Abstract
The Mv1751 gene product is thought to catalyze the first step in the N-glycosylation pathway in Methanococcus voltae. Here, we show that a conditional lethal mutation in the alg7 gene (N-acetylglucosamine-1-phosphate transferase) in Saccharomyces cerevisiae was successfully complemented with Mv1751, highlighting a rare case of cross-domain complementation.
The process of covalently attaching a carbohydrate glycan to asparagine residues within a protein is termed N-linked glycosylation. The vast majority of eukaryotes (fungi, plants, animals, slime molds, and euglenas) synthesize asparagine-linked glycosylation (ALG) by initiation of the synthesis of lipid-linked oligosaccharides in a cyclic pathway, the dolichol cycle (11, 12, 14, 19, 22). Although the body of evidence recognizing that Archaea also N glycosylate proteins has existed for some time (9, 17), only recently have genes responsible for assembling glycans and attaching them to archaeal proteins been experimentally identified (1, 4). These genes, termed agl (archaeal glycosylation) genes, are recognized as possessing many similarities to their counterparts in the eukaryotic and/or bacterial domains. For example, the oligosaccharyl transferase which transfers the complete glycan to its target protein and creates the N-glycosidic bond is a STT3 homolog in all three domains, although in prokaryotes it acts as a single protein and in eukaryotes it acts as part of a multiprotein oligosaccharyl transferase complex (3, 20).
Research into the N-glycosylation pathway in Archaea in our laboratory has used the methanogen Methanococcus voltae as a model organism. The flagella and surface layer proteins in this species are known to possess a trisaccharide composed of N-acetylglucosamine (GlcNAc) (linked to asparagine residues), followed by a diacetylated glucuronic acid and an acetylated mannuronic acid with the amino acid threonine covalently attached at position 6 (21). The genes encoding the glycosyl transferases responsible for the assembly of the second and terminal sugar, as well as for the oligosaccharyl transferase, have been experimentally confirmed (4; B. Chaban and K. F. Jarrell, unpublished results).
The glycosyl transferase gene Mv1751 was identified in M. voltae as the most likely gene to encode the enzyme to carry out the first step in this pathway, i.e., the attachment of a GlcNAc residue to a dolichol lipid carrier in the membrane. This is the only protein in the M. voltae genome that belongs to Pfam family PF00953 (glycosyl transferase family 4), which includes the eukaryotic N-acetylglucosamine-1-phosphate (GlcNAc-1-P) transferase (encoded by the alg7 gene). This enzyme catalyzes the conversion of UDP-N-acetyl-d-glucosamine and dolichyl phosphate to UMP and N-acetyl-d-glucosaminyl-diphosphodolichol. It is known that alg7 is an essential gene that is responsible for the first committed step in eukaryotic N glycosylation (3). Unlike bacteria that assemble N-linked glycans on undecaprenol as the lipid carrier, both archaea and eukaryotes use dolichol as a carrier (9).
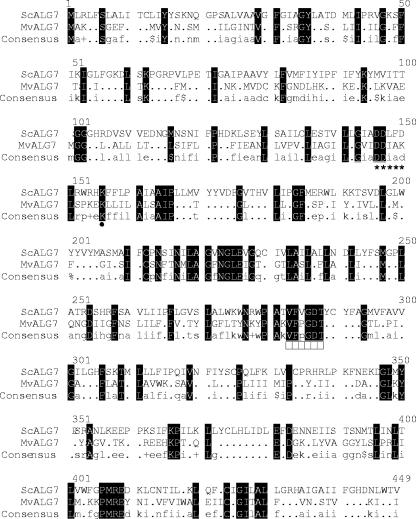
Conceptual translation of Mv1751 (317 amino acids) revealed 25% amino acid sequence identity and 41% similarity with the S. cerevisiae ALG7/GPT protein, where blocks of homology were distributed over the entire length of the protein (Fig. 1). Furthermore, the hydrophobicity plots (13) revealed multiple hydrophobic regions that could function as transmembrane segments. Analysis of the sequence of Mv1751 using the Husar software program package (Unix Sequence Analysis Resources, DKFZ, Heidelberg, Germany) revealed the presence of seven predicted transmembrane segment domains interspersed by hydrophilic/less-hydrophobic areas (data not shown). Two consensus motifs for N-linked glycosylation (N-X-T/S) were also detected. Similar motifs were shown in other GPT proteins, but there is no direct evidence as to whether or not these sites are glycosylated in the native protein. Also, the Mv1751 protein contains a conserved pair of aspartate residues, D-101 and D-102, in a position topologically similar to that of other GPT proteins, which are part of the highly conserved DDXXD motif (Fig. 1) that binds Mg2+ (5, 16). These motifs are predicted to be located on the cytoplasmic loop 2 in yeast GPT as well as Mv1751 (data not shown). As found in most cases, a basic residue (Arg or Lys) (18) is located a few residues downstream of the DDXXD motif (Fig. 1), presumably implicated also in binding of metal ions (2). Furthermore, the archaeal VFPGDT motif is highly conserved, with similarity to the equivalent region around Asp-287 in the yeast GPT (283-VFVGDT-288), and it is believed to be the GPT active-site nucleophile region (18) (Fig. 1).
FIG. 1.
Multiple sequence alignment with hierarchical clustering (7) of S. cerevisiae and M. voltae GPTs. Black boxes indicate identical amino acid residues. The conserved Asp-Asp in the putative DDXXD motif (marked with asterisks) is predicted to be coordinated to the Mg2+ cofactor. The putative active site nucleophilic Asp VFPGDT motif for Mv1751 is indicated with rectangles. •, conserved lysine located downstream of the DDXXD motif.
In view of the sequence similarity to the yeast ALG7 and the Pfam motif, topology, and hydrophobicity comparisons, we reasoned that Mv1751 may be a functional homolog of the yeast gene and might be able to complement a yeast alg7 mutant. Such an approach has already proven successful for identification of the human alg7 gene (8). Therefore, we tested the ability of Mv1751 to suppress the functional loss of a conditional lethal yeast alg7 mutant.
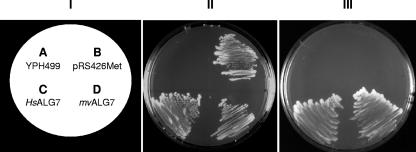
As a first step, we amplified the M. voltae Mv1751 coding region (GenBank accession no. DQ372942.1) with the primers MvALG7_forward (5′GGGGAATTCATGGCTAAAAGCGGTGAATTTATG) and MvALG7_reverse_FLAG (GGGCTCGAGCTTGTCATCGTCGTCCTTGTAGTCAATTATTTTAACCGTCGAATTTAC). The primers not only introduced EcoRI or XhoI sites at either end but also incorporated a FLAG tag on the C-terminal end of the amplification product, which was then digested with EcoRI/XhoI and ligated into a similarly restricted yeast shuttle vector containing ura3+ as a selective marker (carried on pRS426Met) (6), yielding the pRS426Met-Mv1751 plasmid. This plasmid was then transformed into a conditional lethal yeast mutant of the ALG7 gene, YPH499-HIS-GAL-ALG7 (15), as previously described (8), and transformants were streaked on minimal medium lacking histidine and uracil. The conditional lethal mutation was made by replacing the native alg7 promoter with the selection marker/promoter HIS3/GAL1 cassette, thereby eliminating the strain's requirement for histidine supplementation and placing the alg7 gene under the regulation of GAL1. GAL1 has been shown to be induced in the presence of galactose and to be tightly repressed in the presence of glucose. The expression of the endogenous ALG7 can be turned off by shifting the yeast cells from galactose-containing medium (YPGR medium [1% yeast extract, 2% peptone, 4% galactose, 2% raffinose]), in which the promoter is not repressed, to glucose-containing medium (synthetic dextrose minimal medium containing 5% glucose). Mutant cells transformed with plasmids carrying the human ALG7 (Fig. 2, quadrant C) (positive control [8]) or Mv1751 (Fig. 2, quadrant D) genes displayed slow but sustained growth and were able to suppress the functional loss of the ALG7 mutant and grow on glucose. Conversely, cells transformed with the control plasmid pRS426Met failed to grow (Fig. 2, quadrant B). This result indicates that the transformed M. voltae Mv1751 gene is indeed functional in yeast.
FIG. 2.
Rescue of YPH499-HIS-GAL-ALG7 by complementation with either the human ALG7 or Mv1751 gene. The conditional lethal mutant YPH499-HIS-GAL-ALG7 was transformed with the pRS426Met plasmids carrying either the human ALG7 (HsALG7) or the archaeal Mv1751 (MvALG7). The transformed cells were then streaked onto plates containing minimal medium lacking histidine and containing either galactose (II) or glucose (III) and incubated at 30°C. Panel III clearly shows that both the human and archaeal alg7 genes rescue the conditional lethal phenotype of YPH499-HIS-GAL and complement the yeast alg7.
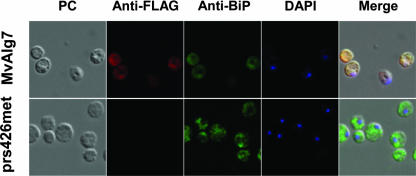
To visualize the Mv1751 protein and its localization within the yeast cells, indirect immunofluorescence microscopy of methanol/acetone-treated yeast spheroplasts using mouse anti-FLAG and goat anti-mouse immunoglobulin G conjugated with rhodamine was employed. ALG7 is an endoplasmic reticulum (ER) membrane-bound enzyme in eukaryotes (10), and Mv1751 also has multiple potential transmembrane domains, suggesting a membrane location. Consistent with the growth data shown in Fig. 2, the immunofluorescence data demonstrate that the yeast cells were synthesizing detectable levels of the Mv1751 protein and the expressed protein displayed predominantly a reticular pattern, including staining of the nuclear rim, a distribution typically observed for proteins localized to the ER. When costained with the ER marker protein BiP (binding protein), a nearly complete overlap was observed (Fig. 3, top).
FIG. 3.
Indirect immunofluorescence microscopy comparing the distributions of the ER marker BiP and the expressed Mv1751 in S. cerevisiae. FLAG-tagged Mv1751 is clearly visible inside cells containing the MvALG7 plasmid, while cells containing the empty pRS426Met plasmid showed no fluorescence. Costaining with BiP showed nearly complete overlapping with Mv1751. Cells were stained with DAPI (4′,6′-diamidino-2-phenylindole) to detect chromosomal DNA. PC, phase contrast.
Interestingly, the percent identity between the entire human and yeast alg7 proteins is 38% (53% similarity), while the percent identity between the archaeal and yeast proteins is only 25% (41% similarity). However, much of the conservation is found within the glycosyl transferase 4 domain, implying that these positions might represent important catalytic or structural residues (Fig. 1, between position markers 127 to 327). The important fact highlighted is that such little direct sequence conservation can result in the complementation of an essential gene.
The result of this complementation demonstrates that Mv1751 is indeed a GlcNAc-1-P transferase that is capable of replacing the essential alg7 gene in S. cerevisiae. For the first time, this allows for an experimentally verified function to be assigned to this gene and offers strong support to the proposal that Mv1751 catalyzes the first step in the N-glycosylation pathway in M. voltae. The linking sugar of both eukaryotic and M. voltae glycans is GlcNAc, and both use dolichol as the anchoring lipid to assemble N-glycans.
It therefore follows that a homologous protein would carry out the enzymatic attachment of nucleotide-activated GlcNAc to dolichol in both domains. As such, in recognition of its place in the archaeal N-glycosylation pathway, we propose to rename the M. voltae Mv1751 gene as aglH and assign it the function of GlcNAc-1-P transferase.
It is interesting that attempts to knock out this gene have proven unsuccessful in M. voltae (3) as well as in the related methanogen, Methanococcus maripaludis (D. VanDyke and K. F. Jarrell, unpublished data), while later steps in the N-linked pathway, including the oligosaccharyl transferase final step, have been disrupted. This suggests that AglH may play an essential role in additional pathways. One possibility is in glycolipid biosynthesis. It is known that M. voltae contains a GlcNAc-1-P diether glycolipid, where the attachment of the sugar is directly to archaeol (10). The pathways for glycolipid biosynthesis are poorly understood and it may be the case that AglH is involved in this process.
Of final note is the fact that the Mv1751 gene was able to complement an essential gene in another domain of life. It is rare to find two genes from different domains of life, especially essential genes, that are interchangeable. Because of the conservation of many aspects of the N-linked glycosylation systems in bacteria, archaea, and eukaryotes, the deciphering of the roles and interchangeability of various components may be advanced by consideration of the use of cross-domain complementation.
Acknowledgments
B.C. was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Post-Graduate Scholarship. This work was supported by a Discovery Grant from NSERC (to K.F.J.) and by the Deutsche Forschungsgemeinschaft (DFG), Bonn, Germany (to R.T.S.).
Footnotes
Published ahead of print on 4 January 2008.
REFERENCES
- 1.Abu-Qarn, M., and J. Eichler. 2006. Protein N-glycosylation in Archaea: defining Haloferax volcanii genes involved in S-layer glycoprotein glycosylation. Mol. Microbiol. 61511-525. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, M. S., S. S. Eveland, and N. P. Price. 2000. Conserved cytoplasmic motifs that distinguish sub-groups of the polyprenol phosphate:N-acetylhexosamine-1-phosphate transferase family. FEMS Microbiol. Lett. 191169-175. [DOI] [PubMed] [Google Scholar]
- 3.Burda, P., and M. Aebi. 1999. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426239-257. [DOI] [PubMed] [Google Scholar]
- 4.Chaban, B., S. Voisin, J. Kelly, S. M. Logan, and K. F. Jarrell. 2006. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol. Microbiol. 61259-268. [DOI] [PubMed] [Google Scholar]
- 5.Chappell, J. 1995. The biochemistry and molecular biology of isoprenoid metabolism. Plant Physiol. 1071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110119-122. [DOI] [PubMed] [Google Scholar]
- 7.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1610881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckert, V., M. Blank, R. Mazhari-Tabrizi, D. Mumberg, M. Funk, and R. T. Schwarz. 1998. Cloning and functional expression of the human GlcNAc-1-P transferase, the enzyme for the committed step of the dolichol cycle, by heterologous complementation in Saccharomyces cerevisiae. Glycobiology 877-85. [DOI] [PubMed] [Google Scholar]
- 9.Eichler, J., and M. W. Adams. 2005. Posttranslational protein modification in Archaea. Microbiol. Mol. Biol Rev. 69393-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrante, G., I. Ekiel, and G. D. Sprott. 1986. Structural characterization of the lipids of Methanococcus voltae, including a novel N-acetylglucosamine 1-phosphate diether. J. Biol. Chem. 26117062-17066. [PubMed] [Google Scholar]
- 11.Helenius, A., and M. Aebi. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 731019-1049. [DOI] [PubMed] [Google Scholar]
- 12.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54631-664. [DOI] [PubMed] [Google Scholar]
- 13.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157105-132. [DOI] [PubMed] [Google Scholar]
- 14.Lehle, L., A. Eiden, K. Lehnert, A. Haselbeck, and E. Kopetzki. 1995. Glycoprotein biosynthesis in Saccharomyces cerevisiae: ngd29, an N-glycosylation mutant allelic to och1 having a defect in the initiation of outer chain formation. FEBS Lett. 37041-45. [DOI] [PubMed] [Google Scholar]
- 15.Mazhari-Tabrizi, R., M. Blank, D. Mumberg, M. Funk, R. T. Schwarz, and V. Eckert. 1999. Chromosomal promoter replacement in Saccharomyces cerevisiae: construction of conditional lethal strains for the cloning of glycosyltransferases from various organisms. Glycoconj. J. 16673-679. [DOI] [PubMed] [Google Scholar]
- 16.McGarvey, D. J., and R. Croteau. 1995. Terpenoid metabolism. Plant Cell 71015-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mescher, M. F., J. L. Strominger, and S. W. Watson. 1974. Protein and carbohydrate composition of the cell envelope of Halobacterium salinarium. J. Bacteriol. 120945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price, N. P., and F. A. Momany. 2005. Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases. Glycobiology 1529R-42R. [DOI] [PubMed] [Google Scholar]
- 19.Schwarz, R. T., and R. Datema. 1982. Inhibition of the dolichol pathway of protein glycosylation. Methods Enzymol. 83432-443. [DOI] [PubMed] [Google Scholar]
- 20.Szymanski, C. M., S. M. Logan, D. Linton, and B. W. Wren. 2003. Campylobacter—a tale of two protein glycosylation systems. Trends Microbiol. 11233-238. [DOI] [PubMed] [Google Scholar]
- 21.Voisin, S., R. S. Houliston, J. Kelly, J. R. Brisson, D. Watson, S. L. Bardy, K. F. Jarrell, and S. M. Logan. 2005. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J. Biol. Chem. 28016586-16593. [DOI] [PubMed] [Google Scholar]
- 22.Wirth, D. F., H. F. Lodish, and P. W. Robbins. 1979. Requirements for the insertion of the Sindbis envelope glycoproteins into the endoplasmic reticulum membrane. J. Cell Biol. 81154-162. [DOI] [PMC free article] [PubMed] [Google Scholar]