attractants. As it swims, the cell must identify and respond to these gradients and at the same time integrate and amplify those signals to coordinate a response that modulates the direction of swimming. The net result is that cells tumble less frequently when they are moving toward higher concentrations of attractants or lower concentrations of repellents, and conversely, they tumble more frequently when moving in disadvantageous directions. Bacterial chemotaxis is achieved, therefore, using a biased random walk.
Bacterial chemotaxis is regulated by a well-studied signal transduction pathway. It is a variant of the two-component system and is easily the best-understood signaling system at the molecular level in any biological system. Transmembrane receptor proteins also known as methyl-accepting chemotaxis proteins (MCPs) or chemoreceptors are the entry point into the signal transduction system that regulates bacterial chemotaxis. MCPs typically have a periplasmic ligand-binding domain for monitoring chemoeffector levels and a highly conserved cytoplasmic signaling domain that communicates with the flagellar rotary motors via protein phosphorylation reactions (50). Chemoreceptors cluster at the poles of the cell in E. coli (39) and form trimers of homodimers that undergo conformational changes upon ligand binding (73). The conformational change in the MCP is transmitted across the membrane, regulating the activity of an associated histidine kinase dimer, CheA, whose autophosphorylation is regulated by receptor occupancy (10, 48). Complexes of chemoreceptors associate with CheA and CheW (a docking protein that promotes the formation of ternary complexes) (11, 55). In enteric bacteria, two response regulators compete for binding to CheA: CheY, a 14-kDa single-domain response regulator, and CheB, a methyl esterase activated via a response regulator domain (24, 25). Phosphorylated CheY (CheY-P) binds to the flagellar switch protein FliM to induce CW rotation and tumbling to change the direction of swimming (68). In the enteric bacteria, CheY-P is dephosphorylated by CheZ, a phosphatase which enhances the speed and coordination of the motor response; e.g., CheY binding to CheZ activates the phosphatase and terminates CW rotation (15, 74). The activity of CheB, another response regulator phosphorylated by CheA, increases about 100-fold on phosphorylation (2). It serves, with the constitutive methyltransferase CheR, to reset the signaling state of the chemoreceptors in a process termed adaptation (36). The rate of spontaneous CheY-P dephosphorylation is increased by the CheZ phosphatase to allow signal termination or decay within the time frame required for the spatial sensing of a gradient (35).
CHEMORECEPTORS: FROM NANODISCS TO TERAFLOPS
The first meeting session was devoted to MCPs or chemoreceptors. Sandy Parkinson (University of Utah) gave an overview on how chemoreceptors function to detect and transduce signals, which mostly came from studies of chemoreceptors in E. coli (50, 57). Two subsequent presentations in the session highlighted the state of the art in chemoreceptor research, with exciting results obtained by applying novel technologies to well-studied systems. The E. coli chemoreceptors are transmembrane dimers that also form higher-order structures—trimers of dimers and extended clusters. Interactions of dimers in higher-order structures are thought to enable receptor cooperativity, their cross-influences, and a multifold gain between ligand binding (signal input) and downstream kinase activity (signal output). An immediate question is which of these properties can be attributed to chemoreceptor dimers and which require a higher-order organization?
Gerald Hazelbauer (University of Missouri—Columbia) reported on the use of an emerging technology for handling MCPs and other membrane proteins through the use of so-called nanodiscs—small, defined-size patches of lipid bilayer (18)—to obtain functional chemoreceptor units consisting of a single dimer or a trimer of dimers (8, 9). Nanodiscs allowed scientists to measure ligand binding (input) and kinase activation (output), as well as transmembrane signaling and adaptational modification of the receptors (methylation and demethylation). Single dimers are capable of binding the ligand, transmembrane signaling, and covalent modification (methylation/demethylation); however, they are hardly able to activate the CheA histidine kinase (Fig. 1). On the other hand, trimers of dimers are capable of full-range control of CheA activation. The significance of this work is that transmembrane signaling occurs without receptor oligomerization beyond homodimers but that downstream signaling requires higher-order oligomerization.
FIG. 1.
Representation of individual bacterial chemoreceptors embedded in lipid bilayers of nanodiscs. These dimeric transmembrane proteins are incorporated in an active state into nanoparticles formed upon the removal of detergent from a solution of membrane scaffold protein (blue cylinders), phospholipid (red ball with white tails), and chemoreceptors (Corey-Pauling-Kolton representation). Chemoreceptors are ∼30 nm long, and nanodiscs are ∼13 nm in diameter.
Another emerging technology that has enabled researchers to achieve substantial progress in studying chemoreceptors is the rapidly accelerating ability to sequence and the ever-increasing number of annotated genomes. Roger Alexander (I. B. Zhulin laboratory, Oak Ridge National Laboratory) reported on the computational structure-guided analysis of more than 2,000 chemoreceptor sequences from more than 150 archaeal and bacterial genomes. Although data mining for this work involved an entry-level, high-performance computer, the exponentially growing number of genome sequences will require that the next round of data mining be carried out on a teraflop scale (computers capable of trillions of mathematical calculations per second). This research revealed several important characteristics about chemoreceptor molecules. First, it is apparent that there are several major classes of chemoreceptor signaling domains that differ in length and sequence conservation. Interestingly, each of the classes has a specific pattern of predicted methylation sites that play a significant role in adaptation, which implies that signaling and adaptation coevolved throughout the natural history of the chemotaxis signal transduction system. Second, this analysis revealed that chemoreceptors maintain the symmetry of their N- and C-terminal helical arms, although the overall lengths of the cytoplasmic domain vary dramatically from more than 400 amino acid residues to less than 170 (Fig. 2). The violation of symmetry was observed in several specific instances, such as in certain types of soluble cytoplasmic receptors, indicating the importance of symmetry in transmembrane signaling. Third, analysis of the chemoreceptor region between the adaptation subdomain (where methylation and demethylation occur) and the signaling subdomain (which interacts with the signaling complex of CheA and CheW) led to the discovery of a new functional region in chemoreceptors, the “flexible bundle subdomain” (1).
FIG. 2.
Cosmological image of chemoreceptor evolution. The figure represents receptor evolution from long to short forms (clockwise). A more realistic view of receptor evolution may be found in reference 1.
The identification of the flexible-bundle subdomain with a conserved glycine hinge, which was previously found to be an important signaling element (16), in the cytoplasmic domain of all known chemoreceptor sequences strongly reinforced the notion that bending of the cytoplasmic domain is central to the signaling mechanism (1). This work also prompted scientists to present a model detailing how trimers of dimers can function based on glycine hinge/flexible-subdomain signaling. This model pulls together a number of recent experimental and computational findings (49).
BLUE LIGHT DISTRICT: NOVEL SIGNALS OF GLOBAL IMPORTANCE
The signals perceived by bacteria were hot topics at BLAST IX, and one of the more interesting aspects discussed involved the perception of light, specifically light in the blue wavelengths, by the nonphotosynthetic bacterium C. crescentus.
The aquatic bacterium C. crescentus, a member of the Alphaproteobacteria group, which includes Sinorhizobium and Rhodobacter, is dimorphic; e.g., each cell division produces two daughter cells that are morphologically and physiologically distinct. The life cycle starts with a motile, chemotactic swarmer cell that has a single polar flagellum used for motility and polar type IV pili that mediate adhesion to biotic and abiotic surfaces. The swarmer cell releases its polar flagellum and retracts its polar pili, causing differentiation into a “stalked” cell (54). The signals and cues perceived by C. crescentus that initiate the differentiation between swarmer and stalked cells are not well understood.
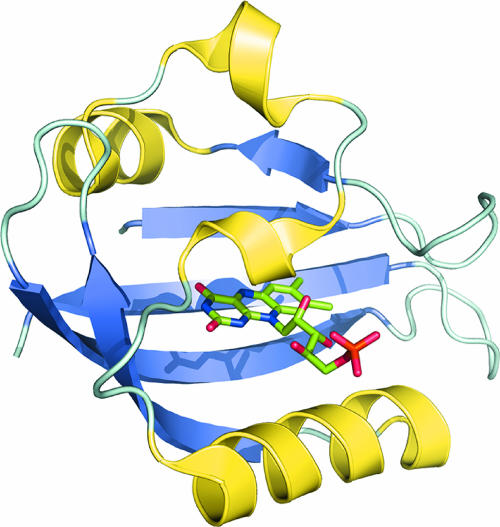
The C. crescentus genome (47) possesses over 64 histidine kinases and 43 response regulators, suggesting that this bacterium has numerous means of sensing its environment and physiological state via two-component signal transduction circuits. These proteins often are modular, containing sensory domains that control signaling outputs, and one of the more common domains found in 20 of the 64 histidine kinases of C. crescentus is the PAS (Per-ARNT-Sim) domain, which is conserved across all kingdoms of life and is capable of specifically binding a wide range of ligands, including heme, flavins, p-coumaric acid, citrate, and other small molecules (61). While PAS domains have low sequence identity, they maintain a conserved structure possessing a fold that is often capable of binding to different ligands. A subclass of PAS domains, called LOV domains (light, oxygen, or voltage) (Fig. 3), commonly bind a flavin cofactor and function to regulate a number of blue-light-dependent processes in plants and fungi (17).
FIG. 3.
Ribbon rendering of the three-dimensional structure of a flavin-binding, photosensory LOV domain (Protein Data Bank identification no. 1G28). In plants and fungi, these domains regulate a number of developmental and circadian responses upon the absorption of blue photons. LOV family sensor histidine kinases have been discovered in a number of bacterial species. The bacterium C. crescentus encodes a LOV photosensory histidine kinase that has been implicated in the light-mediated regulation of cell attachment.
Sean Crosson (University of Chicago) reported on the role of the LovK and LovR two-component system in sensing blue light and in regulating cell activity in C. crescentus. The genes for the LOV histidine kinase LovK (open reading frame CC0285) and the single-domain response regulator LovR (CC0284) are only 17 bp apart (52). LovK is an unorthodox histidine kinase in that its kinase domain is missing an F box and contains a truncated G box and an N box with an E-for-N substitution. The kinase domain of LovK is known as an HWE histidine kinase (29) and is also called COG3920 in the conserved domain database.
The phenotypes of lovR and lovK mutants suggested that this two-component system is involved in light regulation, and a microarray transcriptome analysis showed that the levels of lovK and lovR are modulated by the cell cycle. The results from mutating lovR and lovK came as a surprise. When lovR is knocked out, biofilm and stalk formation are reduced and the reduction is blue light dependent. In either a lovK or lovR null mutant, stalk length decreases, and the overexpression of LovK causes a loss of the stalk in the dark. When both LovK and LovR are overexpressed, stalk length is partially rescued. These data fit into a scenario where LovK is activated by light and acts as a repressor of stalk length in the dark, and LovR can titrate out the LovK effect. In response to light, LovK is activated and is phosphorylated, after which LovK-P transfers the phosphoryl group to LovR. LovR-P acts as a negative regulator of stalk length. However, in the light, knocking out LovK or LovR does not have a strong effect, and long stalks comparable to those observed in the dark are not seen in the light, so there may be other factors controlling stalk length. Nonetheless, these findings are exciting and underscore light as a signal that may influence the physiology of many other nonphotosynthetic bacteria, including the human/animal facultative intracellular pathogen Brucella melitensis, the plant pathogen Pseudomonas syringae, and the marine bacterium Erythrobacter litoralis (32), all of which possess histidine kinases with LOV domains.
How does Caulobacter use blue light to aid in adaptation and survival? Crosson suggests that light is an environmental signal that can cue the Caulobacter cell on where it is positioned in the water column (52). Since the blue region of the visible spectrum penetrates the water column to a greater depth than lower-energy yellow and red light and nutrients are less abundant at the surface than in deeper water, sensing blue light may signal the cells to attach to nutrient-rich particles at the surface and could provide a survival advantage.
LOCATION, LOCATION, LOCATION: GETTING YOUR TRANSDUCER TO WHERE IT DOES THE MOST GOOD
How proteins are localized within the cell was the focus of several talks and posters at BLAST IX, and no system was discussed more than the molecular mechanisms underlying the localization of chemotaxis proteins and receptors. The components of the E. coli chemotaxis pathway predominantly localize to specific regions at the poles of the bacterial cell (39, 58). Localization is dependent upon the presence of CheW (a docking protein), CheA (a histidine protein kinase), and MCPs (also referred to as receptors). As stated earlier, MCPs are dimeric receptors whose periplasmic ligand-binding domains signal across the cell membrane to the cytoplasmic signaling domain. This link is mediated by HAMP domains, which are found after the cytoplasmic end of the transmembrane region of the receptor and are essential for chemotaxis (14). MCP-CheW-CheA form a complex that is predominantly found in clusters at the poles of the cell (37, 55). Both CheA and CheW are required for the formation of tight receptor clusters (55). The formation of tight clusters of chemotaxis proteins has been suggested to be important in the process of signal generation and amplification and receptor adaptation (13, 19).
The purple nonsulfur bacterium Rhodobacter sphaeroides responds by chemotaxis to a wide range of stimuli and provides an interesting twist to the canonical E. coli model of receptor-mediated localization. The R. sphaeroides genome harbors not one but three major chemotaxis operons (cheOp1, cheOp2, and cheOp3), containing multiple homologues of most of the chemotaxis genes found in E. coli. R. sphaeroides has 13 putative chemoreceptors, nine of which are transmembrane MCPs. R. sphaeroides membrane-spanning MCPs localize to the poles of the cell, with most of the chemosensory homologues encoded in cheOp2 (40, 64, 66). The remaining four putative receptors, the transducer-like proteins (Tlps), lack transmembrane domains and localize to a discrete region in the cytoplasm with chemosensory homologues encoded by cheOp3 (65, 66). Genes in both cheOp2 and cheOp3 are essential for chemotaxis (51). It was perhaps not unexpected that some of the chemotaxis proteins, namely, CheA2, CheW2, and CheW3, localize to the poles of the cell in a manner reminiscent of that seen in E. coli. What was surprising was that TlpT along with other components (CheA3, CheA4, CheW4, and TlpC) localizes to a discrete cluster within the cytoplasm of the bacterium (66).
Continuing this story, George Wadhams (Armitage Laboratory, Oxford University) reported on the laboratory's recent results in finding the intrinsic determinants of TlpT controlling its localization in the cytoplasmic cluster. TlpT, homologous to other MCPs, with a conserved signaling domain, lacks transmembrane regions. It contains two putative HAMP domains and a proline-rich N terminus. Wadhams and colleagues examined the role of the HAMP domains in signaling and in cytoplasmic cluster localization using a TlpT-yellow fluorescent protein (YFP) fusion protein (62). As shown in Fig. 4B, TlpT-YFP localizes to a discrete single cluster positioned roughly in the middle of the cell. Prior to cell division, two clusters located at the one-fourth and three-quarter positions are seen (62), which return to one cluster per cell after division. Deletion of either or both HAMP domains (Fig. 4C to E) results in nonchemotactic cells and the loss of the tight clustering of TlpT. Surprisingly, TlpT HAMP domains are not required for the correct cytoplasmic localization of CheA3 or CheA4, as their localization is unaffected by HAMP domain deletion.
FIG. 4.
Fluorescence microscopy images of derivatives of TlpT-YFP in R. sphaeroides. (A) Wild type; (B) TlpT-YFP; (C) TlpT-YFP ΔHAMPA (amino acids 107 to 169); (D) TlpT-YFP ΔHAMPB (amino acids 198 to 253); (E) TlpT-YFP ΔHAMPAB (amino acids 107 to 253); (F) TlpT-YFP in cephalexin-treated cells; (G) TlpT-YFP ΔN terminus (amino acids 11 to 102) in cephalexin-treated cells. Note that the HAMP domains of TlpT are involved in the localization of the protein to the central cytoplasmic chemotaxis protein cluster but that the N-terminal proline-rich region is involved in regulating the number and position of the cytoplasmic cluster in a way similar to that for PpfA (62).
What is the role of the proline-rich N terminus of TlpT? Proline-rich domains of other signaling proteins are important in protein-protein interactions (30). Wadhams treated cells with cephalexin, a drug that inhibits division resulting in long filaments, and examined the localization of TlpT-YFP when its proline-rich N terminus is removed (Fig. 4F and G). A wild-type Tlp-YFP fusion protein localizes to multiple foci in the cephalexin-induced long filaments. Deletion of the proline-rich domain results in a single cluster that is much brighter than wild-type TlpT-YFP, indicating that the loss of the N-terminal proline-rich domain of TlpT is important for the correct localization and partitioning of this cytoplasmic chemoreceptor in R. sphaeroides.
Do the HAMP domains of other chemoreceptor proteins also function in determining localization in the cell? Is the function of the proline-rich N-terminal region of TlpT to interact and bind with other proteins in the cytoplasmic cluster or nearby? The answers to these questions have implications that go beyond Rhodobacter.
MYXOCOCCUS GLIDING: TO BE OR NOT TO BE POLAR—THAT IS THE QUESTION
Dynamic protein localization was at the core of two competing models for the adventurous (A) gliding motility of M. xanthus. It has been known for almost 30 years that M. xanthus possesses two genetically distinct systems for its surface gliding motility (26). Isolated cells use A motility to move about, whereas social (S) motility is functional only when cells are in close proximity or in groups. Recent advances support a model in which S motility is powered by type IV pili that localize at the leading pole of a rod-shaped M. xanthus cell (42), which is similar to the mechanism used in bacterial twitching motility (41). The mechanism for A motility still remains more of a mystery. The talks by David Zusman (Berkeley, CA) and Simone Leonardy (Søgaard-Andersen Group, Marburg, Germany) focused on unraveling this mystery by examining the localization of AglZ and RomR, two proteins essential for A motility (34, 44). Their results led them to almost opposite conclusions regarding the location of the A-motility engine: the lagging pole is where the A engine resides according to Leonardy, whereas Zusman concluded that A engines are distributed periodically along the cell body.
AglZ, indispensable for A motility (71), was found to localize to discrete foci along the cell by the Zusman group (46). This was surprising because FrzS, an S-motility protein with architecture similar to that of AglZ, colocalizes with type IV pili to the leading pole (45). Another of their critical and keen observations was that these AglZ foci remained stationary relative to the surface over which an A-motile cell glides. That is, the cell body moves through these seemingly fixed foci during gliding. As the lagging cell pole moved close to a preexisting AglZ cluster, it was seen to disintegrate, while new clusters appeared near the leading pole. Quantitative analysis indicates that the spacing of AglZ foci is periodic, with an average distance of 466 nm between them. This distance is about the same as the helical pitch of the filaments formed by the bacterial-actin-like protein MreB (22), suggesting the involvement of cytoskeleton-associated structures/complexes in A motility. Observations of moving cells that bend indicated that AglZ clusters mark sites of focal adhesions of the cell to the substratum underneath. Furthermore, examination of the cells that underwent flailing motion (because their leading poles were stuck on the substratum) suggested that the propulsion or force required for A motility is generated at these adhesion sites. They proposed that these adhesion complexes along the cell body power M. xanthus A motility (Fig. 5). Intracellular motors are proposed to be connected to both a helical cytoskeleton and membrane-spanning adhesion complexes. One end of the motor would move along the cytoskeleton, while the other end would be tethered to the immobilized adhesion complex. Such a mechanism, reminiscent of the cytoskeleton and adhesion-based gliding motility in the eukaryotic apicomplexa, would allow forward movement accompanied by rotation of the cell body (31, 44, 46).
FIG. 5.
Focal-adhesion model for M. xanthus A motility. According to Mignot et al. (46), A motility involves multiple transient adhesion complexes (colored ovals on the bottoms of the cells) that are located throughout the length of a cell. Motors within the cytoplasm are part of the large transmembrane adhesion complexes. The motors are proposed to move along an unknown helical cytoskeletal filament. The A-motility motors, which remain stationary relative to the substratum, move along the filament, providing cell locomotion and cell body rotation. The vertical dashed line indicates one stationary adhesion site as the cell moves forward through three different time frames.
The Søgaard-Andersen group discovered an A-motility protein, RomR (34). Using green fluorescent protein (GFP) fusions, RomR was found to localize dynamically in a bipolar but asymmetric pattern. In a cell that moves by A motility, the larger of the two RomR clusters coincides mostly with the lagging pole. Dynamic RomR localization parallels cell reversal. That is, during a directional reversal in gliding, the larger RomR-GFP cluster at the previous lagging pole fades to a smaller one, while the new lagging pole acquires a brighter cluster. This change in RomR localization after a reversal is not due to new protein synthesis since it occurs in the presence of chloramphenicol. The dynamic localization of RomR is likely regulated by the Frz chemosensory system: in a frzE mutant which does not reverse its gliding direction (75), lagging poles predominantly harbor larger RomR clusters. RomR is a protein with an N-terminal receiver domain and a C-terminal proline- and glutamine-rich output domain. It appears that the output domain alone, which localizes asymmetrically to the cell poles, is sufficient to confer A motility but not to correct regulation: cells with only the output domain move in one direction and fail to reverse. Therefore, the receiver domain, which in itself fails to localize to the cell poles, is possibly responsible for the dynamic localization of RomR. What was striking was that a mutated and likely constitutively active fusion protein, RomRD53E-GFP, restores A motility and cell reversal to a romR frzE double mutant. In other words, mutations in romR can suppress frzE mutations. This indicates that the Frz pathway and RomR interact functionally, if not physically, and underscores the importance of RomR as a regulator of A motility. The Søgaard-Andersen group suggested that RomR is a positive regulator of the A engine. If so and if the function of the A-motility engine is sensitive to RomR in a dose-dependent manner, the dynamic localization of RomR would support a model in which the A-motility engine is located at the lagging pole to propel cells forward by slime secretion.
There needs to be a consolidated model for A motility that can accommodate the findings from RomR, AlgZ, and other studies of A motility. There are two main lines of evidence for a slime jet engine at the rear of the cell (69, 70, 72). The first is the observation of increasing numbers of nozzle-like structures at cell poles and their possible association with slime secretion and A motility. The second includes theoretical calculations that studied the feasibility of slime secretion as a propelling force for A motility. The focal adhesion model for A motility is a newcomer (31, 44). It is supported by the studies of AglZ by the Zusman group. In addition, two studies support the notion that A-motility engines are mostly, if not solely, located along the cell body instead of the lagging pole (56, 60). Is it possible that RomR could be a negative regulator of assembly of the focal adhesion and motor complexes proposed by the Zusman group? In a moving cell, these complexes disappear near the lagging pole with the larger RomR cluster and reappear near the leading pole with the smaller RomR cluster after all. An alternative explanation, acknowledged (perhaps reluctantly) by both sides, is that there could be two sets of A engines (34, 46): one set at the lagging pole for slime secretion and another along the cell body as signified by the periodic AglZ clusters. If this were the case, might it be possible to document the A-motility movement of either a romR or an aglZ mutant but not a romR aglZ double mutant? Hopefully there will be a unified model for A motility that is well reasoned and testable as more progress is made.
BLAST AWARDS
In keeping with past meetings, BLAST IX participants were delighted to award two prizes for student poster presentations from the many excellent posters presented. The Robert Macnab prize was awarded to Takanori Hirano, a postdoctoral scientist in Kelly Hughes's (University of Utah) laboratory for his poster “Hook Length versus Rod plus Hook Length: Longer Rod Structure Bypasses Negative Regulation of Flagellar Assembly in Salmonella.” The inaugural Robert Kadner prize was given to Roger Draheim, a graduate student in Michael Manson's (Texas A&M University) lab, who presented his poster “Tuning a Bacterial Chemoreceptor with Protein-Membrane Interactions.”
FUTURE PERSPECTIVES
The last 4 decades have seen amazing advances in our understanding of the mechanics of bacterial motion. During this time, new discoveries of the molecules and mechanisms involved in transmitting environmental and cellular signals in bacteria have been unveiled. The state of the art in flagellum-dependent swimming motility and chemotaxis is moving beyond molecular interactions and, in some cases, is now focused on interactions at the atomic scale. The advent of rapid sequencing and computer analysis has revealed numerous signaling proteins and domains, and new ones are discovered on an almost monthly basis. Yet, these revelations have also brought forth a new set of questions that beg further study and analysis. We have raised some of these questions during our discussion of the highlights from BLAST IX in this meeting review, but many more remain. Questions related to the less-studied forms of motility and their regulation continue to be hot topics in our field. For example, while the mechanisms and regulation of gliding motility in M. xanthus are now becoming clearer, it is apparent that other species use different mechanisms to glide (43). Many of these less-studied bacteria also possess multiple chemosensory systems; why and how do they “isolate” or manage the unavoidable cross talk? As we gaze into our crystal ball, we boldly list the following as some of the areas of breakthroughs and rapid advances in the foreseeable future.
New insights from genomics.
Increased computational power and sequencing speeds will allow greater systematic analysis of signaling proteins and circuits. How many more signaling domains and their combinations are out there? How well does the knowledge derived from model organisms represent the diversity in nature?
Discovery of novel signaling and new signaling molecules.
C. crescentus LovK provides a wonderful example of the unexpected, i.e., the response to light by a nonphotosynthetic bacterium. Other blue-light sensors, e.g., AppA of R. sphaeroides (28), containing BLUF (sensors of blue light by using flavin adenine dinucleotide) are also known and currently being studied (12, 21). We often have little knowledge of the “true” signal sensed by the cell. Even in well-studied models such as C. crescentus, the true identity of the signal inducing (in this case) differentiation remains unknown. What other cues are sensed by bacteria, and how are those signals received and transmitted into the cell?
Better understanding of supermolecular organization of signaling components.
Morphological and functional asymmetry occurs in many bacteria. What purpose may the differential localization of signaling proteins serve in the bacterial cell? What are the mechanisms by which localization and trafficking of signaling molecules occur in bacteria? What controls the localization of proteins in bacteria? How do receptors aggregate into clusters? How many conformational states exist for receptors? How are signals passed from receptors to CheA? Is ligand binding important in clustering? New technologies are enhancing our ability to answer these questions.
Insights into integration and coordination of multiple signals.
Cells are faced with a plethora of diverse signals in nature. How does the cell integrate and coordinate stimulation from multiple environmental stimuli?
Acknowledgments
We thank the organizers, participants, and all the many people who worked to make BLAST IX possible. We especially acknowledge the chairpersons, Ikuro Kawagishi (Nagoya University, Japan), David Zusman (University of California, Berkeley, CA), John Parkinson (University of Utah, Salt Lake City, UT), Nyles Charon (West Virginia University), Bob Bourret (University of North Carolina), Linda Kenney (University of Illinois—Chicago), and Rudy Schmitt (University of Regensburg). Our special thanks go to Phil Matsumura (chairman of BLAST), Peggy O'Neill, and Tarra Bollinger for all of their efforts in organizing the meeting and ensuring its success. We are indebted to Gerry Hazelbauer, Sean Crosson, George Wadhams, and David Zusman for contributing figures and helpful suggestions during the writing of this report. We thank the speakers for permission to cite their work and apologize to those whose work we were unable to fit into this short summary.
The work in our laboratories was supported by grants from the National Science Foundation (MCB0446001 to R.B.) and the National Institutes of Health (GM72285 to I.B.Z. and GM071601 to Z.Y.).
Footnotes
Published ahead of print on 18 January 2008.
REFERENCES
- 1.Alexander, R. P., and I. B. Zhulin. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. USA 1402885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand, G. S., P. N. Goudreau, and A. M. Stock. 1998. Activation of methylesterase CheB: evidence of a dual role for the regulatory domain. Biochemistry 3714038-14047. [DOI] [PubMed] [Google Scholar]
- 3.Balish, M. F., and D. C. Krause. 2006. Mycoplasmas: a distinct cytoskeleton for wall-less bacteria. J. Mol. Microbiol. Biotechnol. 11244-255. [DOI] [PubMed] [Google Scholar]
- 4.Berg, H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 7219-54. [DOI] [PubMed] [Google Scholar]
- 5.Berg, H. C., and D. Brown. 1972. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 239500-504. [DOI] [PubMed] [Google Scholar]
- 6.Bhaya, D. 2004. Light matters: phototaxis and signal transduction in unicellular cyanobacteria. Mol. Microbiol. 53745-754. [DOI] [PubMed] [Google Scholar]
- 7.Blair, D. F., and H. C. Berg. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60439-449. [DOI] [PubMed] [Google Scholar]
- 8.Boldog, T., S. Grimme, M. Li, S. G. Sligar, and G. L. Hazelbauer. 2006. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. USA 10311509-11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boldog, T., M. Li, and G. L. Hazelbauer. 2007. Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 423317-335. [DOI] [PubMed] [Google Scholar]
- 10.Borkovich, K. A., N. Kaplan, J. F. Hess, and M. I. Simon. 1989. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. USA 861208-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boukhvalova, M. S., F. W. Dahlquist, and R. C. Stewart. 2002. CheW binding interactions with CheA and Tar. Importance for chemotaxis signaling in Escherichia coli. J. Biol. Chem. 27722251-22259. [DOI] [PubMed] [Google Scholar]
- 12.Braatsch, S., and G. Klug. 2004. Blue light perception in bacteria. Photosynth. Res. 7945-57. [DOI] [PubMed] [Google Scholar]
- 13.Bray, D., M. D. Levin, and C. J. Morton-Firth. 1998. Receptor clustering as a cellular mechanism to control sensitivity. Nature 39385-88. [DOI] [PubMed] [Google Scholar]
- 14.Butler, S. L., and J. J. Falke. 1998. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry 3710746-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantwell, B. J., R. R. Draheim, R. B. Weart, C. Nguyen, R. C. Stewart, and M. D. Manson. 2003. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J. Bacteriol. 1852354-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coleman, M. D., R. B. Bass, R. S. Mehan, and J. J. Falke. 2005. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching. Biochemistry 447687-7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crosson, S., S. Rajagopal, and K. Moffat. 2003. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry 422-10. [DOI] [PubMed] [Google Scholar]
- 18.Denisov, I. G., Y. V. Grinkova, A. A. Lazarides, and S. G. Sligar. 2004. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 1263477-3487. [DOI] [PubMed] [Google Scholar]
- 19.Duke, T. A. J., and D. Bray. 1999. Heightened sensitivity of a lattice of membrane receptors. Proc. Natl. Acad. Sci. USA 9610104-10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisenbach, M. 2007. A hitchhiker's guide through advances and conceptual changes in chemotaxis. J. Cell. Physiol. 213574-580. [DOI] [PubMed] [Google Scholar]
- 21.Gomelsky, M., and G. Klug. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27497-500. [DOI] [PubMed] [Google Scholar]
- 22.Graumann, P. L. 2007. Cytoskeletal elements in bacteria. Annu. Rev. Microbiol. 61589-618. [DOI] [PubMed] [Google Scholar]
- 23.Harshey, R. 1994. Bees aren't the only ones: swarming in gram-negative bacteria. Mol. Microbiol. 13389-394. [DOI] [PubMed] [Google Scholar]
- 24.Hess, J. F., R. B. Bourret, K. Oosawa, P. Matsumura, and M. I. Simon. 1988. Protein phosphorylation and bacterial chemotaxis. Cold Spring Harbor Symp. Quant. Biol. 141-48. [DOI] [PubMed] [Google Scholar]
- 25.Hess, J. F., R. B. Bourret, and M. I. Simon. 1988. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature 336139-143. [DOI] [PubMed] [Google Scholar]
- 26.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus: two gene systems control movement. Mol. Gen. Genet. 171177-191. [Google Scholar]
- 27.Jacobs-Wagner, C. 2004. Regulatory proteins with a sense of direction: cell cycle signalling network in Caulobacter. Mol. Microbiol. 517-13. [DOI] [PubMed] [Google Scholar]
- 28.Jäger, A., S. Braatsch, K. Haberzettl, S. Metz, L. Osterloh, Y. Han, and G. Klug. 2007. The AppA and PpsR proteins from Rhodobacter sphaeroides can establish a redox-dependent signal chain but fail to transmit blue-light signals in other bacteria. J. Bacteriol. 1892274-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karniol, B., and R. D. Vierstra. 2004. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling. J. Bacteriol. 186445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kay, B. K., M. P. Williamson, and M. Sudol. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14231-241. [PubMed] [Google Scholar]
- 31.Kearns, D. B. 2007. Microbiology. Bright insight into bacterial gliding. Science 315773-774. [DOI] [PubMed] [Google Scholar]
- 32.Kennis, J. T., and S. Crosson. 2007. Microbiology. A bacterial pathogen sees the light. Science 3171041-1042. [DOI] [PubMed] [Google Scholar]
- 33.Kojima, S., and D. F. Blair. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 23393-134. [DOI] [PubMed] [Google Scholar]
- 34.Leonardy, S., G. Freymark, S. Hebener, E. Ellehauge, and L. Sogaard-Andersen. 2007. Coupling of protein localization and cell movements by a dynamically localized response regulator in Myxococcus xanthus. EMBO J. 264433-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukat, G. S., and J. B. Stock. 1993. Response regulation in bacterial chemotaxis. J. Cell. Biochem. 5141-46. [DOI] [PubMed] [Google Scholar]
- 36.Lupas, A., and J. Stock. 1989. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J. Biol. Chem. 26417337-17342. [PubMed] [Google Scholar]
- 37.Lybarger, S. R., and J. R. Maddock. 1999. Clustering of the chemoreceptor complex in Escherichia coli is independent of the methyltransferase CheR and the methylesterase CheB. J. Bacteriol. 1815527-5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macnab, R. M., and D. E. Koshland, Jr. 1972. The gradient-sensing mechanism in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 692509-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 2591717-1723. [DOI] [PubMed] [Google Scholar]
- 40.Martin, A. C., G. H. Wadhams, and J. P. Armitage. 2001. The roles of the multiple CheW and CheA homologues in chemotaxis and in chemoreceptor localization in Rhodobacter sphaeroides. Mol. Microbiol. 401261-1272. [DOI] [PubMed] [Google Scholar]
- 41.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56289-314. [DOI] [PubMed] [Google Scholar]
- 42.Mauriello, E. M., and D. R. Zusman. 2007. Polarity of motility systems in Myxococcus xanthus. Curr. Opin. Microbiol. 10624-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McBride, M. J. 2004. Cytophaga-flavobacterium gliding motility. J. Mol. Microbiol. Biotechnol. 763-71. [DOI] [PubMed] [Google Scholar]
- 44.Mignot, T. 26 July 2007, posting date. The elusive engine in Myxococcus xanthus gliding motility. Cell. Mol. Life Sci. doi: 10.1007/s00018-007-7176-x. [DOI] [PMC free article] [PubMed]
- 45.Mignot, T., J. P. Merlie, Jr., and D. R. Zusman. 2005. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science 310855-857. [DOI] [PubMed] [Google Scholar]
- 46.Mignot, T., J. W. Shaevitz, P. L. Hartzell, and D. R. Zusman. 2007. Evidence that focal adhesion complexes power bacterial gliding motility. Science 315853-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 984136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ninfa, E. G., A. Stock, S. Mowbray, and J. B. Stock. 1991. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J. Biol. Chem. 2669764-9770. [PubMed] [Google Scholar]
- 49.Parkinson, J. S. 2007. Ancient chemoreceptors retain their flexibility. Proc. Natl. Acad. Sci. USA 1042559-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parkinson, J. S., P. Ames, and C. A. Studdert. 2005. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 8116-121. [DOI] [PubMed] [Google Scholar]
- 51.Porter, S. L., and J. P. Armitage. 2002. Phosphotransfer in Rhodobacter sphaeroides chemotaxis. J. Mol. Biol. 32435-45. [DOI] [PubMed] [Google Scholar]
- 52.Purcell, E. B., D. Siegal-Gaskins, D. C. Rawling, A. Fiebig, and S. Crosson. 2007. A photosensory two-component system regulates bacterial cell attachment. Proc. Natl. Acad. Sci. USA 10418241-18246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson, V. L., D. R. Buckler, and A. M. Stock. 2000. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 7626-633. [DOI] [PubMed] [Google Scholar]
- 54.Skerker, J. M., and M. T. Laub. 2004. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat. Rev. Microbiol. 2325-337. [DOI] [PubMed] [Google Scholar]
- 55.Skidmore, J. M., D. D. Ellefson, B. P. McNamara, M. M. P. Couto, A. J. Wolfe, and J. R. Maddock. 2000. Polar clustering of the chemoreceptor complex in Escherichia coli occurs in the absence of complete CheA function. J. Bacteriol. 182967-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sliusarenko, O., D. R. Zusman, and G. Oster. 2007. The motors powering A-motility in Myxococcus xanthus are distributed along the cell body. J. Bacteriol. 1897920-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sourjik, V. 2004. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 12569-576. [DOI] [PubMed] [Google Scholar]
- 58.Sourjik, V., and H. C. Berg. 2000. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37740-751. [DOI] [PubMed] [Google Scholar]
- 59.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 60.Sun, H., Z. Yang, and W. Shi. 1999. Effect of cellular filamentation on adventurous and social gliding motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 9615178-15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson, S. R., G. H. Wadhams, and J. P. Armitage. 2006. The positioning of cytoplasmic protein clusters in bacteria. Proc. Natl. Acad. Sci. USA 1038209-8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turner, L., W. S. Ryu, and H. C. Berg. 2000. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 1822793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wadhams, G. H., A. C. Martin, and J. P. Armitage. 2000. Identification and localization of a methyl-accepting chemotaxis protein in Rhodobacter sphaeroides. Mol. Microbiol. 361222-1233. [DOI] [PubMed] [Google Scholar]
- 65.Wadhams, G. H., A. C. Martin, S. L. Porter, J. R. Maddock, J. C. Mantotta, H. M. King, and J. P. Armitage. 2002. TlpC, a novel chemotaxis protein in Rhodobacter sphaeroides, localizes to a discrete region in the cytoplasm. Mol. Microbiol. 461211-1221. [DOI] [PubMed] [Google Scholar]
- 66.Wadhams, G. H., A. V. Warren, A. C. Martin, and J. P. Armitage. 2003. Targeting of two signal transduction pathways to different regions of the bacterial cell. Mol. Microbiol. 50763-770. [DOI] [PubMed] [Google Scholar]
- 67.Wagner, J. K., and Y. V. Brun. 2007. Out on a limb: how the Caulobacter stalk can boost the study of bacterial cell shape. Mol. Microbiol. 6428-33. [DOI] [PubMed] [Google Scholar]
- 68.Welch, M., K. Oosawa, S. Aizawa, and M. Eisenbach. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. USA 908787-8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolgemuth, C., E. Hoiczyk, D. Kaiser, and G. Oster. 2002. How myxobacteria glide. Curr. Biol. 12369-377. [DOI] [PubMed] [Google Scholar]
- 70.Wolgemuth, C. W. 2005. Force and flexibility of flailing myxobacteria. Biophys. J. 89945-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, R., S. Bartle, R. Otto, A. Stassinopoulos, M. Rogers, L. Plamann, and P. Hartzell. 2004. AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus. J. Bacteriol. 1866168-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu, R., and D. Kaiser. 2007. Gliding motility and polarized slime secretion. Mol. Microbiol. 63454-467. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, P., E. Bos, J. Heymann, H. Gnaegi, M. Kessel, P. J. Peters, and S. Subramaniam. 2004. Direct visualization of receptor arrays in frozen-hydrated sections and plunge-frozen specimens of E. coli engineered to overproduce the chemotaxis receptor Tsr. J. Microsc. 21676-83. [DOI] [PubMed] [Google Scholar]
- 74.Zhao, R., E. J. Collins, R. B. Bourret, and R. E. Silversmith. 2002. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat. Struct. Biol. 9570-575. [DOI] [PubMed] [Google Scholar]
- 75.Zusman, D. R., A. E. Scott, Z. Yang, and J. R. Kirby. 2007. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 5862-872. [DOI] [PubMed] [Google Scholar]